ABSTRACT
This study aimed to explore the role and potential mechanism of the long non-coding (lncRNA) MBNL1-AS1 in human breast cancer. We included 80 patients with breast cancer in this study. Breast cancer cell lines, including MCF7, SKBR3, MDA-MB-231 and MDA-MB-415, and the normal human breast cell line MCF10A were used in this study. MBNL1-AS1, miR-889-3p mimics, si-Krüppel-like factor 9 (KLF9) or their controls were transfected in the cells. Quantitative reverse transcription polymerase chain reaction (qRT-PCR), Western blotting and immunohistochemistry assay were performed to detect the expression of MBNL1-AS1, miR-889-3p and KLF9. Cell proliferation, invasion and migration were detected. Luciferase reporter gene and pull-down assay were performed to verify the target relationship among MBNL1-AS1, miR-889-3p and KLF9. Glycolysis was also detected after transfection. The expression of the lncRNA MBNL1-AS1 was low in the breast cancer tissues and cells. Lower expression levels of the lncRNA MBNL1-AS1 were associated with poor prognosis of breast cancer. Overexpression of the lncRNA MBNL1-AS1 decreased proliferation, invasion, migration and glycolysis of breast cancer cells. The lncRNA MBNL1-AS1 could interact with miR-889-3p, and KLF9 was the downstream target of miR-889-3p. Moreover, miR-889-3p was negatively correlated with KLF9 and lncRNA MBNL1-AS1. Both miR-889-3p and si-KLF9 could reverse the overexpression of lncRNA MBNL1-AS1 in breast cancer development. The lncRNA MBNL1-AS1 decreased proliferation, invasion, migration and glycolysis of breast cancer via the miR-889-3p/KLF9 axis, which might be a potential biomarker for the diagnosis of breast cancer.
KEYWORDS: Breast cancer, lncRNA MBNL1-AS1, miR-889-3p, KLF9
Introduction
Breast cancer is the second most common cancer in the world and the fourth leading cause of cancer-related deaths in women [1]. The incidence and mortality of female breast cancer in China has increased gradually in recent years, and recurrence and distant metastasis of breast cancer are the main causes of cancer-related deaths [2]. Previous studies have shown that hereditary factors and gene mutations (including BRCA1, BRCA2 and other breast cancer susceptibility genes) account for 5–10% of the total cases of breast cancer, which indicates that the treatment approach should focus on targeting the genes [3]. Molecular therapy has become a new biological treatment method for breast cancer in addition to the four traditional treatment methods, including surgery, radiotherapy, chemotherapy and endocrine therapy [4].
The expression of long non-coding RNA (lncRNA) is one of the most common transcriptional changes in cancer [5]. In addition, a previous study has demonstrated that lncRNAs play an important role in tumorigenesis [6] and participates in the occurrence, development, invasion and metastasis of breast cancer through various pathways [7]. Moreover, various lncRNAs related to the development of breast cancer have been studied till date. The lncRNA UCA1 participates in the Wnt/β-catenin signaling pathway, accelerates the epithelial-to-mesenchymal transition (EMT) and facilitates the development of breast cancer [8]. The levels of lncRNA GAS5 were low in patients with breast cancer; additionally, it induced apoptosis of breast cancer cells [9]. However, overexpression of the lncRNA HOTAIR not only improved the proliferation of breast cancer cells but also accelerated tamoxifen resistance [10]. In addition, the lncRNA HOTAIR interacted with FOXM1 and then differentiated between the patients who responded to endocrine therapy [11]. Although the number of new lncRNAs have increased, <1% of the functions of lncRNA have been examined to date [12]. Therefore, in addition to discovering new lncRNAs, their mechanism, function and potential application should be determined.
Previous studies have reported abnormal expression levels of MBNL1-AS1 in various malignant tumors, and MBNL1-AS1 is involved in the malignant progression of human non-small cell lung cancer [13], bladder cancer [14] and colon cancer [15]. However, the biological role of MBNL1-AS1 in human breast cancer remains unclear. This study aimed to explore the role and potential mechanism of action of MBNL1-AS1 in human breast cancer. Our results may provide important insights for improving the prognosis of breast cancer.
Materials and methods
Patients and tissues
From June 2019 to April 2020, breast cancer tissue and paired adjacent tissues of 80 cases were collected in Shanghai Seventh People’s Hospital Affiliated to Shanghai University of Traditional Chinese Medicine. The patient age ranged from 34 to 79 years, with a median age of 56.0 years. The American Cancer Council (AJCC) breast cancer staging system was used for clinical T staging, and these patients included 39 cases of TI/II and 41 cases of TIII/IV. All patients did not receive chemotherapy, endocrine therapy or molecular targeted therapy before surgery. This study was approved by the ethics committee of Shanghai Seventh People’s Hospital Affiliated to Shanghai University of Traditional Chinese Medicine, and all patients signed the informed consent forms. After the tissue sample was obtained, it was immediately stored in the refrigerator at −80°C.
Cell culture and transfection
Human breast cancer cell lines (MCF-7, SKBR3, MDA-MB-231 and MDA-MB-415) and normal breast epithelial cells (MCF-10A) were purchased from the Cell Resource Center, Shanghai Institutes for Biological Sciences and the Chinese Academy of Sciences. The cells were cultured in RPMI1640 medium containing 10% fetal bovine serum. An appropriate amount of penicillin–streptomycin–gentamicin solution was added to the medium to achieve a final concentration of 100-U/mL penicillin, 100-μg/mL streptomycin and 0.05-mg/mL gentamicin. The cells were cultured in an incubator at 37°C with 5% CO2.
One day before transfection, the cells in the logarithmic growth phase were inoculated into a 6-well plate. When the cell density reached 60–80%, transfection was performed using Lipofectamine 2000 (Invitrogen, USA) according to the instructions of the manufacturer. The plasmid overexpressing MBNL1-AS1 and its negative control plasmid were transferred into the cells. Both plasmids were designed and constructed by The Beijing Genomics Institute (BGI). Serum-free and antibiotic-free RPMI1640 medium was used during transfection. After 6 h of transfection, the medium was replaced with RPMI1640 medium containing 10% fetal calf serum. The cells were collected 24 h after transfection, and the expression level of lncRNA XIST in the cells was detected using quantitative reverse transcription-polymerase chain reaction (qRT-PCR) for subsequent experiments.
Immunohistochemistry assay
Frozen breast cancer and adjacent tissue sections were obtained, and then deparaffinized in xylene, absolute ethanol, ethanol and phosphate-buffered saline (PBS) solution. The sections were treated with 3% hydrogen peroxide and washed thrice with PBS solution, and the antigen was retrieved using a microwave retrieval method and placed in sodium citrate buffer. After heating in a microwave oven to boiling, the slices were cooled, washed with PBS and sealed for 10 min. The primary antibody was added to the sectioned tissue and incubated at 4°C for 12 h. After washing, the sample was incubated with the secondary antibody at 4°C for 20 min. 3,3'-diaminodbenzidine (DAB) chromogenic solution was added, and the sample was counterstained with hematoxylin and dehydrated in different gradients of ethanol, mounted with neutral resin and observed under a microscope. Five high-power fields were selected for counting the number of stained cells.
qRT-PCR assay
According to the instructions in the kit, the total RNA in the breast cancer tissue specimens was extracted, and the absorbance value was measured using an ultraviolet spectrophotometer. RNA samples with a ratio (260 nm/280 nm) between 1.8 and 2.0 were used for subsequent experiments. The RNA samples were reverse transcribed into cDNA using the Revertra Ace qRT-PCR Master Mix reverse transcription kit (ToYoBo, Japan) and stored at −80°C. The fluorescence quantitative PCR kit was purchased from TaKaRa Company. PCR detection was performed according to the manufacturer’s instructions. The reaction system included 10-μL SYBR Green Realtime PCR MasterMix, 1 μL of 10-μmol/L upstream and downstream primers, 2.5-μL cDNA templates and 5.5-μL ddH2O. The cycle parameters were 95°C for 60s, 95°C for 15s, 60°C for 34s and 72°C for 45s for a total of 40 cycles. The fluorescence signal was recorded at 60°C, and the purity of the product was analyzed using the melting curve. The final result was expressed using the 2-ΔCt method. The experiment was repeated thrice.
Cell counting kit 8 assay for proliferation
The cell counting kit 8 (CCK-8) assay was used to detect the growth and proliferation status of the cells after transfection. In a 96-well plate, 100 μL of suspension (concentration of 2,000 cells per 100 μL) was added to each well. After overnight incubation, 10 μL of MBNL1-AS1, MBNL1-AS1+ miR-889-3p or MBNL1-AS1-si-Krüppel-like factor 9 (KLF9) and the control transfection solution were added. Three multiple wells were set in each group, and 90 μL of medium was added to each well. After 24, 48 and 72 h of plating, the medium was discarded individually from each well, and 100 μL of the medium containing 10% CCK-8 reagent was added. After incubating for 1 h at 37°C, the optical density (OD) value of each well was measured at a wavelength of 450 nm.
Transwell assay
Pre-cooled Matrigel glue and serum-free RPMI 1640 medium were mixed in a ratio of 1:9, and 80 μL per well of this mixture was evenly coated on the bottom membrane of the Transwell chamber and was placed in an incubator. After 24 h of transfection, the cells were digested and resuspended, and a single cell suspension was prepared in a serum-free medium at a concentration of 3 × 104 cells/mL. Furthermore, 200 μL of the cell suspension was added to the upper chamber of the Transwell chamber, and 500 μL of RPMI1640 medium containing 10% fetal calf serum was added to the lower chamber for culture. After culturing for 24 h, the cells in the upper chamber were carefully wiped off with cotton swabs. The cells were washed thrice with PBS and fixed with 4% paraformaldehyde fixative solution for 20 min at room temperature. The cotton swab was used to suck up the liquid in the upper chamber, and the cells were stained with 0.1% crystal violet at room temperature and protected from light for 10 min. After rinsing the chamber, stained cells were photographed and counted.
Colony formation assay
Cells in each group were seeded in 6-well plates at a density of 300 cells/well, and 2 mL of culture medium was added to each well. After culturing for 10 days, the cells were fixed with 4% paraformaldehyde for 15 minutes and stained with 1% crystal violet for 4 h. After washing and drying, images were taken to observe and count the number of cells. After washing and drying, the cells were observed and the numbers of cells were counted.
Scratch healing assay
Cells in the logarithmic growth phase were seeded on a 6-well plate at 1 × 105 cells/well. After overnight incubation, 10 μL of MBNL1-AS1, MBNL1-AS1+ miR-889-3p or MBNL1-AS1-siKLF9 and the control transfection solution were added. After 24 h of transfection, a pipette tip was used to make a straight line on the cell surface. The cell migration of each group was observed and measured after 24 h.
Glycolysis assay
Glucose uptake
Glucose oxidase catalyses the oxidation of the glucose in the sample to produce gluconic acid and hydrogen peroxide. Peroxidase catalyses the coupling of the reducing 4-aminoantipyrin and phenol and condensation into quinone compounds. Briefly, the process was as follows: after transfection, the cells were plated in a 6-well plate at a density of 1 × 106 cells/well. The final volume of the medium was 3 mL, and the sample was placed in an incubator at 37°C for 48 h. The medium was collected 48 h after the cells were cultured, and the medium collected at 0 h of culture was used to determine the background glucose concentration. Subsequently, 10 μL of the sample was added to 1000 μL of the working solution. The glucose uptake colorimetric assay kit was purchased from Biovision Co. Ltd (USA). After mixing the sample, it was added to a 96-well plate (100 μL/well) and incubated in a water bath at 37°C for 15 min. The absorbance was measured at a wavelength of 505 nm.
Lactose production
Conversion of lactate to pyruvate was catalyzed by lactate dehydrogenase, which converted NAD+ into NADH. Phenazine methosulphate (PMS) mediated the reduction of nitroblue tetrazolium (NBT), which appeared purple and could be detected using a spectrophotometer at 530 nm. Lactate assay kit was purchased from Biovision Co. Ltd. The procedure performed was similar to that of the glucose uptake assay.
Luciferase reporter experiment
The binding site in the 3ʹ-untranslated region (UTR) sequence of the target gene was mutated and cloned into the p-LUC vector. The dual luciferase reporter gene vector (PLU) containing the 3ʹ-UTR sequence of the target gene was synthesized by Guangzhou Ruibo Biotechnology Co., Ltd. Lipofectamine 2000 (Invitrogen) was used to transfect human embryonic kidney (HEK293) cells with the pLUC vector with or without the 3ʹ-UTR sequence of the target gene. The G418 antibiotic was used to screen stable transfected cell lines. Subsequently, the chemically synthesized miR-889-3p mimic or control was transfected into stable cell lines. After 48 h of transfection, the fluorescence intensity was measured according to the instructions of the Dual-luciferase reporter assay system (Promega, USA).
RNA pull-down assay
The pGEM-T-lncRNA MBNL1-AS1 vector was digested with SalI, purified using a gel, added to the in vitro transcription mixture and incubated at 37°C for 3 h. After the colorless liquid became turbid, DNase I was added to digest the DNA template and incubated at 37°C for 15 min. RNA was extracted using the kit to obtain biotin-labeled RNA transcripts. Subsequently, 3 μg of the obtained RNA was heated at 90°C for 2 min to denature the RNA. Thereafter, an equal volume of RNA-binding buffer was added, and the mixture was incubated at room temperature for 30 min to create the RNA fold into a high-level structure. The folded RNA was mixed with 1 mg of total cell protein solution, vortexed at room temperature and incubated for 1 h while leaving a portion of the protein lysate as an input. A total of 40 μL magnetic beads were added to each sample and incubated overnight with vortexing at 4°C. Subsequently, the magnetic beads were washed 8 times, and proteinase K buffer was added and incubated in a molecular hybridization oven at 55°C for 30 min. The supernatant was transferred to a new centrifuge tube, and RNA was extracted via phenol–chloroform extraction. After reverse transcription, qRT-PCR was performed to detect the adsorbed miRNA.
Western blot assay
The cells were collected after 48 h of transfection. Radioimmunoprecipitation assay (RIPA) cell lysate (Beyotime Biotechnology, China) was used to extract the total protein. A BCA kit was used to determine the protein concentration, and the protein content of the sample was adjusted. We added 50 μg of protein sample to each well, and electrophoresis, membrane transfer and sealing were performed. The sample was incubated with the primary antibody (1:1000) and placed in a refrigerator at 4°C overnight. After washing the membrane thrice with PBS Tween 20 (PBST) buffer, secondary antibody was added to the sample and incubated for 1 h. Finally, electrochemiluminescence (ECL) was observed, and the relative expression of KLF9 was calculated.
Statistical analysis
SPSS 17.0 statistical software was used for statistics. The relative expression of genes was expressed as mean ± standard deviation, and comparison between groups was performed by t test. The survival curve was analyzed by Kaplan-Meier. Pearson correlation coefficient was applied to evaluation relevance among lncRNA MBNL1-AS1, miR-889-3p and KLF9. P less than 0.05 were considered statistically significant.
Results
Low expression levels of lncRNA MBNL1-AS1 are associated with poor prognosis of breast cancer
Results of qRT-PCR performed to detect the expression of lncRNA MBNL1-AS1 in 80 pairs of breast cancer tissues and adjacent tissues showed that the expression of MBNL1-AS1 was downregulated in breast cancer tissues (Figure 1(a)). In addition, the results of immunohistochemistry assay were similar to those of qRT-PCR (Figure 1(b)). Moreover, the expression of MBNL1-AS1 was lower in patients with stage III/IV breast cancer than in patients with stage I/II breast cancer (Figure 1(c)). Furthermore, we determined the expression of MBNL1-AS1 in controls relative to that in breast cancer cells. MBNL1-AS1 expression levels were lower in the breast cancer cells than in controls (Figure 1(d)). The lowest expression of MBNL1-AS1 was observed in MCF7 and SKBR3 cell lines; therefore, we used these two cell lines for subsequent experiments. We divided all patients into the high- and low-MBNL1-AS1-expression groups on the basis of the median value of MBNL1-AS1. Clinical data indicated that low MBNL1-AS1 expression levels were associated with a poor prognosis of breast cancer (Figure 1(e)). Additionally, we analyzed the the correlation between MBNL1-AS1 expression and the clinical pathology of breast cancer patients. It was found that MBNL1-AS1 expression was related to the tumor size, stage, and TNM stage of breast cancer patients (Table 1).
Figure 1.
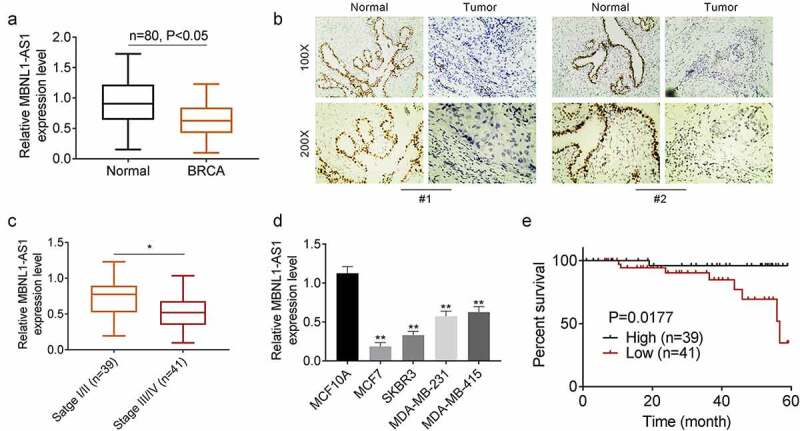
Lower expressed lncRNA MBNL1-AS1 is associated with poor prognosis of breast cancer A.QRT-PCR assay for detecting the expression of lncRNA MBNL1-AS1 in 80 pairs of breast cancer tissues and adjacent tissues. B. Immunohistochemistry assay for lncRNA MBNL1-AS1 expression. C. lncRNA MBNL1-AS1 expression in different stage. D. The expression of lncRNA MBNL1-AS1 in breast cancer and normal cells. E. Survival rate of patients with low and high lncRNA MBNL1-AS1 levels. *P < 0.05, **P < 0.01.
Table 1.
The correlation between MBNL1-AS1 expression and breast cancer clinical pathology
| Characteristics | Number of patients | MBNL1-AS1 high expression (> median) |
MBNL1-AS1 low expression (≤ median) |
P value |
|---|---|---|---|---|
| Number | 80 | 39 | 41 | |
| Ages(years) | 0.371 | |||
| <50 | 38 | 21 | 17 | |
| ≥50 | 42 | 18 | 24 | |
| Tumor size | 0.027 | |||
| ≤5 cm | 43 | 26 | 17 | |
| >5 cm | 37 | 13 | 24 | |
| Stage | 0.043 | |||
| I–II | 39 | 24 | 15 | |
| III–IV | 41 | 15 | 26 | |
| TMN stage | 0.048 | |||
| I–II | 42 | 25 | 17 | |
| III–IV | 38 | 14 | 24 | |
| Lymphatic metastasis | 0.509 | |||
| Yes | 44 | 23 | 21 | |
| No | 36 | 16 | 20 |
Overexpression of lncRNA MBNL1-AS1 inhibits proliferation, migration, and glycolysis of breast cancer cells
A cell line overexpressing MBNL1-AS1 was constructed, and the expression of MBNL1-AS1 was detected using qRT-PCR. The expression levels of MBNL1-AS1 were higher in the MBNL1-AS1 group than in the control group (Figure 2(a)). The CCK8 experiment was used to analyze the effect of MBNL1-AS1 overexpression on the proliferation of MCF7 and SKBR4 cells at 0, 24, 48 and 72 h. At 72 h after MBNL1-AS1 transfection, the proliferation of both MCF7 and SKBR4 decreased significantly (Figure 2(b)). In addition, we examined the effects of MBNL1-AS1 overexpression on the clone formation of MCF7 and SKBR4 cells. The number of cell clones formed decreased significantly, indicating that MBNL1-AS1 overexpression inhibited the proliferation of breast cancer cells (Figure 2(c)). Moreover, invasion and migration were suppressed by MBNL1-AS1 (Figure 2(d,e)). Additionally, the results of the glycolysis assay confirmed that relative glucose uptake and lactate production were inhibited in the group overexpressing MBNL1-AS1 (Figure 2(f)). Therefore, we inferred that overexpression of MBNL1-AS1 inhibited the proliferation, migration and glycolysis of breast cancer cells.
Figure 2.
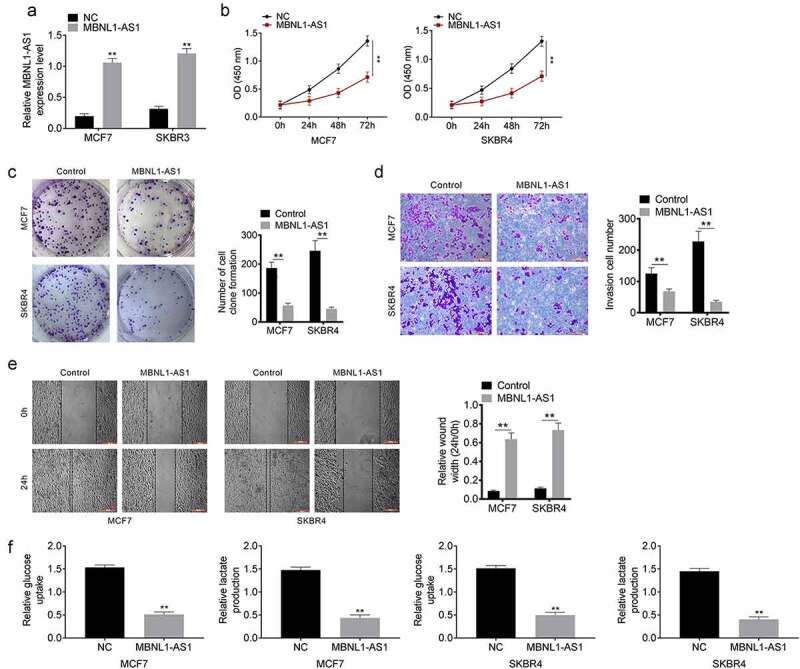
Overexpression of lncRNA MBNL1-AS1 inhibits proliferation, migration and glycolysis of breast cancer cells A. The lncRNA MBNL1-AS1 overexpression cell line was successfully constructed. B. LncRNA MBNL1-AS1 inhibited proliferation of MCF7 and SKBR4 cells. C. Clone formation after lncRNA MBNL1-AS1 transfection. D. Invasion of MCF7 and SKBR4 cells after lncRNA MBNL1-AS1 transfection. E. Migration of MCF7 and SKBR4 cells after lncRNA MBNL1-AS1 transfection. F. Glucose uptake and lactate production. **P < 0.01.
Interaction of lncRNA MBNL1-AS1 with miR-889-3p
Bioinformatic databases Starbase and miRDB were used to predict the candidate target miRNA of MBNL1-AS1. miR-889-3p, which was involved in the progression of various cancers, was selected as a candidate target. The binding site is shown in Figure 3(a). We examined the expression of miR-899-3p in 80 pairs of clinical samples, and the results confirmed that miR-889-3p was upregulated in tumor tissues (Figure 3(b)). Dual-luciferase reporter assay showed the targeting relationship between MBNL1-AS1 and miR-899-3p in HEK293T cells. The miR-889-3p mimic decreased the relative luciferase activity of MBNL1-AS1 in the wild-type (wt) cells, whereas no significant effect was observed in the luciferase activity of MBNL1-AS1 in the mutant (mut) groups (Figure 3(c)). RNA pull-down assay was performed to verify the binding between MBNL1-AS1 and miR-899-3p in MCF7 and SKBR4 cells. The enrichment of miR-889-3p was higher with the biotinylated WT lncRNA (bio-MBNL1-AS1-WT) than with biotinylated mutant lncRNA (bio-MBNL1-AS1-mut) (Figure 3(d)). Additionally, we examined the effect of MBNL1-AS1 overexpression on the expression of miR-889-3p in MCF7 and SKBR4 cells. Overexpression of MBNL1-AS1 significantly decreased miR-899-3p expression in both MCF7 and SKBR4 cells (Figure 3(e)).
Figure 3.
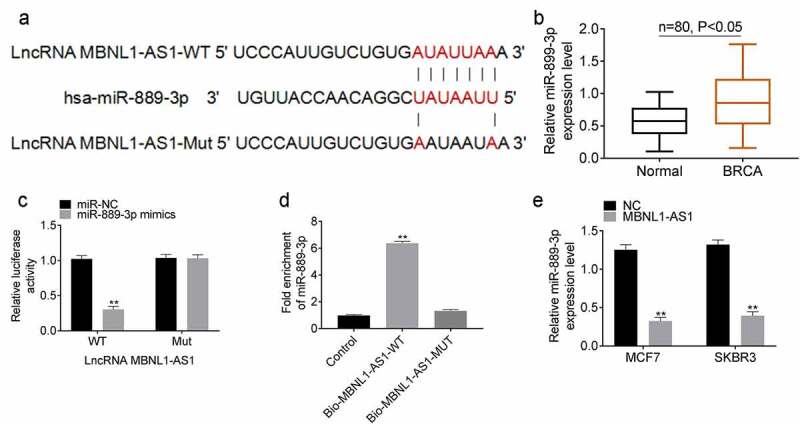
lncRNA MBNL1-AS1 interacted with miR-889-3p A. The binding site of lncRNA MBNL1-AS1 and miR-889-3p. B. The expression of miR-889-3p in breast cancer and normal tissues. C. Dual luciferase reporter experiment verifies the targeting relationship between lncRNA MBNL1-AS1 and miR-899-3p in HEK293T cells. D. The binding relationship between lncRNA MBNL1-AS1 and miR-899-3p in MCF7 and SKBR4 cells was verified by RNA pull down assay. E. The effect of lncRNA MBNL1-AS1 overexpression on miR-889-3p expression in MCF7 and SKBR4 cells. **P < 0.01.
KLF9 is the downstream target of miR-889-3p
The downstream target mRNA of miR-889-3p was predicted using the biological information database TargetSCan and miRanda. KLF9 was selected as a candidate target, which was involved in the progression of multiple cancers. The binding site of KLF9 and miR-889-3p is shown in Figure 4(a). KLF9 expression in 80 pairs of clinical samples was analyzed using qRT-PCR. The expression of KLF9 was downregulated in tumor tissues (Figure 4(b)). The results of Western blotting and immunohistochemical analysis in pairs of clinical samples showed that KLF9 was downregulated in tumor tissues (Figure 4(c,d)). Luciferase reporter experiment was performed to verify the binding of miR-889-3p mimics and KLF9. The miR-889-3p mimic significantly decreased luciferase activity in the KLF9-wt group, whereas it did not affect the KLF9-mut group (Figure 4(e)). qRT-PCR and Western blotting were used to determine the effect of miR-889-3p mimics on the expression of KLF9 in MCF7 and SKBR4 cells. Our results showed that miR-889-3p mimics downregulated KLF9 expression in both MCF7 and SKBR3 cells (Figure 4(f)). These results confirmed that KLF9 was the downstream target of miR-889-3p.
Figure 4.
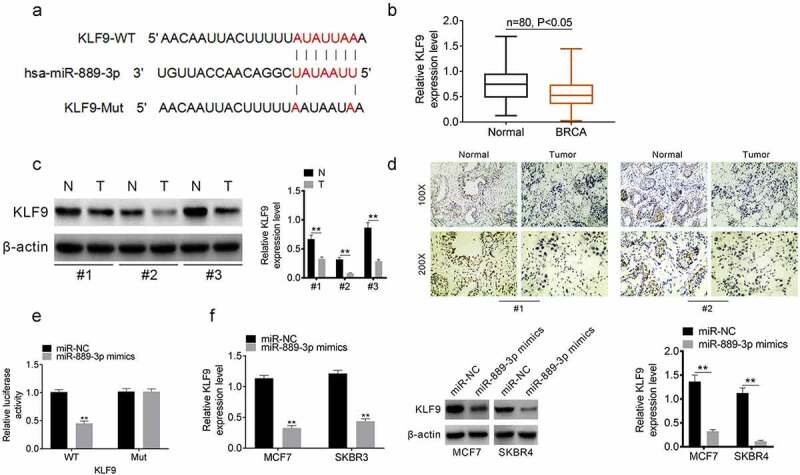
KLF9 is the downstream target of miR-889-3p A. The binding site of KLF9 and miR-889-3p. B. KLF9 expression in 80 pairs of clinical samples. C. Western blot results of KLF9 expression in 3 pairs of clinical samples. D. Immunohistochemical analysis of KLF9 expression in 2 pairs of clinical samples. E. Luciferase reporter experiment for verifing the binding of miR-889-3p mimics and KLF9. F. QRT-PCR and Western blot for analyzing the effect of miR-889-3p mimics on the expression of KLF9 in MCF7 and SKBR4 cells. **P < 0.01.
MiR-889-3p and si-KLF9 rescued the effect of upregulated lncRNA MBNL1-AS1 in breast cancer
We performed a rescue experiment to determine the effect of lncRNA MBNL1-AS1 upregulation. MCF7 cells were divided into MBNL1-AS1, MBNL1-AS1 + miR-889-3p, MBNL1-AS1+ si KLF9, and control groups based on the transfection. Western blotting was performed to determine the effect of miR-889-3p and si-KLF9 transfection on the expression of KLF9. MBNL1-AS1 upregulated KLF9 expression, whereas miR-889-3p and si-KLF9 rescued the effects of MBNL1-AS1 (Figure 5(a)). Moreover, MBNL1-AS1 inhibited invasion, migration and progression of glycolysis, whereas miR-889-3p and si-KLF9 rescued the effects (Figure 5(c-e)).
Figure 5.
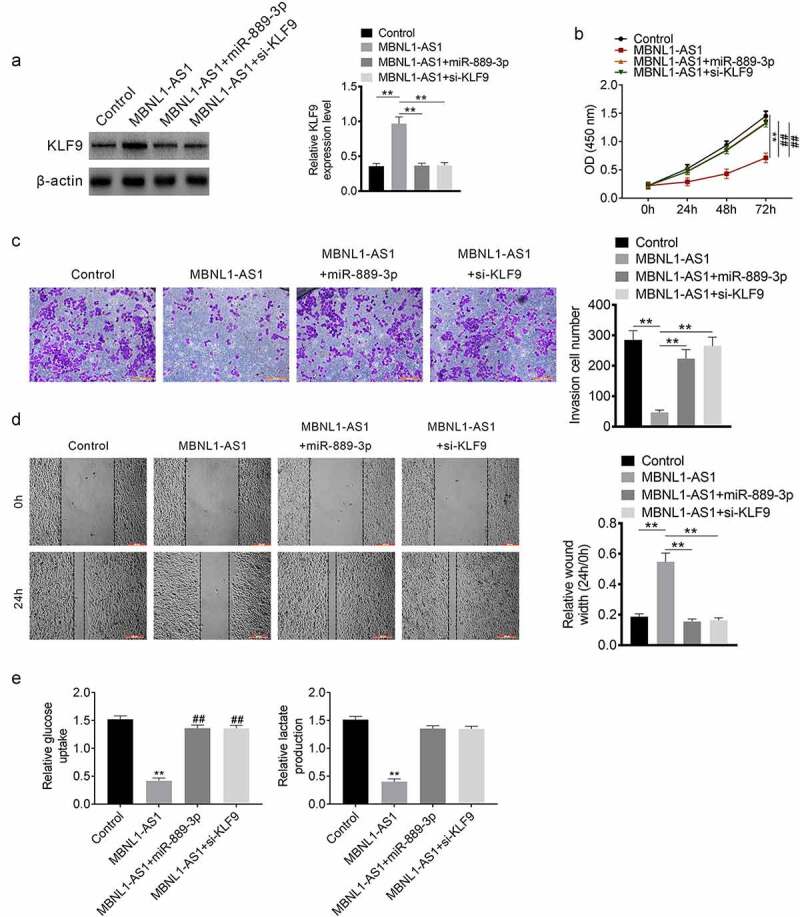
Rescue experiment MCF7 cells were divided into MBNL1-AS1, MBNL1-AS1+ miR-889-3p, MBNL1-AS1+ si KLF9 and control groups based on the transfection. A. Western blot assay. B. Transwell assay was performed for verifying invasion. C. Scratch healing assay was undertaken to detect mgration. E. Glucose uptake and lactate production. **P < 0.01.
Spearman correlation analysis
Spearman correlation analysis was used to evaluate the correlation between the expression of MBNL1-AS1, miR-889-3p and KLF9 in 80 pairs of tumor tissues. MiR-889-3p was negatively correlated with MBNL1-AS1 and KLF9 (Figure 6(a,b)). MBNL1-AS1 was positively correlated with KLF9 (Figure 6(c)).
Figure 6.
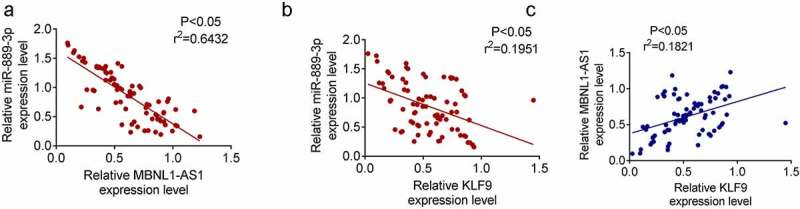
Spearman correlation analysis A. MiR-889-3p was with negative correlation with MBNL1-AS1. B. MiR-889-3p was with negative correlation with KLF9. C. MBNL1-AS1 and KLF9 were with positive correlation.
Discussion
lncRNAs are abnormally expressed in various cancers and play a key role in promoting and maintaining the formation and progression of tumors [16]. Therefore, screening and identifying more effective lncRNAs is important for the diagnosis and treatment of breast cancer. Our results showed that lncRNA MBNL1-AS1 was expressed in low levels in breast cancer tissues and cells. We examined the effect of MBNL1-AS1 on the proliferation, invasion, migration and glycolysis of breast cancer cells to determine the role and understand the mechanism of action of MBNL1-AS1 in human breast cancer. Additionally, we examined the effect of miR-889-3p and KLF9.
MBNL1-AS1 inhibits proliferation, invasion and migration in various cancers. MBNL1-AS1 was downregulated in colon cancer, which regulated the expression of miR-412-3p [15]. Furthermore, upregulation of MBNL1-AS1 accelerated the apoptosis of non-small-cell lung cancer cells via miR-135a-3p [13]. Furthermore, MBNL1-AS1 regulated the miR-135a/PHLPP2/FOXO1 axis, increased apoptosis and inhibited the proliferation of bladder cancer [14]. Among these miRNAs regulated by MBNL1-AS1, miR-135a regulates the expression of HOXA10 and then accelerates the invasion and migration of breast cancer cells [17]. Moreover, miR-135a was involved in the infiltration and progression of breast cancer [[18]]. Our results showed that low expression levels of MBNL1-AS1 were associated with a poor prognosis of breast cancer. Overexpression of MBNL1-AS1 decreases the proliferation, invasion, migration and glycolysis of breast cancer cells and hence plays an important role in the invasion, migration and proliferation of breast cancer.
Furthermore, our results showed that MBNL1-AS1 interacted with miR-889-3p. Results of previous studies showed that miR-889-3p increases cell growth and progression in various cancers by regulating different genes such as ZEB1 [19], E2F7 [20] and DAB2IP [21]. Ge et al. reported that miR-889-3p promoted the proliferation of osteosarcoma through inhibiting the expression of myeloid cell nuclear differentiation antigen (MNDA) [22]. Xu et al. showed that miR-889 overexpression stimulated proliferation of EC109 and EC9706 cells by targeting DAB2IP in esophageal squamous cell carcinomas [21]. Xiao et al. demonstrated that miR-889-3p targeting DAB2IP/ZEB1 regulated the arsenite-induced acquisition of cancer stem cells-like properties by human keratinocytes in carcinogenesis [19]. ZEB1 is an important transcription factor that regulates EMT, and EMT is an important process that promotes the invasion of cancer cells to the adjacent tissues and their infiltration and metastasis [23]. In addition, E2F7 had additional cyclin binding domains, which can simultaneously mediate cell proliferation and p53-dependent/independent apoptosis [24,25]. DAB2IP hypermethylation was detected in breast cancer cell lines and lymph node metastasis samples, which suggested that DAB2IP methylation was related to lymph node metastasis of breast cancer [26]. Previous studies have shown that miR-889-3p-related genes are closely associated with the metastasis and proliferation of cancer. Hayes et al. [27] showed that miR-889 was related to estrogen levels, development of breast cancer and tamoxifen resistance. In the present study, we found that miR-889-3p was overexpressed in breast cancer samples, and miR-889-3p could reverse the anti-tumor effect of MBNL1-AS1 on breast cancer cells. Thus, we proposed that miR-889-3p might act as a tumorigenesis role in breast cancer, and more detailed studies are needed. Therefore, the lncRNA MBNL1-AS1 decreased the proliferation, invasion, and migration of cancer via miR-889-3p.
Our results showed that KLF9 was the downstream target of miR-889-3p. KLF9 was a member of the KLF family. The genes in the family are highly conserved zinc finger proteins that played important roles in the suppression and tumorigenicity of breast cancer tumors [28]. For example, KLF4 was a tumor suppressor, which was downregulated in human tumor cell samples [29]. In addition, KLF4 promoted the expression of CDH1, thereby inhibiting the process of EMT in tumor cells [30]. KLF9 was involved in the E2-mediated signal transduction and estrogen response of breast cancer cells [31]. Additionally, the methylation of KLF9 has been proven to be a potential independent biomarker in breast cancer [32]. Moreover, our results showed that both miR-889-3p and si-KLF9 could rescue the effect of upregulated lncRNA MBNL1-AS1 in the development of breast cancer. Therefore, KLF4 was a critical factor in inhibiting the effects of MBNL1-AS1 in breast cancer.
Our results showed that MBNL1-AS1 decreased the glycolysis of breast cancer. Breast cancer is a highly heterogeneous malignant tumor, and different types of breast cancer have different characteristics of glycolytic metabolism [33]. Previous studies have shown that the metastatic breast cancer cell line MDA shows high levels of glycolysis under aerobic conditions, whereas the non-metastatic breast cancer cell line MCF-7 shows an increase in the levels of glycolysis under low oxygen conditions [34]. High levels of glycolysis can provide tumor cells with sufficient ATP and NADPH to repair DNA damage caused by anti-tumor drugs, promote intracellular drug efflux, repair oxidative damage and promote the drug resistance of breast cancer cells in the high-level glycolysis state [35].
In conclusion, MBNL1-AS1 decreased the proliferation, invasion, migration and glycolysis of breast cancer cells via the miR-889-3p/KLF9 axis and may be a potential biomarker for the diagnosis of breast cancer. However, to date, a limited number of studies have investigated the complex interactions between lncRNA MBNL1-AS1 and related miRNAs and proteins in breast cancer. Additional detailed studies are required to identify the mechanisms underlying the role of MBNL1-AS1 and design potential targeted drugs for breast cancer.
Funding Statement
This study is supported by Shanghai Municipal Health Commission Project[(No. 201940502)]and Pudong New Area Science and Technology Development Fund [(PKJ2019-Y14)].
Highlights
Lower expressed lncRNA MBNL1-AS1 is associated with poor prognosis of breast cancer.
Overexpression of lncRNA MBNL1-AS1 inhibits proliferation and migration of breast cancer cell.
lncRNA MBNL1-AS1 can interact with miR-889-3p.
KLF9 is the downstream target of miR-889-3p.
lncRNA MBNL1-AS1 regulates proliferation and migration of breast cancer cells through miR-889-3p/KLF9
Authors’ contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Availability of data and materials
All data generated and/or analyzed during this study are included in this published article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Ethics approval and consent to participate
The project protocol was authorized by Ethic Committee of Shanghai Seventh People’s Hospital Affiliated to Shanghai University of Traditional Chinese Medicine and carried out in accordance with the Guidelines of Shanghai Seventh People’s Hospital Affiliated to Shanghai University of Traditional Chinese Medicine.
References
- [1].Biersack B, Schobert R. “Current State of Metal-Based Drugs for the Efficient Therapy of Lung Cancers and Lung Metastases.” Lung Cancer and Personalized Medicine Springer. 211–224, 2016. [DOI] [PubMed] [Google Scholar]
- [2].Zeng H, Zheng R, Zhang S, et al. Incidence and mortality of female breast cancer in C hina, 2009. Thorac Cancer. 2013;4(4):400–404. [DOI] [PubMed] [Google Scholar]
- [3].Ford D, Easton D, Stratton M, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Am J Hum Genet. 1998;62(3):676–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lin S-X, Chen J, Mazumdar M, et al. Molecular therapy of breast cancer: progress and future directions. Nat Rev Endocrinol. 2010;6(9):485. [DOI] [PubMed] [Google Scholar]
- [5].Sun T, Ye H, Wu C-L. Lee G-SM and Kantoff PW: emerging players in prostate cancer: long non-coding RNAs. Am J Clin Exp Urol. 2014;2(4):294. [PMC free article] [PubMed] [Google Scholar]
- [6].Wang J, Liu X, Wu H, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38(16):5366–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li N, Gao W, Liu N. LncRNA BCAR4 promotes proliferation, invasion and metastasis of non-small cell lung cancer cells by affecting epithelial-mesenchymal transition. Eur Rev Med Pharmacol Sci. 2017;21(9):2075–2086. [PubMed] [Google Scholar]
- [8].Xiao C, Wu C, Hu H. LncRNA UCA1 promotes epithelial-mesenchymal transition (EMT) of breast cancer cells via enhancing Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2016;20(13):2819–2824. [PubMed] [Google Scholar]
- [9].Pickard MR, Williams GT. The hormone response element mimic sequence of GAS5 lncRNA is sufficient to induce apoptosis in breast cancer cells. Oncotarget. 2016;7(9):10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Xue X, Yang YA, Zhang A, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35(21):2746–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Milevskiy MJ, Al-Ejeh F, Saunus JM, et al. Long-range regulators of the lncRNA HOTAIR enhance its prognostic potential in breast cancer. Hum Mol Genet. 2016;25(15):3269–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang G, Liu C, Deng S, et al. Long noncoding RNAs in regulation of human breast cancer. Brief Funct Genomics. 2016;15(3):222–226. [DOI] [PubMed] [Google Scholar]
- [13].Cao G, Tan B, Wei S, et al. Down-regulation of MBNL1-AS1 contributes to tumorigenesis of NSCLC via sponging miR-135a-5p. Biomed Pharmacother. 2020;125:109856. [DOI] [PubMed] [Google Scholar]
- [14].Wei X, Yang X, Wang B, et al. Lnc RNA MBNL 1- AS 1 represses cell proliferation and enhances cell apoptosis via targeting miR-135a-5p/ PHLPP2/FOXO1 axis in bladder cancer. Cancer Med. 2020;9(2):724–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhu K, Wang Y, Liu L, et al. Long non-coding RNA MBNL1-AS1 regulates proliferation, migration, and invasion of cancer stem cells in colon cancer by interacting with MYL9 via sponging microRNA-412-3p. Clin Res Hepatol Gastroenterol. 2020;44(1):101–114. [DOI] [PubMed] [Google Scholar]
- [16].Liu Y, Zhao M. lnCaNet: pan-cancer co-expression network for human lncRNA and cancer genes. Bioinformatics. 2016;32(10):1595–1597. [DOI] [PubMed] [Google Scholar]
- [17].Chen Y, Zhang J, Wang H, et al. miRNA-135a promotes breast cancer cell migration and invasion by targeting HOXA10. BMC Cancer. 2012;12(1):111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ahmad A, Zhang W, Wu M, Tan S, and Zhu T.. Tumor-suppressive miRNA-135a inhibits breast cancer cell proliferation by targeting ELK1 and ELK3 oncogenes. Genes Genomics. 2018. Mar;40(3):243–251. [DOI] [PubMed] [Google Scholar]
- [19].Xiao T, Xue J, Shi M, et al. Circ008913, via miR-889 regulation of DAB2IP/ZEB1, is involved in the arsenite-induced acquisition of CSC-like properties by human keratinocytes in carcinogenesis. Metallomics. 2018;10(9):1328–1338. [DOI] [PubMed] [Google Scholar]
- [20].Zhou H, Guo R, Wang C. Long non-coding RNA NEAT1 accelerates cell progression in cervical cancer by regulating the miR-889-3p/E2F7 axis through the activation of the PI3K/AKT pathway. RSC Adv. 2019;9(59):34627–34635. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [21].Xu Y, He J, Wang Y, et al. miR-889 promotes proliferation of esophageal squamous cell carcinomas through DAB2IP. FEBS Lett. 2015;589(10):1127–1135. [DOI] [PubMed] [Google Scholar]
- [22].Ge D, Chen H, Zheng S, et al. Hsa-miR-889-3p promotes the proliferation of osteosarcoma through inhibiting myeloid cell nuclear differentiation antigen expression. Biomed Pharmacother. 2019;114:108819. [DOI] [PubMed] [Google Scholar]
- [23].Christiansen JJ, Rajasekaran AK. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res. 2006;66(17):8319–8326. [DOI] [PubMed] [Google Scholar]
- [24].Pandit SK. Novel functions for atypical E2Fs, E2F7 and E2F8, in polyploidization and liver cancer. Utrecht University; 2014. [Google Scholar]
- [25].Schlereth K, Heyl C, Krampitz A-M, et al. Characterization of the p53 cistrome–DNA binding cooperativity dissects p53ʹs tumor suppressor functions. PLoS Genet. 2013;9(8):e1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dote H, Toyooka S, Tsukuda K, et al. Aberrant Promoter Methylation in Human DAB2 Interactive Protein (hDAB2IP) Gene in Breast Cancer. Clin Cancer Res. 2004;10(6):2082–2089. [DOI] [PubMed] [Google Scholar]
- [27].Hayes EL, Knapp J, Lewis-Wambi J. MicroRNA analysis of aromatase inhibitor-resistant breast cancer cells reveals upregulation of a unique miRNA cluster on chromosome 14. AACR; 2015. [Google Scholar]
- [28].Farrugia MK. The Diverse Relationship of Kruppel-Like Factors 4 and 5 in Breast Cancer. 2015.
- [29].Guan H, Xie L, Leithäuser F, et al. KLF4 is a tumor suppressor in B-cell non-Hodgkin lymphoma and in classic Hodgkin lymphoma. Blood J Am Soc Hematol. 2010;116:1469–1478. [DOI] [PubMed] [Google Scholar]
- [30].Tiwari N, Meyer-Schaller N, Arnold P, et al. Klf4 is a transcriptional regulator of genes critical for EMT, including Jnk1 (Mapk8). PloS One. 2013;8(2):e57329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pitch MA, Russell BA, Greene GL. Characterization of the role of KLF9 transcription factor in breast cancer estrogen response. AACR; 2012. [Google Scholar]
- [32].Wang L, Mao Q, Zhou S, et al. Hypermethylated KLF9 Is An Independent Prognostic Factor For Favorable Outcome In Breast Cancer. Onco Targets Ther. 2019;12:9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Laranjo M, Carvalho M, Botas F, et al. 182 Breast Cancer Stem Cells Glycolytic Metabolism and Response to Chemotherapy. Eur J Cancer. 2012;48:S44. [Google Scholar]
- [34].Jiang X-P, RL E, Jf H. Exogenous normal mammary epithelial mitochondria suppress glycolytic metabolism and glucose uptake of human breast cancer cells. Breast Cancer Res Treat. 2015;153(3):519–529. [DOI] [PubMed] [Google Scholar]
- [35].Milane L, Duan Z, Amiji M. Role of hypoxia and glycolysis in the development of multi-drug resistance in human tumor cells and the establishment of an orthotopic multi-drug resistant tumor model in nude mice using hypoxic pre-conditioning. Cancer Cell Int. 2011;11(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated and/or analyzed during this study are included in this published article.


