Abstract
We report a novel phenotype of methicillin resistance, designated “Eagle-type” resistance, which is characteristic in its resistance to high concentrations of methicillin (64 to 512 μg/ml) and susceptibility to low concentrations of methicillin (2 to 16 μg/ml). The type of resistance was expressed in mutant strains selected with high concentrations (e.g., 128 to 512 μg/ml) of methicillin from the pre-methicillin-resistant Staphylococcus aureus strain N315, whose mecA gene transcription is strongly repressed by the mecI gene-encoded repressor protein MecI. The Eagle-type mutant strains harbored no mutation in the mecI gene or in the operator region of mecA gene to which MecI repressor is supposed to bind. In the representative Eagle-type strain h4, repression of mecA gene transcription and penicillin-binding protein 2′ production were found to be released by exposing the cells to a high concentration (128 μg/ml) of methicillin but not to lower concentrations (1 and 8 μg/ml) of methicillin. The strain h4 expressed paradoxical susceptibility (Eagle effect) to the cytokilling activity of methicillin. Experimental deletion of mecI gene from the chromosome of h4 by mecI-specific gene substitution converted its Eagle-type resistance to homogeneously high methicillin resistance. We cloned two novel genes, designated hmrA and hmrB, from genomic library of h4, which conferred Eagle-type resistance to N315 when introduced into the cell in multiple copies. The genes were shown to confer homogeneous methicillin resistance to the heterogeneously methicillin-resistant strain LR5 when they were introduced into on multicopy plasmids. This result strongly indicated that the genetic alteration responsible for the expression of the Eagle phenotype is identical, or equivalent in its effect, to the genetic alteration underlying heterogeneous-to-homogeneous conversion of methicillin resistance in S. aureus.
Beta-lactam resistance of methicillin-resistant Staphylococcus aureus (MRSA) is caused by the expression of penicillin-binding protein 2′ (PBP2′ or PBP2a) encoded by the mecA gene (28), which has low binding affinities to practically all beta-lactam antibiotics so far introduced into clinical use (11, 21, 32). Because of the low binding affinities, PBP2′ is not inhibited by beta-lactam antibiotics and continues synthesizing cell wall in the presence of beta-lactam antibiotics. However, it has been pointed out that production of PBP2′ in Staphylococcus aureus cells per se does not make the entire cell population resistant to methicillin but, instead makes the strain a mixture of cells with different levels of methicillin resistance (23). This peculiar resistance phenotype has long been recognized as heterogeneous (hetero)-methicillin resistance (19, 24). This inability of PBP2′ to confer full or homogeneous (homo) methicillin resistance has also been demonstrated using pre-MRSA strain N315 (12, 17). Pre-MRSA is an S. aureus having a susceptible level of methicillin MIC (<4 mg/liter) because of a strong repression of the mecA gene transcription exerted by mecI gene-encoded repressor protein MecI (13, 17, 30). Inactivation of MecI function in pre-MRSA strain by mecI-specific gene substitution derepresses PBP2′ production but causes the strain express only a hetero type of methicillin resistance (17). It is only after further selection with a high concentration of methicillin (e.g., 128 μg/ml) that the homoresistant strain, whose entire cell population is uniformly resistant to high (126 to 512 μg/ml) concentrations of methicillin, is generated (12, 17). Ryffel et al. have also shown that, besides PBP2′ production, a chromosomal mutation, designated chr∗, is required for the hetero-to-homo phenotypic conversion to occur (23). So far, no phenotypic expression intrinsic to chr∗ mutation (i.e., in the absence of coexpression of PBP2′) has been identified (23). We report here that a mutant strain of pre-MRSA N315 with a novel methicillin resistance phenotype, designated Eagle-type resistance, is considered to manifest the effect of chr∗-equivalent genetic alteration under mecI-mediated repression of PBP2′ production.
MATERIALS AND METHODS
Bacterial strains.
Table 1 lists the bacterial strains used in this study. All of the mutant strains with various methicillin-resistance phenotypes were obtained from N315 and its derivative strains by a one-step selection procedure with methicillin as follows. Portions of overnight culture of N315 and its derivative strains were spread onto heart infusion agar plates containing various concentrations (4 to 512 μg/ml) of methicillin, and the plates were incubated overnight at 37°C. The grown colonies were picked as resistant mutants from each of the selective concentrations of methicillin. The mutants were then streaked onto drug-free heart infusion agar plates, and single-colony isolation (colony purification) was performed before establishing them as strains. A mutant strain, h4, is one of the Eagle-type resistant strains obtained as described above by selecting N315 with 128 μg of methicillin per ml. Strains ΔI and h4ΔI are derivatives of N315 and h4, respectively; they were obtained by substituting their mecI genes with tetL by using the gene substitution procedure described previously (17). Mutant strain ΔI-HR expressing methicillin homoresistance was obtained by one-step selection of ΔI with 128 μg of methicillin per ml. Mutant strain h4-4R is a methicillin homoresistant strain obtained by one-step selection of h4 with 4 μg of methicillin per ml. Mutant strain LR5, in which mecI gene function is inactivated by a 62-bp deletion (from nucleotide positions 130 to 191 of the mecI structure gene [13]), is a derivative of N315 expressing hetero-type methicillin resistance as described previously (1).
TABLE 1.
Bacterial strains used in this study
| Strain | Description and relevant genotypea | Phenotype of methicillin resistanceb | Source or reference |
|---|---|---|---|
| N315 | Japanese clinical isolate of 1982 with a functional mecI gene (mecI+) | Pre-MRSA | 13 |
| h4 | Mutant derived from N315 by selection with 128 mg of methicillin per liter | Eagle-type | This work |
| h4ΔI | Derived from h4 by mecI substitution (mecI::tetL) | Homo-type | This work |
| h4-4R | Mutant derived from h4 by selection with 4 mg of methicillin per liter | Homo-type | This work |
| ΔI | Derived from N315 by mecI substitution (mecI::tetL) | Hetero-type | 17 |
| ΔI-HR | Mutant derived from ΔI by selection with 128 mg of methicillin per liter (mecI::tetL chr∗) | Homo-type | This work |
| LR5 | Mutant derived from N315 by selection with 4 mg of methicillin per liter (ΔmecI) | Hetero-type | 1 |
| N315(pYT3) | Transformant of N315 with vector plasmid pYT3 | Pre-MRSA | This work |
| N315(pHMR-A) | Transformant of N315 with plasmid pHMR-A (multiple hmrA gene copies) | Eagle-type | This work |
| N315(pHMR-B) | Transformant of N315 with plasmid pHMR-B (multiple hmrB gene copies) | Eagle-type | This work |
| LR5(pYT3) | Transformant of LR5 with vector plasmid pYT3 | Hetero-type | This work |
| LR5(pHMR-A) | Transformant of LR5 with plasmid pHMR-A (multiple hmrA copies) | Homo-type | This work |
| LR5(pHMR-B) | Transformant of LR5 with plasmid pHMR-B (multiple hmrB copies) | Hetero-typec | This work |
Strains obtained by methicillin selection are denoted as “mutants.”
Determined by population analysis.
Although classified as hetero-type, the resistance level was higher than that of LR5 and was lower than the typical homo-type resistance (see Fig. 1D and E).
Plasmids.
Escherichia coli-S. aureus shuttle vector pYT3 was used to construct a genomic library of strain h4. The plasmid carries a tetracycline-resistant gene (tetL) and the temperature-sensitive origin of pE194ts and has been described previously (9, 16). Another shuttle vector, pRIT5, was used for the subcloning of cloned DNA fragments (20).
Two recombinant plasmids, pHMR-A and pHMR-B, were obtained by screening the genomic library of strain h4; they had 2,063- and 2,100-base Sau3AI fragments inserted at the BamHI site of pYT3 vector, respectively. The 1.3-kb HindIII-Sau3AI fragment of the insert was removed from pHMR-A by restriction enzyme cleavage, followed by self-ligation, to obtain plasmid pHMR-A1 having the rest of the insert (a 757-bp Sau3AI-HindIII fragment). Plasmid pHMR-A2 was constructed by subcloning the 1.3-kb HindIII-Sau3AI fragment cut out from pHMR-A into the multicloning site of plasmid vector pRIT5 in the same orientation of transcription with spa gene on the vector (20). Plasmid pHMR-A3 was constructed by cloning a PCR-amplified DNA fragment into vector pYT3 as follows. A PCR reaction was performed with pHMR-A plasmid DNA as a template and using two synthetic oligonucleotide primers: primer 1, 5′-AAAAGAGCTCAAAACCCGGGCTTATGTTTACAATTTGA-3′ (the introduced SacI site is underlined), and primer 2, 5′-TTTTTGGTACCTTGATGTTCGTCCGGTTTCA-3′ (the introduced KpnI site is underlined). Amplified DNA was doubly digested with SacI and KpnI and subjected to agarose gel electrophoresis. The restricted DNA fragment was purified from the agarose gel by electroelution and was ligated into the pYT3 vector predigested with SacI and KpnI.
Plasmid pHMR-A4 was constructed by cutting out the 850-base EcoRI DNA fragment from pHMR-A plasmid DNA and then ligating it into the EcoRI site of vector pRIT5 in the same orientation of transcription with the spa gene on the vector. Plasmids pHMR-A5 and pHMR-A5∗ were constructed as follows. The DNA fragment containing the hmrA gene was amplified by PCR with h4 genomic DNA as a template and the following two synthetic oligonucleotides as primers: primer 3, 5′-AAAAGGATCCGATCTAAACTTTCAQCATTCATTT-3′ and 5′-AAAAGGATCCGAATTCCCAATTCTTAATCTCGAA-3′ (the introduced BamHI sites are underlined). The amplified DNA was cut with BamHI and ligated into the BamHI site of pYT3, generating plasmid pHNR-A5. Plasmid pHMR-A5 was digested with NdeI (the NdeI site is located within hmrA gene at bases 201 to 206 from the start codon). The cohesive ends of the DNA were filled in by Klenow fragment, followed by blunt-end ligation. The plasmid pHMR-A5∗ thus generated possessed two additional bases in the hmrA gene, causing a shift of the reading frame and generating a premature termination codon at base 632 from the starting codon.
Plasmids pHMR-B1 to pHMR-B4 were constructed by cutting out the respective restriction fragments from pHMR-B and subcloning them into pYT3 vector; the inserts were Sau3AI-TthI fragment (pHMR-B1), SalI-Sau3AI fragment (pHMR-B2), TthI-Sau3AI fragment (pHMR-B3), and TthI-SalI fragment (pHMR-B4), respectively.
Screening of h4 gene library.
Genomic DNA was extracted from h4 as described previously (13), partially digested with Sau3AI, and ligated with the BamHI-cleaved dephosphorylated pYT3 vector. The plasmid library was established by transforming E. coli JM109 (Takara, Tokyo, Japan) with the recombinant plasmids and by selecting transformants on the heart infusion agar plates containing 10 μg of tetracycline per ml at 37°C. A total of 2,871 transformants were obtained. Recombinant plasmids were then isolated from the transformants, purified by CsCl isopycnic centrifugation, and introduced into N315 by electroporation as described previously (17). A total of 22,580 transformants were obtained by the selection on heart infusion agar plates containing 10 μg of tetracycline per ml at 30°C (a permissive temperature for the replication of pYT3 vector in S. aureus cells). The transformants were then replicated onto two heart infusion agar (Difco, Detroit, Mich.) plates containing 10 and 100 μg of methicillin per ml, respectively. The plates were then incubated at 30°C for 48 h. Transformants were sought that formed greater colonies on the plate containing 100 μg of methicillin per ml than those on the plate containing 10 μg of methicillin per ml (Eagle phenotype). Two transformants carrying plasmids pHMR-A and pHMR-B were thus obtained.
Nucleotide sequencing.
The nucleotide sequence of hmrA and hmrB genes were determined using AmpliTaq Cycle Sequencing Kit (Perkin-Elmer) based on the dideoxynucleotides termination method described by Sanger et al. (26) with synthetic oligonucleotides as primers. Sequences of the mecI genes and mecA gene operator regions of N315 and N315-derived mutants were determined by using the PCR-amplified DNA fragments as templates as described previously (17). The primers used were 5′-GACTTGATTGTTTCCTCTGTT-3′ and 5′-GACTTGATTGTTTCCTCTGTTT-3′ for mecI gene sequencing and 5′-TATACCAAACCCGACAAG-3′ and 5′-CATATCGTGAGCAATGAACTG-3′ for mecA operator region sequencing. The primers corresponded to the nucleotide positions 1923 to 2403, 2403 to 1983 (complementary), 66 to 83, and 287 to 257 (complementary), respectively, of the reported nucleotide sequence of mecI-mecR1-mecA gene complex of N315 (13).
Analysis of nucleotide sequences.
The homology search for the deduced polypeptides encoded by hmrA and hmrB genes was performed with the nucleotide sequences registered in the EMBL and GenBank databases and with the protein sequences of the SWISSPlot and NBRF data libraries. A BLAST search was performed to compare hmrA and hmrB gene sequences in public S. aureus genome databases as follows: strains COL (TIGR [http://www.tigr.org/tdb/mdb /mdbinprogress.html]), NCTC8325 (University of Oklahama [http://www.genome.ou.edu/]), and EMRSA-16 and MSSA strain 476 (The Sanger Centre [http://www.sanger.ac.uk/Projects/S_aureus]). Complete nucleotide sequences of hmrA and hmrB are available under the DDJB accession numbers D85816 and D85817, respectively.
Population analysis.
Analysis of resistant subpopulations was performed with an Autoplate Model 3000 (Spiral Biotech, Inc., Bethesda, Md.). as described previously (17). In brief, 50-μl aliquots of overnight culture and its 1:10 serial dilutions were inoculated onto heart infusion agar plates containing various concentrations of methicillin. The plates were incubated at 37°C for 48 h, and then mature colonies were counted. The number of resistant cells theoretically contained in 50 μl of the undiluted overnight culture was calculated and plotted on a bilogarithmic graph.
Western blotting.
The Western blotting to detect methicillin-induced PBP2′ production in N315-derived strains has been described previously (17). In this study, three different concentrations of methicillin (1, 8, and 128 μg/ml) were used. Densitometry was performed with Master Scan (CSPI, Billerica, Mass.).
Cytokilling assay.
Overnight cultures of test strains in L broth were diluted to 1% (vol/vol) in fresh L broth and further cultivated with gentle shaking until an optical density at 578 nm of 0.3 (ca. 108 CFU/ml) was reached. Then, 0.1 ml of the cell suspension was inoculated into 10 ml of prewarmed L broth containing 0, 1, 8, or 128 μg of methicillin per ml in glass test tubes. The test tubes were incubated with gentle shaking, and 0.1-ml portions of the culture were harvested at 0, 0.5, 1, 2, and 4 h after the initiation of the culture. The sample was serially diluted with saline and spread onto heart infusion agar plates, followed by incubation at 37°C for 18 h. Grown colonies were counted and plotted in the graph. The cultivation temperature of the test strains was 37°C for N315, h4, h4-4R, ΔI, and ΔI-HR and 30°C for the transformant strains N315(pYT3), N315(pHMR-A), and N315(pHMR-B).
RESULTS
Selection of Eagle-type methicillin-resistant strain h4 from pre-MRSA N315.
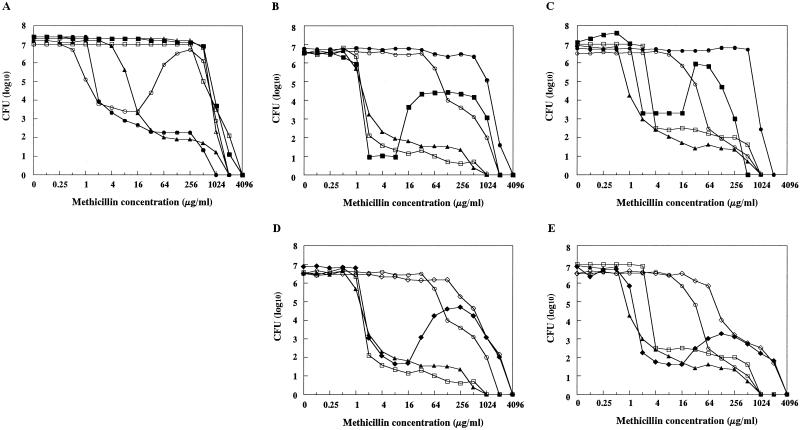
The in vitro mutant strain h4 was obtained from N315 as a methicillin-resistant strain grown on the heart infusion agar plate containing 128 μg of methicillin per ml. Figure 1A shows a population analysis of strain h4 in comparison with those of pre-MRSA strain N315 and its derivative strains with various phenotypes of methicillin resistance, ΔI (hetero-type), ΔI-HR (homo-type), h4-4R (homo-type), and h4-ΔI (homo-type). Strain h4 showed an unusual pattern of distribution of resistant cell subpopulation, i.e., more cells grew in high concentrations than in low concentrations of methicillin (Fig. 1A). Since the pattern reminded us of the “Eagle effect” of penicillin killing (7), we designated the resistance phenotype Eagle-type resistance after him. Nucleotide sequencing of the mecI gene and mecA operator region of h4 revealed no sequence alteration. This was in contrast to the derivatives of N315 with hetero-type methicillin resistance phenotypes, in which mutations are identified either in mecI gene or in the operator region of mecA gene (12, 17).
FIG. 1.
Population analysis of N315-derived strains with various methicillin-resistance phenotypes. The number of colonies grown on agar plates containing various concentrations of methicillin were counted after 48 h incubation at 37°C (A, C, and E) and at 30°C (B and D). (A) Comparison of N315 and its mutant and mecI-gene substituted strains. Symbols: ○, Eagle-type strain h4; ▵, homo-type strain h4ΔI; ▴, hetero-type strain ΔI; ■, homo-type strain ΔI-HR; ●, pre-MRSA strain N315; □, homo-type strain h4-4R. (B to E) Comparison of N315 and its transformants. Symbols: □, N315; ○, LR5; ▴, N315(pYT3); ■, N315(pHMR-A); ⧫, N315(pHMR-B); ●, LR5(pHMR-A); ◊, LR5(pHMR-B).
To see if the Eagle-type phenotype is stably maintained during passage, h4 was serially cultivated for 7 days in drug-free medium as well as in the medium containing 128 μg of methicillin per ml. In both cases, the cultures showed essentially the same population curve, and no mutation was detectable in either mecI gene or in the operator region of mecA gene. Thus, this phenotype was considered stable enough to be analyzed further with various experimental procedures.
There was a clear correlation between the appearance of mutants with Eagle-type resistance and the concentrations of methicillin used for the selection of the mutants. Table 2 shows that the mutants with Eagle-type resistance were predominantly obtained from N315 when selected with a concentration of methicillin of 32 μg/ml or greater. On the other hand, lower concentrations of methicillin (4 to 16 μg/ml) predominantly selected heteroresistant mutants. All of the five Eagle-type mutants obtained with each selective concentration of methicillin (32, 64, 128, 256, and 512 μg/ml; Table 2) retained intact mecI gene and mecA gene operator region. Table 2 also shows that no mutant with homo-type methicillin resistance was obtained in the selection procedures, indicating that the homoresistant mutants do not emerge within the cell population of N315 at a high frequency comparable to those with Eagle-type or hetero-type mutants: the appearance rates for the latter were in the range of 10−4 to 10−5.
TABLE 2.
Phenotypic expression of methicillin resistance of the mutants obtained by selecting N315 with various concentrations of methicillin
| Methicillin concn (μg/ml)a | No. of grown coloniesb | Appearance rate of resistant mutants (10−4) | No. of mutants with each resistance phenotypec
|
||
|---|---|---|---|---|---|
| Hetero | Eagle | Homo | |||
| 4 | 37 | 2.96 | 10 | 0 | 0 |
| 8 | 21 | 1.68 | 9 | 1 | 0 |
| 16 | 6 | 0.48 | 6 | 0 | 0 |
| 32 | 5 | 0.40 | 1 | 4 | 0 |
| 64 | 4 | 0.32 | 1 | 3 | 0 |
| 128 | 6 | 0.48 | 0 | 6 | 0 |
| 256 | 6 | 0.48 | 0 | 6 | 0 |
| 512 | 5 | 0.04 | 0 | 5 | 0 |
Methicillin concentration used for the selection of mutants.
A total of 1.25 × 105 CFU of N315 were inoculated onto the agar plate containing each concentration of methicillin except for that with 512 μg of methicillin per ml, onto which 1.25 × 106 CFU were inoculated.
Each grown colony was picked, colony isolated on a drug-free heart infusion agar plate, grown overnight in drug-free heart infusion broth, and tested for the phenotypic expression of methicillin resistance with population analysis. Ten colonies were arbitrarily picked from each of the plates with 4 and 8 μg of methicillin per ml. Hetero, hetero-type methicillin resistance; Eagle, Eagle-type methicillin resistance; Homo, homo-type methicillin resistance.
Conversion of Eagle-type resistance to homo methicillin resistance by mecI gene inactivation.
Eagle-type strain h4 was plated onto the heart infusion agar containing a low concentration of methicillin (4 μg/ml). After incubation overnight at 37°C, a number of colonies grew at a plating efficiency of 3.2 × 10−4. Ten such colonies were picked, grown in drug-free medium, and subjected to population analysis. All 10 strains had homo-type methicillin resistance. The population curve of representative strain h4-4R is shown in Fig. 1A. Nucleotide sequence determination of the mecI gene of strain h4-4R revealed a point mutation that caused an amino acid substitution from Glu74 to Gly in the MecI protein. This observation suggested that inactivation of MecI was responsible for the Eagle-type-to-homo-type phenotypic conversion. To prove this hypothesis, the mecI gene of h4 was substituted by the tetL gene (17). The resultant strain h4ΔI expressed homo-type resistance, as judged by population analysis (see Fig. 1A).
PBP2′ production in Eagle-type strain in response to various concentrations of methicillin.
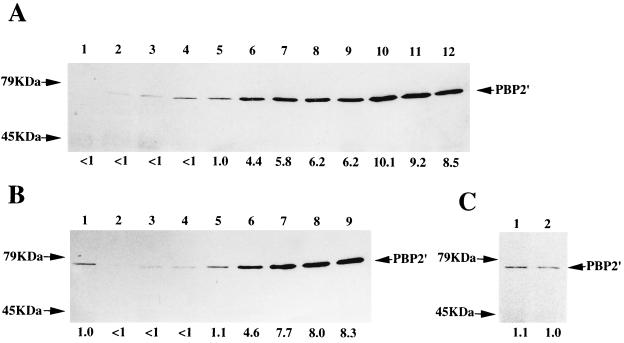
The methicillin-induced production of PBP2′ in the Eagle-type strain h4 was studied in comparison with those in other test strains with various types of methicillin resistance (Fig. 2). The intensity of the PBP2′ band of each lane was measured by densitometry and is shown under the lane as a value relative to that of ΔI without methicillin induction (Fig. 2A, lane 5). The amount of PBP2′ induced by methicillin was measured 1 h after exposure to 1, 8, or 128 μg of methicillin per ml, respectively. Because of the mecI gene-mediated repression of mecA gene transcription (17), N315 produced a very-low-level amount of PBP2′ before induction, which was scarcely visible in the assay (Fig. 2A, lane 1). Induction with 1 and 8 μg of methicillin per ml could not induce PBP2′ production appreciably (Fig. 2A, lanes 2 and 3).
FIG. 2.
PBP2′ production induced by various concentrations of methicillin. (A) Western blot of PBP2′ after 1 h of induction with methicillin. The strains are N315 (lanes 1 to 4), ΔI (lanes 5 to 8), and ΔI-HR (lanes 9 to 12). The methicillin concentrations used for induction were 0, 1, 8, and 128 μg/ml from left to right for each strain. (B) Same experiment as in panel A with strains h4 (lanes 2 to 5) and h4ΔI (lanes 6 to 9). Lane 1 is ΔI without induction (the same sample for the lane 5 of panel A). (C) Comparison of PBP2′ amount in strain h4 during growth in 128 μg of methicillin per ml (lane 1) and that after 1-h induction with 128 μg of methicillin per ml (lane 2). The values under the lanes are relative amounts of PBP2′ as determined by densitometry to that of strain ΔI without induction as a unit of comparison for panels A and B and to that of h4 for panel C.
In contrast, N315 was found to produce detectable amount of PBP2′ when 128 μg of methicillin per ml was used for the induction (Fig. 2A, lane 4). The amount of induced PBP2′ was comparable to that in ΔI without induction (lane 5), but the amount was only 12% of that produced in ΔI after induction with 128 μg of methicillin per ml (compare lanes 4 and 8). This induction profile of N315 greatly contrasted with those of strains ΔI (Fig. 2A, lanes 5 to 8) and ΔI-HR (Fig. 2A, lanes 9 to 12), in which production of PBP2′ was derepressed because of the absence of the mecI gene. The PBP2′ production in ΔI was still partially suppressed, presumably due to the presence of the blaI-blaRI regulatory complex carried by the penicillinase plasmid (8), and was increased by 4.4, 5.8, and 6.2 times upon induction with 1, 8, and 128 μg of methicillin per ml, respectively. Elimination of the penicillinase plasmid from ΔI made it a constitutive producer of PBP2′, whose PBP2′ amount was comparable to those in ΔI after methicillin induction (data not shown).
It was noticed that the amounts of PBP2′ produced in ΔI-HR, a methicillin-selected homoresistant derivative of ΔI, exceeded those of ΔI by 6.2 times before induction (compare lanes 5 and 9) and by 1.4- to 2.3 times upon methicillin induction (compare lanes 6 to 8 and 10 to 12). However, the “induction ratio” (defined by dividing the PBP2′ amount produced after induction by that before induction) of ΔI-HR (1.4 to 1.6) was smaller than that of ΔI (4.4 to 6.2).
PBP2′ production in h4 in response to methicillin was essentially the same as in N315 (Fig. 2B, lanes 2 to 5). PBP2′ production was strongly repressed both before and after induction with 1 or 8 μg of methicillin per ml. However, PBP2′ induction was evident after exposure to 128 μg of methicillin per ml, and an amount of PBP2′ comparable to that of N315 was produced (compare lane 5 in Fig. 2B with lane 4 in Fig. 2A). The amount, however, was still only 24% of that produced by the homoresistant strain h4ΔI before methicillin induction (Fig. 2B, lane 6). Strain h4ΔI had a pattern of PBP2′ induction (lanes 6 to 9) which was quite similar to that of ΔI-HR. PBP2′ production in h4ΔI was derepressed much further than in the heteroresistant strain δI, and, as in ΔI-HR, the “induction ratio” (1.7 to 1.8) was smaller than that in ΔI.
Figure 2C shows the comparison of PBP2′ amounts produced by the strain h4 after cultivation for 1 and 12 h in the heart infusion broth containing 128 μg of methicillin per ml. The amount of PBP2′ produced in h4 after prolonged culture in methicillin-containing medium was comparable to, and only slightly greater than, that produced after 1 h exposure to methicillin.
Paradoxical effect of methicillin in the cytokilling activity against Eagle-type strain h4.
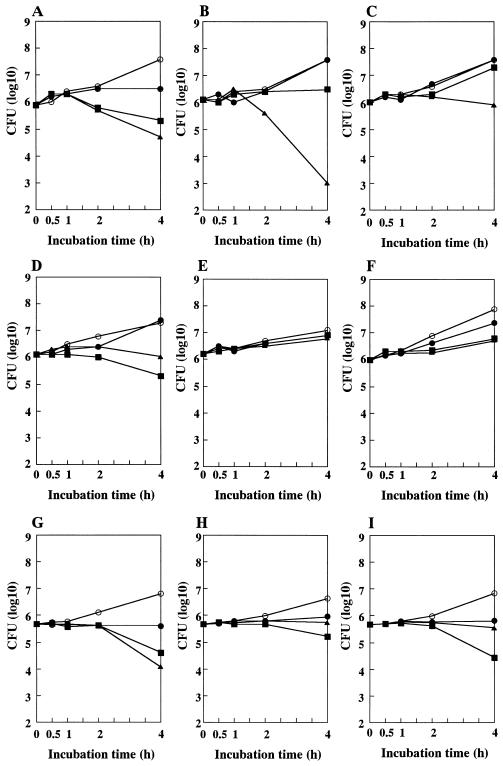
Figure 3 illustrates short-time killing curves of N315 and is derivative strains with various methicillin resistance phenotypes. N315 cells were killed by methicillin in a dose-dependent manner, i.e., the viable cell count decreased from 7.9 × 105 CFU at time zero to 1.9 × 105 CFU at 4 h in the presence of 8 μg of methicillin per ml and to 4.9 × 104 CFU at 4 h in the presence of 128 μg of methicillin per ml (Fig. 3A). The heteroresistant strain, ΔI, showed resistance to 8 μg of methicillin per ml (the cell count increased at 4 h; Fig. 3B). However, it showed an enhanced susceptibility to 128 μg of methicillin per ml: the cell count decreased by 3 log10 units (from 1.1 × 106 CFU at time zero to 1.1 × 103 CFU at 4 h; Fig. 3B). This enhanced susceptibility of ΔI to a high concentration of methicillin disappeared in its homo-converted mutant strain ΔI-HR (Fig. 3C). No apparent decrease in the viable cell count was observed with the strain after a 4-h exposure to 128 μg of methicillin per ml.
FIG. 3.
Methicillin time-kill study of N315 and its derivative strains with various methicillin-resistance phenotypes. The tested strains were N315 (A), ΔI (B), ΔI-HR (C), h4 (D), h4ΔI (E), h4-4R (F), N315(pYT3) (G), N315(pHMR-A) (H), and N315(pHMR-B) (I). Cytokilling was performed in the presence of methicillin at concentrations of 0 (○), 1 (●), 8 (■), and 128 (▴) μg/ml. Viable cell counts were done at 1, 2, and 4 h after the start of culture. The incubation temperature was 37°C for panels A to F and 30°C for panels G to I. Note that the cytokilling activity of methicillin at 128 μg/ml was remarkably decreased with the strains with the Eagle-type or the homo-type methicillin resistance and with the transformant strains harboring multiple copies of hmrA or hmrB genes.
Eagle-type strain h4 (Fig. 3D) showed almost equivalent susceptibility with that of N315 to 8 μg of methicillin per ml (84% decrease; from 1.4 × 106 CFU at time zero to 2.2 × 105 CFU at 4 h; compare closed-square lines in panels A and D). However, h4 exhibited reduced susceptibility to the cytokilling activity of methicillin at 128 μg/ml; no reduction in the cell count was observed at 4 h (1.4 × 106 CFU) compared to that at time zero (1.4 × 106 CFU) (Fig. 3D). The mecI-inactivated strain h4ΔI showed resistance to all the tested concentrations of methicillin as shown in Fig. 3E, which was also the case with h4-4R (Fig. 3F).
Isolation of N315 transformants with Eagle-type methicillin resistance.
To identify a responsible gene involved in the expression of Eagle-type methicillin resistance, in screening 22,580 N315 transformants by replica plating as described in Materials and Methods, we obtained three transformant strains whose colony sizes were greater on the plate containing 100 μg of methicillin per ml than on the plate containing a 10-μg/ml concentration of the antibiotic (Eagle phenotype; see above). By restriction mapping analysis of the recombinant plasmids extracted from the transformants, two of the three had an identical restriction pattern. Therefore, the two different plasmids were designated pHMR-A and pHMR-B, respectively. The plasmids, amplified in E. coli JM109, were then introduced into strain N315 and the hetero-type strain LR5, and their methicillin resistance phenotypes were evaluated by population analysis (Fig. 1B to E). The analysis was performed both at 30°C (Fig. 1B and D; the copy number of plasmid is 14) and at 37°C (Fig. 1C and E; the copy number is 1). When introduced into N315, both plasmids caused Eagle-type methicillin resistance, although the sizes of highly methicillin-resistant subpopulations were different in the two transformants, and they changed depending on the temperature.
Heteroresistant strain LR5 had a higher level of resistance at 30°C than at 37°C but showed typical hetero-type methicillin resistance at both temperature (compare Fig. 1B and C). When pHMR-A was introduced into LR5, homo-type methicillin resistance was expressed at both temperatures (Fig. 1B and C). On the other hand, introduction of plasmid pHMR-B into LR5 could not confer complete homo-type methicillin resistance to LR5, although it did increase the level of methicillin resistance significantly at both temperatures (Fig. 1D and E). Introduction of vector plasmid pYT3 did not change the pattern of resistance of N315 or LR5 (Fig. 1B to E).
Localization of ORFs responsible for Eagle-type resistance.
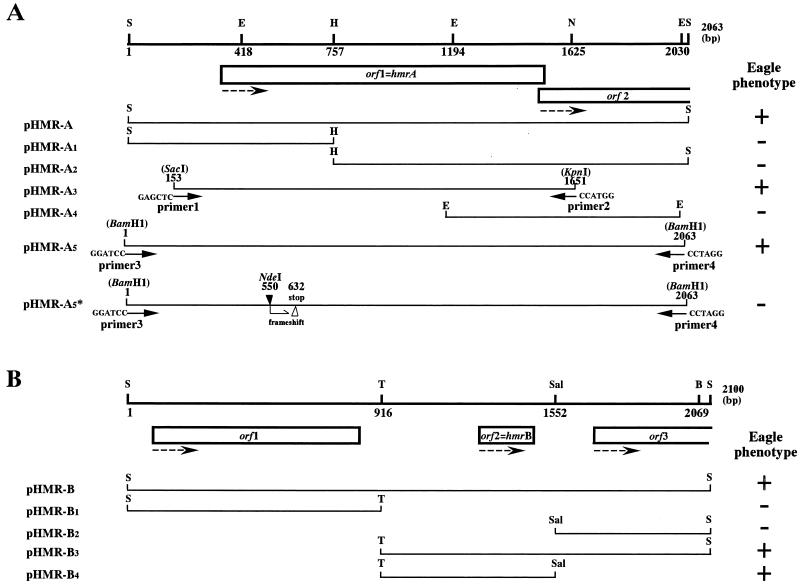
Restriction maps and the location of open reading frames (ORFs) found in the inserted DNAs of pHMR-A and pHMR-B are illustrated in Fig. 4. To localize the function conferring the Eagle phenotype, subcloning of the inserted DNAs was performed either by restriction enzyme cleavage or by PCR amplification of a part of the inserts. The constructed recombinant plasmids were introduced into N315, and their ability to confer Eagle-type resistance to N315 was evaluated by population analysis. The correlation between the tested DNA fragments and their ability to confer Eagle phenotypes to N315 is summarized in Fig. 4. For the orf1 of pHMR-A, we constructed plasmid pHMR-A5∗ that carried a single frameshift mutation at NdeI site. The plasmid did not cause Eagle phenotype when introduced in N315 (Fig. 4A). With these studies, the activity to confer Eagle phenotype to N315 was localized to one ORF of each plasmid; the ORF of pHMR-A was designated hmrA (standing for high methicillin resistance), and that of pHMR-B was designated hmrB.
FIG. 4.
Restriction map and functional assignment of pHMR-A and pHMR-B insert DNAs. Restriction maps and distribution of ORFs of pHMR-A (A) and pHMR-B (B) inserts are shown. Abbreviations for restriction enzymes: S, Sau3AI; E, EcoRI; H, HindIII; N, NcoI; T, Tth111I; Sal, SalI, B, BamHI. The locations of the subcloned fragments and their capability of conferring Eagle phenotype (denoted as +) to N315 are illustrated to the right of the recombinant plasmid names harboring the fragments. See Materials and Methods for the detailed method of constructing each recombinant plasmid. In the case of pHMR-A5∗, a frameshift mutation was introduced at the NdeI site (indicated by a closed arrowhead) of orf1; the location of the premature stop codon generated by this procedure is indicated by an open arrowhead. Dotted arrows show the direction of transcription of the ORFs. The function to confer Eagle-type resistance to strain N315 was confined to orf1 in pHMR-A and orf2 in pHMR-B; they were designated hmrA and hmrB, respectively.
Deduced amino acid sequences encoded by hmrA and hmrB.
The predicted amino acid sequences, HmrA and HmrB, encoded by hmrA and hmrB, respectively, are shown in Fig. 5. The amino acid sequences of HmrA and HmrB were aligned with those of the extant proteins having the highest homology score. The protein with the highest homology score to HmrA was AmiB of Mycobacterium tuberculosis that showed 33% amino acid identity to HmrA over the 380 aligned sequences (Fig. 5A). AmiB is a probable amidohydrolase protein of M40 family with unknown physiological function. Figure 5B shows that HmrB had the highest homology score to constitutive acyl-carrier protein of Rhizobium leguminosarum; the amino acid identity was 66% over 72 aligned sequences. A characteristic prosthetic group attachment site serine of acyl-carrier protein was also present in HmrB (indicated by the arrow in Fig. 5B).
FIG. 5.
Deduced amino acid sequences of HmrA and HmrB. Amino acid sequence of HmrA is aligned with AmiB (a probable aminohydrolase) of M. tuberculosis (A), and that of HmrB is aligned with the ACP of R. leguminosarum (B). Amino acid residues identical and similar between the two peptides are indicated by asterisks and dots, respectively. The boxed sequences, a to d, are parts of sequences that were well conserved among amidohydrolase proteins of the M40 family. The serine residue implicated as the attachment site for prosthetic group of the ACP is indicated by an arrow in panel B.
The nucleotide sequencing of hmrA and hmrB genes in N315 and h4 was performed to reveal that they were identical between the two strains in both coding frames, as well as in promoter regions. A BLAST search was performed to identify the genes in other S. aureus strains. The genes were found as unique genes in all the four strains whose genomic sequence data are available (i.e., strains COL, NCTC8325, E-MRSA-16, and MSSA 146). The amino acid sequences of HmrB of the four S. aureus strains were identical. On the other hand, HmrA of N315 and h4 differed by one amino acid substitution from that of EMRSA-16 (Gly71 to Asp) and from those of COL, NCTC8325, and strain 476 (Pro276 to Arg).
The hmr genes confer high-dose tolerance to methicillin.
Figures 3G to I illustrate the cytokilling curves of the N315-derived transformants harboring plasmids pYT3 (Fig. 3G), pHMR-A (Fig. 3H), and pHMR-B (Fig. 3I). The control transformant N315(pYT3) responded dose dependently to the killing activity of methicillin in the same manner as N315 (Fig. 3A). On the other hand, the transformants N315(pHMR-A) and N315(pHMR-B) had remarkably reduced susceptibility to the killing effect of methicillin at 128 μg/ml, a finding which was comparable to that observed with N315-derived Eagle-type mutant strain h4. Thus, both hmr genes encoded the function that confer N315 high-dose tolerance or the paradoxical phenotype to methicillin-induced killing.
DISCUSSION
It is well established that PBP2′ production is prerequisite for the expression of methicillin resistance (18). Since Eagle-type strain h4 was capable of growth in the presence of high concentrations of methicillin (Fig. 1), it was predictable that PBP2′ was inducible with a high concentration of methicillin. The amount of PBP2′ induced in h4, however, was several times smaller than those expressed in hetero- or homoresistant strains. In contrast, the methicillin-heteroresistant strain ΔI produced a far greater amount of PBP2′ than h4 but did not grow in the presence of 128 μg of methicillin per ml (Fig. 1). This reconfirms the previous reports that high production of PBP2′ alone is not enough for the high-level or homoresistance expression of MRSA, and expression of some factor other than mecA gene referred to as chr∗ is involved in the hetero-to-homo conversion of methicillin resistance (2). Ryffel et al. (23), however, did not look for a phenotypic expression of chr∗ mutation alone, i.e., a phenotypic change caused by chr∗ mutation itself in the absence of the combined function of PBP2′.
In this study, we observed that there was a significant difference between N315 and h4 in terms of vulnerability of cells to methicillin-induced cytokilling (Fig. 3). Pre-MRSA strain N315 was killed by methicillin dose dependently, whereas the Eagle-type strain h4 was paradoxically resistant to high concentrations of methicillin. This resembles the “high-dose tolerance” or “paradoxical effect” observed first by Eagle and Musselman (7). We consider that the tolerance to a high concentration of methicillin is the phenotypic expression of chr∗ mutation in the genetic background of pre-MRSA strain N315, i.e., under mecI gene-mediated mecA gene repression.
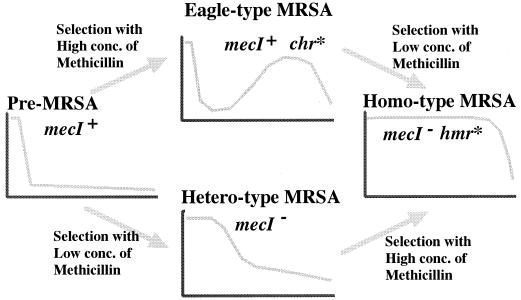
Figure 6 summarizes the proposed evolutionary status of Eagle-type resistance described in this study in correlation with other phenotypes of methicillin resistance. There are two alternate evolutionary pathways for pre-MRSA to acquire the homo-type methicillin resistance, and Eagle-type resistance is assigned to the intermediate stage in one of the evolutionary pathways. Since mecI gene inactivation is common in both pathways, the genetic event underlying the conversion from pre-MRSA to the Eagle phenotype is considered to be equivalent to that underlying the hetero-to-homo phenotypic conversion, the latter having been designated as chr∗ mutation by Ryffel et al. (23). This hypothetical equation was further supported by our successful cloning of hmr genes that conferred both pre-MRSA-to-Eagle and hetero-to-homo conversions to the host strain with respective phenotype.
FIG. 6.
Two pathways of acquisition of homo-type methicillin resistance by pre-MRSA strain N315. Two sequencial genetic alterations, mecI inactivation and chr∗ mutation (see the text) are required for N315 to achieve homo-type methicillin resistance. We propose that the phenotypic expression of chr∗ mutation in the pre-MRSA genetic background corresponds to the Eagle phenotype.
It is noteworthy that the heteroresistant strain ΔI producing a high amount of PBP2′ exhibited much greater susceptibility to 128 μg of methicillin per ml than did N315 (Fig. 3B). The reason for this hypersusceptibility is unknown, but it could be due to deleterious effect of PBP2′ overproduction on the cell wall synthesis of S. aureus cell. PBP2′ is reported to be an inefficient transpeptidase in S. aureus cell (6). Therefore, overexpression of the exogenous PBP2 might have caused a decrease in the cross-linkage of the cell wall, resulting in the increased susceptibility to beta-lactam action. In that sense, the chr∗ mutation may be considered as a “counter mutation” to complement the weakness of the host cell wall produced by the expression of PBP2′. In this hypothesis, derepression production of PBP2′ would be regarded as an “internal selective pressure” for the evolution of host cells.
Many genes, designated fem or aug, have been reported whose genetic alteration affect the expression of homo-type methicillin resistance in MRSA (2). Most of the genes have been identified by transposon mutagenesis of S. aureus strains expressing homo-type methicillin resistance. They are the genes whose partial or complete inactivation lead to the decrease in the methicillin resistance, and most of the genes encode enzymes involved in the cell wall synthesis of S. aureus cell. We approached this problem from a different direction, i.e. we adopted the Eagle phenotype as the assay for cloning the genes responsible for hetero-to-homo conversion of methicillin resistance.
With regard to the fem and aug genes, little is known about how frequently these genes are involved in the hetero-to-homo conversion of methicillin resistance. In the case of hmr genes, we have recently analyzed 10 each of the N315-derived mutant strains with Eagle- and homo-type methicillin resistance. We found that the hmrA overexpression, defined as such when the amount of hmrA transcript in the mutant strain exceeded that in N315(pHMR-A) at 37°C as measured by quantitative reverse transcription-PCR (4), was found in one strain (10%) of Eagle-type and two strains (20%) of homo-type resistance (H. Kuroda-Murakami et al., unpublished results). On the other hand, no strain with hmrB overexpression was found among the mutant strains. Curiously, overexpression of hmrA or hmrB gene was not observed in strain h4. Therefore, it is probable that some other alternate genetic mechanisms exist that cause the expression of homo- and Eagle-type methicillin resistance.
The mechanism how chr∗ mutation causes high-dose tolerance to methicillin will become clear through the study of hmr gene function. HmrA had a considerable homology to the amidohydrolase of M40 family, into which many enzymes of diverse physiological or unknown functions are classified, e.g., AmiB of M. tuberculosis, hippuricase of Campylobacter jejuni (10), N-carbamyl-l-amino acid aminohydrolase of Pseudomonas species (34), N-acyl-l-amino acid amidohydrolase of Bacillus stearothermophilus (25), and thermostable carboxypeptidase of Sulfolobus solfataricus (5). At this moment, we cannot infer any plausible function of HmrA to lead to the tolerance to methicillin caused by its expression.
HmrB, on the other hand, turned out to be the S. aureus homologue of acyl carrier protein (ACP). The hmrB gene homologue found in the Bacillus subtilis genome corresponded to the unique ACP gene acpA (accession no. CAB 13465). hmrB corresponds to the unique ACP gene homologue in N315 chromosome based on a homology search over the complete nucleotide sequence of N315 genome (K. Hiramatsu, unpublished data). ACP is a small acidic protein having multiple functions in the biosynthesis of macromolecules in bacteria. The protein is involved in the synthesis of fatty acids (33), phospholipids (22), aromatic polyketides (14), membrane-derived oligosaccharides (31), and lipopolysaccharides such as lipid A (3). The importance of ACP-dependent protein acylation has also been demonstrated in posttranslational protoxin activation in bacteria, e.g., in the conversion of prohemolysin into hemolysin in pathogenic E. coli (15). It is tempting to speculate that overproduction of ACP in the cell may cause tolerance through the stabilization of the autolytic activity by increasing the synthesis of lipoteichoic acid, which is known as a stabilizer of autolysin in S. aureus (29). Alternatively, ACP may function through enhancing cell wall synthesis of S. aureus, since an increased amount of ACP may lead to the increased production of undecaprenol, the membrane lipid known as the carrier of murein monomer precursors (27). A study is under way to clarify the molecular mechanism how HmrA and HmrB confer high methicillin resistance to S. aureus.
In conclusion, we have described Eagle-type resistance as a novel phenotype of methicillin resistance. We propose that it is the phenotype caused by chr∗ mutation responsible for the hetero-to-homo conversion of methicillin resistance under the mec regulator gene control of mecA gene transcription.
ACKNOWLEDGMENTS
This work was supported by Core University Program under Japan Society for the Promotion of Science, coordinated by the University of Tokyo, Graduate School of Medicine and Universiti Sains Malaysia, School of Medical Sciences, and by Specially Designated Research Promotion of Monbusho, Japan.
REFERENCES
- 1.Asada K, Inaba Y, Tateda-Suzuki E, Kuwahara-Arai K, Ito T, Hiramatsu K. Evolution and resistance expression of MRSA: evaluation of beta-lactam antibiotics against a set of isogenic strains with different types of phenotypic expression. Acta Biochim Polon. 1995;42:517–524. [PubMed] [Google Scholar]
- 2.Berger-Bachi B, Tschierske M. Role of Fem factors in methicillin resistance Drug Resist. Updates. 1998;1:325–335. doi: 10.1016/s1368-7646(98)80048-4. [DOI] [PubMed] [Google Scholar]
- 3.Brozek K A, Raetz C R H. Biosynthesis of lipid A in Escherichia coli:acyl carrier protein-dependent incorporation of laurate and myristate. J Biol Chem. 1990;265:15410–15417. [PubMed] [Google Scholar]
- 4.Christiane G, Silvia C, Manfred G B, Gerd D, Konrad B, Christiane W. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vivo. Infect Immun. 2000;68:1304–1311. doi: 10.1128/iai.68.3.1304-1311.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colombo S, De Auria S, Fusi P, Zecca L, Raia C A, Tortora P. Purification and characterization of a thermostable carboxypeptidase from the extreme thermophilic archaebacterium Sulfolobus solfataricus. Eur J Biochem. 1992;206:349–357. doi: 10.1111/j.1432-1033.1992.tb16934.x. [DOI] [PubMed] [Google Scholar]
- 6.De Jonge B L M, Tomasz A. Abnormal peptidoglycan produced in a methicillin-resistant strain of Staphylococcus aureus grown in the presence of methicillin: functional role for penicillin-binding protein 2A in cell wall synthesis. Antimicrob Agents Chemother. 1993;37:342–346. doi: 10.1128/aac.37.2.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eagle H, Musselman A D. The rate of bactericidal action of penicillin in vitro as a function of its concentration, and its paradoxically reduced activity at high concentrations against certain organisms. J Exp Med. 1948;88:99–131. doi: 10.1084/jem.88.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hackbarth C J, Chambers H F. blaI and blaR1 regulate beta-lactamase and PBP2a production in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1993;37:1144–1149. doi: 10.1128/aac.37.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanaki H, Kuwahara-Arai K, Boyle-Vavra S, Daum R S, Labischinski H, Hiramatsu K. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J Antimicrob Chemother. 1998;42:199–209. doi: 10.1093/jac/42.2.199. [DOI] [PubMed] [Google Scholar]
- 10.Hani E K, Chan V L. Expression and characterization of Campylobacter jejuni benzoylglycine amidohydrolase (Hippuricase) gene in Escherichia coli. J Bacteriol. 1995;177:2396–2402. doi: 10.1128/jb.177.9.2396-2402.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartman B J, Tomasz A. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984;158:513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiramatsu K. Molecular evolution of MRSA. Microbiol Immunol. 1995;39:531–543. doi: 10.1111/j.1348-0421.1995.tb02239.x. [DOI] [PubMed] [Google Scholar]
- 13.Hiramatsu K, Asada K, Suzuki E, Okonogi K, Yokota T. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA) FEBS Lett. 1991;298:133–136. doi: 10.1016/0014-5793(92)80039-j. [DOI] [PubMed] [Google Scholar]
- 14.Hopwood D A, Sherman D H. Molecular genetics of polyketides and its composition to fatty acid biosynthesis. Annu Rev Genet. 1990;24:37–66. doi: 10.1146/annurev.ge.24.120190.000345. [DOI] [PubMed] [Google Scholar]
- 15.Issartel J-P, Koronakis V, Hughes C. Activation of Escherichia coli prohaemolysin to the mature toxin by acyl carrier protein-dependent fatty acylation. Nature. 1991;351:759–761. doi: 10.1038/351759a0. [DOI] [PubMed] [Google Scholar]
- 16.Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, Staphylococcus Cassette Chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44:1549–1555. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuwahara-Arai K, Kondo N, Hori S, Tateda-Suzuki E, Hiramatsu K. Suppression of methicillin resistance in a mecA-containing pre-methicillin-resistant Staphylococcus aureus strain is caused by the mecI-mediated repression of PBP2′ production. Antimicrob Agents Chemother. 1996;40:2680–2685. doi: 10.1128/aac.40.12.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews P, Tomasz A. Insertional inactivation of the mec gene in a transposon mutant of a methicillin-resistant clinical isolate of Staphylococcus aureus. Antimicrob Agents Chemother. 1990;34:1777–1779. doi: 10.1128/aac.34.9.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthews P R, Stewart P R. Resistance heterogeneity in methicillin-resistant Stapylococcus aureus. FEMS Microbiol Lett. 1984;22:161–166. [Google Scholar]
- 20.Nilsson B, Abrahmsen L, Uhlen M. Immobilization and purification of enzymes with staphylococcal protein A gene fusion vectors. EMBO J. 1985;4:1075–1080. doi: 10.1002/j.1460-2075.1985.tb03741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reynolds P E, Fuller C. Methicillin-resistant strains of Staphylococcus aureus: presence of identical additional penicillin-binding protein in all strains examined. FEMS Microbiol Lett. 1986;33:251–254. [Google Scholar]
- 22.Rock C, Jackowski S. Regulation of phospholipid synthesis in Escherichia coli. Composition of the acyl-acyl carrier protein pool in vivo. Biol Chem. 1982;257:10759–10765. [PubMed] [Google Scholar]
- 23.Ryffel C, Strassle A, Kayser F H, Berger-Bachi B. Mechanism of heteroresistance in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:724–728. doi: 10.1128/aac.38.4.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sabath L D, Wallace S J. Factors influencing methicillin resistance in staphylococci. Ann N Y Acad Sci. 1971;182:258–266. doi: 10.1111/j.1749-6632.1971.tb30662.x. [DOI] [PubMed] [Google Scholar]
- 25.Sakanyan V, Desmarez L, Legrain C, Charlier D, Mett I, Kochikyan A, Savchenko A, Falmagne P, Pierard A, et al. Gene cloning, sequence analysis, purification, and characterization of thermostable aminoacylase from Bacillus stearothermophilus. Appl Environ Microbiol. 1993;59:3878–3888. doi: 10.1128/aem.59.11.3878-3888.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siewert G, Strominger J L. Biosynthesis of the peptidoglycan of bacterial cell walls. J Biol Chem. 1968;243:783–790. [PubMed] [Google Scholar]
- 28.Song M D, Wachi M, Doi M, Ishino F, Matsuhashi M. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 1987;221:167–171. doi: 10.1016/0014-5793(87)80373-3. [DOI] [PubMed] [Google Scholar]
- 29.Sugai M. Peptidoglycan hydrolases of the staphylococci J. Infect Chemother. 1997;3:113–127. [Google Scholar]
- 30.Tesch W, Ryffel C, Strassle A, Kayser F H, Berger-Bachi B. Evidence of a novel staphylococcal mec-encoded element (mecR) controlling expression of penicillin-binding protein 2′. Antimicrob Agents Chemother. 1990;34:1703–1706. doi: 10.1128/aac.34.9.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Therisod H, Kennedy E P. The function of acyl carrier protein in the synthesis of membrane-derived oligosaccharides does not require its phosphopanthetheine prosthetic group. Proc Natl Acad Sci USA. 1987;84:8235–8238. doi: 10.1073/pnas.84.23.8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Utsui Y, Yokota T. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1985;28:397–403. doi: 10.1128/aac.28.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van den Boom T, Cronan J E J. Genetics and regulation of bacterial lipid metabolism. Annu Rev Microbiol. 1989;43:317–343. doi: 10.1146/annurev.mi.43.100189.001533. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe K, Ishikawa T, Mukohara Y, Nakamura H. Cloning and sequencing of the genes involved in the conversion of 5-substituted hydantoins to the corresponding l-amino acids from the native plasmid of Pseudomonas sp. strain NS671. J Bacteriol. 1992;174:962–969. doi: 10.1128/jb.174.3.962-969.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]