Abstract
There is growing need for a safe, efficient, specific and non-pathogenic means for delivery of gene therapy materials. Nanomaterials for nucleic acid delivery offer an unprecedented opportunity to overcome these drawbacks; owing to their tunability with diverse physico-chemical properties, they can readily be functionalized with any type of biomolecules/moieties for selective targeting. Nucleic acid therapeutics such as antisense DNA, mRNA, small interfering RNA (siRNA) or microRNA (miRNA) have been widely explored to modulate DNA or RNA expression Strikingly, gene therapies combined with nanoscale delivery systems have broadened the therapeutic and biomedical applications of these molecules, such as bioanalysis, gene silencing, protein replacement and vaccines. Here, we overview how to design smart nucleic acid delivery methods, which provide functionality and efficacy in the layout of molecular diagnostics and therapeutic systems. It is crucial to outline some of the general design considerations of nucleic acid delivery nanoparticles, their extraordinary properties and the structure–function relationships of these nanomaterials with biological systems and diseased cells and tissues.
Gene therapy uses nucleic acids as functional molecules to activate biological treatment for a wide range of diseases, such as cancer1,2, cystic fibrosis3, heart disease4, diabetes5, haemophilia and HIV/AIDS6. Nucleic acids have been attracting increasing attention owing to the global effort in the human genome elucidation together with recent discoveries such as RNA interference (RNAi) and CRISPR-based genome editing7–9. Gene therapy uses genetic material to alter the expression of a target gene or to modify the biological properties of living cells for therapeutic needs. In recent years, multiple gene therapy products have been approved by the regulatory agencies for various applications10. Perhaps the most relevant example is the authorization of mRNA vaccines to fight the COVID-19 outbreak11.
Gene therapy can be divided into three main avenues, as detailed in FIG. 1. First is editing mutated genes using CRISPR–Cas technology to cause gain or loss of function12,13. Second, upregulating gene expression can be achieved through the insertion of a functional gene copy to be expressed by using molecules such as DNA plasmid (pDNA), minicircle DNA (mcDNA), synthetic mRNA, circular RNA and self-amplifying RNA (saRNA)14–16. Last is downregulating gene expression using molecules such as small interfering RNA (siRNA), antisense oligonucleotides (ASOs), short hairpin RNA (shRNA) and microRNA (miRNA)17,18.
Fig. 1 |. Schematic pathways of three gene therapy avenues.
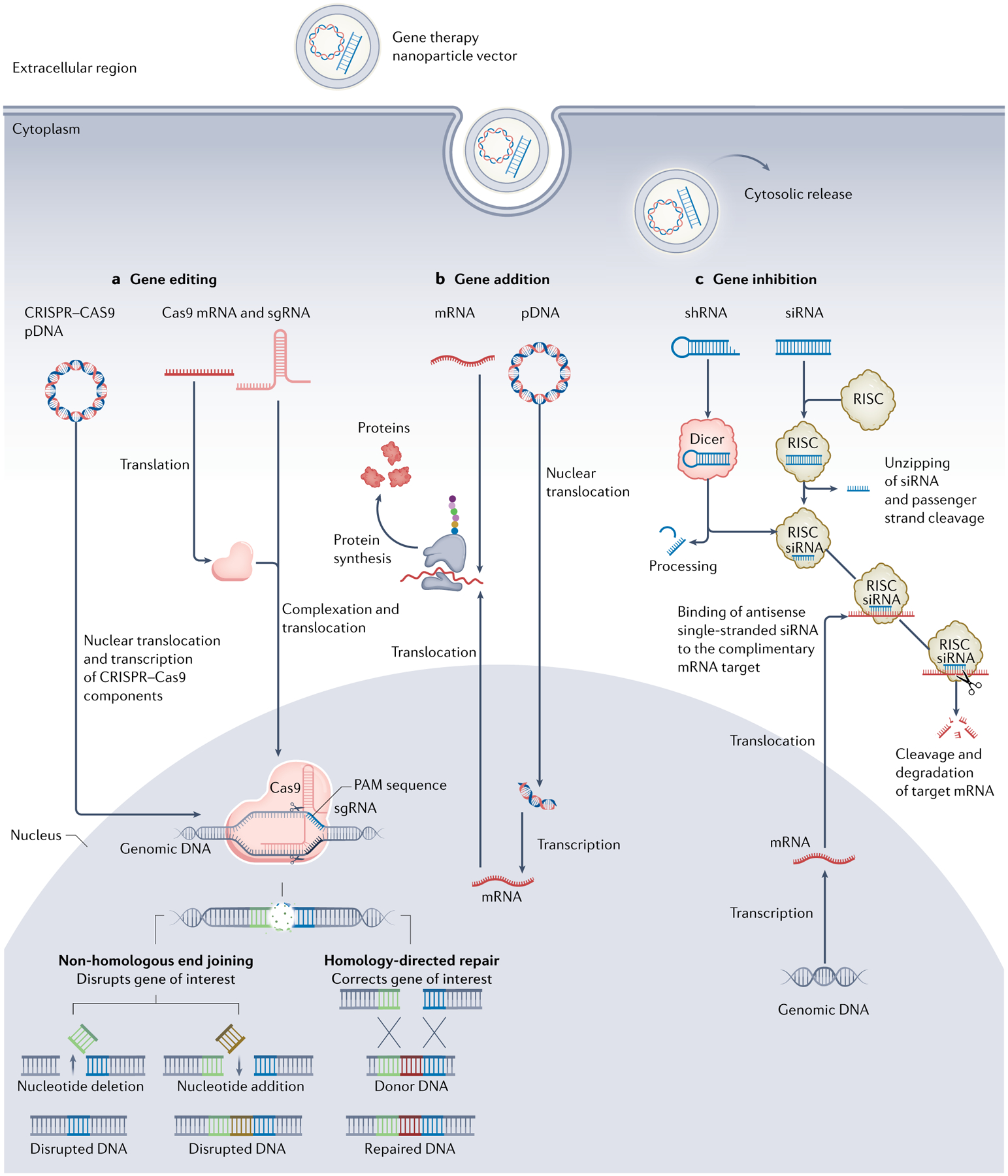
a | Gene editing. CRISPR–Cas9 coding DNA plasmid (pDNA) or Cas9 mRNA and single guide RNA (sgRNA) that contains a targeting sequence are delivered to the cell and enter the cytoplasm. pDNA translocates to the nucleus followed by transcription to Cas9 mRNA and sgRNA. Following translocation to the cytosol, Cas9 mRNA is translated to Cas9 protein and complexes sgRNA. Thereafter, Cas9–sgRNA complex translocates to the nucleus, where it binds the genomic DNA target sequence containing a proto-spacer adjacent motif (PAM) sequence at its 3’ end. Specific binding causes double-stranded DNA breakage resulting in non-homologous end joining or homology-directed repair, leading to mutations or changes within the gene sequence, respectively. b | Gene addition. After entering the cytoplasm, pDNA carrying the desired gene sequence translocates to the nucleus and undergoes transcription to mRNA. Next, transcripted or external mRNA is translated by the ribosome, promoting desired protein synthesis. c | Gene inhibition. After entering the cytoplasm, short hairpin RNA (shRNA) is detected and processed by Dicer protein to generate small interfering RNA (siRNA) later loaded onto RNA-induced silencing complex (RISC). Subsequently, double-stranded siRNA is unzipped, and passenger strand is cleaved. Antisense strand guides RISC towards target mRNA and aligns with it, thus leading to mRNA cleavage and degradation.
Nucleic acids have promising advantages compared with conventional drugs19. Unlike the latter, the mechanism of action and high specificity of nucleic acids present a possible therapy route for viral infections, various cancers and undruggable genetic disorders with unmet clinical need. Moreover, theoretically, a single treatment of the genetic payload can achieve a durable and even curative effect20. However, delivering nucleic acids to reach their active site inside the cell is challenging owing to their low in vivo stability and rapid host clearance outside cells. Additionally, nucleic acids are poorly permeable through the cellular membrane owing to their negative charge, high molecular weight and hydrophilicity21. Nonetheless, few delivery challenges differ between DNA and RNA. For example, the payload and carrier toxicity are of greater concern when delivering RNA molecules usually associated with short-term activity and low retention inside the cell, hence requiring more frequent administration22. Alternatively, DNA activity inside the nucleus adds complexity related to low nuclear transport, thus leading to distinguishing design concepts regarding the delivery system compared with RNA molecules23. Together with specific challenges relevant to the delivered molecule, the fundamental challenge is to develop tailored systems that can facilitate nucleic acid uptake into target cells. The carrier itself needs to overcome extracellular and intracellular barriers, provide protection from nuclease activity in the bloodstream, enhance and assist with cellular uptake, and promote endosomal escape once entered into the cell24.
Gene delivery strategies are usually classified into viral and non-viral24, maturing over the past few decades as presented in Box 1. Viruses can be used to deliver a gene of interest by its insertion into the viral genome, followed by cell infection and gene expression25. Viruses that have high DNA transfection efficiency, such as lentiviruses, retroviruses, adenoviruses, parvoviruses and adeno-associated viruses, have long been used to treat diseases such as HIV, cancer and muscular dystrophy26. Despite the high cellular transfection efficiency of viral gene delivery systems, rapid clearance due to pre-existing antibodies, generation of neutralizing antibodies against the vector, limited vector size capacity (usually below 7 kb) and possible side effects27,28 leave room for complementary gene delivery techniques in biomedical research and in the clinic. Thus, efforts have been made to design non-viral gene delivery systems, based on lipids, polymers, peptides and inorganic compounds.
Box 1 |. Timeline of globally approved gene therapies in the past decade defined by ClinicalTrials.gov identifier.
2010
Rexin-G (retrovirus, plasmid DNA (pDNA), NCT00121745)
2012
Glybera (adeno-associated virus, DNA, NCT03293810)
2013
Kynamro (naked antisense oligonucleotide (ASO), NCT00607373)
2016
Exondys 51 (naked phosphorodiamidate morpholino backbone (PMO), NCT00844597)
Spinraza (naked ASO, NCT01494701)
Imlygic (herpes simplex virus (HSV), DNA, NCT00289016)
Defitelio (naked DNA, NCT00003966)
Strimvelis (retrovirus, cDNA, NCT00599781)
2017
Luxturna (adeno-associated virus, DNA, NCT00643747)
Invossa (retrovirus, DNA, NCT03412864)
Yescarta (retrovirus, DNA, NCT05108805)
Kymriah (lentivirus, DNA, NCT05075603)
2018
Patisiran (lipid nanoparticle (LNP), small interfering RNA (siRNA), NCT01148953)
Tegsedi (naked ASO, NCT04306510)
2019
Zolgensma (adeno-associated virus, DNA, NCT02122952)
Collategene (naked pDNA, NCT04267640)
Waylivra (naked ASO, NCT01529424)
Zynteglo (lentivirus, DNA, NCT01745120)
Givlaari (N-acetylgalactosamine (GalNAc), siRNA, NCT03338816)
2020
Libmeldy (lentivirus, DNA, NCT03771898)
BNT162b2 (LNP, mRNA, NCT04862806)
mRNA-1273 (LNP, mRNA, NCT04470427)
Tecartus (retrovirus, DNA, NCT04880434)
Transfection efficiency.
The percentage of cells successfully inserted with genetic payload from the initial cell population.
Lipid-based structures usually consist of a core-shell arrangement and ligands can be conjugated to their surface for active targeting29,30. Cationic lipids (such as N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium chloride (DOTMA)) are usually used to increase encapsulation efficiency when using liposomes for nucleic acid delivery31. However, the use of cationic liposomes is limited owing to their toxicity at the site of administration32. Ionizable lipids present a positive charge under acidic pH conditions, favouring complexation with nucleic acids and enabling their endosomal escape once inside cells33. Ideally, these lipids should have a pKa value around 6.4 and near-neutral surface charge to prevent rapid sequestration by the mononuclear phagocyte system and a lack of immune stimulation and toxic effects upon in vivo intravenous administration34.
Polymeric nanocarriers generally consist of polycations often containing ionizable amine groups35. These groups interact with negatively charged nucleic acids containing phosphate groups via attractive coulombic forces, thus promoting self-assembly to form condensed nanosized structures termed polyplexes. Choosing an adequate polymer (for example, polyethyleneimine (PEI) and poly-l-lysine (PLL)) should be a consideration when designing the delivery system based on its application36. For instance, branched PEI contains many available amine groups that contribute to its high buffering capacity, and hence facilitate endosomal escape within the cell freeing the RNA to the cytoplasm37; however, it was shown to induce some in vivo and in vitro toxicity38.
Polycations.
Chemical compounds, molecules or complexes obtaining positive charge at several sites.
Polyplexes.
Two-element systems comprising a cationic polymer complexed with an anionic nucleic acid payload (for example, small interfering RNA (siRNA)) through electrostatic interaction.
Peptide-based formulations are another delivery platform introduced in recent years, consisting of 5–30 amino acids typically attributed as cell-penetrating peptides or homing peptides. Both can interact with nucleic acids in a covalent or non-covalent manner, thus facilitating cell internalization39. Inorganic compound-based nano-vectors provide unique multipurpose delivery platforms. This versatile group consists of noble metals (such as gold or silver), carbon nanotubes, cholesterol conjugates, iron oxide, silica and lanthanides as nucleic acid carriers40. Inorganic nano-vectors can be tailored to effectively target and transport therapeutic payloads41. For example, mesoporous silica nanoparticles (MSNs) covered with a polymeric coating were shown to successfully carry siRNA and downregulate osteoporosis-related genes as part of a potential treatment42.
Given the benefits and drawbacks of each delivery class, considerable research showed that a combination of different classes improves therapeutic efficacy43. Gene delivery carriers have made significant steps in clinical implementation alongside the development of new technologies; however, many questions remain unanswered regarding how these carriers are expected to target a given nucleic acid towards a specific cell type. Hence, the quest for a one-size-fits-all formulation platform should be re-evaluated44. This same approach finds itself relevant to the genetic payload, when resembling technologies unexpectedly differ in their outcome. CureVac’s mRNA lipid nanoparticle (LNP) vaccine against COVID-19, CVnCoV, was a promising candidate using a similar LNP formulation to Pfizer and Moderna’s successful vaccines45. Unfortunately, CVnCoV two-dose vaccination showed only 47% effectiveness at preventing the disease. A conjecture to these results is, presumably, the difference between Pfizer and Moderna’s modified versus CureVac’s unmodified mRNA payload and the need to engineer the particle for the specific RNA sequence46.
Therefore, next-generation gene delivery strategies need to integrate material properties, nucleic acid intracellular function and modifications, and disease characteristics. For this purpose, advanced robotic high screening technologies and artificial intelligence algorithms are being developed to process and analyse large data sets of successful delivery vehicles47. This will selectively optimize the process according to an intended target, thereby achieving accuracy, precise gene therapy and better clinical results while saving time and effort.
The purpose of this Primer is to focus on different aspects of nucleic acid delivery using nanomaterials. The Primer outlines the experimentation of various delivery platforms such as LNPs, liposomes and polymeric and inorganic nanoparticles by emphasizing the primary consideration taken when designing delivery platforms as well as possible manufacturing methods. The Primer covers different techniques to characterize nanomaterials’ properties, discusses results analysis and helps deduce downstream biological implications. Key applications of nucleic acid nanoparticles including bioanalysis, nano-barcodes, gene silencing and editing, and vaccines and immunotherapy are described and detailed through relevant examples. Data reproducibility and deposition are reviewed in the context of the field’s limitations and optimization. The future perspective and outlook are deliberated, indicating the major challenges that await the scientific community in the next decade.
Experimentation
Lipid-based nanoparticles, polymeric nanoparticles and inorganic nanoparticles are the most common non-viral nanocarriers for nucleic acid delivery (FIG. 2; TABLE 1). In this section, the key factors involved in nanoparticle design and fabrication as well as the commonly used modification methods to improve nucleic acid stability are described.
Fig. 2 |. Scheme for the main nanoparticle design used in gene delivery.
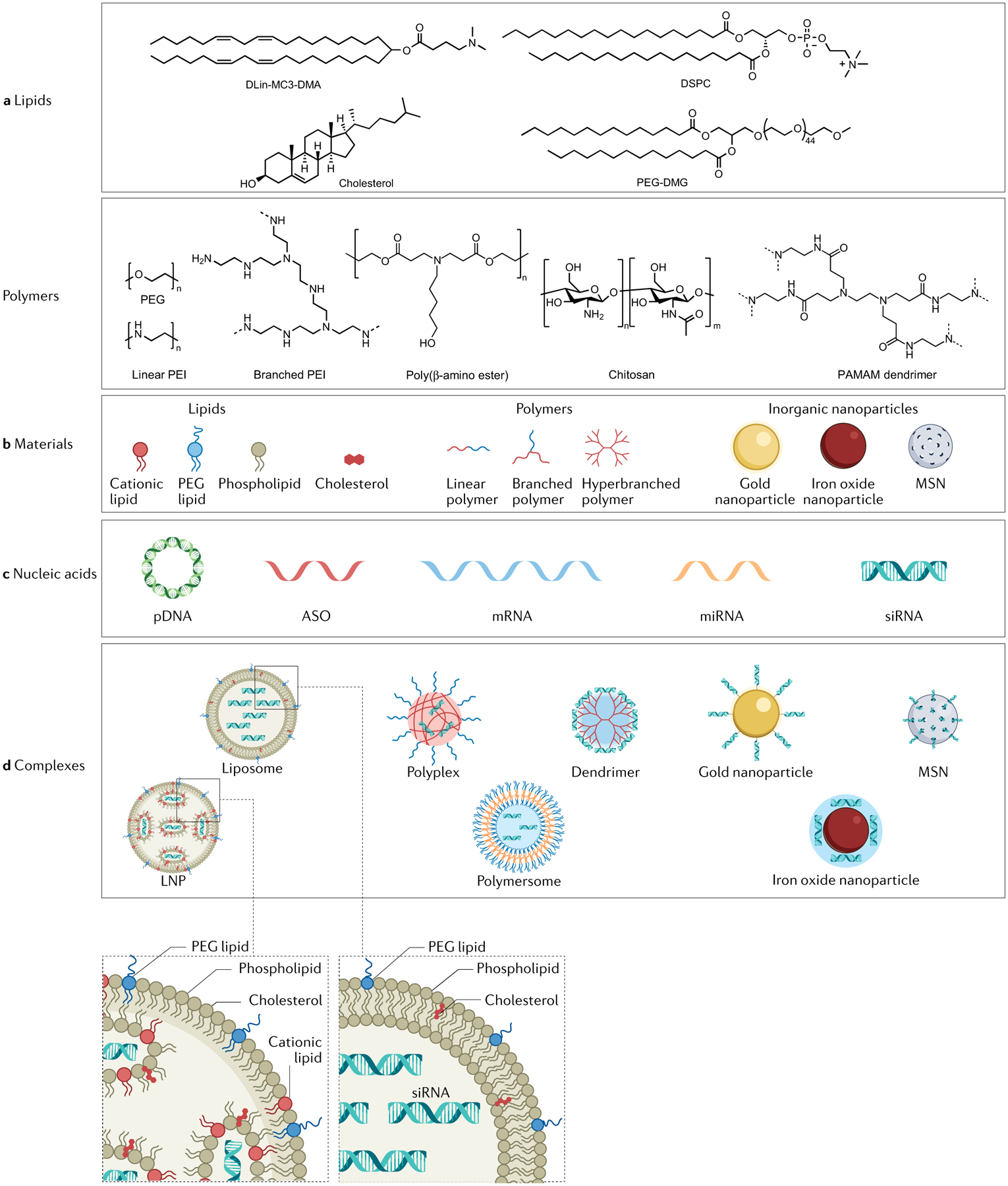
a | Chemical structures of typically used lipids on lipid nanoparticle (LNP) formulations and representative formulation materials for polymeric nanoparticles: cationic lipid (dilinoleylmethyl-4-dimethylaminobutyrate (DLin-MC3-DMA)), phospholipid (1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC)), cholesterol, PEG lipid (1,2-dimyristoyl-sn-glycerol, methoxypolyethylene glycol and polyethylene glycol-dimyristoyl glycerol (PEG-DMG)), PEG, linear and branched polyethylenimine (PEI), poly(β-amino ester), chitosan and polyamidoamine (PAMAM) dendrimer. b | Needed molecular structures for construction of lipid-based nanoparticles (liposomes and LNPs), polymeric nanoparticles (polyplexes, polymersomes and dendrimers) and commonly used inorganic nanoparticles for gene delivery (including gold nanoparticles, iron oxide nanoparticles and mesoporous silica nanoparticles (MSNs)). c | Molecular structures of typical nucleic acids for gene delivery using lipid-based, polymeric and inorganic nanoparticles. d | Structures of commonly used nanoparticle complexes containing nucleic acids. ASO, antisense oligonucleotide; miRNA, microRNA; pDNA, plasmid DNA; PEG, polyethylene glycol; siRNA, small interfering RNA.
Table 1 |.
Synthesis and functionalization methods of common nanocarriers for nucleic acid delivery and their applications in clinical trials
| Nanoparticle type | Synthesis method | Functionalization with nucleic acid | Nucleic acid | Disease type | Clinical trials |
|---|---|---|---|---|---|
| Liposome | Film hydration, solvent injection and reverse phase evaporation | Non-covalent encapsulation | DNA; siRNA | Relapsed or refractory non- Hodgkin lymphoma and diffuse large B cell lymphoma; advanced or metastatic pancreatic cancer | Phase II; phase I/II |
| LNP | Detergent dialysis, ethanol-loading technique, T-mixing technique, microfluidic hydrodynamic focusing and staggered herringbone mixing | Non-covalent encapsulation | siRNA; pDNA | Neuroendocrine tumours, adrenocortical carcinoma and advanced hepatocellular carcinoma; relapsed or refractory leukaemia | Phase I/II; phase I |
| Polyplex | Spontaneous condensation of nucleic acids by polycations | Electrostatic interaction | siRNA; pDNA | Solid tumours and pancreatic ductal adenocarcinomas; colorectal peritoneal carcinomatosis, ovarian cancer, pancreatic adenocarcinoma and non-muscle-invasive bladder cancer | Phase I; phase II |
| Polymersome | Probe sonication, film rehydration, direct dissolution of copolymer, solvent injection, solvent exchange method and phase-guided assembly | Non-covalent encapsulation | NA | NA | NA |
| Dendrimer | Divergent growth method, convergent growth method, double exponential growth technique, double-stage convergent method and hypermonomer method | Non-covalent electrostatic attachment | NA | NA | NA |
| Gold nanoparticle | Turkevich method, the Brust- Schiffrin method, electrochemical method, seeding growth method and others | Gold-thiol conjugation, surface adsorption | siRNA | Recurrent glioblastoma | Phase I |
| MSN | Sol gel, microwave synthesis, hydrothermal synthesis, template synthesis, soft and hard templating methods, and others | Non-covalent encapsulation (hydrogen bonding, physical adsorption, electrostatic interaction or aromatic stacking), surface adsorption, covalent modification | NA | NA | NA |
| Iron oxide nanoparticle | Co-precipitation, thermal decomposition, hydrothermal synthesis, microemulsion, sonochemical synthesis and others | Non-covalent electrostatic attachment, covalent modification | NA | NA | NA |
LNP, lipid nanoparticle; MSN, mesoporous silica nanoparticle; NA, not applicable; pDNA, plasmid DNA; siRNA, small interfering RNA.
Lipid-based nanoparticles
Lipid-based nano-delivery systems are the most widely used non-viral nucleic acid vehicle, mainly due to their stable nanostructure in physiological fluids and their fusion with negatively charged endosomal membranes that allows effective nucleic acid delivery (FIG. 2d). Lipids are amphiphilic molecules that contain a polar head group, a hydrophobic tail region and a linker between the two domains48. Lipid-based nano-delivery systems typically contain other lipid components, such as phospholipids, cholesterol or polyethylene glycol (PEG) (FIG. 2a,b). The main differences between these nanoparticles rely on their lipid constituents, synthesis parameters, the methods used for nucleic acids and encapsulation48. These systems are clinically used or under clinical evaluation to treat genetic disorders (such as cystic fibrosis and propionic acidaemia)48 and solid tumours48 (FIG. 3), being at the forefront of clinical advanced nano-delivery systems for gene therapy following Onpattro approval by the US Food and Drug Administration (FDA)49 and SARS-CoV-2 vaccines50.
Fig. 3 |. Experimental workflow of nucleic acid-loaded LNP delivery in cancer therapies.
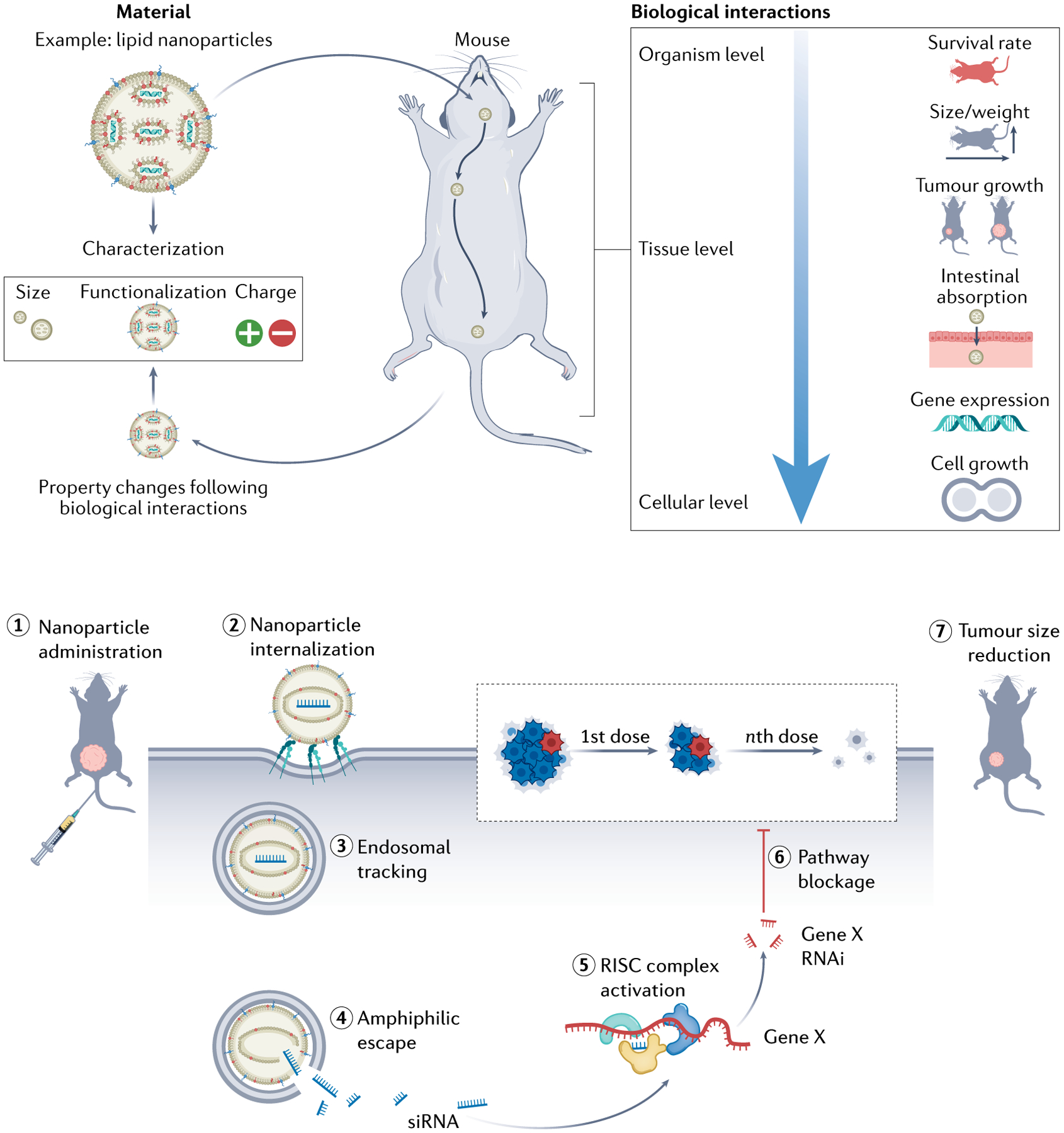
After physico-chemical characterization, nucleic acid-loaded lipid nanoparticle (LNP) formulations are administered in vivo. LNP intrinsic features will determine their biological behaviour at organism, tissue and cellular levels. Upon successful LNP administration (1), internalization (2) and siRNA release/escape (3–4), gene silencing (5–6) and subsequent tumour size reduction (7) are achieved. RISC, RNA-induced silencing complex ; RNAi, RNA interference; siRNA, small interfering RNA.
Liposomes.
Liposomes are composed of phospholipids with polar head groups and non-polar tails along with stabilizers (such as cholesterol), and they spontaneously self-assemble into vesicles owing to their amphiphilic nature. Cationic lipids, such as DOTMA, 1,2-dioleoyl-3-trimethylammonium propane (DOTAP) and zwitterionic 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), are commonly used to form cationic liposomes by exploiting electrostatic interactions between lipids and negatively charged nucleic acids, which increases encapsulation efficiency51. Smaller liposomes have greater likelihood of escaping phagocyte uptake (≤100 nm)52. Strikingly, positive nanoparticle surface charge may lead to non-specific binding of serum proteins and immune stimulation, causing toxiciy32. To this end, a PEGylated cationic liposomal bilayer has been explored as a substitute53. There are three typical methods for liposome preparation: film hydration, solvent injection and reverse phase evaporation54, which are used to achieve effective drug encapsulation, narrow particle size dispersion and long-term stability. Despite the tremendous benefits of liposomes as nucleic acid carriers, they require complex production methods that use organic solvents (for example, solvent injection), which can jeopardize their large-scale production.
Lipid nanoparticles.
LNPs (50–100 nm in diameter) typically comprise ionizable and cationic lipids, cholesterol, phospholipids and PEG lipids (FIG. 2a), among which ionizable lipid plays a major role in protecting nucleic acid from nuclease degradation55. In addition, helper lipids (such as phospholipids and cholesterol) promote formulation stability and membrane fusion34 — approximately 30–40 mol% helper lipid content is required to efficiently entrap siRNA within LNPs56. PEG lipids, which consist of a hydrophilic PEG polymer conjugated to a hydrophobic lipid anchor, can improve the circulatory half-life and stability, preventing LNP clearance57. In particular, PEG lipids of low molecular weights have been shown to decrease non-specific protein adsorption, and re-establish specific binding of receptor-targeting nanoparticles58. In addition, PEG lipid content dictates the particle size59. For example, siRNA-loaded LNPs ranging from 27 to 117 nm in size were obtained by varying the PEG lipid content (0.25–5 mol% lipid), achieving an optimal size of 80 nm for gene silencing potency60.
As opposed to liposomes, LNPs are able to have micellar structures within the particle core44 (FIG. 3). Moreover, LNPs exhibited better kinetic stability and more rigid morphology than liposomes; a large-scale commercial fabrication method can be used to obtain more homogeneous LNPs. LNPs can be subdivided into solid nanoparticles at room temperature or nanostructured lipid carriers, which are a mixture of solid and liquid lipids. Interestingly, ionizable LNPs have a near-neutral charge at physiological pH, but the amine groups on ionizable lipids become protonated and positively charged at low pH, allowing assembly with negatively charged phosphate groups on nucleic acids. After complexation, the pH can be adjusted to a neutral or physiological pH to allow therapeutic administration. After their in vivo injection, ionizable LNPs can extravasate from the bloodstream to target tissues. LNPs can then adsorb onto the cell surface, and enter the cell via endocytosis61. Positively charged ionizable lipids aid in endosomal escape and interact with negative charges on the endosomal lipid membrane, which causes destabilization of the endosome and promotes nucleic acid release62.
The fabrication methods to encapsulate nucleic acids (for example, siRNA, mRNA and pDNA) in LNP systems needs to be fine-tuned in terms of molar ratio and lipid composition, mainly owing to the differences in nucleic acid size and charge that might result in variations of lipid packing and LNP structure63,64. Moreover, targeted LNPs can be fabricated by conjugating surface-attached ligands (such as folate and transferrin) to the surface of LNPs to recognize and bind to specific receptors on cells65.
The first methods of generating LNP–nucleic acid formulations include detergent dialysis and the ethanol-loading technique. Rapid-mixing methods have gained favour for their less laborious requirements as they combine nanoparticle formation and nucleic acid entrapment into a single step while improving nanoparticle homogeneity. For example, a T-mixing technique was used as a fully scalable method for the production of homogeneous nucleic acid LNPs with entrapment efficiencies >90%34. More recently, two microfluidic mixing approaches were designed based on a rapid-mixing technique. Microfluidic hydrodynamic focusing is a continuous-flow technique used to manufacture LNPs in a reproducible and scalable fashion66, and staggered herringbone mixing is the most commercially used67. This technique can improve the control over the mixing process and shorten the mixing time59. All of the previously mentioned methods allow fast mixing of an organic phase containing the lipid components with a liquid phase containing the nucleic acid, leading to high encapsulation of the nucleic acid. LNPs of specific sizes can be accurately fabricated through control of microfluidic operating parameters, such as the flow rate ratio and/or total flow rate68.
Polymeric nanoparticles
Polymeric nanoparticles are produced using natural polymers, namely dextran, chitosan and/or cyclodextrins, or synthetic polymers, such as PLL, PEI, polyamidoamine (PAMAM), and/or poly(lactic-co-glycolic acid) (PLGA) (FIG. 2a), allowing a wide variety of polymeric nanoparticles with different compositions and structures. The most common forms of polymeric nanoparticles are nanocapsules and nanospheres, which have multiple subclasses such as polyplexes, polymersomes and dendrimers44 (FIG. 2d). Polymeric nanoparticles are among the most promising artificial materials for nano-delivery of nucleic acids, mainly owing to their simple synthesis and functionalization, structural versatility, synthetic scalability, high transfection efficiency, gene immunogenicity and good biocompatibility.
Nanocapsules.
Systems with a typical core-shell structure in which the drug is confined to an inner liquid cavity surrounded by a polymeric membrane or shell.
Nanospheres.
Solid matrix systems consisting of a continuous polymeric network in which the drug can be either entrapped in the polymer matrix or adsorbed on the nanoparticle surface.
Polymersomes.
Polymeric vesicles consisting of an aqueous interior surrounded by a polymer shell, which are formed by the self-assembly of amphiphilic block copolymers.
Dendrimers.
A class of dendritic polymers with 3D, nanoscale, hyperbranched, well-defined and monodisperse architectures, comprising a central core, branched repeat units and terminal groups.
Polyplexes contain cationic polymers (for example, PEI, chitosan and biodegradable polyesters) that bind to and condense nucleic acids into small, compact structures through electrostatic interactions. The process of condensation is entropically driven, and upon mixing cationic polymers with nucleic acids, the polyplexes are spontaneously produced. For their preparation, an excess of cationic polymer relative to oligonucleotide is typically used, which generates particles with a positive surface charge and better condenses nucleic acids into smaller-sized nanoparticles35. Nucleic acids are entrapped in the polymer matrix and protected by the polymer chains, which can sterically block the access of nucleolytic enzymes. Additionally, higher packaging stability of polyplexes can be achieved by introducing hydrophobic elements (for example, alkyl groups) to promote particle formation through hydrophobic aggregation61 or by incorporation of covalent cross-linkers within the particle core69.
Polymersomes, also known as polymeric vesicles, are formed by the self-assembly of amphiphilic block copolymers via the hydrophobic effect. The hydrophilic fraction (fhydrophilic) of an amphiphilic block copolymer dictates its self-assembly morphology, and, as a general rule, polymersomes are generated with fhydrophilic of approximately 35 ± 10%70. Polymersome preparation methods are mostly adapted from liposome protocols71, including direct dissolution of copolymer, film rehydration, solvent exchange and probe sonication. Generally, various nucleic acids (such as pDNA, ASOs and siRNA) can be favourably encapsulated into the inner aqueous core of polymeric vesicles owing to the hydrophilic nature of biomacromolecules as block copolymers self-assemble into polymersomes. For example, polymeric vesicles that are formed by film hydration of diblock copolymer poly(1,2-butadiene)-b-poly(ethylene oxide) in aqueous solutions have been applied for the encapsulation of siRNA with an efficiency of about 51%72. Polymersomes with asymmetrical membranes formed from ABC triblock copolymers are applied to increase the volume of the inner aqueous core, improving nucleic acid encapsulation73. Membrane ionizable polymersomes are also used to increase loading efficiency owing to electrostatic interactions between charged membranes and nucleic acids74.
Hydrophobic effect.
The tendency of non-polar molecules and molecular segments in an aqueous medium to minimize their interactions with water molecules and form intermolecular aggregates.
Dendrimers have been synthesized in an iterative sequence of reaction steps by divergent or convergent methods75. Each successive reaction step leads to an additional generation of branching, and the number of repeated cycles is defined as the dendrimer generation. The mass and size of dendrimers can be controlled by tuning the dendrimer generations, whereas the dendrimer surface can be easily functionalized through conjugation of different molecules to the reactive terminal groups, which make them attractive systems for gene delivery applications. Some dendrimers, such as PAMAM, poly(propylene imine) and PLL, have a well-defined number of positive terminal amine groups, allowing the attachment of nucleic acids by ionic interactions. For example, RNA nanoparticles formed by complexation of triple-helix strands of miRNAs with PAMAM G5 dendrimer create a branched sponge-like nanoscopic structure used to regulate key genetic drivers in the breast tumour microenvironment76. Dendrimers with higher generations become more compact and provide a surface with a high density of amine groups for nucleic acid binding77.
For polyplexes and dendrimers, conjugation of hydrophilic polymers (typically PEG chains) to the nanoparticle surface can be applied to improve serum stability and biocompatibility77,78. For all types of polymeric nanoparticle, the attachment of targeting moieties (including antibodies, transferrin, folate and glycosidic moieties) to the nanoparticle surface increases cell uptake and target cell specificity through interactions between targeting moieties and molecules on the target cell’s surface44. Through the incorporation of moieties (for example, PEI, chitosan and imidazole groups) that possess unprotonated amine groups into polymeric nanoparticles, endosomal swelling and rupture may be induced by the proton sponge effect, allowing endosomal escape to be facilitated79.
Inorganic nanoparticles
Inorganic nanoparticles have been studied for nucleic acid delivery and imaging, including gold nanoparticles, silica nanoparticles and iron oxide nanoparticles (FIG. 2d). They can be engineered to exhibit specific sizes, structures and/or geometries44. Among inorganic nanoparticles, gold and iron oxide nanoparticles are generally considered non-toxic nanomaterials40,80. Interestingly, research shows that inorganic nanoparticles can stimulate and/or suppress the immune responses, and their compatibility with the immune system is largely determined by surface chemistry. In fact, the influence of size, solubility and surface modification on the biocompatibility of nanoparticles and their use in biological applications is well known81. Therefore, it is essential to test nanoparticle/biological interactions experimentally and modify the nanoparticles for best biocompatibility with the cell82.
Gold nanoparticles.
Gold nanoparticles exhibit unique optical properties, simple synthesis and surface functionalization, being selectively and cooperatively decorated with nucleic acids via covalent or non-covalent conjugation83. Nucleic acid strands are covalently attached to gold nanoparticle cores (typically 13–15 nm) through thiol moieties84–86. This strategy is used for DNA and siRNA, which can be conjugated directly to gold cores or to polymer-modified gold cores. Gold nanoparticle surfaces can be coated by alternating layers of anionic nucleic acids and polycations (such as PEI or PAMAMs)87 as well as by adding targeting ligands that enable a specific interaction and binding of nanoparticles to cell surface receptors88. Ligand length, density, hydrophobicity and avidity need to be taken into account and optimized for efficient targeting of nanoparticles89.
Mesoporous silica nanoparticles.
MSNs (100–250 nm in diameter) have been used for nucleic acid delivery owing to the good biocompatibility and tunability of the porous architecture40. Typically, nucleic acid molecules are loaded in MSNs through weak non-covalent interactions90. Pore size and surface functionalization play a crucial role in the loading capacity and nucleic acid release rate. MSNs with small pores can provide a tunable release rate for small nucleic acids, whereas larger pores provide higher loading capacity and a faster release rate. For example, MSNs of ~100 nm with small pore size (2.5–5 nm) are suitable for the delivery of small siRNA, and MSNs of ~250 nm with large pore sizes (above 15 nm) are used for loading large pDNA40. Negatively charged surfaces of MSN can be covalently modified into cationic surfaces in order to greatly impact cargo loading, protein adsorption and the release rate91. Approaches such as the addition of different types of cationic macromolecule, including PEI, dendrimers and lipids, have been used to modify MSN surfaces to adsorb and deliver nucleic acids.
Iron oxide nanoparticles.
Iron oxide nanoparticles (composed of Fe3O4 or Fe2O3) possess superparamagnetic properties at certain sizes, demonstrating successful outcomes as delivery vehicles and thermal-based therapeutics. Most of these delivery systems rely on surface-engineered cationic iron oxide nanoparticles that electrostatically interact with anionic nucleic acid cargos. For example, lipidoid-coated iron oxide nanoparticles of 50–100 nm display optimal siRNA delivery activity92. Micellar lipid coatings have been used to adsorb and layer nucleic acids93. Interestingly, iron oxide nanoparticles coated with nucleic acid-loaded lipidoids demonstrated an efficient siRNA and pDNA delivery and in vitro transfection.
Methods to improve stability
One of the greatest challenges of nucleic acid delivery is their poor stability. Indeed, nucleic acids without modification have poor drug-like properties, are unstable in blood and can be quickly degraded by nucleases in extracellular fluids and serum94. Besides the encapsulation of nucleic acids in nanocarriers, chemical modification of nucleotides (including backbone, nucleobase and sugar modifications) can also enhance chemical stability and reduce immunological effects of nucleic acids95–97. For instance, phosphorothioate-modified siRNA drugs have exhibited an improved stability and efficacy by replacing the unstable phosphodiester backbone into a phosphorothioate backbone, which is one of the most used strategies98,99. Unlike unmodified siRNAs, siRNAs with several 2-thiouracil modified units are thermally stable and displayed similar silencing activity100. Both the cap at the 5’ end and the poly(A) tail at the 3’ end of mRNA can enhance the stability and translation efficiency of mRNA101,102. Researchers can also increase the stability of mRNA by modifying the chemical structure of the cap at the 5’ end. In addition, chemical modifications can also be introduced to miRNAs and ASOs to improve their stability.
Results
Even though it is not completely understood how nanoparticle properties interact with biological systems, the pivotal role of these properties on the elicited biological response is widely accepted103,104. In this section, we discuss techniques to characterize the physico-chemical features of nanomaterials and how these properties can modulate the biological response of nanoparticles (FIG. 4).
Fig. 4 |. Overview of the common nucleic acid delivery system characterization methods.
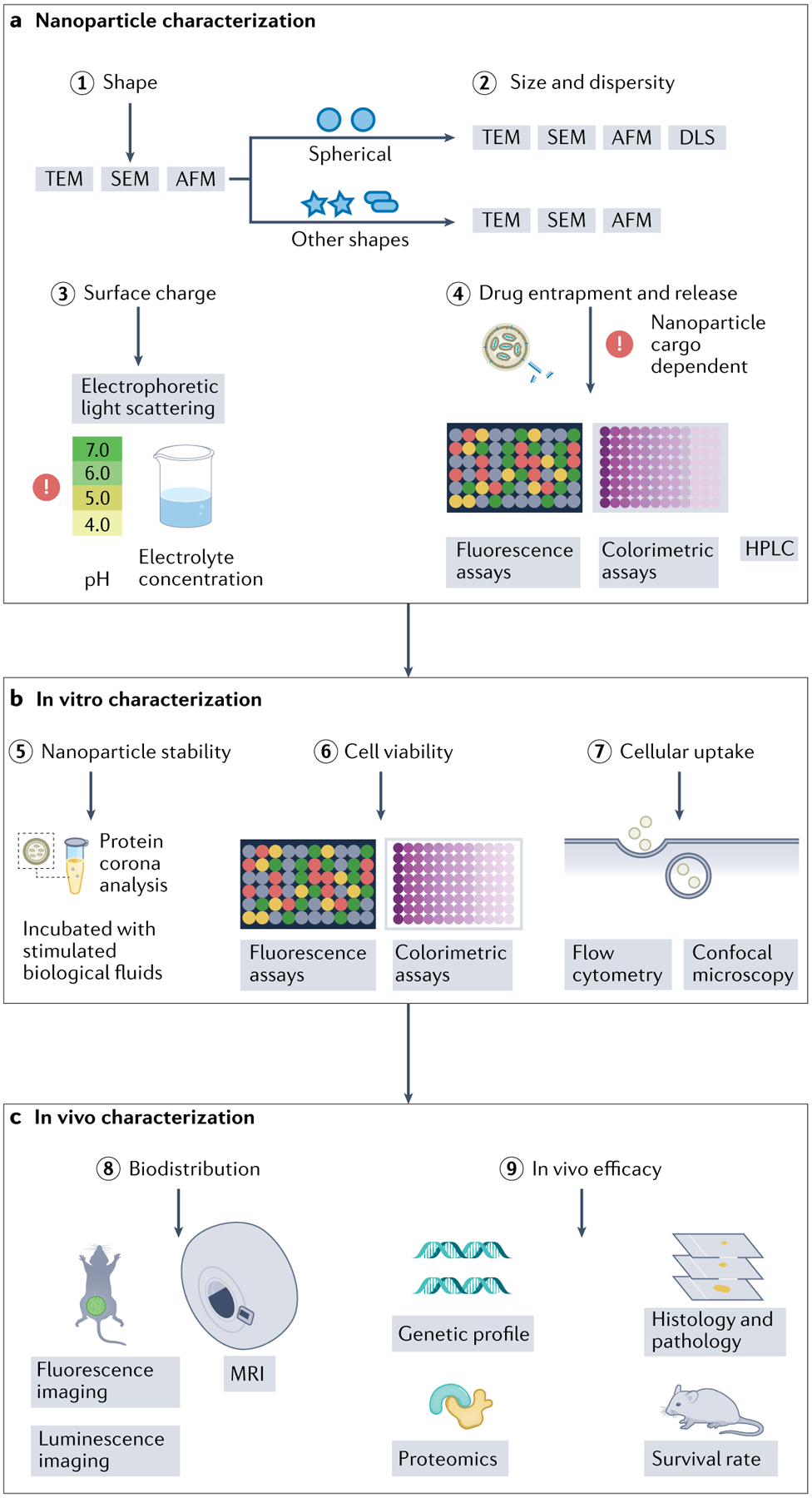
a | Upon nanoparticle synthesis, physical and chemical features are determined, namely morphology (step 1), size (step 2), surface charge (step 3), and nucleic acid loading and profile release (step 4). b | Nanoparticle physico-chemical properties fine-tune their in vitro biological response, which ranges from characterization of the produced protein corona (step 5) to study of nanoparticle cellular uptake mechanisms (steps 6,7). c | Although study of in vivo nanoparticle behaviour will depend on target tissue and/or disease (step 8), some of the most commonly used laboratory procedures are recommended to evaluate nanoparticle efficacy and toxicity in vivo, such as animal survival rate, proteomics, genetic profile, histology and pathology (step 9). AFM, atomic force microscopy; DLS, dynamic light scattering; HPLC, high-performance liquid chromatography; SEM, scanning electron microscopy; TEM, transmission electron microscopy.
Nanomaterial characterization
Nanomaterial composition and synthesis.
As biological interactions are strongly influenced by the composition of nanomaterials, a detailed description of the constituents is very important. However, the protocols for material characterization can be extremely variable and will depend highly on the type of material used. Therefore, it would be too restrictive to impose specific required analysis for some materials, and certainly insufficient for others. There should be a joint effort from academia and the regulators to ensure that novel materials have a detailed synthesis protocol, rigorously assessed by peer-reviewers, and that particularly sensitive or challenging steps are highlighted. Examples of best practices for synthesis and methods have been extensively detailed elsewhere105. Purification methods should also be detailed as the presence of precursor residues in the final nanomaterial may change the biological effect104.
Morphology and size dispersity.
The size and shape of nanomaterials can determine the specific cellular pathways engaged in nanomaterial internalization44,106. Additionally, these properties have influence on the adsorption of biomolecules and biodistribution in vivo107,108. For spherical particles, diameter determination is sufficient; however, for other shapes (for example, cubes or rods), measuring every dimension is fundamental. Moreover, the size of the nanomaterial in vivo should also be considered, as biomolecule adsorption onto the particle surface may alter the particle size109,110. Besides the average size, it is crucial to report size dispersity as samples with the same average size can, in fact, correspond to samples with totally different nanoparticle populations and, thus, have an influence on particle interaction with biological systems111.
Nanomaterial internalization.
By tuning nanoparticle design, their initial recognition at the cell membrane and the further mechanisms of internalization by the cells can be fine-tuned.
Several methods have been used to characterize nanoparticle morphology and size dispersity112 (FIG. 4a). In general, evaluation of the parameters can be performed in suspension (wet) or in a dry state. Unlike inorganic metal nanoparticles, the morphology of organic nanomaterials, in particular their size, can change substantially depending on the wet or dry state. Therefore, the protocol used to assess particle morphology should be provided. Dynamic light scattering (DLS) is the main technique to determine the particle hydrodynamic diameter in suspension112. This method estimates particle size from Brownian motion of the particles in suspension. It can be used in the range from 5 nm to 10 μm, with short acquisition time, requires low sample volume and is compatible with a wide variety of solvents. The DLS size estimation depends on the scattering intensity; thus, for highly polydisperse particle samples, size determination is typically biased towards the larger populations. In this case, size fractionation of the sample is the most effective protocol to prevent biased size estimations113,114. Size characterization can be performed by nanoparticle tracking analysis, based on particle free diffusion in suspension. By means of enhanced contrast microscopy or temporal-resolution video acquisition, diffusion is measured by tracking random movement of single particles115. As nanoparticle tracking analysis relies on single-particle resolution, it is suitable for highly polydisperse populations and can be used to determine hydrodynamic diameters ranging from ≈30 nm to ≈1 μm. However, the wide application of nanoparticle tracking analysis has been limited by the higher costs of equipment compared with DLS and the need for a sample concentration or dilution to around 109 particles/ml for measurement (which may be significantly different from the concentration for the intended application)112.
Brownian motion.
Small particles in a liquid are subjected to random motion in all directions, which is caused by collisions with other molecules. By measuring the particle speed, it is possible to determine the hydrodynamic diameter.
Considering dry state methods for nanoparticle characterization, transmission electron microscopy (TEM) is one of the most important techniques. TEM uses a focused electron beam on a thin sample to produce micrographs of nanomaterials, and can provide high-resolution images with atomic resolution and size diameters spanning from >1 nm to <1 μm (ReFS112,116). TEM also allows particle shape evaluation, single-particle visual inspection and size distribution. Nevertheless, the whole sample preparation, measurement and analysis is laborious, and nanomaterials must be electron transparent and withstand high beam energy and vacuum, which can lead to sample damage, especially in organic, polymeric and hybrid nanoparticles117,118. Cryo electron microscopy enables direct imaging of some nanoparticles, such as protein/RNA complexes, liposomes and viruses that are damaged during the drying process119. Scanning electron microscopy (SEM) imaging is another alternative, which is based on secondary electrons emitted by the surface of the sample upon interaction with the impinging electron beam120. Compared with TEM, SEM uses lower energies beams, which limits the resolution to around 2–3 nm, reducing the chances of beam-induced damage to the sample. For high-resolution images, SEM requires conductive substrates. Therefore, non-conductive samples are typically coated with a thin metallic film (5–10 nm) and this modification of size and surface must be considered during the analysis112,120. Atomic force microscopy (AFM) is a technique based on a scanning probe that enables surface visualization in the atomic range121. A sharp tip at the end of the cantilever is rastered across the surface of the sample and the repulsion and attraction forces between the tip and the atoms of the surface are recorded and used to assess surface topography. AFM can be used for dry and wet samples. Particles in suspension have to be deposited on a flat and smooth surface (such as mica) and dried before analysis. If the nanomaterial is already in a dry state and on a smooth and flat surface, no sample preparation is required112,121.
Zeta potential.
The surface charge of nanomaterials not only influences cellular response but also affects biomolecule adsorption onto the particle surface and the nanoparticle biodistribution44,122,123 (FIG. 4a). Surface charge also plays an important role on particle stability in suspension124. Zeta potential varies with the surrounding environment; thus, pH, electrolyte concentration and the fluid used for the measurements should be detailed111.
Zeta potential.
A physical property that is established on the surface of any particle in a liquid, representing the effective net charge.
Drug entrapment and release.
If the nanomaterial has been developed to carry a molecule, the amount of mole cule that can be entrapped should be calculated and reported as the concentration of molecule/particle or as percentage by mass (FIG. 4a). The amount of carried molecule release from the nanomaterial and its profile release should also be considered and detailed111,125.
Targeting.
Nanomaterials can be designed as targeted systems with increased affinity to specific cells or tissues44. This can be accomplished based on the physico-chemical properties of the nanomaterials, such as size, shape and surface chemistry44, or by covalent126 or electrostatic127 conjugation of targeting moieties onto the surface of the nanoparticles. The targeting efficacy depends on the amount of ligand and the conjugation strategy. Therefore, when nanomaterial is conjugated with ligands on their surface, these must be quantified and reported. Although this task might be challenging, at least (semi-)quantitative analysis should be performed to demonstrate the specificity and the affinity of the nanomaterial for the intended targeted111. More recent methods have been developed to assess the density or the number of biologically functional moieties on the particle’s surface, to provide a deeper understanding of the interaction with their targets. In the future, these assays should be included as reporting standards128.
Biological characterization
In vitro studies.
Nucleic acid-loaded nanoparticles are incubated with primary cells and/or cell lines of interest as the first screening of their toxicity and functionality. Before moving to cell studies, there are several reports that study the stability of nanoparticles in stimulated biological fluids (such as fetal bovine serum) in order to understand the nanoparticle identity interface under physiological conditions129,130 (FIG. 4b). Nanoparticles have a large surface area, which encourages the formation of complex interactions at the bio–nano interface, influencing their reactivity with toxicity targets in the cells.
Bio–nano interface.
The interaction of the nanomaterial surface with the biological microenvironment, which alters their surface characteristics and greatly influences their behaviour in vivo.
In vitro studies are limited to recapitulate biological and physical features of the local microenvironment, therefore adequate cell models should be chosen according to nanoparticle type and composition, nucleic acid category and target disease (FIG. 4b). Experiments involving different cell types (for example, immune cells, patient-derived disease cells and fibroblasts) and recapitulating key features from disease pathophysiology (for example, dynamic culture conditions, biomechanical forces and architecture) have been developed using 3D tissue models, such as organoids131,132 or organs-on-chips133. These models have been shown to better mimic the in vivo scenario, and thus they are now considered key to successfully determine nanoparticle drug efficiency and cytotoxicity134.
The cellular uptake of the nucleic acid-loaded nanoparticle can be studied by TEM, confocal microscopy and/or analytical flow cytometry, which are the methods of choice to determine whether the nanoparticles are taken by the cells135. The impact of nucleic acid delivery on cells of interest can be evaluated by the expression level of the target gene. Moreover, it can also be assessed through the proliferation, migration and survival of the studied cells as a function of nucleic acid-loaded nanoparticle dose by performing light microscopy and/or viability assays (for example, MTT).
Preclinical in vivo models.
The nanomedicine community have been discussing the importance of personalized therapies; thus, the choice of the animal model needs to reflect the biological scenario, which can include gender differences136. Moreover, the administration route to deliver the nanoparticle systems needs to be correctly chosen owing to their impact on nucleic acid-loaded nanoparticle pharmacokinetics, in vivo biodistribution and overall therapeutic efficacy. Interestingly, local administration of hydrogel patches embedded with siRNA–gold nanospheres was demonstrated to improve nanoparticle stability and minimize side effects, while promoting complete colon cancer regression135. Also, it was reported that nanoparticle type, shape, charge and size impact the in vivo biodistribution130,137,138. Whereas smaller nanoparticles (10–20 nm) are rapidly eliminated by renal clearance139, the mononuclear phagocyte system can uptake nanoparticles larger than 100 nm owing to nanoparticle opsonization upon contact with blood, then being filtered by the spleen and sequestered in the liver140,141. In addition, the amount of nanoparticles that reaches the target site is of utmost importance to evaluate the efficacy of the studied nanomedicines111. Therefore, the percentage of injected dose delivered per gram of tissue and data on weight of organs measured need to be reported. After in vivo studies, it is recommended to analyse the sample genetic profile in response to the applied treatment and respective gene pathways (FIG. 4c).
Applications
Nucleic acid-based therapeutics are revolutionizing the pharmaceutical landscape, as they can target the genetic bases of diseases rather than the downstream proteins, offering long-lasting or even potentially curative treatments for many intractable conditions. In this section, we discuss the application of nanoparticle-based nucleic acid delivery in several key areas and offer some representative examples.
Bioanalysis
Nucleic acid–nanoparticle conjugates have found broad applications in ultra-sensitive detection of various analytes, such as nucleic acids142, proteins143, small molecules144 and metal ions145. One prominent example is spherical nucleic acid (SNA) conjugates, which were initially prepared by covalently attaching oligonucleotide strands onto gold nanoparticles. In the presence of oligonucleotide linkers containing complementary sequences at multiple sites, these SNA–gold nanoparticles form agglomerates in solution, altering the optical properties of the gold nanoparticles (such as surface plasma resonance). This principle has been exploited for colorimetric detection of RNA142 and DNA146 targets in solution, enabling diverse bioanalytical applications such as the rapid naked-eye diagnosis of SARS-CoV-2 infection without requiring advanced instruments142 (FIG. 5a). Notably, SNA–gold nanoparticles exhibit very narrow transition (melting) temperatures, which allows differentiation of targets with a single base mismatch146. Array-based gene detection platforms were created by immobilizing oligonucleotides onto a chip and applying the use of SNA–gold nanoparticles, both of which contained sequences complementary to different regions of the target gene. Taking advantage of the gold nanoparticle-mediated catalytic reduction of silver ions onto its surface, the gold nanoparticle signal could be amplified and read out through various means such as light scattering147, surface-enhanced Raman spectroscopy (SERS)148 (FIG. 5d) and electrical conductivity with high sensitivity147 (FIG. 5b). Automated systems incorporating this array-based gene detection technology (such as VERIGENE) have been commercialized for rapid diagnosis of infectious diseases in clinical settings149.
Fig. 5 |. Nanoparticle-based nucleic acid platforms for diagnosis applications.
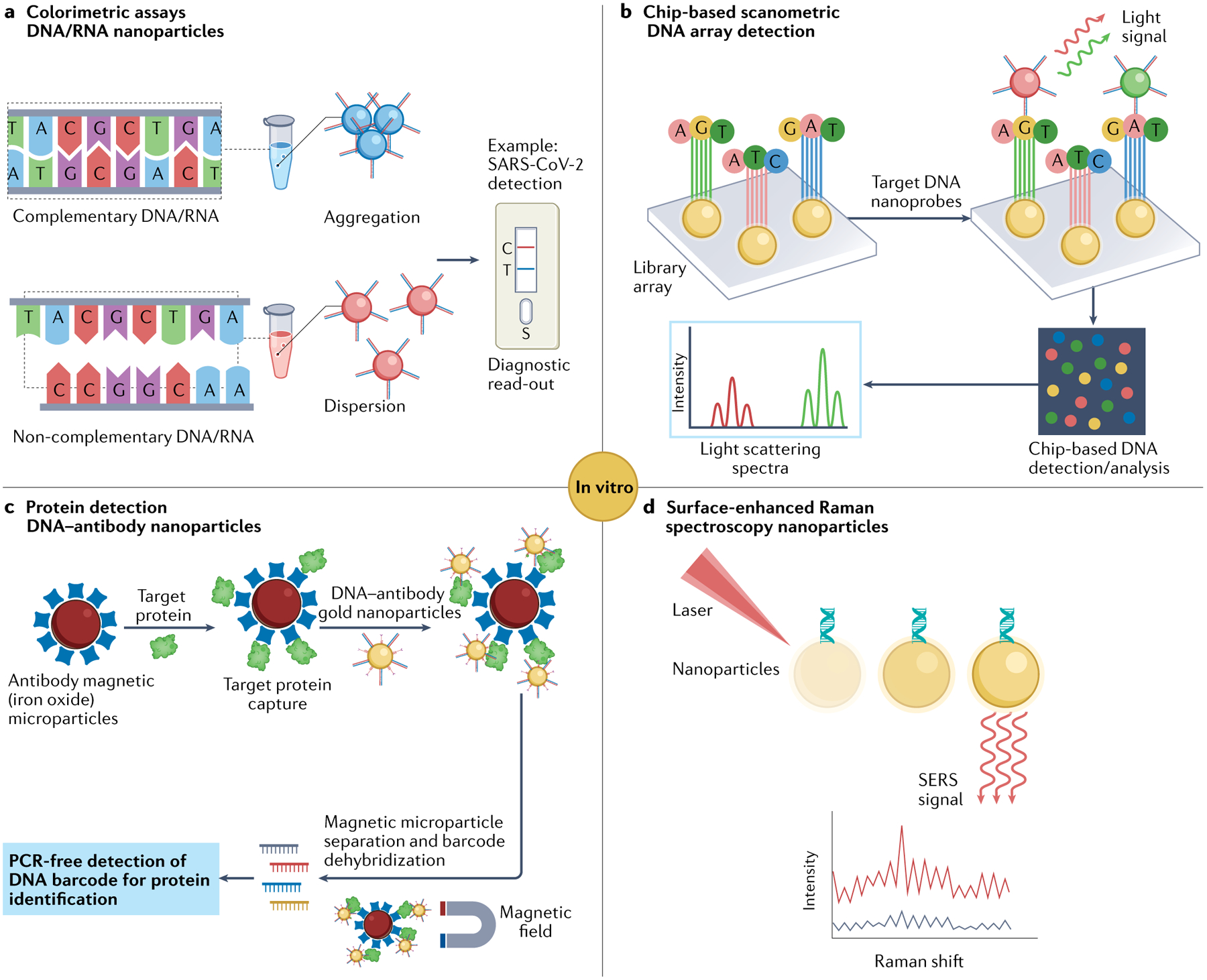
a | Antisense oligonucleotide (ASO)-capped gold nanoparticles aggregate in the presence of target RNA sequence of the virus, leading to change in surface plasmon resonance of gold nanoparticles and, thus, observed colour142. b | Chip-based scanometric DNA array detection using oligonucleotide-modified gold nanoparticles and silver enhancing for amplification of light scattering signal147. c | Ultra-sensitive protein detection using antibody-functionalized magnetic microparticle and DNA–antibody co-functionalized nanoparticle for signal amplification143. d | Surface-enhanced Raman spectroscopy (SERS)148.
Ultra-sensitive detection of proteins by signal amplification was also realized by implementing a two-particle system comprising nanoparticles co-functionalized with DNA duplexes and antibodies as well as magnetic microparticles coated with antibodies143. In the presence of target proteins (for example, antigen), a sandwiched complex forms, in which both the nanoparticle and the microparticle are bound to the antigen through the antibody–antigen interaction. This complex can be easily separated using a magnetic field, followed by dehybridization of DNA duplexes to release the DNA barcodes for detection (FIG. 5c). Because each nanoparticle carries many DNA barcodes per protein binding event, there is substantial signal amplification, and the target protein can be detected with a sensitivity six orders of magnitude higher than conventional protein assays commonly used in the clinic. This method was later successfully applied to detect a soluble protein biomarker (amyloid-β-derived diffusible ligands) of Alzheimer disease in human cerebral spinal fluid, which conventional ELISA or blotting assays failed to detect owing to its extremely low concentration150. Besides nucleic acids and proteins, nucleic acid–nanoparticle conjugates can be used to sensitively detect other analytes such as small molecules and metal ions by leveraging DNA-binding molecules151, aptamer–substrate interactions144 and metal ion-dependent nuclease activity152.
Gene silencing
Gene silencing technologies based on ASOs and RNAi are transforming the pharmaceutical industry, as they allow targeting of virtually any disease-causing genes (FIG. 6a). Nanoparticles for ASOs and RNAi (such as siRNA and miRNA) delivery include LNPs153, polymeric nanoparticles154, lipid–polymer hybrid nanoparticles155 and inorganic nanoparticles156. Systemic delivery of ASOs or RNAi by nanoparticles has been explored for the treatment of many indications such as cancer and cardiovascular, neurological, infectious and genetic diseases157,158. Owing to the preferential hepatic uptake of nanoparticles after systemic administration, the liver has been an important target organ for nanoparticle-mediated ASO and RNAi therapies, which have been used to treat various liver-related disorders such as hyperlipidaemia159 and viral infection160. Nanoparticle accumulation and gene silencing in solid tumours can be achieved by non-targeting nanoparticles through the enhanced permeability and retention effect. Stimuli-responsive nanoparticles were developed to enable more controllable delivery to lesions in response to various endogenous and exogenous triggers125. For example, tumour microenvironment-responsive nanoparticles sensitive to acidic pH, enzymatic and hypoxic conditions in the tumour microenvironment were exploited to promote penetration, cellular uptake and cytosolic delivery of siRNAs in tumours161. ASO/siRNA delivery to specific cells and subsequent gene silencing in tissues of interest can be improved by incorporation of targeting ligands and by precise control of their protein coronas in vivo. Moreover, the local administration of siRNA-loaded nanoparticles has been shown to induce effective and sustained gene silencing in the reproductive mucosa, making them promising microbicides against sexually transmitted diseases (for example, HIV)162. siRNAs and ASOs encapsulated in nanoparticles can also be shielded from the harsh environment of the gastrointestinal tract and protected from premature degradation, allowing them to be delivered orally to treat gastrointestinal diseases163.
Fig. 6 |. Nanoparticle-based nucleic acid delivery systems for therapeutical applications.
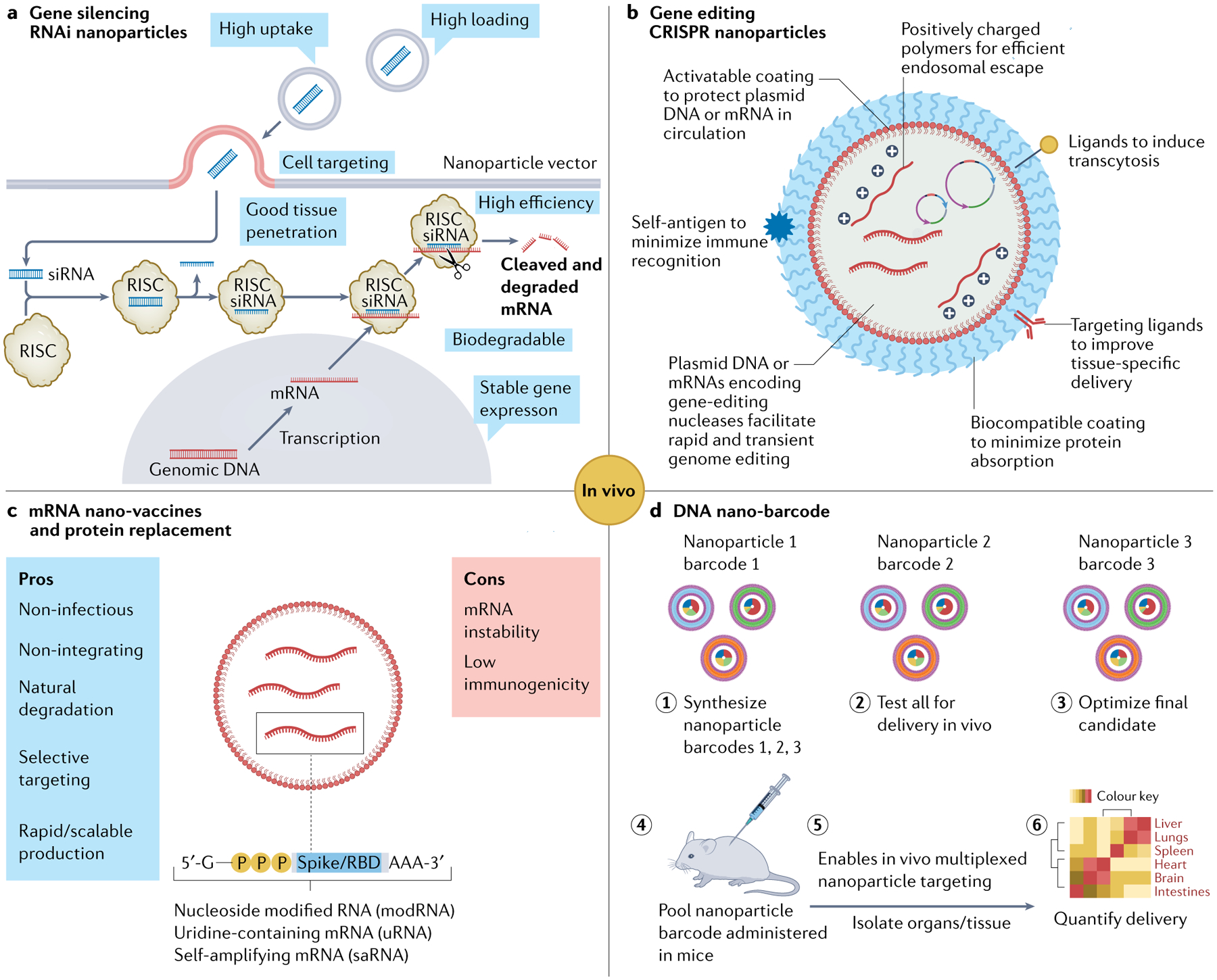
a | Small interfering RNA (siRNA) nanoparticle-mediated gene silencing163. b | Design and assembly of nucleic acid-based CRISPR nanoparticles169.c | Pros and cons of mRNA lipid nanoparticles (LNPs) for vaccination and protein replacement therapy171. For example, Pfizer/BioNTech vaccine or individual LNP encapsulated mRNA vaccines (nucleoside modified RNA, one uridine-containing mRNA, one self-amplifying RNA (saRNA)) encoding spike protein or receptor-binding domain (RBD). d | Specific DNA sequences can be incorporated in many different nanoparticles (1) as DNA barcodes to assess their in vivo delivery profiles (2–3) at once in a single experiment184. DNA barcode system enables multiplexed nanoparticle-targeted studies in vivo (4–6). RISC, RNA-induced silencing complex; RNAi, RNA interference.
The advanced chemical modifications have dramatically improved the enzymatic stability of ASOs and siRNAs. All of the clinically approved ASO/siRNA drugs are chemically modified, and do not require the use of nanoparticles except for Onpattro. In addition, chemical conjugation with targeting ligands such as N-acetylgalactosamine (GalNAc) can increase the specific delivery of nucleic acids to cells and tissues of interest, as exemplified by the FDA approval of three GalNAc–siRNA conjugates for the treatment of acute hepatic porphyria (givosiran), primary hyperoxaluria type 1 (lumasiran) and hypercholesterolaemia or mixed dyslipidaemia (inclisiran)164. Moreover, a robust pipeline of GalNAc-conjugated therapeutics is progressing into the clinic for liver diseases including hepatitis B virus infection, transthyretin-mediated amyloidosis and α1-antitrypsin deficiency. Beyond the success of chemical modification/conjugation strategies, nanoparticle delivery is expected to be complimentary and significantly expand these gene silencing technologies to many other biomedical applications.
Gene editing
Modern gene editing tools rely on zinc-finger nucleases, transcription activator-like effector nucleases and CRISPR–Cas9. Unlike gene silencing (which is usually transient), gene editing technologies allow permanent modification of DNA at specific sites. Nanoparticles were explored to deliver these nuclease proteins directly for gene editing165. Alternatively, pDNAs or mRNAs encoding gene-editing nucleases can be loaded into nanoparticles, which allows delivery of a relatively small number of gene copies to achieve the same editing efficiency. Delivery of pDNA or mRNAs for gene editing may be more challenging than delivery of ASOs or RNAi agents. pDNA and mRNAs are much larger, so they require carriers that can accommodate bulky nucleic acids while providing effective cytosolic trafficking to achieve gene editing efficiency166,167. Currently, LNPs and polymeric nanoparticles are the most widely explored non-viral platforms, owing to the vast tunability of their chemical compositions for screening and optimizing formulations168, and their high gene packaging capacity that is unattainable by conventional viral vectors.
Delivery of gene editing tools in the form of DNA has several potential drawbacks, including the risk of insertional mutagenesis and persistent nuclease expression, which could lead to off-target editing and genome instability. By contrast, delivery of mRNA ensures transient expression of the nucleases and minimizes the risk of off-target effects and genome insertion. In a mouse model of transthyretin-mediated amyloidosis, a single injection of chemically modified single guide RNA (sgRNA) and Cas9 mRNA encapsulated in LNPs induced nearly 70% gene editing efficiency in the liver with >97% decrease in serum transthyretin levels for more than a year. Initial human results (NCT04601051) have revealed profound reductions in serum transthyretin levels after a single dose without safety concerns. Delivery of mRNA encoding genome-editing agents by targeted nanoparticles may allow reprogramming of specific cell subtypes (for example, T cells and macrophages) in a heterogeneous pool, eliminating the elaborate and costly ex vivo cell purification process in conventional cell therapies and enabling applications such as in situ programming of circulating T cells for chimeric antigen receptor (CAR) T cell therapy169 (FIG. 6b). Interestingly, nanoparticle composition can be tuned to deliver organ-selective Cas9 mRNA/sgRNA for gene editing in extra-hepatic tissues, which broaden the development of new treatments for tissue-specific diseases170.
Protein replacement
Protein replacement therapy can correct the dysfunction that produced the disease in the first place, with the delivery of protein-coded mRNA being one of the most promising methods171. Systemic delivery of mRNA utilizing LNPs was evaluated as protein replacement therapy in preclinical models of haemophilia, liver and lung fibrosis, and metabolic diseases171 (FIG. 6c). Successful roduction of human erythropoietin in non-human primates was achieved via systemically administration of mRNA LNPs172. Some mRNA nanoparticles have also been developed for use in producing therapeutic antibodies such as antibodies against HIV-1 in mouse liver. A clinical trial (NCT03829384) by Moderna is testing the systemic administration of mRNA nanoparticles encoding secreted antibodies for rapid protection against chikungunya virus infection. More recently, mRNA nanoparticles have been used to restore tumour suppressors such as PTEN (phosphatase and tensin homo-logue deleted on chromosome 10) and p53, which could inhibit tumour growth and sensitize tumours to other therapies such as chemotherapy and immune checkpoint blockade therapy173–175.
Vaccines and immunotherapy
Nucleic acid-based vaccines have shown remarkable potential, as manifested by recent FDA approval for emergency use of two mRNA vaccines (BNT162b2 by Pfizer/BioNTech and mRNA-1273 by Moderna) for combating the COVID-19 pandemic. Their impressive efficacy can be largely attributed to the successful LNP delivery platforms, which are used extensively for the delivery of other mRNA vaccines176,177. For example, saRNA encoding the SARS-CoV-2 spike protein encapsulated in LNPs achieved high and dose-dependent SARS-CoV-2-specific antibody titres in mouse sera, leading to neutralization of pseudotyped and wild-type virus14. The potency of mRNA vaccines could be boosted by nanoparticle-mediated co-delivery of immunostimulatory adjuvants178. Recently, lipid adjuvant-assisted mRNA vaccines were developed by high-throughput screening of ionizable or cationic lipids, which can simultaneously mediate mRNA delivery and provide adjuvant stimulation via the STING pathway or Toll-like receptor 4 pathway179. Beyond LNPs, polymeric nanoparticles and some inorganic nanoparticles have also been adopted to deliver nucleic acids for vaccination (FIG. 6c).
In addition to vaccine development, nucleic acid nanoparticle delivery has also been explored for immuno-oncology. For instance, mRNA nanoparticle-mediated restoration of tumour suppressor PTEN in PTEN-mutated/null tumours induced autophagy and triggered cell death-associated immune activation175. The combination of PTEN mRNA nanoparticles with an anti-PD1 antibody elicited a highly potent antitumour effect in multiple PTEN-mutated/null cancer models and immunological memory175. T cell-based cancer immunotherapies are emerging as powerful tools for cancer therapy. In vivo nanoparticle co-delivery of plasmids encoding a 194–1BBz CAR and a piggyBac transposase to T cells induced CAR expression and expansion, a system now moving into clinical trials180. Although systemically administered checkpoint inhibitors (such as anti-CTLA4, anti-PD1 and anti-PDL1 antibodies) have shown effectiveness in certain cancers, many other tumours are resistant to them. Agonizing T cell co-stimulators, such as B7 immunoglobulin and tumour necrosis factor receptor family members (for example, OX40), in combination with checkpoint blockade improve patient responses181. Intratumoural administration of IL-23, IL-36γ and OX40L triplet mRNA LNPs elicited durable anticancer immunity, and had good efficacy in combination with checkpoint blockade in models otherwise resistant to systemic immune checkpoint inhibition182. In addition, delivery of nucleic acid-based immunostimulatory adjuvants (such asCpG) via nanoparticles has been considered a promising strategy for inducing a cascading adaptive immune response183.
Nucleic acid nano-barcodes
Rationally designed DNA sequences can be tagged to specific nanoparticles as DNA barcodes to assess their in vivo biodistribution via deep sequencing, allowing hundreds of chemically diverse nanoparticle formulations to be administered as a pool and screened simultaneously in a single animal184 (FIG. 6d). This robust DNA barcode technology is being commercialized by GuideTx (now part of Beam Therapeutics, Inc.) for high-throughput in vivo screening of nanoparticles for targeting different cells. Similarly, mRNA barcodes could also accelerate in vivo screening, particularly for mRNA nanoparticles185. In addition, owing to the strong and highly specific Watson–Crick base pairing, DNA-functionalized nanoparticles could be used as building blocks to construct programmable nanoparticle superstructures to be explored for their unique physical, chemical and biological properties186. DNA is emerging as a promising high-capacity data storage medium, and encapsulation of DNA in nanoparticles has been pursued to prolong the storage time and protect data integrity.
Reproducibility and data deposition
Significant efforts have been undertaken by the community to develop standards for nanomaterial design, experimental report accuracy and data deposition. However, their implementation remains challenging111,187 (FIG. 7). To improve data reproducibility and result transparency, several important features should be carefully discussed.
Fig. 7 |. Key limitations of nucleic acid-based nanoparticle clinical translation and potential strategies to tackle their limitations.
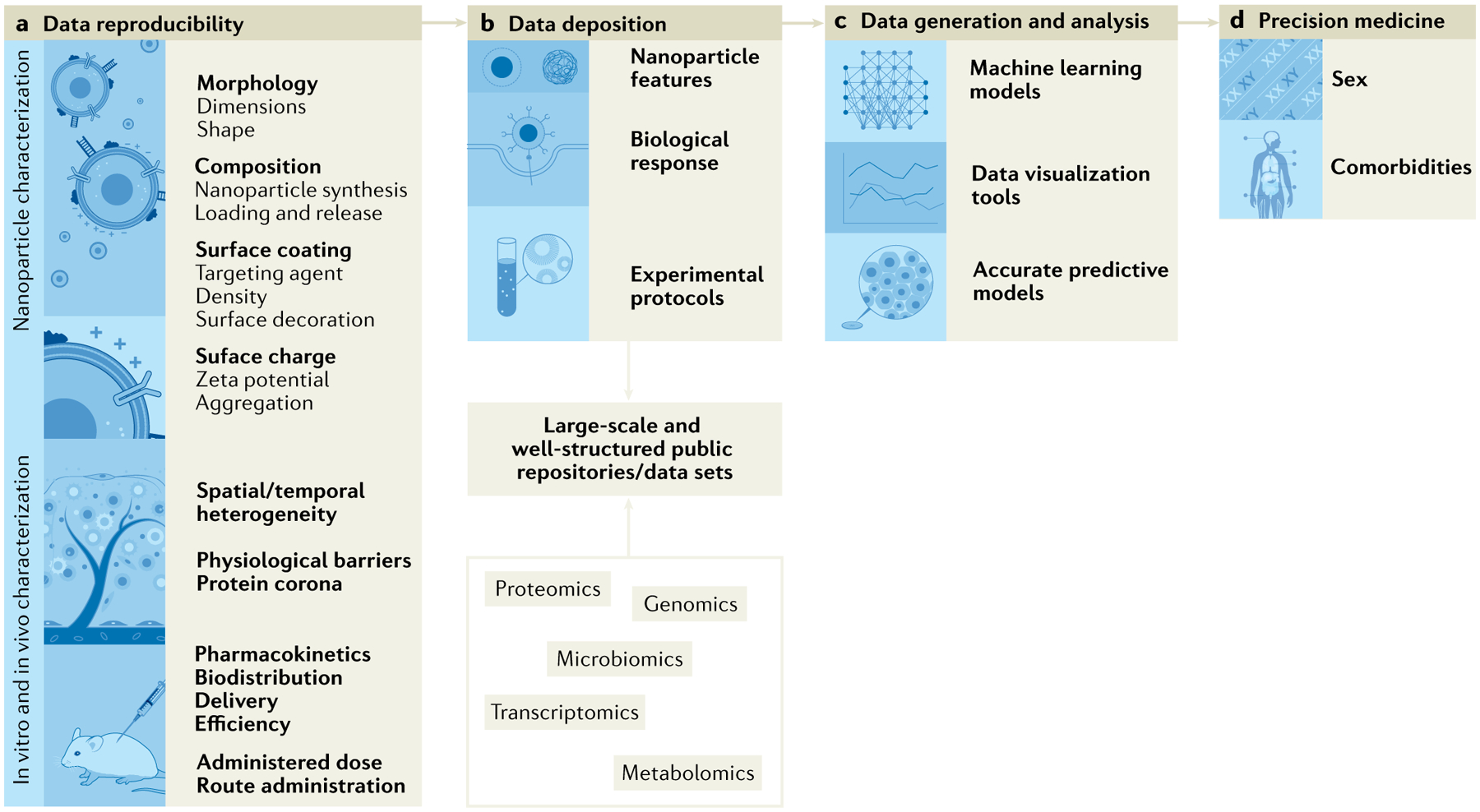
a | Scientific community efforts focus on the importance of standardized nanoparticles and bio–nano interface characterization, and their impact on data reproducibility111. b | Development of large-scale and well-structured repositories is challenging owing to the broad procedures, terms, abbreviations and acronyms of the scientific field111. c | Machine learning will allow generation of predictable models for in silico nanoparticle design taking into account physico-chemical properties (size, zeta potential) and their in vitro and in vivo response47,205. d | Nanoparticle design is envisioned to focus on precision medicine therapies, instead of one-size-fits-all strategies44.
Nano-engineered materials are designed to overcome numerous biophysical barriers as well as physiological barriers inherent to the complex cellular microenvironment188,189. Moreover, the disease genetic and phenotypic variation amongst patients along with the spatial and temporal heterogeneity in the same patient provides additional challenges for the design of such delivery systems190,191. For example, shear forces, nucleic acid degradation and protein adsorption that depend on the previously discussed parameters alter the nanoparticle biodistribution, functionality and drug delivery profile, which makes them hard to isolate and characterize broadly. Indeed, nanoparticle in vivo fate and behaviour are more complex in comparison with conventional drug formulations, hindering their data reproducibility and, thus, their successful clinical translation (FIG. 7a). In spite of the underexplored heterogeneity, basic/translational nanotechnology studies are still focusing on a one-size-fits-all solution instead of designing precision medicine therapies that reflect patient-specific data and particular physiological barrier features44.
In the literature, the lack of physico-chemical data (such as surface charge, coating or loading materials, encapsulation efficiency) and/or preclinical data (for example, biodistribution, off-target tissues, volume tumour reduction that is not normalized to the naked/empty nanoparticles) jeopardizes the comparison between nanomaterial studies and the fundamental understanding of the bio–nano interface. In addition, non-uniformity in the reports by describing the administered dose of nanoparticles as milligrams of material per kilogram of animal weight, drug/nanoparticle/material quantity or, even, drug/nanoparticle/material concentration continues to generate additional experimental inconsistencies. Recently, the community has proposed the use of minimum information reporting in bio–nano experimental literature (MIRIBEL) criteria to ensure that the researchers provide sufficient general information as well as key characterization parameters111. Nevertheless, broader uptake in the field has been widely discussed192. The main concerns of the research community include the practicability and research costs. It is not easy for poorly resourced laboratories in developing countries or young faculty members with limited laboratory funding and interdisciplinary collaborations to meet all of the MIRIBEL criteria, which are often dependent on the research field. There are deep differences between academia and industry settings that greatly affect data reproducibility reports192–195.
The development of large-scale and well-structured repositories are limited by the interdisciplinary nature of the nanotechnology field that ranges from medicine to chemical procedures and terms, as well as the use of abbreviations and acronyms that can have multiple meanings (FIG. 7b). Also, the low number of open software and repositories that describe experimental protocols, methods and biomaterials hampers the usability of biological, experimental and computational data. The support of text mining by defined terminologies and structured data enables the retrieval of relevant information via automated processing, which is crucial to promote data sharing and reuse. After this optimization, machine learning will be able to model the best-fit nanoparticle features, to obtain a specific biological behaviour (FIG. 7c). Finally, it is of utmost importance that software is kept under a public licence that allows the community to use it without further restrictions. The same applies to data sets, which are recommended to be publicly available through version control repository (such as GitHub) so the community is able to validate and contribute to collected data.
Limitations and optimizations
Effective gene delivery for therapeutic purposes remains a translational challenge. In general, oligonucleotides are large and polyanionic molecules that are unable to instantly cross the plasma membrane. Additionally, before reaching their target these nucleic acids must resist nuclease degradation in vivo and avoid the reticuloendothelial system and renal clearance196. Viral and non-viral nanocarriers have been proposed to overcome these limitations. Most of the currently approved cancer gene therapies are based on viral vectors. For instance, Gendicine (2003, China), the first to be approved, is based on recombinant adenovirus to express the tumour suppressor p53 (ReF.197). Although a large number of clinical trials are underway, assessing both viral and non-viral delivery strategies198, non-viral delivery systems have achieved less robust outcomes compared with viral approaches. This has been attributed to challenges in crossing the physiological barriers, entering the cell/nucleus and difficulties in endosomal escape199.
Moreover, oligonucleotide-based therapies do not normally cross the blood–brain barrier, which is an additional obstacle. Thus, the physico-chemical properties (such as dimension, shape, surface charge, targeting agent density and composition) of these non-viral delivery systems have a significant influence on nanomaterial stability, biodistribution and pharmacokinetics, which leads to different therapeutic efficacies and biological responses60,200–202. Strikingly, lyophilization methods have been explored to improve nanoparticle stability for storage and transport, lipid and polymer-based nanoparticles being the most prone to aggregate and to become unstable in solution44,203. So far, most of the approved gene therapies have focused on local delivery, for instance to the spinal cord or the eye, or to the liver, as it is a highly perfused organ with a discontinuous sinusoidal endothelium enabling rapid uptake of the vector and oligonucleotides204.
Although the relevance of physico-chemical properties of non-viral vectors is extremely important, the design of these platforms has become very complex. Not only is their characterization difficult but there is also non-uniformity and often insufficient physico-chemical data in the research reports. The lack of standardization in nanomedicine research reports jeopardizes the comparison of different nanomaterials and the assessment of engineering design approaches, which in turn jeopardizes the comparison with other studies, the fundamental understanding of the bio–nano interface and successful translation to the clinic205. Concerning the biodistribution, it is assumed that non-viral nanocarriers accumulate preferentially in certain tissues, based on the perfusion. For instance, cancer gene therapy has been based mostly on the enhanced permeability and retention effect. However, it is currently known that this phenomenon differs between tumours, and more specific and active delivery strategies should be designed206,207.
Combined efforts from the research community to improve data transparency and reproducibility are considered crucial to overcome some of the limitations, bringing hope for the development of precision genetic therapies for rare diseases or conditions with limited therapeutic options (FIG. 7d).
Outlook
The introduction of consensus in the scientific community, namely on experimental and reporting standards, and the development of ambitious nucleic acid delivery strategies will further push nanomedicine technological clinical barriers. In this section, we look at major challenges that the research community has to face in the next decade.
Challenges
Going beyond the liver.
One of the challenges of nanoparticle delivery of nucleic acids is organ selectivity and/or cell selectivity, mainly owing to the numerous factors that affect the biodistribution of nanoparticles61. The current clinical success of nucleic acid nanotherapeutics has been limited to either liver targeting or local administration. Thus, strategies to improve systemic nanoparticle delivery efficiency to extra-hepatic tissues and organs are highly desired. Such strategies are being actively pursued by both academia and industry. For example, by tuning the composition of LNPs, systemic delivery of Cas9 mRNA/sgRNA for lung-selective gene editing was demonstrated168,170. High-throughput in vivo screening also revealed the important role of lipid component (for example, cholesterol variants) in controlling RNA delivery to hepatic endothelial cells208. More efficient polymer components can be modified on these nucleic acid delivery systems to minimize non-specific protein adsorption, inhibiting the formation of protein corona. Compared with the rapid advance in developing and screening new biomaterials and nanoparticle formulations, the fundamental understanding for nanoparticle-mediated tissue tropism lags. It has been shown that the preferential targeting of the clinically approved siRNA LNPs (Onpattro) to the liver hepatocytes is mediated by adsorbed apolipoprotein E (ApoE) on the nanoparticle surface after intravenous administration49. More efforts for exploring nanoparticle–serum protein interactions are therefore essential, as the protein corona formation on nanoparticles may determine their tissue and/or cell selectivity. It is expected that new understanding of the relationships between nanoparticle physico-chemical properties and their physiological behaviours will facilitate rational development of more effective and selective nucleic acid delivery vehicles to specific tissues/cells.
Lack of standardization and reproducibility in experimental data.
The implementation of standardized reports on nanoparticles and bio–nano interface characterization has been challenging. Although many bottom-up preparation methods enable lipids, polymers and some inorganic nanoparticles to encapsulate or bind with nucleic acids into nucleic acid nanocarriers, these methods are usually variable and not homogeneous, producing a multidispersed population of nanoparticles209. Bottom-up methods can aggravate this inhomogeneity by increasing the scale from that of small-scale bulk mixing environments used in a laboratory to higher volumes, especially as mixing conditions vary with different kinds of processing instruments. To produce more accurate nucleic acid-loaded nanocarriers, increasing interest in the drug delivery field has focused on large-scale continuous production such as microfluidic mixing and extrusion techniques (for LNPs and some polymeric nanoparticles) that can be more easily integrated in a homogeneous way. In addition, to ensure precision in clinical outcomes, homogeneity of nucleic acid nanocarriers is vital and may need improvements in both purification and characterization methods.
Bottom-up preparation methods.
Chemical processes to fabricate nanomaterials from single atoms or molecules.
Exploit gene nanoparticle delivery as a more accessible and affordable clinical strategy.
The approval of SARS-CoV-2 mRNA vaccines by the FDA and their mass usage around the world have paved the way for positioning nucleic acids, and principally synthetic mRNA molecules, as a promising mainstream vaccination therapeutic technology. Nucleic acid vaccines present many advantages compared with conventional vaccines owing to their potential to streamline vaccine discovery and development while possessing rapid development capacity, versatility, efficacy, safety profile, scalable production and cost-effectiveness. For this reason, there is great excitement towards future possibilities of utilizing and expanding nucleic acid vaccines to formulating prophylactic and therapeutic treatments for various diseases, some of which have not been successfully tackled by conventional vaccines. It should be noted, however, that even before the COVID-19 outbreak, several vaccines were already in the pharmaceutical companies’ pipeline210–212. Among the top priorities of global importance to be addressed and developed are cancer vaccines, designed to induce a specific and long-lasting immune response against tumour antigens using precision medicine methodologies. Another essential class are preventive and therapeutic vaccines against infectious viral or bacterial diseases and eukaryotic unicellular parasites such as HIV, malaria, Ebola, rabies, Zika and so on prevalent in developing countries, but with a substantial worldwide potential spreading impact210–212. In this regard, making vaccines more accessible and affordable for developing countries is a necessity, as these countries suffer from low purchasing power and limited dependence on wealthy nations’ donations213. A current example highlighting this issue is being demonstrated by the low SARS-CoV-2 vaccination rates, resulting from scarce vaccine availability, which leads to high infection and morbidity rates in developing countries, posing a global risk of new variant generation214,215. Experts now believe the developing countries’ solution is to obtain technology licensing for self-manufacturing or expanding manufacturing facilities of pharmaceutical companies to these areas215,216.
Strikingly, current patent waiver disagreements may intimidate new players from developing new nanotechnologies, which will require a long development time and many more resources in the gene delivery field. Throughout the course of patent law history, and especially in modern times, inventors are seeking to protect their intellectual property from illegal competition as a means to increase profitability for a limited time217. However, the question of whether patents accelerate or restrict innovation has not been brought up for discussion218. A different, yet no less controversial, debate arises during global emergencies (such as wars or pandemics) over patent waiver. For example, during World War II, Pfizer shared its penicillin production techniques with other US manufacturers to boost antibiotic production to save lives without suing for patent infringement219. Current disagreements regarding COVID-19 vaccine patent reprieve include two main arguments. Advocates state that waivers will help low-income nations vaccinate their population, and therefore help bring the pandemic to an end. Opponents claim raw material shortage is the real bottleneck and such waivers will provide a bypass to competitive companies wishing to achieve expensive technology220. Regardless of the World Trade Organization (WTO) decision in this manner, steps need to be taken to encourage new players to enter the nucleic acid field and develop new delivery technologies which require long development time and many resources.
Artificial intelligence and machine learning for personalized medicine.
The design of nanocarriers and oligonucleotide sequences are essential for controlling the nano-vehicle physiological fate and biological activity221. Furthermore, the broad parameter space of these nanoparticles and the numerous unknown variables affecting their formation and biological interactions make their design process extremely complex. Therefore, rational design efforts of oligonucleotide delivery nanocarriers are often not successful. Wide scans of different compositions, oligonucleotide sequences and component ratios are required to find a formulation with the desired properties and therapeutic efficacy184,222,223. These scans are laborious and resource intensive, and therefore limited in the number of combinations that can be tested. In recent years, computational models and artificial intelligence approaches have been integrated into the design of nanoparticles and oligonucleotide sequences224–226. Nevertheless, further development of machine learning algorithms can provide imperative tools to overcome existing challenges and knowledge gaps in oligonucleotide nanoparticle engineering. For example, development of tools that will assist in the selection and optimization of mRNA sequence elements such as the 5’ and 3’ untranslated regions has potential to substantially improve the mRNA stability and translation level227. On the nanoparticle side, algorithms with predictive capabilities on complexation and encapsulation efficiencies, biodistribution in the body and toxicity will also provide great value. However, wide and reliable data collection and improvements in formulation scanning throughputs are prerequisites in order to make this realizable.
Acknowledgements
This work was supported by the European Research Council (ERC) Starting Grant (ERC-StG-2019–848325 to J. Conde) and the Fundação para a Ciência e a Tecnologia FCT Grant (PTDC/BTM-MAT/4738/2020 to J. Conde). J.S. acknowledges US National Institute of Health (NIH) grants (R01CA200900, R01HL156362 and R01HL159012), the US DoD PRCRP Idea Award with Special Focus (W81XWH1910482), the Lung Cancer Discovery Award from the American Lung Association and the Innovation Discovery Grants award from the Mass General Brigham. H.L., D.Y. and X.Z. were supported by the National Key R&D Program of China (no. 2020YFA0710700), the National Natural Science Foundation of China (nos 21991132, 52003264, 52021002 and 52033010) and the Fundamental Research Funds for the Central Universities (no. WK2060000027).
Footnotes
Competing interests
J. Conde is a co-founder and shareholder of TargTex S.A. The other authors declare no competing interests.
References
- References: 1.Alvarez EM et al. The global burden of adolescent and young adult cancer in 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Oncol . 23, 27–52 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 2.Global Burden of Disease 2019 Cancer Collaboration. Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019: a systematic analysis for the Global Burden of Disease Study 2019. JAMA Oncol . 10.1001/jamaoncol.2021.6987 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 3.Vos T et al. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet 396, 1204–1222 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 4.Roth AG et al. Global burden of cardiovascular diseases and risk factors, 1990–2019. J. Am. Coll. Cardiol . 76, 2982–3021 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 5.Forouzanfar MH et al. Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388, 1659–1724 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 6.Frank TD et al. Global, regional, and national incidence, prevalence, and mortality of HIV, 1980–2017, and forecasts to 2030, for 195 countries and territories: a systematic analysis for the Global Burden of Diseases, Injuries, and Risk Factors Study 2017. Lancet HIV 6, e831–e859 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 7.Watson DJ The human genome project: past, present, and future. Science 248, 44–49 (1990). [DOI] [PubMed] [Google Scholar]
- References: 8.Fire A et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998). [DOI] [PubMed] [Google Scholar]
- References: 9.Martin J et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 10.Shahryari A et al. Development and clinical translation of approved gene therapy products for genetic disorders. Front. Genet 10, 868 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 11.Polack FP et al. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N. Engl. J. Med 383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 12.Gillmore JD et al. CRISPR–Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med 385, 493–502 (2021). [DOI] [PubMed] [Google Scholar]
- References: 13.Ruan G-X et al. CRISPR/Cas9-mediated genome editing as a therapeutic approach for Leber congenital amaurosis 10. Mol. Ther 25, 331–341 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 14.McKay PF et al. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun 11, 3523 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 15.Wesselhoeft RA, Kowalski PS & Anderson DG Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun 9, 2629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 16.Dangouloff T & Servais L Clinical evidence supporting early treatment of patients with spinal muscular atrophy: current perspectives. Ther. Clin. Risk Manag 15, 1153–1161 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 17.Kay MA State-of-the-art gene-based therapies: the road ahead. Nat. Rev. Genet . 12, 316–328 (2011). [DOI] [PubMed] [Google Scholar]
- References: 18.Urits I et al. A review of patisiran (Onpattro®) for the treatment of polyneuropathy in people with hereditary transthyretin amyloidosis. Neurol. Ther 9, 301–315 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 19.Dunbar EC et al. Gene therapy comes of age. Science 359, eaan4672 (2018). [DOI] [PubMed] [Google Scholar]
- References: 20.Deverman BE, Ravina BM, Bankiewicz KS, Paul SM & Sah DWY Gene therapy for neurological disorders: progress and prospects. Nat. Rev. Drug Discov 17, 641–659 (2018). [DOI] [PubMed] [Google Scholar]
- References: 21.Jones CH, Chen C-K, Ravikrishnan A, Rane S & Pfeifer BA Overcoming nonviral gene delivery barriers: perspective and future. Mol. Pharm 10, 4082–4098 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 22.Malaviya M & Shiroya M Systemic gene delivery using lipid envelope systems and its potential in overcoming challenges. Int. J. Pharm. Drug Anal 9, 46–55 (2021). [Google Scholar]
- References: 23.Uchida S & Kataoka K Design concepts of polyplex micelles for in vivo therapeutic delivery of plasmid DNA and messenger RNA. J. Biomed. Mater. Res. A 107, 978–990 (2019). [DOI] [PubMed] [Google Scholar]
- References: 24.Sung YK & Kim SW Recent advances in the development of gene delivery systems. Biomater. Res . 23, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 25.Patil SD, Rhodes DG & Burgess DJ DNA-based therapeutics and DNA delivery systems: a comprehensive review. AAPS J . 7, E61–E77 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 26.Bulcha JT, Wang Y, Ma H, Tai PWL & Gao G Viral vector platforms within the gene therapy landscape. Signal. Transduct. Target. Ther 6, 53 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 27.Mancheño-Corvo P & Martín-Duque P Viral gene therapy. Clin. Transl. Oncol 8, 858–867 (2006). [DOI] [PubMed] [Google Scholar]
- References: 28.Selot RS, Jayandharan GR & Hareendran S Developing immunologically inert adeno-associated virus (AAV) vectors for gene therapy: possibilities and limitations. Curr. Pharm. Biotechnol 14, 1072–1082 (2013). [DOI] [PubMed] [Google Scholar]
- References: 29.Iyer AK, Duan Z & Amiji MM Nanodelivery systems for nucleic acid therapeutics in drug resistant tumors. Mol. Pharm 11, 2511–2526 (2014). [DOI] [PubMed] [Google Scholar]
- References: 30.Rajala A et al. Nanoparticle-assisted targeted delivery of eye-specific genes to eyes significantly improves the vision of blind mice in vivo. Nano Lett . 14, 5257–5263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 31.Ren T, Song YK, Zhang G & Liu D Structural basis of DOTMA for its high intravenous transfection activity in mouse. Gene Ther . 7, 764–768 (2000). [DOI] [PubMed] [Google Scholar]
- References: 32.Wahane A et al. Role of lipid-based and polymer-based non-viral vectors in nucleic acid delivery for next-generation gene therapy. Molecules 25, 2866 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 33.Maugeri M et al. Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells. Nat. Commun 10, 4333 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 34.Witzigmann D et al. Lipid nanoparticle technology for therapeutic gene regulation in the liver. Adv. Drug Deliv. Rev 159, 344–363 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 35.Piotrowski-Daspit AS, Kauffman AC, Bracaglia LG & Saltzman WM Polymeric vehicles for nucleic acid delivery. Adv. Drug Deliv. Rev 156, 119–132 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 36.Rai R, Alwani S & Badea I Polymeric nanoparticles in gene therapy: new avenues of design and optimization for delivery applications. Polymers 11, 745 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 37.Bertschinger M et al. Disassembly of polyethylenimine-DNA particles in vitro: implications for polyethylenimine-mediated DNA delivery. J. Control. Rel 116, 96–104 (2006). [DOI] [PubMed] [Google Scholar]
- References: 38.Wightman L et al. Different behavior of branched and linear polyethylenimine for gene delivery in vitro and in vivo. J. Gene Med 3, 362–372 (2001). [DOI] [PubMed] [Google Scholar]
- References: 39.Hoyer J & Neundorf I Peptide vectors for the nonviral delivery of nucleic acids. Acc. Chem. Res 45, 1048–1056 (2012). [DOI] [PubMed] [Google Scholar]
- References: 40.Luther DC et al. Delivery of drugs, proteins, and nucleic acids using inorganic nanoparticles. Adv. Drug Deliv. Rev 156, 188–213 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 41.Pranatharthiharan S, Patel MD, D’Souza AA & Devarajan PV Inorganic nanovectors for nucleic acid delivery. Drug Deliv. Transl. Res 3, 446–470 (2013). [DOI] [PubMed] [Google Scholar]
- References: 42.Mora-Raimundo P, Lozano D, Manzano M & Vallet-Regí M Nanoparticles to knockdown osteoporosis-related gene and promote osteogenic marker expression for osteoporosis treatment. ACS Nano 13, 5451–5464 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 43.Hadinoto K, Sundaresan A & Cheow WS Lipid–polymer hybrid nanoparticles as a new generation therapeutic delivery platform: a review. Eur. J. Pharm. Biopharm 85, 427–443 (2013). [DOI] [PubMed] [Google Scholar]
- References: 44.Mitchell MJ et al. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov 20, 101–124 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 45.Rauch S et al. mRNA-based SARS-CoV-2 vaccine candidate CVnCoV induces high levels of virus-neutralising antibodies and mediates protection in rodents. NPJ Vaccines 6, 57 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 46.Gebre MS et al. Optimization of non-coding regions improves protective efficacy of an mRNA SARS-CoV-2 vaccine in nonhuman primates. Preprint at bioRxiv 10.1101/2021.08.13.456316 (2021). [DOI] [Google Scholar]
- References: 47.Adir O et al. Integrating artificial intelligence and nanotechnology for precision cancer medicine. Adv. Mater . 32, 1901989 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 48.Hou X, Zaks T, Langer R & Dong Y Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater . 6, 1078–1094 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 49.Akinc A et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol 14, 1084–1087 (2019). [DOI] [PubMed] [Google Scholar]
- References: 50.[No authors listed] Let’s talk about lipid nanoparticles. Nat. Rev. Mater 6, 99 (2021). [Google Scholar]
- References: 51.Mukalel AJ, Riley RS, Zhang R & Mitchell MJ Nanoparticles for nucleic acid delivery: applications in cancer immunotherapy. Cancer Lett . 458, 102–112 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 52.Nagayasu A, Uchiyama K & Kiwada H The size of liposomes: a factor which affects their targeting efficiency to tumors and therapeutic activity of liposomal antitumor drugs. Adv. Drug Deliv. Rev . 40, 75–87 (1999). [DOI] [PubMed] [Google Scholar]
- References: 53.Li W, Huang Z, MacKay JA, Grube S & Szoka FC Jr Low-pH-sensitive poly(ethylene glycol) (PEG)-stabilized plasmid nanolipoparticles: effects of PEG chain length, lipid composition and assembly conditions on gene delivery. J. Gene Med 7, 67–79 (2005). [DOI] [PubMed] [Google Scholar]
- References: 54.Nordling-David MM & Golomb G Gene delivery by liposomes. Isr. J. Chem 53, 737–747 (2013). [Google Scholar]
- References: 55.Han X et al. An ionizable lipid toolbox for RNA delivery. Nat. Commun 12, 7233 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 56.Kulkarni JA, Witzigmann D, Leung J, Tam YYC & Cullis PR On the role of helper lipids in lipid nanoparticle formulations of siRNA. Nanoscale 11, 21733–21739 (2019). [DOI] [PubMed] [Google Scholar]
- References: 57.Ambegia E et al. Stabilized plasmid–lipid particles containing PEG-diacylglycerols exhibit extended circulation lifetimes and tumor selective gene expression. Biochim. Biophys. Act 1669, 155–163 (2005). [DOI] [PubMed] [Google Scholar]
- References: 58.Dai Q, Walkey C & Chan WCW Polyethylene glycol backfilling mitigates the negative impact of the protein corona on nanoparticle cell targeting. Angew. Chem. Int. Ed 53, 5093–5096 (2014). [DOI] [PubMed] [Google Scholar]
- References: 59.Belliveau NM et al. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Mol. Ther. Nucleic Acids 1, e37 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 60.Chen S et al. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J. Control. Rel 235, 236–244 (2016). [DOI] [PubMed] [Google Scholar]
- References: 61.Han X, Mitchell MJ & Nie G Nanomaterials for therapeutic RNA delivery. Matter 3, 1948–1975 (2020). [Google Scholar]
- References: 62.Kowalski PS, Rudra A, Miao L & Anderson DG Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol. Ther 27, 710–728 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 63.Kauffman KJ et al. Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett . 15, 7300–7306 (2015). [DOI] [PubMed] [Google Scholar]
- References: 64.Ramaswamy S et al. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl Acad. Sci. USA 114, E1941–E1950 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 65.Byrne JD, Betancourt T & Brannon-Peppas L Active targeting schemes for nanoparticle systems in cancer therapeutics. Adv. Drug Deliv. Rev 60, 1615–1626 (2008). [DOI] [PubMed] [Google Scholar]
- References: 66.Jahn A, Vreeland WN, Gaitan M & Locascio LE Controlled vesicle self-assembly in microfluidic channels with hydrodynamic focusing. J. Am. Chem. Soc 126, 2674–2675 (2004). [DOI] [PubMed] [Google Scholar]
- References: 67.Zhigaltsev IV et al. Bottom-up design and synthesis of limit size lipid nanoparticle systems with aqueous and triglyceride cores using millisecond microfluidic mixing. Langmuir 28, 3633–3640 (2012). [DOI] [PubMed] [Google Scholar]
- References: 68.Roces CB et al. Manufacturing considerations for the development of lipid nanoparticles using microfluidics. Pharmaceutics 12, 1095 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 69.Lächelt U & Wagner E Nucleic acid therapeutics using polyplexes: a journey of 50 years (and beyond). Chem. Rev 115, 11043–11078 (2015). [DOI] [PubMed] [Google Scholar]
- References: 70.Discher DE & Ahmed F Polymersomes. Annu. Rev. Biomed. Eng 8, 323–341 (2006). [DOI] [PubMed] [Google Scholar]
- References: 71.Iqbal S, Blenner M, Alexander-Bryant A & Larsen J Polymersomes for therapeutic delivery of protein and nucleic acid macromolecules: from design to therapeutic applications. Biomacromolecules 21, 1327–1350 (2020). [DOI] [PubMed] [Google Scholar]
- References: 72.Pangburn TO, Georgiou K, Bates FS & Kokkoli E Targeted polymersome delivery of siRNA induces cell death of breast cancer cells dependent upon orai3 protein expression. Langmuir 28, 12816–12830 (2012). [DOI] [PubMed] [Google Scholar]
- References: 73.Konishcheva EV, Zhumaev UE & Meier WP PEO-b-PCL-b-PMOXA triblock copolymers: from synthesis to microscale polymersomes with asymmetric membrane. Macromolecules 50, 1512–1520 (2017). [Google Scholar]
- References: 74.Li S et al. Biodegradable polymersomes with an ionizable membrane: facile preparation, superior protein loading, and endosomal pH-responsive protein release. Eur. J. Pharm. Biopharm 82, 103–111 (2012). [DOI] [PubMed] [Google Scholar]
- References: 75.Walter MV & Malkoch M Simplifying the synthesis of dendrimers: accelerated approaches. Chem. Soc. Rev 41, 4593–4609 (2012). [DOI] [PubMed] [Google Scholar]
- References: 76.Conde J, Oliva N, Atilano M, Song HS & Artzi N Self-assembled RNA-triple-helix hydrogel scaffold for microRNA modulation in the tumour microenvironment. Nat. Mater 15, 353–363 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 77.Palmerston Mendes L, Pan J & Torchilin VP Dendrimers as nanocarriers for nucleic acid and drug delivery in cancer therapy. Molecules 22, 1401 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 78.Pack DW, Hoffman AS, Pun S & Stayton PS Design and development of polymers for gene delivery. Nat. Rev. Drug Discov 4, 581–593 (2005). [DOI] [PubMed] [Google Scholar]
- References: 79.Lai W-F & Wong W-T Design of polymeric gene carriers for effective intracellular delivery. Trends Biotechnol . 36, 713–728 (2018). [DOI] [PubMed] [Google Scholar]
- References: 80.Arami H, Khandhar A, Liggitt D & Krishnan KM In vivo delivery, pharmacokinetics, biodistribution and toxicity of iron oxide nanoparticles. Chem. Soc. Rev 44, 8576–8607 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 81.Dobrovolskaia MA & McNeil SE Immunological properties of engineered nanomaterials. Nat. Nanotechnol 2, 469–478 (2007). [DOI] [PubMed] [Google Scholar]
- References: 82.Soenen SJ et al. Cellular toxicity of inorganic nanoparticles: common aspects and guidelines for improved nanotoxicity evaluation. Nano Today 6, 446–465 (2011). [Google Scholar]
- References: 83.Cutler JI, Auyeung E & Mirkin CA Spherical nucleic acids. J. Am. Chem. Soc 134, 1376–1391 (2012). [DOI] [PubMed] [Google Scholar]
- References: 84.Conde J et al. Dual targeted immunotherapy via in vivo delivery of biohybrid RNAi–peptide nanoparticles to tumor-associated macrophages and cancer cells. Adv. Funct. Mater 25, 4183–4194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 85.Conde J, Oliva N & Artzi N Implantable hydrogel embedded dark-gold nanoswitch as a theranostic probe to sense and overcome cancer multidrug resistance. Proc. Natl Acad. Sci. USA 112, E1278–E1287 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 86.Gilam A et al. Local microRNA delivery targets Palladin and prevents metastatic breast cancer. Nat. Commun . 7, 12868 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 87.Bishop CJ, Tzeng SY & Green JJ Degradable polymer-coated gold nanoparticles for co-delivery of DNA and siRNA. Acta Biomater . 11, 393–403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 88.Donahue ND, Acar H & Wilhelm S Concepts of nanoparticle cellular uptake, intracellular trafficking, and kinetics in nanomedicine. Adv. Drug Deliv. Rev 143, 68–96 (2019). [DOI] [PubMed] [Google Scholar]
- References: 89.Hak S et al. The effect of nanoparticle polyethylene glycol surface density on ligand-directed tumor targeting studied in vivo by dual modality imaging. ACS Nano 6, 5648–5658 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 90.Tang S, Huang X, Chen X & Zheng N Hollow mesoporous zirconia nanocapsules for drug delivery. Adv. Funct. Mater 20, 2442–2447 (2010). [Google Scholar]
- References: 91.Ahmadi E, Dehghannejad N, Hashemikia S, Ghasemnejad M & Tabebordbar H Synthesis and surface modification of mesoporous silica nanoparticles and its application as carriers for sustained drug delivery. Drug Deliv . 21, 164–172 (2014). [DOI] [PubMed] [Google Scholar]
- References: 92.Li J, Xue S & Mao Z-W Nanoparticle delivery systems for siRNA-based therapeutics. J. Mater. Chem. B 4, 6620–6639 (2016). [DOI] [PubMed] [Google Scholar]
- References: 93.Jiang S, Eltoukhy AA, Love KT, Langer R & Anderson DG Lipidoid-coated iron oxide nanoparticles for efficient DNA and siRNA delivery. Nano Lett . 13, 1059–1064 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 94.Tavitian B et al. In vivo imaging of oligonucleotides with positron emission tomography. Nat. Med 4, 467–471 (1998). [DOI] [PubMed] [Google Scholar]
- References: 95.Khorkova O & Wahlestedt C Oligonucleotide therapies for disorders of the nervous system. Nat. Biotechnol 35, 249–263 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 96.Kanasty R, Dorkin JR, Vegas A & Anderson D Delivery materials for siRNA therapeutics. Nat. Mater . 12, 967–977 (2013). [DOI] [PubMed] [Google Scholar]
- References: 97.Khvorova A & Watts JK The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol 35, 238–248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 98.Dowdy SF Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol 35, 222–229 (2017). [DOI] [PubMed] [Google Scholar]
- References: 99.Sun Y et al. Enhancing the therapeutic delivery of oligonucleotides by chemical modification and nanoparticle encapsulation. Molecules 22, 1724 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 100.Sipa K et al. Effect of base modifications on structure, thermodynamic stability, and gene silencing activity of short interfering RNA. RNA 13, 1301–1316 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 101.Orlandini von Niessen AG et al. Improving mRNA-based therapeutic gene delivery by expression-augmenting 3’ UTRs identified by cellular library screening. Mol. Ther 27, 824–836 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 102.Holtkamp S et al. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood 108, 4009–4017 (2006). [DOI] [PubMed] [Google Scholar]
- References: 103.Jasinski DL, Li H & Guo P The effect of size and shape of RNA nanoparticles on biodistribution. Mol. Ther 26, 784–792 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 104.Albanese A, Tang PS & Chan WCW The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu. Rev. Biomed. Eng 14, 1–16 (2012). [DOI] [PubMed] [Google Scholar]
- References: 105.Buriak JM Preface to the special issue on methods and protocols in materials chemistry. Chem. Mater 29, 1–2 (2017). [Google Scholar]
- References: 106.Behzadi S et al. Cellular uptake of nanoparticles: journey inside the cell. Chem. Soc. Rev 46, 4218–4244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 107.Hadjidemetriou M & Kostarelos K Evolution of the nanoparticle corona. Nat. Nanotechnol 12, 288–290 (2017). [DOI] [PubMed] [Google Scholar]
- References: 108.Satzer P, Svec F, Sekot G & Jungbauer A Protein adsorption onto nanoparticles induces conformational changes: particle size dependency, kinetics, and mechanisms. Eng. Life Sci 16, 238–246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 109.Walkey CD, Olsen JB, Guo H, Emili A & Chan WCW Nanoparticle size and surface chemistry determine serum protein adsorption and macrophage uptake. J. Am. Chem. Soc 134, 2139–2147 (2012). [DOI] [PubMed] [Google Scholar]
- References: 110.Glancy D et al. Characterizing the protein corona of sub-10 nm nanoparticles. J. Control. Rel 304, 102–110 (2019). [DOI] [PubMed] [Google Scholar]
- References: 111.Faria M et al. Minimum information reporting in bio–nano experimental literature. Nat. Nanotechnol 13, 777–785 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 112.Modena MM, Rühle B, Burg TP & Wuttke S Nanoparticle characterization: what to measure? Adv. Mater 31, 1901556 (2019). [DOI] [PubMed] [Google Scholar]
- References: 113.Varenne F, Makky A, Gaucher-Delmas M, Violleau F & Vauthier C Multimodal dispersion of nanoparticles: a comprehensive evaluation of size distribution with 9 size measurement methods. Pharm. Res 33, 1220–1234 (2016). [DOI] [PubMed] [Google Scholar]
- References: 114.Stetefeld J, McKenna SA & Patel TR Dynamic light scattering: a practical guide and applications in biomedical sciences. Biophys. Rev 8, 409–427 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 115.Shen H et al. Single particle tracking: from theory to biophysical applications. Chem. Rev 117, 7331–7376 (2017). [DOI] [PubMed] [Google Scholar]
- References: 116.Dahmen U et al. Background, status and future of the transmission electron aberration-corrected microscope project. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci 367, 3795–3808 (2009). [DOI] [PubMed] [Google Scholar]
- References: 117.Skowron ST et al. Chemical reactions of molecules promoted and simultaneously imaged by the electron beam in transmission electron microscopy. Acc. Chem. Res 50, 1797–1807 (2017). [DOI] [PubMed] [Google Scholar]
- References: 118.Nakamura E, Sommerdijk NAJM & Zheng H Transmission electron microscopy for chemists. Acc. Chem. Res 50, 1795–1796 (2017). [DOI] [PubMed] [Google Scholar]
- References: 119.Stewart PL Cryo-electron microscopy and cryo-electron tomography of nanoparticles. WIREs Nanomed. Nanobiotechnol 9, e1417 (2017). [DOI] [PubMed] [Google Scholar]
- References: 120.Mourdikoudis S, Pallares RM & Thanh NTK Characterization techniques for nanoparticles: comparison and complementarity upon studying nanoparticle properties. Nanoscale 10, 12871–12934 (2018). [DOI] [PubMed] [Google Scholar]
- References: 121.Rao A et al. Characterization of nanoparticles using atomic force microscopy. J. Phys. Conf. Ser 61, 971–976 (2007). [Google Scholar]
- References: 122.Glass JJ et al. Charge has a marked influence on hyperbranched polymer nanoparticle association in whole human blood. ACS Macro Lett . 6, 586–592 (2017). [DOI] [PubMed] [Google Scholar]
- References: 123.Salatin S, Maleki Dizaj S & Yari Khosroushahi A Effect of the surface modification, size, and shape on cellular uptake of nanoparticles. Cell Biol. Int 39, 881–890 (2015). [DOI] [PubMed] [Google Scholar]
- References: 124.Doane TL, Chuang C-H, Hill RJ & Burda C Nanoparticle ζ-potentials. Acc. Chem. Res 45, 317–326 (2012). [DOI] [PubMed] [Google Scholar]
- References: 125.Mura S, Nicolas J & Couvreur P Stimuli-responsive nanocarriers for drug delivery. Nat. Mater 12, 991–1003 (2013). [DOI] [PubMed] [Google Scholar]
- References: 126.Colombo M et al. Tumour homing and therapeutic effect of colloidal nanoparticles depend on the number of attached antibodies. Nat. Commun 7, 13818 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 127.Cheng Y et al. A multifunctional peptide-conjugated AIEgen for efficient and sequential targeted gene delivery into the nucleus. Angew. Chem. Int. Ed 58, 5049–5053 (2019). [DOI] [PubMed] [Google Scholar]
- References: 128.Herda LM, Hristov DR, Lo Giudice MC, Polo E & Dawson KA Mapping of molecular structure of the nanoscale surface in bionanoparticles. J. Am. Chem. Soc 139, 111–114 (2017). [DOI] [PubMed] [Google Scholar]
- References: 129.Conte C et al. Multi-component bioresponsive nanoparticles for synchronous delivery of docetaxel and TUBB3 siRNA to lung cancer cells. Nanoscale 13, 11414–11426 (2021). [DOI] [PubMed] [Google Scholar]
- References: 130.Chen F et al. Complement proteins bind to nanoparticle protein corona and undergo dynamic exchange in vivo. Nat. Nanotechnol 12, 387–393 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 131.Kim J, Koo B-K & Knoblich JA Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol 21, 571–584 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 132.Skylar-Scott MA et al. Biomanufacturing of organ-specific tissues with high cellular density and embedded vascular channels. Sci. Adv 5, eaaw2459 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 133.Low LA, Mummery C, Berridge BR, Austin CP & Tagle DA Organs-on-chips: into the next decade. Nat. Rev. Drug Discov 20, 345–361 (2021). [DOI] [PubMed] [Google Scholar]
- References: 134.Kohl Y et al. Microfluidic in vitro platform for (nano) safety and (nano)drug efficiency screening. Small 17, 2006012 (2021). [DOI] [PubMed] [Google Scholar]
- References: 135.Conde J, Oliva N, Zhang Y & Artzi N Local triple-combination therapy results in tumour regression and prevents recurrence in a colon cancer model. Nat. Mater 15, 1128–1138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 136.Poley M et al. Chemotherapeutic nanoparticles accumulate in the female reproductive system during ovulation affecting fertility and anticancer activity. Preprint at bioRxiv 10.1101/2020.07.22.216168 (2020). [DOI] [Google Scholar]
- References: 137.Duan X & Li Y Physicochemical characteristics of nanoparticles affect circulation, biodistribution, cellular internalization, and trafficking. Small 9, 1521–1532 (2013). [DOI] [PubMed] [Google Scholar]
- References: 138.Elci SG et al. Surface charge controls the suborgan biodistributions of gold nanoparticles. ACS Nano 10, 5536–5542 (2016). [DOI] [PubMed] [Google Scholar]
- References: 139.Soo Choi H et al. Renal clearance of quantum dots. Nat. Biotechnol 25, 1165–1170 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 140.Hoshyar N, Gray S, Han H & Bao G The effect of nanoparticle size on in vivo pharmacokinetics and cellular interaction. Nanomedicine 11, 673–692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 141.García KP et al. Zwitterionic-coated “stealth” nanoparticles for biomedical applications: recent advances in countering biomolecular corona formation and uptake by the mononuclear phagocyte system. Small 10, 2516–2529 (2014). [DOI] [PubMed] [Google Scholar]
- References: 142.Moitra P, Alafeef M, Dighe K, Frieman MB & Pan D Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano 14, 7617–7627 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 143.Jwa-Min N, Shad TC & Mirkin CA Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science 301, 1884–1886 (2003). [DOI] [PubMed] [Google Scholar]
- References: 144.Liu J & Lu Y Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angew. Chem. Int. Ed 45, 90–94 (2006). [DOI] [PubMed] [Google Scholar]
- References: 145.Liu J & Lu Y A colorimetric lead biosensor using DNAzyme-directed assembly of gold nanoparticles. J. Am. Chem. Soc 125, 6642–6643 (2003). [DOI] [PubMed] [Google Scholar]
- References: 146.Robert E, Storhoff JJ, Mucic RC, Letsinger RL & Mirkin CA Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science 277, 1078–1081 (1997). [DOI] [PubMed] [Google Scholar]
- References: 147.Andrew TT, Mirkin CA & Letsinger RL Scanometric DNA array detection with nanoparticle probes. Science 289, 1757–1760 (2000). [DOI] [PubMed] [Google Scholar]
- References: 148.Charles CY, Rongchao J & Mirkin CA Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science 297, 1536–1540 (2002). [DOI] [PubMed] [Google Scholar]
- References: 149.Walker T et al. Clinical Impact of laboratory implementation of verigene BC-GN microarray-based assay for detection of Gram-negative bacteria in positive blood cultures. J. Clin. Microbiol 54, 1789–1796 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 150.Georganopoulou DG et al. Nanoparticle-based detection in cerebral spinal fluid of a soluble pathogenic biomarker for Alzheimer’s disease. Proc. Natl Acad. Sci. USA 102, 2273–2276 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 151.Han MS, Lytton-Jean AKR, Oh B-K, Heo J & Mirkin CA Colorimetric screening of DNA-binding molecules with gold nanoparticle probes. Angew. Chem. Int. Ed 45, 1807–1810 (2006). [DOI] [PubMed] [Google Scholar]
- References: 152.Liu J & Lu Y Accelerated color change of gold nanoparticles assembled by DNAzymes for simple and fast colorimetric Pb2+ detection. J. Am. Chem. Soc 126, 12298–12305 (2004). [DOI] [PubMed] [Google Scholar]
- References: 153.Yang L et al. Efficient delivery of antisense oligonucleotides using bioreducible lipid nanoparticles in vitro and in vivo. Mol. Ther. Nucleic Acids 19, 1357–1367 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 154.Khan FO et al. Endothelial siRNA delivery in nonhuman primates using ionizable low-molecular weight polymeric nanoparticles. Sci. Adv 4, eaar8409 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 155.Wei T et al. siRNA nanoparticles targeting CaMKIIγ in lesional macrophages improve atherosclerotic plaque stability in mice. Sci. Transl. Med 12, eaay1063 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 156.Conde J et al. Design of multifunctional gold nanoparticles for in vitro and in vivo gene silencing. ACS Nano 6, 8316–8324 (2012). [DOI] [PubMed] [Google Scholar]
- References: 157.Wen L et al. BBB pathophysiology-independent delivery of siRNA in traumatic brain injury. Sci. Adv 7, eabd6889 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 158.Zhao W, Hou X, Vick OG & Dong Y RNA delivery biomaterials for the treatment of genetic and rare diseases. Biomaterials 217, 119291 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 159.Zimmermann TS et al. RNAi-mediated gene silencing in non-human primates. Nature 441, 111–114 (2006). [DOI] [PubMed] [Google Scholar]
- References: 160.Morrissey DV et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol 23, 1002–1007 (2005). [DOI] [PubMed] [Google Scholar]
- References: 161.Mendes BB, Sousa DP, Conniot J & Conde J Nanomedicine-based strategies to target and modulate the tumor microenvironment. Trends Cancer 7, 847–862 (2021). [DOI] [PubMed] [Google Scholar]
- References: 162.Woodrow KA et al. Intravaginal gene silencing using biodegradable polymer nanoparticles densely loaded with small-interfering RNA. Nat. Mater 8, 526–533 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 163.Wilson DS et al. Orally delivered thioketal nanoparticles loaded with TNF-α–siRNA target inflammation and inhibit gene expression in the intestines. Nat. Mater 9, 923–928 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 164.Debacker AJ, Voutila J, Catley M, Blakey D & Habib N Delivery of oligonucleotides to the liver with GalNAc: from research to registered therapeutic drug. Mol. Ther 28, 1759–1771 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 165.Zuris JA et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol 33, 73–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 166.Zhang H-X, Zhang Y & Yin H Genome editing with mRNA encoding ZFN, TALEN, and Cas9. Mol. Ther . 27, 735–746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 167.Xu C-F et al. Rational designs of in vivo CRISPR–Cas delivery systems. Adv. Drug Deliv. Rev 168, 3–29 (2021). [DOI] [PubMed] [Google Scholar]
- References: 168.Liu S et al. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR–Cas gene editing. Nat. Mater 20, 701–710 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 169.Parayath NN, Stephan SB, Koehne AL, Nelson PS & Stephan MT In vitro-transcribed antigen receptor mRNA nanocarriers for transient expression in circulating T cells in vivo. Nat. Commun 11, 6080 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 170.Cheng Q et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol . 15, 313–320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 171.Wolff AJ et al. Direct gene transfer into mouse muscle in vivo. Science 247, 1465–1468 (1990). [DOI] [PubMed] [Google Scholar]
- References: 172.Sabnis S et al. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther 26, 1509–1519 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 173.Islam MA et al. Restoration of tumour-growth suppression in vivo via systemic nanoparticle-mediated delivery of PTEN mRNA. Nat. Biomed. Eng 2, 850–864 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 174.Kong N et al. Synthetic mRNA nanoparticle-mediated restoration of p53 tumor suppressor sensitizes p53-deficient cancers to mTOR inhibition. Sci. Transl. Med 11, eaaw1565 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 175.Yao-Xin L et al. Reactivation of the tumor suppressor PTEN by mRNA nanoparticles enhances antitumor immunity in preclinical models. Sci. Transl. Med . 13, eaba9772 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 176.Chaudhary N, Weissman D & Whitehead KA mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat. Rev. Drug Discov 20, 817–838 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 177.Pardi N, Hogan MJ, Porter FW & Weissman D mRNA vaccines — a new era in vaccinology. Nat. Rev. Drug Discov 17, 261–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 178.Islam MA et al. Adjuvant-pulsed mRNA vaccine nanoparticle for immunoprophylactic and therapeutic tumor suppression in mice. Biomaterials 266, 120431 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 179.Zhang H et al. Delivery of mRNA vaccine with a lipid-like material potentiates antitumor efficacy through Toll-like receptor 4 signaling. Proc. Natl Acad. Sci. USA 118, e2005191118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 180.Smith TT et al. In situ programming of leukaemia-specific T cells using synthetic DNA nanocarriers. Nat. Nanotechnol 12, 813–820 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 181.Capece D, Verzella D, Fischietti M, Zazzeroni F & Alesse E Targeting costimulatory molecules to improve antitumor immunity. J. Biomed. Biotechnol 2012, 926321 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 182.Hewitt LS et al. Durable anticancer immunity from intratumoral administration of IL-23, IL-36γ, and OX40L mRNAs. Sci. Transl. Med 11, eaat9143 (2019). [DOI] [PubMed] [Google Scholar]
- References: 183.Conniot J et al. Immunization with mannosylated nanovaccines and inhibition of the immune-suppressing microenvironment sensitizes melanoma to immune checkpoint modulators. Nat. Nanotechnol 14, 891–901 (2019). [DOI] [PubMed] [Google Scholar]
- References: 184.Sago CD et al. High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing. Proc. Natl Acad. Sci. USA 115, E9944–E9952 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 185.Guimaraes PPG et al. Ionizable lipid nanoparticles encapsulating barcoded mRNA for accelerated in vivo delivery screening. J. Control. Rel 316, 404–417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 186.Seiichi O, Dylan G & Chan WCW DNA-controlled dynamic colloidal nanoparticle systems for mediating cellular interaction. Science 351, 841–845 (2016). [DOI] [PubMed] [Google Scholar]
- References: 187.Schrurs F & Lison D Focusing the research efforts. Nat. Nanotechnol . 7, 546–548 (2012). [DOI] [PubMed] [Google Scholar]
- References: 188.Cheng Y, Morshed RA, Auffinger B, Tobias AL & Lesniak MS Multifunctional nanoparticles for brain tumor imaging and therapy. Adv. Drug Deliv. Rev 66, 42–57 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 189.Jain RK & Stylianopoulos T Delivering nanomedicine to solid tumors. Nat. Rev. Clin. Oncol 7, 653–664 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 190.Junttila MR & de Sauvage FJ Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501, 346–354 (2013). [DOI] [PubMed] [Google Scholar]
- References: 191.Bedard PL, Hansen AR, Ratain MJ & Siu LL Tumour heterogeneity in the clinic. Nature 501, 355–364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 192.Leong HS et al. On the issue of transparency and reproducibility in nanomedicine. Nat. Nanotechnol 14, 629–635 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 193.Lammers T & Storm G Setting standards to promote progress in bio–nano science. Nat. Nanotechnol 14, 626 (2019). [DOI] [PubMed] [Google Scholar]
- References: 194.Florindo HF, Madi A & Satchi-Fainaro R Challenges in the implementation of MIRIBEL criteria on nanobiomed manuscripts. Nat. Nanotechnol 14, 627–628 (2019). [DOI] [PubMed] [Google Scholar]
- References: 195.[No authors listed] Voices from the community. Nat. Nanotechnol 14, 625 (2019). [DOI] [PubMed] [Google Scholar]
- References: 196.Zu H & Gao D Non-viral vectors in gene therapy: recent development, challenges, and prospects. AAPS J 23, 78 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 197.Hill AB, Chen M, Chen C-K, Pfeifer BA & Jones CH Overcoming gene-delivery hurdles: physiological considerations for nonviral vectors. Trends Biotechnol . 34, 91–105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 198.Ginn SL, Amaya AK, Alexander IE, Edelstein M & Abedi MR Gene therapy clinical trials worldwide to 2017: an update. J. Gene Med 20, e3015 (2018). [DOI] [PubMed] [Google Scholar]
- References: 199.Ramamoorth M & Narvekar A Non viral vectors in gene therapy — an overview. J. Clin. Diagn. Res 9, GE01–GE06 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 200.Wei Y, Quan L, Zhou C & Zhan Q Factors relating to the biodistribution & clearance of nanoparticles & their effects on in vivo application. Nanomedicine 13, 1495–1512 (2018). [DOI] [PubMed] [Google Scholar]
- References: 201.Sato Y et al. Elucidation of the physicochemical properties and potency of siRNA-loaded small-sized lipid nanoparticles for siRNA delivery. J. Control. Rel 229, 48–57 (2016). [DOI] [PubMed] [Google Scholar]
- References: 202.Cabral H et al. Accumulation of sub-100 nm polymeric micelles in poorly permeable tumours depends on size. Nat. Nanotechnol 6, 815–823 (2011). [DOI] [PubMed] [Google Scholar]
- References: 203.Phan HT & Haes AJ What does nanoparticle stability mean? J. Phys. Chem C 123, 16495–16507 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 204.Roberts TC, Langer R & Wood MJA Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov 19, 673–694 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 205.Lorenc A et al. Machine learning for next-generation nanotechnology in healthcare. Matter 4, 3078–3080 (2021). [Google Scholar]
- References: 206.Shi Y, van der Meel R, Chen X & Lammers T The EPR effect and beyond: strategies to improve tumor targeting and cancer nanomedicine treatment efficacy. Theranostics 10, 7921–7924 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 207.Park J et al. Alliance with EPR effect: combined strategies to improve the EPR effect in the tumor microenvironment. Theranostics 9, 8073–8090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 208.Paunovska K et al. Analyzing 2000 in vivo drug delivery data points reveals cholesterol structure impacts nanoparticle delivery. ACS Nano 12, 8341–8349 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 209.Vaughan HJ, Green JJ & Tzeng SY Cancer-targeting nanoparticles for combinatorial nucleic acid delivery. Adv. Mater 32, 1901081 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 210.Chakraborty C, Sharma AR, Bhattacharya M & Lee S-S From COVID-19 to cancer mRNA vaccines: moving from bench to clinic in the vaccine landscape. Front. Immunol 12, 2648 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 211.Tombácz I, Weissman D & Pardi N in Vaccination with Messenger RNA: A Promising Alternative to DNA Vaccination BT - DNA Vaccines: Methods and Protocols (ed. Sousa Â) 13–31 (Springer US, 2021). [DOI] [PubMed] [Google Scholar]
- References: 212.Maruggi G, Zhang C, Li J, Ulmer JB & Yu D mRNA as a transformative technology for vaccine development to control infectious diseases. Mol. Ther 27, 757–772 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 213.Fundytus A et al. Access to cancer medicines deemed essential by oncologists in 82 countries: an international, cross-sectional survey. Lancet Oncol 22, 1367–1377 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 214.Excler J-L, Privor-Dumm L & Kim JH Supply and delivery of vaccines for global health. Curr. Opin. Immunol 71, 13–20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 215.Eccleston-Turner M & Upton H International collaboration to ensure equitable access to vaccines for COVID-19: the ACT-Accelerator and the COVAX Facility. Milbank Q . 99, 426–449 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 216.Plotkin S, Robinson JM, Cunningham G, Iqbal R & Larsen S The complexity and cost of vaccine manufacturing — an overview. Vaccine 35, 4064–4071 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 217.Prager FD Historic background and foundation of american patent law. Am. J. Leg. Hist 5, 309–325 (1961). [Google Scholar]
- References: 218.Lévêque F & Ménière Y Patents and innovation: friends or foes? SSRN 10.2139/ssrn.958830 (2006). [DOI] [Google Scholar]
- References: 219.Braithwaite J Corporate Crime in the Pharmaceutical Industry (Routledge Revivals) (Routledge, 2013). [Google Scholar]
- References: 220.Okereke M Towards vaccine equity: should big pharma waive intellectual property rights for COVID-19 vaccines? Public Heal. Pract 2, 100165 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 221.Kon E, Elia U & Peer D Principles for designing an optimal mRNA lipid nanoparticle vaccine. Curr. Opin. Biotechnol 73, 329–336 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 222.Ramishetti S et al. A combinatorial library of lipid nanoparticles for RNA delivery to leukocytes. Adv. Mater 32, 1906128 (2020). [DOI] [PubMed] [Google Scholar]
- References: 223.Billingsley MM et al. Ionizable lipid nanoparticle-ediated mRNA delivery for human CAR T cell engineering. Nano Lett 20, 1578–1589 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 224.Yamankurt G et al. Exploration of the nanomedicine-design space with high-throughput screening and machine learning. Nat. Biomed. Eng 3, 318–327 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 225.Shamay Y et al. Quantitative self-assembly prediction yields targeted nanomedicines. Nat. Mater 17, 361–368 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 226.Yesselman JD et al. Computational design of three-dimensional RNA structure and function. Nat. Nanotechnol 14, 866–873 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- References: 227.Leppek K et al. Combinatorial optimization of mRNA structure, stability, and translation for RNA-based therapeutics. Preprint at bioRxiv 10.1101/2021.03.29.437587 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]


