Abstract
We have developed a novel, high-throughput scintillation proximity assay to measure the membrane-associated steps (stages 2 and 3) of peptidoglycan synthesis in Escherichia coli. At least five enzymes are involved in these two stages, all of which are thought to be essential for the survival of the cell. The individual enzymes are difficult to assay since the substrates are lipidic and difficult to isolate in large quantities and analysis is done by paper chromatography. We have assayed all five enzymes in a single mixture by monitoring synthesis of cross-linked peptidoglycan, which is the final product of the pathway. E. coli membranes are incubated with the two sugar precursors, UDP–N-acetyl muramylpentapeptide and UDP–[3H]-N-acetylglucosamine. The radiolabel is incorporated into peptidoglycan, which is captured using wheat germ agglutinin-coated scintillation proximity assay beads. The assay monitors the activity of the translocase (MraY), the transferase (MurG), the lipid pyrophosphorylase, and the transglycosylase and transpeptidase activities of the penicillin-binding proteins. Vancomyin, tunicamycin, nisin, moenomycin, bacitracin, and penicillin inhibit the assay, and these inhibitors have been used to validate the assay. The search for new antimicrobial agents that act via the late stages of peptidoglycan biosynthesis can now be performed in high throughput in a microtiter plate.
Peptidoglycan is the major structural component of the bacterial cell wall. It is a polymer of a repeating disaccharide-peptide unit, where the pentapeptide chains attached to adjacent sugar molecules are cross-linked. For convenience, the synthesis of peptidoglycan can be divided into three stages. In the first stage, in the cytoplasm, the two nucleotide-linked sugar precursors are synthesised: UDP–N-acetylglucosamine (UDP-GlcNAc) and UDP–N-acetylmuramylpentapeptide (UDP–MurNAc-pp). In the second stage, in the membrane, the disaccharide precursor is built up on a lipid carrier molecule; in the third stage, at the extracellular surface of the membrane, the sugars are polymerized, and the peptide chains are cross-linked.
Peptidoglycan is present in most bacteria and has no mammalian counterpart, making its synthesis an attractive target for the development of new antimicrobials. In particular, the membrane-associated enzymes (see Fig. 1) are much-favored targets because of their accessibility to drug molecules. This precludes problems associated with the permeability of the cell wall to the drug and resistance due to drug efflux. In addition, all five enzymes have been shown to be essential for the survival of the bacterial cell, either by genetic or biochemical means (3, 9, 11, 19, 30, 34). However, these targets have largely been underexploited because the assays are difficult to perform and are not amenable to high-throughput screening.
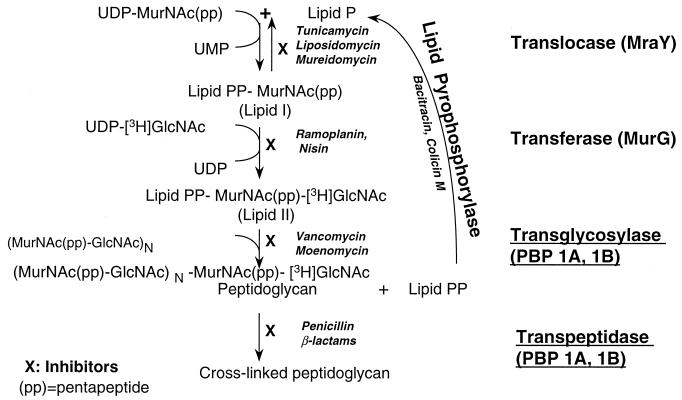
FIG. 1.
Enzymes of stages 2 and 3 of the peptidoglycan biosynthesis pathway and their inhibitors; enzymes of stage 3 are underlined. The enzymes are often referred to by their gene names in E. coli, which are indicated in parentheses.
Five enzymes are involved in stages 2 and 3 of peptidoglycan synthesis (Fig. 1). The translocase (MraY protein) catalyzes the transfer of MurNAc-pp to the lipid carrier, undecaprenol phosphate (2, 12, 14, 37). The transferase (MurG protein) catalyzes transfer of GlcNAc to lipid I to form lipid II (19, 29). The transglycosylase catalyzes transfer of the disaccharide (from the lipid) and polymerization to a preexisting peptidoglycan chain (38). The lipid pyrophosphate, which is released, is hydrolyzed to the monophosphate by a lipid pyrophosphorylase, so it can reenter the cycle (30, 34, 35). The transpeptidase catalyzes the cross-linking of adjacent peptide chains (14, 23) (Fig. 1). Both the transglycosylase and the transpeptidase activities may be present on the same polypeptide, e.g., PBP1a or PBP1b of Escherichia coli (13, 22, 38, 40).
The first two enzymes, MraY and MurG, can be assayed by using the respective radiolabeled sugar precursor as substrate and analyzing the product by paper chromatography or by extracting the lipid product in butanol (4, 31, 39). Alternatively, the translocase (MraY) can be assayed by fluorimetric means, although the sensitivity of this assay is rather low (4, 5, 44). A high-throughput assay for the transferase (MurG) has been described, but this requires synthesis of a complex artificial substrate (10, 18); a simpler assay measures MurG and MraY together, but it has many washing steps (6). The transglycosylase is one of the most difficult enzymes to assay. Radioactive lipid II (the substrate) has to be made in vitro, and incorporation of the radiolabel into peptidoglycan is monitored by paper chromatography (22, 23, 38, 42). Very recently, a moenomycin-binding and filtration assay has been described for detecting inhibitors of the transglycosylase (43), but this is not an enzyme assay and it detects only a specific type of inhibitor. The alternative to assaying these three enzymes individually is to assay part of the biosynthetic pathway by starting with the nucleotide-sugar precursors, which are incorporated into peptidoglycan by the action of the membrane-associated enzymes described above (2, 8, 14). Permeabilized bacteria can be used as a source of the enzymes, in which case the paper chromatography can be replaced by an acid precipitation step (17, 20). However, since a filtration step is required, this assay is also not very convenient for high-throughput screening.
Several assays have been described for the transpeptidase activity of the penicillin-binding proteins (PBPs), but most of these are binding assays and are not truly reflective of the enzyme activity (1, 14, 15, 16, 26, 36). The most commonly used is the binding of a β-lactam to the PBPs to screen for agents that compete with this binding (27, 33, 45). The lipid pyrophosphorylase has been assayed by radiolabeling the phosphate moiety of undecaprenol pyrophosphate and monitoring the release of the radiolabel by the enzyme (30, 34, 35). Thus, for all these membrane-associated enzymes of peptidoglycan synthesis there has been no convenient enzyme assay that can be performed in high throughput to screen for inhibitors.
We have measured here stages 2 and 3 of peptidoglycan synthesis in a single step which measures part of the peptidoglycan synthesis pathway in E. coli. We have used the principle of the scintillation proximity assay (SPA), a powerful technology that allows reactions to be performed in a single tube without any separation steps. Paper chromatography has been used since the 1960s to analyze the products of some of these enzymes, but with this development the assay for these enzymes is made much simpler. In this assay lectin-coated SPA beads are used to measure the radiolabel incorporated into peptidoglycan. The product measured by the SPA beads was shown to be peptidoglycan by comparing the results with the classical assay, i.e., analyzing peptidoglycan by paper chromatography. A number of inhibitors of these two stages of peptidoglycan synthesis were used to validate the assay. Tunicamycin, nisin, vancomycin, moenomycin, and bacitracin, as well as penicillin, inhibit the reaction. The assay can be used to screen, in high-throughput mode, for inhibitors of all five enzymes involved in the late stages of peptidoglycan synthesis, and it could lead to the discovery of novel antibacterial agents.
(Part of this work is presented in a pending patent [S. deSousa and D. Prahlad, A scintillation proximity assay for the detection of peptidoglycan synthesis, International Patent WO9960155, 25 November 1999.])
MATERIALS AND METHODS
Materials.
Wheat germ agglutinin-coated SPA (WGA-SPA) beads (RPNQ0001, PVT beads) were purchased from Amersham International Plc. UDP-[3H]GlcNAc was purchased from NEN Dupont. UDP-GlcNAc, tunicamycin, nisin, vancomycin, penicillin G, ampicillin, bacitracin, chloramphenicol, erythromycin, novobiocin, cerulenin, rifampin, and Triton X-100 were from Sigma Chemical Co. Flavomycin (moenomycin) was a gift from Hoechst. Antibiotic Medium 3 was from Difco Laboratories. BioGel chromatography materials were from Bio-Rad. DEAE-cellulose was from Whatman.
Substrates.
UDP–MurNac-pp was purified from Bacillus cereus 6A1 as described earlier (8). Briefly, cells were grown in Antibiotic Medium 3 to an A578 of 0.7. Chloramphenicol was added to 130 μg/ml, and 15 min later vancomycin was added to a concentration of 5 μg/ml. The cells were harvested 60 min after the addition of vancomycin. The bacterial pellet was resuspended in water, and a hot-water extract was made by adding this suspension dropwise to a flask of boiling water. The hot-water extract was ultracentrifuged, and the supernatant was purified by chromatography on a Bio-Gel P6 column followed by a Bio-Gel P2 column and ion-exchange chromatography on DEAE-cellulose eluted with a gradient of 0 to 0.35 M LiCl in 10 mM Tris-HCl (pH 7.5). Fractions that were positive for hexosamine (21) were pooled at each stage for purification on the next column. The eluate of the DEAE-cellulose column was used as a source of UDP–MurNAc-pp. The concentration of the precursor was estimated from its A262 value using a molar extinction coefficient of 10,000.
Enzyme preparation.
Membranes were prepared from E. coli AMA1004 as follows. The cells were grown in Luria-Bertani broth (6 liters) and harvested at an A600 of ∼1.8. The cells were washed, resuspended in a minimal volume (∼20 ml) of Buffer A (50 mM Tris-HCl pH7.5; 0.1 mM MgCl2), and lysed in a French pressure cell. The lysate was diluted to 100 ml with Buffer A and spun at 3,500 × g for 45 min, and the supernatant was ultra centrifuged at 150,000 × g for 45 min. The pellet of this spin was gently resuspended in ∼100 ml of Buffer A and recentrifuged at 150,000 × g for 45 min. The pellet was resuspended in a minimal volume (∼10 ml) of Buffer A, stored in aliquots at −70°C, and used as the enzyme preparation. The protein content was estimated by using the Coomassie blue dye binding reagent from Pierce Chemical Co. The quality of each membrane batch was monitored by determining the the quantity of peptidoglycan synthesized by different quantities of protein (under standard assay conditions), as well as by determining the counts per minute (cpm) obtained in the blank reaction (see below). Little variation was observed; occasional batches with poor activity or high blank values were discarded. The 50% inhibitory concentration (IC50) for the various inhibitors was similar with different batches of membrane.
Enzyme assay.
Membranes (4 μg of protein) were incubated for 90 min at 37°C with 100 μM UDP–MurNAc-pp and 2.5 μM UDP-[3H]GlcNAc (0.5 μCi) in a buffer of 100 mM Tris-HCl (pH 7.5)–10 mM MgCl2–4% dimethyl sulfoxide (DMSO) in a final volume of 25 μl (unless specified otherwise). The reaction was stopped by adding 5 μl of 90 mM EDTA. The product of the reaction was analyzed either by paper chromatography or by the addition of WGA-SPA beads. All reactions were carried out in triplicate.
For the paper chromatography, 20 μl of the reaction was spotted on Whatman no. 3 paper, which was then dried and chromatographed overnight in isobutyric acid– 1M ammonia (5:3 [vol/vol]). Peptidoglycan stays at the origin, which was cut out, and the radioactivity was measured in a liquid scintillation counter (using Optiphase HiSafe2 Scintillation Fluid; Wallac) after the paper had dried; lipid I and lipid II have Rf values of ∼0.9 (2).
For the SPA the enzyme reaction was done in flexible plates (no. 1450-401) from Wallac. The product was captured by the addition of 170 μl of a suspension of WGA-SPA beads in Triton X-100–Tris buffer such that the final concentration (in 200 μl) was 0.05% Triton X-100 and 100 mM Tris-HCl (pH 7.5). Radioactivity was measured in a Microbeta Trilux 3 h after addition of the beads (unless otherwise specified). The signal was quite stable, and samples may be counted from 3 to 48 h after bead addition.
A reaction without the first sugar nucleotide (UDP–MurNAc-pp) was run in parallel. This was treated as a blank and, for both types of analysis, the cpm obtained in this reaction was subtracted from that of reactions containing both sugar precursors as a measure of peptidoglycan synthesis.
Radioactive GlcNAc was incorporated into peptidoglycan, and the quantity of peptidoglycan formed in the reaction was measured as the cpm or picomoles of GlcNAc incorporated into the product. For the SPA it is difficult to determine the counting efficiency, so all results are represented as cpm. For the paper chromatography analysis, the counting efficiency was low and, for the data shown here, resulted in 350 to 500 cpm per pmol of GlcNAc.
Graphs were plotted using the GraphPad Prism software; error bars are used to indicate the standard error of mean, but in some figures these may not be obvious since they are smaller than the symbols. Where the purity of the starting material is not defined, compound concentrations are expressed in units other than micromolar concentrations.
RESULTS
Assay principle.
All five enzymes involved in stages 2 and 3 of peptidoglycan synthesis are membrane associated (Fig. 1); the membrane also provides the lipid substrate, undecaprenol phosphate. Thus, by incubating membranes with the two UDP-linked sugar precursors, UDP–MurNAc-pp and UDP-GlcNAc, the part of the peptidoglycan synthetic pathway shown in Fig. 1 can be reproduced in a cell-free system (2, 14). The five enzymes work in sequence to incorporate the sugars into cross-linked peptidoglycan and if one of the sugar precursors is radiolabeled (e.g., UDP-[3H]GlcNAc), peptidoglycan that is synthesized is radioactive and can be easily monitored.
The classical way of monitoring peptidoglycan is by paper chromatography, which separates peptidoglycan from the radioactive sugar precursor and lipid intermediate. In the assay described here we have, instead, used the SPA technology to monitor the synthesis of peptidoglycan; the beads are added at the end of the reaction to capture the peptidoglycan synthesized.
SPA beads are microspheres impregnated with a scintillant, and any radioactive product that is brought into the proximity of the bead can thus be directly monitored in a scintillation counter (7). If the radioactive substrate is not captured, then this results in an easy, homogenous assay with no separation steps. In this assay we have used WGA-SPA beads.
WGA binds GlcNAc and polymers containing GlcNAc, and WGA-SPA beads are widely used to capture mammalian membranes (7, 24). In the bacterial cell wall, GlcNAc is associated with peptidoglycan and with the lipopolysaccharides attached to the outer membrane. Peptidoglycan is localized to the periplasm, the region between the inner and outer membranes, so if the beads bind to any of these components, the radioactive peptidoglycan will be brought into the proximity of the bead.
Assay development.
The reaction conditions and substrate concentrations for peptidoglycan synthesis were initially worked out by analyzing the reaction products by paper chromatography. The assay was subsequently converted to a microtiter format, and the product was monitored by the addition of WGA-SPA beads.
Since the product captured by the WGA-SPA beads was not defined, to ensure the scintillation proximity assay was measuring peptidoglycan a series of comparative experiments was done. Two sets of enzyme reactions were run in parallel in a microtiter plate and, after stopping the enzyme reaction with EDTA, the products were analyzed either by paper chromatography or by adding SPA beads. While the radioactivity measured in the two sets of analyses could be different, due to the different counting efficiencies of the two systems, the kinetics of both assays should be similar if the SPA monitors peptidoglycan synthesis.
In both analyses, the quantity of radioactive product formed was insignificant when the reaction was stopped with EDTA at time zero, when the protein was left out of the reaction, or when the protein was heat inactivated (Table 1). Also, incorporation of the radiolabel into the product was insignificant when the first sugar precursor, UDP–MurNAc-pp, was left out of the reaction. This is particularly important in the SPA, in which there is no separation step and peptidoglycan cannot be distinguished from other radioactive products that may be captured by the beads.
TABLE 1.
Comparison of peptidoglycan synthesis, as measured by paper chromatography, with the SPAa
| Reaction conditions | Peptidoglycan synthesis (cpm)
|
|
|---|---|---|
| SPA | Paper chromatography | |
| Complete reaction | 9,805 | 10,198 |
| No UDP-MurNAc-pp | 878 | 190 |
| No protein | 530 | 154 |
| Heat-inactivated protein | 558 | 140 |
| EDTA at time zero | 609 | 183 |
| UDP-[3H]GlcNAc plus beads (NPE)b | 517 | NAc |
Enzyme reactions were performed in parallel, and the products were analyzed either by paper chromatography or with SPA beads.
NPE, nonproximity effect.
NA, not applicable.
The SPA beads do not capture the substrate, UDP-[3H]GlcNAc; the signal due to the nonproximity effect is ∼5% of the radioactivity captured in the enzymatic reaction (Table 1). Triton X-100 was added along with the beads at the capture step, at a final concentration of 0.05%, since this reduced the background and thus increased the signal-to-noise ratio (data not shown).
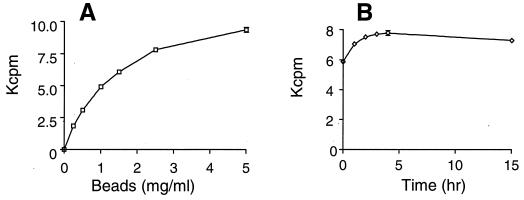
In the SPA format the concentration of beads per well was varied (Fig. 2A), and a concentration of 2.5 mg/ml (or 500 μg of beads per well) was chosen as the quantity for all further assays. The radioactivity was measured at various times after the addition of the WGA-SPA beads (Fig. 2B). Significant counts were observed immediately after bead addition, indicating the capture was instantaneous. The counts increased for up to 3 h and were fairly stable for up to 48 h; in most of the figures shown here the samples were counted ∼3 h after bead addition. DMSO stimulated peptidoglycan synthesis, as measured by the conventional paper chromatography analysis, as well as the radioactivity monitored by the SPA (data not shown), and was included in all assays at a concentration of 4%. With 4 or 10% DMSO the peptidoglycan synthesized was at a ∼1.5 or ∼3 times higher level, respectively, than if no DMSO was present during the assay.
FIG. 2.
SPA. (A) Product captured as a function of the SPA bead concentration. (B) SPA signal at different times after bead addition. Enzyme reactions were carried out in triplicate under standard assay conditions.
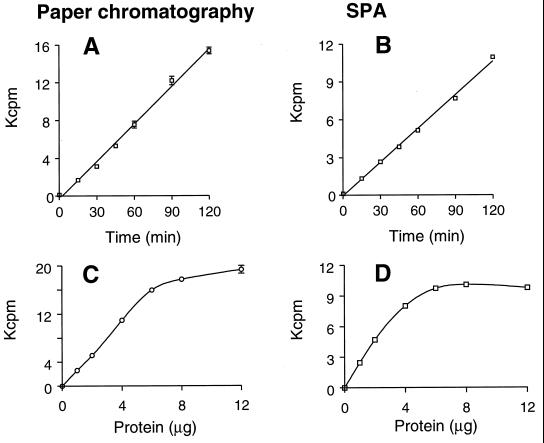
The time course of the reaction was similar for both peptidoglycan synthesis (monitored by paper chromatography) and the SPA analysis (Fig. 3A and B), as was the dependence of the two assays on protein quantity (Fig. 3C and D). Based on these results, 4 μg of protein and an incubation time of 90 min were chosen for all further experiments. Also, the dependence of both peptidoglycan synthesis and the SPA bead-captured product on concentrations of the first substrate, UDP–MurNAc-pp, or the second substrate, UDP-GlcNAc (Fig. 4) was very similar. These experiments strongly suggested that the product being monitored by the addition of WGA-SPA beads was the same as that monitored at the origin of the paper chromatogram, i.e., peptidoglycan.
FIG. 3.
Effect of various incubation times (A and B) and various protein concentrations (C and D; expressed in micrograms per reaction) on the synthesis of peptidoglycan (A and C) or on the product captured in the SPA (B and D).
FIG. 4.
Effect of various concentrations of substrates, UDP–MurNAc-pp (A and B) and UDP-GlcNAc (C and D) on the synthesis of peptidoglycan (A and C) or on the product captured in the SPA (B and D). In panels A and B the concentration of UDP-GlcNAc was 2.5 μM, and in panels C and D the concentration of UDP–MurNAc-pp was 100 μM. In panels C and D, for concentrations of UDP-GlcNAc of 0.25 to 5 μM a specific activity of 8 Ci/mmol was used, and for 10 to 25 μM a specific activity of 1.5 Ci/mmol was used. Because of the varying specific activity, the paper chromatography results are plotted as picomoles of GlcNAc incorporated, and for the SPA a correction was made for the two higher concentrations of UDP-GlcNAc so that the cpm are what would be expected if a specific activity of 8 Ci/mmol was used.
Assay validation. (i) Lysozyme.
Lysozyme cleaves peptidoglycan at the β1-4 bond between muramic acid and GlcNAc. Thus, if it is added during the synthesis of peptidoglycan, it should inhibit the formation of peptidoglycan polymers.
When lysozyme was added to the enzyme reaction it inhibited the incorporation of radiolabel into the SPA-captured product and also into peptidoglycan, as monitored by paper chromatography. At a concentration of 10 μg/ml lysozyme caused 70% inhibition of the SPA, and at 100 μg/ml it caused >90% inhibition of both the SPA and peptidoglycan synthesis. This further suggested that the SPA-captured product was peptidoglycan.
(ii) Effect of inhibitors.
A number of inhibitors are available for the individual enzymes in the late stages of the peptidoglycan biosynthesis pathway (Fig. 1), and these were used to validate the new assay. Tunicamycin is a known inhibitor of the first enzyme, the translocase (or mraY gene product), and thus is expected to inhibit peptidoglycan synthesis in this pathway assay (5). The effect of tunicamycin on peptidoglycan synthesis was compared with its effect on the SPA product. The IC50s in the two systems are very similar (∼0.3 μg/ml). The transferase enzyme (murG gene product) is inhibited by ramoplanin and nisin (28, 31). Nisin inhibited both the synthesis of peptidoglycan and the SPA with similar IC50s (0.9 and 3.9 μM, respectively; Table 2).
TABLE 2.
Comparison of the IC50s of inhibitors on peptidoglycan synthesis, as measured by paper chromatography, with the SPA
| Inhibitor | Enzyme inhibited | IC50
|
|
|---|---|---|---|
| Peptidoglycan (Paper chromatography) | SPA | ||
| Tunicamycin | Translocase | 0.27 μg/ml | 0.28 μg/ml |
| Nisin | Transferase | 0.9 μM | 3.9 μM |
| Vancomycin | Translocase, transferase, and transglycosylase | 13 μM | 22 μM |
| Moenomycin | Transglycosylase | 0.009 μM | 0.01 μM |
| Penicillin | Transpeptidase | No inhibition | 3 μM |
| Ampicillin | Transpeptidase | No inhibition | 8 μM |
| Bacitracin | Lipid pyrophosphorylase | 0.075 U/ml | 0.065 U/ml |
For the nisin IC50 determinations, 75 μM UDP–MurNAc-pp was used. Where the purity of the starting material is not defined, the concentrations are expressed in units other than micromolar concentrations. For tunicamycin, the IC50 approximates to ∼0.3 μM.
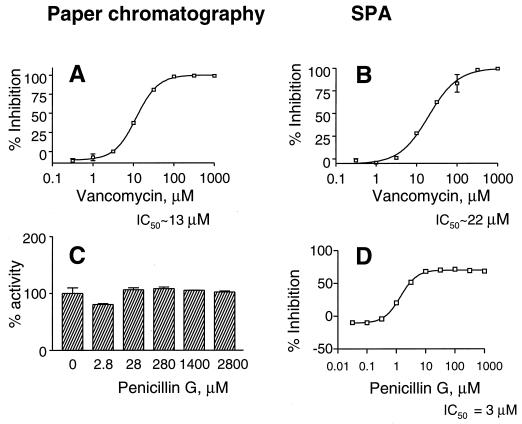
Vancomycin is a glycopeptide antibiotic that is known to bind to the terminal part of the peptide chain attached to muramic acid. As a consequence, it inhibits the transglycosylase enzyme; at higher concentrations it is reported to inhibit the transferase and translocase as well. The effect of vancomycin on the SPA (IC50 ∼22 μM) is similar to its effect on peptidoglycan synthesis (IC50 ∼ 13 μM; Fig. 5A and B). Moenomycin, also called flavomycin, is known as an inhibitor of the transglycosylase. The IC50 for the SPA is very similar to that for peptidoglycan synthesis (∼10 nM). The SPA was also inhibited by bacitracin, the lipid pyrophosphorylase inhibitor, showing an IC50 for the SPA (∼0.07 U/ml) that is very similar to that seen for peptidoglycan synthesis. For all of the inhibitors discussed so far, the inhibition levels were very similar for both the paper chromatography and the SPA analyses. Hence, only the vancomycin graphs are shown; for other inhibitors the IC50 values are summarized in Table 2.
FIG. 5.
Effect of vancomycin (A and B) and penicillin G (C and D) on peptidoglycan synthesis as analyzed by paper chromatography (A and C) and SPA (B and D). Note that, in panel C, the y axis is the percent activity (and not inhibition), since penicillin does not show inhibition in this assay.
We next tested the effect of the β-lactam antibiotics on the SPA and paper chromatography analysis. The β-lactams are one of the most successful antibiotics in the clinic and are known as inhibitors of the transpeptidase activity of the PBPs; they do not, however, affect the transglycosylase or polymerizing activity. Thus, in the presence of β-lactam antibiotics peptidoglycan is formed, but it is not cross-linked. It is very difficult to distinguish cross-linked peptidoglycan from that which is not cross-linked: both run at the origin in the paper chromatogram (14), and thus the β-lactams do not inhibit peptidoglycan synthesis as monitored by the paper chromatography assay (Fig. 5C).
Interestingly, the β-lactams did inhibit the SPA (Fig. 5D) with IC50s in the micromolar range: ∼3 μM for penicillin G and ∼8 μM for ampicillin (Table 2). If penicillin (300 μM) was added after the reaction was stopped with EDTA (and incubated for 30 min before addition of the beads), no inhibition was observed. Under these conditions we expect penicillin would have bound to the PBPs, suggesting that mere binding of the β-lactam to the PBPs in the membranes is insufficient to cause inhibition. Also, if the SPA beads were preincubated with the equivalent quantity of penicillin, no inhibition was observed, from which we conclude that penicillin does not prevent the capture of peptidoglycan by the beads. The β-lactams only inhibited when they were present during the reaction, when peptidoglycan was being synthesized; under these conditions we expect the peptidoglycan that was synthesized was not cross-linked, as has been shown earlier (23). This is the only situation in which the SPA results are markedly different from those of the paper chromatography assay. The data obtained with the β-lactams suggest that the product captured by the WGA-SPA beads is cross-linked peptidoglycan. It appears the SPA can monitor not only the polymerization but also the cross-linking step of peptidoglycan synthesis. This is a powerful advantage over the classical paper chromatography assay, which cannot distinguish cross-linked peptidoglycan from that which is not cross-linked.
The negative inhibition observed in the SPA with the β-lactams (Fig 5D), and sometimes with other inhibitors, occurs when the signal (cpm) in the treated sample is greater than that of the untreated control. This effect of the β-lactams on peptidoglycan synthesis (measured by paper chromatography) has been reported in the literature; the reason given is that the β-lactams also inhibit carboxypeptidases, which degrade the substrate and thus decrease the quantity of peptidoglycan synthesized. When the negative inhibition is observed only in the SPA and not in the paper chromatography analysis, we think this is a result of the complexity of the SPA capture. It could be that in the presence of the inhibitor more of the product is captured by the beads or that the efficiency of counting of the captured product is higher under this condition.
Inhibitors of enzymes unrelated to peptidoglycan synthesis, such as erythromycin, novobiocin, cerulenin, and rifampin, showed no inhibition of the SPA at concentrations of 100 μM. However, Triton X-100, a detergent, inhibited the assay (as well as peptidoglycan synthesis [data not shown]). Since the target enzymes are all membrane associated, other membrane-perturbing agents are also expected to inhibit the assay.
DISCUSSION
Peptidoglycan synthesis has for a long while been a favorite target for the development of antibacterial drugs because of the success of antibiotics (e.g., the β-lactams and glycopeptides) that act on these targets. However, research in this area has been hampered by the lack of assays amenable to high-throughput screening. In particular, there has been no easy assay described thus far that can measure the enzyme activities of the PBPs (transglycosylase or transpeptidase) or the lipid pyrophosphorylase. Because of this, several indirect assays have been used to screen for antibacterial compounds that act via inhibition of cell wall synthesis. For example, one screen looked for agents that inhibit incorporation of radioactive diaminopimelic acid into acid-insoluble fractions of B. subtilis but that do not inhibit the growth of Mycoplasma (which lacks a cell wall) (25, 32).
We have described here a scintillation proximity enzyme assay to measure peptidoglycan synthesis in E. coli. The SPA measures the combined activities of the translocase (MraY), the transferase (MurG), and the lipid pyrophosphorylase and the transglycosylase and transpeptidase activities of the PBPs, leading to peptidoglycan synthesis. The observation that the SPA was inhibited by lysozyme, as well as by inhibitors of each of the five enzymes involved in these stages of peptidoglycan synthesis, validates the assay as a monitor of peptidoglycan synthesis. Moreover, the IC50s observed in the SPA were similar to those obtained by the paper chromatography analysis and ranged from 10 nM to 20 μM for the compounds tested (Table 2).
The SPA can be used as a convenient, high-throughput screen for inhibitors of any of these five enzymes. Because it screens for the inhibitors of five enzymes in a single reaction, this results in a considerable saving of test compounds and cost when performing a large screen. An added advantage of the system is that, since wild-type membranes are used as a source of the enzymes, the pathway closely resembles the physiological situation with respect to the ratio of the five enzymes, as well as the concentration of the carrier lipid, undecaprenol phosphate. Also, since the assay does not measure binding of a β-lactam, we think it represents a true enzyme assay for the transpeptidase and could pick up inhibitors of a different chemical class and those with a mechanism of inhibition distinct from that of the β-lactams.
However, the assay does have a disadvantage in that it does not distinguish between inhibitors of the five enzymes or the inhibition caused by detergents. Thus, for an unknown compound, secondary assays will have to be carried out to determine which of the five enzymes is the target of the inhibitor. Also, the IC50 value must be interpreted with caution, particularly with compounds that are expected to inhibit more than one enzyme in the cascade (e.g., vancomycin). While the IC50 is a convenient way of measuring the potency of an inhibitor, compounds with similar IC50s may be targeting two different enzymes.
It is noticeable that, for some of the inhibitors we tested, the IC50s are in the micromolar range, which appears to be high for an antibacterial compound. This is possibly due to the mechanism by which these compounds inhibit, since many of them bind to the substrates or enzymes in this pathway. For example, the β-lactams bind covalently to the PBPs, of which there are several in the cell, while only a few of these probably contribute to the transpeptidase activity measured in this assay. Nisin binds to the lipid intermediates (28), and bacitracin binds to undecaprenol pyrophosphate (28, 34, 35). Similarly, in the whole cell vancomycin is thought to bind to lipid II (which is not abundant), thus inhibiting the transglycosylase. However, in this assay it probably has access to and binds UDP–MurNAc-pp, as well as lipid I, thus giving it a high IC50; if the assay is performed with a lower concentration of UDP–MurNAc-pp, the IC50 of vancomycin falls considerably (data not shown). In contrast, the IC50 of moenomycin, which was thought of as a competitive inhibitor of the transglycosylase (41), is between 1 and 10 nM; however, a recent report claims it, too, binds to the PBPs (43).
We have converted a laborious paper chromatography assay into one that can be performed with high throughput in a microtiter plate. By comparison with the classical paper chromatography assay, where the Rf of peptidoglycan is defined, we have validated the SPA as a way of monitoring peptidoglycan synthesis. Because of the convenience and ease of the assay, it is now possible to look at kinetic parameters that are very tedious to monitor by the classical assay. The most powerful aspect of the SPA is the ability to detect, in a single reaction, the inhibitors of the five enzymes in the late stages of peptidoglycan synthesis that are very popular targets for the development of antibacterial drugs.
ACKNOWLEDGMENTS
We thank Noel deSouza, formerly of Hoechst, India, for the gift of flavomycin and Tanneke den Blaauwen for discussions on the purification of UDP–MurNAc-pp.
REFERENCES
- 1.Adam M, Damblon C, Jamin M, Zorzi W, Dusart V, Galleni M, El Kharroubi A, Piras G, Spratt B G, Keck W, Coyette J, Ghuysen J-M, Nguyen-Disteche M, Frere J-M. Acyltransferase activities of the high-molecular-mass essential penicillin-binding proteins. Eur J Biochem. 1991;279:601–604. doi: 10.1042/bj2790601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson J S, Meadow P M, Haskin M A, Strominger J L. Biosynthesis of the peptidoglycan of bacterial cell walls. Arch Biochem Biophys. 1966;116:487–515. doi: 10.1016/0003-9861(66)90056-7. [DOI] [PubMed] [Google Scholar]
- 3.Boyle D S, Donachie W D. mraY is an essential gene for cell growth in Escherichia coli. J Bacteriol. 1998;180:6429–6432. doi: 10.1128/jb.180.23.6429-6432.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandish P E, Burnham M K, Lonsdale J T, Southgate R, Inukai M, Bugg T D H. Slow binding inhibition of phospho-N-acetylmuramyl-pentapeptide-translocase (Escherichia coli) by mureidomycin A. J Biol Chem. 1996;271:7609–7614. doi: 10.1074/jbc.271.13.7609. [DOI] [PubMed] [Google Scholar]
- 5.Brandish P E, Kimura K I, Inukai M, Southgate R, Lonsdale J T, Bugg T D H. Modes of action of tunicamycin, liposidomycin B, and mureidomycin A: inhibition of phospho-N-acetylmuramyl-pentapeptide-translocase from Escherichia coli. Antimicrob Agents Chemother. 1996;40:1640–1644. doi: 10.1128/aac.40.7.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branstrom A A, Midha S, Longley C B, Han K, Baizman E R, Axelrod H R. Assay for identification of inhibitors for bacterial MraY translocase or MurG transferase. Anal Biochem. 2000;280:315–319. doi: 10.1006/abio.2000.4530. [DOI] [PubMed] [Google Scholar]
- 7.Cook N D. Scintillation proximity assay: a versatile high-throughput screening technology. Drug Discovery Today. 1996;1:287–294. [Google Scholar]
- 8.Den Blaauwen T, Aarsman M, Nanninga N. Interaction of monoclonal antibodies with the enzymatic domains of penicillin-binding protein 1b of Escherichia coli. J Bacteriol. 1990;172:63–70. doi: 10.1128/jb.172.1.63-70.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denome S A, Elf P K, Henderson T A, Nelson D E, Young K D. Escherichia coli mutants lacking all possible combinations of eight penicillin-binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J Bacteriol. 1999;181:3981–3993. doi: 10.1128/jb.181.13.3981-3993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha S, Chang E, Lo M-C, Men H, Park P, Ge M, Walker S. The kinetic characterization of Escherichia coli MurG using synthetic substrate analogues. J Am Chem Soc. 1999;121:8415–8426. [Google Scholar]
- 11.Harkness R E, Braun V. Colicin M inhibits peptidoglycan biosynthesis by interfering with lipid carrier recycling. J Biol Chem. 1989;264:6177–6182. [PubMed] [Google Scholar]
- 12.Ikeda M, Wachi M, Jung H K, Ishino F, Matsuhashi M. The Escherichia coli mraY gene encoding UDP-N-acetylmuramoyl-pentapeptide: undecaprenyl-phosphate phospho-N-acetylmuramoyl-pentapeptide transferase. J Bacteriol. 1991;173:1021–1026. doi: 10.1128/jb.173.3.1021-1026.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishino F, Mitsui K, Tamaki S, Matsuhashi M. Dual enzyme activities of cell wall peptidoglycan synthesis, peptidoglycan transglycosylase and penicillin-sensitive transpeptidase in purified preparations of Escherichia coli penicillin-binding protein 1A. Biochem Biophys Res Commun. 1980;97:287–293. doi: 10.1016/s0006-291x(80)80166-5. [DOI] [PubMed] [Google Scholar]
- 14.Izaki K, Matsuhashi M, Strominger J L. Biosynthesis of the peptidoglycan of bacterial cell walls. XIII. Peptidoglycan transpeptidase and d-alanine carboxypeptidase-sensitive enzymatic reaction in strains of Escherichia coli. J Biol Chem. 1968;246:3180–3192. [PubMed] [Google Scholar]
- 15.Jamin M, Damblon C, Millier S, Hakenbeck R, Frere J-M. Penicillin-binding protein 2x of Streptooccus pneumoniae: enzymic activities and interaction with β-lactams. Biochem J. 1993;292:735–741. doi: 10.1042/bj2920735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laible G, Keck W, Lurz R, Mottl H, Frere J-M, Jamin M, Hakenbeck R. Penicillin-binding protein 2× of Streptococcus pneumoniae: expression in Escherichia coli and purification of a soluble enzymatically active derivative. Eur J Biochem. 1992;207:943–949. doi: 10.1111/j.1432-1033.1992.tb17128.x. [DOI] [PubMed] [Google Scholar]
- 17.Maas D, Pelzer H. Murein biosynthesis in ether permeabilized Escherichia coli starting from early peptidoglycan precursors. Arch Microbiol. 1981;130:301–306. doi: 10.1007/BF00425944. [DOI] [PubMed] [Google Scholar]
- 18.Men H, Park P, Walker S. Substrate synthesis and activity assay for MurG. J Am Chem Soc. 1998;120:2484–2485. [Google Scholar]
- 19.Mengin-Lecreulx D, Texier L, Rousseau M, van Heijenoort J. The murG gene of Escherichia coli codes for the UDP-N-acetylglucosamine: N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase involved in the membrane steps of peptidoglycan synthesis. J Bacteriol. 1991;173:4625–4636. doi: 10.1128/jb.173.15.4625-4636.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirelman D, Yashouv-Gan Y, Schwarz U. Peptidoglycan biosynthesis in a thermosensitive division mutant of Escherichia coli. Biochemistry. 1976;15:1781–1790. doi: 10.1021/bi00654a001. [DOI] [PubMed] [Google Scholar]
- 21.Morgan W T J, Elson L A. A colorimetric method for the determination of N-acetylglucosamine and N-acetyl chondrosamine. Biochem J. 1934;28:988–995. doi: 10.1042/bj0280988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakagawa J, Tamaki S, Matsuhashi M. Purified penicillin binding proteins 1Bs from Escherichia coli membrane showing activities of both peptidoglycan polymerase and peptidoglycan crosslinking enzyme. Agric Biol Chem. 1979;43:1379–1380. [Google Scholar]
- 23.Nakagawa J, Tamaki S, Tomioka S, Matsuhashi M. Functional biosynthesis of cell wall peptidoglycan by polymorphic bifunctional polypeptides. J Biol Chem. 1984;259:13937–13946. [PubMed] [Google Scholar]
- 24.Nelson N. A novel method for the detection of receptors and membrane proteins by scintillation proximity radioassay. Anal Biochem. 1987;165:287–293. doi: 10.1016/0003-2697(87)90271-5. [DOI] [PubMed] [Google Scholar]
- 25.Omura S, Tanaka H, Oiwa R, Nagai T, Koyama Y, Takahashi Y. Studies on bacterial cell wall inhibitors. VI. Screening method for the specific inhibitors of peptidoglycan synthesis. J Antibiot. 1979;32:978–984. doi: 10.7164/antibiotics.32.978. [DOI] [PubMed] [Google Scholar]
- 26.Presslitz J E, Ray V. dd-Carboxypeptidase and peptidoglycan transpeptidase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1975;7:578–581. doi: 10.1128/aac.7.5.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roychoudhury S, Kaiser R E, Brems D N, Yeh W-K. Specific interaction between β-lactams and soluble penicillin-binding protein 2a from methicillin-resistant Staphylococcus aureus: development of a chromogenic assay. Antimicrob Agents Chemother. 1996;40:2075–2079. doi: 10.1128/aac.40.9.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reisinger P, Seidel H, Tschesche H, Hammes W P. Effect of nisin on murein synthesis. Arch Microbiol. 1980;127:187–193. doi: 10.1007/BF00427192. [DOI] [PubMed] [Google Scholar]
- 29.Salmond G P C, Lutkenhaus J F, Donachie W D. Identification of new genes in a cell envelope-cell division gene cluster of Escherichia coli: cell envelope gene murG. J Bacteriol. 1980;144:438–440. doi: 10.1128/jb.144.1.438-440.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siewert G, Strominger J L. Bacitracin: an inhibitor of the dephosphorylateon of lipid pyrophosphate, an intermediate in biosynthesis of the peptidoglycan of bacterial cell walls. Proc Natl Acad Sci USA. 1967;54:767–773. doi: 10.1073/pnas.57.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Somner E A, Reynolds P E. Inhibition of peptidoglycan biosynthesis by ramoplanin. Antimicrob Agents Chemother. 1990;34:413–419. doi: 10.1128/aac.34.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spiri-Nakagawa P, Fukushi Y, Maebashi K, Imamura N, Takahashi Y, Tanaka Y, Tanaka H, Omura S. Izupeptins A and B, new glycopeptide antibiotics produced by an actinomycete. J Antibiot. 1986;39:1719–1723. doi: 10.7164/antibiotics.39.1719. [DOI] [PubMed] [Google Scholar]
- 33.Spratt B G. Properties of the penicillin-binding proteins of Escherichia coli K12. Eur J Biochem. 1977;72:341–352. doi: 10.1111/j.1432-1033.1977.tb11258.x. [DOI] [PubMed] [Google Scholar]
- 34.Stone K J, Strominger J L. Mechanism of action of bacitracin: complexation with metal ion and C55-isoprenyl pyrophosphate. Proc Natl Acad Sci USA. 1971;68:3223–2327. doi: 10.1073/pnas.68.12.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Storm D R, Strominger J L. Complex formation between bacitracin peptides and isoprenyl pyrophosphates. J Biol Chem. 1973;248:3940–3945. [PubMed] [Google Scholar]
- 36.Strominger J L, Izaki K, Matsuhashi M, Tipper D J. Peptidoglycan transpeptidase and d-alanine carboxypeptidase: penicillin-sensitive enzymatic reactions. Fed Proc. 1967;26:9–22. [PubMed] [Google Scholar]
- 37.Struve W G, Sinha R K, Neuhaus F C. On the initial stage in peptidoglycan synthesis: phospho-N-acetylmuramyl-pentapeptide translocase (uridine monophosphate) Biochemistry. 1966;5:82–93. doi: 10.1021/bi00865a012. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki H, van Heijenoort Y, Tamura T, Mizoguchi J, Hirota Y, van Heijenoort J. In vitro peptidoglycan polymerization catalysed by penicillin binding protein 1b of Escherichia coli. FEBS Lett. 1980;110:245–249. doi: 10.1016/0014-5793(80)80083-4. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka H, Oiwa R, Matsukura S, Omura S. Amphomycin inhibits phospho-N-acetylmuramyl-pentapeptide translocase in peptidoglycan synthesis of bacillus. Biochem Biophys Res Commun. 1979;86:902–908. doi: 10.1016/0006-291x(79)91797-2. [DOI] [PubMed] [Google Scholar]
- 40.Terrak M, Ghosh T K, van Heijenoort J, van Beeumen J, Lampilas M, Aszodi J, Ayala J A, Ghuysen J-M, Nguyen-Disteche M. The catalytic, glycosyl transferase and acyl transferase modules of the cell wall peptidoglycan polymerizing penicillin-binding protein 1b of Escherichia coli. Mol Microbiol. 1999;34:350–364. doi: 10.1046/j.1365-2958.1999.01612.x. [DOI] [PubMed] [Google Scholar]
- 41.van Heijenoort Y, Leduc M, Singer H, van Heijenoort J. Effect of moenomycin on Escherichia coli. J Gen Microbiol. 1987;133:667–674. doi: 10.1099/00221287-133-3-667. [DOI] [PubMed] [Google Scholar]
- 42.van Heijenoort Y, Derrien M, van Heijenoort J. Polymerisation by transglycosylation in the biosynthesis of the peptidoglycan of Escherichia coli K12 and its inhibition by antibiotics. FEBS Lett. 1978;89:141–144. doi: 10.1016/0014-5793(78)80540-7. [DOI] [PubMed] [Google Scholar]
- 43.Vollmer W, Holtje J-V. A simple screen for murein transglycosylase inhibitors. Antimicrob Agents Chemother. 2000;44:1181–1185. doi: 10.1128/aac.44.5.1181-1185.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weppner W A, Neuhaus F C. Fluorescent substrate for nascent peptidoglycan synthesis. J Biol Chem. 1977;252:2296–2303. [PubMed] [Google Scholar]
- 45.Zhao G, Meier T I, Kahl S D, Gee K R, Blaszczak L C. BOCILLIN FL, a sensitive and commercially available reagent for the detection of penicillin-binding proteins. Antimicrob Agents Chemother. 1999;43:1124–1128. doi: 10.1128/aac.43.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]