Abstract
Xerophyta retinervis has great ornamental value and numerous traditional uses. This study reported its first complete chloroplast genome sequence, which was 155,109 bp in length, including a pair of inverted repeat regions (IRs) (27,093 bp), a small single-copy (SSC) region (17,385 bp), and a large single-copy (LSC) region (83,538bp). The chloroplast genome encoded 133 genes, including 87 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. The total GC content of the chloroplast genome was 37.55%. The phylogenetic tree showed that X. retinervis was closely related to X. spekei.
Keywords: Chloroplast genome, phylogeny, Xerophyta retinervis
The species of genus Xerophyta Juss. (Velloziaceae) are known to be drought-tolerant plants (Farrant 2000). X. retinervis Baker 1875 is a perennial shrub up to 1.8 m tall and widely distributed through south Africa (Gibbs et al. 1987). Locally, X. retinervis is an extensively applied medicinal plant, with smoke from roots used to relieve asthma and smoke from the whole plant to stop nosebleeds (Van Wyk et al. 1997). Its stems are widely used to make ropes for hut and screen building, brushes, or mats in traditional home crafts (Dyer 1942).
The fresh leaves of X. retinervis were collected from Beijing Botanical Garden (N 39.9920, E 116.2137), Institute of Botany, Chinese Academy of Sciences, kept in silica gel, and stored at the Herbarium of Chengdu Institute of Biology (Bo Xu, xubo@cib.ac.cn) under the voucher number S1091. Total genomic DNA was extracted from dry leaves through Plant DNA Isolation Kit (Cat.No.DE-06111) and sequenced via Illumina pair-end technology. Cleaned reads were assembled using GetOrganelle v1.7.2 (Jin et al. 2020). The assembled chloroplast genome was annotated using PGA (Qu et al. 2019) and manually corrected for the start and stop codons. The annotated chloroplast genome was deposited to GenBank under the accession number MW580856.
The chloroplast genome of X. retinervis was 155,109 bp in length with a typical quadripartite structure, including a pair of inverted repeat regions (IRs) of 27,093 bp, a single-copy (SSC) region of 17,385 bp, and a large single-copy (LSC) region of 83,538bp. The chloroplast genome contained 133 genes, including 87 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. 62% of the genes were located in the single-copy regions, and 19% were duplicated in the IR regions. The total GC content of the chloroplast genome was 37.55%.
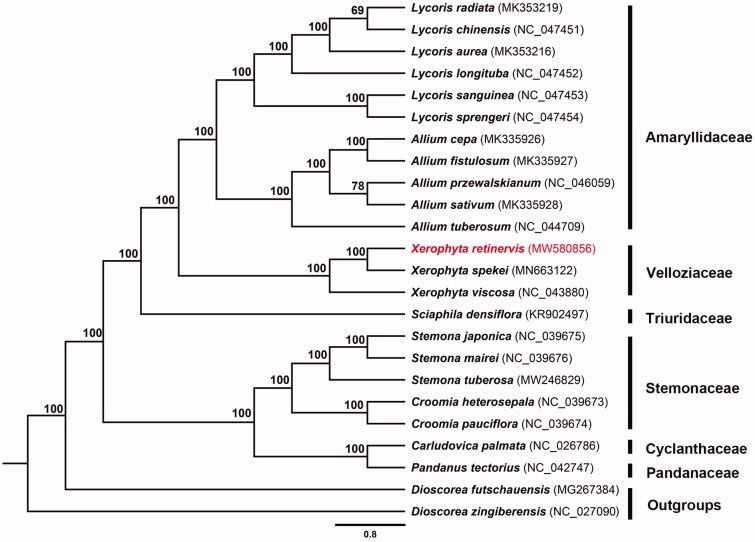
Based on a previous study (Wanga et al. 2019), we included 21 sequences from Pandanales and two sequences from Dioscoreaceae for phylogenetic analysis. Sequences were aligned via MAFFT v7.475 (Katoh and Standley 2013). The phylogenetic tree was reconstructed using maximum-likelihood (ML) method via IQ-Tree v1.6.10 (Nguyen et al. 2015) and visualized in Figtree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree). We found that X. retinervis was closely related to X. spekei Baker 1875 in Velloziaceae clade with strong bootstrap support (Figure 1).
Figure 1.
The maximum-likelihood phylogeny obtained from 24 complete chloroplast sequences.
Acknowledgments
The authors thank Dr. Bing Liu, Institute of Botany, Chinese Academy of Sciences for assistance in obtaining material. The authors also thank Dr. Jianjun Jin, Kunming Institute of Botany, Chinese Academy of Sciences for help and advice on plastid genomes assembly.
Funding Statement
This work was supported by Wild Plants Sharing and Service Platform of Sichuan Province and Vegetation Restoration Techniques for Hydropower Stations in Alpine Regions [HNKJ20-H23].
Ethical approval
The material used in this study is widely distributed in the field and does not belong to the IUCN Red List, the collection area is not a protected area. Moreover, this article was conducted in compliance with the regulations of Chengdu Institute of Biology, Chinese Academy of Sciences.
Disclosure statement
The authors declare there is no conflicts of interest and are responsible for the content.
Data availability statement
The data that support the findings of this study are openly available in GenBank number MW580856 (https://www.ncbi.nlm.nih.gov/nuccore/MW580856) and the related BioProject, raw sequencing files in SRA, and the Bio-Sample number are PRJNA810740, SRR18153545, and SAMN26278338 respectively.
Author contributions
BX and JYZ designed the study. ML and XL performed data analysis. JYZ drafted and BX revised the manuscript. All authors reviewed and approved the final manuscript.
References
- Baker JG. 1875. Synopsis of the African species of Xerophyta. J Bot. 13:231–236. [Google Scholar]
- Dyer RA. 1942. Vellozia retinervis. Fl Pl Africa. 22:856. [Google Scholar]
- Farrant JM. 2000. A comparison of mechanisms of dessication tolerance among three angiosperm resurrection plant species. Plant Ecol. 151(1):29–39. [Google Scholar]
- Gibbs RGE, Welman WGM, Retief E, Immelman KL, Germishuizen G, Pienaar BJ, Van Wyk M, Nicholas A.. 1987. List of species of southern African plants. Mem Bot Surv South Africa. 2(1–2):1. [Google Scholar]
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ.. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu XJ, Moore MJ, Li DZ, Yi TS.. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(50):50–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wyk BE, Van Oudtshoorn B, Gericke N.. 1997. Medicinal plants of South Africa. Pretoria: Briza Publications. [Google Scholar]
- Wanga VO, Dong X, Oulo MA, Munyao JN, Mkala EM, Kirika P, Gituru RW, Hu G-W.. 2019. The complete chloroplast genome sequence of Xerophyta spekei (Velloziaceae). Mitochondrial DNA B Resour. 5(1):100–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in GenBank number MW580856 (https://www.ncbi.nlm.nih.gov/nuccore/MW580856) and the related BioProject, raw sequencing files in SRA, and the Bio-Sample number are PRJNA810740, SRR18153545, and SAMN26278338 respectively.