Summary
The response of vital organs to different types of nutrition or diet is a fundamental question in physiology. We examined the cardiac response to 4 weeks of high-fat diet in mice, measuring cardiac metabolites and mRNA. Metabolomics showed dramatic differences after a high-fat diet, including increases in several acyl-carnitine species. The RNA-seq data showed changes consistent with adaptations to use more fatty acid as substrate and an increase in the antioxidant protein catalase. Changes in mRNA were correlated with changes in protein level for several highly responsive genes. We also found significant sex differences in both metabolomics and RNA-seq datasets, both at baseline and after high fat diet. This work reveals the response of a vital organ to dietary intervention at both metabolomic and transcriptomic levels, which is a fundamental question in physiology. This work also reveals significant sex differences in cardiac metabolites and gene expression.
Subject areas: Pathophysiology, Human metabolism, Metabolomics, Transcriptomics
Graphical abstract

Highlights
-
•
Cardiac gene expression shows adaptive changes after short-term high-fat diet
-
•
Cardiac metabolomics indicate mitochondrial dysfunction after high-fat diet
-
•
Metabolites and gene expression show significant sex differences at baseline
-
•
There are significant sex differences in the changes caused by high-fat diet
Pathophysiology; Human metabolism; Metabolomics; Transcriptomics
Introduction
The requirement for continuous mechanical work from the heart results in an intense metabolic demand on cardiac myocytes. Although it is clear from epidemiologic studies that nutrition can modulate the risk of heart disease, it is controversial how different diets can alter cardiac metabolism and function. Our primary analysis was to determine how a relatively short duration of high-fat diet (4 weeks) changes cardiac biology in wild-type (WT) mice, using unbiased multi-omic methods such as metabolomics and RNA sequencing. We have previously shown that this duration of diet does not cause obesity and abnormalities in either contractile function or cardiac histology but does promote oxidative stress and arrhythmias in rodents (Joseph et al., 2019). Additional experiments were performed to evaluate mitochondrial morphology, cardiac protein levels, and promoter-luciferase activity. In secondary analyses, sex differences were examined; we used equal an number of male and female hearts for metabolomics and RNA sequencing, revealing a complex interplay between sex and diet. This work reveals the response of a vital organ to dietary intervention, which is a fundamental question in physiology that may have relevance for cardiovascular diseases. In addition, this work adds to our understanding of sex differences in cardiac biology.
Results
High-fat diet causes multiple changes in cardiac metabolomics
We fed WT mice a high-fat diet (HFD, with 60% calories from palm oil, which is high in saturated fat) for 4 weeks to determine the effects on cardiac metabolism. This relatively short duration does not result in obesity (Table S1) but has been shown previously to promote electrophysiologic abnormalities and arrhythmias mediated by increased oxidative stress (Joseph et al., 2019). We divided the tissue from each heart into samples for metabolomics and RNA extraction for RNA sequencing (RNA-seq). Principal component analysis (PCA) of the metabolomic dataset indicated that the samples cluster into four groups, with principal component 1 separating chow from HFD cardiac samples and principal component 2 separating male from female cardiac samples (Figure 1A).
Figure 1.
Metabolomic changes after HFD; electron microscopy shows changes in mitochondrial morphology
(A) Principal component analysis (loading) of metabolites demonstrates that the first two principal components (PC) cluster the samples into four groups. PC1 + PC2 account for 85.47% of the proportion of variance in this metabolomic dataset.
(B) Graph of acyl-carnitine species, dots with lines indicated mean ± SEM “C#” refers to the number of carbon atoms in the lipid, that is C16 is palmitoylcarnitine, C14 is myristoylcarnitine, etc. N = 10 hearts for each group. The comparison of control (chow) to HFD is highly statistically significant by two-way ANOVA, p < 0.0001.
(C) Representative examples of electron microscopy from cardiac ventricular samples.
(D) Graphs of average mitochondrial area (um2) and average number of mitochondria per high-power image. N = 4 hearts for each group, ∗ indicates significantly different by t test.
Comparing cardiac tissue metabolites from mice on HFD versus regular chow revealed there were a large number of metabolites with significant differences. With the false discovery rate (FDR) set to 0.05, 275 out of 640 identified metabolites — 43% of the total metabolites — had significant differences. Lipid and carnitine species comprised the majority of the metabolites with the greatest differences. Notably, the saturated fats palmitate (C16) and myristate (C14) were significantly increased after HFD, consistent with palmitate being the most abundant fatty acid in the palm oil used to make this HFD (Table 1). Metabolomic pathway analysis identified several lipid pathways with significant changes after HFD (Table 2). Notably, acyl-carnitine species from C6-C16 were increased after HFD compared to regular chow controls (Figure 1B). This indicates that acyl-carnitine production is occurring faster than consumption by the mitochondria. Pyruvate was almost doubled in HFD hearts, which could indicate that pyruvate is accumulating because fatty acids are being used preferentially as a substrate, rather than glucose (Table 1). In summary, HFD caused significant changes in cardiac metabolites, including increased lipid accumulation and increased acyl-carnitine species, which could indicate mitochondrial dysfunction.
Table 1.
Cardiac metabolomics with greatest differences comparing chow or HFD, n = 10 heart samples for each group
| av chow | av HFD | ratio HFD/chow | FDR | Biochemical |
|---|---|---|---|---|
| 1.37 | 0.80 | 0.58 | 6.276E-07 | deoxycarnitine |
| 0.64 | 1.20 | 1.86 | 6.901E-06 | pyruvate |
| 0.65 | 3.03 | 4.69 | 0.0027 | laurylcarnitine (C12) |
| 0.85 | 1.23 | 1.44 | 0.0027 | myristate (C14:0) |
| 0.83 | 1.22 | 1.46 | 0.0038 | 3-hydroxyhexanoylcarnitine |
| 1.11 | 0.95 | 0.85 | 0.0047 | carnitine |
| 1.11 | 0.89 | 0.81 | 0.0048 | malonylcarnitine |
| 0.74 | 1.20 | 1.63 | 0.0051 | palmitate (16:0) |
| 0.70 | 1.68 | 2.39 | 0.0052 | octanoylcarnitine (C8) |
| 0.58 | 3.74 | 6.41 | 0.0060 | myristoylcarnitine (C14) |
| 0.73 | 2.72 | 3.71 | 0.0065 | myristoleoylcarnitine (C14:1) |
| 0.65 | 1.51 | 2.32 | 0.0066 | 3-hydroxypalmitate |
| 0.73 | 1.75 | 2.40 | 0.0069 | hexanoylcarnitine (C6) |
| 0.72 | 1.79 | 2.47 | 0.0077 | decanoylcarnitine (C10) |
Table 2.
Metabolomic pathway analysis, top hits comparing chow or HFD
| Pathway | Enrichment | Count |
|---|---|---|
| Phosphatidylinositol (PI) | 2.19 | 6 |
| Phosphatidylethanolamine (PE) | 1.83 | 13 |
| Long Chain Saturated Fatty Acid | 1.75 | 6 |
| Long Chain Polyunsaturated Fatty Acid (n3 and n6) | 1.73 | 20 |
| Lysophospholipid | 1.64 | 26 |
| Fatty Acid Metabolism | 1.57 | 5 |
| Fatty Acid Metabolism (Acyl Carnitine, Hydroxy) | 1.49 | 7 |
| Fatty Acid Metabolism (Acyl Carnitine, Polyunsaturated) | 1.43 | 11 |
| Fatty Acid Metabolism (Acyl Carnitine, Long Chain Saturated) | 1.31 | 8 |
Male and female mice have significant differences in cardiac metabolites at baseline, on regular chow
By using an equal number of male and female mouse hearts, we were able to examine sex differences in cardiac metabolites. Comparing cardiac metabolites from male and female mice on regular chow revealed significant differences in approximately 15% of detected metabolites, indicating that there are important sex differences at baseline (Table 3). A diverse range of metabolites were found to be significantly different, comparing male to female hearts on chow diet. Metabolic pathway analysis indicated that the most prominent sex differences involved phospholipid metabolism, ceramides, and several amino acid pathways (Table 4).
Table 3.
Cardiac metabolomics with greatest differences comparing male vs female mice on regular chow, n = 5 heart samples each group
| av male | av female | ratio female/male | FDR | |
|---|---|---|---|---|
| 1.21 | 0.66 | 0.55 | 0.0010 | hypotaurine |
| 1.5 | 0.43 | 0.28 | 0.0011 | carnosine |
| 1.73 | 0.45 | 0.26 | 0.0013 | anserine |
| 1.74 | 0.96 | 0.55 | 0.0014 | gulonate |
| 1.44 | 2.03 | 1.41 | 0.0016 | N6-methyllysine |
| 2.82 | 0.65 | 0.23 | 0.0018 | cinnamoylglycine |
| 0.8 | 1.17 | 1.46 | 0.0018 | sphingosine |
| 0.65 | 1.08 | 1.65 | 0.0024 | homoarginine |
| 1.12 | 0.64 | 0.57 | 0.0026 | N-glycolylneuraminate |
| 0.82 | 1.48 | 1.81 | 0.0028 | pantothenate |
| 0.98 | 0.29 | 0.3 | 0.0030 | alpha-hydroxyisovalerate |
| 1.09 | 2.33 | 2.14 | 0.0036 | sphingomyelin (d18:1/18:1, d18:2/18:0) |
| 1.09 | 0.44 | 0.4 | 0.0044 | imidazole lactate |
| 1.48 | 0.78 | 0.53 | 0.0045 | N-acetylglutamine |
| 1.13 | 0.6 | 0.53 | 0.0045 | glycerophosphoserine |
Table 4.
Metabolomic pathway analysis of comparison of male vs female mice on regular chow
| Pathway | Enrichment | Count |
|---|---|---|
| Phospholipid Metabolism | 2.04 | 10 |
| Ceramides | 2.03 | 5 |
| Plasmalogen (ether phospholipids) | 1.88 | 9 |
| Gamma-glutamyl Amino Acid | 1.69 | 6 |
| Ascorbate and Aldarate Metabolism | 1.69 | 6 |
| Tryptophan Metabolism | 1.69 | 8 |
| Leucine, Isoleucine and Valine Metabolism | 1.63 | 21 |
| Nicotinate and Nicotinamide Metabolism | 1.54 | 11 |
| Histidine Metabolism | 1.54 | 11 |
| Urea cycle; Arginine and Proline Metabolism | 1.48 | 16 |
| Sphingomyelins | 1.46 | 21 |
| Lysine Metabolism | 1.45 | 14 |
| Nucleotide Sugar | 1.34 | 5 |
Male and female hearts have different metabolic responses to HFD
Because there were significant differences in metabolites in male and female hearts at baseline (on regular chow), it was not surprising to find that there were significant differences in metabolites in male and female hearts on HFD. However, the metabolites with significant differences after HFD had moderate overlap with the metabolites with significant differences at baseline. Alpha-hydroxyisovalerate, N6-methyllsyine, and anserine were metabolites that had significant differences after both baseline and HFD (Tables 3 and 5). Similarly, metabolomic pathway analysis shows moderate overlap of the pathways identified as different, comparing male to female hearts on regular chow, with those identified as the top differences comparing male to female on HFD. For example, tryptophan, lysine, leucine/isoleucine, and fatty acid metabolism were among the top sex differences for both chow and HFD comparisons (Tables 4 and 6). In contrast, long-chain saturated fatty acid and hydroxy-acylcarnitine pathways were only identified as having sex differences in the HFD samples (Table 6). Thus, sex differences in cardiac metabolites in response to HFD cannot be reliably predicted from sex differences at baseline.
Table 5.
Cardiac metabolomics with greatest differences comparing male vs female hearts on HFD, n=5 heart samples each group
| av M palm | av F palm | ratio F/M | FDR | |
|---|---|---|---|---|
| 1.21 | 0.34 | 0.29 | 0.0005 | 2,3-dihydroxyisovalerate |
| 0.37 | 1.07 | 2.93 | 0.0008 | linoleoyl-arachidonoyl-glycerol (18:2/20:4) |
| 0.67 | 0.96 | 1.43 | 0.0043 | betaine |
| 0.56 | 1.27 | 2.28 | 0.0042 | dimethylglycine |
| 0.61 | 1.34 | 2.20 | 0.0055 | indolelactate |
| 1.83 | 0.42 | 0.23 | 0.0057 | alpha-hydroxyisovalerate |
| 0.88 | 1.41 | 1.61 | 0.0091 | glycerophosphoglycerol |
| 0.67 | 1.28 | 1.91 | 0.0092 | N6-methyllysine |
| 0.92 | 1.63 | 1.77 | 0.0102 | ascorbic acid 3-sulfate |
| 1.78 | 0.37 | 0.21 | 0.0093 | anserine |
| 0.99 | 1.31 | 1.33 | 0.0098 | glycerophosphorylcholine (GPC) |
| 0.97 | 3.50 | 3.59 | 0.0097 | ophthalmate |
| 0.20 | 0.50 | 2.46 | 0.0100 | N,N,N-trimethyl-5-aminovalerate |
| 0.95 | 1.32 | 1.39 | 0.0105 | glycerophosphoethanolamine |
Table 6.
Metabolomic pathway analysis of comparison of male vs female hearts on HFD
| Pathway | Enrichment | Count |
|---|---|---|
| Tyrosine Metabolism | 2.78 | 6 |
| Fatty Acid Metabolism (also BCAA Metabolism) | 2.66 | 5 |
| Tryptophan Metabolism | 2.08 | 8 |
| Fatty Acid Metabolism (Long Chain Saturated) | 2.08 | 8 |
| Fatty Acid Metabolism (Acyl Carnitine, Hydroxy) | 1.89 | 7 |
| Urea cycle; Arginine and Proline Metabolism | 1.89 | 16 |
| Purine Metabolism, (Hypo)Xanthine/Inosine containing | 1.85 | 9 |
| Glycine, Serine, and Threonine Metabolism | 1.85 | 9 |
| Leucine, Isoleucine, and Valine Metabolism | 1.76 | 21 |
| Phospholipid Metabolism | 1.66 | 10 |
| Lysine Metabolism | 1.66 | 14 |
| Gamma-glutamyl Amino Acid | 1.65 | 6 |
| Pantothenate and CoA Metabolism | 1.65 | 6 |
| Phosphatidylethanolamine (PE) | 1.53 | 13 |
| Fatty Acid Metabolism (Acyl Carnitine, Polyunsaturated) | 1.50 | 11 |
| Phosphatidylcholine (PC) | 1.36 | 17 |
| Long Chain Polyunsaturated Fatty Acid (n3 and n6) | 1.33 | 20 |
To achieve a better understanding of the sex differences on HFD, we compared the values of acyl-carnitine species. We found that female hearts on HFD had significantly higher levels of several acyl-carnitines compared to male hearts on HFD (Figure S1). The female hearts on HFD did not have a significantly greater amount of palmitate compared to male hearts on HFD, implying that sex differences in the processing of fatty acids is more likely to explain this observation rather than a sex difference in uptake. In addition, there was no significant sex difference in pyruvate after HFD (Figure S1).
Electron microscopy shows changes in cardiac mitochondrial size and number after HFD
To determine if there were significant abnormalities in the structure of cardiac mitochondria after HFD, we performed electron microscopy on cardiac ventricular samples (Figure 1C). Although there were no obvious signs of severe mitochondrial damage, quantitative analysis showed significant differences in mitochondrial number and size (as measured by cross-sectional area) after HFD (Figure 1D). HFD hearts had significantly fewer mitochondria, and these mitochondria had significantly larger average size, probably because of shift in mitochondrial fission/fusion dynamics. However, the magnitude of these effects is modest.
RNA-seq shows metabolic reprogramming of the heart after HFD
After finding that HFD causes dramatic changes in the cardiac metabolomic profile, we set out to examine how the cardiac transcriptome is altered by HFD. To determine the transcriptomic profile of heart tissue at baseline and in response to HFD in both male and female mice, we collected ventricular samples from the same cohort of mice as the metabolomic study for RNA-sequencing.
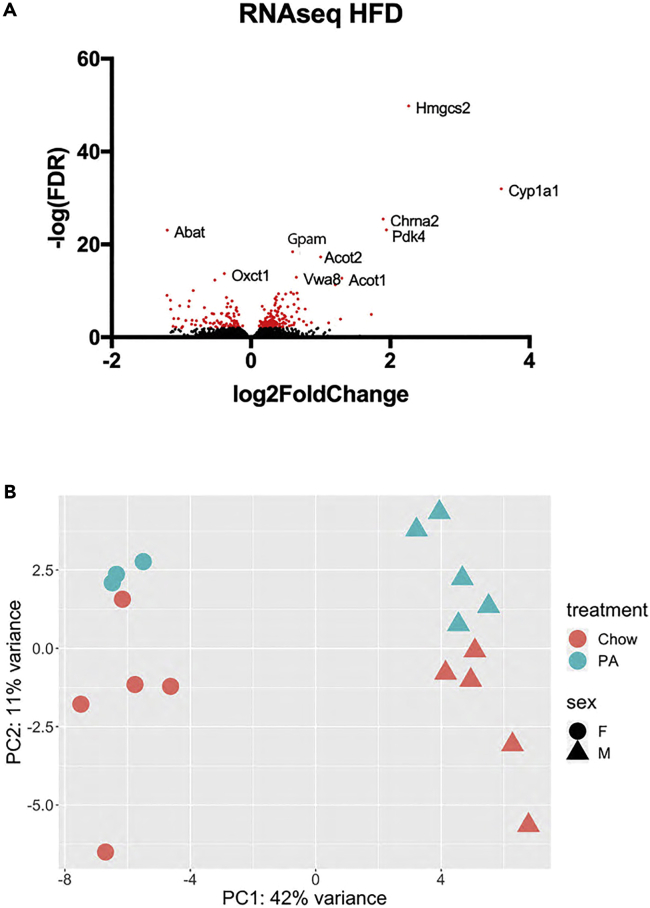
RNA-seq from heart tissue revealed significant differences in expression of a large number of transcripts; 292 mRNA had significantly different expression with FDR <0.05 (Figure 2A, Table 7). Overall, the pattern displayed a shift toward upregulation of genes coding for enzymes used in fatty acid metabolism. There was also a decrease in some enzymes that are important for glucose metabolism. Glucose transporter (Glut) expression was significantly decreased at the RNA level in HFD samples. Abat (4-aminobutyrate aminotransferase, mitochondrial) and Bdh1 (D-beta-hydroxybutyrate dehydrogenase, mitochondrial), which are important for ketone metabolism, were significantly decreased at the mRNA level in HFD samples. Overall, these data indicate that the heart uses less glucose and less ketone as substrate during HFD, which could be adaptive, because there is an abundance of fatty acids during HFD.
Figure 2.
Visualizing RNAseq results
(A) Volcano plot of cardiac RNAseq data comparing HFD to regular chow, with red color indicating statistical significance.
(B) PCA plot illustrates the clustering of RNA-seq samples.
Table 7.
HFD RNAseq highlights comparing HFD to chow
| Gene name | av chow | av HFD | ratio HFD:chow | FDR | notes |
|---|---|---|---|---|---|
| Hmgcs2 | 466.45 | 2243.77 | 4.81 | 2.08E-07 | This enzyme condenses acetyl-CoA with acetoacetyl-CoA to form HMG-CoA |
| Abat | 512.85 | 222.85 | 0.43 | 9.80E-06 | 4-aminobutyrate aminotransferase, mitochondrial |
| Chrna2 | 73.27 | 272.67 | 3.72 | 2.67E-05 | Neuronal acetylcholine receptor subunit alpha-2 |
| Gpam | 5477.43 | 8292.39 | 1.51 | 2.97E-05 | Glycerol-3-phosphate acyltransferase 1, mitochondrial |
| Oxct1 | 40499.83 | 31120.16 | 0.77 | 0.0001 | protein SCOT (succinyl-CoA:3-oxoacid CoA transferase), mitochondrial |
| Vwa8 | 10666.38 | 16767.07 | 1.57 | 0.0002 | mitochondrial: Exhibits ATPase activity in vitro. Increased in liver by HFD |
| Ptpn3 | 2040.2 | 2684.23 | 1.32 | 0.0005 | Tyrosine-protein phosphatase non-receptor type 3:L |
| Acot2 | 2948.08 | 5801.6 | 1.97 | 0.0008 | mitochondrial: Acyl-CoA thioesterases |
| Hsd17b11 | 927.09 | 1468.15 | 1.58 | 0.0008 | Estradiol 17-beta-dehydrogenase 11 |
| Decr1 | 14063.98 | 19875.08 | 1.41 | 0.0011 | Auxiliary enzyme of beta-oxidation |
| Ephx2 | 7777.01 | 11592.17 | 1.49 | 0.0016 | Bifunctional enzyme |
| Acot1 | 586.15 | 1446.88 | 2.47 | 0.0019 | Acyl-CoA thioesterase |
| Hadha | 55932.89 | 72107.1 | 1.29 | 0.0019 | Trifunctional enzyme subunit alpha, mitochondrial |
| Cat | 7041.04 | 8982.66 | 1.28 | 0.0021 | Catalase |
| Mthfd2 | 70.03 | 161.98 | 2.31 | 0.0026 | mitochondrial bifunctional enzyme |
| Slc2a4 | 11871.31 | 9525.91 | 0.8 | 0.0037 | aka Glut4, Solute carrier family 2, facilitated glucose transporter member 4 |
| Bdh1 | 2123.81 | 956.14 | 0.45 | 0.0039 | D-beta-hydroxybutyrate dehydrogenase |
| Pdk4 | 11762.44 | 45314.11 | 3.85 | 0.0045 | Kinase that plays a key role in regulation of glucose and fatty acid metabolism |
| Impa2 | 570.38 | 871.88 | 1.53 | 0.0046 | Inositol monophosphatase 2 |
| Cyp1a1 | 10.18 | 123.51 | 12.13 | 0.0289 | Cytochromes P450 are a group of heme-thiolate monooxygenases |
Hydroxymethylglutaryl-CoA synthase (HMGcs2) showed one of the greatest increases in expression after HFD (Figure 2A). This mitochondrial enzyme condenses acetyl-CoA with acetoacetyl-CoA to form HMG-CoA. In the liver, this enzyme is important for ketogenesis (Wang et al., 2019). The role of HMGcs2 in cardiac metabolism is not well-defined.
We performed PCA to define the overall similarity of the samples. The first principal component separated male and female mice, suggesting significant sex differences in chow-fed mice. The second principal component separated HFD-fed vs. chow-fed samples for both male and female mice (Figure 2B).
We next performed differential expression (DE) analysis to determine the genes showing significantly differential expression using FDR-adjusted p-value < 0.05 as the cutoff and further filtered by using absolute fold-change (FC) > 1.5. DE analysis revealed that 46 genes were significantly upregulated in HFD-fed mice, whereas 35 genes were significantly downregulated in HFD-fed mice (Figure 3A, Table S2). As an additional method to analyze the RNAseq data, we performed Gene Set Enrichment Analysis (GSEA). GSEA analysis identified 753 gene sets that were significantly enriched (FDR <0.25) for DE genes in HFD-fed vs. chow-fed mice (Tables S3 and S4). Of these, 213 gene sets were significantly upregulated, and 540 were significantly downregulated in HFD-fed mice. The top enriched gene sets are visualized in Figure 3. The most significant gene sets enriched for upregulated DE genes in HDF-fed mice include PPAR-alpha activates gene expression and long-chain fatty acid metabolism (Figure 3B), consistent with effects expected for HFD. The most significant gene sets enriched for downregulated DE genes in PA-fed mice include ATP metabolic process, mitochondrial translation, and oxidative phosphorylation (Figure 3C), which could result in impaired mitochondrial function.
Figure 3.
RNA-seq results for HFD vs chow
(A) Results of differential expression (DE) analysis.
(B) The dot plot visualizes the top eight Gene Ontology (GO) Biological Process terms enriched for upregulated genes in HFD-fed vs. chow-fed mice. The two representative GSEA plots for Human Reactome Pathway “PPARa activates gene expression” and GO term “long-chain fatty acid metabolic process” are visualized. The complete GSEA report is available in Table S3.
(C) The dot plot visualizes the top eight GO Biological Process terms enriched for downregulated genes in HFD-fed vs. chow-fed mice. The two representative GSEA plots for GO terms “ATP metabolic process” and “oxidative phosphorylation” are visualized. The complete GSEA report is available in Table S4.
To gain a more detailed understanding of the pathways responsible for the transcriptomic changes caused by HFD, we performed upstream regulator analysis using Ingenuity Pathway Analysis (IPA). IPA indicated that the differences in mRNA between chow and HFD were most likely caused by activation of several pathways: KLF, PPAR, PGC1a, RXRa (which can form heterodimers with PPARs), and FOXO transcription factors (Table 8). The identification of these pathways is not particularly surprising given their known role in cardiac metabolism (Son et al., 2010; Battiprolu et al., 2012). Pathway analysis also indicates activation of p53. Although p53 is best known as a tumor suppressor, it also has roles in regulating mitochondrial metabolism (Matoba et al., 2006) and cardiac hypertrophy remodeling (Sano et al., 2007).
Table 8.
Upstream ingenuity pathway analysis of RNA seq data comparing HFD to chow
| Upstream Regulator | Molecule Type | Activation z-score | p-value |
|---|---|---|---|
| KLF15 | transcription regulator | 4.105 | 1.26E-28 |
| PPARG | ligand-dependent nuclear receptor | 3.407 | 2.75E-27 |
| PPARA | ligand-dependent nuclear receptor | 5.445 | 7.89E-24 |
| PPARD | ligand-dependent nuclear receptor | 2.858 | 1.39E-19 |
| PPARGC1A | transcription regulator | 2.779 | 3.63E-17 |
| RXRA | ligand-dependent nuclear receptor | 1.896 | 1.66E-14 |
| FOXO1 | transcription regulator | 3.232 | 6.61E-14 |
| TP53 | transcription regulator | 1.571 | 7.47E-13 |
Male and female mice have significant differences in cardiac mRNA at baseline
Comparison of RNA-seq data from males and female hearts on chow revealed several statistically significant differences: 25 transcripts had statistically significant differences by FDR (Table 9). Several of the top hits are from genes known to be located on sex chromosomes (e.g., Kdm5d and Uty); therefore, differential sex expression is expected. Some of the differentially expressed genes could be relevant to cardiac physiology. Tcap, also called telethonin, is muscle assembly regulating factor that regulates assembly of titin (TTN) molecules. This gene had significantly greater mRNA levels in female hearts compared to male. UCP3 is a mitochondrial uncoupler protein which showed significantly greater mRNA levels in female hearts compared to male hearts on regular chow. We conclude that there are significant sex differences in the cardiac transcriptome at baseline, and the differences would not necessarily predict the differences in the metabolomic dataset, emphasizing the value of a multi-omic approach.
Table 9.
RNAseq highlights of sex differences on chow diet
| Gene name | av F | av M | ratio | FDR | notes |
|---|---|---|---|---|---|
| Kdm5d | 0.00 | 516.77 | 0.00 | 7.28427E-06 | Lysine-specific demethylase 5D |
| Uty | 1.01 | 660.54 | 0.00 | 1.02269E-05 | Male-specific histone demethylase |
| Eif2s3y | 0.40 | 1622.39 | 0.00 | 2.70527E-05 | As a subunit of eukaryotic initiation factor 2 (eIF2), involved in the early steps of protein synthesis |
| Xist | 7427.97 | 1374.10 | 5.41 | 3.09696E-05 | RNA-binding protein that acts as a key regulator of N6-methyladenosine (m6A) methylation of RNAs |
| Ddx3y | 1.22 | 2417.80 | 0.00 | 0.0001 | Probable ATP-dependent RNA helicase. May play a role in spermatogenesis |
| Arntl | 302.76 | 822.06 | 0.37 | 0.0005 | Transcriptional activator which forms a core component of the circadian clock |
| Tcap | 38893.51 | 19691.70 | 1.98 | 0.0004 | Muscle assembly regulating factor. Mediates the antiparallel assembly of titin (TTN) |
| Dbp | 2711.67 | 731.38 | 3.71 | 0.0008 | D site-binding protein |
| Nr1d1 | 4370.40 | 2177.92 | 2.01 | 0.0009 | Transcriptional repressor which coordinates circadian rhythm and metabolic pathways |
| Kdm6a | 1247.16 | 727.20 | 1.72 | 0.0020 | Lysine-specific demethylase 6A aka UTX |
| C7 | 75.89 | 360.38 | 0.21 | 0.0054 | May be involved in protection from oxidative damage |
| Nr1d2 | 4624.64 | 2686.70 | 1.72 | 0.0090 | Transcriptional repressor which coordinates circadian rhythm and metabolic pathways |
| Cth | 94.28 | 42.13 | 2.24 | 0.0094 | Catalyzes the last step in the trans-sulfuration pathway from methionine to cysteine |
| Urah | 9.63 | 21.77 | 0.44 | 0.0098 | 5-hydroxyisourate hydrolase |
| Cry1 | 247.00 | 450.13 | 0.55 | 0.0092 | Transcriptional repressor which forms a core component of the circadian clock |
| Hsd11b1 | 215.68 | 405.08 | 0.53 | 0.0115 | Catalyzes reversibly the conversion of cortisol to the inactive metabolite cortisone |
| Serpine1 | 1426.33 | 743.36 | 1.92 | 0.0122 | Serine protease inhibitor |
| Adam19 | 1195.23 | 2507.78 | 0.48 | 0.0164 | Disintegrin and metalloproteinase domain-containing protein 19 |
| Ucp3 | 3041.30 | 1400.19 | 2.17 | 0.0401 | Uncoupling protein 3 |
| Npas2 | 129.50 | 220.19 | 0.59 | 0.0482 | Transcriptional activator which forms a core component of the circadian clock |
DE analysis revealed that 154 genes were significantly upregulated in chow-fed females, whereas 130 genes significantly downregulated in chow-fed females compared to chow-fed males (Table S5). Using GSEA analysis and an FDR cutoff at 0.25 revealed that no gene sets were significantly enriched for DE genes. Using nominal p value < 0.01 as the cutoff, 50 gene sets were found to be significantly enriched for DE genes. Of these, 17 were upregulated and 23 were downregulated in chow-fed females (Tables S6 and S7). Thus, although there was a relatively large number of DE genes observed in chow-fed female vs chow-fed male, there was modest enrichment in specific pathways. This is to some extent expected because sex difference is relevant to homeostasis and physiological regulation, thus may involve many pathways with modest effect size. By looking into top enrichment pathways based on nominal p value, GSEA results indicated that female hearts had greater expression of genes involved in placenta development, genetic imprinting, and regulation of circadian cycles. Male hearts had greater expression of genes associated with steroid hormone signaling, androgen receptor signaling, protein demethylation, and leukocyte immunity.
Male and female heart mRNA response to HFD
We next asked if there were sex-specific transcriptomic responses to HFD. For HFD-fed females vs. chow-fed females, DE analysis revealed 77 significantly upregulated genes, whereas 17 were significantly downregulated (Table S8). For HFD-fed males vs. chow-fed males, DE analysis revealed that 41 genes were significantly upregulated, whereas 27 genes significantly downregulated (Table S11). The upregulated genes showed minimal overlap between females and males, i.e., eight genes (Figure 4A). There were fewer genes downregulated by HFD, with greater overlap between females and males (Figure 4B).
Figure 4.
RNA-seq results of sex-specific responses to HFD
(A) The Venn diagram illustrates the overlap of differentially expressed genes upregulated in HFD-fed vs. chow-fed mice for female and male mice, respectively.
(B) The Venn diagram illustrates the overlap of differentially expressed genes downregulated in HFD-fed vs. chow-fed for female and male mice, respectively.
(C) The dot plot visualizes the top 10 GO Biological Process terms enriched for upregulated genes in chow-fed males vs. HFD males. The representative GSEA plots are also included. The complete GSEA report is available in Table S12.
(D) The dot plot visualizes the top 10 GO Biological Process terms enriched for upregulated genes in chow-fed females vs HFD females. The representative GSEA plots are also included. The complete GSEA report is available in Table S13.
(E) Upstream Regulator analysis is performed by IPA for HFD vs. chow-fed for female mice only (female), male mice only (male), and female and male mice with sex as a covariant (all). The activation and inhibition z-score for the top upstream regulators are visualized by heatmap.
Consistent with the observation of sex-specific DE genes in response to HFD, GSEA analysis also revealed gene sets showing specific or more robust enrichment in a sex-specific manner. GSEA analysis for DE genes in HFD-fed females vs. chow-fed females identified 1529 gene sets to be significantly upregulated and 504 gene sets to be downregulated in HFD-fed females (Tables S9 and S10). For DE genes in HFD-fed males vs. chow-fed males, GSEA analyses identified 55 gene sets to be significantly upregulated and 317 gene sets to be downregulated in HFD-fed males (Tables S12 and S13). The analyses imply that female mice showed more substantial responses to HFD, which induced more upregulated genes and pathways. Male mice showed gene sets supplemented with upregulated genes related to fatty acid metabolism and regulation of lipid metabolism by PPARa (Figure 4C). Female mice showed gene sets enriched for upregulated genes related to collagen chain trimerization and beta1 integrin cell surface interaction (Figure 4D). In addition, upstream regulator analysis performed by IPA for HFD-fed vs. chow-fed for female mice and male mice revealed a female-response to HFD that was seen to involve a high activation Z score for a larger number of top upstream regulators, compared to the male response to HFD (Figure 4E). Some of these enriched gene sets were not observed in male mice, indicating that female mice are more subject to HFD-induced cardiac remodeling. Consistent with the relatively higher percentage of overlap in the downregulated DE genes between females and males, the gene sets enriched for downregulated DE genes were similar comparing females and males.
Nuclear western blots show activation of multiple transcription factors after HFD
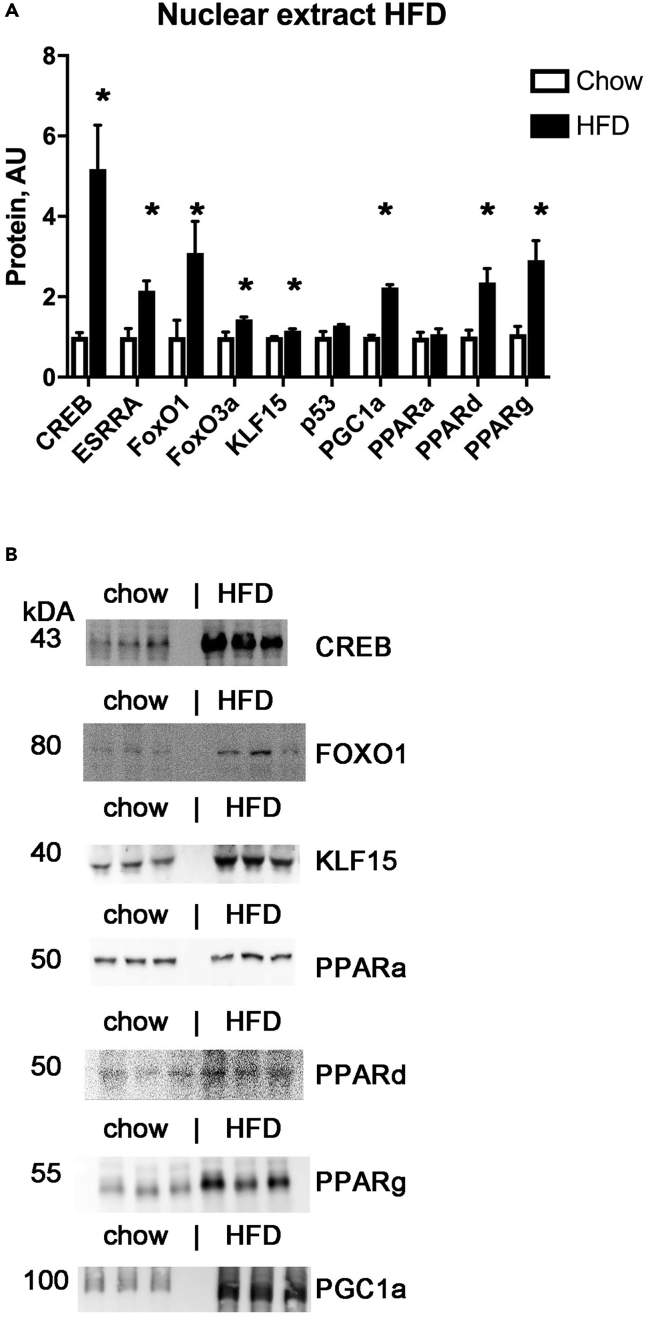
Because the RNA-seq data showed a reprogramming of cardiac metabolic gene expression, we performed nuclear western blots to determine which transcription factors were activated by HFD, with a focus on transcription factors identified by upstream regulator analysis and others known to have a prominent role in metabolic regulation. We confirmed that the nuclear lysates were enriched for nuclear proteins (Figure S2).
Nuclear western blots showed a significant increase in the nuclear levels of the transcription factor CREB (cAMP-response element binding) (Figure 5), which has an important role in the transcription of both mitochondrial genes and some voltage-gated ion channels (Wu et al., 2006; Huang et al., 2013). ESRRa, which regulates cardiac energy production (Vega and Kelly, 2017), was significantly increased in HFD samples. PGC1a, a transcriptional coactivator that has been found to work with CREB, ESRRa, and PPARs, was also found to be significantly higher in cardiac nuclei after HFD. FOXO1 and FOXO3a were significantly increased in HFD samples, which may be maladaptive after longer duration of HFD by promoting insulin resistance (Battiprolu et al., 2012). KLF15, which was identified by IPA and has a known role in the circadian regulation of cardiac metabolism (Zhang et al., 2015), showed a moderate but statistically significant increase at the nuclear protein level.
Figure 5.
Nuclear lysate western blots from mouse heart tissue
(A) Graph of nuclear lysate western blot results. Mean + SEM, ∗ = p < 0.05 by t-test.
(B) Representative examples of nuclear lysate western blots.
There was also a significant increase in nuclear levels of PPARg and PPARd, which are known to have important roles in regulating cardiac metabolism (Figure 5) (Son et al., 2010; Palomer et al., 2016). PPARa is known to have a prominent role in regulation of fatty acid oxidation and diabetic cardiomyopathy (Nakamura et al., 2019), but we did not find increased levels of this protein in the nuclear lysates at this 4 week HFD time point, which is before the onset of obesity. Although p53 was identified by RNAseq upstream regulator analysis, we did not detect any significant differences in cardiac nuclear protein level (Figure 5). In summary, the changes in gene expression observed in the RNA-seq data can be explained by upregulation of multiple transcription factors that regulate mitochondrial genes and/or metabolic enzymes.
Cardiac protein expression after HFD
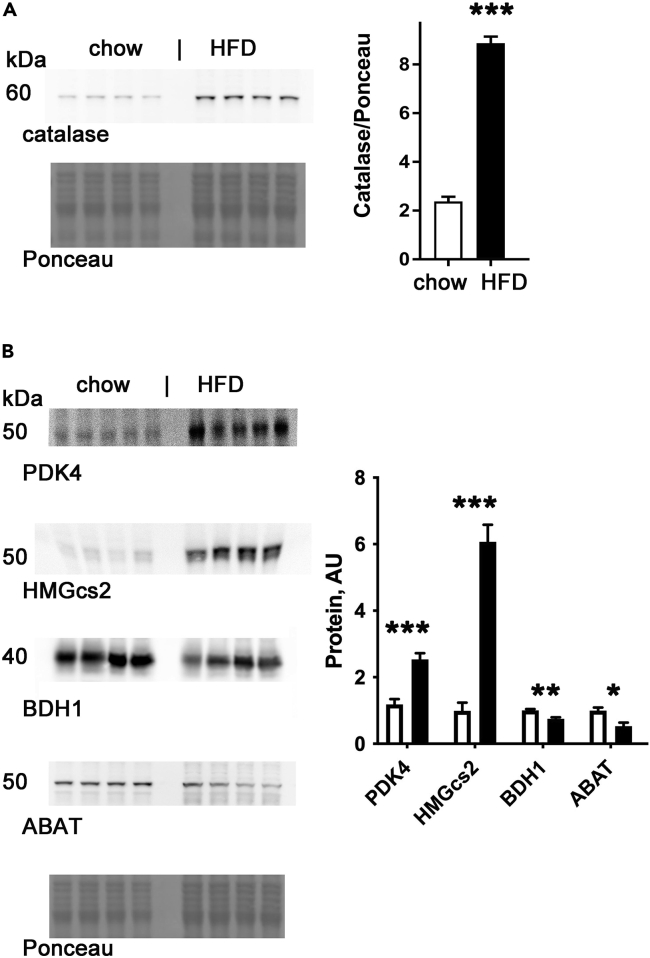
To determine if the RNA-seq data correlated with protein levels, we performed western blots from cardiac tissue for several of the proteins with significant changes in the corresponding mRNA. We found that catalase is increased in the heart at both the mRNA and protein level after 4 weeks of HFD (Figure 6A). This is most likely an adaptive response to the increase in oxidative stress caused by HFD. PDK4 (which is located in the mitochondrial matrix) is increased at both the mRNA and protein levels (Figure 6B). PDK4 inhibits pyruvate dehydrogenase which decreases glucose utilization, consistent with increased use of fatty acids as substrate.
Figure 6.
Western blots of catalase and metabolic proteins after 4-weeks of HFD
(A) Catalase blot, loading control, and graph of quantification.
(B) Blots of PDK4, HMGcs2, BDH1, ABAT, Ponceau loading control, and graph of quantifications adjusted for loading. Mean + SEM, ∗ = p < 0.05 by ∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.0001.
HMGcs2 is increased at both the mRNA and protein level. In contrast, BDH1, an enzyme involved in ketogenesis in the liver and ketolysis in other tissues, is decreased at both the mRNA and the protein level (Figure 6B). ABAT, which is best known for its role in ketone metabolism but may also have a role in regulating mitochondrial nucleoside metabolism (Besse et al., 2015), is decreased at both the mRNA and the protein level (Figure 6B). In summary, changes in protein levels correspond to changes in mRNA for several important metabolic enzymes after HFD, indicating an adaptation to reduce oxidative stress and to use increased levels of fatty acids as a substrate.
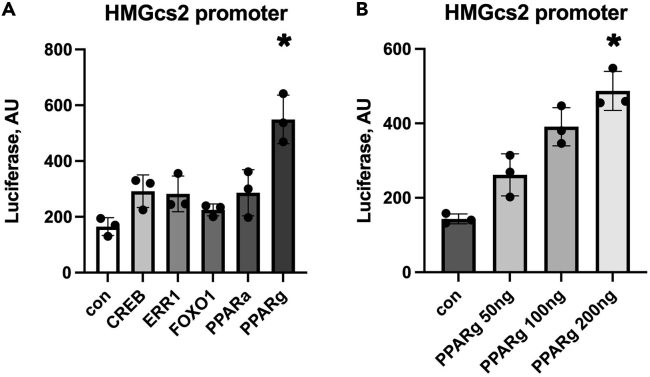
The HMGcs2 promoter is upregulated in response to PPARg
Because HMGcs2 was highly upregulated at the mRNA and protein level, we created a promoter-luciferase construct to examine the response to the transcription factors activated by HFD. We co-transfected the cardiomyoblast H9c2 cell line with the HMGcs2 promoter luciferase plasmid and expression vectors for several of the metabolic transcription factors that appeared to be upregulated by HFD. We found that PPARg had the greatest effect (Figures 7A and 7B). PPARa, in contrast, did not cause a significant increase in promoter activity for HMGcs2.
Figure 7.
HMGcs2 promoter luciferase
(A) Comparison of transcription factors co-transfected with the HMGcs2 promoter luciferase plasmid, n = triplicate wells, each experiment was performed at least 3 times. Mean ± SEM, the means are significantly different by ANOVA, ∗ indicates statistically significant difference from control by Tukey’s post-hoc test.
(B) Dose-response of PPARg expression plasmid with the HMGcs2 promoter luciferase plasmid.
Discussion
To sustain life, the heart must perform rhythmic mechanical work, which results in an intense metabolic demand for cardiomyocytes to produce ATP. The heart has been called a metabolic omnivore in that it can use multiple substrates as fuel (Bertero and Maack, 2018). By using a multi-omic approach, we investigated the response of the heart to short-term HFD, before the onset of obesity. Our metabolomic data indicate that HFD causes cardiac lipid accumulation, a change in substrate preference to use lipids as substrate, and an increase in acyl-carnitines, which could indicate early-stage mitochondrial dysfunction. In fact, increased serum levels of acyl-carnitines are used to diagnose genetic mitochondrial myopathy. The RNA-seq dataset from the same hearts reveals a somewhat different picture. There is a complex response in cardiac mRNA after 4 weeks of HFD, consistent with an adaptive response to use lipids as fuel and reduce oxidative stress. Notably, our data showed large increases in Pdk4, HMGcs2, and catalase mRNA, and the corresponding protein levels increased significantly. Both Pdk4 and HMGcs2 are differentially expressed in obese human hearts with hypertrophy (Newman et al., 2017), which underscores the potential relevance of our findings to some forms of human heart disease. Another group has recently reported metabolomic changes in mouse atrial tissue in response to 16 weeks of HFD (Suffee et al., 2021). Similar to our results from ventricular tissue, this study showed accumulation of some fatty acids and increased use of fatty acid as a substrate.
How could these changes in protein levels alter cardiac metabolism? Pdk4 is located in the mitochondrial matrix and it inhibits pyruvate dehydrogenase, which in turn decreases glucose utilization and increases fat utilization. Pdk4 is known to be a direct transcriptional target of Foxo1 (Gopal et al., 2017) and is also upregulated by PGC1a (Ma et al., 2005). Is the increase in PDK4 in the heart after HFD adaptive? There is prior literature investigating Pdk4 using genetic approaches. A whole-body Pdk4 KO mouse exists. Homozygous Pdk4 KO mice were created and are viable. Interestingly, these mice did better on long-term HFD, with less weight gain and better glucose tolerance (Hwang et al., 2009). This publication did not include data examining the heart. The Pdk4 KO mice have lower blood glucose with starvation, possibly because of a relative inability to change substrates (Jeoung et al., 2006). In addition, cardiac overexpression Pdk4 mice have been created. Although were found to be normal at baseline, when crossed with the calcineurin-induced cardiomyopathy line, they had increased mortality and more fibrosis (Zhao et al., 2008). These cardiac-overexpression-Pdk4 mice had normal contractility, increased fatty acid oxidation, and significantly less cardiac triglyceride accumulation after 4 weeks of HFD (Chambers et al., 2011). In summary, there has been a large amount of prior work examining Pdk4 in the heart. It is known to be upregulated by HFD, and transgenic overexpression does not cause cardiomyopathy, at least in young adult animals. Because the global Pdk4 KO does better with long-term HFD, the increase that we observe may be adaptive in the short term but perhaps would cause cardiac pathology in the long term.
HMGcs2 is best known as a ketogenic enzyme. HMG-CoA is also involved as an intermediate in branch chain amino acid metabolism and mevalonate-cholesterol synthesis. In a rodent model, hepatic KO of HMGcs2 by antisense oligos made HFD-induced liver injury worse, with extensive hepatocyte injury and inflammation (Cotter et al., 2014). Less is known about HMGcs2 in the heart. Our data show that 4 weeks of palm diet causes a large increase in HMGcs2 protein in the mouse heart. This is somewhat puzzling because this enzyme makes ketone bodies, and the heart is thought to use ketones rather than synthesize ketones. Extrahepatic ketone synthesis is controversial (Puchalska and Crawford, 2017). There is one older report of perfused hearts making ketones (Opie and Owen, 1975). Other groups have shown that HMG CoA synthase is upregulated in the mouse heart after HFD or Type 1 DM (Sikder et al., 2018; Cook et al., 2017). One possible physiological explanation is that the heart is using HMGcs2 to shunt excess acetyl-CoA to prevent inhibition of pyruvate metabolism.
We found that catalase protein level is significantly increased after HFD, which is most likely an adaptive response to increased oxidative stress. Prior work showed that HFD increases catalase in cardiac mitochondria and just three days of HFD can cause a significant increase (Rindler et al., 2013). Transcriptional upregulation of catalase is known to involve PPARg (Okuno et al., 2010) and FOXO transcription factors (Tan et al., 2008).
Both RNAseq upstream regulator analysis and nuclear western blots indicate that multiple transcription factors are activated in the heart by HFD, resulting in changes in mRNA expression for a wide range of genes. Some of these findings confirm prior work; for example, KLF15 is known to be an important regulator of cardiac lipid metabolism (Prosdocimo et al., 2014). It is also established that the PPAR transcription factors are important regulators of cardiac metabolism. Despite the large amount of research focused on PPARa in the heart, for some genes (such as HMGcs2), PPARg could be more important for transcriptional regulation than PPARa, as shown by the promoter-luciferase experiments. Model systems that only focus on one transcription factor or pathway are unlikely to capture the complex response of cardiac metabolism to HFD.
The results regarding the transcription factor CREB are interesting for several reasons. The RNA-seq pathway analysis did not identify CREB as one of the top hits; yet, the nuclear western blots indicate that CREB may play an important role in the cardiac response to short-term HFD. CREB is a central regulator of metabolism (Altarejos and Montminy, 2011) and regulates mitochondrial gene expression in part by upregulating PGC1a (which was one of the top hits in the pathway analysis) (Herzig et al., 2001; Wu et al., 2006). We have previously shown that CREB is decreased in WT diet-induced obese mouse hearts (Huang et al., 2013), so it appears that CREB has a biphasic response depending on the duration of HFD. It is also worth noting that CREB has been found to be decreased in animal models of cardiac hypertrophy (Watson et al., 2007), which could involve mitochondrial dysfunction as part of the pathophysiology.
Sex differences
There is increasing interest in sex differences in cardiac biology (Murphy et al., 2017; Walker et al., 2021). Several prior studies have examined sex differences in cardiac metabolism. A human study showed that male hearts have greater glucose uptake than female (Peterson et al., 2015). Prior works based on animal experiments have also shown sex differences in lipid metabolism, with male diabetic rats having greater oxidative metabolism of triglycerides (Devanathan et al., 2016). Although it is established that male and female hearts have significant differences in metabolism and gene expression, our work advances the field by combining metabolomic and RNAsequencing approaches to gain a more detailed understanding of sex differences at baseline and in response to HFD. To the best of our knowledge, this is the first demonstration of sex differences in cardiac metabolomics. One notable finding is that the most prominent sex differences at baseline (on regular chow) in the multi-omic dataset did not necessarily predict the most prominent sex differences after HFD. Thus, there can be important interactions between biological sex and the cardiac response to different forms of nutrition.
Limitations
A limitation of this work is that we measured metabolite levels rather than quantifying metabolic flux. Future work could focus on evaluating metabolic flux in male and female hearts. Another limitation is that this work used mice as a model system, which may not correspond to human cardiac biology. However, rodents have evolved as omnivores, similar to humans, so some metabolic pathways are probably conserved. The experiments with the promoter-luciferase construct in the H9c2 cell line employs a reductionist approach that may not correspond to the complexity of in vivo gene regulation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Monoclonal anti-CREB (48H2) | Cell Signaling | Cat #9197 |
| Rabbit Polyclonal anti-ESRRA | Abclonal | Cat #A14184 |
| Rabbit Polyclonal anti-p53 | Abclonal | Cat #A3185 |
| Rabbit Polyclonal anti-FoxO1 | Abclonal | Cat #A2934 |
| Rabbit Polyclonal anti-FoxO3a | Abclonal | Cat #A0102 |
| Goat Polyclonal anti-KLF15 | Abcam | Cat #ab2647 |
| Rabbit Polyclonal anti-PGC1 alpha+beta | Abcam | Cat #ab72230 |
| Rabbit Polyclonal anti-PPAR alpha | Abcam | Cat #ab8934 |
| Rabbit Polyclonal anti-PPAR delta | Abcam | Cat #ab23673 |
| Mouse Monoclonal [A3409A] anti-PPAR gamma 2 + PPAR gamma | Abcam | Cat #ab41928 |
| Rabbit Polyclonal anti-Catalase – Peroxisome Marker | Abcam | Cat #ab52477 |
| Rabbit Polyclonal anti-PDK4 | Abcam | Cat #ab89295 |
| Rabbit Monoclonal [EPR8642] anti-HMGCS2 | Abcam | Cat #ab137043 |
| Rabbit Monoclonal [EPR4433] anti-ABAT/GABA-T | Abcam | Cat #ab108249 |
| Alkaline Phosphatase-Conjugated Goat anti-Rabbit IgG | Santa Cruz | sc-2307 |
| Critical commercial assays | ||
| Cellytic Nuclear Extraction Kit | Sigma-Aldrich | Product Code: NXTRACT |
| Lipofectamine 2000 Transfection Reagent | Thermo Fisher Scientific | Cat #11668027 |
| RIPA Lysis and Extraction Buffer | Thermo Fisher Scientific | Cat #89900 |
| Luciferase Assay System | Promega | Cat #E1500 |
| Halt Protease Inhibitor Cocktail (100x) | Thermo Fisher Scientific | Cat #78430 |
| Halt Phosphatase Inhibitor Cocktail (100x) | Thermo Fisher Scientific | Cat #78420 |
| Supersignal West Femto Maximum Sensitivity Substrate | Thermo Fisher Scientific | Cat #34094 |
| Deposited data | ||
| RNA Seq Data – Combined metabolomic and transcriptomic profiling approaches reveal the cardiac response to high-fat diet | Morrow Lab | GEO: GSE198546 |
| Experimental models: Cell lines | ||
| Rat Cardiomyocyte H9c2 | ATCC | CRL-1446 |
| Experimental models: Organisms/Strains | ||
| Mouse: C57BL/6J | Jackson Laboratory | JAX: 000664 |
| Recombinant DNA | ||
| Plasmid: RSV CREB | Addgene | #22394 |
| Plasmid: pCMV Flag ERR1 | Addgene | #10975 |
| Plasmid: HA-FOXO1 (pCMV5) | Addgene | #12142 |
| Plasmid: pSG5 PPARa | Addgene | #22751 |
| Plasmid: pSV Sport PPARg | Addgene | #8886 |
| Software and algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Mouse_GOBP_AllPathways_no_GO_iea_February_05_2021_symbol.gmt” | Bader Lab, University of Toronto | http://baderlab.org/GeneSets |
| Gene Set Enrichment Analysis (GSEA) | (Subramanian et al., 2005) | Version 4.1.0 |
| Salmon (version 1.5.1) | (Patro et al., 2017) | https://combine-lab.github.io/salmon/ |
| tximport (version 1.20.0) | (Soneson et al., 2015) | https://combine-lab.github.io/salmon/ |
| DESeq2 (version 1.32.0) | (Love et al., 2018) | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| BioVenn | Tim Hulsen | https://www.biovenn.nl |
| Prism v9 | GraphPad Software Inc | v9 |
| Other | ||
| High Fat Diet; 60 kcal% Palm Oil | Research Diets Inc | D04051707 |
| Picolab Rodent Diet 20 | LabDiet | Cat #3005740-220 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to, and will be fulfilled by the lead contact, John P. Morrow (jpm46@cumc.columbia.edu).
Material availability
This study did not generate new unique reagents.
Experimental model and subject details
Animal care
Animal protocols were approved by the Columbia University Institutional Animal Care and Use Committee and were carried out in accordance with the NIH guidelines for the care and use of laboratory animals. Wild type (WT) C57BL6J mice were purchased from Jackson Labs. Male and female mice were used. Mice were randomized to HFD or regular chow at 12 weeks of age. The high fat diet was a special order from Research Diets Inc: D04051707, with 60 kcal% from palm oil, which is high in saturated fat. Control chow was Picolab Rodent diet 20 from LabDiet, which has 13% kcal from fat. Animals were fed ad libitum.
Cell lines and luciferase assay
The rat cardiomyocyte H9c2 cell line was purchased from ATCC and cultured in DMEM with 10% FBS, penicillin, and streptomycin. This cell line is derived from female tissue (PMID: 23085378). Vectors were co-transfected into H9c2 cells using Lipofectamine 2000 (Invitrogen). We transfected 200 ng of luciferase construct and 100 ng of protein expression vector per well; control transfection wells received empty vector pcDNA3 to equalize the amount of DNA. Standard 12-well tissue culture plates were used. Cell lysates were harvested 36–48 h after transfection. After cell lysis, luciferase substrate (Promega, E1501) was added to lysates in a 96-well plate and luminescence was measured with a Molecular Devices SpectraMax ID3 microplate reader.
Method details
Metabolomic methodology
Sample Preparation: Cardiac samples were flash-frozen and shipped on dry ice to Metabolon (Morrisville, NC). Samples were prepared at Metabolon using the automated Micro-Lab STAR system. Several recovery standards were added prior to the first step in the extraction process for QC purposes. To remove protein and to recover chemically diverse metabolites, proteins were precipitated with methanol under vigorous shaking for 2 min (Glen Mills GenoGrinder, 2000) followed by centrifugation. The resulting extract was divided into five fractions: two for analysis by two separate reverse phase (RP)/UPLC-MS/MS methods with positive ion mode electrospray ionization (ESI), one for analysis by RP/UPLC-MS/MS with negative ion mode ESI, one for analysis by HILIC/UPLC-MS/MS with negative ion mode ESI, and one sample was reserved for backup. Samples were placed briefly on a TurboVap (Zymark) to remove the organic solvent. The sample extracts were stored overnight under nitrogen before preparation for analysis.
QA/QC: Several types of controls were analyzed in concert with the experimental samples. A pooled matrix sample generated by taking a small volume of each experimental sample served as a technical replicate throughout the dataset; extracted water samples served as process blanks; and a cocktail of QC standards that were carefully chosen not to interfere with the measurement of endogenous compounds were spiked into every analyzed sample, allowed instrument performance monitoring and aided chromatographic alignment. Instrument variability was determined by calculating the median relative standard deviation (RSD) for the standards that were added to each sample prior to injection into the mass spectrometers. Overall process variability was determined by calculating the median RSD for all endogenous metabolites (i.e., non-instrument standards) present in 100% of the pooled matrix samples. Experimental samples were randomized across the platform run with QC samples spaced evenly among the injections.
Ultrahigh Performance Liquid Chromatography-Tandem Mass Spectroscopy (UPLC-MS/MS): All methods utilized a Waters ACQUITY ultra-performance liquid chromatography (UPLC) and a Thermo Scientific Q-Exactive high resolution/accurate mass spectrometer interfaced with a heated electrospray ionization (HESI-II) source and Orbitrap mass analyzer operated at 35,000 mass resolution. The sample extract was dried then reconstituted in solvents compatible to each of the four methods. Each reconstitution solvent contained a series of standards at fixed concentrations to ensure injection and chromatographic consistency. One aliquot was analyzed using acidic positive ion conditions, chromatographically optimized for more hydrophilic compounds. In this method, the extract was gradient eluted from a C18 column (Waters UPLC BEH C18-2.1 × 100 mm, 1.7 μm) using water and methanol, containing 0.05% perfluoropentanoic acid (PFPA) and 0.1% formic acid (FA). Another aliquot was also analyzed using acidic positive ion conditions, however it was chromatographically optimized for more hydrophobic compounds. In this method, the extract was gradient eluted from the same afore mentioned C18 column using methanol, acetonitrile, water, 0.05% PFPA and 0.01% FA and was operated at an overall higher organic content. Another aliquot was analyzed using basic negative ion optimized conditions using a separate dedicated C18 column. The basic extracts were gradient eluted from the column using methanol and water, however with 6.5mM Ammonium Bicarbonate at pH 8. The fourth aliquot was analyzed via negative ionization following elution from a HILIC column (Waters UPLC BEH Amide 2.1 × 150 mm, 1.7 μm) using a gradient consisting of water and acetonitrile with 10mM Ammonium Formate, pH 10.8. The MS analysis alternated between MS and data-dependent MSn scans using dynamic exclusion. The scan range varied slighted between methods but covered 70–1000 m/z. Raw data files are archived and extracted as described below.
Bioinformatics: The informatics system consisted of four major components, the Laboratory Information Management System (LIMS), the data extraction and peak-identification software, data processing tools for QC and compound identification, and a collection of information interpretation and visualization tools for use by data analysts. The hardware and software foundations for these informatics components were the LAN backbone, and a database server running Oracle 10.2.0.1 Enterprise Edition.
Electron microscopy
Heart tissue was fixed with 2.5% glutaraldehyde and 2% paraformaldehyde in 100 mM Sodium Cacodylate buffer (pH 7.4) overnight at 4C. Samples were then treated with 1% osmium tetroxide for 1 h, washed in distilled water four times and then treated with 2% aqueous uranyl acetate overnight at 4°C in the dark. Samples were then washed and sequentially dehydrated with increasing concentrations of ethanol (20, 30, 50, 70, 90, and 100%) for 30 min each, followed by three additional treatments with 100% ethanol for 20 min each. Samples were then infiltrated with increasing concentrations of Spurr’s resin (25% for 1 h, 50% for 1 h, 75% for 1 h, 100% for 1 h, 100% overnight at room temperature), and then incubated overnight at 70°C in a resin mold. Sections of 80 nm were cut on a Leica ultramicrotome with a diamond knife. Imaging then took place using an ThermoFisher Talos L120C transmission electron microscope operating at 120 kV.
RNA-seq
Cardiac samples were preserved in RNAlater, frozen, and shipped to Genewiz (South Plainfield, NJ) on dry ice. RNA was extracted and reverse-transcribed after polyA selection to complementary DNA for sequencing on an Illumina NovaSeq platform.
Preparation of protein extracts and western blots
Mouse ventricular samples were homogenized in RIPA buffer (Thermo Scientific) containing protease and phosphatase inhibitors (Roche) using a mini bead-beater (Biospec, Bartlesville, OK). The nuclear fraction was extracted from ventricular lysates using the Cellytic Nuclear Extraction Kit (Product Code: NXTRACT, Sigma-Aldrich) according to the manufacturer’s instructions. Samples were subjected to 10% SDS-PAGE. Proteins were transferred onto a nitrocellulose membrane and probed with the appropriate antibodies. Primary antibody was used at a 1:1000 dilution. Alkaline phosphatase-conjugated goat anti-rabbit IgG (Santa Cruz) was used as secondary antibody (1:5000 dilution), and immunoreactive bands were detected with ECL solution (Thermo Scientific), according to the manufacturer’s instructions.
Quantification and statistical analysis
Data extraction and compound identification
Raw data was extracted, peak-identified and QC processed using Metabolon’s hardware and software. Compounds were identified by comparison to library entries of purified standards or recurrent unknown entities. Metabolon maintains a library based on authenticated standards that contains the retention time/index (RI), mass to charge ratio (m/z), and chromatographic data (including MS/MS spectral data) on all molecules present in the library. Furthermore, biochemical identifications are based on three criteria: retention index within a narrow RI window of the proposed identification, accurate mass match to the library +/− 10 ppm, and the MS/MS forward and reverse scores between the experimental data and authentic standards. The MS/MS scores are based on a comparison of the ions present in the experimental spectrum to the ions present in the library spectrum. While there may be similarities between these molecules based on one of these factors, the use of all three data points can be utilized to distinguish and differentiate biochemicals. The formula for metabolic pathway enrichment is (k/m)/((n-k)/(N-m)), where k = # of significant metabolites in pathway, m = total # of detected metabolites in pathway, n = total # of significant metabolites, N = total # of detected metabolites.
RNA-seq data processing and differential expression analysis
Raw sequencing reads were mapped to the mouse genome version GRCm39 (M27) using Salmon (version 1.5.1) to obtain transcript abundance counts (39). The resulting counts were summarized to the gene level using tximport (vevison 1.20.0) (40). Subsequently, genes with more than ten counts were retained for differential gene expression analysis using DESeq2 (version 1.32.0) and apeglm log fold change shrinkage (41, 42). The Venn diagrams were generated using BioVenn (43). Outlier identification: To identify and confirm outliers, we calculated each gene’s coefficient of variation (CV) and ranked genes from high CV to low CV. We then performed PCA using the top 5000 genes with large CV, including 10 dimensions. For each PC, we detect outliers by identifying samples with >6 median absolute deviations from the median. This rigorous approach detected two HFD-fed female mice samples (4 and 5) as outliers from PC1 (Figure S1). The two samples had a good quality of RNAs and RNA-seq data. The facility (GENEWIZ) processing and sequencing the libraries did not note specific QC issues. Yet, based on the PCA, we determine that the two samples have quality issues and thus excluded the two samples from further analyses.
Gene set enrichment analysis
Pre-ranked gene list for Gene Set Enrichment Analysis (GSEA) (version 4.1.0) was prepared using ranking metrics “−log10 p value multiplied by the sign of log-transformed fold-change” (44). The pathway definition file “Mouse_GOBP_AllPathways_no_GO_iea_February_05_2021_symbol.gmt” downloaded from http://baderlab.org/GeneSets.
According to the GSEA User Guide, the nominal p value estimates the significance of the observed enrichment score for a single gene set. The FDR is the estimated probability that a gene set with a given enrichment score (normalized for gene set size) represents a false positive finding. A reported p value of zero (0.0) indicates an actual p value of less than 1/number-of-permutations, which was set up as 2,000 in our analysis as recommended.
Ingenuity pathway analysis
Ingenuity pathway analysis (IPA) software using build-in scientific literature based database was used to identify canonical pathways, diseases and functions, and gene networks overrepresented in differentially expressed genes using threshold log2(Fold Change) > 0.58 or < −0.58 and FDR-adjusted p < 0.1.
Western blot analysis
Quantification of bands was performed using ImageJ software and normalized to Ponceau.
Statistical analysis
Results are given as mean ± SEM. The unpaired t test was used for comparisons of 2 means with a 2-tailed value of p <.05 was considered statistically significant. For groups of 2 or more, one-way analysis of variance (ANOVA) was used with Tukey post hoc testing or non-parametric methods (Prism v7, GraphPad Software Inc., La Jolla, CA).
Acknowledgments
The authors thank Chenyi Xue for technical advice.
Sources of Funding: This work was supported in part by T32 HL007343 to L.C.J. J.P.M. is supported by NIH R01 HL136758. H.Z. is supported by R00HL130574, R01HL151611, and the Irving Scholar award by the National Center for Advancing Translational Sciences (NCATS) at NIH through UL1TR001873. R.C.B. is supported by R01HL141745.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author contributions
L.C.J. and J.P.M. designed experiments. L.C.J., Q.N., V.P., C.G., and J.P.M. carried out experiments. L.C.J., Q.N., V.P., J.S., C.G., R.C.B., H.Z, and J.P.M. performed data analysis. J.P.M. conceived the study and wrote the manuscript. L.C.J., H.Z., and R.C.B. contributed to editing and writing sections of the manuscript. All authors reviewed and edited the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: May 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104184.
Supplemental information
Data and code availability
-
•
The metabolomic datasets and RNAseq datasets generated during the current study will be available in the public repository.
-
•
All original code has been deposited at GitHub repository and is publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Altarejos J.Y., Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battiprolu P.K., Hojayev B., Jiang N., Wang Z.V., Luo X., Iglewski M., Shelton J.M., Gerard R.D., Rothermel B.A., Gillette T.G., et al. Metabolic stress-induced activation of Foxo1 triggers diabetic cardiomyopathy in mice. J. Clin. Invest. 2012;122:1109–1118. doi: 10.1172/JCI60329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertero E., Maack C. Metabolic remodelling in heart failure. Nat. Rev. Cardiol. 2018;15:457–470. doi: 10.1038/s41569-018-0044-6. [DOI] [PubMed] [Google Scholar]
- Besse A., Wu P., Bruni F., Donti T., Graham B.H., Craigen W.J., Mcfarland R., Moretti P., Lalani S., Scott K.L., et al. The GABA transaminase, ABAT, is essential for mitochondrial nucleoside metabolism. Cell Metab. 2015;21:417–427. doi: 10.1016/j.cmet.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers K.T., Leone T.C., Sambandam N., Kovacs A., Wagg C.S., Lopaschuk G.D., Finck B.N., Kelly D.P. Chronic inhibition of pyruvate dehydrogenase in heart triggers an adaptive metabolic response. J. Biol. Chem. 2011;286:11155–11162. doi: 10.1074/jbc.M110.217349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook G.A., Lavrentyev E.N., Pham K., Park E.A. Streptozotocin diabetes increases mRNA expression of ketogenic enzymes in the rat heart. Biochim. Biophys. Acta Gen. Subj. 2017;1861:307–312. doi: 10.1016/j.bbagen.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter D.G., Ercal B., Huang X., Leid J.M., D'avignon D.A., Graham M.J., Dietzen D.J., Brunt E.M., Patti G.J., Crawford P.A. Ketogenesis prevents diet-induced fatty liver injury and hyperglycemia. J. Clin. Invest. 2014;124:5175–5190. doi: 10.1172/JCI76388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devanathan S., Whitehead T.D., Fettig N., Gropler R.J., Nemanich S., Shoghi K.I. Sexual dimorphism in myocardial acylcarnitine and triglyceride metabolism. Biol. Sex Differ. 2016;7:25. doi: 10.1186/s13293-016-0077-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal K., Saleme B., Al Batran R., Aburasayn H., Eshreif A., Ho K.L., Ma W.K., Almutairi M., Eaton F., Gandhi M., et al. Foxo1 regulates myocardial glucose oxidation rates via transcriptional control of pyruvate dehydrogenase kinase 4 expression. Am. J. Physiol. Heart Circ. Physiol. 2017;313:H479–H490. doi: 10.1152/ajpheart.00191.2017. [DOI] [PubMed] [Google Scholar]
- Herzig S., Long F., Jhala U.S., Hedrick S., Quinn R., Bauer A., Rudolph D., Schutz G., Yoon C., Puigserver P., et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- Huang H., Amin V., Gurin M., Wan E., Thorp E., Homma S., Morrow J.P. Diet-induced obesity causes long qt and reduces transcription of voltage-gated potassium channels. J. Mol. Cell. Cardiol. 2013;59:151–158. doi: 10.1016/j.yjmcc.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B., Jeoung N.H., Harris R.A. Pyruvate dehydrogenase kinase isoenzyme 4 (Pdhk4) deficiency attenuates the long-term negative effects of a high-saturated fat diet. Biochem. J. 2009;423:243–252. doi: 10.1042/BJ20090390. [DOI] [PubMed] [Google Scholar]
- Jeoung N.H., Wu P., Joshi M.A., Jaskiewicz J., Bock C.B., Depaoli-Roach A.A., Harris R.A. Role of pyruvate dehydrogenase kinase isoenzyme 4 (Pdhk4) in glucose homoeostasis during starvation. Biochem. J. 2006;397:417–425. doi: 10.1042/BJ20060125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph L.C., Avula U.M.R., Wan E.Y., Reyes M.V., Lakkadi K.R., Subramanyam P., Nakanishi K., Homma S., Muchir A., Pajvani U.B., et al. Dietary saturated fat promotes arrhythmia by activating Nox2 (NADPH oxidase 2) Circ. Arrhythm. Electrophysiol. 2019;12:E007573. doi: 10.1161/CIRCEP.119.007573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Soneson C., Patro R. Swimming downstream: statistical analysis of differential transcript usage following Salmon quantification. F1000Research. 2018;7:952. doi: 10.12688/f1000research.15398.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K., Zhang Y., Elam M.B., Cook G.A., Park E.A. Cloning of the rat pyruvate dehydrogenase kinase 4 gene promoter: activation of pyruvate dehydrogenase kinase 4 by the peroxisome proliferator-activated receptor gamma coactivator. J. Biol. Chem. 2005;280:29525–29532. doi: 10.1074/jbc.M502236200. [DOI] [PubMed] [Google Scholar]
- Matoba S., Kang J.G., Patino W.D., Wragg A., Boehm M., Gavrilova O., Hurley P.J., Bunz F., Hwang P.M. P53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Murphy E., Amanakis G., Fillmore N., Parks R.J., Sun J. Sex differences in metabolic cardiomyopathy. Cardiovasc. Res. 2017;113:370–377. doi: 10.1093/cvr/cvx008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Liu T., Husain S., Zhai P., Warren J.S., Hsu C.P., Matsuda T., Phiel C.J., Cox J.E., Tian B., et al. Glycogen synthase kinase-3alpha promotes fatty acid uptake and lipotoxic cardiomyopathy. Cell Metab. 2019;29:1119–1134.e12. doi: 10.1016/j.cmet.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M.S., Nguyen T., Watson M.J., Hull R.W., Yu H.G. Transcriptome profiling reveals novel BMI- and sex-specific gene expression signatures for human cardiac hypertrophy. Physiol. Genomics. 2017;49:355–367. doi: 10.1152/physiolgenomics.00122.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y., Matsuda M., Miyata Y., Fukuhara A., Komuro R., Shimabukuro M., Shimomura I. Human catalase gene is regulated by peroxisome proliferator activated receptor-gamma through a response element distinct from that of mouse. Endocr. J. 2010;57:303–309. doi: 10.1507/endocrj.k09e-113. [DOI] [PubMed] [Google Scholar]
- Opie L.H., Owen P. Effects of increased mechanical work by isolated perfused rat heart during production or uptake of ketone bodies. Assessment of mitochondrial oxidized to reduced free nicotinamide-adenine dinucleotide ratios and oxaloacetate concentrations. Biochem. J. 1975;148:403–415. doi: 10.1042/bj1480403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomer X., Barroso E., Zarei M., Botteri G., Vazquez-Carrera M. Pparbeta/delta and lipid metabolism in the heart. Biochim. Biophys. Acta. 2016;1861:1569–1578. doi: 10.1016/j.bbalip.2016.01.019. [DOI] [PubMed] [Google Scholar]
- Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson L.R., Herrero P., Coggan A.R., Kisrieva-Ware Z., Saeed I., Dence C., Koudelis D., Mcgill J.B., Lyons M.R., Novak E., et al. Type 2 diabetes, obesity, and sex difference affect the fate of glucose in the human heart. Am. J. Physiol. Heart Circ. Physiol. 2015;308:H1510–H1516. doi: 10.1152/ajpheart.00722.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosdocimo D.A., Anand P., Liao X., Zhu H., Shelkay S., Artero-Calderon P., Zhang L., Kirsh J., Moore D., Rosca M.G., et al. Kruppel-like factor 15 is a critical regulator of cardiac lipid metabolism. J. Biol. Chem. 2014;289:5914–5924. doi: 10.1074/jbc.M113.531384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchalska P., Crawford P.A. Multi-dimensional roles of ketone bodies in fuel metabolism, signaling, and therapeutics. Cell Metab. 2017;25:262–284. doi: 10.1016/j.cmet.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rindler P.M., Plafker S.M., Szweda L.I., Kinter M. High dietary fat selectively increases catalase expression within cardiac mitochondria. J. Biol. Chem. 2013;288:1979–1990. doi: 10.1074/jbc.M112.412890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano M., Minamino T., Toko H., Miyauchi H., Orimo M., Qin Y., Akazawa H., Tateno K., Kayama Y., Harada M., et al. P53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature. 2007;446:444–448. doi: 10.1038/nature05602. [DOI] [PubMed] [Google Scholar]
- Sikder K., Shukla S.K., Patel N., Singh H., Rafiq K. High fat diet upregulates fatty acid oxidation and ketogenesis via intervention of PPAR-gamma. Cell Physiol. Biochem. 2018;48:1317–1331. doi: 10.1159/000492091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son N.H., Yu S., Tuinei J., Arai K., Hamai H., Homma S., Shulman G.I., Abel E.D., Goldberg I.J. Ppargamma-induced cardiolipotoxicity in mice is ameliorated by pparalpha deficiency despite increases in fatty acid oxidation. J. Clin. Invest. 2010;120:3443–3454. doi: 10.1172/JCI40905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C., Love M.I., Robinson M.D. Differential analyses for rna-seq: transcript-level estimates improve gene-level inferences. F1000Research. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suffee N., Baptista E., Piquereau J., Ponnaiah M., Doisne N., Ichou F., Lhomme M., Pichard C., Galand V., Mougenot N., et al. Impacts of a high fat diet on the metabolic profile and the phenotype of atrial myocardium in mice. Cardiovasc. Res. 2021 doi: 10.1093/cvr/cvab367. [DOI] [PubMed] [Google Scholar]
- Tan W.Q., Wang K., Lv D.Y., Li P.F. Foxo3a inhibits cardiomyocyte hypertrophy through transactivating catalase. J. Biol. Chem. 2008;283:29730–29739. doi: 10.1074/jbc.M805514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega R.B., Kelly D.P. Cardiac nuclear receptors: architects of mitochondrial structure and function. J. Clin. Invest. 2017;127:1155–1164. doi: 10.1172/JCI88888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C.J., Schroeder M.E., Aguado B.A., Anseth K.S., Leinwand L.A. Matters of the heart: cellular sex differences. J. Mol. Cell. Cardiol. 2021;160:42–55. doi: 10.1016/j.yjmcc.2021.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.H., Liu C.L., Chiu W.C., Twu Y.C., Liao Y.J. HMGCS2 mediates ketone production and regulates the proliferation and metastasis of hepatocellular carcinoma. Cancers (Basel) 2019;11:1876. doi: 10.3390/cancers11121876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P.A., Reusch J.E., Mccune S.A., Leinwand L.A., Luckey S.W., Konhilas J.P., Brown D.A., Chicco A.J., Sparagna G.C., Long C.S., Moore R.L. Restoration of CREB function is linked to completion and stabilization of adaptive cardiac hypertrophy in response to exercise. Am. J. Physiol. Heart Circ. Physiol. 2007;293:H246–H259. doi: 10.1152/ajpheart.00734.2006. [DOI] [PubMed] [Google Scholar]
- Wu Z., Huang X., Feng Y., Handschin C., Feng Y., Gullicksen P.S., Bare O., Labow M., Spiegelman B., Stevenson S.C. Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1alpha transcription and mitochondrial biogenesis in muscle cells. Proc. Natl. Acad. Sci. U S A. 2006;103:14379–14384. doi: 10.1073/pnas.0606714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Prosdocimo D.A., Bai X., Fu C., Zhang R., Campbell F., Liao X., Coller J., Jain M.K. Klf15 establishes the landscape of diurnal expression in the heart. Cell Rep. 2015;13:2368–2375. doi: 10.1016/j.celrep.2015.11.038. [DOI] [PubMed] [Google Scholar]
- Zhao G., Jeoung N.H., Burgess S.C., Rosaaen-Stowe K.A., Inagaki T., Latif S., Shelton J.M., Mcanally J., Bassel-Duby R., Harris R.A., et al. Overexpression of pyruvate dehydrogenase kinase 4 in heart perturbs metabolism and exacerbates calcineurin-induced cardiomyopathy. Am. J. Physiol. Heart Circ. Physiol. 2008;294:H936–H943. doi: 10.1152/ajpheart.00870.2007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The metabolomic datasets and RNAseq datasets generated during the current study will be available in the public repository.
-
•
All original code has been deposited at GitHub repository and is publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.