ABSTRACT
Failed recognition and clearance of damaged mitochondria contributes to memory loss as well as Aβ and MAPT/Tau pathologies in Alzheimer disease (AD), for which there is an unmet therapeutic need. Restoring mitophagy to eliminate damaged mitochondria could abrogate metabolic dysfunction, neurodegeneration and may subsequently inhibit or slow down cognitive decline in AD models. We have developed a high-throughput machine-learning approach combined with a cross-species screening platform to discover novel mitophagy-inducing compounds from a natural product library and further experimentally validated the potential candidates. Two lead compounds, kaempferol and rhapontigenin, induce neuronal mitophagy and reduce Aβ and MAPT/Tau pathologies in a PINK1-dependent manner in both C. elegans and mouse models of AD. Our combinational approach provides a fast, cost-effective, and highly accurate method for identification of potent mitophagy inducers to maintain brain health.
KEYWORDS: Aging, Alzheimer’s disease, autophagy, machine learning, mitophagy
Drug development is a costly, time-consuming, and uncertain process. It has been estimated that research and development (R&D) for a single new drug costs approximately $1-2 billion per drug and involves timelines that typically run between 10–20 years. Even so, only 12% of drugs making it to clinical trials can be approved by the FDA in the end. Artificial intelligence (AI) has emerged as a cost-effective and fast approach for the identification of novel compounds and is especially useful for new drug development. With increasingly impressive developments in artificial intelligence and accompanying increases in computational power, AI will effect an improvement in the efficiency of compound screening, disease model establishment, new target discovery, lead compound discovery, and lead drug optimization, among other tasks. Further combinations with cross-species wet lab validation, such as the use of mammalian cells, nematodes, and mice will provide biological validation and assist with elimination of “false positive candidates”.
Alzheimer disease (AD) is the most common form of dementia. Approximately 50 million people are suffering from dementia, where 70% of the afflicted have AD. Over 250 AD drug candidates have been developed during the last 15 years, but almost all of them failed. Recently, it has been proposed that the clearance capacity of impaired mitochondria via mitophagy, a selective form of macroautophagy, influences AD progression. Restoring mitophagy using genetic and/or pharmacological methods can inhibit the development and propagation of AD pathology in preclinical models. However, robust neuronal mitophagy inducers that work on mechanisms other than those that induce mitochondrial damage are sparse. We thus developed a workflow combining machine learning (a subdiscipline of AI, dry lab) and a cross-species platform (wet lab) to rapidly, accurately, and cost-effectively identify mitophagy inducers [1].
To create the screening technology, molecular representation approaches including Mol2vec, pharmacophore fingerprint, and 3D conformers fingerprint, were used for modeling. We started by creating a large pre-training dataset using compounds retrieved from the ChEMBL and ZINC natural products databases, with 19.9 million potential compounds included after filtering for relevance. SMILES representations were created for these compounds using RDKit and Word2vec. Pharmacore (2D) and conformers (3D) fingerprints were generated, and the resulting dataset used for training the natural language processing model. Structural distances between the targets (known mitophagy inducers) and each of the compounds in the database were calculated. The trained model was used to filter a dataset consisting of 3,274 plant-based natural products used in traditional Chinese medicine. Similarity scores were calculated, and the top compounds were selected. From these 3,274 compounds, 18 scored over the cutoff threshold for similarity and were selected for further study. Although 18 may seem like a small number when compared to the original 3,274 possibilities, this method was able to increase the success rate from 0.01%-10.00% to 44% with 8 of the 18 compounds showing mitophagy induction capacity at 10 µM in HeLa-mt-Keima cells (Figure 1).
Figure 1.
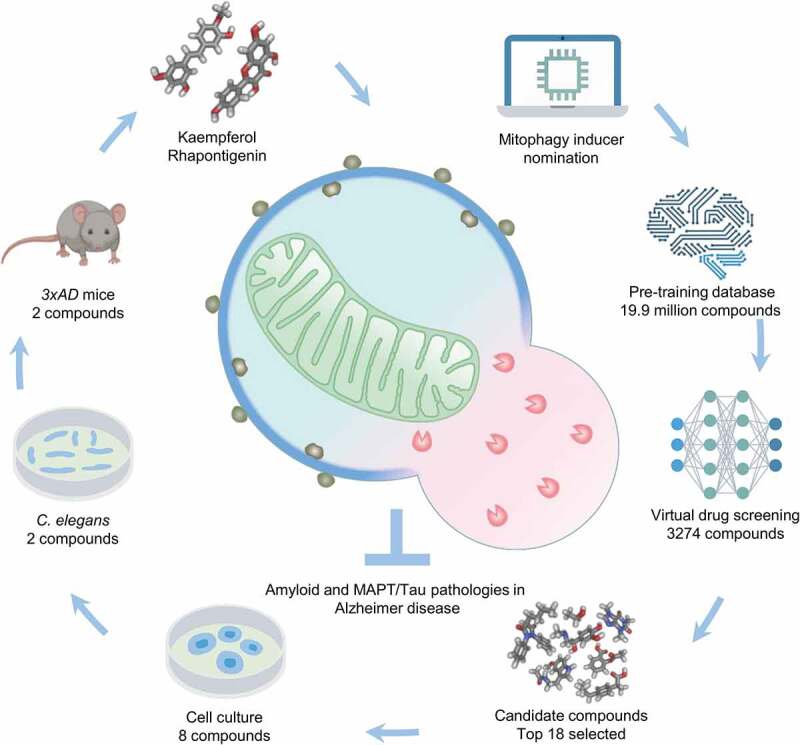
A summary of the workflow. The workflow comprises chemical structure nomination, pre-training model set-up, virtual screening, top scoring compound selection, and biological evaluation in vitro and in vivo. “#” denotes the numbers of compounds left in each step.
In order to validate these in silico data, we performed in vitro studies in cells and in vivo studies in nematodes and mice. In the first round using HeLa cells, 8 molecules from among the 18 AI top-ranked compounds were verified to induce mitophagy at 10 μM (quercetin [Macau library ID: T-2174], quercetin dihydrate [T-6630], tacrolimus [T-2144], ascomycin [T-2481], isorhamnetin [T-2836], pinostilbene [T-3755], kaempferol [Kaem, T-2177], and rhapontigenin [Rhap, T-3776]). In a second-pass study, pan-neuronal expression of mitochondria-targeted Rosella (mt-Rosella, a dual color-emission biosensor) transgenic nematodes were used to detect neuronal mitophagy in vivo. In total, three compounds (quercetin, Kaempferol, Rhapontigenin) were validated as mitophagy stimulating molecules; however only Kaem and Rhap improved memory in the AD nematodes at either of the doses (0.2 mM, 1 mM) used. Kaeml and Rhap were finally chosen to validate their capacities to retain memory in the 3xTg AD mice. These two compounds can restore memory deficit and ameliorate pathologies and were our lead compounds. Mechanistically, Kaem (and similarly Rhap) induces mitophagy via upregulation of key mitophagy/autophagy-related proteins, such as PINK1, PRKN/parkin, p-DNM1L/DRP1 (Ser616), BECN1, AMBRA1, and LC3B-II. Molecular docking suggests Kaem and Rhap can bind PINK1 (unpublished data), supporting the evidence that PINK1 knockdown almost completely annuls the anti-MAPT/Tau pathology effect of the two lead compounds. A summary of our workflow is shown in Figure 1.
In total, two robust mitophagy inducers were successfully identified through the combination of machine learning-based virtual screening and cross-species platform-supported wet lab validation. This illustrates that the combination of AI and preclinical drug research can indeed improve the efficiency of lead compound screening, reducing the time and labor consumption, and greatly speed up the development of novel treatments for AD.
Acknowledgments
We acknowledge contributions from the other co-authors in this project. We are grateful for the support of HELSE SØR-ØST (#2017056, #2020001, #2021021), the Research Council of Norway (#262175), the National Natural Science Foundation of China (#81971327), Akershus University Hospital (#269901, #261973), the Civitan Norges Forskningsfond for Alzheimers sykdom (#281931), the Czech Republic-Norway KAPPA programme (with Martin Vyhnálek, #TO01000215), and the Rosa sløyfe/Norwegian Cancer Society & Norwegian Breast Cancer Society (#207819) to E.F.F. We thank the Science and Technology Development Fund, Macau SAR (Grants No. 0128/2019/A3, 024/2017/AMJ), the University of Macau grants (Grants No. MYRG2019-00129-ICMS) awarded to J.H.L. R.A. were funded by the China Scholarship Council (http://www.csc.edu.cn/). We thank Thale Dawn Patrick-Brown for reading of the paper and Dr. Feixiong Cheng (Cleveland Clinic) for figure editing. Some of the cartons used in Figure 1 were from BioRender.
Funding Statement
This work was supported by the macau government [Grants No. 0128/2019/A3, 024/2017/AMJ].
Disclosure statement
E.F.F. has a CRADA arrangement with ChromaDex (USA) and is consultant to Aladdin Healthcare Technologies (UK and Germany), the Vancouver Dementia Prevention Centre (Canada), Intellectual Labs (Norway), and MindRank AI (China).
Reference
- [1].Xie C, Zhuang -X-X, Niu Z, et al. Amelioration of Alzheimer’s disease pathology by mitophagy inducers identified via machine learning and a cross-species workflow. Nat Biomed Eng. 2022;6:76–93. [DOI] [PMC free article] [PubMed] [Google Scholar]


