Abstract
Background
Schizophrenia is a multifactorial disease in which genetic factors play a greater role than other factors. The genes of importance in schizophrenia patients are the genes that encode for neurotransmitters associated with low minor allele frequency (MAF) scores. This study was aimed to determine the association of genetic variations in catechol-O-methyl transferase (COMT), Ras association domain family member 1 (RASSF1) and glycoprotein M6A (GPM6A) with the risk of paranoid schizophrenia (PS) in patients admitted to Prof HB Saanin Psychiatric Hospital, West Sumatra, Indonesia.
Methods
Genotyping analysis through polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and PCR-amplification refractory mutation system (ARMS) was performed in 100 PS patients and 100 healthy controls. Chi-square and Fisher’s exact tests were used to compare the frequencies of genotype and allotype between the PS and control groups. Odds ratio (OR) with 95% confidence interval (95% CI) were calculated to determine the relative risk of PS with respect to genetic variations.
Results
Polymorphism rs13142920 in GPM6A was associated with significantly elevated risk of PS (P = 0.020; OR = 1.60 [95% CI: 1.08, 2.39]). However, COMT rs4680 and RASSF1 rs2073499 polymorphisms were not significantly associated with PS.
Conclusion
The GPM6A rs13142920 polymorphism holds great potential as a genetic marker in PS patients.
Keywords: paranoid schizophrenia, polymorphism, GPM6A, RASSF1, COMT
Introduction
Schizophrenia is a psychiatric disorder with complex pathophysiology related to the development neurons (1). Early symptoms of this disorder usually appear in late adolescence or early adulthood. These disorders are distinguished by common psychotic symptoms, such as delusions and hallucinations, loss of interest and encouragement, changes in emotional reactivity and disorganised behaviour (2). In 2016, the World Health Organization (WHO) reported that more than 21 million people worldwide experience schizophrenia. According to Ministry of Health, Indonesia data from 2018, the prevalence of severe mental disorders, such as schizophrenia, in Indonesia was estimated to be around 1.6 million people or 7 per 1,000 inhabitants. This number has increased four-fold since the previous Ministry of Health consensus in 2013. West Sumatra ranked seventh out of 33 provinces with an increase in the number of schizophrenia cases between 2013 and 2018 (3). Schizophrenia affects 0.5%–1% of individuals across distinct ethnic populations (4).
Schizophrenia is classified into five types based on the predominant symptoms: i) paranoid schizophrenia (PS); ii) hebephrenic schizophrenia (HS); iii) catatonic schizophrenia (CS); iv) residual schizophrenia (RS) and v) unclassified schizophrenia (US). PS is characterised by delusions, hallucinations, uncertain anxiety, contentiousness and arguing and violent behaviour. HS is also known as disorganised schizophrenia or chaos. CS is characterised by symptoms of decreased reactivity to the environment. RS is characterised by flat feeling, withdrawal from social interactions, eccentric behaviour, illogical and irrational thoughts. US is characterised by general clinical features, such as delusions, hallucinations, incoherence and chaotic behaviour (5). The prevalence of PS is higher than other types of schizophrenia. Therefore, we focused on PS cases in this study. Furthermore, focusing on only one type of schizophrenia reduces the genetic bias that can arise when studying multiple types of schizophrenia (6–7).
Schizophrenia is a multifactorial disease in which both genetic and environmental factors interact (8). A high heritability of schizophrenia (about 80%) shows that genetic factors are more influential in schizophrenia than environmental ones (9). The candidate-gene approach has been used in several molecular studies on schizophrenia, which focus on genes that encode for proteins associated with schizophrenia pathophysiology, such as neurotransmitter dopamine (catechol-O-methyl transferase [COMT]), Ras association domain family member 1 (RASSF1) and glycoprotein M6A (GPM6A) genes (10).
Three candidate genes were focused upon in this study: COMT, RASSF1 and GPM6A. The COMT gene has been widely studied in different populations. It encodes the enzyme COMT, which is the main enzyme involved in the metabolism of the neurotransmitter dopamine (13). The rs4680, which is located in exon 4, is a variant in the COMT that has been linked to an increased risk of schizophrenia. In this variation, the gene undergoes a transversion, which leads to conversion of the amino acid valine to methionine. This alteration results in decrease in the activity of enzymes associated with the activity of the neurotransmitter dopamine, which is linked to symptoms of schizophrenia (14–15). The minor allele frequency (MAF) of rs4680 polymorphism in the general population is less than 10%, with 4% in the Japanese population, 6.9% in the Han Chinese population and 8% in the sub-Saharan African population (16). A genetic variant with a low MAF, especially one with MAF < 10%, might be considered as a candidate gene for disease.
The RASSF1 and GPM6A genes were selected for this study based on the findings of genome-wide association study (GWAS), which revealed rs2073499 (A/G) and rs13142920 (A/C) polymorphisms in these genes, respectively, which were found to be associated with schizophrenia and had a MAF < 10% in East Asian population (12, 17–18).
The RASSF1 rs2073499 polymorphism had an MAF 9.3% in the Han Chinese population and 9.7% in the Chinese population (17–18). The RASSF1 gene encodes a protein that is associated with the RAS protein that modulates several growth-inhibiting responses and also acts as a tumour suppressor gene (19). It is hypothesised that increased expression of these tumour suppressor genes will result in excessive nerve cell death, which might impair the nervous system function associated with schizophrenia (20–21). rs13142920 is a polymorphism in GPM6A with an MAF of 5.8% in the Japanese population, 6.6% in the Chinese population and 7.1% in the Han Chinese population (18). This gene encodes the glycoprotein M6A protein, which is a transmembrane protein and a member of the proteolipid protein family that is most commonly expressed during nerve cell differentiation and development (22).
In Indonesia, several studies have been conducted to elucidate the relationship between genetic variation and schizophrenia. According to Rudianto et al. (23), the 8NRG433E1006 variant in the NRG1 (Neuregulin-1) was not associated with Javanese schizophrenic patients. Sutrisna and Yulianti (24) found that the rs3213207 polymorphism is not significantly associated with schizophrenia among Javanese patients. So far, no candidate genetic markers for schizophrenia have been discovered in Indonesia. Hence, this study was aimed to determine the association of genetic variations in COMT, RASSF1 and GPM6A with the risk of PS in patients admitted to Prof HB Saanin Psychiatric Hospital, West Sumatra, Indonesia.
Methods
Patient Characteristics and Ethical Approval
In this study, we used a case-control design with a total of 200 people, including 100 patients with PS at the Prof HB Saanin Psychiatric Hospital and 100 people as the controls who had no history of schizophrenia or any other mental disorders for at least three generations. The affected group included 83 males and 17 females aged 19 years old–64 years old, while the healthy controls were selected from the general public using a questionnaire and consisted of 26 males and 72 females aged 18 years old–39 years old.
Blood Sample Collection
Two to three millilitres of whole blood was collected from every participant and stored in a storage tube containing the anticoagulant ethylenediaminetetraacetic acid (EDTA).
DNA Extraction
DNA was isolated from whole blood using the salting out method that has been validated by Molecular Biology Laboratory of Biology Department, Faculty of Medicine, Universitas Indonesia. Briefly, 3 mL blood is added to a 15-mL tube and filled with red blood lysis solution in a ratio of 1:3. The mixture was incubated at room temperature for 10 min. Then, it was centrifuged at 1,500 rpm for 10 min and the supernatant was discarded. This step was repeated until the pellet was white. Next, 2 mL cell lysis solution (1 M Tris hydrocloric acid, 0.5 M EDTA and 10% sodium dodecyl sulfate (SDS) was added to the pellet and mixed until homogeneous. The mixture was incubated at 37 °C for 30 min. Then, 1.3 mL protein precipitation solution (5 M ammonium acetate) was added to the mixture and vortexed for 15 s–20 s. The mixture was centrifuged at 3000 rpm for 15 min. The supernatant was transferred into a tube that already contained 2.3 mL cold isopropanol. The tube was then inverted until DNA chromatin was visible. Then, the mixture was incubated at 37 °C for 2 h on water bath, followed by centrifugation at 3,000 rpm for 5 min. Next, 1.3 mL alcohol (70%) was added and then the tube was inverted. The supernatant was discarded and the tube was dried in an inverted position for 1 h. Finally, 300 μL Tris-EDTA (TE) buffer was added and the tube was incubated at 37 °C for 2 h on a water bath. The solution was then transferred to a 1.5-mL tube and refrigerated at −20 °C.
DNA Amplification
The PCR technique was used to amplify the DNA target using the Gotaq™ PCR Core System Kit (Promega) according to the manufacturer’s protocol. The RFLP-PCR primer for the RASSF1 was designed using the PrimerQuest online software, whereas the PCR-amplification refractory mutation system (ARMS) primers for the COMT and GPM6A were designed using the Primer1 online software. In order to prevent hairpins and dimers, the following primer criteria was selected: number of bases, 18 bp −30 bp; melting temperature (Tm), 52 °C– 58 °C; and guanine-cytosine (GC) percentage, 45%–60% (25). The National Centre for Biotechnology Information (NCBI)’s basic local alignment tool (BLAST) was used to confirm primer specificity. Table 1 contains detailed information about the PCR primers used in this study.
Table 1.
PCR primer sequences corresponding to each candidate gene
| Gene/Polymorphism | Primer sequences | PCR product (abp) | Annealing temperature (°C) | GC content (%) |
|---|---|---|---|---|
| RASSF1/rs2043799 | F: 5′ GCTGGCTCCATACAGGAGTG 3′ | 233 | 60 | 60 |
| R: 5′ GGCTTGTGGTAGACCTGAGC 3′ | 60 | |||
|
| ||||
| COMT/rs4680 | FI (A) : 5′ CCAGCGGATGGTGGATTTCGCTGTCA 3′ | 216 | 69 | 62 |
| RO: 5′ CTGAGCTGCTGGGGGGGTCTTTCCTCAG 3′ | 418 | |||
| FO: 5′ TCTCTCCACCTGTGCTCACCTCTCCTCCG 3′ | ||||
| RI (G): 5′CGGGTCAGGCATGCACACCTTGTCCTTAAC3′ | 258 | 64 | ||
|
| ||||
| GPM6A/rs13142920 | FI (C): 5′ TCTTTCGATTGCAAAGAATAGAGATTTAC 3′ | 201 | 56 | 38 |
| RO: 5′ AGCAATCTACGACTTGTAAGTCGTGAAT 3′ | 316 | |||
| FO: 5′ AATATACAGTTGATTCAGCTTCGACTCAC 3′ | ||||
| RI(A): 5′ CTGCCCCATCTTTCAGCTACTCTAGT 3′ | 170 | 39 | ||
Note:
Base pair
DNA samples were amplified for up to 30–35 cycles, beginning with a 5-min pre-denaturation temperature of 94 °C, followed by a cycle of denaturation at 95 °C for 30 s, annealing at 56 °C–69 °C for 30 s (Table 1) and elongation at 72 °C for 30 s. The extension phase comprised 72 °C for 7 min at the end of the cycle. The PCR product was verified using electrophoresis using 1.5%–2% agarose gel. Ethidium bromide was added to agarose gel for DNA visualisation with ultraviolet (UV) on UV longlife™ Filter Spectroline and photographed. The size of PCR product for each gene is listed on Table 1.
Genotyping
Restriction Fragment Length Polymorphism
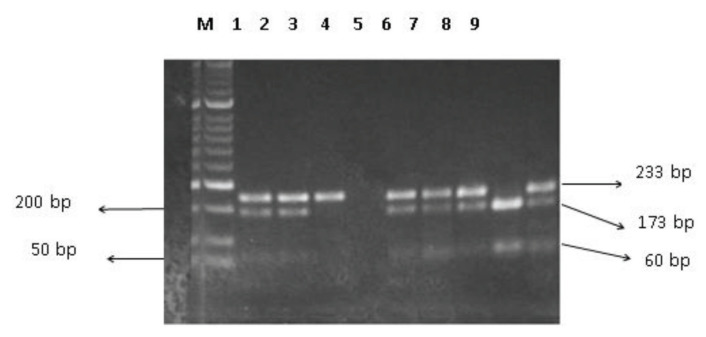
DrdI enzyme (New England BioLabs) was used to perform RFLP for rs2073499 (G/A). The restriction reaction was performed in a 20-μL system, containing 1 unit of DrdI, 1 μg DNA fragment, 2 μL restriction enzyme (10×) buffer solution and remaining ddH2O. The DNA fragment was detected for their quality using 2% agarose. The restriction fragments produced by restriction enzymes were 173 bp and 60 bp long (Figure 1).
Figure 1.
Agarose gel electrophoresis of PCR-RFLP products for the RASSF1 rs2073499. Lane M represents 50 bp DNA ladder marker; lanes 1, 2, 5, 6, 7 and 9 represent GA genotype; lane 3 represents AA genotype; lane 8 represents GG genotype; and lane 4 represents negative control
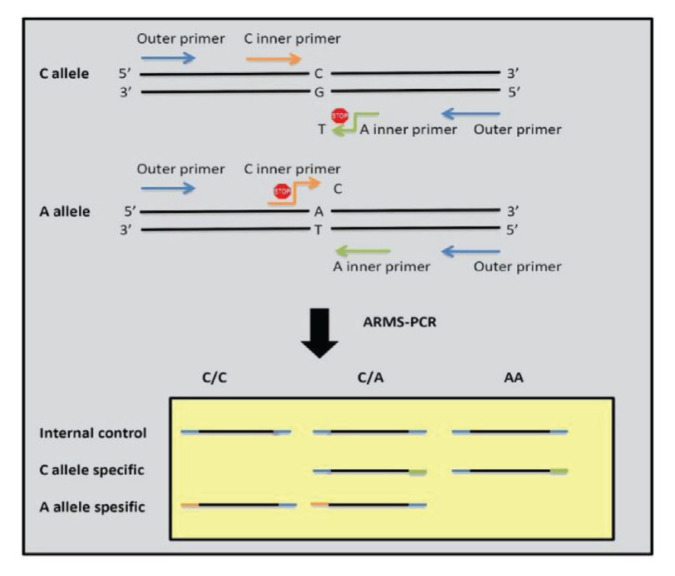
Amplification Refractory Mutation System
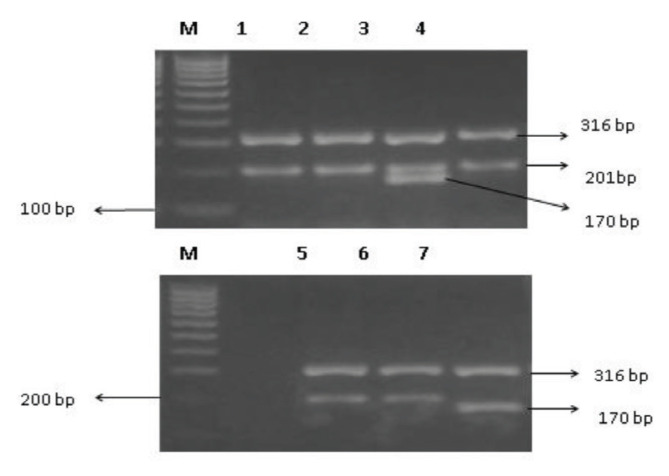
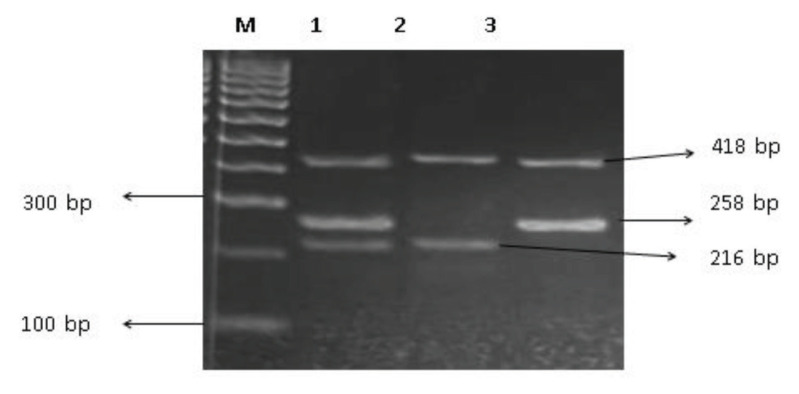
In ARMS, four primers were used to detect the rs4680 (G/A) and the rs13142920 (C/A) polymorphisms (Table 1). Each gene has one pair of internal control primers known as forward outer and reverse outer primers. For the detection of genetic variations in the COMT gene, reverse inner and forward outer primers were used for the G allele and forward inner and reverse outer primers were used for the A allele. Figure 2 depicts the PCR-ARMS method scheme (26). The PCR product of the GPM6A gene contained three DNA fragments with sizes of 316 bp for internal control, 201 bp for C allele-specific primers and 170 bp for A allele-specific primers (Figure 3). The PCR product of the COMT gene contained three DNA fragments with sizes of 418 bp for internal control, 258 bp for G allele-specific primers and 216 bp for A allele-specific primers (Figure 4).
Figure 2.
Tetra-primer PCR-ARMS method (26) (with modification). Different colours represent different primers involved in the PCR reaction. Blue: outer primer, orange: inner specific primer for C allele, and green: inner specific primer for A allele
Figure 3.
Agarose gel electrophoresis of ARMS-PCR products for the GPM6A rs13142920. Lane M represents 100 bp DNA ladder marker; lanes 1, 2 and 4–6 represent CC genotype; lane 3 represents CA genotype; and lane 7 represents AA genotype
Figure 4.
Agarose gel electrophoresis of ARMS-PCR products for the COMT rs4680. Lane M represents 100 bp DNA ladder marker; lane 1 represents GA genotype; lane 2 represents AA genotype; and lane 3 represents GG genotype
Statistical Analysis
Statistical analysis was performed to determine the association between genotype and allotype frequency in PS group using the Chi-square or the Fisher’s exact tests, while the odds ratio (OR) was used to determine the significance of genetic variations in schizophrenia disease manifestations. Statistical Package for the Social Science (SPSS) software version 25 was used for statistical analysis.
Results
Table 2 shows the genotype distribution of the COMT, RASSF1 and GPM6A genes. Statistical analysis revealed that the GPM6A polymorphism was significantly related to the risk of PS (P < 0.05; OR (CA and AA) = 2.92; CI 95%: 1.47, 5.81). To determine which genotypes were risk factors for PS, additional analysis was performed using dominant genetic modeling (Table 3). Individuals with the CA and AA genotypes had a P-value < 0.05, an OR > 1, and a CI > 1, indicating that they were at a higher risk of schizophrenia than those with the CC genotype. However, there was no significant association between rs4680 and rs2073499 genotypes and PS.
Table 2.
The genotype frequencies of polymorphisms in RASSF1, GPM6A and COMT in PS and control groups
| Polymorphisms | Ref/Alt | Genotype | Frequency | Hardy-Weinberg equilibrium | P-value | cOR (95% CI) | ||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| PS (n = 100) | Control (n = 100) | Patients P-value | Control P-value | |||||
| RASSF1 rs2073499 | G/A | GG | 0.13 | 0.17 | 0.458 | 0.752 | 0.358a | |
| GA | 0.54 | 0.44 | ||||||
| AA | 0.33 | 0.39 | ||||||
| GPM6A rs13142920 | C/A | CC | 0.15 | 0.34 | 0.000* | 0.051 | 0.007 a * | 2.92 (1.47, 5.81) |
| CA | 0.73 | 0.58 | ||||||
| AA | 0.12 | 0.08 | ||||||
| COMT rs4680 | G/A | GG | 0.59 | 0.51 | 0.831 | 0.509 | 0.508b | |
| GA | 0.37 | 0.44 | ||||||
| AA | 0.04 | 0.05 | ||||||
Notes: ref/alt = reference/alteration;
Chi-square test;
Fisher’s exact test;
Odds ratio;
indicates significant difference
Table 3.
Combined genotype frequencies of the RASFF1, GPM6A and COMT in PS and control groups
| No | Gene | Polymorphism | Genotype | Genotype Frequency | P-value | aOR (CI 95%) | |||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Patients | Control | ||||||||
|
| |||||||||
| Genotype I | Genotype II | Genotype I | Genotype II | ||||||
| 1 | RASSF1 | rs2073499 | GG versus GA + AA | 0.07 | 0.43 | 0.09 | 0.41 | 0.428b | 2.92 (1.47, 5.81) |
| AA versus GG + GA | 0.17 | 0.33 | 0.20 | 0.30 | 0.377b | ||||
| 2 | GPM6A | rs13142920 | CC versus CA + AA | 0.07 | 0.42 | 0.17 | 0.33 | 0.002 b * | |
| AA versus CA + CC | 0.06 | 0.45 | 0.04 | 0.46 | 0.346b | ||||
| 3 | COMT | rs4680 | GG versus AA + GA | 0.30 | 0.20 | 0.23 | 0.25 | 0.256b | |
| AA versus GG + GA | 0.02 | 0.48 | 0.03 | 0.49 | 1b | ||||
Notes:
Odds ratio;
Chi-square test;
indicates significant difference
The Hardy-Weinberg analyses for the RASSF1, GPM6A and COMT genes in both the case and control groups are shown in Table 2. In two groups, the RASSF1 and COMT genes exhibited non-significant P-values for Hardy-Weinberg equilibrium. This result indicated that the observed versus expected genotype distribution are comparable, or that the population is in Hardy-Weinberg equilibrium. Surprisingly, the affected group exhibited a significant P-value for the GPM6A genotype distribution, which was not in Hardy-Weinberg equilibrium.
Table 4 also shows the allotype distribution of the COMT, RASSF1 and GPM6A genes. The statistical test revealed that the allotype of the GPM6A gene exhibited a significant association with PS (P < 0.05; OR (A allele) = 1.60; 95% CI: 1.08, 2.39). In the West Sumatran population, the A allele was found to be more likely to be associated with schizophrenia than the C allele. COMT and RASSF1 allotypes, on the other hand, exhibited no significant association with PS.
Table 4.
The allotype frequencies of polymorphisms in RASSF1, GPM6A and COMT in PS and control groups
| Gene polymorphisms | Ref/Alt | Allotype | Frequency | P-value | aOR (CI 95%) | |
|---|---|---|---|---|---|---|
|
| ||||||
| PS (n = 200) | Control (n = 200) | |||||
| RASSF1 rs2073499 | G/A | G | 0.40 | 0.39 | 0.838 | |
| A | 0.60 | 0.61 | ||||
| GPM6A rs13142920 | C/A | C | 0.52 | 0.63 | 0.020b* | 0.62 (0.42, 0.93) |
| A | 0.49 | 0.37 | 1.60 (1.08, 2.39) | |||
| COMT rs4680 | G/A | G | 0.78 | 0.73 | 0.297 | |
| A | 0.23 | 0.27 | ||||
Notes: ref/alt = reference/alteration;
Odds ratio;
Chi-square test;
indicates significant difference
Discussion
Our findings revealed that there was no significant association between the rs4680 and rs2073499 genotypes and allotypes and the risk of PS. The results pertaining to COMT gene variant were similar to those of the studies done on Asian populations reported in Korea (27), Taiwan (28) and France (13). However, in other ethnic Asian populations, such as Saudi Arabia (31), India (32), China (30) and Turkey (29), a significant association has been reported between genotype and allotype frequencies in schizophrenia group compared to the control group.
Previous studies have also shown that the rs4680 polymorphism is associated with schizophrenia risk (33–34), which warrants the need to assess the other populations belonging to different ethnicities. Furthermore, the rs4680 variant in exon 4 is linked to the amino acid change from valine to methionine, with the val/val genotype associated with higher enzyme activity and less dopamine than the met/met genotype. As a result, the val/met heterozygous genotype exhibits more stable enzyme activity and dopamine levels (35). Changes in dopamine levels in the prefrontal cortex are associated with both positive and negative symptoms of schizophrenia (36–37), with dopamine hyperactivity associated with positive PS symptoms and vice versa (30, 38).
Previous studies on the association between rs4680 and schizophrenia in some populations have yielded conflicting results. This variation in the results could be attributed to schizophrenia being a polygene disease (35). Our findings showed that rs4680 is not a candidate genetic marker of schizophrenia in the population from West Sumatra, Indonesia.
Our results also showed that rs2073499, similar to rs4680, was not significantly associated with PS. This finding differs from previous GWAS studies in East Asia, including Indonesia (12) and China (39), and is associated with ethnic divergence. In this study, we focused on a more specific population in West Sumatra, with Minang being the dominant ethnic group.
The RASSF1 gene encodes a protein that belongs to the RAS family and plays a role in the apoptotic pathway. This protein functions as a tumor suppressor, suppressing proliferation and increasing apoptosis. According to Catts and Catts (20), an increased risk of schizophrenia is associated with increased expression of tumor suppressor genes, such as p53. It is assumed that variation in the RASSF1 gene increases apoptosis of excess nerve cells, resulting in decreased nerve cell function and association with schizophrenia.
In this study, we observed that rs13142920 was significantly associated with PS (P-value = 0.007; 95% CI: 0.17, 0.68). When compared to the CA and AA genotypes, the OR of the CC genotype was 0.34, indicating that it had a low probability of being associated with schizophrenia manifestations (Table 2). This result was related to the OR of the C allele, which was lower than that of A allele. These finding indicated that the A allele had dominant effect on schizophrenia manifestations. The Hardy-Weinberg equilibrium analysis revealed that the rs13142920 genotype for the patient group has a significant value. Our findings suggested that the GPM6A variant might contribute to schizophrenia risk.
These findings are also supported by GWAS studies conducted by Lam et al. (12) and Ma et al. (40), which demonstrated that the GPM6A could be a candidate gene worth further research for mental illnesses, such as schizophrenia. Ma et al. (40) discovered a decrease in GPM6A expression in the hippocampal area in the schizophrenic patients. An association study on the rs10520303 polymorphism has also been conducted by Boks et al. (41). They reported that there was a significant association between this polymorphism and schizophrenia with depressive sub-phenotypes. This finding could be attributed to stressor regulatory activity of the GPM6A, which might result in a change of expression, thus affecting the function of M6A proteins in nerve cell differentiation and development. As a result, changes in the function of nerve cells in the hippocampus are associated with the emergence of schizophrenia symptoms (42–43). As previously report, the rs13142920 polymorphism is located in an intronic area of GPM6A that has an associated to schizophrenia and is likely to be linked to causative variants that can directly affect the appearance of schizophrenia by acting as an enhancer in the transcription process (44). Furthermore, variation in the intronic region directly adjacent to the exon can affect the accuracy of the splicing process (45).
Conclusion
In conclusion, the rs13142920 polymorphism of GPM6A is a candidate genetic marker for PS patients in West Sumatra, Indonesia. Meanwhile, the rs4680 polymorphism in COMT and rs2073499 polymorphism in RASSF1 do not hold great potential candidates for genetic marker for PS in West Sumatra. As each ethnicity has different genotype and allotype frequencies, it is suggested that the ethnicity of our cohort might influence our findings. Furthermore, schizophrenia is a polygenic disease with many genes influencing the pathogenesis of the disease as well as a number of genotype and allotype frequencies in each population. Therefore, further investigations on COMT, RASSF1 and GPM6A on larger population are warranted to validate our findings and further elucidate the association between schizophrenia and variations in these genes.
Acknowledgements
Many thanks to the Director and staff of Prof HB Saanin Psychiatric Hospital, Indonesia, and Dr Pradiptajati Kusuma for their contributions and assistance in completion of this research.
Footnotes
Ethics of Study
This study was approved by the University of Indonesia’s Ethical Committee on Medical Research (No. KTE-323/UN2.F1/ETIK/PPM.00.02/2019), and the participants and their families were informed of the study’s purpose. Meanwhile, each participant also signed written informed consent forms.
Conflict of Interest
None.
Funds
This research was supported by the Universitas Indonesia’s PUTI Proceeding Grant 2020 (No. NKB-3504/UN2.RST/HKP.05.00/2020).
Authors’ Contributions
Conception and design: LY
Analysis and interpretation of the data: EK
Drafting of the article: EK
Critical revision of the article for important intellectual content: LY
Final approval of the article: LY
Provision of study materials or patients: EK
Statistical expertise: LY, EK
Obtaining of funding: LY
References
- 1.Yamada K, Iwayama Y, Toyota T, Ohnishi T, Ohba H, Maekawa M, et al. Association study of the KCNJ3 gene as a susceptibility candidate for schizophrenia in the Chinese population. Hum Genet. 2012;131(3):443–451. doi: 10.1007/s00439-011-1089-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davidson GC, Neale JM, Kring AM. Psikologi abnormal. 2nd ed. Jakarta: Raja Grafindo Persada; 2010. [Google Scholar]
- 3.Kementerian Kesehatan Republik Indonesia. Riskesdas. 2018. [Retrieved 2019 Feb 5]. https://kesmas.kemkes.go.id/assets/upload/dir_519d41d8cd98f00/files/Hasil-riskesdas-2018_1274.pdf.
- 4.Ohi K, Shimada T, Yasuyama T, Uehara T, Kawasaki Y. Variability of 128 schizophrenia-associated gene variants across distinct ethnic populations. Transl Psychiatry. 2017;7(1):e988. doi: 10.1038/tp.2016.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hawari D. Pendekatan holistik pada gangguan jiwa skizofrenia. Jakarta: Balai Penerbitan FKUI; 2006. [Google Scholar]
- 6.Zahnia S, Wulan Sumekar D. Kajian epidemiologis skizofrenia. Majority. 2016;5(5):160–166. [Google Scholar]
- 7.Muhyi A. Prevalensi penderita skiofrenia paranoid dengan gejala depresi di RSJ Dr. Soeharto Heerdjan Jakarta. [Bachelor thesis] UIN Syarif Hidayatullah; Jakarta: 2010. [Google Scholar]
- 8.Gejman PV, Sanders AR, Duan J. The role of genetics in the etiology of schizophrenia. Psychiatr Clin North Am. 2010;33(1):35–66. doi: 10.016/j.psc.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riley B, Kendler KS. Molecular genetic studies of schizophrenia. Eur J Hum Genet. 2006;14(6):669–680. doi: 10.1038/sj.ejhg.5201571. [DOI] [PubMed] [Google Scholar]
- 10.Allen NC, Bagade S, McQueen MB, Ioannidis JPA, Kavvoura FK, Khoury MJ, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40(7):827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 11.Farrell MS, Werge T, Sklar P, Owen MJ, Ophoff RA, O’donovan MC, et al. Evaluating historical candidate genes for schizophrenia. Mol Psychiatry. 2015;20(5):555–562. doi: 10.1038/mp.2015.16. https://doi.org/10/1038/mp.2015.16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lam M, Chen CY, Li Z, Martin AR, Bryois J, Ma X, et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat Genet. 2019;51(12):1670–1678. doi: 10.1038/s41588-019-0512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Chaldée M, Corbex M, Campion D, Jay M, Samolyk D, Petit M, et al. No evidence for linkage between COMT and schizophrenia in a French population. Psychiatry Res. 2001;102(1):87–90. doi: 10.1016/s-165-1781901000237-2. [DOI] [PubMed] [Google Scholar]
- 14.Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, et al. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci U S A. 2001;98(12):6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bilder RM, Volavka J, Czobor P, Malhotra AK, Kennedy JL, Ni X, et al. Neurocognitive correlates of the COMT Val158Met polymorphism in chronic schizophrenia. Biol Psychiatry. 2002;52(7):701–707. doi: 10.1016/s0006-3223(02)01416-6. [DOI] [PubMed] [Google Scholar]
- 16.National Centre for Biotechnology Information (NCBI) db SNP short genetic variation of rs4680 [Internet] [Retrieved 2020 Sep]. Available at: https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?do_not_redirect&rs=rs4680.
- 17.National Centre for Biotechnology Information (NCBI) db SNP short genetic variation of rs2073499 [Internet] [Retrieved 2020 Sep]. Available at: https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?do_not_redirect&rs=rs2073499.
- 18.National Centre for Biotechnology Information (NCBI) db SNP short genetic variation of rs13142920 [Internet] [Retrieved 2020 Sep]. Available at: https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?do_not_redirect&rs=rs13142920.
- 19.van der Weyden L, Adams DJ. The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochim Biophys Acta - Rev Cancer. 2007;1776(1):58–85. doi: 10.1016/j.bbcan.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Catts VS, Catts SV. Apoptosis and schizophrenia: is the tumour suppressor gene p53, a candidate susceptibility gene? Schizophr Res. 2000;41(3):405–415. doi: 10.1016/s0920-9964(99)00077. [DOI] [PubMed] [Google Scholar]
- 21.Zhuo C, Wang D, Zhou C, Chen C, Li J, Tian H, et al. Double-edged sword of tumour suppressor genes in schizophrenia. Front Mol Neurosci. 2019;12(1):1–9. doi: 10.3389/fnmol.2019.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michibata H, Okuno T, Konishi N, Wakimoto K, Kyono K, Aoki K, et al. Inhibition of mouse GPM6A expression leads to decreased differentiation of neurons derived from mouse embryonic stem cells. Stem Cells Dev. 2008;17(4):641–651. doi: 10.1089/scd.2008.0088. [DOI] [PubMed] [Google Scholar]
- 23.Rudianto CC, Brajadenta GS, Fitrikasari A, Winarni TI. Correlation of SNP8NRG433E1006 polymorphism Neuregulin 1 (NRG1) gene with schizophrenia in Java ethnic. Glob Med Health Comm. 2018;6(1):49–56. doi: 10.29313/gmhc.v6i1.2658. [DOI] [Google Scholar]
- 24.Sutrisna E, Yulianti R. Polymorphism of DTNBP1 gene P 1635 (Rs3213207) in schizophrenic Javanese, Indonesia. Bangladesh J Med Sci. 2019;18(4):703–705. doi: 10.3329/bjms.v18i4.42872. [DOI] [Google Scholar]
- 25.Elsalam KAA. Bioinformatic tools and guideline for PCR primer design. African J Biotechnol. 2003;2(5):91–95. doi: 10.5897/AJB2003.000-1019. [DOI] [Google Scholar]
- 26.Peng BY, Wang Q, Luo YH, He JF, Tan T, Zhu H. A novel and quick PCR-based method to genotype mice with a leptin receptor mutation (db/db mice) Acta Pharmacol Sin. 2018;39(1):117–123. doi: 10.1038/aps.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang HJ, Choe BM, Kim SH, Son S-R, Lee K-M, Kim BG, et al. No association between functional polymorphisms in COMT and MTHFR and schizophrenia risk in Korean population. epiH. 2010;32:e2010011. doi: 10.4178/epiH/e2010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen CY, Lu RB, Yeh YW, Shih MC, Huang SY. Association study of catechol-O-methyltransferase gene polymorphisms with schizophrenia and psychopathological symptoms in Han Chinese. Genes Brain Behav. 2011;10(3):316–324. doi: 10.1111/j.1601-183X.2010.00670. [DOI] [PubMed] [Google Scholar]
- 29.Altintas N, Sengoz S, Karamil S. Investigation of catechol-o-methyltransferase (COMT) val158met polymorphism in chronic schizophrenia diagnosed individuals in Manisa. JABS. 2019;13(3):128–132. https://jabsonline.org/index.php/jabs/article/view/626 . [Google Scholar]
- 30.Wang Y, Fang Y, Shen Y, Xu Q. Analysis of association between the catechol-O-methyltransferase (COMT) gene and negative symptoms in chronic schizophrenia. Psychiatry Res. 2010;179(2):147–150. doi: 10.1016/j.psychres.2009.03.029. [DOI] [PubMed] [Google Scholar]
- 31.Al-Asmary S, Kadasah S, Arfin M, Tariq M, Al-Asmari A. Genetic association of catechol-O-methyltransferase val(158)met polymorphism in Saudi schizophrenia patients. Genet Mol Res. 2014;13(2):3079–3088. doi: 10.4238/2014.April.17.4. [DOI] [PubMed] [Google Scholar]
- 32.Gupta M, Bhatnagar P, Grover S, Kaur H, Baghel R, Bhasin Y, et al. Association studies of catechol-O-methyltransferase (COMT) gene with schizophrenia and response to antipsychotic treatment. Pharmacogenics. 2009;10(3):385–397. doi: 10.2217/14622416.10.3.385. [DOI] [PubMed] [Google Scholar]
- 33.Baron M. Genetics of schizophrenia and the new millennium: Progress and pitfalls. Am J Hum Genet. 2001;68(2):299–312. doi: 10.1086/318212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenwood TA, Light GA, Swerdlow NR, Radant AD, Braff DL. Association analysis of 94 candidate genes and schizophrenia-related endophenotypes. PLoS ONE. 2012;7(1):e29630. doi: 10.1371/journal.pone.0029630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen X, Wang X, O’Neill AF, Walsh D, Kendler KS. Variants in the catechol-o-methyltransferase (COMT) gene are associated with schizophrenia in Irish high-density families. Mol Psychiatry. 2004;9(10):962–967. doi: 10.1038/sj.mp.4001519. [DOI] [PubMed] [Google Scholar]
- 36.De Castro-Catala M, Barrantes-Vidal N, Sheinbaum T, Moreno-Fortuny A, Kwapil TR, Rosa A. COMT-by-sex interaction effect on psychosis proneness. Biomed Res Int. 2015;2015:829237. doi: 10.1155/2015/829237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis KL, Kahn RS, Ko G, Davidson M. Dopamine in schizophrenia: a review and reconceptualization. Am J Psychiatry. 1991;148(11):1474–1486. doi: 10.1176/ajp.148.11.1474. [DOI] [PubMed] [Google Scholar]
- 38.Laruelle M. Schizophrenia: from dopaminergic to glutamatergic interventions. Curr Opin Pharmacol. 2014;14(1):97–102. doi: 10.1016/j.coph.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Liu D, Cen H, Jiang K, Xu Y. Research progress in biological studies of schizophrenia in China in 2017. Shanghai Arch Psychiatry. 2018;30(3):147–153. doi: 10.11919/j.issn.1002-0829.218041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma C, Gu C, Huo Y, Li X, Luo XJ. The integrated landscape of causal genes and pathways in schizophrenia. Transl Psychiatry. 2018;8(1):67. doi: 10.1038/s41398-018-0114-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boks MPM, Hoogendoorn M, Jungerius BJ, Bakker SC, Sommer IE, Sinke RJ, et al. Do mood symptoms subdivide the schizophrenia phenotype? Association of the GMP6A gene with a depression subgroup. Am J Med Genet Part B Neuropsychiatr Genet. 2008;147(6):707–711. doi: 10.1002/ajmg.b.30667. [DOI] [PubMed] [Google Scholar]
- 42.Alfonso J, Fernández ME, Cooper B, Flugge G, Frasch AC. The stress-regulated protein M6a is a key modulator for neurite outgrowth and filopodium/spine formation. Proc Natl Acad Sci U S A. 2005;102(47):17196–17201. doi: 10.1073/pnas.0504262102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alfonso J, Frick LR, Silberman DM, Palumbo ML, Genaro AM, Frasch AC. Regulation of hippocampal gene expression is conserved in two species subjected to different stressors and antidepressant treatments. Biol Psychiatry. 2006;59(3):244–251. doi: 10.1016/j.biopsych.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 44.Siekmann TE, Gerber MM, Amanda ET. Variants in an Hdac9 intronic enhancer plasmid impact Twist1 expression in vitro. Mamm Genome. 2017;27(3–4):99–110. doi: 10.1007/s00335-015-9618-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faried A, Halim D, Achmad TH. Biologi molekuler dasar dalam bidang kesehatan. Jakarta: Erlangga Medical Series; 2019. pp. 62–63. [Google Scholar]