Abstract
Glycosaminoglycans (GAGs) are long, linear polysaccharides that are ubiquitously expressed on the cell surface and in the extracellular matrix of all animal cells. These complex carbohydrates play important roles in many cellular processes and have been implicated in many disease states, including cancer, inflammation, and genetic disorders. GAGs are among the most complex molecules in biology with enormous information content and extensive structural and functional heterogeneity. GAG biosynthesis is a nontemplate-driven process facilitated by a large group of biosynthetic enzymes that have been extensively characterized over the past few decades. Interestingly, the expression of the enzymes and the consequent structure and function of the polysaccharide chains can vary temporally and spatially during development and under certain pathophysiological conditions, suggesting their assembly is tightly regulated in cells. Due to their many key roles in cell homeostasis and disease, there is much interest in targeting the assembly and function of GAGs as a therapeutic approach. Recent advances in genomics and GAG analytical techniques have pushed the field and generated new perspectives on the regulation of mammalian glycosylation. This review highlights the spatiotemporal diversity of GAGs and the mechanisms guiding their assembly and function in human biology and disease.
Keywords: chondroitin sulfate, glycosaminoglycans, heparan sulfate, regulation, transcription factors
INTRODUCTION
The glycocalyx is a diverse network of complex carbohydrates that coats the surface of all animal cells. The major components of the glycocalyx, including glycolipids and glycoproteins, serve not only to protect the cell, but also function as the primary mediators of a cell’s interaction with its environment (1, 2). Glycosaminoglycans (GAGs) are long, linear polysaccharides and key members of the glycocalyx that play important roles in a variety of normal and pathophysiological cellular processes (3). These complex carbohydrates are composed of repeating disaccharide units containing amino sugar (e.g., glucosamine) and uronic acid (e.g., glucuronic acid) building blocks. These sugar moieties can be O- or N-sulfated at different positions, which give them a high overall negative charge. There are four major families of GAGs that are classified based on their disaccharide composition and biosynthesis: heparin/heparan sulfate (HP/HS), chondroitin/dermatan sulfate (CS/DS), keratan sulfate (KS), and hyaluronan (HA). In the interest of space, here we have focused on HS and CS/DS and their roles in cell homeostasis and human disease (4, 5). For further reading on the other GAG classes, we refer the reader to reviews by Karamanos et al. (6) and Caterson and Melrose (7).
HS and CS/DS are ubiquitously expressed in all animal cells and are attached to core proteins, known as proteoglycans, on the cell surface and extracellular matrix (ECM). These sulfated GAGs profoundly impact the biology of cells and tissues and act as structural components of connective tissue and the basement membrane (8). The biosynthesis of HS and CS/DS occurs in the endoplasmic reticulum (ER) and the Golgi and is catalyzed by a large family of biosynthetic enzymes (Fig. 1). As the chains are assembled, they undergo a series of modification reactions including sulfation and epimerization, which generates sulfated regions/domains that encode binding sites for ligands, such as growth factors and chemokines (9). Interestingly, various mammalian tissues and cell types exhibit distinct expression profiles for the biosynthetic enzymes and proteoglycans and, as a result, exhibit characteristic patterns of HS and CS/DS chain modifications. This structural heterogeneity is found temporally and spatially during development (10–12) and is altered when cells undergo transformation (13). The immense structural diversity and anionic nature of these polysaccharides enable them to interact with many protein ligands at the cell surface and ECM interface, thus impacting many facets of cell biology, including differentiation, cell signaling, angiogenesis, and coagulation (14). Despite the breadth of published studies detailing their biosynthesis in cells, the regulatory mechanisms that dictate tissue-specific patterns and resultant functions of HS and CS/DS remain poorly understood. Recent advances in genomics, chemical biology, and bioinformatics have led to novel insights into the regulation of GAG assembly in cells. In this review, we highlight GAG structure and function, biosynthesis, and their regulation in the context of cell biology and human disorders.
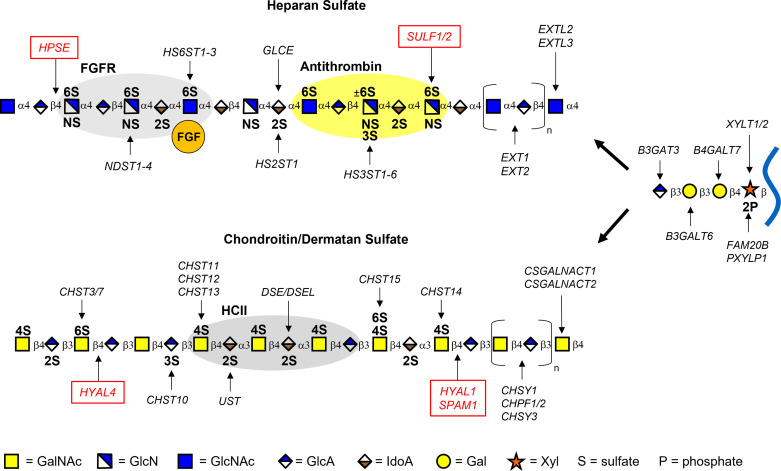
Figure 1.
Heparan and chondroitin/dermatan sulfate biosynthesis. Assembly of the chains is performed by a large group of biosynthetic enzymes located in the Golgi. Heparan sulfate (HS) and chondroitin sulfate (CS)/dermatan sulfate (DS) share a common tetrasaccharide linker (Xyl-Gal-Gal-GlcA) that is attached to a serine residue of the core protein. HS and CS/DS assembly is initiated by EXTL3 (HS) or CSGALNACT1/2 (CS/DS), respectively, followed by polymerization and sulfation at specific sites. The gray and yellow ovals signify binding sites for ligands/proteins. FGF, fibroblast growth factor; FGFR, fibroblast growth factor receptor; HCII, heparin cofactor 2; GalNAc, N-acetylgalactosamine; GlcN, glucosamine; Gal, galactose; GlcA, glucuronic acid; GlcNAc, N-acetylglucosamine; IdoA, iduronic acid; Xyl, xylose.
GLYCOSAMINOGLYCAN STRUCTURE AND BIOSYNTHESIS
HS and CS/DS are linear polysaccharides that are assembled while attached to one of ∼77 known proteoglycan core proteins. These proteoglycans are differentially expressed in various tissues and cell types, and certain core proteins can also function as part-time proteoglycans, meaning they may exist with or without the GAG chains (15, 16). These heterogeneous polysaccharides are biosynthesized in a nontemplate-driven manner by a family of enzymes encoded by nearly 40 genes (17). The process of HS and CS/DS biosynthesis begins in the cytoplasm with the synthesis of 5-uridine-diphosphate (UDP) sugars that are then transported to the Golgi apparatus (18). The assembly of both HS and CS/DS is initiated by the formation of a common tetrasaccharide structure catalyzed by xylosyltransferase I/II (XYLT1/XYLT2), B4GALT7, B3GALT6, and B3GAT3. This linker region contains xylose (Xyl), two galactose (Gal) molecules, and d-glucuronic acid (GlcA), with xylose covalently bound to the serine residue of the proteoglycan core protein (Fig. 1). The xylose sugar can be phosphorylated or dephosphorylated at the C2 hydroxyl group by FAM20B and PXYLP1, respectively, which regulates the rate of formation of the linker region and subsequent chain elongation (19–21). HS synthesis is initiated once a N-acetyl-d-glucosamine (GlcNAc) residue is added by EXTL3 (22, 23), and CS/DS assembly begins upon addition of N-acetyl-d-galactosamine (GalNAc) by CSGALNACT1 or CSGALNACT2 (24, 25). The N-acetylhexosaminyltransferase, EXTL2, can transfer GalNAc and GlcNAc to the common GAG linkage region (26), although the exact function of this enzyme in GAG assembly remains unclear (27). Interestingly, this enzyme has been shown to specifically control HS biosynthesis via addition of a phosphorylated GlcNAc residue to the tetrasaccharide linkage, which thereby terminates chain elongation (28). HS polymerization occurs through the addition of alternating GlcNAc and GlcA residues by the heterodimeric co-polymerase complex containing exostosin-1 (EXT1) and exostosin-2 (EXT2) (29, 30). CS polysaccharides consist of alternating units of N-acetyl-d-galactosamine (GalNAc) and GlcA and are synthesized by a series of seemingly redundant glycosyl- and sulfotransferases. DS is structurally related to CS; however, DS chains uniquely contain GalNAc and d-iduronic acid (IdoA) repeating disaccharide units. CS/DS elongation occurs upon addition of repeating GalNAc and GlcA repeating units by any combination of a hetero-oligomer complex of chondroitin sulfate synthase 1 (CHSY1), chondroitin polymerizing factor 1/2 (CHPF1/CHPF2), and chondroitin sulfate synthase 3 (CHSY3), depending on tissue-specific expression of the enzymes (31–33).
After elongation, HS and CS/DS chains are sulfated at specific sites by multiple sulfotransferase enzymes, which creates highly sulfated domains that can interact with protein binding partners (9, 34). HS sulfation is thought to proceed in a processive manner driven by enzyme substrate specificities. First, GlcNAc residues are N-deacetylated and subsequently N-sulfated by N-deacetylase/N-sulfotransferases (NDST1-4), followed by addition of a 2-O-sulfate group to C2 of GlcA or IdoA (HS2ST1), which is generated by the C5-epimerase GLCE, and subsequent 6-O-sulfation of C6 on GlcN/GlcNAc residues (HS6ST1-3) (35). In addition, relatively rare 3-O sulfate groups can be added to the C3 position of specific N-sulfoglucosamine residues by seven 3-O sulfotransferases (HS3ST1–6) (36). CS chains can be O-sulfated at the C4 (CHST11-14) and C6 (CHST3/7/15) of GalNAc residues and at C2 (UST) and more rarely at C3 (CHST10) of GlcA (37). In addition, GlcA residues may be epimerized into IdoA residues via DSE/DSEL and 2-O sulfated by UST, resulting in structurally related chondroitin/dermatan sulfate (CS/DS) hybrid chains (38) (Fig. 1). Overall, the variation in polymer length and sulfation patterns of HS and CS/DS regulate their interactions with GAG-binding proteins and ligands in the ECM. Since sulfation is not uniform across the polysaccharide chain and most modification reactions do not go to completion, GAG chains are variably sulfated at structurally diverse, containing both highly sulfated regions and regions lacking all or most of these modifications (39–41).
After assembly and presentation on the cell surface and ECM, both HS and CS chains can be further processed by extracellular sulfatase and/or glycosidase enzymes. These secreted factors are implicated in remodeling of the ECM and regulating GAG-protein interactions via tuning GAG assembly in the extracellular space. A classic example is heparanase-1 (HPSE), which is an endo-β-d-glucuronidase encoded by the gene HPSE. This enzyme degrades HS chains through cleavage of the β-1,4 glycosidic bond between GlcA and GlcN at sites of low sulfation (NA domains) (42). HS degradation by HPSE results in the release of 5–10 kDa fragments of HS, which facilitates structural alterations to the ECM and basement membrane and is associated with disease states, such as tumor metastasis and sepsis (43). In fact, it has been shown that degradation of HS chains by HPSE enhances the accessibility of the proteoglycan core protein to enzymatic cleavage by matrix metalloproteases and other proteases (44). Heparanase-2 (HPSE2), a secreted protein that shares 40% homology to HPSE, interestingly does not exhibit glycosidase activity but can block HPSE activity and act as a tumor suppressor (45).
The endo-6-O-sulfatases, SULF1 and SULF2, are secreted proteins that regulate the binding and activity of growth factors through removal of a subset of 6-O sulfate groups from highly sulfated regions (NS domains) of HS (46, 47). Consequently, cell signaling can be attenuated (fibroblast growth factors) or promoted (canonical WNT) by SULF-mediated HS desulfation (48). For example, studies have implicated the SULFs acting as both a tumor suppressor (49) and promoter of tumorigenesis (50–52) depending on cancer type. In vitro and in vivo models have revealed that SULF1 and SULF2 are functionally redundant in certain contexts, such as during skeletal development (53). Interestingly, the isoforms display distinct expression patterns among various adult tissues (54) and brain regions (55) yet exhibit overlapping expression patterns during embryonic development (53). Together, this suggests that SULF1 and SULF2 dynamically influence HS-protein interactions by regulating tissue-specific HS fine structure. In contrast, CS endosulfatases have not been discovered (56), yet there is some evidence that the lysosomal exosulfatase, arylsulfatase B (ARSB), can localize at the cell membrane and impact CS levels under certain pathophysiological conditions (57, 58). Three mammalian hydrolases capable of cleaving CS chains have been identified, including PH20 (SPAM1), hyaluronidase 1 (HYAL1), and hyaluronidase 4 (HYAL4). Although all three can also cleave hyaluronic acid glycosaminoglycans, HYAL4 is predominantly an endo-β-N-acetylgalactosaminidase that degrades CS motifs containing 2-O and 6-O sulfated subunits (CS-D). HYAL4 has been shown to cleave CS chains on multiple proteoglycans, including serglycin, aggrecan, and CD44, and its expression is altered in certain disease states, such as cancer (59). Altogether, the activity and expression of biosynthetic enzymes and secreted remodeling factors discussed here have a significant impact on the huge chemical diversity of HS and CS/DS chains and their resultant functions across tissue/cell types and disease states.
SPATIAL AND TEMPORAL DIVERSITY OF GLYCOSAMINOGLYCANS
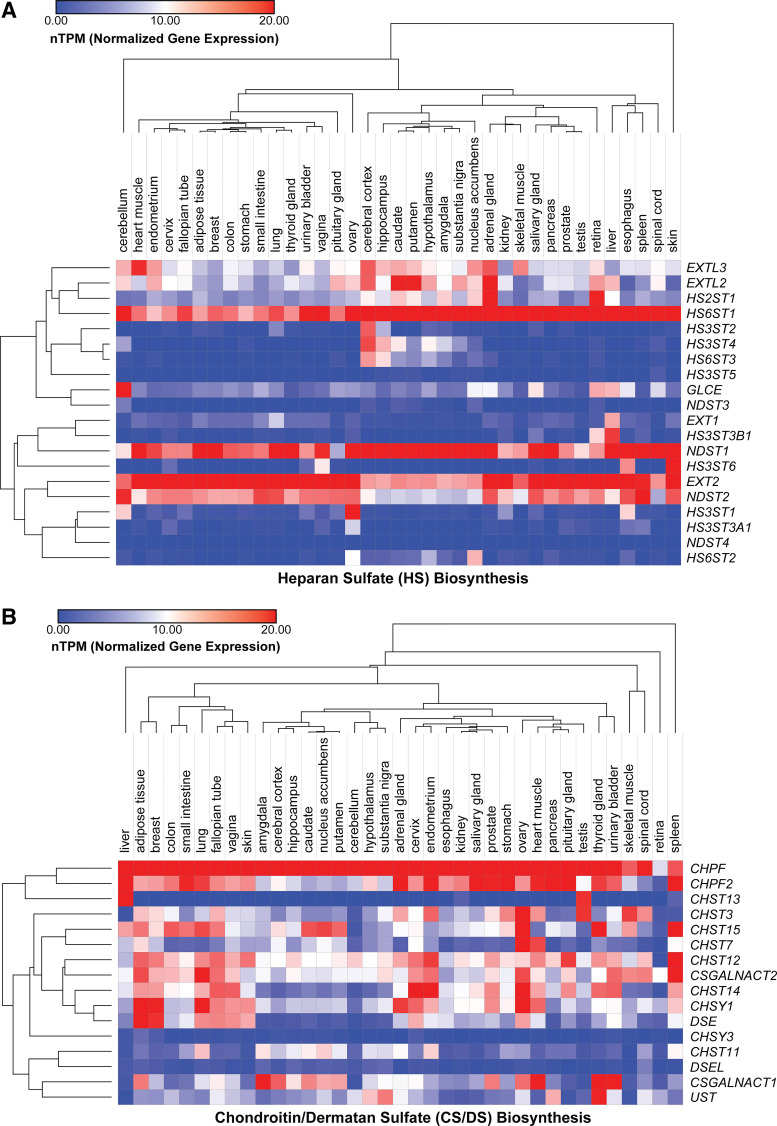
The structural diversity of GAG polysaccharides is vast among different species and can vary temporally during development and spatially in tissues. Consequently, GAGs contribute significantly to cell- and organ-specificity, cell lineage (60–62), tissue/organ function (63–65), and pathophysiological conditions, such as tumorigenesis (66) and inflammation (67). The development of pattern-specific antibodies for CS (68) and HS (69, 70) facilitated the initial discovery of cellular and tissue distribution of GAGs during development. Recent advances in next generation sequencing and state-of-the-art analytical tools have enabled detailed profiling of the spatiotemporal expression of GAG enzymes (10, 71) and their diverse structure/composition (72) across various organisms, cells, and tissues. In Fig. 2, we present tissue-specific gene expression of all the enzymes involved in HS and CS biosynthesis across 37 human tissues obtained from the Human Protein Atlas (73, 74). This data emphasizes differential gene expression of individual HS and CS biosynthetic enzymes across various human tissues and highlights distinct clustering of certain enzymes based on similar expression signatures. Generally, genes encoding for enzymes involved in elongation of the GAG chains, such as EXT2 and CHPF1/2, are highly expressed across most tissues, whereas sulfotransferase enzymes, such as HS3STs and HS6STs, tend to cluster with localized expression patterns, especially in regions of the brain (75). Clearly, there are dominant isoforms across all tissues for a given enzyme family (NDST1, HS6ST1, EXT2, and SULF2), and certain genes have low expression across all 37 tissues (e.g., NDST3 and CHSY3). Overall, CS genes are more highly expressed, whereas HS genes show more tissue specificity, depending on the enzyme subclass. Although it is obvious that GAG expression is regulated at the transcriptional level, the mechanisms controlling this spatiotemporal regulation in cells and tissues remain still unclear.
Figure 2.
Expression profiles of glycosaminoglycan (GAG) biosynthetic enzymes in human tissues. Heatmaps show transcript expression levels for heparan sulfate (HS, A) and chondroitin sulfate/dermatan sulfate (CS/DS, B) biosynthetic enzymes in 37 human tissues based on RNA sequencing (normalized transcripts per million, nTPM). Hierarchical clustering was performed based on the Pearson correlation. Data were obtained from GTEx and is based on The Human Protein Atlas version 21.0 and Ensembl version 103.38.
Many studies up to this point have utilized the availability of animal models to explore the temporal and spatial diversity of GAG assembly. For example, it has been shown that total HS amounts vary widely when isolated from different mouse tissues (76). Along with changes in overall amounts of HS, their composition and sulfation patterns are diverse across different cell types, allowing for different capacities to bind ligands and receptors involved in cell signaling (54). Furthermore, multiple diverse forms of CS/DS (CS A–E) with distinct sulfated disaccharide repeats are present across various species and tissues. Depending on sulfation pattern and IdoA content, CS/DS chains can be classified into different subtypes: CS-A [GlcA-GalNAc(4S)], CS-B/DS [IdoA(2S)-GalNAc(4S)], CS-C [GlcA-GalNAc(6S)], CS-D [GlcA(2S)-GalNAc(6S)], CS-E [GlcA-GalNAc(4S,6S)], and CS-iE [IdoA-GalNAc(4S,6S)] (77, 78). Interestingly, these different types of CS exhibit distinct spatiotemporal expression across tissues, particularly in the brain (79, 80). In the human brain, CS is highly expressed and experiences a significant shift from CS-C to CS-A during development (81). In addition, CS gene expression and composition is altered in the postnatal mouse cerebellum during development, with a significant shift from CS-E to CS-D/DS (82). Finally, HS and CS composition were also found to differ significantly between the model organisms Hydra vulgaris, Drosophila melanogaster, and Caenorhabditis elegans, which represents a potential for evolutionary distinct functional motifs across metazoans (83).
Recently, publicly available RNA sequencing data from various mouse tissues and across developmental stages were comprehensively analyzed and integrated to profile the spatial, temporal expression for genes involved in HS biosynthesis (84). Similar to the human expression data (Fig. 2), many of the enzymes involved in HS sulfation in mice show distinct spatial expression patterns, particularly among different isoforms. Within the family of NDST enzymes, for example, Ndst1 and Ndst2 expression were detected at significant levels across all 21 mouse tissue types, whereas Ndst3 and Ndst4 were both only detected in the cerebral cortex and testis. In the same study, temporal expression of certain enzymes was shown to vary significantly across different time points from embryonic to postnatal development in the mouse forebrain. For instance, certain 3-O sulfotransferases (Hs3st1 and Hs3st5) steadily increase in expression during embryonic development but are attenuated at the postnatal stage. Moreover, Sulf1 expression decreases throughout the embryonic stage and maintains low expression throughout the postnatal stage, whereas Sulf2 exhibits a general decrease in expression throughout the embryonic stage but fluctuates with no discernible pattern of expression in the postnatal stage. Other studies have revealed significant alterations in GAG composition during stem cell differentiation and fate commitment (85). Interestingly, ∼80% of GAGs produced by undifferentiated mouse embryonic stem cells (mESCs) contain a low-sulfated form of HS (with low overall CS levels), but upon transition to committed cell types, their HS chains become increasingly sulfated with a concurrent increase in CS synthesis (86, 87). Comprehensive profiling of the glycome in human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs) also revealed overall low sulfation of HS and predominantly 4-mono-sulfated CS disaccharides in both populations, and total amounts of these “immature” GAGs were significantly higher when compared with other nonstem human cell lines (62). Differentiation of mESCs along the mesodermal lineage to the hemangioblast stage results in transient expression of a unique 3-O sulfated HS epitope that is a marker for distinct cell populations along the lineage (88). These transient changes in GAG expression during differentiation presumably impacts GAG-growth factor interactions and downstream cell signaling pathways. The mechanisms that control these changes in GAG composition during different developmental stages are still unknown. Presumably, epigenetic factors (e.g., chromatin remodeling complexes), which play key well-studied roles in mammalian development (89), may also tune GAG gene expression as a way to respond to extracellular cues and impact stem cell differentiation and pluripotency.
REGULATORY MECHANISMS OF GLYCOSAMINOGLYCAN ASSEMBLY
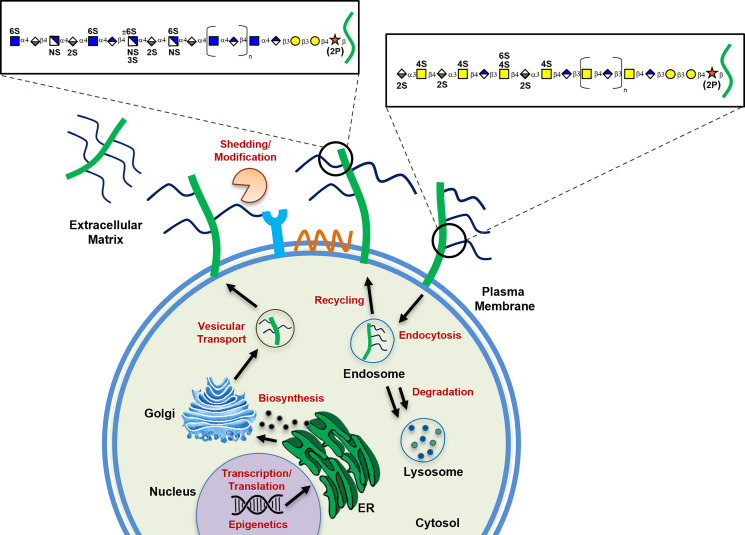
Over the past few decades, GAG biosynthetic enzymes have been identified, cloned, biochemically characterized, and mutated in cells and model organisms (17). Although much is known about the enzymes and their impact on HS/CS composition, little is known about the mechanisms that dictate spatiotemporal patterning of the chains and trigger their dysregulation in human disease (5, 90, 91). The organization of sulfated residues along the chains, in part, reflects the specificity of the enzymes (9); however, other nonenzymatic factors could regulate patterning of the chains and expression of the biosynthetic enzymes and proteoglycan core proteins. Before HS/CS chains are presented as proteoglycans on the cell surface and in the ECM, it is conceivable that many steps along the pathway could be regulated and thus impact GAG structure/function in the extracellular space, including: 1) the expression of the core proteins or biosynthetic enzymes by transcriptional, translational, or epigenetic factors, 2) the organization and activity of enzymes in the ER and Golgi, 3) nucleotide sugar and 3′-phosphoadenosine-5′-phosphosulfate (PAPS) levels, 4) chaperone and/or scaffolding proteins in the ER and Golgi during biosynthesis, 5) rate of turnover and secretion, and 6) other unknown factors (Fig. 3). Here, we will discuss potential regulatory mechanisms and highlight studies that have shed light on the regulation of GAG assembly.
Figure 3.
Regulation of glycosaminoglycan (GAG) biosynthesis. Illustration depicting regulatory pathways controlling heparan sulfate (HS) and chondroitin sulfate (CS)/dermatan sulfate (DS) biosynthesis in the cell. Red lettering indicates potential steps of regulation before presentation of GAG chains in the extracellular space.
Regulation of Enzyme Expression and Activity
As aforementioned, the expression of GAG biosynthetic enzymes and the proteoglycan core proteins varies across different tissues and is tightly regulated during development. Early studies described the impact of growth factors on the regulation of proteoglycan expression and GAG chain length. The family of fibroblast growth factors (FGFs) was shown to activate expression of the proteoglycan, syndecan-1 (Sdc1) (92, 93). In a related study, the activation of Sdc1 expression in mouse keratinocytes and fibroblasts was growth factor-dependent and initiated by transcription factors and response elements upstream of Sdc1 (94). In addition, treatment of arterial smooth muscle cells with platelet-derived growth factor (PDGF) led to increases in CS/DS chain length, whereas transforming growth factor-β1 (TGFβ1) treatment resulted in enhanced mRNA levels of biglycan, a CS proteoglycan (CSPG) (95). TGFβ1 also was shown to augment CS biosynthesis by upregulating CHSY1 expression via the MAPK signaling pathway (96). Another report showed a role for TGFβ1 in regulating CS/DS chain polymerization (97).
Other genetic factors, such as 5′ untranslated region (UTR) and 3′ UTR mRNA sequences (98–100), microRNAs (101–103), and epigenetic factors (104–106) have been shown to affect expression of the enzymes and core proteins, which in turn affects the assembly of the chains, their binding properties, and their downstream biological activity. Multiple transcriptional regulatory elements have been reported to play a role in controlling GAG assembly and core protein expression (107–119). For example, an astrocyte-specific basic leucine zipper transcription factor, OASIS, was recently shown to positively regulate CHST3 expression during spinal cord injury (120). In addition, alternative splice forms of certain enzymes can be expressed in a spatiotemporal manner and impact biosynthesis. For example, a shorter alternative splice form of NDST1 (“NDST1B”) is expressed at a low level in normal tissue but is overexpressed in certain cancers, where it might function in a dominant negative manner by replacing active endogenous NDST1 in the HS enzyme complexes during biosynthesis (121).
Recent advances in genomic technologies and bioinformatic tools have enhanced our ability to explore the genome to identify novel regulatory factors involved in GAG assembly. A recent data mining effort utilizing publicly available chromatin immunoprecipitation sequencing (ChIP-Seq) and RNA sequencing data uncovered the transcription factor, MEF2C, as a potential regulator of HS and CS enzyme expression (122). Bioinformatic analyses examining the promoter region of all HS enzymes for enrichment of transcription factor binding motifs led to the discovery of ZNF263 as a novel transcriptional repressor of HS3ST1 and HS3ST3A1 expression (123). The advent of CRISPR/Cas9 as a genetic tool (124, 125) and associated screening technologies (126, 127) has enabled unbiased genome-wide searches for nonenzymatic factors regulating GAG assembly. Our group recently reported the development of a genome-wide CRISPR screen to uncover novel factors regulating HS assembly in human cells. This genetic screen uncovered many potential regulatory pathways for HS biosynthesis and, importantly, revealed that the histone demethylase, KDM2B, controls HS-protein interactions via epigenetic regulation of SULF1 expression (128). Another CRISPR screen designed to identify host factors of Sindbis virus infection found that mutating COG4, a member of the conserved Golgi complex, results in a defect in HS biosynthesis in human colorectal carcinoma cells (129).
Enzyme Localization and Golgi Homeostasis
The Golgi can be divided into cis, medial, and trans compartments. Interestingly, the majority of HS enzymes localize to the cis/medial stacks, whereas CS enzymes are located in the trans-Golgi network (130, 131). Disruption of the Golgi network with the inhibitor brefeldin-A results in mis-localization of multiple CS enzymes and defects in CS polymerization (132). In addition, deletion of the stem region of HS6ST isoforms inhibits their localization to the Golgi and reduces sulfotransferase activity (133). β-Secretase was also found to indirectly regulate 6-O sulfation of HS through truncation of HS6ST3 thus altering its cellular localization (134). Moreover, knockout mice with mutations in the Golgi-resident bisphosphate nucleotidase 2 (BPNT2), which catalyzes the breakdown of 3′-phosphoadenosine-5′-phosphate in the Golgi, led to impaired 4-O sulfation of CS (135). A genome-wide screen recently revealed that the transmembrane protein, TM9SF2, is a novel host factor for chikungunya virus infection and a critical regulator of N-sulfation of HS and localization and stability of NDST1 in the Golgi (136). Also, the Golgi-resident transmembrane protein 165 (TMEM165) was discovered to be a manganese transporter and crucial regulator of GAG biosynthesis. Knockout of TMEM165 resulted in a defect in Mn2+ availability in the Golgi and a profound decrease in HS/CS chain elongation. Supplementation with Mn2+ rescued the GAG polymerization process, suggesting that the availability of metal ion cofactors is also important for GAG synthesis in the Golgi (137). Other factors, such as the Drosophila GOLPH3 homolog, have been shown to regulate the retrograde trafficking of EXT1 and EXT2 (138).
The “GAGosome”
The “GAGosome” hypothesis was first proposed by Esko and Selleck (139) suggesting that a physical complex of the biosynthetic enzymes exists in the Golgi. This theory was built upon early evidence suggesting that rapid biosynthesis of the chains is due to the cooperative activity of the enzymes (140, 141). Another early report showed evidence of specific interactions between XYLT1 and B4GALT7 during CS biosynthesis (142). Multiple studies since then have provided additional evidence to support this concept. The first example is the seminal discovery that heterooligomeric complexes form to polymerize both HS (EXT1/EXT2) (30) and CS chains (CHSY1/CHSY3/CHPF1/CHPF2) (31). In addition, the HS C5-epimerase GLCE was also shown to directly interact with the 2-O sulfotransferase HS2ST1 in vivo, which impacts epimerase stability and translocation to the Golgi (143). More recently, immunoprecipitation experiments showed that EXT1 and EXT2 can directly interact with NDST1, which affects HS sulfation levels and NDST1 expression (144). Further studies and new methods are needed to elucidate other nonenzymatic factors, such as chaperones and/or scaffolding proteins that may regulate enzyme activity and impact GAG synthesis in the Golgi.
Proteoglycan Structure and HS/CS Attachment Sites
In certain contexts, the proteoglycan core protein may compete with the biosynthetic enzymes and may be the limiting step in GAG biosynthesis. Certain protein domains of proteoglycans have been shown to influence CS or HS utilization of their GAG attachment sites and sulfation patterns. In an earlier study, the globular domain of glypican-1 (GPC1), a major glycosylphosphatidylinositol-anchored cell surface HS proteoglycan, was found to be a key structural motif that influences whether HS or CS chains are added to the core protein. Removal of this domain resulted in a shift from ∼90% HS to ∼90% CS chains on the protein (145). Another study investigated whether the organization of non-GAG bearing domains from two matrix proteoglycans, perlecan and aggrecan, can impact HS/CS occupancy. Intriguingly, expression of perlecan/aggrecan chimeras with different non-GAG bearing domains resulted in significant changes to the HS/CS ratio on the core proteins, depending on which domains were present. This suggested that utilization of attachment sites for HS and CS may be influenced, at least in part, by non-GAG-bearing domains (146). Kokenyesi and Bernfield discovered that mouse Sdc1 contains five GAG attachment sites containing either HS or CS chains, depending on the protein domain. HS was found solely at the N-terminus while CS chains were attached to the C-terminus sites, suggesting that core protein structure regulates HS/CS occupancy (147). A more recent study compared the sulfation levels of HS attached to mouse syndecan-2 (Sdc2) and syndecan-4 (Sdc4). Interestingly, they found that insertion of the N-terminal domain sequence of Sdc2 into Sdc4 drove a significant increase in 6-O sulfation of its HS chains compared with the native Sdc4 sequence. These results provide further evidence of the existence of core protein-determined HS sulfation patterns (148). Newer analytical approaches have helped address these questions of site-specific glycosylation by identifying HS/CS attachment sites of proteoglycans (15, 149, 150).
The activity of certain enzymes can regulate HS/CS assembly and act as a quality control mechanism. As discussed earlier, transient phosphorylation of the C2 position of xylose in the linker region functions as a molecular switch to regulate GAG synthesis. In fact, loss of FAM20B-dependent phosphorylation leads to incomplete linkage regions that are capped with sialic acid, thus halting elongation (151). Interestingly, sulfation of the linkage region at various positions can function as a regulatory signal in chain initiation. For instance, 6-O sulfation of the two galactoses in the linkage region influences B3GAT3 (152) and B4GALT7 activity (20), whereas 4- or 6-O sulfation can significantly enhance N-acetylgalactosaminyltransferase-1 (CSGALNACT1) activity (25). In addition, sulfation of the GlcA residue in the tetrasaccharide linker by HNK-1 (CHST10) can suppress CS assembly in certain core proteins (153). EXTL2 can function as both a terminator and activator of GAG synthesis through addition of GlcNAc and GalNAc to the linker region. Addition of α-GalNAc to the linker region terminates CS biosynthesis, whereas adding GlcNAc initiates HS biosynthesis (26). Suppression of EXTL2 and EXTL3 leads to reduced HS biosynthesis (154), yet knockdown of EXTL3 also results in increased chain length, indicating a complex regulatory mechanism (23). The HS enzyme NDST2 exhibits high N-deacetylase but weaker N-sulfotransferase activity compared with NDST1 (155), yet a recent study showed that this enzyme could also stimulate HS chain elongation through an unknown mechanism (156). Intriguingly, the addition of a 4-O sulfate group to the nonreducing terminal N-acetylgalactosamine during CS synthesis by CHST11 was found to facilitate chain elongation by CHSY1/CHPF in cooperation with the N-acetylgalactosaminyltransferase, CSGALNACT2 (157). Overall, these findings have exposed novel mechanisms dictating CS or HS chain attachment and elongation at certain serine residues of proteoglycans; however, there is still much unknown on this topic.
GLYCOSAMINOGLYCAN REGULATION IN HUMAN DISEASE
Dysregulation of GAG biosynthesis has been implicated in a variety of human disorders, and alterations in GAG structure have a direct impact on their function in disease states. Such defects can occur within the biosynthetic machinery, altering the polymerization or modifications of the chains, which affects their ability to bind proteins, such as growth factors and chemokines, to impact tissue function and physiology. Depending on the underlying alterations in GAG assembly, diverse pathophysiological phenotypes have been observed within a given disorder (e.g., congenital disorders). Here, we have outlined how the dysregulation of GAG assembly impacts human disease.
Genetic Disorders
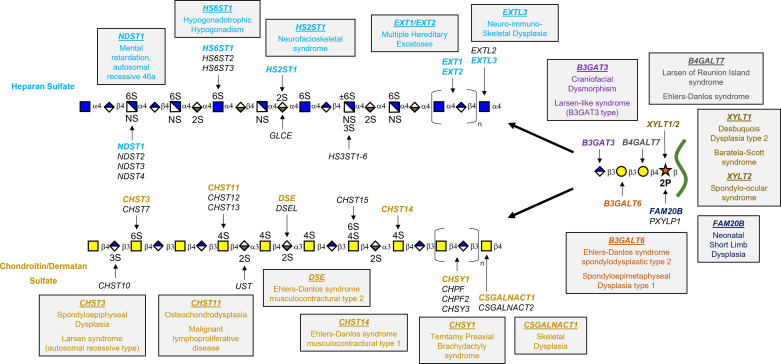
A variety of mutations in genes encoding GAG biosynthesis enzymes have been implicated in several relatively rare human diseases (Fig. 4). Interestingly, defects in GAG biosynthesis usually portray clinical features in bone or cartilage, ligaments, or subepithelial layers (158). For example, dysregulation of HS chain polymerization is a key feature in multiple hereditary exostoses (MHE), an autosomal dominant skeletal disorder hallmarked by the formation of benign cartilage-capped tumors. This disorder is caused by a heterozygous loss-of-function mutation in EXT1 or EXT2 (159, 160). HS synthesized by chondrocytes at the growth plate of long bones is crucial for growth factor signaling. Mutations altering HS chain polymerization leads to truncation of the HS chains and a consequential decrease in HS levels, which results in disruption of signaling pathways [e.g., FGF, bone morphogenetic protein (BMP), and Hedgehog], leading to exostoses formation (161, 162).
Figure 4.
Congenital disorders of glycosaminoglycan biosynthesis. Diagram highlighting genetic disorders caused by mutations in key biosynthetic enzymes for heparan sulfate (HS) (blue text) and chondroitin sulfate (CS)/dermatan sulfate (DS) (gold text) assembly and the shared tetrasaccharide linker region. A complete reference list is available in Ref. 158.
A variety of mutations in genes involved in GAG biosynthesis have been shown to cause Ehlers–Danlos syndromes (EDS), a heterogeneous group of hereditary connective tissue disorders that are characterized by hypermobility of joints, hyperextensible skin, and weakened or fragile connective tissue (163). One variant of the disease, EDS musculocontractural type 2, is caused by homozygous mutations in DSE, which impedes DS formation via a reduction in GlcA epimerization to IdoA (164). In addition, mutations in CHST14, which encodes for dermatan 4-O-sulfotransferase 1 (D4ST1), results in the absence of DS and excess CS synthesis, causing EDS musculocontractural type 1 (165). We have highlighted multiple human disorders caused by genetic mutations in GAG enzymes in Fig. 4. Currently, there are limited therapies for these patients, therefore understanding the regulation of the genes and proteins involved in GAG biosynthesis will help identify targets for new treatment strategies.
Neurodegenerative Diseases
Many neurological disorders can be traced to the dysregulation of GAGs, including tauopathies (166–168), Parkinson’s disease (169), multiple sclerosis (170), and neuroinflammation during spinal cord injury (171). HS has been repeatedly shown to play a key role in forming protein aggregates in various neurodegenerative disorders (172). It was originally reported that highly sulfated GAGs such as heparin, a highly sulfated form of HS produced in mast cells, promote aggregations of tau by competing with microtubules as binding sites (173). Heparin binding of tau induces a conformational change that consequently exposes previously masked phosphorylation sites. Different protein kinases can phosphorylate these newly exposed sites and lead to tau hyperphosphorylation, which is a hallmark of a variety of tauopathies, including Alzheimer’s disease, Pick’s disease, and progressive supranuclear palsy (174, 175). Recently, a rare 3-O-sulfation modification of HS was implicated in the spread of tau due to this modification’s enhancement of tau-HS binding (176). Furthermore, 6-O-sulfation of HS was also revealed to bind to and compete with cellular intake of tau (177). These findings present potential new therapeutic avenues to treat Alzheimer’s progression.
HS has also been suggested to play a role in forming protein aggregates of α-synuclein in Parkinson’s disease. Different sulfation patterns of HS allow for N-terminal domain interactions of the protein to promote the formation of α-synuclein fibrils and promotes their aggregation. On the other hand, the role of HS in multiple sclerosis (MS) etiology and pathology is less understood. Multiple studies have highlighted the beneficial and deleterious effects of the HS degrading enzyme, HPSE, in MS. In a mouse model of experimental autoimmune encephalitis (EAE), inhibition of HPSE resulted in reduced EAE (178). It is thought that increased expression of HPSE in the central nervous system (CNS) of EAE mice promotes MS disease progression and T cell invasiveness. Other limited studies have suggested that HPSE plays a beneficial role in reducing inflammation and blocking T cell migration into the central nervous system during MS pathogenesis (179). Future work will need to fully elucidate the effects of HPSE expression and activity on MS progression. Contrary to HS, CSPGs have been highly implicated in MS progression and neuroinflammation during spinal cord injury. CS is the main component of “glial scars” that form after spinal cord injury and act as inhibitors of axon regrowth (180). Therefore, there is much interest in targeting CSPGs during spinal cord injury (181). CSPGs are upregulated in plaques in patients with MS, particularly in white matter (182) and play a proinflammatory role during MS progression (183, 184).
Inflammation
The varying sulfation patterns of HS allow for binding interactions with a wide array of ligands and receptors. Chemokines are a family of chemoattractant signaling proteins known to bind to HS across sulfated domains and are involved in inflammatory and immune responses via the recruitment of leukocytes (185). This binding results in a phenomenon called chemotaxis in which a concentration gradient of immobilized chemokines is established at the cell surface. Almost all chemokines have been shown to bind HS/heparin, and this process is thought to be the mechanism by which circulating blood leukocytes roll to positions along the endothelium to sites of inflammation (186).
Heparin is one of the most widely prescribed drugs in the world. Its potent anticoagulant activity makes it the drug of choice for treating deep vein thrombosis and pulmonary embolism in the acute care setting. The molecular basis of the anticoagulant action of heparin is linked to its ability to bind to the serine protease inhibitor antithrombin, leading to inhibition of the coagulation cascade (187). Of the 12 million patients treated with heparin in the USA annually, up to 5% of patients can develop a life-threatening autoimmune response triggered by the interaction between heparin and a platelet-derived chemokine, platelet factor 4 (PF4), known as heparin-induced thrombocytopenia (HIT). Heparin binds to PF4 and forms a supramolecular complex that is recognized by IgG antibodies, which results in promoting thrombin generation and other complications (188). Interestingly, prior studies have shown that PF4 binding to heparin depends on distinct sulfated sites compared with AT, suggesting that a safer form of anticoagulant heparin could be developed (189, 190).
Cancer
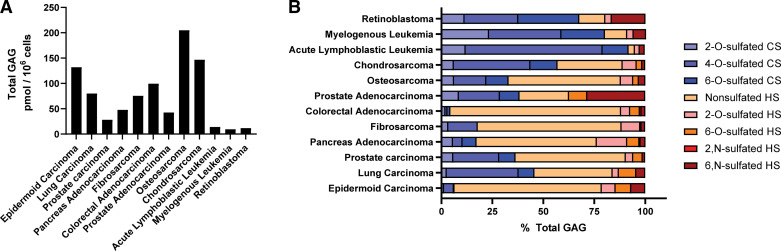
Structural heterogeneity of the GAG chains also varies when cells undergo transformation (191). Alterations in GAG structure and function have been implicated in cancer growth, progression, and tumor metastasis (37). Cancer cells can dynamically regulate the structure and sequence of cell-surface and extracellular matrix HS, and many cancer cells secrete proangiogenic factors (e.g., vascular endothelial growth factors) that bind to HS on the cell surface and stimulate tumor angiogenesis and metastasis (13). Certain cancers display an increase in overall sulfation levels causing defects in growth factor signaling pathways that contribute to tumorigenesis. In fact, a recent study profiled HS/CS across various human cancer lines by reverse-phase HPLC (192). This study revealed immense structural diversity and differences in total amount of HS/CS, depending on the tissue of origin (see Fig. 5). FGF2 is a growth factor that binds to such sites and can increase cell proliferation in various cancers (193, 194). Since HS can promote cell-cell and cell-ECM adhesion (195, 196), many cancers show reduced HS levels (197), thereby promoting metastasis. The secreted 6-O-endosulfatase, SULF1, is downregulated in breast cancer, which results in high basal 6-O-sulfation levels and increased cell migration and invasion (198). HS has also been shown to play a role in epithelial to mesenchymal transition by binding growth factors in the tumor microenvironment (199). In the same way, CS has been associated with almost all the hallmarks of cancer, including proliferation, migration, invasion, angiogenesis, and metastasis (200). Many cancers show CS-E overexpression (201, 202), which allows for increased binding of vascular endothelial growth factor (VEGF) in ovarian adenocarcinomas (203), negative regulation of Wnt/β-catenin signaling in breast cancer (204), and increased metastasis of Lewis Lung carcinoma (201). Increased prevalence of 6-O-sulfation and nonsulfated disaccharides of CS are also implicated in the malignant phenotype of pancreatic cancer (205).
Figure 5.
Glycosaminoglycan (GAG) profiling across various human cancer cell lines. Total heparan sulfate (HS)/chondroitin sulfate (CS) amounts (A) and disaccharide composition (B) vary between human cancer cells, as determined by RP-HPLC (Data obtained from Ref. 192). Sulfation patterns are grouped for CS (blue colors) and HS (red/orange colors).
CONCLUSIONS AND PERSPECTIVE
GAGs play many roles in essential cellular processes, including cell proliferation, signaling, and development, and interact with a diverse set of proteins and ligands in the extracellular space. Their distinct functions in biology are directly linked to their inherent structural diversity. As detailed above, there is ample evidence that GAG biosynthesis is spatiotemporally regulated in cells. Advances in genomics have enabled genome-wide profiling of GAG expression across tissues, and the development of effective analytical tools has supported the detailed structural analysis and distribution of GAG structures across cell types. Although a significant amount of research has been done to understand GAG structure and function, further investigation is needed to elucidate the distinct regulatory mechanisms controlling GAG assembly. One of the current challenges in the field is the analysis of intact GAG chains and longer oligosaccharides at high resolution. Classically, GAG structural information has been obtained via profiling the composition of the chains after enzymatic/chemical depolymerization of the chains (83, 149, 206) or sequencing intact purified oligosaccharides (“top-down”) by NMR (207) and high-resolution MS techniques, such as tandem MS (208, 209) and ion mobility MS (210). Sequencing intact GAG chains is still under development, and new technologies, such as solid-state nanopores (211, 212), are also being tested for this application.
New genomic technologies have been described to help link gene expression to functional changes in glycan structure at the cell surface. Single-cell RNA sequencing and spatially resolved transcriptomics have revolutionized our understanding of the positional context of gene expression in tissues (213). New tools were recently developed for detailed glycan analysis at the single-cell level (214, 215). These tools will enhance our ability to measure glycan diversity across cell types and understand their dysregulation in human disorders. One could imagine combining spatial transcriptomics with new mass spectrometry imaging techniques (216) to obtain detailed GAG profiles in tissues. Innovations in the field of epigenetics have allowed genome-wide profiling of the localization and function of transcription factors, chromatin remodeling complexes, and chromatin status in diverse cell types and pathophysiological conditions. The role of epigenetics in controlling GAG assembly is still unclear (106), despite obvious applications for these tools to understand the genetic factors controlling the dynamic process of glycosylation. Certainly, understanding the regulation of GAG assembly would provide avenues for controlling their synthesis in different biological contexts, exploiting novel therapeutic targets for treating human disorders, and developing bioengineered-defined structures for GAG-based therapeutics.
GRANTS
Funding provided by University of Georgia Research Foundation (to R. J. Weiss) and NIH T32 training Grant GM107004 (to N. G. Patel).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
This article is part of the special collection “Deciphering the Role of Proteoglycans and Glycosaminoglycans in Health and Disease.“ Liliana Schaefer, MD, served as Guest Editor of this collection.
AUTHOR CONTRIBUTIONS
R.J.W. conceived and designed research; E.D.N. and R.J.W. analyzed data; A.B., N.G.P., and R.J.W. prepared figures; A.B., N.G.P., and R.J.W. drafted manuscript; A.B., N.G.P., E.D.N., and R.J.W. edited and revised manuscript; A.B., N.G.P., E.D.N., and R.J.W. approved final version of manuscript.
ACKNOWLEDGMENTS
We apologize to all authors of relevant and important references we were unable to include in this review due to space limitations. We thank Dr. Marissa Maciej-Hulme for careful review and feedback on this manuscript.
REFERENCES
- 1.Möckl L. The emerging role of the mammalian glycocalyx in functional membrane organization and immune system regulation. Front Cell Dev Biol 8: 253, 2020. doi: 10.3389/fcell.2020.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinbaum S, Tarbell JM, Damiano ER. The structure and function of the endothelial glycocalyx layer. Annu Rev Biomed Eng 9: 121–167, 2007. doi: 10.1146/annurev.bioeng.9.060906.151959. [DOI] [PubMed] [Google Scholar]
- 3.Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH (Editors). Essentials of Glycobiology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2015. –2017. [PubMed] [Google Scholar]
- 4.Sasarman F, Maftei C, Campeau PM, Brunel-Guitton C, Mitchell GA, Allard P. Biosynthesis of glycosaminoglycans: associated disorders and biochemical tests. J Inherit Metab Dis 39: 173–188, 2016. doi: 10.1007/s10545-015-9903-z. [DOI] [PubMed] [Google Scholar]
- 5.Hacker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: the sweet side of development. Nat Rev Mol Cell Biol 6: 530–541, 2005. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- 6.Karamanos NK, Piperigkou Z, Theocharis AD, Watanabe H, Franchi M, Baud S, Brezillon S, Gotte M, Passi A, Vigetti D, Ricard-Blum S, Sanderson RD, Neill T, Iozzo RV. Proteoglycan chemical diversity drives multifunctional cell regulation and therapeutics. Chem Rev 118: 9152–9232, 2018. doi: 10.1021/acs.chemrev.8b00354. [DOI] [PubMed] [Google Scholar]
- 7.Caterson B, Melrose J. Keratan sulfate, a complex glycosaminoglycan with unique functional capability. Glycobiology 28: 182–206, 2018. doi: 10.1093/glycob/cwy003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss RJ, Esko JD, Tor Y. Targeting heparin and heparan sulfate protein interactions. Org Biomol Chem 15: 5656–5668, 2017. doi: 10.1039/c7ob01058c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu D, Esko JD. Demystifying heparan sulfate-protein interactions. Annu Rev Biochem 83: 129–157, 2014. doi: 10.1146/annurev-biochem-060713-035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y-F, Mizumoto S, Fujita M. Novel insight into glycosaminoglycan biosynthesis based on gene expression profiles. Front Cell Dev Biol 9: 709018, 2021. doi: 10.3389/fcell.2021.709018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sjöberg I, Fransson LA. Synthesis of glycosaminoglycans by human embryonic lung fibroblasts. Different distribution of heparan sulphate, chondroitin sulphate and dermatan sulphate in various fractions of cell culture. Biochem J 167: 383–392, 1977. doi: 10.1042/bj1670383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caniggia I, Tanswell K, Post M. Temporal and spatial differences in glycosaminoglycan synthesis by fetal lung fibroblasts. Exp Cell Res 202: 252–258, 1992. doi: 10.1016/0014-4827(92)90072-g. [DOI] [PubMed] [Google Scholar]
- 13.Wei J, Hu M, Huang K, Lin S, Du H. Roles of proteoglycans and glycosaminoglycans in cancer development and progression. Int J Mol Sci 21: 5983, 2020. doi: 10.3390/ijms21175983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi D, Sheng A, Chi L. Glycosaminoglycan-protein interactions and their roles in human disease. Front Mol Biosci 8: 639666, 2021. doi: 10.3389/fmolb.2021.639666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toledo AG, Pihl J, Spliid CB, Persson A, Nilsson J, Pereira MA, Gustavsson T, Choudhary S, Oo HZ, Black PC, Daugaard M, Esko JD, Larson G, Salanti A, Clausen TA. Affinity chromatography and glycoproteomics workflow to profile the chondroitin sulfate proteoglycans that interact with malarial VAR2CSA in the placenta and in cancer. Glycobiology 30: 989–1002, 2020. doi: 10.1093/glycob/cwaa039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noborn F, Nikpour M, Persson A, Nilsson J, Larson G. Expanding the chondroitin sulfate glycoproteome—but how far? Front Cell Dev Biol 9: 695970, 2021. doi: 10.3389/fcell.2021.695970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L. Glycosaminoglycan (GAG) biosynthesis and GAG-binding proteins. In: Progress in Molecular Biology and Translational Science, edited by Zhang L. London: Academic Press, 2010, p. 1–17. [DOI] [PubMed] [Google Scholar]
- 18.Ghiselli G. Drug-mediated regulation of glycosaminoglycan biosynthesis. Med Res Rev 37: 1051–1094, 2017. doi: 10.1002/med.21429. [DOI] [PubMed] [Google Scholar]
- 19.Koike T, Izumikawa T, Tamura J-I, Kitagawa H. FAM20B is a kinase that phosphorylates xylose in the glycosaminoglycan–protein linkage region. Biochem J 421: 157–162, 2009. doi: 10.1042/BJ20090474. [DOI] [PubMed] [Google Scholar]
- 20.Gulberti S, Lattard V, Fondeur M, Jacquinet JC, Mulliert G, Netter P, Magdalou J, Ouzzine M, Fournel-Gigleux S. Phosphorylation and sulfation of oligosaccharide substrates critically influence the activity of human beta1,4-galactosyltransferase 7 (GalT-I) and beta1,3-glucuronosyltransferase I (GlcAT-I) involved in the biosynthesis of the glycosaminoglycan-protein linkage region of proteoglycans. J Biol Chem 280: 1417–1425, 2005. doi: 10.1074/jbc.M411552200. [DOI] [PubMed] [Google Scholar]
- 21.Koike T, Izumikawa T, Sato B, Kitagawa H. Identification of phosphatase that dephosphorylates xylose in the glycosaminoglycan-protein linkage region of proteoglycans. J Biol Chem 289: 6695–6708, 2014. doi: 10.1074/jbc.M113.520536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamada S. Specific functions of Exostosin-like 3 (EXTL3) gene products. Cell Mol Biol Lett 25: 39, 2020. doi: 10.1186/s11658-020-00231-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Busse M, Feta A, Presto J, Wilen M, Gronning M, Kjellen L, Kusche-Gullberg M. Contribution of EXT1, EXT2, and EXTL3 to heparan sulfate chain elongation. J Biol Chem 282: 32802–32810, 2007. doi: 10.1074/jbc.M703560200. [DOI] [PubMed] [Google Scholar]
- 24.Sato T, Gotoh M, Kiyohara K, Akashima T, Iwasaki H, Kameyama A, Mochizuki H, Yada T, Inaba N, Togayachi A, Kudo T, Asada M, Watanabe H, Imamura T, Kimata K, Narimatsu H. Differential roles of two N-acetylgalactosaminyltransferases, CSGalNAcT-1, and a novel enzyme, CSGalNAcT-2—initiation and elongation in synthesis of chondroitin sulfate. J Biol Chem 278: 3063–3071, 2003. doi: 10.1074/jbc.M208886200. [DOI] [PubMed] [Google Scholar]
- 25.Gulberti S, Jacquinet JC, Chabel M, Ramalanjaona N, Magdalou J, Netter P, Coughtrie MW, Ouzzine M, Fournel-Gigleux S. Chondroitin sulfate N-acetylgalactosaminyltransferase-1 (CSGalNAcT-1) involved in chondroitin sulfate initiation: Impact of sulfation on activity and specificity. Glycobiology 22: 561–571, 2012. doi: 10.1093/glycob/cwr172. [DOI] [PubMed] [Google Scholar]
- 26.Kitagawa H, Shimakawa H, Sugahara K. The tumor suppressor EXT-like gene EXTL2 encodes an alpha1, 4-N-acetylhexosaminyltransferase that transfers N-acetylgalactosamine and N-acetylglucosamine to the common glycosaminoglycan-protein linkage region. The key enzyme for the chain initiation of heparan sulfate. J Biol Chem 274: 13933–13937, 1999. doi: 10.1074/jbc.274.20.13933. [DOI] [PubMed] [Google Scholar]
- 27.Busse-Wicher M, Wicher KB, Kusche-Gullberg M. The extostosin family: proteins with many functions. Matrix Biol 35: 25–33, 2014. doi: 10.1016/j.matbio.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Nadanaka S, Zhou S, Kagiyama S, Shoji N, Sugahara K, Sugihara K, Asano M, Kitagawa H. EXTL2, a member of the EXT family of tumor suppressors, controls glycosaminoglycan biosynthesis in a xylose kinase-dependent manner. J Biol Chem 288: 9321–9333, 2013. doi: 10.1074/jbc.M112.416909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lind T, Tufaro F, McCormick C, Lindahl U, Lidholt K. The putative tumor suppressors EXT1 and EXT2 are glycosyltransferases required for the biosynthesis of heparan sulfate. J Biol Chem 273: 26265–26268, 1998. doi: 10.1074/jbc.273.41.26265. [DOI] [PubMed] [Google Scholar]
- 30.McCormick C, Duncan G, Goutsos KT, Tufaro F. The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proc Natl Acad Sci USA 97: 668–673, 2000. doi: 10.1073/pnas.97.2.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izumikawa T, Uyama T, Okuura Y, Sugahara K, Kitagawa H. Involvement of chondroitin sulfate synthase-3 (chondroitin synthase-2) in chondroitin polymerization through its interaction with chondroitin synthase-1 or chondroitin-polymerizing factor. Biochem J 403: 545–552, 2007. doi: 10.1042/BJ20061876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitagawa H, Izumikawa T, Uyama T, Sugahara K. Molecular cloning of a chondroitin polymerizing factor that cooperates with chondroitin synthase for chondroitin polymerization. J Biol Chem 278: 23666–23671, 2003. doi: 10.1074/jbc.M302493200. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa H, Shionyu M, Sugiura N, Hatano S, Nagai N, Kubota Y, Nishiwaki K, Sato T, Gotoh M, Narimatsu H, Shimizu K, Kimata K, Watanabe H. Chondroitin sulfate synthase-2/chondroitin polymerizing factor has two variants with distinct function. J Biol Chem 285: 34155–34167, 2010. doi: 10.1074/jbc.M110.109553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang B, Chi L. Chondroitin sulfate/dermatan sulfate-protein interactions and their biological functions in human diseases: implications and analytical tools. Front Cell Dev Biol 9: 693563, 2021. doi: 10.3389/fcell.2021.693563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarrazin S, Lamanna WC, Esko JD. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol 3: a004952, 2011. doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thacker BE, Xu D, Lawrence R, Esko JD. Heparan sulfate 3-O-sulfation: a rare modification in search of a function. Matrix Biol 35: 60–72, 2014. doi: 10.1016/j.matbio.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Afratis N, Gialeli C, Nikitovic D, Tsegenidis T, Karousou E, Theocharis AD, Pavao MS, Tzanakakis GN, Karamanos NK. Glycosaminoglycans: key players in cancer cell biology and treatment. FEBS J 279: 1177–1197, 2012. doi: 10.1111/j.1742-4658.2012.08529.x. [DOI] [PubMed] [Google Scholar]
- 38.Silbert JE, Sugumaran G. Biosynthesis of chondroitin/dermatan sulfate. IUBMB Life 54: 177–186, 2002. doi: 10.1080/15216540214923. [DOI] [PubMed] [Google Scholar]
- 39.Esko JD, Lindahl U. Molecular diversity of heparan sulfate. J Clin Invest 108: 169–173, 2001. doi: 10.1172/JCI200113530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Persson A, Vorontsov E, Larson G, Nilsson J. Glycosaminoglycan domain mapping of cellular chondroitin/dermatan sulfates. Sci Rep 10: 3506, 2020. doi: 10.1038/s41598-020-60526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schultz V, Suflita M, Liu X, Zhang X, Yu Y, Li L, Green DE, Xu Y, Zhang F, DeAngelis PL, Liu J, Linhardt RJ. Heparan sulfate domains required for fibroblast growth factor 1 and 2 signaling through fibroblast growth factor receptor 1c*. J Biol Chem 292: 2495–2509, 2017. doi: 10.1074/jbc.M116.761585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fux L, Ilan N, Sanderson RD, Vlodavsky I. Heparanase: busy at the cell surface. Trends Biochem Sci 34: 511–519, 2009. doi: 10.1016/j.tibs.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masola V, Greco N, Gambaro G, Franchi M, Onisto M. Heparanase as active player in endothelial glycocalyx remodeling. Matrix Biol Plus 13: 100097, 2022. doi: 10.1016/j.mbplus.2021.100097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramani VC, Pruett PS, Thompson CA, Delucas LD, Sanderson RD. Heparan sulfate chains of syndecan-1 regulate ectodomain shedding. J Biol Chem 287: 9952–9961, 2012. doi: 10.1074/jbc.M111.330803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKenzie E, Tyson K, Stamps A, Smith P, Turner P, Barry R, Hircock M, Patel S, Barry E, Stubberfield C, Terrett J, Page M. Cloning and expression profiling of Hpa2, a novel mammalian heparanase family member. Biochem Biophys Res Commun 276: 1170–1177, 2000. doi: 10.1006/bbrc.2000.3586. [DOI] [PubMed] [Google Scholar]
- 46.Morimoto-Tomita M, Uchimura K, Werb Z, Hemmerich S, Rosen SD. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J Biol Chem 277: 49175–49185, 2002. doi: 10.1074/jbc.M205131200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamanna WC, Frese MA, Balleininger M, Dierks T. Sulf loss influences N-, 2-O-, and 6-O-sulfation of multiple heparan sulfate proteoglycans and modulates fibroblast growth factor signaling. J Biol Chem 283: 27724–27735, 2008. doi: 10.1074/jbc.M802130200. [DOI] [PubMed] [Google Scholar]
- 48.Vives RR, Seffouh A, Lortat-Jacob H. Post-synthetic regulation of HS structure: the Yin and Yang of the sulfs in cancer. Front Oncol 3: 331, 2014. doi: 10.3389/fonc.2013.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narita K, Staub J, Chien J, Meyer K, Bauer M, Friedl A, Ramakrishnan S, Shridhar V. HSulf-1 inhibits angiogenesis and tumorigenesis in vivo. Cancer Res 66: 6025–6032, 2006. doi: 10.1158/0008-5472.CAN-05-3582. [DOI] [PubMed] [Google Scholar]
- 50.Lai JP, Sandhu DS, Yu C, Han T, Moser CD, Jackson KK, Guerrero RB, Aderca I, Isomoto H, Garrity-Park MM, Zou H, Shire AM, Nagorney DM, Sanderson SO, Adjei AA, Lee JS, Thorgeirsson SS, Roberts LR. Sulfatase 2 up-regulates glypican 3, promotes fibroblast growth factor signaling, and decreases survival in hepatocellular carcinoma. Hepatology 47: 1211–1222, 2008. doi: 10.1002/hep.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morimoto-Tomita M, Uchimura K, Bistrup A, Lum DH, Egeblad M, Boudreau N, Werb Z, Rosen SD. Sulf-2, a proangiogenic heparan sulfate endosulfatase, is upregulated in breast cancer. Neoplasia 7: 1001–1010, 2005. doi: 10.1593/neo.05496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lemjabbar-Alaoui H, van Zante A, Singer MS, Xue Q, Wang YQ, Tsay D, He B, Jablons DM, Rosen SD. Sulf-2, a heparan sulfate endosulfatase, promotes human lung carcinogenesis. Oncogene 29: 635–646, 2010. doi: 10.1038/onc.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ratzka A, Kalus I, Moser M, Dierks T, Mundlos S, Vortkamp A. Redundant function of the heparan sulfate 6-O-endosulfatases Sulf1 and Sulf2 during skeletal development. Dev Dyn 237: 339–353, 2008. doi: 10.1002/dvdy.21423. [DOI] [PubMed] [Google Scholar]
- 54.Nagamine S, Tamba M, Ishimine H, Araki K, Shiomi K, Okada T, Ohto T, Kunita S, Takahashi S, Wismans RG, van Kuppevelt TH, Masu M, Keino-Masu K. Organ-specific sulfation patterns of heparan sulfate generated by extracellular sulfatases Sulf1 and Sulf2 in mice. J Biol Chem 287: 9579–9590, 2012. doi: 10.1074/jbc.M111.290262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miya K, Keino-Masu K, Okada T, Kobayashi K, Masu M. Expression of heparan sulfate endosulfatases in the adult mouse brain: co-expression of sulf1 and dopamine D1/D2 receptors. Front Neuroanat 15: 726718, 2021. doi: 10.3389/fnana.2021.726718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang S, Sugahara K, Li F. Chondroitin sulfate/dermatan sulfate sulfatases from mammals and bacteria. Glycoconj J 33: 841–851, 2016. doi: 10.1007/s10719-016-9720-0. [DOI] [PubMed] [Google Scholar]
- 57.Bhattacharyya S, Zhang X, Feferman L, Johnson D, Tortella FC, Guizzetti M, Tobacman JK. Decline in arylsulfatase B and Increase in chondroitin 4-sulfotransferase combine to increase chondroitin 4-sulfate in traumatic brain injury. J Neurochem 134: 728–739, 2015. doi: 10.1111/jnc.13156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prabhu SV, Bhattacharyya S, Guzman-Hartman G, Macias V, Kajdacsy-Balla A, Tobacman JK. Extra-lysosomal localization of arylsulfatase B in human colonic epithelium. J Histochem Cytochem 59: 328–335, 2011. doi: 10.1369/0022155410395511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maciej-Hulme ML. New insights into human hyaluronidase 4/chondroitin sulphate hydrolase. Front Cell Dev Biol 9: 767924, 2021. doi: 10.3389/fcell.2021.767924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith RA, Meade K, Pickford CE, Holley RJ, Merry CL. Glycosaminoglycans as regulators of stem cell differentiation. Biochem Soc Trans 39: 383–387, 2011. doi: 10.1042/BST0390383. [DOI] [PubMed] [Google Scholar]
- 61.Izumikawa T, Sato B, Kitagawa H. Chondroitin sulfate is indispensable for pluripotency and differentiation of mouse embryonic stem cells. Sci Rep 4: 3701, 2014. doi: 10.1038/srep03701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fujitani N, Furukawa J-I, Araki K, Fujioka T, Takegawa Y, Piao J, Nishioka T, Tamura T, Nikaido T, Ito M, Nakamura Y, Shinohara Y. Total cellular glycomics allows characterizing cells and streamlining the discovery process for cellular biomarkers. Proc Natl Acad Sci USA 110: 2105–2110, 2013. doi: 10.1073/pnas.1214233110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clark SJ, Bishop PN, Day AJ. The proteoglycan glycomatrix: a sugar microenvironment essential for complement regulation. Front Immunol 4: 412, 2013. doi: 10.3389/fimmu.2013.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Puri S, Coulson-Thomas YM, Gesteira TF, Coulson-Thomas VJ. Distribution and function of glycosaminoglycans and proteoglycans in the development, homeostasis and pathology of the ocular surface. Front Cell Dev Biol 8: 731, 2020. doi: 10.3389/fcell.2020.00731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Melrose J. Glycosaminoglycans in wound healing. Bone Tissue Regen Insights 7: BTRI.S38670, 2016. doi: 10.4137/BTRI.S38670. [DOI] [Google Scholar]
- 66.Chakraborty S, Njah K, Hong W. Agrin mediates angiogenesis in the tumor microenvironment. Trends Cancer 6: 81–85, 2020. doi: 10.1016/j.trecan.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 67.Mousavi S, Moradi M, Khorshidahmad T, Motamedi M. Anti-inflammatory effects of heparin and its derivatives: a systematic review. Adv Pharmacol Sci 2015: 507151, 2015. doi: 10.1155/2015/507151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caterson B. Fell-Muir lecture: chondroitin sulphate glycosaminoglycans: fun for some and confusion for others. Int J Exp Pathol 93: 1–10, 2012. doi: 10.1111/j.1365-2613.2011.00807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Kuppevelt TH, Dennissen MABA, Van Venrooij WJ, Hoet RMA, Veerkamp JH. Generation and application of type-specific anti-heparan sulfate antibodies using phage display technology—further evidence for heparan sulfate heterogeneity in the kidney. J Biol Chem 273: 12960–12966, 1998. doi: 10.1074/jbc.273.21.12960. [DOI] [PubMed] [Google Scholar]
- 70.David G, Bai XM, Van der Schueren B, Cassiman JJ, Van den Berghe H. Developmental changes in heparan sulfate expression: in situ detection with mAbs. J Cell Biol 119: 961–975, 1992. doi: 10.1083/jcb.119.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee J, Ha S, Kim M, Kim S-W, Yun J, Ozcan S, Hwang H, Ji IJ, Yin D, Webster MJ, Shannon Weickert C, Kim J-H, Yoo JS, Grimm R, Bahn S, Shin H-S, An HJ. Spatial and temporal diversity of glycome expression in mammalian brain. Proc Natl Acad Sci USA 117: 28743–28753, 2020. doi: 10.1073/pnas.2014207117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song Y, Zhang F, Linhardt RJ. Analysis of the glycosaminoglycan chains of proteoglycans. J Histochem Cytochem 69: 121–135, 2021. doi: 10.1369/0022155420937154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Tissue-based map of the human proteome. Science 347: 1260419, 2015. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 74.Karlsson M, Zhang C, Méar L, Zhong W, Digre A, Katona B, Sjöstedt E, Butler L, Odeberg J, Dusart P, Edfors F, Oksvold P, Kv F, Zwahlen M, Arif M, Altay O, Li X, Ozcan M, Mardinoglu A, Fagerberg L, Mulder J, Luo Y, Ponten F, Uhlén M, Lindskog C. A single-cell type transcriptomics map of human tissues. Sci Adv 7: eabh2169, 2021. doi: 10.1126/sciadv.abh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Denys A, Allain F. The emerging roles of heparan sulfate 3-O-sulfotransferases in cancer. Front Oncol 9: 507, 2019. doi: 10.3389/fonc.2019.00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ledin J, Staatz W, Li JP, Gotte M, Selleck S, Kjellen L, Spillmann D. Heparan sulfate structure in mice with genetically modified heparan sulfate production. J Biol Chem 279: 42732–42741, 2004. doi: 10.1074/jbc.M405382200. [DOI] [PubMed] [Google Scholar]
- 77.Sugahara K, Mikami T, Uyama T, Mizuguchi S, Nomura K, Kitagawa H. Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate. Curr Opin Struct Biol 13: 612–620, 2003. doi: 10.1016/j.sbi.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 78.Nakanishi K, Higashi K, Toida T, Asai M. Characterization of chondroitin sulfate in stem cells derived from umbilical cord blood in rats. PLoS One 17: e0262854, 2022. doi: 10.1371/journal.pone.0262854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miyata S, Nadanaka S, Igarashi M, Kitagawa H. Structural variation of chondroitin sulfate chains contributes to the molecular heterogeneity of perineuronal nets. Front Integr Neurosci 12: 3, 2018. doi: 10.3389/fnint.2018.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kubota Y, Morita T, Kusakabe M, Sakakura T, Ito K. Spatial and temporal changes in chondroitin sulfate distribution in the sclerotome play an essential role in the formation of migration patterns of mouse neural crest cells. Dev Dyn 214: 55–65, 1999. doi:. [DOI] [PubMed] [Google Scholar]
- 81.Bandtlow CE, Zimmermann DR. Proteoglycans in the developing brain: new conceptual insights for old proteins. Physiol Rev 80: 1267–1290, 2000. doi: 10.1152/physrev.2000.80.4.1267. [DOI] [PubMed] [Google Scholar]
- 82.Ishii M, Maeda N. Spatiotemporal expression of chondroitin sulfate sulfotransferases in the postnatal developing mouse cerebellum. Glycobiology 18: 602–614, 2008. doi: 10.1093/glycob/cwn040. [DOI] [PubMed] [Google Scholar]
- 83.Lawrence R, Olson SK, Steele RE, Wang L, Warrior R, Cummings RD, Esko JD. Evolutionary differences in glycosaminoglycan fine structure detected by quantitative glycan reductive isotope labeling. J Biol Chem 283: 33674–33684, 2008. doi: 10.1074/jbc.M804288200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moon S, Zhao Y-T. Spatial, temporal and cell-type-specific expression profiles of genes encoding heparan sulfate biosynthesis enzymes and proteoglycan core proteins. Glycobiology 31: 1308–1318, 2021. doi: 10.1093/glycob/cwab054. [DOI] [PubMed] [Google Scholar]
- 85.Kraushaar DC, Dalton S, Wang L. Heparan sulfate: a key regulator of embryonic stem cell fate. Biol Chem 394: 741–751, 2013. doi: 10.1515/hsz-2012-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nairn AV, Kinoshita-Toyoda A, Toyoda H, Xie J, Harris K, Dalton S, Kulik M, Pierce JM, Toida T, Moremen KW, Linhardt RJ. Glycomics of proteoglycan biosynthesis in murine embryonic stem cell differentiation. J Proteome Res 6: 4374–4387, 2007. doi: 10.1021/pr070446f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Johnson CE, Crawford BE, Stavridis M, Ten Dam G, Wat AL, Rushton G, Ward CM, Wilson V, van Kuppevelt TH, Esko JD, Smith A, Gallagher JT, Merry CL. Essential alterations of heparan sulfate during the differentiation of embryonic stem cells to Sox1-enhanced green fluorescent protein-expressing neural progenitor cells. Stem Cells 25: 1913–1923, 2007. [Erratum in Stem Cells 25: 2389, 2007]. doi: 10.1634/stemcells.2006-0445. [DOI] [PubMed] [Google Scholar]
- 88.Baldwin RJ, ten Dam GB, van Kuppevelt TH, Lacaud G, Gallagher JT, Kouskoff V, Merry CL. A developmentally regulated heparan sulfate epitope defines a subpopulation with increased blood potential during mesodermal differentiation. Stem Cells 26: 3108–3118, 2008. doi: 10.1634/stemcells.2008-0311. [DOI] [PubMed] [Google Scholar]
- 89.Piunti A, Shilatifard A. The roles of Polycomb repressive complexes in mammalian development and cancer. Nat Rev Mol Cell Biol 22: 326–345, 2021. doi: 10.1038/s41580-021-00341-1. [DOI] [PubMed] [Google Scholar]
- 90.Cool SM, Nurcombe V. Heparan sulfate regulation of progenitor cell fate. J Cell Biochem 99: 1040–1051, 2006. doi: 10.1002/jcb.20936. [DOI] [PubMed] [Google Scholar]
- 91.Bulow HE, Hobert O. The molecular diversity of glycosaminoglycans shapes animal development. Annu Rev Cell Dev Biol 22: 375–407, 2006. doi: 10.1146/annurev.cellbio.22.010605.093433. [DOI] [PubMed] [Google Scholar]
- 92.Jaakkola P, Vihinen T, Maatta A, Jalkanen M. Activation of an enhancer on the syndecan-1 gene is restricted to fibroblast growth factor family members in mesenchymal cells. Mol Cell Biol 17: 3210–3219, 1997. doi: 10.1128/MCB.17.6.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jaakkola P, Jalkanen M. Transcriptional regulation of Syndecan-1 expression by growth factors. Prog Nucleic Acid Res Mol Biol 63: 109–138, 1999. doi: 10.1016/s0079-6603(08)60721-7. [DOI] [PubMed] [Google Scholar]
- 94.Jaakkola P, Maatta A, Jalkanen M. The activation and composition of FiRE (an FGF-inducible response element) differ in a cell type- and growth factor-specific manner. Oncogene 17: 1279–1286, 1998. doi: 10.1038/sj.onc.1202002. [DOI] [PubMed] [Google Scholar]
- 95.Schönherr E, Järveläinen HT, Kinsella MG, Sandell LJ, Wight TN. Platelet-derived growth factor and transforming growth factor-β1 differentially affect the synthesis of biglycan and decorin by monkey arterial smooth muscle cells. Arterioscler Thromb 13: 1026–1036, 1993. doi: 10.1161/01.atv.13.7.1026. [DOI] [PubMed] [Google Scholar]
- 96.Hu B, Shi C, Tian Y, Zhang Y, Xu C, Chen H, Cao P, Yuan W. TGF-β induces up-regulation of chondroitin sulfate synthase 1 (CHSY1) in nucleus pulposus cells through MAPK signaling. Cell Physiol Biochem 37: 793–804, 2015. doi: 10.1159/000430396. [DOI] [PubMed] [Google Scholar]
- 97.Tiedemann K, Olander B, Eklund E, Todorova L, Bengtsson M, Maccarana M, Westergren-Thorsson G, Malmstrom A. Regulation of the chondroitin/dermatan fine structure by transforming growth factor-β1 through effects on polymer-modifying enzymes. Glycobiology 15: 1277–1285, 2005. doi: 10.1093/glycob/cwj027. [DOI] [PubMed] [Google Scholar]
- 98.Grobe K, Esko JD. Regulated translation of heparan sulfate N-acetylglucosamine N-deacetylase/N-sulfotransferase isozymes by structured 5'-untranslated regions and internal ribosome entry sites. J Biol Chem 277: 30699–30706, 2002. doi: 10.1074/jbc.M111904200. [DOI] [PubMed] [Google Scholar]
- 99.Bornemann DJ, Park S, Phin S, Warrior R. A translational block to HSPG synthesis permits BMP signaling in the early Drosophila embryo. Development 135: 1039–1047, 2008. doi: 10.1242/dev.017061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Morii E, Ogihara H, Oboki K, Sawa C, Sakuma T, Nomura S, Esko JD, Handa H, Kitamura Y. Inhibitory effect of the mi transcription factor encoded by the mutant mi allele on GA binding protein-mediated transcript expression in mouse mast cells. Blood 97: 3032–3039, 2001. doi: 10.1182/blood.v97.10.3032. [DOI] [PubMed] [Google Scholar]
- 101.Small EM, Sutherland LB, Rajagopalan KN, Wang S, Olson EN. MicroRNA-218 regulates vascular patterning by modulation of Slit-Robo signaling. Circ Res 107: 1336–1344, 2010. doi: 10.1161/CIRCRESAHA.110.227926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ibrahim SA, Hassan H, Gotte M. MicroRNA regulation of proteoglycan function in cancer. FEBS J 281: 5009–5022, 2014. doi: 10.1111/febs.13026. [DOI] [PubMed] [Google Scholar]
- 103.Kasza Z, Fredlund Fuchs P, Tamm C, Eriksson AS, O'Callaghan P, Heindryckx F, Spillmann D, Larsson E, Le Jan S, Eriksson I, Gerwins P, Kjellen L, Kreuger J. MicroRNA-24 suppression of N-deacetylase/n-sulfotransferase-1 (NDST1) reduces endothelial cell responsiveness to vascular endothelial growth factor A (VEGFA). J Biol Chem 288: 25956–25963, 2013. doi: 10.1074/jbc.M113.484360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Haupt LM, Murali S, Mun FK, Teplyuk N, Mei LF, Stein GS, van Wijnen AJ, Nurcombe V, Cool SM. The heparan sulfate proteoglycan (HSPG) glypican-3 mediates commitment of MC3T3-E1 cells toward osteogenesis. J Cell Physiol 220: 780–791, 2009. doi: 10.1002/jcp.21825. [DOI] [PubMed] [Google Scholar]
- 105.Ropero S, Setien F, Espada J, Fraga MF, Herranz M, Asp J, Benassi MS, Franchi A, Patino A, Ward LS, Bovee J, Cigudosa JC, Wim W, Esteller M. Epigenetic loss of the familial tumor-suppressor gene exostosin-1 (EXT1) disrupts heparan sulfate synthesis in cancer cells. Hum Mol Genet 13: 2753–2765, 2004. doi: 10.1093/hmg/ddh298. [DOI] [PubMed] [Google Scholar]
- 106.Hull EE, Montgomery MR, Leyva KJ. Epigenetic regulation of the biosynthesis & enzymatic modification of heparan sulfate proteoglycans: implications for tumorigenesis and cancer biomarkers. Int J Mol Sci 18: 1361, 2017. doi: 10.3390/ijms18071361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pursiheimo JP, Jalkanen M, Tasken K, Jaakkola P. Involvement of protein kinase A in fibroblast growth factor-2-activated transcription. Proc Natl Acad Sci USA 97: 168–173, 2000. doi: 10.1073/pnas.97.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sun H, Berquin IM, Owens RT, O'Flaherty JT, Edwards IJ. Peroxisome proliferator-activated receptor gamma-mediated up-regulation of syndecan-1 by n − 3 fatty acids promotes apoptosis of human breast cancer cells. Cancer Res 68: 2912–2919, 2008. doi: 10.1158/0008-5472.CAN-07-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li H, Melford K, Judson A, Bensadoun A. Murine glypican-4 gene structure and expression; Sp1 and Sp3 play a major role in glypican-4 expression in 3T3-F442A cells. Biochim Biophys Acta 1679: 141–155, 2004. doi: 10.1016/S0167-4781(04)00110-1. [DOI] [PubMed] [Google Scholar]
- 110.Huber R, Schlessinger D, Pilia G. Multiple Sp1 sites efficiently drive transcription of the TATA-less promoter of the human glypican 3 (GPC3) gene. Gene 214: 35–44, 1998. doi: 10.1016/S0378-1119(98)00233-9. [DOI] [PubMed] [Google Scholar]
- 111.Asundi VK, Keister BF, Carey DJ. Organization, 5'-flanking sequence and promoter activity of the rat GPC1 gene. Gene 206: 255–261, 1998. doi: 10.1016/S0378-1119(97)00594-5. [DOI] [PubMed] [Google Scholar]
- 112.Takagi A, Kojima T, Tsuzuki S, Katsumi A, Yamazaki T, Sugiura I, Hamaguchi M, Saito H. Structural organization and promoter activity of the human ryudocan gene. J Biochem 119: 979–984, 1996. doi: 10.1093/oxfordjournals.jbchem.a021338. [DOI] [PubMed] [Google Scholar]
- 113.Armelin-Correa LM, Lin CJ, Barbosa A, Bagatini K, Winnischofer SMB, Sogayar MC, Passos-Bueno MR. Characterization of human collagen XVIII promoter 2: interaction of Sp1, Sp3 and YY1 with the regulatory region and a SNP that increases transcription in hepatocytes. Matrix Biol 24: 550–559, 2005. doi: 10.1016/j.matbio.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 114.Iozzo RV, Pillarisetti J, Sharma B, Murdoch AD, Danielson KG, Uitto J, Mauviel A. Structural and functional characterization of the human perlecan gene promoter. Transcriptional activation by transforming growth factor-β via a nuclear factor 1-binding element. J Biol Chem 272: 5219–5228, 1997. doi: 10.1074/jbc.272.8.5219. [DOI] [PubMed] [Google Scholar]
- 115.Shigemoto K, Kubo S, Maruyama N, Yamada S, Obata K, Kikuchi K, Kondo I. Identification and characterization of 5' extension of mammalian agrin cDNA, the exons and the promoter sequences. Biochim Biophys Acta 1494: 170–174, 2000. doi: 10.1016/s0167-4781(00)00214-1. [DOI] [PubMed] [Google Scholar]
- 116.Schick BP, Petrushina I, Brodbeck KC, Castronuevo P. Promoter regulatory elements and DNase I-hypersensitive sites involved in serglycin proteoglycan gene expression in human erythroleukemia, CHRF 288-11, and HL-60 cells. J Biol Chem 276: 24726–24735, 2001. doi: 10.1074/jbc.M102958200. [DOI] [PubMed] [Google Scholar]
- 117.Ghiselli G, Agrawal A. The human d-glucuronyl C5-epimerase gene is transcriptionally activated through the β-catenin-TCF4 pathway. Biochem J 390: 493–499, 2005. doi: 10.1042/BJ20050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Barre L, Venkatesan N, Magdalou J, Netter P, Fournel-Gigleux S, Ouzzine M. Evidence of calcium-dependent pathway in the regulation of human beta1,3-glucuronosyltransferase-1 (GlcAT-I) gene expression: a key enzyme in proteoglycan synthesis. FASEB J 20: 1692–1694, 2006. doi: 10.1096/fj.05-5073fje. [DOI] [PubMed] [Google Scholar]
- 119.Hiyama A, Gajghate S, Sakai D, Mochida J, Shapiro IM, Risbud MV. Activation of TonEBP by calcium controls β1,3-glucuronosyltransferase-I expression, a key regulator of glycosaminoglycan synthesis in cells of the intervertebral disc. J Biol Chem 284: 9824–9834, 2009. doi: 10.1074/jbc.M807081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Okuda H, Tatsumi K, Horii-Hayashi N, Morita S, Okuda-Yamamoto A, Imaizumi K, Wanaka A. OASIS regulates chondroitin 6-O-sulfotransferase 1 gene transcription in the injured adult mouse cerebral cortex. J Neurochem 130: 612–625, 2014. doi: 10.1111/jnc.12736. [DOI] [PubMed] [Google Scholar]
- 121.Missaghian P, Dierker T, Khosrowabadi E, Axling F, Eriksson I, Ghanem A, Kusche-Gullberg M, Kellokumpu S, Kjellén L. A dominant negative splice variant of the heparan sulfate biosynthesis enzyme NDST1 reduces heparan sulfate sulfation. Glycobiology 2022. doi: 10.1093/glycob/cwac004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Groth T, Gunawan R, Neelamegham S. A systems-based framework to computationally describe putative transcription factors and signaling pathways regulating glycan biosynthesis. Beilstein J Org Chem 17: 1712–1724, 2021. doi: 10.3762/bjoc.17.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Weiss RJ, Spahn PN, Toledo AG, Chiang AWT, Kellman BP, Li J, Benner C, Glass CK, Gordts P, Lewis NE, Esko JD. ZNF263 is a transcriptional regulator of heparin and heparan sulfate biosynthesis. Proc Natl Acad Sci USA 117: 9311–9317, 2020. doi: 10.1073/pnas.1920880117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823, 2013. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science 339: 823–826, 2013. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343: 84–87, 2014. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34: 184–191, 2016. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Weiss RJ, Spahn PN, Chiang AWT, Liu Q, Li J, Hamill KM, Rother S, Clausen TM, Hoeksema MA, Timm BM, Godula K, Glass CK, Tor Y, Gordts P, Lewis NE, Esko JD. Genome-wide screens uncover KDM2B as a modifier of protein binding to heparan sulfate. Nat Chem Biol 17: 684–692, 2021. doi: 10.1038/s41589-021-00776-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Petitjean O, Girardi E, Ngondo RP, Lupashin V, Pfeffer S. Genome-wide CRISPR-Cas9 screen reveals the importance of the heparan sulfate pathway and the conserved oligomeric Golgi complex for synthetic double-stranded RNA uptake and Sindbis virus infection. mSphere 5, 2020. doi: 10.1128/mSphere.00914-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Multhaupt HA, Couchman JR. Heparan sulfate biosynthesis: methods for investigation of the heparanosome. J Histochem Cytochem 60: 908–915, 2012. doi: 10.1369/0022155412460056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Velasco A, Hidalgo J. Ultrastructural demonstration of proteoglycans in adult rat cornea. Tissue Cell 20: 567–575, 1988. doi: 10.1016/0040-8166(88)90058-4. [DOI] [PubMed] [Google Scholar]
- 132.Sugumaran G, Katsman M, Silbert J. Effects of Brefeldin A on the synthesis of chondroitin 4-sulfate by cultures of mouse mastocytoma cells. Biochem Biophys Res Commun 183: 357–361, 1992. doi: 10.1016/0006-291x(92)90488-7. [DOI] [PubMed] [Google Scholar]
- 133.Nagai N, Habuchi H, Esko JD, Kimata K. Stem domains of heparan sulfate 6-O-sulfotransferase are required for Golgi localization, oligomer formation and enzyme activity. J Cell Sci 117: 3331–3341, 2004. doi: 10.1242/jcs.01191. [DOI] [PubMed] [Google Scholar]
- 134.Nagai N, Habuchi H, Kitazume S, Toyoda H, Hashimoto Y, Kimata K. Regulation of heparan sulfate 6-O-sulfation by beta-secretase activity. J Biol Chem 282: 14942–14951, 2007. doi: 10.1074/jbc.M610691200. [DOI] [PubMed] [Google Scholar]
- 135.Eisele BS, Luka Z, Wu AJ, Yang F, Hale AT, York JD. Sulfation of glycosaminoglycans depends on the catalytic activity of lithium-inhibited phosphatase BPNT2 in vitro. J Biol Chem 297: 101293, 2021. doi: 10.1016/j.jbc.2021.101293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tanaka A, Tumkosit U, Nakamura S, Motooka D, Kishishita N, Priengprom T, Sa-Ngasang A, Kinoshita T, Takeda N, Maeda Y. Genome-wide screening uncovers the significance of n-sulfation of heparan sulfate as a host cell factor for chikungunya virus infection. J Virol 91: e00432-17, 2017. doi: 10.1128/JVI.00432-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Khan S, Sbeity M, Foulquier F, Barré L, Ouzzine M. TMEM165 a new player in proteoglycan synthesis: loss of TMEM165 impairs elongation of chondroitin- and heparan-sulfate glycosaminoglycan chains of proteoglycans and triggers early chondrocyte differentiation and hypertrophy. Cell Death Dis 13: 11, 2021. doi: 10.1038/s41419-021-04458-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chang WL, Chang CW, Chang YY, Sung HH, Lin MD, Chang SC, Chen CH, Huang CW, Tung KS, Chou TB. The Drosophila GOLPH3 homolog regulates the biosynthesis of heparan sulfate proteoglycans by modulating the retrograde trafficking of exostosins. Development 140: 2798–2807, 2013. doi: 10.1242/dev.087171. [DOI] [PubMed] [Google Scholar]
- 139.Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem 71: 435–471, 2002. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 140.Höök M, Lindahl U, Hallén A, Bäckström G. Biosynthesis of heparin. Studies on the microsomal sulfation process. J Biol Chem 250: 6065–6071, 1975. [PubMed] [Google Scholar]
- 141.Forsee WT, Rodén L. Biosynthesis of heparin. Transfer of N-acetylglucosamine to heparan sulfate oligosaccharides. J Biol Chem 256: 7240–7247, 1981. [PubMed] [Google Scholar]
- 142.Schwartz NB. Biosynthesis of chondroitin sulfate: Immunoprecipitation of interacting xylosyltransferase and galactosyltransferase. FEBS Lett 49: 342–345, 1975. doi: 10.1016/0014-5793(75)80781-2. [DOI] [PubMed] [Google Scholar]
- 143.Pinhal MAS, Smith B, Olson S, Aikawa J, Kimata K, Esko JD. Enzyme interactions in heparan sulfate biosynthesis: uronosyl 5-epimerase and 2-O-sulfotransferase interact in vivo. Proc Natl Acad Sci USA 98: 12984–12989, 2001. doi: 10.1073/pnas.241175798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Presto J, Thuveson M, Carlsson P, Busse M, Wilen M, Eriksson I, Kusche-Gullberg M, Kjellen L. Heparan sulfate biosynthesis enzymes EXT1 and EXT2 affect NDST1 expression and heparan sulfate sulfation. Proc Natl Acad Sci USA 105: 4751–4756, 2008. doi: 10.1073/pnas.0705807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen RL, Lander AD. Mechanisms underlying preferential assembly of heparan sulfate on glypican-1. J Biol Chem 276: 7507–7517, 2001. doi: 10.1074/jbc.M008283200. [DOI] [PubMed] [Google Scholar]
- 146.Doege K, Chen XC, Cornuet PK, Hassell J. Non-glycosaminoglycan bearing domains of perlecan and aggrecan influence the utilization of sites for heparan and chondroitin sulfate synthesis. Matrix Biol 16: 211–221, 1997. doi: 10.1016/S0945-053X(97)90010-X. [DOI] [PubMed] [Google Scholar]
- 147.Kokenyesi R, Bernfield M. Core protein structure and sequence determine the site and presence of heparan sulfate and chondroitin sulfate on syndecan-1. J Biol Chem 269: 12304–12309, 1994. [PubMed] [Google Scholar]
- 148.Corti F, Wang Y, Rhodes JM, Atri D, Archer-Hartmann S, Zhang J, Zhuang ZW, Chen D, Wang T, Wang Z, Azadi P, Simons M. N-terminal syndecan-2 domain selectively enhances 6-O heparan sulfate chains sulfation and promotes VEGFA165-dependent neovascularization. Nat Commun 10: 1562, 2019. [Erratum in Nat Commun 10: 2124, 2019]. doi: 10.1038/s41467-019-09605-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Noborn F, Gomez Toledo A, Green A, Nasir W, Sihlbom C, Nilsson J, Larson G. Site-specific identification of heparan and chondroitin sulfate glycosaminoglycans in hybrid proteoglycans. Sci Rep 6: 34537, 2016. doi: 10.1038/srep34537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Klein JA, Meng L, Zaia J. Deep sequencing of complex proteoglycans: a novel strategy for high coverage and site-specific identification of glycosaminoglycan-linked peptides. Mol Cell Proteomics 17: 1578–1590, 2018. doi: 10.1074/mcp.RA118.000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wen J, Xiao J, Rahdar M, Choudhury BP, Cui J, Taylor GS, Esko JD, Dixon JE. Xylose phosphorylation functions as a molecular switch to regulate proteoglycan biosynthesis. Proc Natl Acad Sci USA 111: 15723–15728, 2014. doi: 10.1073/pnas.1417993111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tone Y, Pedersen LC, Yamamoto T, Izumikawa T, Kitagawa H, Nishihara J, Tamura J, Negishi M, Sugahara K. 2-O-phosphorylation of xylose and 6-o-sulfation of galactose in the protein linkage region of glycosaminoglycans influence the glucuronyltransferase-I activity involved in the linkage region synthesis. J Biol Chem 283: 16801–16807, 2008. doi: 10.1074/jbc.M709556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hashiguchi T, Mizumoto S, Nishimura Y, Tamura J, Yamada S, Sugahara K. Involvement of human natural killer-1 (HNK-1) sulfotransferase in the biosynthesis of the GlcUA(3-O-sulfate)-Gal-Gal-Xyl tetrasaccharide found in {α}-thrombomodulin from human urine. J Biol Chem 286: 33003–33011, 2011. doi: 10.1074/jbc.M111.279174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kaidonis X, Liaw WC, Roberts AD, Ly M, Anson D, Byers S. Gene silencing of EXTL2 and EXTL3 as a substrate deprivation therapy for heparan sulphate storing mucopolysaccharidoses. Eur J Hum Genet 18: 194–199, 2010. doi: 10.1038/ejhg.2009.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Aikawa J, Grobe K, Tsujimoto M, Esko JD. Multiple isozymes of heparan sulfate/heparin GlcNAc N-deacetylase/N-sulfotransferase: structure and activity of the fourth member, NDST4. J Biol Chem 276: 5876–5882, 2001. doi: 10.1074/jbc.M009606200. [DOI] [PubMed] [Google Scholar]
- 156.Deligny A, Dierker T, Dagalv A, Lundequist A, Eriksson I, Nairn AV, Moremen KW, Merry CL, Kjellen L. NDST2 (N-deacetylase/N-sulfotransferase-2) enzyme regulates heparan sulfate chain length. J Biol Chem 291: 18600–18607, 2016. doi: 10.1074/jbc.M116.744433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Izumikawa T, Okuura Y, Koike T, Sakoda N, Kitagawa H. Chondroitin 4-O-sulfotransferase-1 regulates the chain length of chondroitin sulfate in co-operation with chondroitin N-acetylgalactosaminyltransferase-2. Biochem J 434: 321–331, 2011. doi: 10.1042/BJ20101456. [DOI] [PubMed] [Google Scholar]
- 158.Mizumoto S, Yamada S. Congenital disorders of deficiency in glycosaminoglycan biosynthesis. Front Genet 12: 717535, 2021. doi: 10.3389/fgene.2021.717535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cheung PK, McCormick C, Crawford BE, Esko JD, Tufaro F, Duncan G. Etiological point mutations in the hereditary multiple exostoses gene EXT1: a functional analysis of heparan sulfate polymerase activity. Am J Hum Genet 69: 55–66, 2001. doi: 10.1086/321278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Faiyaz-Ul-Haque M, Ahmad W, Zaidi S, Hussain S, Haque S, Ahmad M, Cohn D, Tsui LC. Novel mutations in the EXT1 gene in two consanguineous families affected with multiple hereditary exostoses (familial osteochondromatosis). Clin Genet 66: 144–151, 2004. doi: 10.1111/j.1399-0004.2004.00275.x. [DOI] [PubMed] [Google Scholar]
- 161.D'Arienzo A, Andreani L, Sacchetti F, Colangeli S, Capanna R. Hereditary multiple exostoses: current insights. Orthop Res Rev 11: 199–211, 2019. doi: 10.2147/ORR.S183979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pacifici M. The pathogenic roles of heparan sulfate deficiency in hereditary multiple exostoses. Matrix Biol 71–72: 28–39, 2018. doi: 10.1016/j.matbio.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Malfait F, Castori M, Francomano CA, Giunta C, Kosho T, Byers PH. The Ehlers–Danlos syndromes. Nat Rev Dis Primers 6: 64, 2020. doi: 10.1038/s41572-020-0194-9. [DOI] [PubMed] [Google Scholar]
- 164.Syx D, Van Damme T, Symoens S, Maiburg MC, van de Laar I, Morton J, Suri M, Del Campo M, Hausser I, Hermanns-Lê T, De Paepe A, Malfait F. Genetic heterogeneity and clinical variability in musculocontractural Ehlers-Danlos syndrome caused by impaired dermatan sulfate biosynthesis. Hum Mutat 36: 535–547, 2015. doi: 10.1002/humu.22774. [DOI] [PubMed] [Google Scholar]
- 165.Miyake N, Kosho T, Mizumoto S, Furuichi T, Hatamochi A, Nagashima Y, Arai E, Takahashi K, Kawamura R, Wakui K, Takahashi J, Kato H, Yasui H, Ishida T, Ohashi H, Nishimura G, Shiina M, Saitsu H, Tsurusaki Y, Doi H, Fukushima Y, Ikegawa S, Yamada S, Sugahara K, Matsumoto N. Loss-of-function mutations of CHST14 in a new type of Ehlers-Danlos syndrome. Hum Mutat 31: 966–974, 2010. doi: 10.1002/humu.21300. [DOI] [PubMed] [Google Scholar]
- 166.Alavi Naini SM, Soussi-Yanicostas N. Heparan sulfate as a therapeutic target in tauopathies: insights from zebrafish. Front Cell Dev Biol 6: 163, 2018. doi: 10.3389/fcell.2018.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Naini SMA, Yanicostas C, Hassan-Abdi R, Blondeel S, Bennis M, Weiss RJ, Tor Y, Esko JD, Soussi-Yanicostas N. Surfen and oxalyl surfen decrease tau hyperphosphorylation and mitigate neuron deficits in vivo in a zebrafish model of tauopathy. Transl Neurodegener 7: 6, 2018. [Erratum in Transl Neurodegener 9: 45, 2020]. doi: 10.1186/s40035-018-0111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lin J-Z, Duan M-R, Lin N, Zhao W-J. The emerging role of the chondroitin sulfate proteoglycan family in neurodegenerative diseases. Rev Neurosci 32: 737–750, 2021. doi: 10.1515/revneuro-2020-0146. [DOI] [PubMed] [Google Scholar]
- 169.Mehra S, Ghosh D, Kumar R, Mondal M, Gadhe LG, Das S, Anoop A, Jha NN, Jacob RS, Chatterjee D, Ray S, Singh N, Kumar A, Maji SK. Glycosaminoglycans have variable effects on alpha-synuclein aggregation and differentially affect the activities of the resulting amyloid fibrils. J Biol Chem 293: 12975–12991, 2018. doi: 10.1074/jbc.RA118.004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cohlberg JA, Li J, Uversky VN, Fink AL. Heparin and other glycosaminoglycans stimulate the formation of amyloid fibrils from alpha-synuclein in vitro. Biochemistry 41: 1502–1511, 2002. doi: 10.1021/bi011711s. [DOI] [PubMed] [Google Scholar]
- 171.Dyck SM, Karimi-Abdolrezaee S. Role of chondroitin sulfate proteoglycan signaling in regulating neuroinflammation following spinal cord injury. Neural Regen Res 13: 2080–2082, 2018. doi: 10.4103/1673-5374.241452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Maiza A, Chantepie S, Vera C, Fifre A, Huynh MB, Stettler O, Ouidja MO, Papy-Garcia D. The role of heparan sulfates in protein aggregation and their potential impact on neurodegeneration. FEBS Lett 592: 3806–3818, 2018. doi: 10.1002/1873-3468.13082. [DOI] [PubMed] [Google Scholar]
- 173.Goedert M, Jakes R, Spillantini MG, Hasegawa M, Smith MJ, Crowther RA. Assembly of microtubule-associated protein tau into Alzheimer-like filaments induced by sulphated glycosaminoglycans. Nature 383: 550–553, 1996. doi: 10.1038/383550a0. [DOI] [PubMed] [Google Scholar]
- 174.Zheng-Fischhöfer Q, Biernat J, Mandelkow EM, Illenberger S, Godemann R, Mandelkow E. Sequential phosphorylation of Tau by glycogen synthase kinase-3β and protein kinase A at Thr212 and Ser214 generates the Alzheimer-specific epitope of antibody AT100 and requires a paired-helical-filament-like conformation. Eur J Biochem 252: 542–552, 1998. doi: 10.1046/j.1432-1327.1998.2520542.x. [DOI] [PubMed] [Google Scholar]
- 175.Paudel HK, Li W. Heparin-induced conformational change in microtubule-associated protein Tau as detected by chemical cross-linking and phosphopeptide mapping. J Biol Chem 274: 8029–8038, 1999. doi: 10.1074/jbc.274.12.8029. [DOI] [PubMed] [Google Scholar]
- 176.Zhao J, Zhu Y, Song X, Xiao Y, Su G, Liu X, Wang Z, Xu Y, Liu J, Eliezer D, Ramlall TF, Lippens G, Gibson J, Zhang F, Linhardt RJ, Wang L, Wang C. 3-O-Sulfation of heparan sulfate enhances tau interaction and cellular uptake. Angew Chem Int Ed Engl 59: 1818–1827, 2020. doi: 10.1002/anie.201913029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rauch JN, Chen JJ, Sorum AW, Miller GM, Sharf T, See SK, Hsieh-Wilson LC, Kampmann M, Kosik KS. Tau internalization is regulated by 6-O sulfation on heparan sulfate proteoglycans (HSPGs). Sci Rep 8: 6382, 2018. doi: 10.1038/s41598-018-24904-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Willenborg DO, Parish CR. Inhibition of allergic encephalomyelitis in rats by treatment with sulfated polysaccharides. J Immunol 140: 3401–3405, 1988. [PubMed] [Google Scholar]
- 179.Bitan M, Weiss L, Reibstein I, Zeira M, Fellig Y, Slavin S, Zcharia E, Nagler A, Vlodavsky I. Heparanase upregulates Th2 cytokines, ameliorating experimental autoimmune encephalitis. Mol Immunol 47: 1890–1898, 2010. doi: 10.1016/j.molimm.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hussein RK, Mencio CP, Katagiri Y, Brake AM, Geller HM. Role of chondroitin sulfation following spinal cord injury. Front Cell Neurosci 14: 208, 2020. doi: 10.3389/fncel.2020.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hryciw T, Geremia NM, Walker MA, Xu X, Brown A. Anti-chondroitin sulfate proteoglycan strategies in spinal cord injury: temporal and spatial considerations explain the balance between neuroplasticity and neuroprotection. J Neurotrauma 35: 1958–1969, 2018. doi: 10.1089/neu.2018.5928. [DOI] [Google Scholar]
- 182.Sobel RA, Ahmed AS. White matter extracellular matrix chondroitin sulfate/dermatan sulfate proteoglycans in multiple sclerosis. J Neuropathol Exp Neurol 60: 1198–1207, 2001. doi: 10.1093/jnen/60.12.1198. [DOI] [PubMed] [Google Scholar]
- 183.Stephenson EL, Zhang P, Ghorbani S, Wang A, Gu J, Keough MB, Rawji KS, Silva C, Yong VW, Ling CC. Targeting the chondroitin sulfate proteoglycans: evaluating fluorinated glucosamines and xylosides in screens pertinent to multiple sclerosis. ACS Cent Sci 5: 1223–1234, 2019. doi: 10.1021/acscentsci.9b00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Stephenson EL, Mishra MK, Moussienko D, Laflamme N, Rivest S, Ling CC, Yong VW. Chondroitin sulfate proteoglycans as novel drivers of leucocyte infiltration in multiple sclerosis. Brain 141: 1094–1110, 2018. doi: 10.1093/brain/awy033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Crijns H, Vanheule V, Proost P. Targeting chemokine-glycosaminoglycan interactions to inhibit inflammation. Front Immunol 11: 483, 2020. doi: 10.3389/fimmu.2020.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Massena S, Christoffersson G, Hjertstrom E, Zcharia E, Vlodavsky I, Ausmees N, Rolny C, Li JP, Phillipson M. A chemotactic gradient sequestered on endothelial heparan sulfate induces directional intraluminal crawling of neutrophils. Blood 116: 1924–1931, 2010. doi: 10.1182/blood-2010-01-266072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Jin L, Abrahams JP, Skinner R, Petitou M, Pike RN, Carrell RW. The anticoagulant activation of antithrombin by heparin. Proc Natl Acad Sci USA 94: 14683–14688, 1997. doi: 10.1073/pnas.94.26.14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Arepally GM, Kamei S, Park KS, Kamei K, Li ZQ, Liu W, Siegel DL, Kisiel W, Cines DB, Poncz M. Characterization of a murine monoclonal antibody that mimics heparin-induced thrombocytopenia antibodies. Blood 95: 1533–1540, 2000. [PubMed] [Google Scholar]
- 189.Stringer SE, Gallagher JT. Specific binding of the chemokine platelet factor 4 to heparan sulfate. J Biol Chem 272: 20508–20514, 1997. doi: 10.1074/jbc.272.33.20508. [DOI] [PubMed] [Google Scholar]
- 190.Pempe EH, Burch TC, Law CJ, Liu J. Substrate specificity of 6-O-endosulfatase (Sulf-2) and its implications in synthesizing anticoagulant heparan sulfate. Glycobiology 22: 1353–1362, 2012. doi: 10.1093/glycob/cws092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Marques C, Reis CA, Vivès RR, Magalhães A. Heparan sulfate biosynthesis and sulfation profiles as modulators of cancer signalling and progression. Front Oncol 11: 778752, 2021. doi: 10.3389/fonc.2021.778752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ambrosius M, Kleesiek K, Gotting C. Quantitative determination and comparison of the glycosaminoglycan delta-disaccharide composition in 22 different human cell lines. Cell Biol Int 33: 848–852, 2009. doi: 10.1016/j.cellbi.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 193.Lundin L, Larsson H, Kreuger J, Kanda S, Lindahl U, Salmivirta M, Claesson-Welsh L. Selectively desulfated heparin inhibits fibroblast growth factor-induced mitogenicity and angiogenesis. J Biol Chem 275: 24653–24660, 2000. doi: 10.1074/jbc.M908930199. [DOI] [PubMed] [Google Scholar]
- 194.Maeda T, Desouky J, Friedl A. Syndecan-1 expression by stromal fibroblasts promotes breast carcinoma growth in vivo and stimulates tumor angiogenesis. Oncogene 25: 1408–1412, 2006. doi: 10.1038/sj.onc.1209168. [DOI] [PubMed] [Google Scholar]
- 195.Lim HC, Multhaupt HA, Couchman JR. Cell surface heparan sulfate proteoglycans control adhesion and invasion of breast carcinoma cells. Mol Cancer 14: 15, 2015. doi: 10.1186/s12943-014-0279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Levy-Adam F, Feld S, Suss-Toby E, Vlodavsky I, Ilan N. Heparanase facilitates cell adhesion and spreading by clustering of cell surface heparan sulfate proteoglycans. PLoS One 3: e2319, 2008. doi: 10.1371/journal.pone.0002319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Morla S. Glycosaminoglycans and glycosaminoglycan mimetics in cancer and inflammation. Int J Mol Sci 20: 1963, 2019. doi: 10.3390/ijms20081963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lai J-P, Sandhu DS, Shire AM, Roberts LR. The tumor suppressor function of human sulfatase 1 (SULF1) in carcinogenesis. J Gastrointest Cancer 39: 149–158, 2008. doi: 10.1007/s12029-009-9058-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Elgundi Z, Papanicolaou M, Major G, Cox TR, Melrose J, Whitelock JM, Farrugia BL. Cancer metastasis: the role of the extracellular matrix and the heparan sulfate proteoglycan perlecan. Front Oncol 9: 1482, 2019. doi: 10.3389/fonc.2019.01482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Mikami T, Kitagawa H. Biosynthesis and function of chondroitin sulfate. Biochim Biophys Acta 1830: 4719–4733, 2013. doi: 10.1016/j.bbagen.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 201.Li F, Ten Dam GB, Murugan S, Yamada S, Hashiguchi T, Mizumoto S, Oguri K, Okayama M, van Kuppevelt TH, Sugahara K. Involvement of highly sulfated chondroitin sulfate in the metastasis of the lewis lung carcinoma cells. J Biol Chem 283: 34294–34304, 2008. doi: 10.1074/jbc.M806015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Basappa Rangappa KS, Sugahara K. Roles of glycosaminoglycans and glycanmimetics in tumor progression and metastasis. Glycoconj J 31: 461–467, 2014. doi: 10.1007/s10719-014-9551-9. [DOI] [PubMed] [Google Scholar]
- 203.ten Dam GB, van de Westerlo EM, Purushothaman A, Stan RV, Bulten J, Sweep FC, Massuger LF, Sugahara K, van Kuppevelt TH. Antibody GD3G7 selected against embryonic glycosaminoglycans defines chondroitin sulfate-E domains highly up-regulated in ovarian cancer and involved in vascular endothelial growth factor binding. Am J Pathol 171: 1324–1333, 2007. doi: 10.2353/ajpath.2007.070111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Willis ND, Cox TR, Rahman-Casañs SF, Smits K, Przyborski SA, van den Brandt P, van Engeland M, Weijenberg M, Wilson RG, de Bruïne A, Hutchison CJ. Lamin A/C is a risk biomarker in colorectal cancer. PLoS One 3: e2988, 2008. doi: 10.1371/journal.pone.0002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Skandalis SS, Kletsas D, Kyriakopoulou D, Stavropoulos M, Theocharis DA. The greatly increased amounts of accumulated versican and decorin with specific post-translational modifications may be closely associated with the malignant phenotype of pancreatic cancer. Biochim Biophys Acta 1760: 1217–1225, 2006. doi: 10.1016/j.bbagen.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 206.Persson A, Tykesson E, Westergren-Thorsson G, Malmstrom A, Ellervik U, Mani K. Xyloside-primed chondroitin sulfate/dermatan sulfate from breast carcinoma cells with a defined disaccharide composition has cytotoxic effects in vitro. J Biol Chem 291: 14871–14882, 2016. doi: 10.1074/jbc.M116.716829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Soares PA, Queiroz IN, Pomin VH. NMR structural biology of sulfated glycans. J Biomol Struct 35: 1069–1084, 2017. doi: 10.1080/07391102.2016.1171165. [DOI] [PubMed] [Google Scholar]
- 208.Ly M, Leach FE 3rd, Laremore TN, Toida T, Amster IJ, Linhardt RJ. The proteoglycan bikunin has a defined sequence. Nat Chem Biol 7: 827–833, 2011. doi: 10.1038/nchembio.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Yu Y, Duan J, Leach FE III, Toida T, Higashi K, Zhang H, Zhang F, Amster IJ, Linhardt RJ. Sequencing the dermatan sulfate chain of decorin. J Am Chem Soc 139: 16986–16995, 2017. doi: 10.1021/jacs.7b10164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Miller RL, Guimond SE, Schworer R, Zubkova OV, Tyler PC, Xu Y, Liu J, Chopra P, Boons GJ, Grabarics M, Manz C, Hofmann J, Karlsson NG, Turnbull JE, Struwe WB, Pagel K. Shotgun ion mobility mass spectrometry sequencing of heparan sulfate saccharides. Nat Commun 11: 1481, 2020. doi: 10.1038/s41467-020-15284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Xia K, Hagan JT, Fu L, Sheetz BS, Bhattacharya S, Zhang F, Dwyer JR, Linhardt RJ. Synthetic heparan sulfate standards and machine learning facilitate the development of solid-state nanopore analysis. Proc Natl Acad Sci USA 118: e2022806118, 2021. doi: 10.1073/pnas.2022806118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Im J, Lindsay S, Wang X, Zhang P. Single molecule identification and quantification of glycosaminoglycans using solid-state nanopores. ACS Nano 13: 6308–6318, 2019. doi: 10.1021/acsnano.9b00618. [DOI] [PubMed] [Google Scholar]
- 213.Crosetto N, Bienko M, van Oudenaarden A. Spatially resolved transcriptomics and beyond. Nat Rev Genet 16: 57–66, 2015. doi: 10.1038/nrg3832. [DOI] [PubMed] [Google Scholar]
- 214.Kearney CJ, Vervoort SJ, Ramsbottom KM, Todorovski I, Lelliott EJ, Zethoven M, Pijpers L, Martin BP, Semple T, Martelotto L, Trapani JA, Parish IA, Scott NE, Oliaro J, Johnstone RW. SUGAR-seq enables simultaneous detection of glycans, epitopes, and the transcriptome in single cells. Sci Adv 7: eabe3610, 2021. doi: 10.1126/sciadv.abe3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Minoshima F, Ozaki H, Odaka H, Tateno H. Integrated analysis of glycan and RNA in single cells. iScience 24: 102882, 2021. doi: 10.1016/j.isci.2021.102882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kunzke T, Balluff B, Feuchtinger A, Buck A, Langer R, Luber B, Lordick F, Zitzelsberger H, Aichler M, Walch A. Native glycan fragments detected by MALDI-FT-ICR mass spectrometry imaging impact gastric cancer biology and patient outcome. Oncotarget 8: 68012–68025, 2017. doi: 10.18632/oncotarget.19137. [DOI] [PMC free article] [PubMed] [Google Scholar]