ABSTRACT
Recent rodent microbiome experiments suggest that besides Akkermansia, Parasutterella sp. are important in type 2 diabetes and obesity development. In the present translational human study, we aimed to characterize Parasutterella in our European cross-sectional FoCus cohort (n = 1,544) followed by validation of the major results in an independent Canadian cohort (n = 438). In addition, we examined Parasutterella abundance in response to a weight loss intervention (n = 55). Parasutterella was positively associated with BMI and type 2 diabetes independently of the reduced microbiome α/β diversity and low-grade inflammation commonly found in obesity. Nutritional analysis revealed a positive association with the dietary intake of carbohydrates but not with fat or protein consumption. Out of 126 serum metabolites differentially detectable by untargeted HPLC-based MS-metabolomics, L-cysteine showed the strongest reduction in subjects with high Parasutterella abundance. This is of interest, since Parasutterella is a known high L-cysteine consumer and L-cysteine is known to improve blood glucose levels in rodents. Furthermore, metabolic network enrichment analysis identified an association of high Parasutterella abundance with the activation of the human fatty acid biosynthesis pathway suggesting a mechanism for body weight gain. This is supported by a significant reduction of the Parasutterella abundance during our weight loss intervention. Together, these data indicate a role for Parasutterella in human type 2 diabetes and obesity, whereby the link to L-cysteine might be relevant in type 2 diabetes development and the link to the fatty acid biosynthesis pathway for body weight gain in response to a carbohydrate-rich diet in obesity development.
KEYWORDS: Gut microbiome, L-cysteine, obesity, Parasutterella, fatty acid biosynthesis
Introduction
Scientific evidence indicates that obesity and obesity-related comorbidities such as type 2 diabetes and cardiovascular diseases are associated with a dysbiosis of the gut microbiome.1 Several studies of the last decade imply that environmental factors influence the composition of the gut microbiome2 and that the gut microbiome plays a crucial role in the regulation of host metabolism via specific host-microbiome interactions.1
Mechanistically, it is thought that a significant elevation in host metabolism-related microbial communities might be associated with an increased capacity to harvest energy from the diet.3 In addition, it is now known that an obesity-associated microbiome can alter multiple host factors including inflammation, intestinal permeability, hormonal regulation, bile acids, and lipid metabolism. In the past, nutritional components were thought to be predominant factors inducing alterations of the immune system in obesity (e.g. unsaturated fatty acids4), while current evidence highlights the gut microbiome as a key mediator of metabolic inflammation.5
Several studies investigated the relation of gut microbial composition at the phylum level to metabolic parameters and dietary components.6,7 Recently, however, several pilot projects have targeted specific microbial species, like Akkermansia muciniphila, which have been shown to be independently associated with beneficial health traits in obese subjects.8–10
In the present study, we followed this more specific approach by focusing on Parasutterella sp. at the genus level (and Parasutterella excrementihominis at the species level) which belong to the family of Sutterellaceae, the order of Burkholderiales, the class of Betaproteobacteria, and the phylum of Proteobacteria.11 This microbe is linked to metabolic abnormalities in rodents12 and responds to a high fat diet (HFD) intervention in obesity-prone mice.13 In addition, recent evidence suggests Parasutterella is involved in the mediation of ω3-fatty acid effects on host physiology.14 From a mechanistic point of view, Parasutterella is associated with both, inflammatory reactions in the intestinal mucosa15 and systemic metabolic abnormalities12 which might suggest a role in the development of systemic low-grade metabolic inflammation due to dysbiosis. In fact, in a recent MRI (magnetic resonance imaging) study by our group, we found Parasutterella associated with hypothalamic inflammation in obese humans6 which is thought to interfere with appetite- and satiety regulation contributing to the development of obesity.
Since most published data are on rodent models, the aim of this present, explorative study was to characterize Parasutterella more deeply with respect to normal human physiology as well as human metabolic and chronic inflammatory diseases and nutrient intake in order to understand the development of obesity and to identify novel therapeutic approaches for future treatments.16 For this purpose, we analyzed 16s amplicon microbiome data in 1,544 subjects of our large FoCus cohort in Kiel, Germany.17 The subjects were extensively geno- and phenotyped. In addition, HPLC metabolomics and detailed dietary phenotyping were performed (EPIC protocol18) and analyzed with regard to Parasutterella in the form of metabolic pathway over-representation analysis and microbial metabolic networks. In a previous analysis using the FoCus cohort, we found the overall gut microbiota composition to be associated with single nucleotide polymorphisms (SNPs) in the human vitamin D receptor (VDR) gene locus and the Proopiomelanocortin (POMC) gene locus. Therefore, we aimed to analyze as well if Parasutterella abundance is determined by these two human genetic factors.
To increase reliability and interpretability, we also performed a human intervention study in order to show that Parasutterella is not only associated with an obesity phenotype but also responds to a weight loss therapy indicating a potential functional relevance. In addition, in the analysis of the intervention study, we aimed to quantify Parasutterella abundance levels by qPCR not to rely solely on 16S rRNA data.
Research design and methods
Study cohorts and study design
Cross-sectional cohort (FoCus cohort)
The present investigation included n = 1,544 subjects (63% females) of the northern German Food Chain Plus (FoCus) cohort (established from 2011 to 2015), which has been previously reported.19 Subjects were stratified into five groups according to BMI and the presence of type 2 diabetes (Table 1). 422 subjects were enrolled from the obesity outpatient clinic of the Department of Internal Medicine I of Kiel University. The remaining 1,122 subjects were recruited from regional registration offices as cross-sectional controls. Fasted serum and -stool samples as well as anthropometric values were collected at the study center. In addition to these measurements, subjects completed a 12-month retrospective food frequency questionnaire used by the European Prospective Investigation into Cancer Nutrition (EPIC) study. In addition, for validation purpose in 10% of the study probands, an additional 24-h nutrition profile was obtained by a phone interview. The median age of the whole cohort was 52 ± 14.2 years. The study population was generally overweight in Body Mass Index (BMI 27.8 kg/m2), with median height being 1.72 m and the median weight being 85 kg. Metabolic parameters including HOMA-IR index indicated low level of insulin resistance (2.42). The fasting glucose level was 95 mg/dL. Serum triglyceride levels appeared to be within a normal range of 108 mg/dL. Inflammatory parameters such as Interleukin-6 (IL-6) (3.7 pg/mL) and C-reactive protein (CRP) (3.3 mg/L) were within the normal range in the study population. This FoCus subset had prevalence for diabetes of 14.3%, whereas 11.9% of the whole subset was affected by type 2 diabetes.
Table 1.
Descriptive statistics of the three cohorts included in the study showing variables such as age, sex, and BMI. Further division of the cohorts was based on five groups: underweighted, normal weighted, overweight, obese with T2D, and obese without T2D
| |
FoCus cohort (n = 1,544) |
ATP cohort (n = 438) |
Intervention cohort (n = 55) |
|---|---|---|---|
| Parameter | Median (25th and 75th percentile) or Mean ± |
Median (25th and 75th percentile) or Mean ± |
Median (25th and 75th percentile) or Mean ± |
| Age (years) | 51.62 ± 14.22 | 56.9 ± 6.27 | 45.79 ± 10.89 |
| Sex (% female) | 63 | 72 | 69 |
| BMI (kg/m2) | 27.8 (23.68; 35.9) | 30.62 (26.55; 36.88) | 45.25 (43.36; 48.13) |
| Underweighted (<20 kg/m2) (%) | 4.5 | - | - |
| Normal weighted (20–25 kg/m2) (%) | 28.6 | 2.9 | - |
| Overweighted (25–30 kg/m2) (%) | 27 | 46 | - |
| Obese (>30 kg/m2) with type 2 diabetes (%) | 10.6 | 5.2 | 17.3 |
| Obese (>30 kg/m2) without type 2 diabetes (%) | 29.2 | 45.8 | 82.7 |
The study was conducted according to the guidelines laid down in the Declaration of Helsinki and was approved by the ethic committee of the Medical Faculty of the University of Kiel (Germany). All subjects gave their informed consent for study participation and data usage.
Validation cohort
To validate the FoCus results, a study population of n = 438 subjects from Alberta’s Tomorrow Project (ATP), a cross-sectional cohort from Alberta Canada was included for comparison20 (Table 1). These included 242 healthy controls (with a BMI between 20 and 25 kg/m2), 120 subjects with cardiovascular disease, 36 with chronic metabolic disease, and 44 with chronic inflammatory disease. Fecal samples from all n = 438 ATP subjects were available for the present analysis and 16S rRNA gene sequencing was performed in the same laboratory and using the same methodology as for the FoCus samples.
Intervention cohort
For validation of 16S rRNA gene sequencing data, we performed quantitative polymerase chain reaction (qPCR) for Parasutterella excrementihominis of n = 55 patients of an intervention cohort described elsewhere21 (Table 1). In brief, this group of severely overweight patients underwent a dietary intervention that consisted of a very low-calorie formula diet for the duration of 12 weeks, followed by a stabilization period of another 14 weeks. It is important to mention that the formula diet used was enriched in fish oil according to the EU regulations.
Patients received extensive medical, dietary, and psychological monitoring during the intervention and gave blood and stool samples at the beginning, mid-point, and end of the study. All patients of the intervention study gave their informed consent.
16S rRNA gene sequencing of the FoCus cohort
Stool samples of the study populations were immediately stored at −80°C and were passed forward to the Institute of Clinical Molecular Biology (IKMB) in Kiel (Germany) for microbiome sequencing. 16S rRNA gene sequencing was performed as explained previously.22 Bioinformatic analysis was based on amplicon sequence variants (ASVs) in R. For the FoCus cohort, the median sequencing depth was 36,048 reads, with an IQR from 23,444 to 52,786 reads. The sequencing depth of the Canadian ATP cohort was slightly lower with a median of 22,931 and an IQR of 18,301–28,758. Samples of the FoCus and ATP cohorts were sequenced in the same lab, on the same platform. Samples with a sequencing depth below 10,000 were removed as part of the quality control offered by the sequencing lab. Complete sets of genetic, food questionnaire (EPIC), and microbiome data were available for n = 1,443 individuals.
Metabolomics sample preparation in the FoCus cohort
Serum samples were thawed on ice and extracted by a modified SIMPLEX approach according to Matyash et al.23 From 100 µL blood samples, a lipophilic methyl-tert-butyl ether (MTBE) phase, a hydrophilic methanol-water phase, and a protein pellet were obtained; dried under vacuum (speed-vac from Thermo Fisher, Germany); and resuspended with the following solvent: the lipophilic phase with a mixture of isopropanol/chloroform (3/1, v/v) with 0.1% acetic acid; the hydrophilic phase with water/methanol (50/50, v/v) with 0.1% acetic acid.
To each sample, 4 µL of an internal standard mixture was added. The hydrophilic standard contains a mixture of 13C-labeled tyrosine and tryptophan. The lipophilic standard contains synthetic lipids PC 5:0, PC 11:0, PC 19:0, and PG 17:0. All samples were stored at −80°C until the day of measurements.
Mass spectrometry was conducted using a FT-ICR-MS (7 Tesla, SolariXR, Bruker, Bremen, Germany) in the flow-injection mode. The injection was facilitated by a HPLC autosampler (1260 Infinity, Agilent, Waldbronn, Germany). The eluent for the hydrophilic samples was water/methanol (50/50, v/v) with 0.1% acetic acid and for the lipophilic samples, isopropanol/chloroform (3/1, v/v) with 0.1% acetic acid, respectively. The samples were ionized with an electrospray ionization source (in both modes). Different methods were used, each optimized to the respective detection range (in total, the range was from 65 to 3000 m/z). The average resolution at 400 m/z was 600,000. Evaluation of mass features was conducted with DataAnalysis 5.0 and MetaboScape 4.0.1 both from Bruker (Bremen, Germany). Sum formulas were calculated based on the mass error and isotopic fine structure of mass features. To reduce false-positive results, the seven golden rules of Kind and Fiehn were applied.24
Metabolomics pathway analysis
Based on the results of mass spectrometry metabolite analysis, we performed an enrichment analysis in MetaboAnalyst 5.0 in order to find common pathways of all the metabolites that were significantly associated with Parasutterella. Out of 256 nominally significant metabolites, we made use of n = 126 metabolites that were significantly associated with Parasutterella abundance after FDR correction of p-values in the count part of the Hurdle model. Of the 126, a total of 76 metabolites could be matched in terms of The Small Metabolites Pathway Database (SMPDB), whereas the other 40 were unidentified compounds.
Microbial community metabolism
We used metabolic network modeling to elucidate to which extent Parasutterella and associated gut microbes are involved in the consumption or production of certain metabolites, like L-cysteine. In order to predict L-cysteine consumption by bacteria of the gut microbiota, we employed gapseq (v1.2) to reconstruct metabolic models25 based on genomic data originating from the reference set of 820 bacteria and archaea belonging to the human gut microbiota (AGORA collection).(25) Default settings of gapseq were used. For gap filling and modeling of in silico growth, we assumed a nutritional environment corresponding to the average dietary input recorded for a cohort of human participants as described previously (“Kiel cohort”).26 For each individual bacterial model, we used flux balance analysis27 implemented in the R-package Sybil version 2.2.028 with biomass production as objective and unconstrained L-cysteine uptake as input to predict maximal L-cysteine consumption during optimal growth. We used information about the relative abundances of bacterial species from the FoCus cohort as references to determine the average abundance of each bacterial species across human participants. Relative cysteine uptake was then determined by multiplying the maximal L-cysteine uptake of each bacterial species by the average relative abundance of the corresponding bacterial species in the FoCus cohort.
Quantitative polymerase chain reaction
Genomic DNA (gDNA) was extracted from stool samples using the QIA amp Fast DNA Stool Mini Kit according to the manufacturer’s protocol. We decided for measuring Parasutterella excrementihominis since the DNA for the genus of Parasutterella was not available at DSMZ, Germany. Furthermore, we found appropriate primers for Parasutterella excrementihominis in the literature. This species is known to be the primary species of intestinal Parasutterella at least in our 16S rRNA sequencing data of the FoCus cohort (shotgun analysis). For quantifying the amount of DNA in each fecal sample, we used a Thermo NanoDrop 2000 spectrophotometer. A concentration of 2.5 ng/µL was used of each sample. For qPCR, a tenfold serial dilution of Parasutterella excrementihominis was generated to produce a standard curve (ordered from DSMZ, Germany). The qPCR system contained a PowerUpTM SYBRTM Green Master Mix (5 µL), the forward primer and reverse primer (2.5 µM each), and DNAse free water (4.5 µL). The primers were selected according to a study of Chen and coworkers: GGAAGTACGGTCGCAAGA (forward) and TGTCAAGGGTTGGGTAAGACA (reverse).29 Melt curves and relative quantification of Parasutterella excrementihominis were generated with Bio-Rad CFX ConnectTM Real-Time System PCR instrument with the help of the following template: 50°C for 2 min, preliminary denaturation at 95°C for 2 min, 40 cycles at 95°C (15 s), 40 cycles annealing at 60°C (30 s), and 40 cycles extension at 60°C (1 min). Melt curve analysis was performed between 65°C and 95°C (increment: 0.5).
Bio-Rad CFX Manager 3.0 was used to analyze melt curves and copy numbers of the target gene concentrations. Ct-values of the target gene and the standard curve indicated the final concentration through the following formula: y = −1.48lg(x) + 13.854 (R2 = 0.9979).
Genotyping
FoCus probands were genotyped using the Iscan Immunochip Opticall and the Iscan Omniexpress Exome Chip. Quality control and preprocessing of genetic data are described in detail elsewhere.30 For the purpose of this study, the VDR and POMC genes were defined as genes of interest. Quality control of genetic data was done in PLINK v. 1.9.
57 SNPs in the VDR locus (chr 12, 48.22–48.32 Mb) and 26 SNPs in the POMC locus (chr2, 25.36–25.46Mb) passed quality control and filtering.
Kruskal-Wallis Test was used in R to test for associations between SNP genotypes and Parasutterella sp. abundance groups.
Statistical analysis
Statistical significance was set at P < .05 P-values are shown as FDR (False Discovery Rate) adjusted to correct for multiple hypotheses testing.
Univariate analysis
The statistical and graphical data analysis was done in Rv3.6.0. Data were tested for normality by Shapiro-Wilk-Test and are presented as means ± SDs (standard deviation) for normally distributed variables and as median for non-normally distributed variables. In the intervention study, paired Wilcoxon signed-rank tests (for non-normally distributed variables) have been used for metric variables to determine differences between two time points in one group.
Multivariate analysis
Since Parasutterella abundances in the explorative part of this study were derived from 16s rRNA amplicon sequence data, Hurdle models were chosen in order to handle the excess number of zeros and overdispersion in the data. Hurdle models are two-part (negative) binomial regression models in which probabilities for the non-zero and zero abundances of Parasutterella are handled separately. In the first part of the model (henceforth called “count part”), the model is truncated at zero and a negative-binomial linear regression is fitted to the remaining abundance data. In the second part of the model (henceforth called “zero part”), a logistic regression is fitted to determine the binary probability of Parasutterella abundance being zero vs. non-zero.
ASV abundances of Parasutterella sp. lower than 10 counts were set to zero before using the Hurdle algorithm to adjust for the error in determining low counts of 16S rRNA data. A multivariate Hurdle algorithm was applied to model the association of Parasutterella and the independent variable of interest for dietary data, phenotypes linked to obesity, and metabolic health and clinical biomarkers. Since inflammatory comorbidities (inflammatory bowel disease (IBD) and psoriasis) other than metabolic diseases that are known to affect the microbial composition, the presence of such diseases and the proband age were included as covariates.31,32 In some analysis when marked accordingly, we additionally used BMI as a covariate.
Metabolomics data was either K-nearest neighbor (KNN) (for samples with < 50% missingness) or limit of detection (LOD) (for samples with > 50% missingness) imputed and then log-transformed. For the metabolomics analysis, once again the Hurdle models were used as previously described, since the resulting data matrix consists of zero-inflated integers, which can statistically be treated similarly to gene-based count data. For the peak annotation, a customized and local database was established, which contained sum formula and names of all metabolites of interest. The database was used for annotation with MetaboScape and the resulting data frame contained information about each mass error and isotopic fine structure (as the mSigma value). By means of these both values, the resulting metabolites were validated (only Metabolites with an error below 2 ppm and a “good” mSigma (< 200) were accepted for further statistical evaluation).
For microbiomics analysis, subjects were grouped into high (>10) or low (≤10) counts of Parasutterella and Mann-Whitney-U test were used to find differences between the two groups. Microbiome β- and α- diversity was characterized by the Bray-Curtis dissimilarity index and the Shannon, Chao1, and Species Richness indices, respectively, and compared between the groups. Differential abundance between BMI groups was tested with the DESEQ2 integration for phyloseq data in R. To evaluate classifier performance in human obesity of Parasutterella in relation to Akkermansia, prediction models were built using random forests and ROC curves and AUC values were determined. For some of these analyses, the FoCus cohort was stratified into five groups concerning BMI and diabetes status (see Table 1).
Results
To generate a comprehensive overview of Parasutterella on the development of obesity and type 2 diabetes, we decided to investigate the bacterium on metabolic, inflammatory, dietary, microbiome, metabolome, and genetic level. Additionally, we examined qPCR-determined Parasutterella abundances in a weight loss intervention study to further validate bioinformatical calculations and to gain insight into the functional capabilities of this species.
As described in the method section, we used two-part Hurdle models to test for association between our independent variables of interest and Parasutterella abundance. Since we did not observe any significant correlation in the zero part of each model (logistic regression) except of microbiomics calculations, the results described in the following pages refer solely to the count part of the Hurdle models.
Parasutterella and measures of obesity
We first examined 1,544 subjects of the FoCus cohort regarding the obesity phenotype. Therefore, we used Parasutterella sp. > 10 counts (n = 1132 samples that met the threshold of Parasutterella sp. > 10 counts) for multivariate modeling in regard to different metric variables such as BMI and body weight. This threshold was recommended by QIIME pipeline (https://docs.qiime2.org/2021.4/) in order to only incorporate values of Parasutterella sp. that were truly measured through 16S rRNA gene sequencing. We found a significant positive correlation between positive counts of Parasutterella sp. abundance and BMI (3.71e−2, P = 3.11e−3) (Figure 1a), meaning that there was an increased Parasutterella sp. abundance in subjects with higher BMI. The results were in line with data regarding weight measurements: there was a nominally significantly positive association between Parasutterella sp. abundance and weight of the subjects (7.95e−3, P = 2.0e−2).
Figure 1.
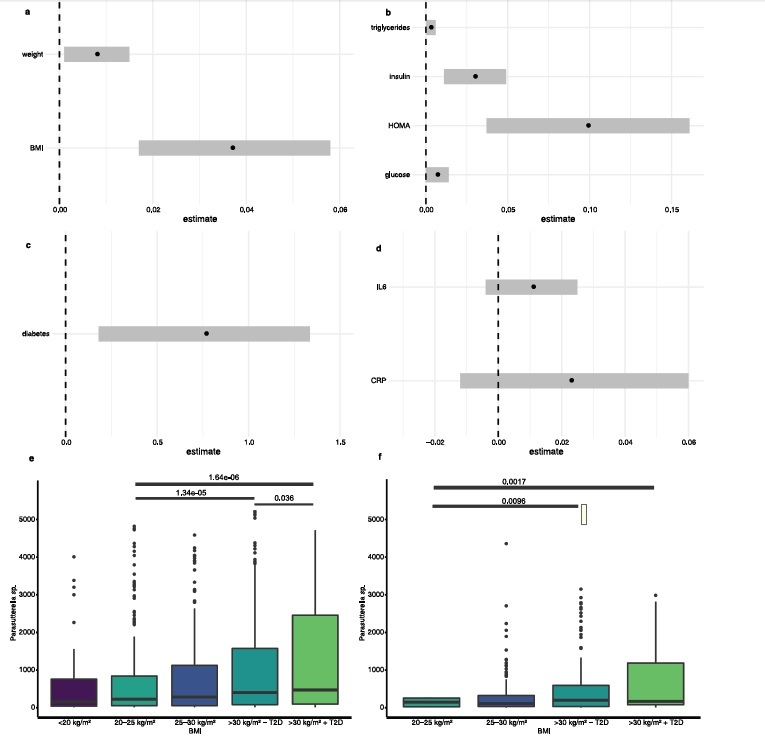
Parasutterella and obesity, glucose and lipid abnormalities as well as metabolic inflammation. (a) Association of Parasutterella sp. with weight and BMI in n = 1,544 subjects reported through estimate and standard error (Hurdle count model). (b) Association of Parasutterella sp. with metabolic parameter in n = 1,544 subjects reported through estimate and standard error (Hurdle count model). (c) Association of Parasutterella sp. with diabetes in n = 1,544 subjects reported through estimate and standard error (Hurdle count model). (d) Association of Parasutterella sp. with inflammatory parameters in n = 1,544 subjects reported through estimate and standard error (Hurdle count model). (e) Parasutterella sp. abundance in relation to BMI and T2D groups in the FoCus cohort. (f) Parasutterella sp. abundance in relation to BMI and T2D groups in the ATP cohort. While the sequencing depth of the ATP cohort was slightly lower overall than in FoCus (median FoCus: 36,048; median ATP: 22,931), Figures 1e and f demonstrate that the distribution of Parasutterella abundance in relation to BMI is comparable in both cohorts and that Parasutterella is a highly abundant microbe in ATP as well, although the height of box plots differs slightly between cohorts.
The significant positive association of Parasutterella and BMI found in the European FoCus cohort was validated in the independent Canadian ATP cohort (n = 305 samples that met the threshold of Parasutterella sp. > 10 counts). Of interest, we found that Parasutterella sp. abundance was increased in probands with higher BMI (P = 2.0e−3) in the Canadian cohort as well.
Parasutterella and diabetes phenotypes
In the next step, we analyzed glucose metabolism and found Parasutterella sp. abundance to be positively associated with elevated HOMA (9.88e−2, P = 1.5e−2) and fasting insulin (3.02e−2, P = 1.38e−2) (Figure 1b). In addition, we found a positive association between Parasutterella sp. abundance and fasting glucose (6.81e−3, P = 5.38e−2). Furthermore, there was a significantly positive association between Parasutterella sp. abundance and the presence of diabetes type 2 (7.58e−1, nominal significant, P = 1.02e−2) (Figure 1c). With regard to lipid metabolism, Parasutterella sp. was nominally positively associated with serum triglyceride levels (2.61e−3, P = 3.34e−2) (Figure 1b). However, when adapting for BMI, the significant association to the measures of insulin sensitivity diminished, suggesting the association of Parasutterella with insulin resistance to be indirectly, mediated via its effect on body weight gain.
In a subsequent analysis, we stratified the FoCus cohort into five groups concerning BMI and diabetes status (see Table 1) and found a significant difference between the lean and the obese group concerning Parasutterella abundance (Figure 1e)). Of interest, a significant difference in Parasutterella abundance was further identified between obese probands without type 2 diabetes compared to obese probands with type 2 diabetes, whereby diabetics had around 70% higher Parasutterella abundance (Figure 1e). In the ATP cohort, a similar pattern was observed, whereby the difference between obese individuals with and without type 2 diabetes did not reach statistical significance, which might be due to the smaller number of subjects in ATP (n = 438, 5.2% diabetics) compared to FoCus (n = 1,544, 10.6% diabetics) (Figure 1f).
Parasutterella and inflammatory phenotypes
In order to gain insights into the inflammatory properties of Parasutterella sp., we examined two biomarkers that mirror the inflammatory state of the subjects. Parasutterella sp. abundance was neither associated with CRP nor with IL-6 (2.39e−2, P = 1.9e−1; 1.07e−2, P = 1.5e−1) (Figure 1d). This finding indicates no major association of Parasutterella sp. to systemic metabolic inflammation.
Parasutterella and dietary phenotypes
Dietary data were derived from 12-month food frequency questionnaires (n = 1,443). Analysis of 19 dietary components revealed that the intake of total carbohydrates showed a nominal significant positive correlation with Parasutterella sp. abundance (4.51e−2, P = 4.24e−3) (Table 2) falling in line with the data on diabetes phenotypes. The composition of total carbohydrate intake consisted of 22.09% monosaccharides, 31.56% disaccharides, 44.22% polysaccharides, and minor parts of sugar alcohol and oligosaccharides. Appropriately, monosaccharides showed a nominal significant positive association with Parasutterella sp. abundance (1.115e−2P = 1.19e−3). In contrast, the total fat intake of the subjects was nominally significantly negatively associated with Parasutterella sp. abundance (−5.13e−2, P = 5.97e−3). In more detail, we found the polyunsaturated fatty acid (PUFA) linolenic acid (ω3-fatty acid) nominally significantly negatively associated with Parasutterella sp. abundance (−3.3e−1, P = 4.99e−2). Furthermore, eicosenoic acid was nominally significantly negatively associated with Parasutterella sp. (−3.25, P = 3.04e−3). The mean percentage of the intake of total fiber was 22.3%. Dietary data were additionally adjusted for BMI.
Table 2.
Dietary parameters regarding the abundance of Parasutterella sp. (two-part Hurdle model), truncated linear model considering only counts of Parasutterella sp. (count part). Dependencies of parameters and the abundance of Parasutterella sp. reported through estimate, confidence intervals, and p-values in the respective model. First part of the Hurdle model considers only counts of Parasutterella sp. using a negative binomial regression. After FDR-correction, p-values were not significant
| Parameter | Estimate | Confidence interval [2.5%, 97.5%] | p-value |
|---|---|---|---|
| Carbohydrates (g/day) | 4.51e−2 | [1.42e−2, 7.60e−2] | 4.24e−3 |
| Monosaccharides (g/day) | 1.15e−2 | [4.53e−3, 1.88e−2] | 1.19e−3 |
| Protein (g/day) | 3.76e−2 | [8.03e−2, 1.56e−1] | 5.31e−1 |
| Fat (g/day) | −5.13e−2 | [−8.78e−2, 1.47e−2] | 5.97e−3 |
| Linolenic acid (g/day) | −3.3e−1 | [−6.61e−1, −2.63e−5] | 4.99e−2 |
| Eicosenoic acid (g/day) | −3.25 | [−6.21, −3.07e−1] | 3.04e−2 |
| Butanoic acid (g/day) | 3.07e−1 | [−7.99e−1, 1.84e−1] | 2.2e−1 |
| Hexanoic acid (g/day) | −4.84e−1 | [−1.25, 2.85e−1] | 2.17e−1 |
| Vitamin D (mg/day) | −72.75 | [−1.58e+2, 13.13] | 9.68e−2 |
| Vitamin B9 (mg/day) | 2.61 | [−6,24, 11.46] | 5.62e−1 |
| Vitamin B6 (mg/day) | −7.51e−1 | [−1.48, −2.14e−2] | 4.36e−2 |
| Vitamin C (mg/day) | 2.04e−3 | [−2.14e−3, 6.21e−3] | 3.39e−1 |
| Vitamin B12 (mg/day) | −45.81 | [−1.75e+2, 83.74] | 4.88e−1 |
| Vitamin E (mg/day) | −3.65e−2 | [−1.01e−1, 2.8e−2] | 2.68e−1 |
| Iodine (mg/day) | 4.16 | [−1.37, 9.7] | 1.41e−1 |
| Iron (mg/day) | 2.28e−1 | [7.99e−2, 3.77e−1] | 2.55e−3 |
| Calcium (g/day) | 3.55e−1 | [−7.35e−1, 1.44] | 5.23e−1 |
| Magnesium (g/day) | −5.35e−1 | [−4.35, 3.28] | 7.83e−1 |
| Zinc (mg/day) | −3.98e−2 | [−1.52e−1, 7.21e−2] | 4.85e−1 |
Besides macronutrients, we examined several micronutrients, whereby iron was nominally significantly positively associated with Parasutterella sp. abundance (2.28e−1, P = 2.55e−3). Regarding vitamins, a nominal significant negative association was found for vitamin B6 and Parasutterella sp. (−7.51e−1, P = 4.36e−2).
Having found an association of Parasutterella sp. abundance with carbohydrate intake we also examined carbohydrate intake independently of the Parasutterella abundance and found a significant higher intake in obese individuals compared to lean controls (P = 3.2e−2). Importantly, the association of Parasutterella sp. and the carbohydrate intake remained significant when adjusting for BMI.
Parasutterella and microbiomics
A comparison of Parasutterella sp. levels with the general microbiome composition revealed significant differences in β-diversity measures (Bray-Curtis distance: P = 9.0e−3, Jaccard index: P = 8.0e−4) between groups with low versus high Parasutterella sp. abundance (Figure 2a). Likewise, α-diversity was significantly reduced in individuals with low Parasutterella sp. abundance (zero part of the Hurdle model: Shannon index: P = 2.4e−23, Chao Index: P = 2.8e−25, Species richness: P = 2.6e−26) (Figure 2b). In the count part of the Hurdle model, no significant correlation with Parasutterella sp. abundances was detected.
Figure 2.
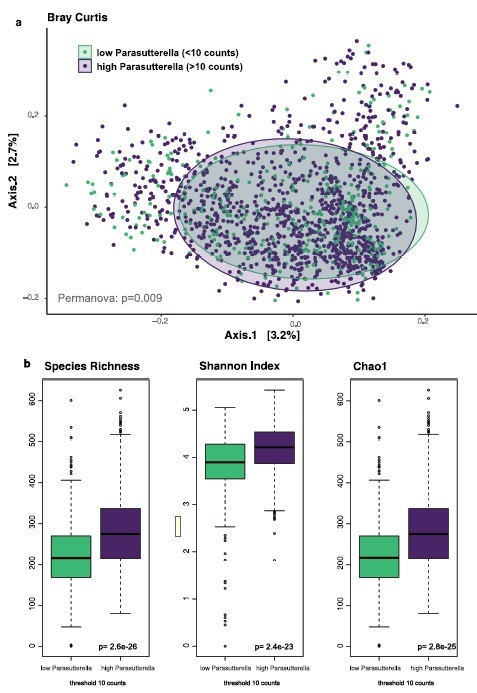
Parasutterella and gut microbiome diversity measures. Beta diversity was assessed by Bray-Curtis distance and PERMANOVA. Alpha diversity was assessed by species richness, Chao1 Index and Shannon Index. Statistical significance between high and low Parasutterella groups was tested by Wilcoxon tests.
The association of obesity and the gut microbiome was evaluated by differential Fold Change analysis. In this analysis, Parasutterella was among the top 3% differentially abundant species (placed 18th out of 665 species tested) and consistently exhibited higher fold changes than Akkermansia sp., a species that is an already established marker for obesity (Figure 3a). We have also performed a differential abundance analysis on the genus level (Figure 3b), which revealed that subjects with high Parasutterella abundance are characterized by nominally higher abundances of Alistepes (r2 = 0.43, P = .02), Faecalibacterium (r2 = 0.58, P = .01), and Roseburia (r2 = 0.47, P = .02). Probands with low counts of Parasutterella are in turn characterized by higher abundances of Bacteroides and Subdoligranulum.
Figure 3.
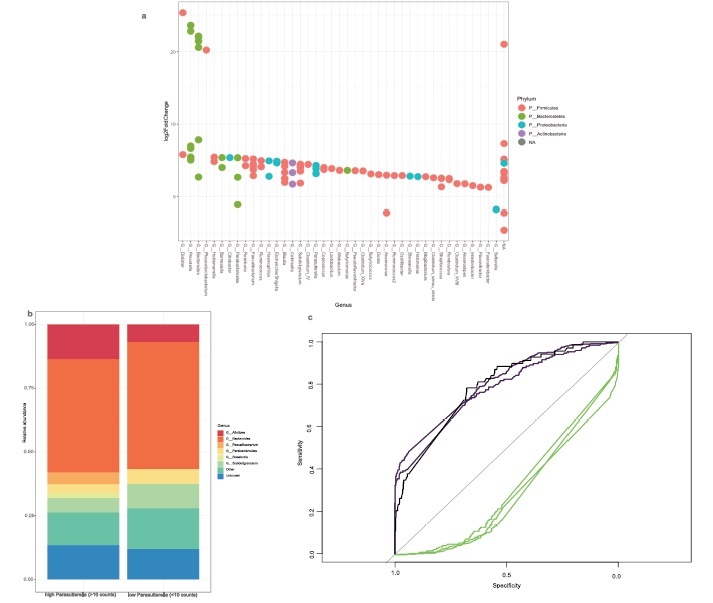
Parasutterella in relation to other gut microbiome species in human obesity. (a) LogFold Changes of differentially abundant microbes in human obesity in the FoCus cohort. Comparison was made between the normal weight group (BMI < 25) and the obese group (BMI > 30, without T2D). Parasutterella sp. is among the top 20 (=3%) differentially abundant species out of 665 species tested. Plot shows the top 50 differentially expressed species, eight species could not be assigned to a genus (marked NA in the plot). Parasutterella sp. is placed 18th among the top 50. (b) Composition plots of probands with high and low (threshold <10 counts) Parasutterella sp. (c) ROC curves for prediction models (random forests) of BMI and T2D groups by Akkermansia (green, AUC: 0.22–0.28) and Parasutterella (purple, AUC: 0.83–0.87) abundance. Comparisons were made between the normal weight group and the (1) obese group without diabetes, (2) obese group with diabetes, and (3) the overweight group according to Table 1. Classifier performance was tested using bootstrapping of AUC results and revealed significantly better performance of Parasutterella compared to Akkermansia in all comparison groups (P = 5.2e-191;-2.8e-185).
Finally, an ROC analysis revealed that in the FoCus cohort, increased abundance of Parasutterella is an even better classifier for obesity compared to a reduced abundance of Akkermansia. (Figure 3c). AUCs of Parasutterella ranged from 0.83 to 0.87, while Akkermansia had AUCs ranging from 0.22–0.28. Differences in AUC for Parasutterella and Akkermansia were significant for each comparison between normal weight, obese, and diabetes groups (PNW:OBS-T2D = 5.2e-191; PNW:OBS+T2D = 1.4e-171; PNW:Overweught = 2.8e-185, according to the BMI groups shown in Table 1).
Parasutterella and host metabolomics
Presently, MS-metabolomics analysis data are available of n = 470 subjects of the FoCus cohort, consisting of n = 178 metabolically diseased subjects and 292 healthy controls. This analysis took n = 955 detectable metabolites into account. 269 metabolites were nominally associated with Parasutterella abundance, of which 126 remained significant after BH adjustment for multiple comparisons. Of interest, we identified L-cysteine (LC) as a metabolite that showed a significant inverse correlation with Parasutterella sp. abundance in the count part of the Hurdle model (−13.00, P = 1.78e−3) (Table 3). Furthermore, 6-Hydroxynicotinic acid was positively correlated with Parasutterella sp. abundance (1.57, P = 1.22e−2) (Table 4). Tables 3 and 4 show the five highest estimates either negative or positive out of in total n = 126 significant metabolites in association with Parasutterella sp. abundance in the count part. There were no significant associations in the zero part of the Hurdle model for all metabolites analyzed.
Table 3.
Metabolomic parameters regarding the abundance of Parasutterella sp. (two-part Hurdle model) showing negative associations by using a truncated linear model considering only counts of Parasutterella sp. (count part). Dependencies of parameters and the abundance of Parasutterella sp. reported through estimate, confidence intervals, and FDR- adjusted p-values in the respective model. First part of the Hurdle model considers only counts of Parasutterella sp. using a negative binomial regression. Parameters are chosen considering the five highest estimates
| Parameter | Estimate | Confidence interval [2.5%, 97.5%] | p-adjusted |
|---|---|---|---|
| L-Cysteine | −13.00 | [−18.89, −7.11] | 1.78e−3 |
| 19,20-DiHDPA | −6.74 | [−10.45, −3.02] | 1.11e−2 |
| Hydroxycholesterol | −4.60 | [−7.43, −1.78] | 1.98e−2 |
| Tetrahydrocortisone | −3.93 | [−6.62, −1.24] | 3.94e−2 |
| Tetracosatetraenoic acid | −3.62 | [−5.34, −1.89] | 3.04e−3 |
Table 4.
Metabolomic parameters regarding the abundance of Parasutterella sp. (two-part Hurdle model) showing positive associations by using a truncated linear model considering only counts of Parasutterella sp. (count part). Dependencies of parameters and the abundance of Parasutterella sp. reported through estimate, confidence intervals, and BH- adjusted p-values in the respective model. First part of the Hurdle model considers only counts of Parasutterella sp. using a negative binomial regression. Parameters are chosen considering the five highest estimates
| Parameter | Estimate | Confidence interval [2.5%, 97.5%] | p-adjusted |
|---|---|---|---|
| [(2 R)-1-[2-aminoethoxy(hydroxy)phosphoryl]oxy-3-[(1Z,11Z)-octadeca-1,11-dienoxy]propan-2-yl] (9Z,12Z,15Z)-octadeca-9,12,15-trienoate | 3.53 | [1.11, 5.95] | 3.99e−2 |
| Oxoglutaric acid | 1.99 | [0.55, 3.44] | 5.36e−2 |
| 6-Hydroxynicotinic acid | 1.57 | [0.68, 2.46] | 1.22e−2 |
| Prostaglandin f1 alpha | 1.33 | [0.80, 1.87] | 1.57e−4 |
| Isocaproic acid | 1.25 | [0.61, 1.88] | 5.52e−3 |
Second, the pathway enrichment analysis of 76 metabolites that were successfully matched to terms of the SMPDB database resulted in two nominally significant pathways: fatty acid biosynthesis and α-linolenic acid metabolism (P = 1.16e−2, P = 4.62e−2) (Figure 4). All other pathways in the figure were not significantly enriched.
Figure 4.
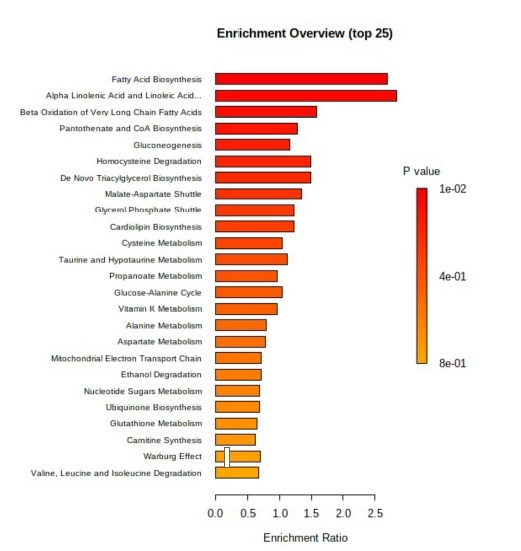
Parasutterella and metabolic pathway enrichment analysis.
Enrichment ratio and related p-value of the pathway enrichment analysis (SMPDB database) displaying fatty acid biosynthesis and α-linolenic acid metabolism as nominally significant (P < .05).
Microbial metabolic network analysis
We used metabolic network modeling to investigate maximal L-cysteine consumption by member species of the human gut microbiota. This analysis revealed that Parasutterella is among the top five consumers, together with Dialister invisus, Enterobacter cancerogenus, Prevotella copri, and Escherichia coli K12. Hence, the fact that Parasutterella is a high L-cysteine consumer fits with the findings of reduced L-cysteine levels in the serum of human subjects with high Parasutterella abundance. Table 5 gives an overview of the top L-cysteine consumers in the gut microbiome.
Table 5.
Gut bacteria on species level with the highest relative L-cysteine consumption in human samples. The second column indicates the maximal predicted L-cysteine consumption of each strain during optimal growth, the third column the average abundance in the FoCus cohort and the fourth column the abundance-weighted cystein consumption. The fifth column shows the relative L-cysteine consumption as a sum of 1 regarding microbiome abundance in the FoCus cohort
| Species (strain designation in the AGORA collection) | Maximal L-cysteine consumption (mmol/gDW/h) |
Average relative abundance |
Abundance-weighted cystein consumption | Relative L-cysteine consumption |
|---|---|---|---|---|
| Dialister invisus DSM 15470 | 40.8 | 0.078 | 3.20 | 13.72% |
| Enterobacter cancerogenus ATCC 35316 | 97.7 | 0.027 | 2.68 | 11.53% |
| Escherichia coli K12 MG1655 | 47.1 | 0.052 | 2.47 | 10.59% |
| Prevotella copri CB7 DSM 18205 | 38.9 | 0.058 | 2.27 | 9.77% |
| Parasutterella excrementihominis YIT 11859 | 81.9 | 0.026 | 2.12 | 9.10% |
| Sutterella wadsworthensis 3145B | 96.3 | 0.008 | 0.81 | 3.48% |
| Roseburia hominis A2 183 | 89.4 | 0.008 | 0.70 | 3.02% |
| Hafnia alvei BIDMC 31 | 98.1 | 0.007 | 0.70 | 3.01% |
| Bilophila wadsworthia 316 | 96.3 | 0.005 | 0.51 | 2.17% |
| Acidaminococcus intestini RyC MR95 | 88.2 | 0.004 | 0.39 | 1.68% |
Parasutterella and host genomics
In total, we examined 83 different SNPs. None of the SNPs at the VDR locus were significantly associated with Parasutterella abundance. At the POMC locus, wild-type homozygosity of the SNP rs7565877 was nominally significantly associated (P = 3.0e−2) with abundance of Parasutterella.
Parasutterella and weight loss intervention
The patients of the weight loss intervention lost a significant amount of weight (mean of 23 kg; Wilcoxon paired, P = 5.30e−10). BMI dropped from 45.2 kg/m2 to 38.5 kg/m2 (Figure 5a). Of interest, Parasutterella excrementihominis was significantly reduced during the intervention (Figure 5b). The median value of relative Parasutterella excrementihominis abundance at baseline was 1.62% in comparison to 0.62% of Parasutterella excrementihominis after the intervention. These data indicate that Parasutterella is not only associated to the obesity phenotype in a steady state but also reacts significantly to variations in body weight. However, it has to be mentioned that the formula diet used within the human intervention study has a low carbohydrate content which also might have affected the change in Parasuterella abundance.
Figure 5.
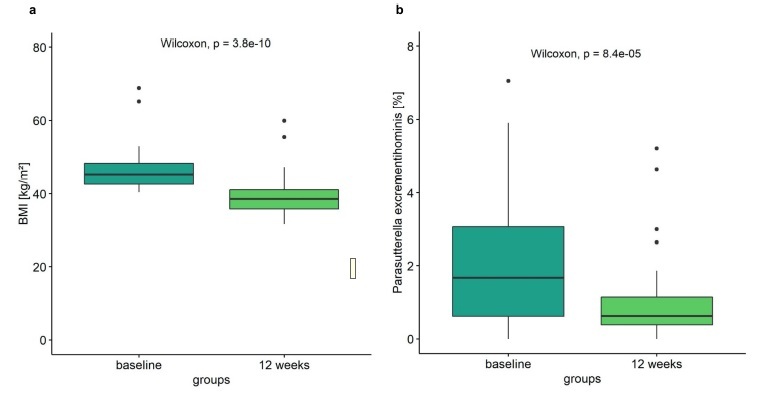
Parasutterella and human weight loss intervention. (a) Difference of BMI in subjects of intervention at baseline compared with 12 weeks (Wilcoxon signed-rank test, P < .05), (n = 55). (b) Abundance of Parasutterella excrementihominis in subjects of intervention at baseline compared with 12 weeks (Wilcoxon signed-rank test, P < .05), (n = 55).
Comparability of cohorts used in this study
We performed the following analysis to ensure that the ethnically and obesity-status diverse cohorts in this study allow for comparable results. Figure 1e and 1f provide an overview of the distribution of Parasutterella abundances in relation to BMI for both the FoCus cohort and the Canadian ATP cohort. Furthermore, we compared the dietary patterns in both cohorts by calculating the Mediterranean Diet Score, a score that is commonly used to evaluate healthy dietary patterns. There was no significant difference in adherence to the Mediterranean diet between the German and Canadian cohort (Supp. Figure 1). Lastly, we compared the microbial composition of the population-based FoCus cohort with the intervention cohort at baseline (Supp. Figure 2) and found no significant difference (P = .872, multivariate comparison of abundance at phylum level). Overall, we conclude that the cohorts in this study can justifiably be compared to each other.
Discussion
In recent years, several studies have investigated the relation of gut microbial composition at the phylum level to obesity and its metabolic comorbidities. Due to previous findings in various rodent models, we further characterized a specific bacterium, Parasutterella, at the genus level using more than 1.500 subjects from the FoCus cohort in Kiel. We observed a positive association between Parasutterella abundance and obesity, as well as type 2 diabetes and carbohydrate-rich food intake. Additionally, Parasutterella was significantly reduced after a sustained weight loss intervention in subjects with obesity and associated with L-cysteine among other metabolites. Metabolite pathway analysis revealed an enrichment of the fatty acid biosynthesis pathway.
Our data indicate a positive association between Parasutterella and obesity, which we were able to validate in the independent Canadian ATP cohort. These results are in line with research of mechanistic studies in animal models from independent groups. For example, Gu et al. found that Parasutterella sp. was significantly enriched in obesity-prone mice in comparison to obesity-resistant mice both fed a HFD.13 Furthermore, bacteria of Proteobacteria phylum, to which Parasutterella sp. belongs, are known to be highly abundant in obesity and other metabolic diseases.33,34 In addition, several authors classify Parasutterella sp. as pro-diabetic in animal studies. For instance, in a study by Cheng et al., the authors reported that mice fed with galacto-oligosaccharides (GOS) exhibit a change in their microbiota with a significant enrichment in Parasutterella sp. and at the same time a significantly increased blood glucose level compared to control animals.35 This is in line with our finding that Parasutterella sp. is elevated in patients with type 2 diabetes.
Dietary patterns are a key factor in determining gut microbiota composition in humans as well as the pathogenesis of obesity.36 As such, we examined the role of diet in Parasutterella sp. abundance. Therefore, in the present analysis, we made use of the detailed EPIC dietary data available for all FoCus subjects. Our data indicate that carbohydrate (especially monosaccharides) and ω3 linolenic acid intake are associated with the abundance of our target bacterium. A higher intake of carbohydrates was associated with a higher abundance of Parasutterella sp., whereas a higher intake of linolenic acid was associated with a lower abundance of Parasutterella sp. In a previous study using the FoCus cohort, we found that polyunsaturated fatty acids are positively associated with the β-diversity of microbial composition.30 Hence, the negative association of Parasutterella with ω3 linolenic acid fits into the concept that this bacterium is related to negative cardiovascular effects.37 This theory is further supported by the metabolic pathway enrichment results, which showed that many of the metabolites that are associated with Parasutterella are involved in the fatty acids synthesis pathways.
By using our untargeted HPLC-based mass spectrometry metabolomics data set, L-cysteine revealed an inverse correlation with Parasutterella sp. displaying a convincing estimate that underlines the fact that L-cysteine is linked to blood glucose control which was described in the literature.38 Intriguingly, metabolic network modeling revealed Parasutterella sp. as a potential top consumer of L-cysteine within the human gut microbiota. Furthermore, a study of Ju et al. investigated mice colonized with Parasutterella which had significantly reduced concentrations of taurine (degradation product of cysteine) and N-Acetyl-DL-methionine that implies Parasutterella to be able to reduce also cysteine associated metabolites.39
So far, it is known that supplementation of L-cysteine improves glycemia and also lowers vascular inflammation in diabetic rodents38 meaning L-cysteine reveals anti-diabetic characteristics. In rats that were fed a high-sucrose diet and a whey concentrate rich in L-cysteine, the data showed that an increase in L-cysteine limited the impairment of glucose homeostasis through lowering oxidative stress.40 Of interest, especially pancreatic β cells are extremely susceptible to oxidative stress due to a high endogenous production of reactive oxygen species (ROS) and a low expression of antioxidative enzymes.41 Since we found that Parasutterella is associated with type 2 diabetes but not with (BMI-independent) measures of insulin resistance, this finding might point toward abnormalities in insulin secretion due to Parasutterella promoting type 2 diabetes development.
In a second analysis, we examined the enrichment of metabolic sets and found an association of Parasutterella sp. abundance with the fatty acid biosynthesis pathway. This finding falls in line with the fact that elevated BMI and body weight linked to an obese phenotype have elevated fatty acid biosynthesis systemically. Of interest, it is known that Parasutterella secunda does not produce significant amounts of propionate11 and Parasutterella excrementihominis is producing only traces of propionate.42 Since SCFA are known to inhibit human fatty acid biosynthesis and induce human fatty acid oxidation,43 this might suggest a mechanism on how Parasutterella induces human fatty acid biosynthesis as found in our analysis. However, we have to admit that (1) we did not measure SCFA fecal concentrations in the present analysis in relation to Parasutterella abundance and (2) that in our differential abundance analysis Parasutterella is correlated with bacteria that are SCFA producers, e. g. Faecalibacterium and Roseburia. Hence, while our human cohort study can only identify association, the data presented here might serve as a basis for future intervention studies in model systems of obesity and type 2 diabetes to identify the exact molecular and cellular mechanisms on how Parasutterella affects fatty acid biosynthesis.
Due to our previous findings on SNPs in the human VDR influencing gut microbiome composition, we decided to include human genetic factors in our present analysis. However, none of the SNPs at the VDR locus were significantly associated with Parasutterella sp. abundance. Furthermore, we examined common SNPs in the POMC locus, since these were also found to influence the gut microbiome in our previous study30 and SNPs in the POMC gene are associated with body weight dysregulation.44 In that analysis, we found one nominally significant SNP at the POMC locus, in which lower Parasutterella abundance was associated with the wild-type homozygous allele. Taken together, our data suggest that Parasutterella abundance is more related to environmental (nutrition) compared to host-genetic factors at least in the VDR and POMC regions.
Most research data regarding the role of the gut microbiome in the development of obesity and type 2 diabetes are on phylum level, showing that both diseases are associated with a reduced α- and/or β-diversity. It has to be mentioned that this reduction in diversity measures is not specific for metabolic diseases but is also found in chronic IBD,45 chronic heart failure46 and diverse oncological diseases.47 Thus, from our point of view, the present data and the published data on Akkermansia indicate that future research should also examine single bacteria or networks of defined bacteria on genus level as indicators or even pathogenic factors of specific diseases rather than solely relying on phylum data. In addition, the finding of higher Parasutterella abundance in obese subjects known to have lower overall diversity might underline the specificity of the novel nutrition-Parasutterella-host metabolic axis.
In our previous project regarding hypothalamic inflammation, we found an association of Parasutterella with the MRI density in the hypothalamus in severe obese subjects.6 This is of interest since in the present study Parasutterella was associated with obesity and type 2 diabetes, but not with systemic inflammatory markers like IL-6 and CRP in the periphery. As IL-6 and CRP reflect the degree of metabolic inflammation in obesity,48 our data might suggest that Parasutterella mediates its negative effects on host glucose metabolism – at least in the periphery – via direct metabolic effects rather by influencing the innate immune system. In this respect, L-cysteine, identified in our metabolomics analysis, might be an interesting candidate for future investigations.
Conclusion
Our data indicate that Parasutterella sp. is associated with human obesity and type 2 diabetes and might be implicated in a novel dietary carbohydrate – microbiome – host metabolic axis. Thereby, the link to the fatty acid biosynthesis pathway might be important for body weight gain in obesity in response to a carbohydrate-rich diet, whereas the link to L-cysteine could be relevant for type 2 diabetes development (Figure 6). In addition, the inverse relationship of Parasutterella with the dietary ω3 fatty intake should be of interest for future studies, given the convincing data of the recent REDUCE-IT trial.49
Figure 6.
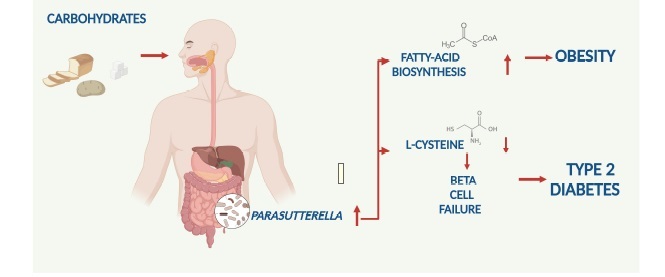
Summary figure on the proposed dietary Carbohydrate – Gut Parasutterella – Human Fatty Acid Biosynthesis metabolic axis.
Supplementary Material
Funding Statement
This work was supported by a grant of the Bundesministerium für Bildung und Forschung (BMBF, Germany, FKZ: 0315540) and by a grant of the ERA-HDHL Initiative “Gut metabotypes as Biomarkers for Nutrition and Health” (BLE, Germany, FKZ: 2816ERA14E & 2816ERA13E and CIHR, Canada, FRN #150364)
Acknowledgments
The authors gratefully thank all participants of the study. Matthias Laudes acts as guarantor in this project.
Author Contributions
ML and DP conceived the research idea and acted as PIs. LH and KS performed data analysis, laboratory experiments, and wrote the manuscript with support from ML, TH. CK and NR performed additional data analysis. CK and JZ performed microbial metabolic network analysis. NA, TD, JJ, CG, DMS, KH, US, KT, JB, JS, SS, GST, KSCHW, and SS were involved in clinical studies and laboratory experiments. AF provided bioinformatic expertise for 16S rRNA gene sequencing.
Role of Study Funder
The study funder was not involved in the design of the study; the collection, analysis, and interpretation of data; writing the report; and did not impose any restrictions regarding the publication of the report.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data Availability
Samples and data are stored in the PopGen Biobank (Schleswig-Holstein, Germany) and can be accessed via a structured application procedure (https://www.uksh.de/p2n/Information+for+Researchers.html).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
References
- 1.Cox AJ, West NP, Cripps AW.. Obesity, inflammation, and the gut microbiota. Lancet Diabetes Endo. 2015. Mar;3(3):207–18. [DOI] [PubMed] [Google Scholar]
- 2.Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med [Internet]. 2016 Apr 27 [cited 2020 Dec 9];8. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4848870/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006. Dec;444(7122):1027–1031. [DOI] [PubMed] [Google Scholar]
- 4.de MAH, Uberti MF, de FBX, de SNAR, Rezin GT. n-3 PUFA and obesity: from peripheral tissues to the central nervous system. Br J Nutr. 2018. Jun;119(11):1312–1323. [DOI] [PubMed] [Google Scholar]
- 5.Boulangé CL, Neves AL, Chilloux J, Nicholson JK, Dumas M-E. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med [Internet]. 2016 Apr 20 [cited 2019 Jan 20];8. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4839080/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kreutzer C, Peters S, Schulte DM, Fangmann D, Türk K, Wolff S, van Eimeren T, Ahrens M, Beckmann J, Schafmayer C, et al. Hypothalamic inflammation in human obesity is mediated by environmental and genetic factors. Diabetes. 2017. Sep;66(9):2407–2415. [DOI] [PubMed] [Google Scholar]
- 7.Mandaşescu S, Mocanu V, Dăscaliţa A-M, Haliga R, Nestian I, Stitt PA, et al. Flaxseed supplementation in hyperlipidemic patients. Rev Med Chir Soc Med Nat Iasi. 2005. Sep;109(3):502–506. [PubMed] [Google Scholar]
- 8.Depommier C, Everard A, Druart C, Plovier H, Van Hul M, Vieira-Silva S, Falony G, Raes J, Maiter D, Delzenne NM, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med. 2019 Jul 1;25(7):1096–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anhê FF, Schertzer JD, Marette A. Bacteria to alleviate metabolic syndrome. Nat Med. 2019 Jul 1;25(7):1031–1033. [DOI] [PubMed] [Google Scholar]
- 10.Basolo A, Hohenadel M, Ang QY, Piaggi P, Heinitz S, Walter M, Parrington S, Trinidad DD, von Schwartzenberg RJ, et al. Effects of underfeeding and oral vancomycin on gut microbiome and nutrient absorption in humans. Nat Med. 2020 Apr 1;26(4):589–598. [DOI] [PubMed] [Google Scholar]
- 11.Morotomi M, Nagai F, Watanabe Y. Parasutterella secunda sp. nov., isolated from human faeces and proposal of Sutterellaceae fam. nov. in the order Burkholderiales. Int J Syst Evol Microbiol. 2011 Mar 1;61(3):637–643. [DOI] [PubMed] [Google Scholar]
- 12.Xiao S, Liu C, Chen M, Zou J, Zhang Z, Cui X, Jiang S, Shang E, Qian D, Duan J. Scutellariae radix and coptidis rhizoma ameliorate glycolipid metabolism of type 2 diabetic rats by modulating gut microbiota and its metabolites. Appl Microbiol Biotechnol. 2020. Jan;104(1):303–317. [DOI] [PubMed] [Google Scholar]
- 13.Gu Y, Liu C, Zheng N, Jia W, Zhang W, Li H. Metabolic and gut microbial characterization of obesity-prone mice under a high-fat diet. J Proteome Res. 2019 Apr 5;18(4):1703–1714. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Wang H, Yin P, Fan H, Sun L, Liu Y. Flaxseed oil ameliorates alcoholic liver disease via anti-inflammation and modulating gut microbiota in mice. Lipids Health Dis. 2017 Feb 22;16(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiodini RJ, Dowd SE, Chamberlin WM, Galandiuk S, Davis B, Glassing A. Microbial population differentials between mucosal and submucosal intestinal tissues in advanced Crohn’s disease of the Ileum. PLoS ONE. 2015;10:e0134382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. BMJ. 2018 Jun 13;361:k2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thingholm LB, Rühlemann MC, Koch M, Fuqua B, Laucke G, Boehm R, Bang C, Franzosa EA, Hübenthal M, Rahnavard A, et al. Obese individuals with and without Type 2 diabetes show different gut microbial functional capacity and composition. Cell Host Microbe. 2019 Aug 14;26(2):252–264.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroke A, Klipstein-Grobusch K, Voss S, Möseneder J, Thielecke F, Noack R, Boeing H. Validation of a self-administered food-frequency questionnaire administered in the European Prospective Investigation into Cancer and Nutrition (EPIC) study: comparison of energy, protein, and macronutrient intakes estimated with the doubly labeled water, urinary nitrogen, and repeated 24-h dietary recall methods. Am J Clin Nutr. 1999. Oct;70(4):439–447. [DOI] [PubMed] [Google Scholar]
- 19.Fangmann D, Theismann E-M, Türk K, Schulte DM, Relling I, Hartmann K, Keppler JK, Knipp JR, Rehman A, Heinsen FA, et al. Targeted microbiome intervention by microencapsulated delayed-release niacin beneficially affects insulin sensitivity in humans. Diabetes Care. 2018;41(3):398–405. [DOI] [PubMed] [Google Scholar]
- 20.Ye M, Robson PJ, Eurich DT, Vena JE, Xu J-Y, Johnson JA. Cohort profile: Alberta’s tomorrow project. Int J Epidemiol. 2017 Aug 1;46(4):1097–1098l. [DOI] [PubMed] [Google Scholar]
- 21.Rohmann N, Schlicht K, Geisler C, Hollstein T, Knappe C, Krause L, Hagen S, Beckmann A, Seoudy AK, Wietzke-Braun P , et al. Circulating sDPP-4 is increased in obesity and insulin resistance but is not related to systemic metabolic inflammation. J Clin Endocrinol Metab. 2020 Oct. 21;106(2):e592–e601. [DOI] [PubMed] [Google Scholar]
- 22.Heinsen F-A, Fangmann D, Müller N, Schulte DM, Rühlemann MC, Türk K, Settgast U, Lieb W, Baines JF, Schreiber S, et al. Beneficial effects of a dietary weight loss intervention on human gut microbiome diversity and metabolism are not sustained during weight maintenance. Obes Facts. 2016;9(6):379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008. May;49(5):1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kind T, Fiehn O. Seven golden rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinform. 2007 Mar 27;8:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmermann J, Kaleta C, Waschina S. gapseq: informed prediction of bacterial metabolic pathways and reconstruction of accurate metabolic models. Genome Biol. 2021 Mar 10;22:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pryor R, Norvaisas P, Marinos G, Best L, Thingholm LB, Quintaneiro LM, De Haes W, Esser D, Waschina S, Lujan C, et al. Host-microbe-drug-nutrient screen identifies bacterial effectors of metformin therapy. Cell. 2019 Sep 5;178(6):1299–1312.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orth JD, Thiele I, Bø P. What is flux balance analysis? Nat Biotechnol. 2010. Mar;28(3):245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gelius-Dietrich G, Desouki AA, Fritzemeier CJ, Lercher MJ. Sybil – efficient constraint-based modelling in R. BMC Syst Biol. 2013 Nov 13;7:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y-J, Wu H, S-D W, Lu N, Wang Y-T, Liu H-N, Dong L, Liu TT, Shen XZ. Parasutterella, in association with irritable bowel syndrome and intestinal chronic inflammation: parasutterella may be related with IBS. J Gastroenterol Hepatol. 2018. Nov;33(11):1844–1852. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Thingholm LB, Skiecevičienė J, Rausch P, Kummen M, Hov JR, Degenhardt F, Heinsen FA, Rühlemann MC, Szymczak S, et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet. 2016. Nov;48(11):1396–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delday M, Mulder I, Logan ET, Grant G. Bacteroides thetaiotaomicron Ameliorates colon inflammation in preclinical models of Crohn’s disease. Inflamm Bowel Dis. 2019. Jan;25(1):85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, Li J, Zhu W, Kuang Y, Liu T, Zhang W, Chen X, Peng C. Skin and gut microbiome in psoriasis: gaining insight into the pathophysiology of it and finding novel therapeutic strategies. Front Microbiol [Internet]. 2020 Dec 15 [cited 2021 Mar 23];11. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7769758/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Zhao Y, Zhang M, Pang X, Xu J, Kang C, Li M, Zhang C, Zhang Z, Zhang Y, et al. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PLoS One [Internet]. 2012 Aug 3 [cited 2020 Dec 16];7(8). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3411811/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM, Gibson GR, Delzenne NM. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxemia. Diabetologia. 2007. Nov;50(11):2374–2383. [DOI] [PubMed] [Google Scholar]
- 35.Cheng W, Lu J, Lin W, Wei X, Li H, Zhao X, Jiang A, Yuan J. Effects of a galacto-oligosaccharide-rich diet on fecal microbiota and metabolite profiles in mice. Food Funct. 2018;9(3):1612–1620. [DOI] [PubMed] [Google Scholar]
- 36.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen -Y-Y, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011 Oct 7;334(6052):105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamada T, Takahashi D, Hase K. The diet-microbiota-metabolite axis regulates the host physiology. J Biochem. 2016. Jul;160(1):1–10. [DOI] [PubMed] [Google Scholar]
- 38.Jain SK, Velusamy T, Croad JL, Rains JL, Bull R. L-cysteine supplementation lowers blood glucose, glycated hemoglobin, CRP, MCP-1, oxidative stress and inhibits NFkB activation in the livers of Zucker diabetic rats. Free Radic Biol Med. 2009 Jun 15;46(12):1633–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ju T, Kong JY, Stothard P, Willing BP. Defining the role of Parasutterella, a previously uncharacterized member of the core gut microbiota. ISME J. 2019. Jun;13(6):1520–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blouet C, Mariotti F, Azzout-Marniche D, Mathé V, Mikogami T, Tomé D, Huneau JF. Dietary cysteine alleviates sucrose-induced oxidative stress and insulin resistance. Free Radic Biol Med. 2007 Apr 1;42(7):1089–1097. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Wang H. Oxidative stress in pancreatic beta cell regeneration. Oxid Med Cell Longev. 2017;2017:1930261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagai F, Morotomi M, Sakon H, Tanaka R. Parasutterella excrementihominis gen. nov., sp. nov., a member of the family Alcaligenaceae isolated from human faeces. Int J Syst Evol Microbiol. 2009;59:1793–1797. [DOI] [PubMed] [Google Scholar]
- 43.He J, Zhang P, Shen L, Niu L, Tan Y, Chen L, Zhao Y, Bai L, Hao X, Li X, et al. Short-chain fatty acids and their association with signalling pathways in inflammation, glucose and lipid metabolism. Int J Mol Sci. 2020 Sep 2;21(17):6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farooqi IS, Drop S, Clements A, Keogh JM, Biernacka J, Lowenbein S, Challis BG, O’Rahilly S. Heterozygosity for a POMC-null mutation and increased obesity risk in humans. Diabetes. 2006 Sep 1;55(9):2549–2553. [DOI] [PubMed] [Google Scholar]
- 45.Matsuoka K, Kanai T. The gut microbiota and inflammatory bowel disease. Semin Immunopathol. 2015;37:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kitai T, Tang WHW. GUT MICROBIOTA IN CARDIOVASCULAR DISEASE AND HEART FAILURE. Clin Sci (Lond). 2018 Jan 16;132(1):85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lauka L, Reitano E, Carra MC, Gaiani F, Gavriilidis P, Brunetti F, de’Angelis GL, Sobhani I, de’Angelis N. Role of the intestinal microbiome in colorectal cancer surgery outcomes. World J Surg Oncol [Internet]. 2019 Dec 2 [cited 2021 Mar 26];17. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6889350/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park HS, Park JY, Yu R. Relationship of obesity and visceral adiposity with serum concentrations of CRP, TNF-α and IL-6. Diabetes Res Clin Pract. 2005 Jul 1;69(1):29–35. [DOI] [PubMed] [Google Scholar]
- 49.Bhatt DL, Steg PG, Miller M, Brinton EA, Jacobson TA, Ketchum SB, Doyle Jr RT, Juliano RA, Jiao L, Granowitz C, et al. Cardiovascular risk reduction with icosapent ethyl for hypertriglyceridemia. N Engl J Med. 2019 Jan 3;380(1):11–22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Samples and data are stored in the PopGen Biobank (Schleswig-Holstein, Germany) and can be accessed via a structured application procedure (https://www.uksh.de/p2n/Information+for+Researchers.html).


