Abstract
The complete chloroplast genome of an important medicinal plant, Convallaria majalis Linnaeus, was sequenced for the first time. The entire circular genome is 162,218 bp in length, with 37.9% GC contents. The genome has consisted of a large single-copy region (LSC) with a length of 85,417 bp, a small single-copy region (SSC) with a length of 18,495 bp, and two inverted repeat regions (IRs) with a length of 29,153 bp each. The genome harbored 133 genes, including 87 protein coding genes, 38 tRNA genes, and eight rRNA genes. The phylogenetic tree of 24 plant species was constructed based on the maximum-likelihood method. This study will provide theoretical basis for further study on plant genetics phylogenetic research.
Keywords: Asparagaceae, chloroplast genome, Convallaria majalis, phylogenetic analysis
Convallaria majalis Linnaeus (1753), a perennial herb of Asparagaceae, is widely distributed in Asia, Europe and North America, growing in wet places such as shady forest or ditch according to the record of Flora Reipublicae Popularis Sinicae (FRPS). It contains many cardiac glycoside components, which provide cardiotonic and diuretic effects. Furthermore, one study in 2014 showed that odorous components derived from C. majalis constituted more than 20% of perfume raw material market (Dörrich et al. 2014). High content of aromatic oil in C. majalis possesses sweet and elegant fragrance, making it widely used in the production of soap and cosmetics. In addition to its ornamental and pharmaceutical values, C. majalis is known as a toxic plant. Tissue factor expression induced by saponins and various cardiac glycosides in C. majalis contributes to the development of a hypercoagulable state, which often led to plant poisoning among children in Finland (Lamminpää and Kinos 1996; Morimoto et al. 2021). Current studies show that steroidal glycosides derived from C. majalis possessed cytotoxicity to human lung adenocarcinoma cells and thus can be a potential agent for anti-lung cancer (Matsuo et al. 2017).
According to the Regulations of the People’s Republic of China on Wild Plants Protection, C. majalis is not in the list of national key protection of wild plants. On-site and ex-situ protection of wild plants and scientific research on wild plants are supported in article five of the regulations. With the permission of Pharmacy College in Liaoning University of traditional Chinese Medicine, C. majalis was identified by professor Ting-guo Kang and transplanted in the University herbal garden (E 121°53′14″, N 39°4′12″). The voucher specimen and genomic DNA were deposited at the herbarium of Liaoning University of Chinese Medicine (Liang Xu 861364054@qq.com, C. majalis number: 10162210515067LY). All operations are carried out in accordance with guidelines in Specification on Good Agriculture and Collection Practices for Medicinal Plants (GACP; Number: T/CCCMHPIE 2.1-2018). The extraction of total genomic DNA from fresh leaves was achieved by Magbead Plant DNA Kit (CWBIO China) and sequenced on Illumina Novaseq 6000 platform. Data were edited and assembled by NGS QC toolkit (Patel and Jain 2012) and SPAdes v3.11.0 (Bankevich et al. 2012), respectively. The protein coding sequences of chloroplast (CP) genome were compared with NR protein databases for protein-coding gene prediction and annotation.
The chloroplast genome length of C. majalis was 162,218 bp. The genome harbored 133 genes, including 87 protein coding genes, 38 tRNA genes, and eight rRNA genes, with a GC content of 37.9%. The genome has consisted of a large single-copy region (LSC) with a length of 85,417 bp, a small single-copy region (SSC) with a length of 18,495 bp and two inverted repeat regions (IRs) with a length of 29,153 bp each. Additionally, we find that 15 genes, including trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC and ndhA, each of which contain one intron, clpP and ycf3 genes contain two introns, and rps12 gene has trans splicing.
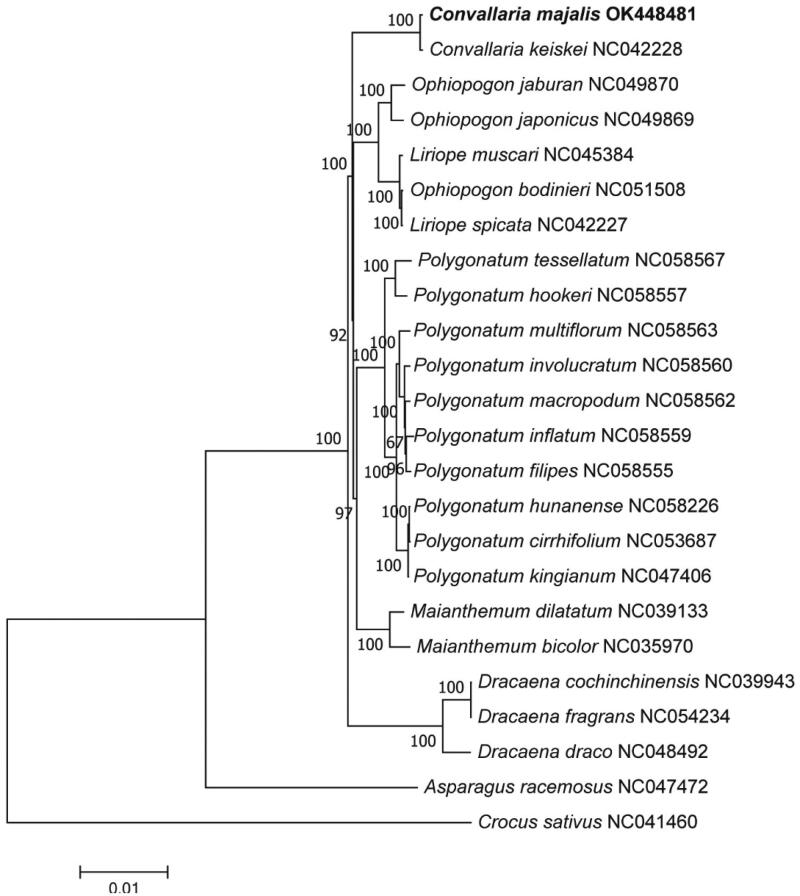
Twenty-three species of plant and the outgroup Crocus sativus were selected for complete CP genome phylogenetic analysis according to the Bayesian information criterion (BIC). Maximum likelihood (ML) tree was constructed with the model TVM + F+R3 (Figure 1) by IQ-TREE 1.6.12 software (Nguyen et al. 2015) (bootstrap value 1000). The phylogenetic tree shows that C. majalis and Convallaria keiskei are sister groups. Crocus sativus, as an outgroup, is far from the other species. The species of each genus are clearly distinguished, except for Ophiopogon bodinieri, which is closer to the genus Liriope, since Liriope spicata and Liriope muscari were once separated from the genus Ophiopogon.
Figure 1.
Maximum-likelihood (ML) phylogenetic tree based on the complete CP genome of C. majalis and 23 other species.
In conclusion, the complete CP genome of C. majalis was determined in this study, which provides theoretical foundation for further study on the phylogenetic relationship of Asparagaceae family.
Funding Statement
This research was funded by 2019 Liaoning Provincial Department of Education Scientific Research Project, China [No. L201942], National Key Research and Development in the 13th Five-Year Plan [No. 2018YFC1708200], Major Special Fund for Science and Technology of Inner Mongolia Autonomous Region [No. 2019ZD004], Natural Science Fund Project of Liaoning Province [No. 2020-MS-224] Mongolian Medicine R & D National Local Joint Engineering Research Center Open Fund Project, China [No. MDK2019047], and Liaoning BaiQianWan Talents Program.
Author contributions
W.X.M., Y.Y.S., C.B., and Y.P.X. carried out the sampling and analyses; T.G.K. and M.X. were involved in validation and supervision; All authors contributed to the design of the work, the analysis of data for the work and draft revising. All authors finally approved this version.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession NO. OK448481. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA769782, SRX12547454 and SAMN22169498, respectively.
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. . 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörrich S, Mahler C, Tacke R, Kraft P.. 2014. Synthesis and olfactory characterization of silicon-containing derivatives of the acyclic lily-of-the-valley odorant 5,7,7-trimethyl-4-methylideneoctanal. Chem Biodivers. 11(11):1675–1687. [DOI] [PubMed] [Google Scholar]
- Lamminpää A, Kinos M.. 1996. Plant poisonings in children. Hum Exp Toxicol. 15(3):245–249. [DOI] [PubMed] [Google Scholar]
- Matsuo Y, Shinoda D, Nakamaru A, Kamohara K, Sakagami H, Mimaki Y.. 2017. Steroidal Glycosides from Convallaria majalis Whole Plants and Their Cytotoxic Activity. IJMS. 18(11):2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto M, Tatsumi K, Yuui K, Terazawa I, Kudo R, Kasuda S.. 2021. Convallatoxin, the primary cardiac glycoside in lily of the valley (Convallaria majalis), induces tissue factor expression in endothelial cells. Vet Med Sci. 7(6):2440–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RK, Jain M.. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7(2):e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession NO. OK448481. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA769782, SRX12547454 and SAMN22169498, respectively.