Abstract
Recent studies have shown that point mutations in the dihydropteroate synthase (DHPS) gene of human-derived Pneumocystis carinii are related to exposure to sulfa drugs and possibly represent the emergence of sulfa resistance. We developed a simple single-strand conformation polymorphism (SSCP) method to permit rapid detection of these mutations. With plasmid constructs, SSCP was able to detect as little as 10% of a minority population. The SSCP assay was compared to direct sequencing for typing the DHPS gene by examining 37 clinical isolates with known DHPS sequences and 41 clinical isolates with unknown DHPS sequences. The typing results were consistent between these two methods for all isolates except 11 in which mutations were detected by SSCP but not by direct sequencing. Sequencing of individual clones after subcloning confirmed the presence of mutations in a minority population as determined by SSCP. SSCP is a very simple and sensitive method for rapid identification of P. camii DHPS mutations.
An increasing number of studies have demonstrated that mutations at amino acids 55 (Thr→Ala) and 57 (Pro→Ser) in the human-derived Pneumocystis carinii dihydropteroate synthase (DHPS) gene are associated with prior exposure to sulfa or dapsone, which target the DHPS (7, 10, 11, 13, 17; Q. Mei, S. Gurunathan, H. Masur, and J. A. Kovacs, Letter, Lancet 351:1631, 1998). These mutations likely represent emergence of sulfa resistance in P. carinii, since they are present in one of the active sites of the enzyme (1) and correlate with mutations that are shown to confer sulfa resistance in other organisms, such as Streptococcus pneumoniae (16), Plasmodium falciparum (4), and Mycobacterium leprae (9). Because no reliable culture system for human-derived P. carinii is currently available, direct proof of sulfa resistance in P. carinii isolates with DHPS mutations cannot be determined by traditional in vitro drug susceptibility testing. Evaluation of the relationship between these mutations and clinical resistance to sulfa drugs would be facilitated by a rapid method for identifying the mutations. To date, detection of the P. carinii DHPS mutations has relied on direct DNA sequencing of PCR-amplified products. Although sequencing has the advantage of high accuracy and possibly of identifying other mutations, it is not amenable to rapid screening of large numbers of samples due to its technical complexity and high cost. Furthermore, minority populations can be detected only if they represent 20 to 30% of the population.
Our goal in the present study was to develop a rapid, simple method for detecting these mutations in clinical isolates that not only could be utilized by other investigators but in the future could also be used by clinical microbiology laboratories. Single-strand conformation polymorphism (SSCP) is a relatively recently developed method for detecting mutations that relies upon the ability of one or more nucleotide changes to alter the electrophoretic mobility of single-stranded DNA molecules under nondenaturing conditions (18). Because of its technical simplicity and relatively high sensitivity, SSCP has become one of the most popular strategies for detection of genetic variations and mutations. Hauser et al. (6) have previously shown this method to be a promising option for molecular typing of human-derived P. carinii isolates. Here we describe a simple SSCP protocol using a compact electrophoresis unit, small gel format, and nonradioactive staining to detect the previously described point mutations (codons 55 and 57) in the DHPS gene of human-derived P. carinii.
(This work was presented in part at the 40th Interscience Conference on Antimicrobial Agents and Chemotherapy, Toronto, Ontario, Canada, 17 to 20 September 2000 [L. Ma and J. A. Kovacs, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1951, 2000].)
MATERIALS AND METHODS
Clinical P. carinii isolates and DNA extraction.
Two sets of clinical P. carinii isolates were used in this study. The first consisted of 37 isolates for which the P. carinii DHPS nucleotide sequences had been determined previously by direct sequencing and/or subcloning (17). The second set consisted of 41 isolates (from 14 sputum samples and 27 bronchoalveolar lavage fluid samples) which were obtained from patients diagnosed with P. carinii pneumonia between 1991 and 1995 but for which the DHPS sequence had not been previously determined. For the first set of isolates, genomic DNA was extracted by treatment with proteinase K followed by phenol-chloroform extraction as described previously (17). For the second set, genomic DNA was extracted either by that method or by use of the NucliSens isolation kit (Organon Teknika, Durham, N.C.).
Experimentation guidelines of the U.S. Department of Health and Human Services and the National Institutes of Health were followed in the conduct of this research.
PCR.
On the basis of the full-length human-derived P. carinii DHPS gene (17), we designed two primers, A2 (5′-TTACTCCTGATTCTTTTTTCGATGGG-3′) and PS308 (5′-GCCTTAATTGCTTGTTCTGCAACC-3′), which amplify a 259-bp fragment spanning the mutation sites (codons 55 and 57). For determination of the basic SSCP patterns of the human-derived P. carinii DHPS variants and for optimization of the assay conditions, we used plasmids containing wild-type or mutant (either single or double mutation) sequences of the human-derived P. carinii DHPS gene. The wild-type plasmid contained a DHPS sequence with nucleotides A and C at positions 163 and 169, respectively (17). In the single mutant plasmids, a G was at position 163 instead of A (resulting in amino acid change Thr-55→Ala-55) or a T was at position 169 instead of C (Pro-57→Ser-57), and in the double mutant plasmid, nucleotides G and T were present at positions 163 and 169 (Thr-55→Ala-55 and Pro-57→Ser-57, respectively). These plasmids were obtained by subcloning of the PCR products from several clinical isolates in our previous study (17) and in the present study. PCR amplification with plasmids was performed in a 50-μl reaction volume containing 10 ng of plasmid DNA, 0.4 μM (each) primers A2 and PS308, 0.2 mM deoxynucleoside triphosphates, 1× native Pfu buffer [20 mM Tris-HCl (pH 8.0), 10 mM KCl, 2 mM MgCl2, 6 mM (NH4)2SO4, 0.1% Triton X-100, and 10 μg of bovine serum albumin per ml), and 2.5 U of native Pfu DNA polymerase (Stratagene, La Jolla, Calif.). The thermal cycling conditions included an initial incubation at 95°C for 45 s, followed by 35 cycles of 95°C for 45 s, 60°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 10 min. PCR products were examined on a 3% agarose gel and then used in the SSCP assay described below.
To amplify the DHPS fragments from clinical isolates, a nested PCR protocol was performed using the previously described primer pair PK95 and PS876 (17) for the first round and the primer pair A2 and PS308 (described above) for the second round. All PCRs were carried out in a total volume of 50 μl. The first round of PCR was carried out with a touchdown protocol, which consisted of 10 cycles of 45 s at 94°C and 2 min at 65°C, with a decrease by 1°C every cycle to reach 55°C in the last cycle of the period, and 2 min at 72°C, followed by 25 cycles of 45 s at 94°C, 1 min at 55°C, and 2 min at 72°C. The thermal cycling parameters for the second round of PCR were the same as for the PCR with the plasmid DNA as described above. Each experiment included a negative control without template DNA and a positive control containing 100 ng of human-derived P. carinii genomic DNA. The amplified products from the second-round PCR were used in the SSCP assay after examination by 3% agarose electrophoresis.
SSCP.
The SSCP conditions were optimized using plasmids containing either wild-type or mutant human-derived P. carinii DHPS sequences and the GeneGel SSCP Starter Kit according to the instructions of the manufacturer (Amersham Pharmacia Biotech, San Francisco, Calif.). Four-microliter aliquots of PCR products were mixed with 4 μl of loading buffer containing 90% formamide, 0.025% (wt/vol) bromophenol blue, 0.025% (wt/vol) xylene cyanol, 3% (vol/vol) glycerol, and 0.5 μM (each) primers A2 and PS308. The addition of the primers has been shown to improve the resolution of the single-stranded DNA bands in the SSCP gel, presumably by preventing reannealing of denatured DNA fragments, thus decreasing the amount of double-stranded DNA and increasing the concentration of single-stranded DNA (2). This mixture was heated at 95°C for 5 min and then chilled in an ice water bath. Five microliters was loaded on a precast GeneGel SSCP gel (122 by 110 by 0.5 mm; Amersham Pharmacia Biotech). Electrophoresis was performed using the temperature-controlled GenePhor Electrophoresis System (Amersham Pharmacia Biotech) under the conditions recommended by the manufacturer. The gels were stained by using the PlusOne DNA Silver Staining Kit (Amersham Pharmacia Biotech).
DNA sequencing.
PCR products were purified by use of the StrataPrep PCR Purification Kit (Stratagene) and sequenced either by direct sequencing or after subcloning. Subcloning was performed using the PCR-Script Amp Cloning Kit (Stratagene) according to the manufacturer's instructions. Inserts were screened by PCR with primers A2 and PS308 followed by SSCP. Inserts which showed SSCP patterns different from the wild-type pattern were sequenced using universal or sequence-specific primers. DNA sequencing was carried out by the dideoxy chain termination reaction method using the ABI PRISM 377 automated DNA sequencer (Perkin-Elmer, Foster City, Calif.) as described previously (17). Nucleic acid sequences were analyzed using MacVector 6.5.3 software (Oxford Molecular Ltd., Oxford, England).
RESULTS
Optimization of assay conditions.
We initially performed the SSCP analysis using plasmids containing either wild-type or mutant sequences of the human-derived P. carinii DHPS gene and the GeneGel SSCP Starter Kit, and we examined the four key factors, i.e., fragment length, electrophoresis buffer, temperature, and gel matrix, which can affect the resolution of SSCP. Although PCR products ranging in size from 189 to 596 bp were evaluated, optimal separation of wild type (T-55 and P-57), single mutation (A-55 or S-57), and double mutation (A-55 and S-57) was obtained with a 259-bp fragment generated by primers A2 and PS308. Additionally, use of the GeneGel SSCP Clean gel, a running temperature of 12°C, running buffer C (pH 8.3), and a running voltage at 80 V for 20 min followed by 510 V for 140 min provided optimal conditions. Moreover, the addition of primers to PCR products before loading onto the gel increased the yield of single strands (data not shown), as has been described previously (2).
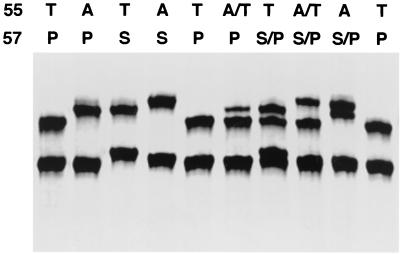
Under these optimal assay conditions, we could distinguish the SSCP patterns of different types of P. carinii DHPS sequences. As shown in Fig. 1, mobility differences among different genotypes can be seen in at least one strand. Compared to the wild type (T-55 and P-57), the single A-55 mutation and the double mutation (A-55 and S-57) cause mobility shifts in the upper band, and the single S-57 mutation causes a shift in both bands. The mobility shift of the upper band in the single A-55 mutation is different from that in the double mutation as clearly shown in the sample containing a mixture of these two types of mutation. In the samples containing a mixture of two different sequences, there are three or four bands corresponding to individual bands seen in the samples with one homozygous sequence. SSCP is capable of distinguishing different mixed forms, including the mixture of wild type and single A-55 mutant (A/T-55, P-57), the mixture of wild type and single S-57 mutant (T-55, S/P-57), the mixture of wild type and double mutant (A/T-55, S/P-57), and the mixture of single A-55 mutant and double mutant (A-55, S/P-57). Except for the mixed form of wild type and single S-57 mutant sequences, all forms shown have been detected in clinical samples in previous studies from our group (17) and/or others (3, 7, 10, 11).
FIG. 1.
SSCP patterns of the human-derived P. carinii DHPS gene. The templates used were plasmid DNAs containing different human-derived P. carinii DHPS sequences. Above each lane is the amino acid sequence at codons 55 and 57. The genotype A/T-55 plus S/P-57 represents a mixture of wild-type (T-55, P-57) and double mutant (A-55, S-57) sequences. The mobility difference is clearly seen in at least one strand among the different sequences.
Sensitivity of SSCP for detection of one allele in a mixture of two alleles.
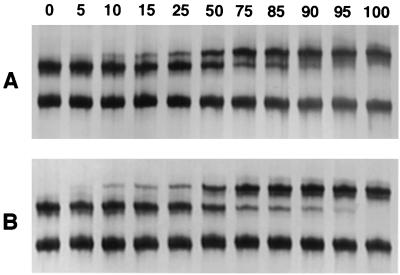
Because multiple alleles may be present in a single clinical isolate, we wanted to examine the ability of SSCP to detect a minority population. For these studies, plasmids containing wild-type or mutant (either a single mutation at codon 55 or double mutations at codons 55 and 57) P. carinii DHPS sequences were mixed, with mutant concentrations ranging from 5 to 95%, and evaluated by SSCP. Under the previously optimized conditions, SSCP was able to detect the minority population (either wild type or mutant) when it was present in as little as 10% of the total population (Fig. 2).
FIG. 2.
Sensitivity of SSCP for detection of minority populations of the P. carinii DHPS gene. Plasmids containing wild-type or mutant DHPS sequences were mixed in various concentrations, amplified by PCR, and analyzed by SSCP. The percentage of mutant DNA template is indicated above each lane. (A) Serial mixtures of wild-type (T-55, P-57) and single A-55 mutant sequences. (B) Serial mixtures of wild-type (T-55, P-57) and double mutant (A-55 and S-57) sequences.
Evaluation of clinical isolates by SSCP.
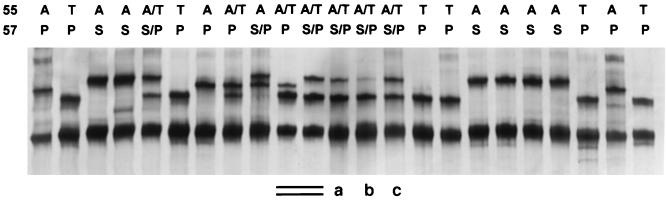
To validate the SSCP method for detection of DHPS mutations, we first examined 37 clinical isolates for which the DHPS sequences had been determined previously by direct sequencing (17). The results of one representative SSCP assay are shown in Fig. 3. The typing results of the SSCP assay were very consistent with those of direct sequencing (Table 1). However, in three samples the results of these two methods were discordant. By direct sequencing of the PCR product (without subcloning), two samples (samples a and b in Table 2) were determined to be wild type (T-55 and P-57) and one sample (sample c in Table 2) was a mixture of wild-type and single A-55 mutant (A/T-55 and P-57) sequences. Based on the SSCP patterns, all three samples (Fig. 3) appeared to be a mixture of wild-type and double mutant sequences (A/T-55 and S/P-57). To clarify the discrepant results in these three samples, we performed subcloning of the PCR products followed by sequencing of multiple clones per sample. As shown in Table 2, in all three samples the presence of double mutant clones as a minority population (9.5 to 36.4%) was verified, although the majority of the clones were wild type. These findings confirmed that the SSCP typing results were correct and suggested that SSCP is more sensitive than direct sequencing for detection of a minority population.
FIG. 3.
Representative SSCP analysis of P. carinii DHPS mutations in clinical isolates. Above each lane is the amino acid sequence at codons 55 and 57. The genotype A/T-55 plus S/P-57 represents a mixture of wild-type (T-55, P-57) and double mutant (A-55, S-57) sequences. Plasmids containing DHPS sequences were used as the standard control (double-underlined lanes). Samples a, b, and c showed discordant results between SSCP and direct sequencing but were confirmed by sequencing of individual clones to be mixtures of wild type and double mutation as determined by SSCP.
TABLE 1.
Comparison of SSCP assay and direct sequencing for detection of P. carinii DHPS mutations in 37 clinical isolates
| No. of isolates | DHPS codons 55 and 57a determined by:
|
|
|---|---|---|
| Direct sequencing | SSCP | |
| 19 | T-55, P-57 | T-55, P-57 |
| 2 | T-55, P-57 | A/T-55, S/P-57 |
| 1 | A/T-55, P-57 | A/T-55, S/P-57 |
| 3 | A/T-55, P-57 | A/T-55, P-57 |
| 2 | A-55, P-57 | A-55, P-57 |
| 7 | A-55, S-57 | A-55, S-57 |
| 2 | A-55, S/P-57 | A-55, S/P-57 |
| 1 | A/T-55, S/P-57 | A/T-55, S/P-57 |
Amino acids that differ from those in the wild-type sequence are underlined; the genotype A/T-55, S/P-57 represents a mixture of wild-type (T-55, P-57) and double mutant (A-55, S-57) sequences.
TABLE 2.
Subcloning of PCR products from five P. carinii isolates which showed different DHPS genotypes in direct sequencing and SSCP
| Sample | Codons determined bya:
|
No. (%) of wild-type clones (T-55, P-57) | No. (%) of mutant clonesa:
|
|||
|---|---|---|---|---|---|---|
| Direct sequencing | SSCP | A-55, S-57 | A-55, P-57 | T-55, S-57 | ||
| a | T-55, P-57 | A/T-55, S/P-57 | 14 (63.6) | 8 (36.4) | 0 | 0 |
| b | T-55, P-57 | A/T-55, S/P-57 | 19 (90.5) | 2 (9.5) | 0 | 0 |
| c | A/T-55, P-57 | A/T-55, S/P-57 | 11 (64.7) | 4 (23.5) | 1 (5.9) | 1 (5.9) |
| TH | A/T-55, P-57 | A/T-55, S/P-57 | 13 (86.7) | 2 (13.3) | 0 | 0 |
| KL | T-55, P-57 | A/T-55, S/P-57 | 10 (71.4) | 4 (28.6) | 0 | 0 |
Amino acids that differ from those in the wild-type sequence are underlined; the genotype A/T-55, S/P-57 represents a mixture of wild-type (T-55, P-57) and double mutant (A-55, S-57) sequences.
Notably, sample c contained four different DHPS sequences, including wild type, double mutation, and single mutation at codon 55 or 57. The single mutation at either codon 55 or 57 was not detected by SSCP or direct sequencing, presumably because each of them accounted for only 5.9% of the total population and were beyond the sensitivity of these two methods.
Following analysis of the 37 clinical samples described above, an additional 41 samples with unknown DHPS sequence were examined by the SSCP assay. Based on the SSCP patterns, 22 samples had wild-type sequence, 17 samples contained either a homozygous or heterozygous mutation(s) at codon 55 alone or both codons 55 and 57, and 2 samples showed unique SSCP patterns not previously identified. To confirm the SSCP typing results, direct sequencing was performed on 7 samples with a wild-type SSCP pattern and all 19 samples showing patterns different from those of wild-type samples. As shown in Table 3, the SSCP typing results were verified by direct sequencing in most samples. However, six samples, which were determined by SSCP to be mixed forms of wild-type and mutant sequences, either at codon 55 alone (three samples) or at both codons 55 and 57 (three samples), were pure wild type by direct sequencing, and two samples, which were determined to be a mixture of wild-type and double mutant sequences by SSCP, were a mixture of wild-type and single A-55 mutant sequences by direct sequencing. To determine which was correct, we performed subcloning of the PCR products from two samples (samples TH and KL in Table 2) which showed inconsistent results between SSCP and direct sequencing. Subsequent sequencing of multiple individual clones confirmed the presence of both wild-type and double mutant sequences in each sample, as determined by SSCP.
TABLE 3.
SSCP analysis of P. carinii DHPS mutations in 41 clinical isolates
| No. of isolates | DHPS codons 55 and 57a determined by:
|
|
|---|---|---|
| SSCP | Direct sequencing | |
| 24b | T-55, P-57 | T-55, P-57 (9)c |
| 2 | A-55, P-57 | A-55, P-57 |
| 3 | A-55, S-57 | A-55, S-57 |
| 2 | A/T-55, P-57 | A/T-55, P-57 |
| 3 | A/T-55, P-57 | T-55, P-57 |
| 2 | A/T-55, S/P-57 | A/T-55, S/P-57 |
| 2 | A/T-55, S/P-57 | A/T-55, P-57 |
| 3 | A/T-55, S/P-57 | T-55, P-57 |
Amino acids that differ from those in the wild-type sequence are underlined; the genotype A/T-55, S/P-57 represents a mixture of wild-type (T-55, P-57) and double mutant (A-55, S-57) sequences.
Including two isolates containing a synonymous change at codon 51 or 73, respectively.
Only nine isolates were examined by direct sequencing.
For the two samples with unique SSCP patterns, direct sequencing showed that both samples had wild-type sequence at both codons 55 and 57 but contained novel synonymous nucleotide changes at other codons that did not result in a change in the predicted amino acid. One sample showed a mixture of nucleotides T and C at position 153 (codon 51), and the other had a mixture of nucleotides T and C at position 219 (codon 73). The latter sample was further examined by subcloning of the PCR product, and we obtained six clones containing T and six clones containing C at nucleotide 219.
DISCUSSION
This study has shown that single or double mutations in the DHPS gene can be easily detected by SSCP, even when they represent as little as 10% of the population. The reliability of SSCP for detection of the DHPS mutations was confirmed by examining 78 clinical isolates of human-derived P. carinii. The results from the present study suggest that the SSCP assay we have developed will prove to be highly useful for detection of the DHPS mutations in clinical P. carinii isolates.
The system we have described has advantages in simplicity and speed, thus permitting rapid detection of DHPS mutations. In this study, we used the small precast gel (12.2 by 11 cm) and semidry running system, which requires no liquid running buffer and allows fast and easy electrophoresis set-up and clean-up. For sample processing, the only step after PCR is a heat denaturation in formamide. For detection of the DNA bands, we used silver staining instead of radioactive labeling. The silver staining method is easy, fast, and highly sensitive. The total time needed for performing this assay starting from PCR was about 10 h, including 6 h for nested PCR, 2.5 h for electrophoresis, and 1.5 h for silver staining.
Another major advantage of the SSCP method is its high sensitivity. Under optimized conditions, a mixed population was detected when the minority sequence represented as little as 10% of the total population. When clinical isolates were studied, SSCP detected heterozygous sequences in 11 isolates, which were not detected by direct sequencing but were documented by sequencing of individual clones after subcloning. These findings indicate that the SSCP assay has higher sensitivity than direct sequencing. In automated direct sequencing with fluorescent terminators, a heterozygous sequence is identified as a mixture of two signals at one position, and it is sometimes difficult to differentiate a weak signal from background noise. More recently, another method based on restriction fragment length polymorphism analysis has been shown to be useful for detection of the P. carinii DHPS mutations (8). However, in preliminary experiments evaluating this methodology, we found that, like direct sequencing, this method has low sensitivity for detection of a minority population.
The ability to detect a minor population in mixed populations will be important to help understand the prevalence as well as the kinetics of the development of DHPS mutations in patients. There have been a number of reports demonstrating a high rate of coinfection with multiple P. carinii strains in clinical isolates. Studies using DNA sequencing-based methods for typing human-derived P. carinii isolates revealed that coinfection rates are seen in 10 to 30% of isolates at several different genetic loci (12, 14, 15). A much higher coinfection rate (69%) has been reported with SSCP analysis of similar loci (5). The reported coinfection rates at the DHPS locus varied from 1 to 11% in studies utilizing direct sequencing of the PCR products (3, 7, 10, 11, 17). The relatively low sensitivity of direct sequencing as shown in the present study suggests that the coinfection rate has been substantially underestimated. In the present study, the coinfection rate determined by SSCP assay was 28% (21 of 78).
Another advantage of the method is its high reproducibility. Once the optimal conditions have been established, this method gives reproducible SSCP patterns. Since all of the materials, including the electrophoresis unit, the gel, and the buffer are commercially available, the results should be reproducible in different laboratories.
SSCP does have disadvantages compared to sequencing. First, this method is unable to detect other mutations that have been reported at codons 23, 111, and 248 (13), because they are located outside the region being investigated. Second, in a very few cases representing novel nucleotide changes, the SSCP patterns may be difficult to interpret. In this situation, sequencing is needed to identify the base change. In the present study, novel nucleotide changes were detected at positions 153 and 219 in two clinical samples. These changes are synonymous and do not result in amino acid changes; thus, it is unlikely that they represent a response to antibiotic pressure. Third, there is a need to perform nested PCR before SSCP, since less than half of clinical samples are positive after one round of PCR. Nested PCR is time-consuming, taking about 6 h. However, nested PCR is also needed for other mutation detection methods, including DNA sequencing.
In conclusion, the SSCP assay we have developed is a very simple and highly sensitive method for rapid identification of P. carnii DHPS mutations.
ACKNOWLEDGMENTS
We thank Philippe Hauser of the Centre Hospitalier Universitaire Vaudois, Division Autonome de Medecine Preventive Hospitaliere, Lausanne, Switzerland, for advice on SSCP methodology.
REFERENCES
- 1.Achari A, Somers D O, Champness J N, Bryant P K, Rosemond J, Stammers D K. Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nat Struct Biol. 1997;4:490–497. doi: 10.1038/nsb0697-490. [DOI] [PubMed] [Google Scholar]
- 2.Almeida T A, Cabrera V M, Miranda J G. Improved detection and characterization of mutations by primer addition in nonradioisotopic SSCP and direct PCR sequencing. BioTechniques. 1998;24:220–221. doi: 10.2144/98242bm10. [DOI] [PubMed] [Google Scholar]
- 3.Beard C B, Carter J L, Keely S P, Huang L, Pieniazek N J, Moura I N, Roberts J M, Hightower A W, Bens M S, Freeman A R, Lee S, Stringer J R, Duchin J S, del Rio C, Rimland D, Baughman R P, Levy D A, Dietz V J, Simon P, Navin T R. Genetic variation in Pneumocystis carinii isolates from different geographic regions: implications for transmission. Emerg Infect Dis. 2000;6:265–272. doi: 10.3201/eid0603.000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks D R, Wang P, Read M, Watkins W M, Sims P F, Hyde J E. Sequence variation of the hydroxymethyldihydropterin pyrophosphokinase: dihydropteroate synthase gene in lines of the human malaria parasite, Plasmodium falciparum, with differing resistance to sulfadoxine. Eur J Biochem. 1994;224:397–405. doi: 10.1111/j.1432-1033.1994.00397.x. [DOI] [PubMed] [Google Scholar]
- 5.Hauser P M, Blanc D S, Bille J, Francioli P. Typing methods to approach Pneumocystis carinii genetic heterogeneity. FEMS Immunol Med Microbiol. 1998;22:27–35. doi: 10.1111/j.1574-695X.1998.tb01184.x. [DOI] [PubMed] [Google Scholar]
- 6.Hauser P M, Francioli P, Bille J, Telenti A, Blanc D S. Typing of Pneumocystis carinii f. sp. hominis by single-strand conformation polymorphism of four genomic regions. J Clin Microbiol. 1997;35:3086–3091. doi: 10.1128/jcm.35.12.3086-3091.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helweg-Larsen J, Benfield T L, Eugen-Olsen J, Lundgren J D, Lundgren B. Effects of mutations in Pneumocystis carinii dihydropteroate synthase gene on outcome of AIDS-associated P. carinii pneumonia. Lancet. 1999;354:1347–1351. doi: 10.1016/S0140-6736(99)03320-6. [DOI] [PubMed] [Google Scholar]
- 8.Helweg-Larsen J, Eugen-Olsen J, Lundgren B. Rapid detection of dihydropteroate polymorphism in AIDS-related Pneumocystis carinii pneumonia by restriction fragment length polymorphism. Scand J Infect Dis. 2000;32:481–483. doi: 10.1080/003655400458730. [DOI] [PubMed] [Google Scholar]
- 9.Kai M, Matsuoka M, Nakata N, Maeda S, Gidoh M, Maeda Y, Hashimoto K, Kobayashi K, Kashiwabara Y. Diaminodiphenylsulfone resistance of Mycobacterium leprae due to mutations in the dihydropteroate synthase gene. FEMS Microbiol Lett. 1999;177:231–235. doi: 10.1111/j.1574-6968.1999.tb13737.x. [DOI] [PubMed] [Google Scholar]
- 10.Kazanjian P, Armstrong W, Hossler P A, Burman W, Richardson J, Lee C H, Crane L, Katz J, Meshnick S R. Pneumocystis carinii mutations are associated with duration of sulfa or sulfone prophylaxis exposure in AIDS patients. J Infect Dis. 2000;182:551–557. doi: 10.1086/315719. [DOI] [PubMed] [Google Scholar]
- 11.Kazanjian P, Locke A B, Hossler P A, Lane B R, Bartlett M S, Smith J W, Cannon M, Meshnick S R. Pneumocystis carinii mutations associated with sulfa and sulfone prophylaxis failures in AIDS patients. AIDS. 1998;12:873–878. doi: 10.1097/00002030-199808000-00009. [DOI] [PubMed] [Google Scholar]
- 12.Keely S P, Stringer J R, Baughman R P, Linke M J, Walzer P D, Smulian A G. Genetic variation among Pneumocystis carinii hominis isolates in recurrent pneumocystosis. J Infect Dis. 1995;172:595–598. doi: 10.1093/infdis/172.2.595. [DOI] [PubMed] [Google Scholar]
- 13.Lane B R, Ast J C, Hossler P A, Mindell D P, Bartlett M S, Smith J W, Meshnick S R. Dihydropteroate synthase polymorphisms in Pneumocystis carinii. J Infect Dis. 1997;175:482–485. doi: 10.1093/infdis/175.2.482. [DOI] [PubMed] [Google Scholar]
- 14.Latouche S, Ortona E, Mazars E, Margutti P, Tamburrini E, Siracusano A, Guyot K, Nigou M, Roux P. Biodiversity of Pneumocystis carinii hominis: typing with different DNA regions. J Clin Microbiol. 1997;35:383–387. doi: 10.1128/jcm.35.2.383-387.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee C H, Helweg-Larsen J, Tang X, Jin S, Li B, Bartlett M S, Lu J J, Lundgren B, Lundgren J D, Olsson M, Lucas S B, Roux P, Cargnel A, Atzori C, Matos O, Smith J W. Update on Pneumocystis carinii f. sp. hominis typing based on nucleotide sequence variations in internal transcribed spacer regions of rRNA genes. J Clin Microbiol. 1998;36:734–741. doi: 10.1128/jcm.36.3.734-741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez P, Espinosa M, Greenberg B, Lacks S A. Sulfonamide resistance in Streptococcus pneumoniae: DNA sequence of the gene encoding dihydropteroate synthase and characterization of the enzyme. J Bacteriol. 1987;169:4320–4326. doi: 10.1128/jb.169.9.4320-4326.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma L, Borio L, Masur H, Kovacs J A. Pneumocystis carinii dihydropteroate synthase but not dihydrofolate reductase gene mutations correlate with prior trimethoprim-sulfamethoxazole or dapsone use. J Infect Dis. 1999;180:1969–1978. doi: 10.1086/315148. [DOI] [PubMed] [Google Scholar]
- 18.Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci USA. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]