Summary
Background
The multiplicity, heterogeneity, and dynamic nature of human immunodeficiency virus type-1 (HIV-1) latency mechanisms are reflected in the current lack of functional cure for HIV-1. Accordingly, all classes of latency-reversing agents (LRAs) have been reported to present variable ex vivo potencies. Here, we investigated the molecular mechanisms underlying the potency variability of one LRA: the DNA methylation inhibitor 5-aza-2’-deoxycytidine (5-AzadC).
Methods
We employed epigenetic interrogation methods (electrophoretic mobility shift assays, chromatin immunoprecipitation, Infinium array) in complementary HIV-1 infection models (latently-infected T-cell line models, primary CD4+ T-cell models and ex vivo cultures of PBMCs from HIV+ individuals). Extracellular staining of cell surface receptors and intracellular metabolic activity were measured in drug-treated cells. HIV-1 expression in reactivation studies was explored by combining the measures of capsid p24Gag protein, green fluorescence protein signal, intracellular and extracellular viral RNA and viral DNA.
Findings
We uncovered specific demethylation CpG signatures induced by 5-AzadC in the HIV-1 promoter. By analyzing the binding modalities to these CpG, we revealed the recruitment of the epigenetic integrator Ubiquitin-like with PHD and RING finger domain 1 (UHRF1) to the HIV-1 promoter. We showed that UHRF1 redundantly binds to the HIV-1 promoter with different binding modalities where DNA methylation was either non-essential, essential or enhancing UHRF1 binding. We further demonstrated the role of UHRF1 in the epigenetic repression of the latent viral promoter by a concerted control of DNA and histone methylations.
Interpretation
A better understanding of the molecular mechanisms of HIV-1 latency allows for the development of innovative antiviral strategies. As a proof-of-concept, we showed that pharmacological inhibition of UHRF1 in ex vivo HIV+ patient cell cultures resulted in potent viral reactivation from latency. Together, we identify UHRF1 as a novel actor in HIV-1 epigenetic silencing and highlight that it constitutes a new molecular target for HIV-1 cure strategies.
Funding
Funding was provided by the Belgian National Fund for Scientific Research (F.R.S.-FNRS, Belgium), the « Fondation Roi Baudouin », the NEAT (European AIDS Treatment Network) program, the Internationale Brachet Stiftung, ViiV Healthcare, the Télévie, the Walloon Region (« Fonds de Maturation »), « Les Amis des Instituts Pasteur à Bruxelles, asbl », the University of Brussels (Action de Recherche Concertée ULB grant), the Marie Skodowska Curie COFUND action, the European Union's Horizon 2020 research and innovation program under grant agreement No 691119-EU4HIVCURE-H2020-MSCA-RISE-2015, the French Agency for Research on AIDS and Viral Hepatitis (ANRS), the Sidaction and the “Alsace contre le Cancer” Foundation. This work is supported by 1UM1AI164562-01, co-funded by National Heart, Lung and Blood Institute, National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, National Institute on Drug Abuse and the National Institute of Allergy and Infectious Diseases.
Keywords: HIV-1 latency, Reactivation, UHRF1, Epigenetics, EGCG
Research in context.
Evidence before this study
Accumulating data highlight the intrinsically dynamic and heterogeneous nature of latent HIV-1 cellular reservoirs within and between infected individuals. This heterogeneity and the multiplicity of the silencing mechanisms underlying HIV-1 latency, rather than latency itself, are now considered as the major barrier to HIV-1 eradication. In agreement, all classes of HIV-1 latency-reversing agents (LRAs) have been reported to present variable ex vivo potencies. In particular, we have previously shown that latency reversal with the DNA methylation inhibitor 5-aza-2’-deoxycytidine (5-AzadC or decitabine) is associated with patient-specific qualitative and quantitative variations in HIV-1 reactivation from latency. However, the underlying molecular mechanisms of LRA various potencies, specifically those of 5-AzadC, had never been studied before.
Added value of this study
For the first time, we explored the molecular mechanisms underlying the heterogeneity of 5-AzadC-induced reversal of HIV-1 latency. We evidenced the existence of non-random and reproducible DNA methylation signatures in response to 5-AzadC treatment at the level of the single CpG dinucleotide in the HIV-1 promoter. We showed that these preferentially differentially-demethylated positions (DDMPs) correspond to binding sites for the epigenetic integrator UHRF1. Characterization of UHRF1’s function in HIV-1 latency revealed a novel role for this factor in the epigenetic silencing of viral gene expression. We further showed that UHRF1 is an interesting target for innovative anti-HIV therapeutic strategies and demonstrated that the main polyphenolic compound of green tea, epigallocatechin-3-gallate (EGCG), presented a new latency reversal activity, in addition to its known antiviral activity.
Implications of all the available evidence
Collectively, these findings demonstrate that the understanding of HIV-1 latency heterogeneity is crucial to define new anti-HIV curative strategies. In particular, we showed that UHRF1 is redundantly recruited to the latent HIV-1 promoter where it controls the cross-talk of epigenetic repressive mechanisms (DNA and histone methylations) to ensure a concurrent silencing of viral gene expression. As a proof of concept, we demonstrated the relevance of deciphering new basic mechanisms of HIV-1 latency for the development of anti-HIV therapeutic approaches.
Alt-text: Unlabelled box
Introduction
Combination antiretroviral therapy (cART) is currently the only therapeutic option available for HIV-1 infected individuals. If cART is efficient in suppressing viral replication and in prolonging the lifespan of infected individuals, the persistence of transcriptionally-silent proviruses, particularly in latently-infected resting memory CD4+ T cells, still prevents HIV-1 eradication.1, 2, 3 As such, much effort has been put into understanding the multiple molecular factors involved in viral latency to develop new anti-HIV therapeutic strategies. One such strategy relies on the use of latency-reversing agents (LRAs) that target repressors of HIV-1 gene expression, thereby inducing a controlled activation of the latent reservoirs of the virus.4, 5, 6
The multifactorial process of HIV-1 silencing during latency is controlled, in part, by several interrelated epigenetic mechanisms that collectively govern the chromatin architecture of the provirus.7,8 In particular, the latent HIV-1 promoter, located within the 5’Long Terminal Region (5’LTR), is maintained in a tight heterochromatic state by the concurrent recruitment of multiple cellular epigenetic machineries. In addition to the precise nucleosome positioning, as well as the accumulation of specific inhibitory histone modifications,8 the HIV-1 promoter contains two CpG islands (CGIs) surrounding the transcriptional start site.9 Both CGIs have been reported to be hypermethylated in latently-infected T-cell line models, thus participating in the 5’LTR heterochromatinization during latency.9, 10, 11, 12 Methylation of the HIV-1 promoter in patient cells has been reported in some studies10,11,13,14 but other reports denied the implication of 5’LTR methylation ex vivo.15, 16, 17, 18 Explaining these apparently contradictory results, recent studies demonstrated that clinical characteristics of HIV+ individuals, such as duration of the infection13 or duration of the antiretroviral treatment,19,20 influence the accumulation of DNA methylation on the 5’LTR. In this regard, an elegant study from the group of Mathias Lichterfeld has recently reported a progressive longitudinal accumulation of proviruses integrated in DNA regions with hypermethylated cytosine residues upstream of the proviral 5’LTR promoter, suggesting a role of DNA methylation in silencing proviral transcriptional activity during prolonged ART.21 In addition, several latency factors have been proposed for the recruitment of the cellular DNA methyltransferases (DNMTs) at the HIV-1 promoter,14,22 indicating an additional heterogeneity in the mechanisms responsible for the accumulation of DNA methylation on the viral promoter. In agreement, we have previously shown that latency reversal with the DNA methylation inhibitor 5-aza-2’-deoxycytidine (5-AzadC or decitabine) is associated with patient-specific qualitative and quantitative variations in HIV-1 reactivation from latency.23
Here, we studied the molecular basis underlying the variations in the HIV-1 reactivation potency of 5-AzadC in terms of proviral DNA demethylation. By highlighting the presence of specific epigenetic signatures in the HIV-1 promoter following 5-AzadC reactivation, we uncovered the role of UHRF1 (Ubiquitin-like with PHD and RING finger domain 1) in the epigenetic control of HIV-1 latency. UHRF1 is known to maintain a heterochromatic environment via its combined action on both DNA and histone methylations, coordinated through its recruitment of the enzymes catalyzing these epigenetic modifications.24, 25, 26 While the role of UHRF1 in the epigenetic silencing of endogenous retroviruses has been reported,27, 28, 29, 30 our data report here for the first time the role of UHRF1 in the epigenetic repression of an exogenous retrovirus. Considering this and as a proof-of-concept, we investigated the relevance of the pharmacological inhibition of UHRF1 and provided evidence that it constitutes a novel molecular target for anti-HIV-1 curative strategies.
Methods
Cell culture
The Jurkat (RRID:CVCL_0065), J-Lat 6.3 (RRID:CVCL_8280), J-Lat 8.4 (RRID:CVCL_8284), J-Lat 9.2 (RRID:CVCL_8285), J-Lat 15.4 (RRID:CVCL_8282) and HEK293T (RRID:CVCL_0063) cell lines were obtained from the AIDS Research and Reference Reagent Program (NIAID, NIH). Cells were grown in RPMI 1640 medium (Gibco-BRL) supplemented with 10 % fetal bovine serum, 50 U/ml of penicillin and 50 µg/mL of streptomycin and were cultivated at 37 °C in a 5% CO2 atmosphere. All cells were negative for mycoplasma (Sigma-Aldrich, MP0040A kit).
Primary models for HIV-1
CD4+ T cells were isolated from buffy coats of healthy donors, obtained from the Belgian Red Cross, by negative magnetic bead selection (StemCell #19052), according to the manufacturer's instructions. Following isolation, cells were stimulated with anti-CD3 + anti-CD28 for three days then infected by spinoculation (2 h, 800 g and 32 °C) with VSV-G pseudotyped HIVGKO particles at an MOI of 3000, where GFP and mKO2 expression were used to distinguish between productively-infected and latently-infected cells.31 After spinoculation, cells were let recover for five days with conditioned medium replacement every 24 h to promote cell survival and quiescence. Five days post-infection, HIV-1 infected cells were sorted using the SH800 cell sorter (Sony) and used for further applications.
Reagents and antibodies
5-aza-2’-deoxycytidine (5-AzadC, A3656), TNF-α (GF314) and epigallocatechin-3-gallate (EGCG, E4143) were purchased from Sigma Aldrich. NSC232003 was purchased from MedChemExpress (HY-103236). Antibodies against CREB (sc-186, RRID:AB_2086021), CREM (sc-440, RRID:AB_673599), ATF-1 (sc-28673, RRID:AB_2274416), MBD4 (sc-10753, RRID:AB_2250279) and purified rabbit IgG (sc-2027, RRID:AB_737197) were purchased from Santa Cruz Biochemical. Antibodies against MBD1 (pAb-078-050) and UHRF1 for ChIP applications (C154110258-100) were purchased from Diagenode. Antibodies against MBD2/3 (07-199, RRID:AB_310423), Kaiso (05-659, RRID:AB_309884) and UHRF1 for EMSA and western blotting (MABE308) were purchased from Upstate/Millipore. Antibodies against MeCP2 (ab2828, RRID:AB_2143853) and RBP-JK (ab33065, RRID:AB_778156) were purchased from Abcam. Antibody against RNA polymerase II (RNAPII) (14958, RRID:AB_2687876) was purchased from Cell Signaling Technology. Secondary antibodies (7074, RRID:AB_2099233 and 7076, RRID:AB_330924) were purchased from Cell Signaling Technology.
Virus production assays
HIV-1 production was measured in cell culture supernatants by ELISA assays on p24Gag using the INNOTEST HIV Antigen mAb kit according to the manufacturer's instructions (Fujirebio).
Sodium bisulfite-mediated mapping of methylcytosines
Genomic DNA was isolated using the DNeasy Blood and Tissue kit (Qiagen), then sodium bisulfite-converted (EpiTect Bisulfite kit, Qiagen). The 5’LTR or the rev regions were amplified by (semi)nested PCR (primer sequences are available upon request). At least 12 clones from each condition were sequenced, and clones with sodium bisulfite conversion higher than 95% were aligned on the reference sequence HIV-1 NL4.3 using ClustalΩ. MethTools32 and Inkscape were used for graphical representations.
Electrophoretic mobility shift assays (EMSAs)
Nuclear extracts were prepared using a protocol described by Dignam and colleagues.33 EMSAs, competition EMSAs and supershift assays were performed as described previously.34 Oligonucleotides sequences used for the probes are available upon request.
Chromatin immunoprecipitation assays
ChIP assays were performed as previously described.35 Relative quantification using the standard curve method on the input was performed for each primer pair and 96-well Optical Reaction plates were read in a StepOnePlus PCR instrument (Applied Biosystems). Fold enrichments were calculated as fold inductions relative to the values measured with IgG. Primer sequences used for quantification are available upon request.
Western blot
Western blotting was performed using 15 μg of total protein extracts. The immunodetection was assessed using primary antibodies targeting UHRF1 and β-actin or GAPDH as a loading control. Horseradish peroxidase (HRP)-conjugated secondary antibodies were used for chemiluminescence detection (Cell Signaling Technology).
RNA extraction and analysis of transcripts
Total RNA samples were isolated using the Tri-Reagent (TRC-118, MRC), according to the manufacturer's protocol. Following DNAse treatment (AM1907, Invitrogen), reverse transcription was performed with the PrimeScript RT reagent kit (RR037A, TaKaRa).
Lentiviral production and transduction assays
TRC Lentiviral shRNA plasmids (pLKO.1) MISSION shRNA were obtained from Sigma-Aldrich (SHC002, TRCN0000273315, TRCN0000273256, TRCN0000273317 and TRC0000004352). The pMD2.G and the psPAX2 packaging system were obtained from Addgene. VSV-G pseudotyped particles were produced by transfection of HEK 293T cells as described previously.36 J-Lat cells were transduced as described previously.37 For primary cell models for HIV-1 infection, the HIVGKO vector was kindly provided by Dr Emilie Battivelli (Buck Institute for Research on Aging, Novato, California, USA).
siRNA transfections
Cells were transfected with 1µM of two different Accell siRNAs targeting UHRF1 (siUHRF1#1: GUAUUAGGGAAGAAUGAGA, siUHRF1#2: CCUCCUUAUUCUUAGAUUA) or a control siRNA (siNT: UGGUUUACAUGUUUUCUGA, Dharmacon) and cultured in Accell delivery media for 96 h.
Cellular proliferation assays and viability
Cellular proliferation was evaluated by the colourimetric test WST-1 according to the manufacturer's instructions (Roche). Cellular viability was assessed by staining the cells with the LIVE/DEAD Fixable Near-IR Dead Cell Stain (Thermo Fisher) and analysis by flow cytometry on a FACSCantoII (Becton-Dickinson), using the FACSDiva software (Becton-Dickinson).
Plasmid constructs and reporter assays
We generated an expression vector for human UHRF1 (pCMV-HA-UHRF1, termed pUHRF1). The non-episomal pLTR-Fluc vector was described previously.38 To obtain the pLTRme-Fluc vector, where only the LTR CpGs are methylated, we methylated in vitro the whole pLTR-Fluc construct using the SssI methyltransferase (New England Biolabs, M0226). The LTR fragment was then purified and cloned back in the parental reporter vector and the resulting pLTRme-Fluc vector was directly transfected without bacterial amplification.
Infinium 850K Human Methylation arrays
Genomic DNA was extracted with the DNeasy Blood and Tissue Kit (QIAGEN) and converted with sodium bisulfite (EZ DNA Methylation Kit, Zymo Research). The quality of each analyzed sample was first evaluated by inspection of the control probes’ intensity level. Raw data (uncorrected probe intensity values) from the Infinium Methylation arrays were processed according to the recommended steps of Dedeurwaerder et al.39 Beta-values were computed using the following formula: Beta-value = M/[U + M] where M and U are the raw “methylated” and “unmethylated” signals, respectively. Beta-values were corrected for type I and type II bias using the peak-based correction.40 Infinium HumanMethylation850K raw data were submitted to the NCBI's Gene Expression Omnibus (GEO) database (GSE139320, token: wnqfssiudxubtgx). Principal component analysis and hierarchical clustering were performed via an in-house R script using the most variable Infinium probes (standard deviation ≥ 0.27). For differential analysis, probes showing an absolute difference between case and control Beta-values higher than 0.3 were assumed significant. Infinium probes located in promoter regions were first associated with their corresponding genes for pathway analysis. Then, a « delta-Beta » was defined for each gene as the difference between case and control Beta-values of its promoter Infinium probe showing the highest absolute difference. Finally, genes were ranked according to their delta-Beta and submitted to the GSEA tool41 to search for significant enrichments among the HALLMARK gene sets from MSigDB (http://software.broadinstitute.org/gsea/msigdb/).
Study subjects
We selected 22 HIV-1-infected individuals at the Saint-Pierre Hospital (Brussels, Belgium) based on the following criteria: all volunteers were treated with cART for at least 1 year, had an undetectable plasma HIV-1 RNA level (20 copies/ml) for at least 1 year, and had a level of CD4+ T lymphocytes higher than 300 cells/mm3 of blood. Characteristics (age, CD4+ T cell count, CD4+ nadir, antiviral regimens, duration of therapy, duration with undetectable plasma HIV-1 RNA level, and HIV-1 subtypes) of HIV+ individuals from the Saint-Pierre Hospital were well documented and are presented in Table S1A. Buffy coats from healthy donors were obtained at the Belgian Red Cross.
Ethical statement
Ethical approval was granted by the Human Subject Ethics Committee of the Saint-Pierre Hospital (Brussels, Belgium). All individuals enrolled in the study provided written informed consent for donating blood.
Isolation of CD8+-depleted PBMCs
CD8+-depleted PBMCs used in HIV-1 reactivation assays were isolated from fresh whole blood of HIV+ individuals as previously described.23
Quantitation of cell-associated HIV-1 unspliced RNA
Total nucleic acids were extracted from pellets of CD8+-depleted PBMCs according to the Boom isolation method.42 Extracted cellular RNA was treated with DNase (DNA-free kit; Thermo Fisher Scientific) and reverse transcribed using the SuperScript III reverse transcriptase (Thermo Fisher Scientific). cDNA was used for the qPCR-based quantification of cell-associated HIV-1 unspliced RNA (amplicon in the gag region), as previously reported.43 HIV-1 RNA copy numbers were normalized to the total cellular RNA (by measurement of 18S ribosomal RNA) inputs as described previously.44 Non-template control wells were included in every qPCR run and were consistently negative.
Quantification of HIV-1 extracellular RNA
Total RNA was extracted from CD8+-depleted PBMCs ex vivo culture supernatants using the QIA amp Viral RNA Mini kit (Qiagen). HIV-1 RNA levels were quantified using the Generic HIV Viral Charge kit (Biocentric) according to the manufacturer's instructions.
Quantification of total HIV-1 DNA
Total cellular DNA was extracted from HIV-1+ individuals CD8+-depleted PBMCs ex vivo cultures using the QIAamp DNA Mini kit (Qiagen). The total cell-associated HIV-1 DNA was then quantified by ultra-sensitive real-time PCR (Generic HIV DNA cell kit, Biocentric) according to the manufacturer's instruction.45
Cell activation analysis by flow cytometry
For cell activation analysis, CD8+-depleted PBMCs from the blood of healthy donors were used to establish ex vivo cell cultures. Cells were collected 24 h after stimulation with EGCG and were stained with relevant antibodies as previously described.23
Probability calculation and statistical analyses
The demethylation probability following 5-AzadC treatment was calculated as follows:
where P(demeth) corresponds to the probability of demethylation following treatment, #D(treated) the number of demethylated CpGs in the treated conditions, #C(total) the number of clones (12 for each condition in the present study) and where #M(mock) corresponds to the number of methylated CpGs in the mock-treated conditions. Sodium bisulfite sequencing data sets were analyzed using Fisher's exact test. For all the analyses, the threshold of statistical significance was set at 0.05. p-values ≤ 0.05 (*: p-value ≤ 0.05, **: p-value ≤ 0.01, ***: p-value ≤ 0.001) were considered statistically significant. All tests were two-sided. All analyses were performed using Prism version 6.0 (GraphPad software, RRID:SCR 002798) and Microsoft Excel. Statistical tests are indicated in the corresponding figure legends.
Results
5-AzadC treatment provokes specific demethylation signatures within the HIV-1 promoter
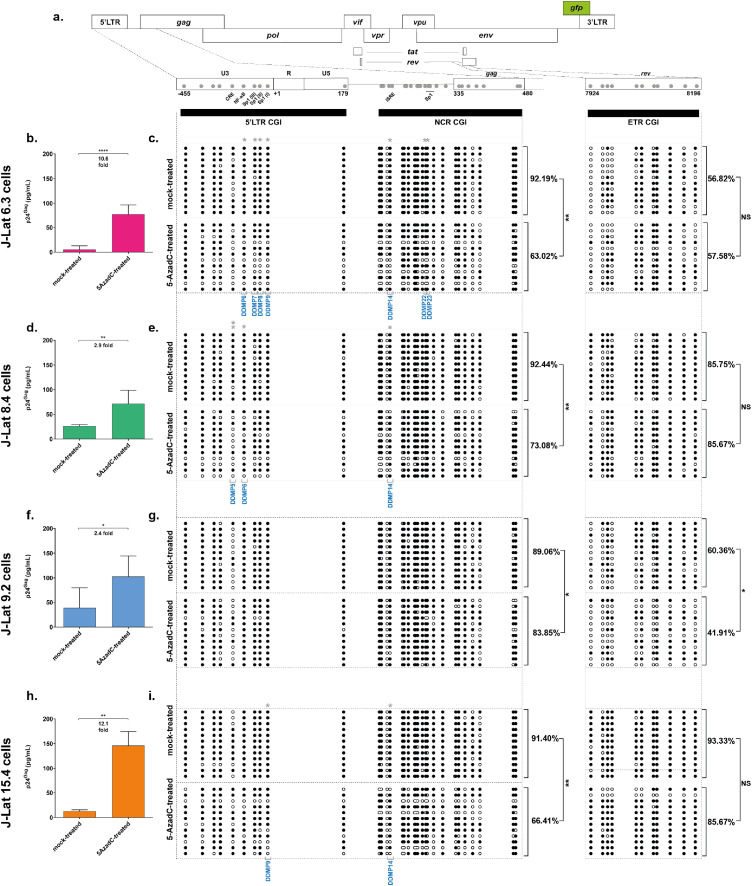
We previously reported that treatment with the DNA methylation inhibitor 5-AzadC provoked reactivation of HIV-1 gene expression in ex vivo patient cell cultures and several T-cell line models for HIV-1 post-integration latency, although to variable extents.23 To investigate how this variability translated at the DNA methylation level in the HIV-1 promoter CGIs, we mock-treated or treated four clones of the CD4+ T-lymphoid J-Lat cell line model for HIV-1 latency with 5-AzadC (J-Lat 6.3 cells, J-Lat 8.4 cells, J-Lat 9.2 cells and J-Lat 15.4 cells).46 First, quantification of the viral progeny particles capsid protein p24Gag in the treated culture supernatants by ELISA confirmed the variations in 5-AzadC reactivation potency in vitro (Figure 1b,d,f,h, indicating a 10.6-fold, a 2.9-fold, a 2.4-fold and a 12.1-fold reactivation, for J-Lat 6.3, J-Lat 8.4, J-Lat 9.2 and J-Lat 15.4 cells, respectively), consistent with the extents of HIV-1 reactivation previously reported following 5-AzadC treatment.10,11 Reactivation by 5-AzadC was associated with a decreased metabolic activity ranging from 58.18% to 67.95% (Fig. S1a), consistent with the tolerable effect of 5-AzadC treatment that we have previously reported.23 We next assessed the methylation status of the two promoter CGIs termed 5’LTR and NCR (from nt -455 to nt 179 and from nt 183 to nt 470, respectively, where nt+1 is the U3/R junction in the 5’LTR, Figure 1a) and of a control intragenic CGI located within the rev gene termed ETR (nt 7924 to nt 8196, Figure 1a).9 Because the two viral LTRs have identical sequences and because we wanted to specifically obtain the methylation profile of the 5’LTR, both 5’LTR and NCR CGIs were analyzed in a single amplicon. We confirmed that promoter CGIs were hypermethylated to similar levels in the four J-Lat clones in mock-treated conditions (Figure 1c,e,g,i, 92.19%, 92.44%, 89.06% and 91.4% of 5mC, respectively, for J-Lat 6.3, J-Lat 8.4, J-Lat 9.2 and J-Lat 15.4 cells), consistent with previous observations.10,11 Treatment with 5-AzadC provoked a global demethylation in the two promoter CGIs, though to various extents in each J-Lat clone allowing the following ranking: J-Lat 6.3 cells (29.17% of 5-AzadC-induced demethylation) > J-Lat 15.4 cells (24.99%) > J-Lat 8.4 cells (19.36%) > J-Lat 9.2 cells (5.21%) (Figure 1, respective p-values of 0.0057; 0.0020; 0.0085; 0.0142; [unpaired T-tests]). This ranking was similar to the one we observed with the fold reactivation levels of HIV-1 production, indicating that 5-AzadC reactivation is dependent on the demethylation of specific sites in the HIV-1 promoter but with a heterogeneous profile. In addition, treatment with 5-AzadC did not alter the methylation profile of the HIV-1 ETR CGI in J-Lat 6.3 cells, J-Lat 8.4 cells and J-Lat 15.4 cells (Figure 1c,e,i, respectively), showing that 5-AzadC-induced HIV-1 provirus demethylation specifically occurs at the 5’LTR in these clones. Of note and in agreement with lower basal promoter CGIs methylation level, 5-AzadC fold reactivation of HIV-1 production was the lowest in the J-Lat 9.2 clone (Figure 1f), in which the ETR CGI was also demethylated following 5-AzadC treatment, suggesting a non-specific action of 5-AzadC on the HIV-1 promoter in this clone (Figure 1g).
Figure 1.
5-AzadC-induced reactivation of HIV-1 gene expression from latency is associated with 5’LTR CGIs demethylation. (a) Schematic presentation of the three CpG islands studied along the HIV-1 provirus, in the HIV-1 promoter (5’LTR and NCR CGIs) and rev (ETR CGI). The reactivation of HIV-1 production following 72 h treatment with 400 nM of 5-AzadC, quantified by ELISA on p24Gag capsid protein in culture supernatants, and the DNA methylation profile, established by sodium bisulfite sequencing for the three CGIs, are respectively presented for the J-Lat 6.3 cells (b and c), the J-Lat 8.4 cells (d and e), the J-Lat 9.2 cells (f and g) and the J-Lat 15.4 cells (h and i). ELISA results are representative of the means ± SD of three independent 5-AzadC treatments. Reactivation folds are indicated. Unmethylated and methylated CpG dinucleotides are respectively represented with open and closed circles, where each line corresponds to individual sequenced molecules. The global methylation level presented correspond to mean percentages of methylated CpGs for the twelve clones of each condition, either on the promoter CGIs (5’LTR + NCR CGIs considered together) or on the ETR CGI. Statistical significance was determined by [unpaired T-tests].
To tease out for specific regulatory mechanisms underlying the heterogeneity of 5-AzadC reactivation potency, we next mapped the probability of demethylation following 5-AzadC treatment at individual CpG positions in the HIV-1 promoter CGIs. This probabilistic analysis highlighted that some CpGs were more prone to 5-AzadC-induced demethylation (Fig. S1 and Methods section). The most statistically significant 5-AzadC-induced differentially-demethylated positions (termed “DDMPs”) are listed in Table 1. Despite some similarities, the position of statistically significant DDMPs varied among the J-Lat clones, illustrating the heterogeneity of the 5-AzadC-induced latency reversal mechanisms recapitulated by the different clones. Some DDMPs were present in sequences giving rise to viral RNA features (in the primer binding site or the packaging signal sequence ψ), suggesting a potential link between DNA methylation deposition and RNA secondary structures (Table 1). Importantly, several DDMPs were positioned within transcription factor binding sites known to be involved in HIV-1 transcriptional regulation (Figure 1 and Table 1). For instance, DDMP5 is located within the cAMP-Responsive Element (CRE),47 DDMP6 within the NF-κB binding sites,47 DDMP7, DDMP8 and DDMP9 within the Sp1 binding sites,48,49 and DDMP14 is located in the interferon-stimulated response element49 (highlighted in Figure 1a and Table 1).
Table 1.
Most statistically significant 5-AzadC-induced differentially-demethylated CpG dinucleotides (DDMPs).
| Cell line | CpG positiona | Probability of 5-AzadC-induced demethylation | p-valueb | Statistical-significance | Locationc | DDMPd |
|---|---|---|---|---|---|---|
|
J-Lat 6.3 cells |
-[158,159] | 0,33 | 0,046 | * | N/A | DDMP3 |
| -[96, 97] | 0,33 | 0,0466 | * | NF-κB site | DDMP6 | |
| -[74,75] | 0,45 | 0,32 | * | Sp1 site III | DDMP7 | |
| -[63,64] | 0,42 | 0,0186 | * | Sp1 site II | DDMP8 | |
| -[47, 48] | 0,42 | 0,0186 | * | Sp1 site I | DDMP9 | |
| +[109, 110] | 0,42 | 0,0186 | * | U5 interacting with ψ | DDMP10 | |
| +[183, 184] | 0,42 | 0,0186 | * | PBS | DDMP11 | |
| +[186, 187] | 0,33 | 0,0466 | * | PBS | DDMP12 | |
| +[205, 206] | 0,42 | 0,0186 | * | Interferon-Stimulated Response Element | DDMP14 | |
| +[243, 244] | 0,33 | 0,0466 | * | Zinc Knuckles in p7Gag binding to SL1 | DDMP17 | |
| +[261, 262] | 0,42 | 0,0186 | * | SL2 of ψ | DDMP20 | |
| +[278, 279] | 0,42 | 0,0186 | * | Sp1 site of HSIV | DDMP22 | |
| +[282, 283] | 0,33 | 0,0466 | * | Sp1 site of HSIV | DDMP23 | |
| +[295, 296] | 0,33 | 0,0466 | * | SL2 of ψ | DDMP24 | |
| +[314, 315] | 0,42 | 0,0186 | * | SL3 of ψ | DDMP25 | |
| +[341, 342] | 0,5 | 0,0069 | ** | SL4 of ψ | DDMP26 | |
| +[360, 361] | 0,33 | 0,0466 | * | Coding sequence of p17Gag | DDMP28 | |
| +[390, 391] | 0,6 | 0,0167 | * | Coding sequence of p17Gag | DDMP30 | |
|
J-Lat 8.4 cells |
-[119, 120] | 0.64 | 0.0045 | ** | CRE site | DDMP5 |
| -[96, 97] | 0.33 | 0.0466 | * | NF-κB site | DDMP6 | |
| +[183, 184] | 0.33 | 0.0466 | * | N/A | DDMP11 | |
| +[205, 206] | 0.33 | 0.0466 | * | Interferon-Stimulated Response Element | DDMP14 | |
| +[231, 232] | 0.45 | 0.0320 | * | N/A | DDMP15 | |
| +[360, 361] | 0.33 | 0.0466 | * | Coding sequence of p17Gag | DDMP28 | |
| J-Lat 15.4 cells | -[217, 218] | 0.33 | 0.0466 | * | N/A | DDMP1 |
| -[47, 48] | 0.42 | 0.0186 | * | Sp1 site | DDMP9 | |
| +[109, 110] | 0.33 | 0.0466 | * | U5 interacting with ψ | DDMP10 | |
| +[205, 206] | 0.42 | 0.0186 | * | Interferon-Stimulated Response Element | DDMP14 | |
| +[231, 232] | 0.42 | 0.0186 | * | N/A | DDMP15 | |
| +[234, 235] | 0.33 | 0.0466 | * | SL1 of ψ | DDMP16 | |
| +[243, 244] | 0.33 | 0.0466 | * | Zinc Knuckles in p7Gag binding to SL1 | DDMP17 | |
| +[295, 296] | 0.50 | 0.0069 | * | SL2 of ψ | DDMP24 | |
| +[314, 315] | 0.33 | 0.0466 | * | SL3 of ψ | DDMP25 | |
| +[341, 342] | 0.42 | 0.0186 | * | SL4 of ψ | DDMP26 | |
| +[347, 348] | 0.33 | 0.0466 | * | SL4 of ψ | DDMP27 | |
| +[360, 361] | 0.36 | 0.0129 | * | Coding sequence of p17Gag | DDMP28 | |
Position given in coordinates where nt+1 is located at the junction U3/R in the 5’LTR.
Statistical significance attributed with * for p ≤ 0.05, ** for p ≤ 0.01 and *** for p ≤ 0.001 by [Fisher's exact test].
Location of CpG within regulatory or structural elements according to the HIV-1 Database.47,92 N/A refers to no known features.
Differentially-Demethylated Position.
DDMPs located in transcription factor binding sites are in bold.
UHRF1 redundantly binds to the HIV-1 promoter
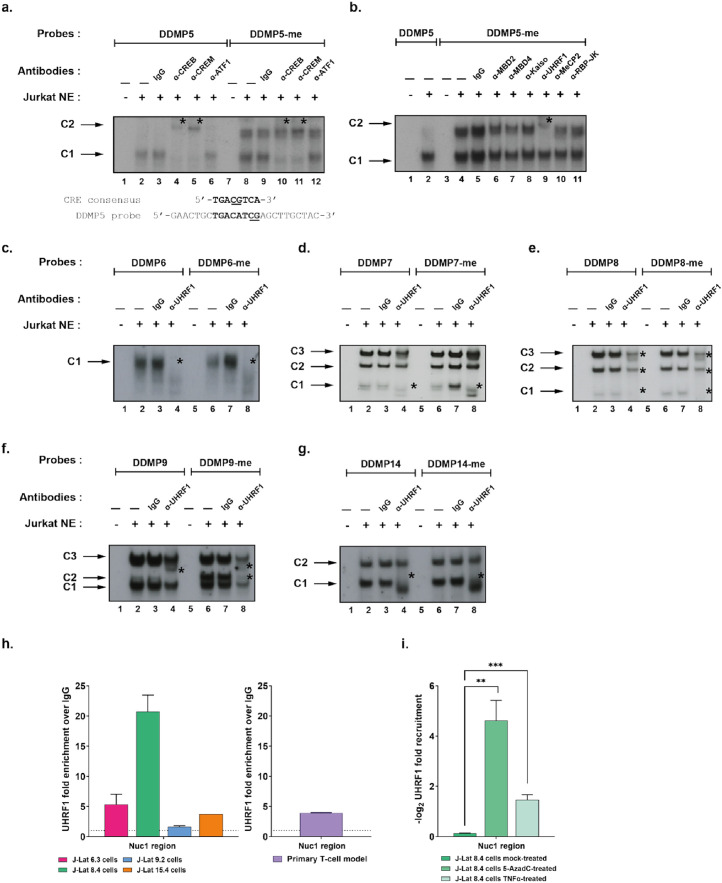
We thus demonstrated that treatment with 5-AzadC provoked the reactivation of HIV-1 production through the non-random DNA demethylation of specific CpG dinucleotides in the 5’LTR. We next postulated that studying transcription factor binding modalities to the DDMPs could provide mechanistic insights into the variations of 5-AzadC reactivation potency. To do so, we first focused on DDMP5, as it presented the highest demethylation probability and the highest statistical significance among all identified DDMPs in all clones (Table 1, 5-AzadC-induced demethylation probability = 0.64 and p-value = 0.005, [Fisher's exact test]). DDMP5 is located within a known HIV-1 promoter CRE.47 Since genome-wide studies have shown that DNA methylation generally affects negatively the binding of CRE factors,50 we hypothesized that DDMP5 methylation would decrease or prevent the binding of cognate transcriptional activators to the HIV-1 promoter CRE. Electrophoretic mobility shift assays (EMSAs) using probes containing the HIV-1NL4.3 DDMP5 sequence in an unmethylated form (“DDMP5” probe) confirmed the presence of a single retarded DNA-protein complex, termed C1 (Figure 2a, lane 2, indicated by an arrow). In supershift experiments, the addition of antibodies raised against CREB and CREM provoked a decrease in the formation of the complex C1 (Figure 2a, lane 4 and lane 5, indicated by an asterisk), whereas the addition of the IgG control or antibodies raised against ATF1 did not affect complex formation (Figure 2a, lane 3 and lane 6, respectively). These supershift experiments thus confirmed the binding of CREB and CREM proteins to the CRE region containing the DDMP5.47,51 Surprisingly, the C1 complex was still observed when the DDMP5 probe was methylated (Figure 2a, lane 8) and supershift experiments showed that CREB and CREM factors could bind to the same extent to the methylated and unmethylated probes (Figure 2a, lane 10 and lane 11, respectively). We further confirmed that the binding of proteins in the C1 complex was independent of the DDMP5 methylation status, since molar excesses of both methylated and unmethylated DDMP5 competed out complex C1 formation (Fig. S2a, compare lanes 3–5 with lanes 11–13 and Fig. S2b). Together, these data indicate that DNA methylation in the HIV-1 promoter CRE site neither prevented nor decreased the binding of its cognate factors. The discrepancy between our results and the reported genome-wide DNA methylation-induced inhibition of CRE factors binding50 could be explained by sequence differences in this transcription factor binding motif (Figure 2a).
Figure 2.
UHRF1 binds in vitro and in vivo to the latent HIV-1 promoter. (a) The radiolabelled unmethylated or the methylated HIV-1 DDMP5 probe (respectively indicated as “DDMP5” and “DDMP5-me”) were incubated with 10µg of nuclear extracts from Jurkat T cells (“Jurkat NE”) and either with a purified rabbit IgG as a negative control (lane 3 and lane 9) or with an antibody directed against CREB/CREM family members including CREB (lane 4 and lane 10), CREM (lane 5 and lane 11) or ATF1 (lane 6 and lane 12). The figure shows the specific retarded bands of interest indicated by arrows. Supershifted complexes are indicated by asterisks. One representative experiment out of three is presented. (b) The “DDMP5” or “DDMP5-me” probes were incubated with 10 µg of Jurkat cells NE, and either with purified rabbit IgG as a negative control (lane 5) or with an antibody directed against methylcytosines-recognizing proteins, including MBD2 (lane 6), MBD4 (lane 7), Kaiso (lane 8), UHRF1 (lane 9), MeCP2 (lane 10) and RBP-JK (lane 11). The figure shows the specific retarded bands of interest indicated by arrows. Supershifted complexes are indicated by asterisks. One representative experiment out of three is presented. (c, d, e, f, g) The “DDMP6” or “DDMP6-me” probes (c), “DDMP7” or “DDMP7-me” probes (d), “DDMP8” or “DDMP8-me” probes (e), “DDMP9” or “DDMP9-me” probes (f) and “DDMP14” or “DDMP14-me” probes (g) were incubated with 10µg of Jurkat cells NE, and either with purified rabbit IgG as a negative control (lanes 3 and 7) or antibodies directed against UHRF1 (lanes 4 and 8). The figure shows the specific retarded bands of interest indicated by arrows. Supershifted complexes are indicated by asterisks. One representative experiment out of three is presented. (h) Chromatin preparations of J-Lat cells and primary CD4+ T cell models for HIV-1 latency were immunoprecipitated with an anti-UHRF1 antibody or with purified rabbit IgG, serving as a negative control. qPCRs were performed with primers hybridizing specifically to the 5’LTR, in the Nuc-1 region. Folds relative to IgG are presented, where fold enrichments for each immunoprecipitated DNA were calculated by the relative standard curve on input DNA. Values represent the means of duplicate samples ± SD. One representative experiment out of three is presented, except for the primary CD4+ T cell model which is representative of two independent infections of healthy donors. (i) Chromatin preparations of J-Lat 8.4 cells, either mock-treated or treated with 400 nM of 5-AzadC or with 10 ng/mL of TNF-α, were immunoprecipitated with an anti-UHRF1 antibody or with purified rabbit IgG, serving as a negative control. Folds relative to IgG were first calculated as above, then normalized to the mock condition and -log2(fold to mock) were plotted. Statistical significance was calculated with an [unpaired T-test].
Interestingly, we observed by EMSAs the formation of an additional retarded complex, termed C2, with the methylated HIV-1 DDMP5 probe (Figure 2a, lanes 8 to 12). The C2 complex was formed when the DDMP5 probe was methylated and was competed out by the methylated DDMP5 probe (Fig. S2a, lanes 7–9). Furthermore, the formation of the C2 complex was not competed out by molar excesses of the methylated consensus for methyl-binding domain (MBD) proteins, or of the methylated consensus for Sp1, indicating that the proteins contained within the C2 complex were specific to the methylated HIV-1 DDMP5 probe, and not to any 5mC-containing sequence (Fig. S2c). Because our in vitro experiments showed that, rather than preventing the binding of transcriptional activators, DDMP5 methylation allowed the binding of methylCpG-recognizing proteins to the HIV-1 promoter, we investigated the nature of the proteins present in the C2 complex. To do so, we performed additional supershift experiments using antibodies raised against proteins known to bind methylcytosines (MBD2, MBD4, MeCP2, Kaiso, UHRF1 and RBP-JK).52,53 Addition of an antibody raised against UHRF1, but not against the other proteins, altered the formation of the C2 complex, concomitantly with the appearance of a supershifted complex of lower mobility (Figure 2b, lane 9, indicated with an asterisk), while the addition of IgG did not affect complex C2 formation (Figure 2b, lane 5). These results thus indicate that the C2 complex contains UHRF1 that binds in vitro to the methylated HIV-1 DDMP5.
Beyond DDMP5, our probabilistic approach identified other CpGs that were prone to demethylation following 5-AzadC-induced reactivation (see Figure. 1, S1 and Table 1). In particular, DDMP6, DDMP7, DDMP8, DDMP9 and DDMP14 appeared to be affected by 5-AzadC in most J-Lat clones and correspond to binding sites for positive regulators of HIV-1 gene expression.47, 48, 49 Similarly to our approach with DDMP5, we first investigated the impact of DNA methylation on complexes formation for these DDMPs (Figure 2c, 2d, 2e, 2f and 2g for DDMP6, DDMP7, DDMP8, DDMP9 and DDMP14, respectively). These experiments showed that DDMP9 DNA methylation enhanced the appearance of a complex termed C2, already observed when we used the unmethylated DDMP9 probe (complex C2, Figure 2f, lanes 6 and 2 for the methylated and unmethylated probes, respectively). Supershift experiments indicated that this C2 complex contained UHRF1 (Figure 2f, lane 8). Thus, in addition to the UHRF1 binding modality to DDMP5 that was DNA methylation-dependent, the observation that UHRF1 binding was increased by DNA methylation for the DDMP9 C2 complex revealed another binding modality to the HIV-1 DDMPs, where DNA methylation enhanced UHRF1 binding. The careful examination of the DDMP9 supershift profile indicated that the formation of the other complexes (C1 and C3 complexes), that were also present when we used the unmethylated DDMP9 probe, was altered following the addition of the anti-UHRF1 antibody (Figure 2f, lane 8), suggesting that UHRF1 could bind to the unmethylated probe. Indeed, supershift experiments using the unmethylated DDMP9 probe showed that the C1 and C3 complexes contained UHRF1 (Figure 2f, lane 4). Contrarily to our observations with DDMP5 and DDMP9, DNA methylation did not affect the formation of the DNA-protein complexes we obtained with the probes containing DDMP6, DDMP7, DDMP8 or DDMP14 (compare lanes 2 and 6 in Figure 2c, 2d, 2e or 2g, respectively). Therefore, we investigated by supershift experiments if the complexes observed with these unmethylated DDMP probes also contained UHRF1. The addition of the anti-UHRF1 antibody caused the decreased formation of the C1 complex for DDMP6 (Figure 2c, lane 4) and DDMP7 (Figure 2d, lane 4), the decreased formation of all three complexes for DDMP8 (Figure 2e, lane 4) and the shift of the C1 complex for DDMP14 (Figure 2g, lane 4), thereby indicating the binding of UHRF1 in vitro to these unmethylated probes. Methylation of the DDMP6, DDMP7, DDMP8 and DDMP14 positions did not affect the supershifts observed with the corresponding methylated probes (lanes 8 in Figure 2c, Figure 2d, Figure 2e and Figure 2g, respectively), demonstrating that UHRF1 bound to the DDMP6, DDMP7, DDMP8 and DDMP14 probes independently of their DNA methylation status. Together, these data indicated that UHRF1 bound to multiple DDMPs within the HIV-1 promoter with different binding modalities where DNA methylation was either non-essential, essential or enhancing UHRF1 binding.
Finally, to garner more insights into the DNA methylation-independent binding of UHRF1 to the HIV-1 promoter and because UHRF1 has been reported to bind to CCAAT DNA sequences of the human Topoisomerase IIa promoter,54 we explored whether UHRF1 could bind to previously characterized CCAAT motifs located within the HIV-1 promoter region.55 Indeed, the HIV-1 promoter contains four binding sites for proteins recognizing the CCAAT motif termed C/EBPs (CCAAT/enhancer-binding proteins), that regulate HIV-1 transcription55 (Fig. S3a). Our data indicated that UHRF1 bound to the C/EBPUS3 site (Fig. S3b, lane 4), the C/EBP US2 site Fig. S3c, lane 4), the C/EBP US1 site overlapping the DDMP5 (Fig. S3d, lane 4) and the C/EBP DS3 site (Fig. S3e).
Altogether, our in vitro studies indicate a redundant binding of UHRF1 to the HIV-1 promoter. UHRF1 binds to multiple DDMPs within the HIV-1 promoter with three different binding modalities: a DNA methylation-dependent binding (for DDMP5), a DNA methylation-dependent enhanced binding (for DDMP9) and a DNA methylation-independent binding (for DDMP6, DDMP7, DDMP8 and DDMP14). Moreover, we showed that UHRF1 binds to the HIV-1 promoter through specific recognition of four CCAAT DNA motifs.
UHRF1 is recruited to the latent HIV-1 5’LTR
We next sought to evaluate the binding of UHRF1 within the context of the latent proviral chromatin structure. To do so, we performed chromatin immunoprecipitation (ChIP) assays in all J-Lat clones using primers hybridizing specifically to the HIV-1 5’LTR, in the Nuc-1 region (Figure 2h). These experiments demonstrated that UHRF1 was recruited to the HIV-1 latent promoter in all J-Lat clones, though at different levels depending on the clone (Figure 2h, recruitment of 5.35-fold, 20.74-fold, 1.61-fold and 3.75-fold in J-Lat 6.3 cells, J-Lat 8.4 cells, J-Lat 9.2 cells and J-Lat 15.4 cells, respectively). UHRF1 presence on the 5’LTR is thus a common feature of HIV-1 latency in J-Lat cells, irrespective of the degree of promoter DNA methylation, although higher methylation of the DDMP5 appeared to be positively associated with higher UHRF1 recruitment (Fig. S3f).
Although latently-infected cell lines constitute model systems highly valuable for mechanistic studies, they might not capture accurately the full spectrum and the heterogeneity of HIV-1 latency mechanisms taking place in vivo. Therefore, we next explored whether UHRF1 recruitment to the viral promoter was a hallmark of HIV-1 latency in T cells by performing ChIP experiments in primary CD4+ T cell models for HIV-1 infection.31 Importantly, these experiments showed that UHRF1 was recruited to the HIV-1 promoter in the primary CD4+ T cell models (Figure 2h, recruitment of 3.90-fold for a representative primary CD4+ T cell model).
Finally, as we originally revealed UHRF1 binding by the characterization of 5-AzadC-induced DDMPs, we studied the recruitment of UHRF1 to the HIV-1 promoter following reactivation with 5-AzadC. To do so, we worked in the J-Lat 8.4 cell line in which UHRF1 recruitment was the highest. We observed a significant reduction in UHRF1 recruitment to the viral promoter upon reactivation with 5-AzadC in J-Lat 8.4 cells (Figure 2i, 26.5-fold decrease, p = 0.01, [unpaired T-test]). As a control, we quantified UHRF1 mRNA and protein levels in J-Lat 8.4 cells in response to 5-AzadC (Fig. S3g and Fig. S3h, respectively) and we showed that 5-AzadC did not alter UHRF1 expression. Furthermore, reactivation of HIV-1 from latency with TNF-α also provoked a decreased recruitment of UHRF1 to the viral promoter (Figure 2i, 2.76-fold decrease, p = 0.001, [unpaired T-test]), further supporting the importance for HIV-1 latency of UHRF1 presence on the heterochromatic viral promoter.
Taken together, we demonstrated the recruitment of UHRF1 to the HIV-1 5’LTR in J-Lat T-cell line models as well as in primary CD4+ T cell models for HIV-1 infection. Furthermore, we showed that reactivation of viral production following 5-AzadC or TNF-α treatment was accompanied by a decreased UHRF1 recruitment to the viral promoter, suggesting a role of UHRF1 in the silencing of HIV-1 gene expression during latency.
UHRF1 downregulation releases HIV-1 from latency
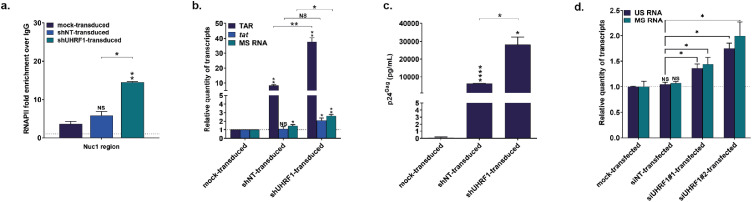
To determine the involvement of UHRF1 in the establishment or the maintenance of transcriptional silencing at the HIV-1 promoter during latency, we induced the downregulation of endogenous UHRF1 by RNA interference using shRNAs. J-Lat 8.4 cells were mock-transduced or stably transduced with lentiviral vectors expressing the puromycin-resistance gene and containing one out of four different shRNAs targeting UHRF1 mRNA (shUHRF1#1-4) or a control non-targeting shRNA (shNT). UHRF1 knockdown at both the protein and mRNA levels in selected puromycin-resistant clones was confirmed by both western blot and RT-qPCR analyses (Fig. S4a and S4b, respectively). Because UHRF1 is essential in the cell cycle control and its downregulation is associated with cellular mortality,56,57 we selected for further analyses one shUHRF1 that did not provoke the most efficient downregulation of UHRF1 and that would, therefore, not be counter-selected (Fig. S4a,b, pLV shUHRF1#4). First, by ChIP experiments, we showed that RNAPII recruitment to the Nuc-1 region of the HIV-1 promoter was statistically higher in UHRF1-knocked down J-Lat 8.4 cells than in shNT-transduced cells, consistent with a release of viral transcriptional blocks from latency (Figure 3a, 2.48-fold increase, p = 0.008, [unpaired T-test]). Reactivation of HIV-1 gene expression from latency in J-Lat 8.4 cells following UHRF1 knockdown was further studied by quantifying by RT-qPCR initiated (TAR region) and elongated (tat region) HIV-1 transcripts (Figure 3b). A statistically higher number of initiated and elongated transcripts was observed when UHRF1 was knocked down in J-Lat 8.4 cells, compared to the amount measured in shNT-transduced cells (Figure 3b, 3.29-fold increase, p = 0.0011 and 1.7-fold increase, p = 0.034, for TAR and tat, respectively, [unpaired T-test]). This indicated that the observed increase in RNAPII recruitment was accompanied by an increased transcription initiation and elongation from the HIV-1 promoter. In addition, we observed a statistically significant increase in multiply spliced (MS) HIV-1 RNA58 in shUHRF1-transduced cells compared to shNT-transduced cells (Figure 3b, 2.99-fold increase, p = 0.0004, [unpaired T-test]). Finally, quantification of p24Gag capsid protein by ELISA in culture supernatants from puromycin-resistant clones showed that UHRF1 knockdown was accompanied by an increased HIV-1 production, as demonstrated by the higher HIV-1 production in shUHRF1-transduced cells compared to shNT-transduced cells (Figure 3c, 4.5-fold increase, p = 0.02, [unpaired T-test]). We observed this statistically significant increase in HIV-1 production upon UHRF1 knockdown with the four different shUHRF1 tested (Fig. S4c). Of note, J-Lat 8.4 cells transduction with the control, non-targeting shRNA, also caused reactivation of HIV-1 gene expression and production, though to lower levels than transduction of the shUHRF1, as seen by increased levels of HIV-1 initiated, gag and multiply spliced transcripts (Figure 3b) and by increased HIV-1 production (Figure. 3c and S4c). This was consistent with the use of lentiviral shRNA vectors and was in agreement with a previous report.36 Nevertheless, we observed the reactivation of HIV-1 gene expression and production when shUHRF1-transduced conditions were normalized to shNT-transduced conditions, and a fortiori when normalized to mock-transduced conditions (Figure 3a–c).
Figure 3.
UHRF1 silences HIV-1 transcription during latency. (a) Chromatin was prepared from J-Lat 8.4 cells, either mock-transduced, control-transduced (with non-targeting shRNA, indicated as “shNT-transduced”) or shUHRF1-transduced (indicated as “shUHRF1-transduced”). Immunoprecipitations were performed using anti-RNAPII or purified rabbit IgG, as a negative control. qPCRs were performed with primers hybridizing specifically to the 5’LTR, in the Nuc-1 region. Folds relative to IgG are presented, where fold enrichments for each immunoprecipitated DNA were calculated by the relative standard curve on input DNA. Values represent the means of duplicate samples ± SD. (b) Total RNA preparations from mock-transduced, shNT-transduced or shUHRF1-transduced J-Lat 8.4 cells were reverse transcribed. Initiated (TAR region), elongated (tat region) transcripts, or HIV-1 multiply spliced RNA (MS RNA) were quantified by RT-qPCR using GAPDH as a first normalizer and the mock-transduced condition as a second normalizer. Means from duplicate ± SD are indicated. Statistical significance was calculated with an unpaired T test. (c) Cultures supernatants from mock-transduced, shNT-transduced or shUHRF1-transduced J-Lat 8.4 cells were probed for viral production as measured by ELISA on p24Gag capsid protein. (d) Total RNA preparations from mock-transfected, siNT-transfected, siUHRF1#1-transfected or siUHRF1#1 primary CD4+ T cell models were reverse transcribed. Unspliced HIV-1 RNA (US RNA) or multiply spliced HIV-1 RNA (MS RNA) were quantified by RT-qPCR using TBP as a first normalizer and the mock-transfected condition as a second normalizer. Means from duplicate ± SD are indicated. One representative model out of two is shown. Statistical significance was calculated with a Mann-Whitney test.
We next sought to confirm these effects in the more physiological context of primary CD4+ T cell models for HIV-1 infection. To do so, we induced the downregulation of endogenous UHRF1 by RNA interference using siRNAs. Primary CD4+ T cell models were mock-transfected, or transiently-transfected with siRNAs targeting UHRF1 mRNA (siUHRF1#1-2) or a control non-targeting siRNA (siNT). UHRF1 knockdown was confirmed by both western blot and RT-qPCR analyses (Fig. S4D and S4E, respectively). Quantification of HIV-1 unspliced RNA levels (US RNA) or multiply-spliced RNA levels (MS RNA) showed a statistically significant increase in both siUHRF1-transfected conditions compared to the control siNT-transfected condition (Figure 3D, 1.34-fold increase, p = 0.029 and 1.76-fold increase, p = 0.029, for siUHRF1#1 and siUHRF1#2 and US RNA and 1.46-fold increase, p = 0.029 and 1.99-fold increase, p = 0.029, for siUHRF1#1 and siUHRF1#2 and MS RNA, respectively, [Mann Whitney test]).
Together, these data demonstrate that UHRF1 knockdown enables a release of the transcriptional blocks from HIV-1 latency, in both cell line models and primary cell models, thereby indicating the role of UHRF1 in the maintenance of HIV-1 latency.
UHRF1 is a novel epigenetic repressor of HIV-1 gene expression
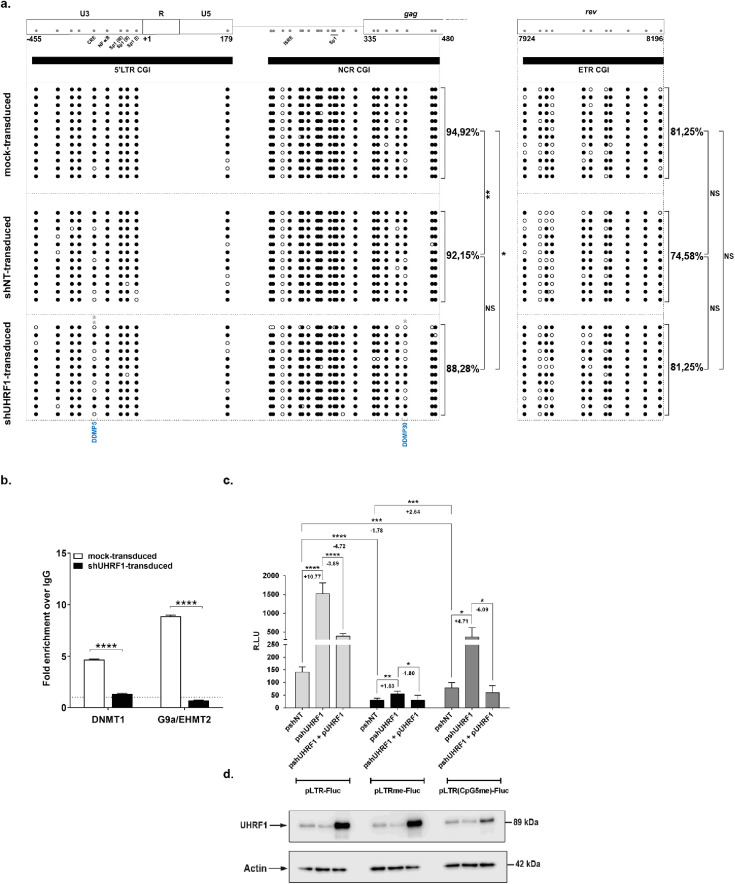
Because UHRF1 is an important epigenetic integrator in the heterochromatinization of cis-regulatory sequences,24, 25, 26 we next studied how UHRF1 knockdown and subsequent reactivation of HIV-1 gene expression translated in terms of DNA methylation modifications in the HIV-1 promoter. UHRF1 knockdown was associated with significant decrease in global HIV-1 promoter DNA methylation compared to the control conditions (Figure 4a, Figure S5), whereas no DNA demethylation was observed on the control ETR CGI. These results thus indicated that UHRF1 downregulation led to HIV-1 transcriptional reactivation through specific 5’LTR demethylation. UHRF1 has been shown to interact with multiple epigenetic enzymes, including the DNA methyltransferase DNMT156,59,60 and the histone methyltransferase G9a/EHMT2,61,62 that are important actors in the HIV-1 promoter heterochromatinization during latency (reviewed in Verdikt et al.8). Thus, we next assessed by ChIP the effect of UHRF1 downregulation on the recruitment of DNMT1 and G9a/EHMT2 to the HIV-1 promoter. Following downregulation of UHRF1 and reactivation of HIV-1 gene expression and production, a significant decrease in DNMT1 and G9a/EHMT2 recruitment was observed on the viral promoter (Figure 4b, 3.61-fold decrease, p = 0.0003 and 12.71-fold decrease, p = 0.00005, [unpaired T-test], for DNMT1 and G9a, respectively). These results thus indicate that UHRF1 silences HIV-1 gene expression during latency by actively promoting the accumulation of DNA methylation on the viral promoter via the recruitment of DNMT1. Furthermore, we demonstrate that UHRF1 also recruits G9a/EHMT2 to the latent promoter, thereby linking DNA methylation with the accumulation of inhibitory histone methylation.
Figure 4.
UHRF1 is a novel epigenetic repressor of HIV-1 latency. (a) DNA methylation mapping performed by sodium bisulfite sequencing is presented for the promoter CGIs or the intragenic ETR CGI in J-Lat 8.4 cells mock-transduced, shNT-transduced or shUHRF1-transduced, as indicated. Unmethylated and methylated CpG dinucleotides are respectively represented with open and closed circles, where each line corresponds to individual sequenced molecules. The global methylation level presented correspond to mean percentages of methylated CpGs for the twelve clones of each condition, either for the promoter, CGIs considered together (5’LTR + NCR CGIs) or for the ETR CGI. Panel (a) to (d) originates from the same representative experiment out of three. (b) Chromatin was prepared from J-Lat 8.4 cells, either mock-transduced or shUHRF1-transduced. Immunoprecipitations were performed using anti-DNMT1, anti-G9a or purified rabbit IgG, as a negative control. qPCRs were performed with primers hybridizing specifically to the 5’LTR, in the Nuc-1 region. Folds relative to IgG are presented, where fold enrichments for each immunoprecipitated DNA were calculated by the relative standard curve on input DNA. Values represent the means of duplicate samples ± SD. (c) HEK293T cells were transfected either with 600 ng of the pshNT vector or with 600 ng of the pshUHRF1#4. Twenty-four hours after this initial transfection, 400 ng of the pLTR-Fluc, the pLTRme-Fluc or the pLTR(CpG5me)-Fluc reporter constructs together with or without 200 ng of the plasmid overexpressing UHRF1 (pUHRF1) were co-transfected. Luciferase activities were measured in the cell lysates 24 h post-transfection. Results are presented as histograms of “relative luciferase units” (R.L.U.), corresponding to the Fluc activity normalized to the total levels of proteins. Means and standard errors of triplicate samples are represented. An experiment representative of three independent experiments is shown. Statistical significance was assessed by an [unpaired T-test]. (d) UHRF1 and β-actin, serving as a loading control, protein levels were assessed by immunoblot in cell lysates of the corresponding transfection points.
UHRF1 possesses multiple repression mechanisms of gene expression and our data indicate that it can be recruited to the 5’LTR independently of DNA methylation. We thus further dissected the dependency of UHRF1 to DNA methylation and, in particular, to the methylation status of DDMP5 for its role in HIV-1 transcriptional repression. To do so, we subcloned the HIV-1 5’LTR region in a reporter construct, where the LTR controls the firefly luciferase gene and is either unmethylated (referred to as the “pLTR-FLuc” vector), in a hypermethylated state (i.e. where each CpG of the 5’LTR has been artificially methylated, the resulting construct being termed “pLTRme-Fluc”), or where only the 5th CpG dinucleotide corresponding to the DDMP5 of the LTR is methylated (referred to as the “pLTR(CpG5me)-Fluc” vector). First, as shown in Figure 4c, these three reporter vectors were transiently transfected in HEK293T cells along with the control non-targeting shRNA vector (referred to as the “pshNT”). These transfection experiments showed that the pLTRme-Fluc vector presented a statistically significant decrease in luciferase activity compared to the pLTR-Fluc vector (Figure 4c, 4.72-fold decrease, p < 0.0001, [unpaired T-test]), thereby confirming that DNA methylation of the HIV-1 LTR provoked a decrease of its promoter activity. Second, we transiently co-transfected the reporter LTR constructs along with the UHRF1-targeting shRNA vector (“pshUHRF1”). Downregulation of endogenous UHRF1 in HEK293T cells was confirmed by western blot (Figure 4d) and was associated with statistically significant increased luciferase activities for all three constructs (Figure 4c, 10.77-fold increase, p < 0.0001; 1.83-fold increase, p = 0.0014 and 4.71-fold increase, p = 0.0152, for the pLTR-Fluc, pLTRme-Fluc and pLTR(CpG5me)-Fluc constructs, respectively and according to an [unpaired T-test]), confirming the repressive role of UHRF1 in the control of HIV-1 gene expression. Interestingly, in this reporter system, UHRF1-mediated repression of the HIV-1 promoter activity was proportionally lower when the LTR was totally methylated or methylated on the DDMP5 in comparison to the repression observed when the LTR was unmethylated (Figure 4c, compare the 1.83-fold and 4.71-fold increases with the 10.77-fold increase, for the fully-methylated, DDMP5-methylated and unmethylated LTRs, respectively). These results indicated that UHRF1 repressive activity was dependent on the HIV-1 promoter DNA methylation status, and, in particular, that this repression was partially, but not totally, dependent on the DDMP5 methylation status. To confirm the dependency of UHRF1 on DNA methylation for its role in HIV-1 transcriptional repression, we performed a rescue experiment in which we transiently co-transfected reporter constructs along with the UHRF1-targeting shRNA vector then, after twenty-four hours, we added a UHRF1 expression vector (“pUHRF1”) before assaying the luciferase activities after another twenty-four hours. Overexpression of UHRF1 was confirmed by western blot (Figure 4d) and decreased the luciferase activities of all three reporter vectors in comparison to the conditions of UHRF1 downregulation (Figure 4c, 3.88-fold decrease, p < 0.0001; 1.80-fold decrease p = 0.0217 and 6.09-fold decrease, p = 0.0114 for the pLTR-Fluc, pLTRme-Fluc and pLTR(CpG5me)-Fluc constructs, respectively), thereby confirming the specific role of UHRF1 in the repression of HIV-1 promoter activity. Together, these results indicated that in the context of an in vitro HIV-1 5’LTR reporter system, UHRF1 repression of HIV-1 transcription depended in part but not exclusively on DNA methylation of the viral promoter.
Altogether, our results highlight the role of UHRF1 in HIV-1 latency through both DNA and histone methylations, thereby demonstrating that UHRF1 participates in the heterochromatinization of the viral promoter during latency.
Pharmacological inhibition of UHRF1 promotes HIV-1 reactivation from latency
Understanding the molecular mechanisms of HIV-1 latency has allowed the development of several classes of LRAs.63, 64, 65 Our results on the role of UHRF1 positioned this cellular factor as an attractive pharmacological target for HIV-1 latency reversal strategies. Epigallocatechin-3-gallate (EGCG), the major polyphenolic compound of green tea, has been shown to inhibit UHRF1 expression.66 Accordingly, we showed here in J-Lat 8.4 cells that increasing concentrations of EGCG steadily decreased UHRF1 protein levels starting from 30µg/mL of EGCG (Fig. S6a). This protein level decrease was not accompanied by a decrease in UHRF1 mRNA level, as quantified by RT-qPCR (Fig. S6b), consistent with a previous report showing that EGCG targets UHRF1 proteins but not UHRF1 transcripts.66
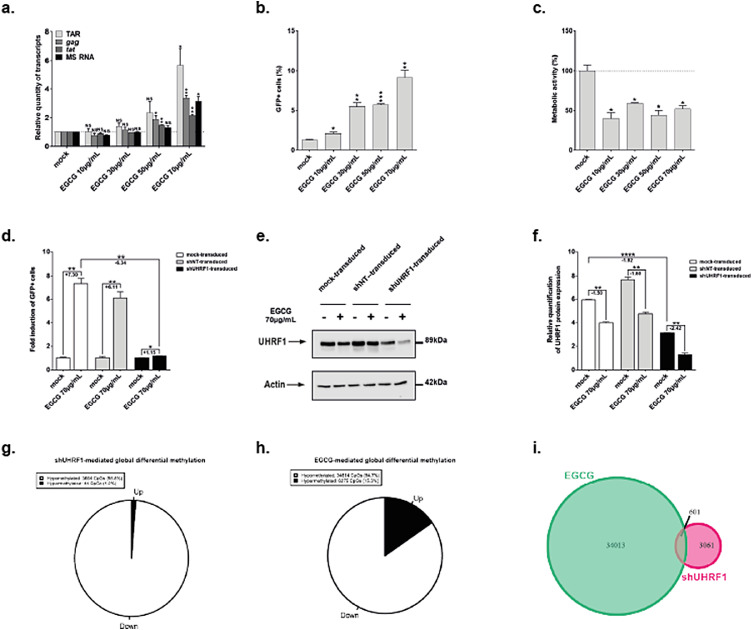
To assess the LRA potential of EGCG, we quantified HIV-1 transcripts by RT-qPCR in EGCG-treated J-Lat 8.4 cells (Figure 5a). We observed statistically significant increases in initiated (TAR region), elongated (gag and tat regions) and MS HIV-1 transcript levels in EGCG-treated compared to mock-treated cells (Figure 5a, 5.70-fold increase and p = 0.03, 3.35-fold increase and p = 0.003, 2.15-fold increase and p = 0.003, and 3.11-fold increase and p = 0.01 for TAR, tat, gag and MS RNAs, respectively at 70µg/mL of EGCG). Furthermore, quantification of GFP+ cells by flow cytometry showed that starting from 10µg/mL of EGCG, a release of the post-transcriptional blocks to the production of HIV-1 was observed (Figure 5b). In addition, the cellular metabolic activity in J-Lat 8.4 cells after treatment with increasing EGCG doses was decreased in a statistically relevant manner, with a metabolic activity of 36% observed at the highest EGCG dose (Figure 5c). Despite increased levels of gag transcripts (Figure 5a), EGCG did not reactivate HIV-1 particles production in J-Lat 8.4 cells, as measured by p24Gag capsid protein ELISA in cell supernatants (Fig. S6c). These data were consistent with a previous report indicating that EGCG destabilizes HIV-1 particles by binding to envelope phospholipids, thereby inducing their degradation.67 Thus, while EGCG reactivation capacity might be lower than the capacities of known LRAs (Fig. S6d,e), EGCG anti-viral activity allows for the degradation of virions produced from HIV-1 reactivation from latency. Furthermore, consistent with UHRF1 being unequivocally recruited to the HIV-1 promoter in latent conditions (Figure 2h), EGCG treatment reactivated HIV-1 gene expression from latency in all J-Lat clones (Fig. S6f,g) as well as in primary CD4+ T cell models for HIV-1 infection (Fig. S6h,i).
Figure 5.
HIV-1 transcription is reactivated from latency upon UHRF1 inhibition by epigallocatechin-3-gallate. (a) Total RNA preparations from J-Lat 8.4 cells mock-treated or treated with increasing doses of EGCG for 24 h were used in RT-qPCR to quantify initiated (TAR region), gag, elongated (tat region) transcripts, or HIV-1 MS RNA, using GAPDH as normalizer. (b) After 24 h of EGCG increasing doses treatment, J-Lat 8.4 cells were analyzed by flow cytometry to quantify the percentage of GFP+ cells. (c) WST-1 proliferation assay, reflective of metabolic activity, was performed on J-Lat 8.4 cells treated for 24 h with increasing doses of EGCG. The result obtained with mock-treated cells was set at a value of 100%. (a), (b) and (c) originate from the same representative experiment out of three. Means ± SD of duplicates are presented. Statistical significance was calculated with an [unpaired T-test] and corresponds to comparisons to the mock-treated condition. (d) J-Lat 8.4 cells, either mock-transduced or shUHRF1-transduced were treated for 24 h with 70 µg/mL of EGCG. The percentage of GFP positive cells was assessed by flow cytometry and was normalized to the respective mock conditions. Means ± SD of duplicates representative of three independent experiments are presented. Statistical significance was assessed by an unpaired T test. (e) Whole protein levels of the experiments presented in Figure 5d were loaded and probed for the presence of UHRF1 and β-actin, serving as a loading control. (f) Quantification of the western blot presented in Figure 5e was performed in ImageJ. Means ± SD of two independent quantifications are presented. Statistical significance was assessed by an [unpaired T-test]. (g) An Infinium assay with gDNA from above was conducted. Differential methylated CpGs upon EGCG treatment were plotted on a pie chart, where the black slice represents hypermethylated CpGs and the white slice represents hypomethylated CpGs. (h) An Infinium assay with gDNA from Figure 3 was conducted. Differential methylated CpGs upon shUHRF1 transduction were plotted on a pie chart, where the black slice represents hypermethylated CpGs and the white slice represents hypomethylated CpGs. (i) A Venn diagram representing the number of hypomethylated CpGs in both conditions is presented.
Since EGCG is a broad-acting compound,68 we next investigated EGCG modes of action in HIV-1 latency reversal, specifically, their dependency on UHRF1 inhibition. First, we performed EGCG reactivation assays in latently-infected J-Lat 8.4 cells in which UHRF1 expression had been downregulated. By normalizing each EGCG treatment to its respective mock control (Fig. S6j), we observed a statistically relevant decrease in EGCG reactivation potency in shUHRF1-transduced versus mock-transduced J-Lat 8.4 cells (Figure 5d, 6.34-fold decrease, p = 0.0032, [unpaired T-test]), whereas UHRF1 expression was further inhibited in EGCG-treated shUHRF1-transduced cells in comparison to mock-treated, mock-transduced cells (Figure 5e,f). Second, we assessed the DNA methylation signatures occurring at genome-scale while knocking down UHRF1 or treating latently-infected J-Lat 8.4 cells with EGCG and compared these signatures. To do so, we performed an Infinium Human Methylation 850 K array.69 By using unsupervised analyses, such as hierarchical clustering and principal component analyses, we revealed a strong effect of both EGCG treatment and UHRF1 knockdown on the cellular methylome, with treated samples being distinctly different from control samples (Fig. S7a,b). In addition, EGCG treatment and UHRF1 knockdown methylation profiles partially clustered together, suggesting that the effect of the two conditions on the DNA methylome was only partly similar. We identified 3664 hypomethylated CpGs through UHRF1 knockdown and 34614 hypomethylated CpGs through EGCG treatment (Figure 5g and Figure 5h, respectively). As already suggested by the unsupervised analyses, the overlap between differential CpGs through EGCG treatment and UHRF1 knockdown, while small, was statistically significant (Figure 5i, 601 sites, hypergeometrical p-value < 1e-170), suggesting that some but not all mechanisms involved in the two processes were similar. We further assessed which pathways were affected at the DNA methylation level in EGCG-treated and shUHRF1-transduced conditions using a Gene Set Enrichment Analysis (GSEA).41 This analysis further confirmed the partial but not total overlap between the two conditions (Fig. S7c,d).
Together, these experiments indicate that EGCG reactivates HIV-1 from latency in part via the inhibition of UHRF1, although this compound has a broader reactivation capacity on HIV-1 gene expression, in line with its pleiotropic action on the HIV-1 replication cycle.67,70 Of note, because of its important role in epigenetic silencing during oncogenesis, UHRF1 has been the target of intense drug development in recent years.71 The search for a specific inhibitor of UHRF1 being still in its early stages, we nevertheless tested the uracil derivative NSC23200372 in HIV-1 reactivation assays. These experiments showed that, at a concentration of 30 µM, NSC232003 provoked a statistically relevant HIV-1 reactivation from latency (Fig. S8a, reactivation of 3.5-fold, p = 0.037 according to an [unpaired T-test]) with no notable effect on cell metabolic activity (Fig. S8b). These results reinforce the role of UHRF1 in HIV-1 latency and highlight the relevance of anti-UHRF1 approaches in latency reversal strategies.
EGCG induces HIV-1 expression in CD8+-depleted PBMCS from HIV-1+ aviremic individuals
The development of LRAs has been guided by deciphering HIV-1 latency molecular mechanisms in in vitro cell models.5 However, these models do not completely recapitulate the biological properties of in vivo latent reservoirs.73 Therefore, we next evaluated the LRA potency of EGCG ex vivo, using cultures of CD8+-depleted PBMCs isolated from the blood of cART-treated aviremic HIV-1+ individuals.
We first assessed cellular viability and metabolic activity in ex vivo cultures of CD8+-depleted PBMCs from six healthy donors in response to EGCG treatments (Fig. S9). Neither TCR stimulation, serving as a positive control, nor increasing EGCG doses affected cellular viability (Fig. S9a). However, consistent with our observations in J-Lat cells, metabolic activity was affected by EGCG, although the median of metabolic activity remained tolerable (Fig. S9b). Suitable LRA candidates for HIV-1 latency reversal strategies in vivo should limit non-specific or strong immune T-cell activation. Therefore, we assessed the level of cell surface activation markers HLA-DR (late activation marker), CD25 (intermediate activation marker), CD69 (early activation marker) and CD38 (late activation marker and predictor of HIV-1 progression) in EGCG-treated cells compared to mock-treated cells (Fig. S9c–f). TCR stimulation consistently and statistically increased the levels of each marker, while EGCG treatment increased slightly but statistically the surface expression of CD69 (Fig. S9e). When EGCG stimulation was sustained for 6 days, this increase in CD69 expression was not maintained (data not shown). Therefore, we attributed it to an indirect epigenetic effect of EGCG.74 Indeed, considering the wide DNA demethylation observed following EGCG treatment (Figure 5h), changes in gene expression were expected. Flow cytometry analyses also highlighted that increasing doses of EGCG were associated with a statistically significant decrease in CD4 expression on the surface of the treated CD8+-depleted PBMCs (Fig. S9g), which would, in the context of HIV-1 infection, reduce the number of target cells and therefore limit HIV-1 dissemination.
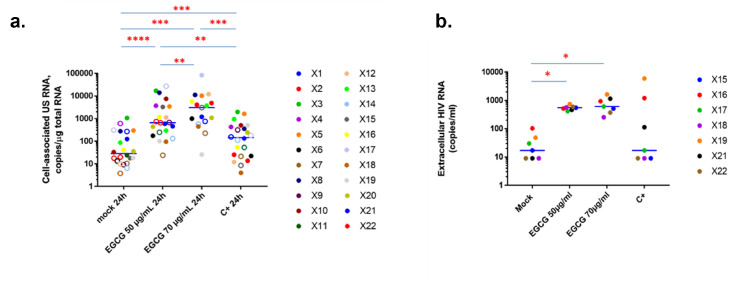
Based on these observations of tolerable cytotoxic, metabolic and immune effects, we next investigated HIV-1 recovery in CD8+-depleted PBMCs in response to EGCG treatments. To do so, we purified CD8+-depleted PBMCs from 22 HIV+ aviremic cART-treated individuals (Table S1a) and evaluated the frequency of infected cells during plating by quantification of cell-associated total HIV-1 DNA (Table S1b). Ex vivo cultures were then mock-treated, treated with 50 µg/mL or 70 µg/mL of EGCG or activated with anti-CD3 + anti-CD28 antibodies as a positive control. Because of EGCG's effect in degrading viral particles, reactivation of HIV-1 gene expression was measured unequivocally by the quantification of intracellular HIV-1 RNA. These quantifications showed that EGCG potently increased HIV-1 unspliced RNA levels in patient cells, even to higher levels than TCR activation (Figure 6a). Moreover, HIV-1 US RNA/DNA ratios were statistically increased in EGCG-treated patient cells (Fig. S10a,c), indicating that proviruses were more transcriptionally active. Quantification of HIV-1 extracellular RNA in supernatants, serving as a surrogate for the completion of the viral replication cycle, further indicated a statistically significant increase in HIV-1 extracellular RNA when CD8+-depleted PBMCs from HIV+ individuals were submitted to EGCG treatments (Figure 6b). Thus, our results showed that, by destabilizing HIV-1 particles in reactivated ex vivo cell cultures,67 EGCG treatment released HIV-1 RNA from virions. Accordingly, we observed a statistically significant increase in HIV-1 extracellular RNA/DNA ratios in EGCG-treated compared to mock-treated ex vivo patient cell cultures (Fig. S10b,c), indicating that not only transcriptional but also post-transcriptional latency blocks were overcome by EGCG treatment. Altogether, our results highlight the strong potency of EGCG as a new LRA ex vivo allowing to reactivate HIV-1 transcription to the completion of the replication cycle, while maintaining low immune activation level, and even allowing to prevent de novo infections by decreasing cell surface CD4 marker expression and by degrading reactivated HIV-1 particles.
Figure 6.
EGCG reactivates the expression of viral RNA in ex vivo cultures of CD8+-depleted PBMCs isolated from cART-treated aviremic HIV-1+ individuals. Ex vivo cultures of CD8+-depleted PBMCs isolated from 22 cART-treated aviremic HIV+ individuals were mock-treated or treated for 24 h with EGCG, at the indicated concentrations, or with anti-CD3+anti-CD28 antibodies serving as positive control stimulation. (a) Total intracellular RNA was extracted and cell-associated HIV-1 US RNA was quantified. Medians are represented. Open circles depict undetectable values, censored to the assay detection limits. The latter depended on the amounts of input cellular RNA and therefore differed between samples. Statistical significance was determined by paired Wilcoxon tests, where pairs were included in the analysis only when either (i) both values in a pair were detectable, or (ii) one value in a pair was undetectable and the other detectable, and the maximal value of the undetectable (the assay detection limit) was lower than the detectable. (b) In 7 out of 22 HIV+ individuals, the concentration of HIV-1 extracellular genomic RNA in culture supernatants was also determined (in copies/ml).
Finally, because our in vitro data pointed to the pleiotropic action of EGCG in terms of HIV-1 reactivation capacity, we further assessed the contribution of HIV-1 promoter methylation ex vivo to EGCG reactivation potency. A dynamical increase in the degree of viral promoter methylation has been shown in HIV+ individuals in response to the duration of the antiretroviral treatment or the time of viral suppression.19,20 Accordingly, strong statistically significant positive correlations were observed between the EGCG-mediated HIV-1 reactivation potency and either the time on cART (Fig. S11a) or the time of virological suppression (Fig. S11b), while no correlation was observed for the positive control (Fig. S11c). These results demonstrate that EGCG modes of action ex vivo are time-dependent in HIV+ individuals, suggesting that EGCG acts, at least in part, through HIV-1 promoter demethylation. To refine this observation, we next assessed the DNA methylation level of the viral promoter in our cohort of HIV+ individuals by sodium bisulfite sequencing. As suggested by Blazkova and colleagues,11 we found that the assessment of DNA methylation in aviremic individuals, who have small size reservoirs that prevent the analysis of proviral DNA, was technically challenging. Nevertheless, we obtained the DNA methylation profile of the HIV-1 promoter for 8 out of the 22 enrolled HIV+ individuals. Out of these, five individuals had no detectable DNA methylation, while three individuals presented a median methylation level of 7.41% mCpG on the HIV-1 promoter, which corresponded to levels reported in other studies.11,13,20 To determine the relationship between EGCG reactivation potency and in vivo HIV-1 promoter methylation, we clustered these 8 individuals in groups of non-methylated or methylated 5’LTR and plotted the cell-associated HIV-1 US RNA for 50µg/mL of EGCG (Fig. S11d). Without reaching statistical significance, a trend towards a higher reactivation could be observed for patients accumulating more DNA methylation on the viral promoter. The lack of statistical significance can be attributed to the low number of patients but it also reveals the pleiotropic reactivation capacity of EGCG. We propose that the mechanism of HIV-1 reactivation from latency by EGCG is through DNA demethylation of the viral promoter, or, when it is not methylated, through indirect demethylation of cellular genes.
Altogether, our ex vivo data indicate, in line with the heterogeneity of the mechanisms responsible for HIV-1 latency, that EGCG, in addition to its antiviral properties, is a heterogeneous LRA, capable of reversing HIV-1 latency through several modes of action depending on the different clinical characteristics of infected individuals.
Discussion
Accumulating data highlight the intrinsically dynamic and heterogeneous nature of latent HIV-1 cellular reservoirs within and between infected individuals. This heterogeneity and the multiplicity of the silencing mechanisms underlying HIV-1 latency rather than latency itself are now considered as the major barrier to HIV-1 eradication.8 In agreement, LRAs have been found to present various reactivation potencies in vitro and ex vivo.63,75,76 In the context of DNA methylation, our previous study has highlighted that the DNA methylation inhibitor 5-AzadC exhibits different ex vivo reactivation potencies in terms of HIV-1 latency reversal.23 Here, we investigated the molecular basis of this potency heterogeneity at the level of proviral DNA methylation.
We first evidenced the existence of non-random and reproducible DNA methylation signatures in response to 5-AzadC treatment at the level of the single CpG dinucleotide in the HIV-1 promoter. Thus, rather than reactivating HIV-1 in a non-specific manner, 5-AzadC acts through specific molecular mechanisms within the viral promoter. Correspondingly, in epigenetic cancer therapies, 5-AzadC was shown to induce non-random DNA demethylation at some specific CpG dinucleotides.77 To tease out the underlying regulatory mechanisms, we mapped preferentially-demethylated positions in the HIV-1 5’LTR. Among all identified DDMPs, DDMP5 presented the highest demethylation probability and the highest statistical significance and DDMP5 is located in a CRE binding site. Interestingly, DDMP5 had been previously identified in response to LPS activation of HIV-1 in humanized mice models,78 pointing towards the importance of this specific DNA sequence in the control of HIV-1 gene expression. We demonstrated that DDMP5 methylation allowed the in vitro binding of an additional DNA-protein complex, containing UHRF1. The exploration of binding modalities to the other DDMPs in our dataset revealed that UHRF1 possessed multiple binding sites along the HIV-1 promoter, where DNA methylation was either non-essential, essential or enhancing UHRF1 binding. The presence of multiple binding sites for the same cellular factor is characteristic of HIV-1 and allows the virus to adapt dynamically to changing cellular environments of infection.8,47,79 Redundant recruitment also indicates the importance of the factor for viral gene regulation. In this regard, we report here the in vivo chromatin recruitment of UHRF1 to the latent HIV-1 promoter in all J-Lat T-cell line models tested and, importantly, in primary CD4+ T cell models for HIV-1 infection.
UHRF1 is an epigenetic integrator as it both reads the chromatin (5mC, H3K9me3 and H3R2) and recruits epigenetic enzymes (DNMT1, G9a/EHMT2, …). UHRF1 is an important regulator of endogenous retroviruses27, 28, 29, 30 but its role in the epigenetic repression of exogenous retroviruses had not been explored so far. Here, we report for the first time the role of UHRF1 in HIV-1 silencing through DNA and histone methylations. Mechanistically, UHRF1 knockdown led to statistically significant decreases in the global DNA methylation level of the 5’LTR and DNMT1 recruitment. We also showed that G9a/EHMT2 recruitment to the 5’LTR was decreased upon UHRF1 knockdown, supporting the model that UHRF1 branches several repressive epigenetic mechanisms to maintain HIV-1 latency. We further sought to determine how much of the UHRF1 repressive action on the 5’LTR was dependent on DNA methylation, and in particular on the methylation status of DDMP5. To answer unambiguously to this question, we decided to work in an artificial system of transient transfection experiments and showed that, even in this system, UHRF1 repression of HIV-1 transcription depended strongly, but not exclusively, on the DNA methylation status of the 5’LTR. Thus, in agreement with the multiple modes of recruitment of UHRF1 to the viral promoter, UHRF1 likely represses HIV-1 gene expression during latency through several mechanisms. In this regard, during the revision of the current manuscript, T. Liang and colleagues have reported a non-epigenetic role of UHRF1 in the control of HIV-1 gene expression through the ubiquitinylation of Tat,80 pointing towards the importance of UHRF1 ubiquitin E3 ligase activity in HIV-1 gene expression. Indirectly, recruitment of UHRF1 to the 5’LTR and the exploitation of its ubiquitin ligase activity might also be used in host:pathogen interplays, to selectively downregulate certain cellular factors in the vicinity of the epigenetically-silent HIV-1 5’LTR.81, 82, 83 UHRF1 also targets several lysine residues of the H3 histone (H3K14, H3K18, H3K23 and H3K27) for mono-ubiquitination, which has an important role in DNMT1 recruitment.84,85 Whether H3 ubiquitination occurs on the 5’LTR during HIV-1 latency and its potential role in viral gene expression still needs to be addressed. Furthermore, the observation that HIV-1 actively controls its own gene expression by hijacking the epigenetic cellular machinery also poses the question of HIV-1-induced epigenetic alterations in the host cells. Indeed, several studies have shown the extent of HIV-1-induced DNA methylation alterations in cellular loci.86,87 Finally, a recent report has shown that UHRF1 and TRIM28/KAP1 overlap in the recruitment of DNMT1 to endogenous retroviruses.30 As our laboratory has recently demonstrated that TRIM28/KAP1 is important for the control of HIV-1 expression in microglial reservoirs88 and as we observed the recruitment of UHRF1 to the 5’LTR in myeloid models for HIV-1 persistence (data not shown), an exciting avenue of research is how these two factors UHRF1 and TRIM28/KAP1 are co-recruited to the latent viral promoter and how they concertedly regulate the epigenetic environment of the 5’LTR. From a larger perspective, how the different epigenetic machineries are recruited in a coordinated manner to the latent HIV-1 promoter remains a challenging open question.8
As a proof-of-concept that the molecular characterization of HIV-1 latency mechanisms leads to the identification of new targets for therapeutic strategies, we studied the latency reversal potency of UHRF1 pharmacological inhibition. Because of the important roles of UHRF1 in cancer, multiple drugs are being tested for their capacity to inhibit UHRF1 expression or functions.71 The uracil derivative NSC232003 is one such inhibitory drug72 and we report here that NSC232003 exhibits a novel HIV-1 latency reversal activity. EGCG, the major phenolic compound of green tea, has also been reported, among other functions, to affect UHRF1 expression.66 Contrarily to NSC232003, EGCG can be safely administered to humans and because of its reported antiviral effects,67 EGCG is currently evaluated in anti-HIV clinical trials (NCT01433289 and NCT03141918, ClinicalTrials.gov). Using complementary models – latently-infected T-cell lines, primary CD4+ T cell models for HIV-1 infection and ex vivo cell cultures from cART-treated HIV+ aviremic individuals – our results showed the potency of EGCG as a new LRA. Indeed, EGCG reactivated HIV-1 from latency up to the completion of the viral replication cycle, in a short time frame, and in all the latency models we tested. In agreement with the EGCG antiviral activity on HIV-1,67 we detected little p24Gag capsid protein in the cell culture supernatants in our reactivation experiments. Nevertheless, in our ex vivo patient cell cultures, we observed that EGCG augmented the recovery of intracellular and extracellular HIV-1 RNA. Thus, these results suggest that reactivated virions were indeed destabilized by EGCG in cell culture supernatants. Furthermore, we showed that EGCG treatment was associated with a decreased expression of the cellular surface marker CD4 on target cells, thereby preventing subsequent new infections following reactivation. Of note, a recent report has also tested the effect of EGCG in ex vivo patient cell cultures, but in reactivated conditions.89 In these reactivated conditions, these authors have demonstrated that EGCG decreases HIV-1 RNA and protein expressions by sequestering NF-κB in the cytoplasm. In agreement, we obtained similar results when working in reactivated and not latent conditions (data not shown). This differential effect of EGCG in unstimulated versus reactivated conditions is likely due to the different subcellular localization of NF-κB in unreactivated versus reactivated cells.90,91 Together, we showed the promising use of EGCG in HIV-1 eradication strategies due to its complementary and synergistic anti-HIV modes of action. Our results indicated, however, that EGCG potency was not uniquely dependent on UHRF1 expression, rather this compound presented a pleiotropic function in HIV-1 reactivation, balancing the heterogeneity observed in the cellular reservoirs of the virus.
In conclusion, we developed a probabilistic methodology to decipher the DNA methylation-mediated mechanisms underlying the heterogeneous capacity of 5-AzadC to reactivate HIV-1 from latency. This approach enabled us to uncover the role of UHRF1, an epigenetic integrator, in HIV-1 promoter silencing via DNA and histone methylations. Our work provides a demonstration that the understanding of the molecular basis of the heterogeneous effects of LRAs, even in in vitro HIV-1 latency models, can bring to light new factors involved in HIV-1 silencing and hence, new targets to devise anti-HIV therapeutic approaches. As a proof-of-concept, we showed that EGCG, a known inhibitor of UHRF1, presents, in addition to its broad antiviral activities, a pleiotropic anti-latency activity, allowing its potential use in HIV-1 cure strategies.
Role of funding sources
Funding agencies providing financial support did not participate in the design, data analyses, interpretation, or writing of this study.
Contributors
Conceived and designed the study: CVL. Conceived and designed the experiments: CVL, OR, RV, MBD, SB and LN. Performed the experiments: RV, MBD, SB, LN, GD, CV, AA-A, MS, EP, ND. Performed the HIV-1 DNA and RNA quantifications: AP, GD, VAF and AC. Performed the activation analyses: FC. Performed the primary cell models of HIV-1 infection: MBD and LN. Performed the Infinium analyses: MB and FF. Performed HIV+ individuals selection: CN, SDW. Analyzed and interpretated the data: RV, MBD, SB, LN, GD, CV, AA-A, MS, EP, ND, OR and CVL. Contributed with reagents/materials/analysis tools: CS, CPBS, AS-C, CR, BB, VG, OR. Wrote the manuscript: RV and CVL. Supervised the study and acquired funding: CVL and OR. All authors read or provided comments on the manuscript. All data were validated by CVL, RV, MBD,SB and LN.
Data sharing statement
All the data/analyses presented in the current publication will be made available upon request to the corresponding author.
Declaration of interests
The authors declare no conflicts of interest.
Acknowledgments
We thank the members of the ANRS (French National Agency for Research on AIDS and Viral Hepatitis) RHIVIERA (Remission of HIV Era) Consortium for helpful discussions. We thank the HIV-1+ individuals for their willingness to participate in this study. We thank the nursing team of CHU Saint-Pierre Hospital (Elodie Goudeseune, Joëlle Cailleau and Annick Caestecker) who cared for the HIV+ individuals. We thank Jacqueline Pineau from the transfusion centre of Charleroi (Belgium) for providing blood from healthy donors. We thank Hilde Vereertbrugghen from Francis Corazza's laboratory for excellent technical assistance. We thank Motoko Unoki for her precious advice regarding UHRF1 RNA interference. We thank Mitia Duerinckx and Benoît Van Driessche for their support in the probabilistic and statistical analyses.
CVL acknowledges funding from the Belgian National Fund for Scientific Research (F.R.S.-FNRS, Belgium), the « Fondation Roi Baudouin », the NEAT (European AIDS Treatment Network) program, the Internationale Brachet Stiftung, ViiV Healthcare, the Walloon Region (« Fonds de Maturation »), « Les Amis des Instituts Pasteur à Bruxelles, asbl », and the University of Brussels (Action de Recherche Concertée ULB grant) related to her work on HIV latency. The laboratory of CVL is part of the ULB-Cancer Research Center (U-CRC). RV was funded by an “Aspirant” fellowship (F.R.S-FNRS), a fellowship from “Les Amis des Instituts Pasteur à Bruxelles, asbl” and is a Belgian American Educational Foundation (BAEF) fellow and a scientific collaborator of the ULB. LN is supported by a “PDR” grant from the F.R.S-FNRS. GD is a postdoctoral clinical master specialist for the F.R.S-FNRS. A A-A is a fellow of the “Wallonie-Bruxelles International” Program and the Marie Skłodowska Curie COFUND action. EP is a fellow of the “Télévie Program” (F.R.S-FNRS). MBD and MS are funded by FRIA fellowships (F.R.S.-FNRS). CVL is "Directeur de Recherches" at the F.R.S-FNRS. Work in OR's laboratory was supported by grants from the French Agency for Research on AIDS and Viral Hepatitis (ANRS), the Sidaction and the “Alsace contre le Cancer” Foundation. This work was supported by the European Union's Horizon 2020 research and innovation program under grant agreement No 691119-EU4HIVCURE-H2020-MSCA-RISE-2015. This work is supported by 1UM1AI164562-01, co-funded by National Heart, Lung and Blood Institute, National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, National Institute on Drug Abuse and the National Institute of Allergy and Infectious Diseases.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103985.
Appendix. Supplementary materials
References
- 1.Deeks S., Autran B., Berkhout B., et al. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol. 2012;12:607–614. doi: 10.1038/nri3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trono D., Van Lint C., Rouzioux C., et al. HIV persistence and the prospect of long-term drug-free remissions for hiv-infected individuals. Science. 2010;329:174–180. doi: 10.1126/science.1191047. [DOI] [PubMed] [Google Scholar]
- 3.Chun T.W.W., Fauci AS. HIV reservoirs: pathogenesis and obstacles to viral eradication and cure. AIDS. 2012;26:1261–1268. doi: 10.1097/QAD.0b013e328353f3f1. [DOI] [PubMed] [Google Scholar]
- 4.Darcis G., Van Driessche B., Van Lint C. HIV latency: should we shock or lock? Trends Immunol. 2017;38:217–228. doi: 10.1016/j.it.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Spivak A., Planelles V. Novel latency reversal agents for HIV-1 cure. Annu Rev Med. 2018;69:421–436. doi: 10.1146/annurev-med-052716-031710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herbein G. Shock and kill, but don't miss the target. EBioMedicine. 2020;58 doi: 10.1016/j.ebiom.2020.102906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lange U.C., Verdikt R., Ait-Ammar A., Van Lint C. Epigenetic crosstalk in chronic infection with HIV-1. Semin Immunopathol. 2020;42:187–200. doi: 10.1007/s00281-020-00783-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verdikt R., Hernalsteens O., Van LintC. Epigenetic mechanisms of HIV-1 persistence. Vaccines. 2021;9:1–23. doi: 10.3390/vaccines9050514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chavez L., Kauder S., Verdin E. In vivo, in vitro, and in silico analysis of methylation of the HIV-1 provirus. Methods. 2011;53:47–53. doi: 10.1016/j.ymeth.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kauder S., Bosque A., Lindqvist A., Planelles V., Verdin E. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 2009;5:1–15. doi: 10.1371/journal.ppat.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blazkova J., Trejbalova K., Gondois-Rey F., et al. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 2009;5:1–14. doi: 10.1371/journal.ppat.1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamann M.V., Ehmele P., Verdikt R., et al. Transcriptional behavior of the HIV-1 promoter in context of the BACH2 prominent proviral integration gene. Virus Res. 2021;293:1–12. doi: 10.1016/j.virusres.2020.198260. [DOI] [PubMed] [Google Scholar]
- 13.Palacios J.A., Pérez-Piñar T., Toro C., et al. Long-term nonprogressor and elite controller patients who control viremia have a higher percentage of methylation in their HIV-1 proviral promoters than aviremic patients receiving highly active antiretroviral therapy. J Virol. 2012;86:13081–13084. doi: 10.1128/JVI.01741-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen K., Dobrowolski C., Shukla M., Cho W.K., Luttge B., Karn J. Inhibition of the H3K27 demethylase UTX enhances the epigenetic silencing of HIV proviruses and induces HIV-1 DNA hypermethylation but fails to permanently block HIV reactivation. PLoS Pathog. 2021;17:1–30. doi: 10.1371/journal.ppat.1010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blazkova J., Murray D., Justement J.S., et al. Paucity of HIV DNA methylation in latently infected, resting CD4+ T cells from infected individuals receiving antiretroviral therapy. J Virol. 2012:1–10. doi: 10.1128/JVI.00040-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho Y.C., Shan L., Hosmane N., et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber S., Weiser B., Kemal K.S., et al. Epigenetic analysis of HIV-1 proviral genomes from infected individuals: predominance of unmethylated CpG's. Virology. 2014;449:181–189. doi: 10.1016/j.virol.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boltz V.F., Ceriani C., Rausch J.W., et al. Cpg methylation profiles of HIV-1 proviral dna in individuals on art. Viruses. 2021;13 doi: 10.3390/v13050799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trejbalova K., Kovarova D., Blazkova J., et al. Development of 5‘ LTR DNA methylation of latent HIV-1 provirus in cell line models and in long-term-infected individuals. Clin Epigenet. 2016;8:1–20. doi: 10.1186/s13148-016-0185-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cortés-Rubio C.N., Salgado-Montes de Oca G., Prado-Galbarro F.J., et al. Longitudinal variation in human immunodeficiency virus long terminal repeat methylation in individuals on suppressive antiretroviral therapy. Clin Epigenet. 2019;11:1–17. doi: 10.1186/s13148-019-0735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Einkauf K.B., Osborn M.R., Gao C., et al. Parallel analysis of transcription, integration, and sequence of single HIV-1 proviruses. Cell. 2022;185:266–282.e15. doi: 10.1016/j.cell.2021.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zapata J.C., Campilongo F., Barclay R.A., et al. The Human immunodeficiency virus 1 ASP RNA promotes viral latency by recruiting the polycomb repressor complex 2 and promoting nucleosome assembly. Virology. 2017;506:34–44. doi: 10.1016/j.virol.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouchat S., Delacourt N., Kula A., et al. Sequential treatment with 5-aza-2’-deoxycytidine and deacetylase inhibitors reactivates HIV- 1. EMBO Mol Med. 2016;8:117–138. doi: 10.15252/emmm.201505557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hashimoto H., Horton J.R., Zhang X., Cheng X. UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications. Epigenetics. 2009;4:8–14. doi: 10.4161/epi.4.1.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bianchi C., Zangi R. UHRF1 discriminates against binding to fully-methylated CpG-Sites by steric repulsion. Biophys Chem. 2013;171:38–45. doi: 10.1016/j.bpc.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Bashtrykov P., Jankevicius G., Jurkowska R., Ragozin S., Jeltsch A. The Uhrf1 protein stimulates the activity and specificity of the maintenance DNA methyltransferase Dnmt1 by an allosteric mechanism. J Biol Chem. 2013:0–18. doi: 10.1074/jbc.M113.528893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meilinger D., Fellinger K., Bultmann S., et al. Np95 interacts with de novo DNA methyltransferases, Dnmt3a and Dnmt3b,and mediates epigenetic silencing of the viral CMV promoter in embryonic stem cells. EMBO Rep. 2009;10:1259–1264. doi: 10.1038/embor.2009.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharif J., Endo T.A., Nakayama M., et al. Activation of endogenous retroviruses in Dnmt1(−/−) ESCs involves disruption of SETDB1-mediated repression by NP95 binding to hemimethylated DNA. Cell Stem Cell. 2016;19:81–94. doi: 10.1016/j.stem.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Dong J., Wang X., Cao C. UHRF1 suppresses retrotransposons and cooperates with PRMT5 and PIWI proteins in male germ cells. Nat Commun. 2019;10:1–14. doi: 10.1038/s41467-019-12455-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haggerty C., Kretzmer H., Riemenschneider C., et al. Dnmt1 has de novo activity targeted to transposable elements. Nat Struct Mol Biol. 2021 doi: 10.1038/s41594-021-00603-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Battivelli E., Dahabieh M., Abdel-Mohsen M., et al. Distinct chromatin functional states correlate with HIV latency reactivation in infected primary CD4+ T cells. Elife. 2018;7:1–22. doi: 10.7554/eLife.34655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grunau C., Schattevoy R., Mache N., Rosenthal A. MethTools: a toolbox to visualize and analyze DNA methylation data. Nucleic Acids Res. 2000;28:1053–1058. doi: 10.1093/nar/28.5.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dignam J.D., Lebovitz R.M., Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierard V., Guiguen A., Colin L., et al. DNA cytosine methylation in the bovine leukemia virus promoter is associated with latency in a lymphoma-derived B-cell line: potential involvement of direct inhibition of cAMP-responsive element (CRE)-binding protein/CRE modulator/activation transcription. J Biol Chem. 2010;285:19434–19449. doi: 10.1074/jbc.M110.107607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colin L., Vandenhoudt N., de Walque S., et al. The AP-1 binding sites located in the pol gene intragenic regulatory region of HIV-1 are important for viral replication. PLoS One. 2011;6:1–19. doi: 10.1371/journal.pone.0019084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lusic M., Marini B., Ali H., Lucic B., Luzzati R., Giacca M. Proximity to PML nuclear bodies regulates HIV-1 latency in CD4+ T cells. Cell Host Microbe. 2013;13:665–677. doi: 10.1016/j.chom.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 37.Naldini L., Blomer U., Gallay P., et al. In vivo gene delivery and stable transduction of post mitotic cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 38.Marban C., Suzanne S., Dequiedt F., et al. Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J. 2007;26:412–423. doi: 10.1038/sj.emboj.7601516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dedeurwaerder S., Defrance M., Bizet M., Calonne E., Bontempi G., Fuks F. A comprehensive overview of infinium human methylation450 data processing. Brief Bioinform. 2013;15:929–941. doi: 10.1093/bib/bbt054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dedeurwaerder S., Defrance M., Calonne E. Evaluation of the infinium methylation 450K technology report. Future Med. 2011;3:771–784. doi: 10.2217/epi.11.105. [DOI] [PubMed] [Google Scholar]
- 41.Subramanian A., Tamayo P., Mootha V.K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boom R., Sol C., Salimans M., Jansen C., Wertheim-Van Dillen P., Van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malnati M.S., Scarlatti G., Gatto F., et al. A universal real-time PCR assay for the quantification of group-M HIV-1 proviral load. Nat Protoc. 2008;3:1240–1248. doi: 10.1038/nprot.2008.108. [DOI] [PubMed] [Google Scholar]
- 44.Pasternak A.O., Jurriaans S., Bakker M., Prins J., Berkhout B., Lukashov V. Cellular levels of HIV unspliced RNA from patients on combination antiretroviral therapy with undetectable plasma viremia predict the therapy outcome. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Avettand-Fenoel V., Chaix M.L., Blanche S. LTR real-time PCR for HIV-1 DNA quantitation in blood cells for early diagnosis in infants born to seropositive mothers treated in HAART area. J Med Virol. 2009;81:217–223. doi: 10.1002/jmv.21390. [DOI] [PubMed] [Google Scholar]
- 46.Jordan A., Bisgrove D., Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003;22:1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pereira L., Bentley K., Peeters A., Churchill M., Deacon N. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000;28:663–668. doi: 10.1093/nar/28.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones C., Kadonaga J.T., Luciw P., Tjian R. Activation of the AIDS retrovirus promoter by the cellular transcription factor, Sp1. Science. 1986;232:755–759. doi: 10.1126/science.3008338. [DOI] [PubMed] [Google Scholar]
- 49.Van Lint C., Amella C.A., Emiliani S.S., John M., Jie T., Verdin E. Transcription factor binding sites downstream of the human immunodeficiency virus type 1 transcription start site are important for virus infectivity. J Virol. 1997;71:6113–6127. doi: 10.1128/jvi.71.8.6113-6127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X., Odom D.T., Koo S.H.H., et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ross H.L., Nonnemacher M.R., Hogan T.H. Interaction between CCAAT /enhancer binding protein and cyclic AMP response element binding protein 1 regulates human immunodeficiency virus type 1 transcription in cells of the monocyte / macrophage lineage. J Virol. 2001;75:1842–1856. doi: 10.1128/JVI.75.4.1842-1856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bartels S., Spruijt C., Brinkman A., Jansen P., Vermeulen M., Stunnenberg H. A SILAC-based screen for methyl-CpG binding proteins identifies RBP-J as a DNA methylation and sequence- specific binding protein. PLoS One. 2011;6:1–10. doi: 10.1371/journal.pone.0025884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Defossez P.A., Stancheva I. Biological functions of methyl-CpG-binding proteins. Prog Mol Biol Transl Sci. 2011:377–398. doi: 10.1016/B978-0-12-387685-0.00012-3. [DOI] [PubMed] [Google Scholar]
- 54.Hopfner R., Mousli M., Jeltsch J.M., et al. ICBP90, a novel human CCAAT binding protein, involved in the regulation of topoisomerase IIα expression. Cancer Res. 2000;60:121–128. [PubMed] [Google Scholar]
- 55.Dahiya S., Liu Y., Nonnemacher M.R., Dampier W., Wigdahl B. CCAAT enhancer binding protein and nuclear factor of activated T cells regulate HIV-1 LTR via a novel conserved downstream site in cells of the monocyte-macrophage lineage. PLoS One. 2014;9:1–14. doi: 10.1371/journal.pone.0088116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharif J., Muto M., Takebayashi S. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 57.Ashraf W., Ibrahim A., Alhosin M. The epigenetic integrator UHRF1: on the road to become a universal biomarker for cancer. Oncotarget. 2017;8:51946–51962. doi: 10.18632/oncotarget.17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pasternak A.O., Adema K.W., Bakker M., et al. Highly sensitive methods based on seminested real-time reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 unspliced and multiply spliced RNA and proviral DNA. J Clin Microbiol. 2008;46:2206–2211. doi: 10.1128/JCM.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bostick M., Kim J.K., Estève P.O., Clark A., Pradhan S., Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 60.Xie S., Qian C. The growing complexity of UHRF1-mediated maintenance DNA methylation. Genes. 2018;9:1–12. doi: 10.3390/genes9120600. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Unoki M., Nishidate T., Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601–7610. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- 62.Kim J.K., Estève P.O., Jacobsen S.E., Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res. 2009;37:493–505. doi: 10.1093/nar/gkn961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ait-Ammar A., Kula A., Darcis G. Current status of latency reversing agents facing the heterogeneity of HIV-1 cellular and tissue reservoirs. Front Microbiol. 2019;10:3060. doi: 10.3389/fmicb.2019.03060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodari A., Darcis G., Van Lint C.M. The current status of latency reversing agents for HIV-1 Remission. Annu Rev Virol. 2021;8:491–514. doi: 10.1146/annurev-virology-091919-103029. [DOI] [PubMed] [Google Scholar]
- 65.Kula-Pacurar A., Rodari A., Darcis G., Van Lint C. Seminars in immunology shocking HIV-1 with immunomodulatory latency reversing agents. Semin Immunol. 2021;51 doi: 10.1016/j.smim.2021.101478. [DOI] [PubMed] [Google Scholar]
- 66.Achour M., Mousli M., Alhosin M. Epigallocatechin-3-gallate up-regulates tumor suppressor gene expression via a reactive oxygen species-dependent down-regulation of UHRF1. Biochem Biophys Res Commun. 2013;430:208–212. doi: 10.1016/j.bbrc.2012.11.087. [DOI] [PubMed] [Google Scholar]
- 67.Yamaguchi K., Honda M., Ikigai H., Hara Y., Shimamura T. Inhibitory effects of (−)-epigallocatechin gallate on the life cycle of human immunodeficiency virus type 1 (HIV-1) Antiviral Res. 2002;53:19–34. doi: 10.1016/s0166-3542(01)00189-9. [DOI] [PubMed] [Google Scholar]
- 68.Singh B.N., Shankar S., Srivastava RK. Green tea catechin, epigallocatechin-3-gallate (EGCG): mechanisms, perspectives and clinical applications. Biochem Pharmacol. 2011;82:1807–1821. doi: 10.1016/j.bcp.2011.07.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moran S., Arribas C., Esteller M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics. 2016;8:389–399. doi: 10.2217/epi.15.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Williamson M.P., McCormick T.G., Nance C.L., Shearer WT. Epigallocatechin gallate, the main polyphenol in green tea, binds to the T-cell receptor, CD4: potential for HIV-1 therapy. J Allergy Clin Immunol. 2006;118:1369–1374. doi: 10.1016/j.jaci.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 71.Patnaik D., Estève P.O., Pradhan S. Targeting the SET and RING-associated (SRA) domain of ubiquitin-like, PHD and ring finger – containing 1 (UHRF1) for anti-cancer drug development. Oncotarget. 2018;9:26243–26258. doi: 10.18632/oncotarget.25425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Myrianthopoulos V., Cartron P.F., Liutkeviciute Z., et al. Tandem virtual screening targeting the SRA domain of UHRF1 identifies a novel chemical tool modulating DNA methylation. Eur J Med Chem. 2016;114:390–396. doi: 10.1016/j.ejmech.2016.02.043. [DOI] [PubMed] [Google Scholar]
- 73.Spina C.A., Anderson J., Archin N.M., et al. An In-Depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4 + T cells from aviremic patients. PLoS Pathog. 2013;9:1–15. doi: 10.1371/journal.ppat.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vazquez B., Laguna T., Carabana J., Krangel M., Lauzurica P. CD69 gene is differentially regulated in T and B cells by evolutionary conserved promoter-distal elements. J Immunol. 2009:1–10. doi: 10.4049/jimmunol.0900839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Darcis G., Kula A., Bouchat S., et al. An in-depth comparison of latency-reversing agent combinations in various in vitro and ex vivo HIV-1 latency models identified bryostatin-1+JQ1 and ingenol-B+JQ1 to potently reactivate viral gene expression. PLoS Pathog. 2015;11:1–36. doi: 10.1371/journal.ppat.1005063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kula A., Delacourt N., Bouchat S., et al. Heterogeneous HIV-1 reactivation patterns of disulfiram and combined disulfiram + romidepsin treatments. J Acquir Immune Defic Syndr. 2019;80:605–613. doi: 10.1097/QAI.0000000000001958. [DOI] [PubMed] [Google Scholar]
- 77.Hagemann S., Heil O., Lyko F., Brueckner B. Azacytidine and decitabine induce gene-specific and non-random DNA demethylation in human cancer cell lines. PLoS One. 2011;6:1–11. doi: 10.1371/journal.pone.0017388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tanaka J., Ishida T., Choi B.I., Yasuda J., Watanabe T., Iwakura Y. Latent HIV-1 reactivation in transgenic mice requires cell cycle -dependent demethylation of CREB/ATF sites in the LTR. AIDS. 2003;17:167–175. doi: 10.1097/00002030-200301240-00005. [DOI] [PubMed] [Google Scholar]
- 79.Colin L., Van Lint C. Molecular control of HIV-1 postintegration latency: implications for the development of new therapeutic strategies. Retrovirology. 2009;6:111. doi: 10.1186/1742-4690-6-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liang T., Zhang Q., Wu Z., et al. Uhrf1 suppresses HIV-1 transcription and promotes HIV-1 latency by competing with p-tefb for ubiquitination-proteasomal degradation of tat. MBio. 2021;12 doi: 10.1128/mBio.01625-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miller L.K., Kobayashi Y., Chen C.C., Russnak T.A., Ron Y., Dougherty J.P. Proteasome inhibitors act as bifunctional antagonists of human immunodeficiency virus type 1 latency and replication. Retrovirology. 2013;10:120. doi: 10.1186/1742-4690-10-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li Z., Wu J., Chavez L., et al. Reiterative enrichment and authentication of CRISPRi targets (REACT) identifies the proteasome as a key contributor to HIV-1 latency. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Verdikt R., Darcis G., Ait-Ammar A., Van Lint C. Applications of CRISPR/Cas9 tools in deciphering the mechanisms of HIV-1 persistence. Curr Opin Virol. 2019;38:63–69. doi: 10.1016/j.coviro.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 84.Qin W., Wolf P., Liu N., et al. DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res. 2015;25:911–929. doi: 10.1038/cr.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.DaRosa P.A., Harrison J.S., Zelter A., et al. A bifunctional role for the UHRF1 UBL domain in the control of hemi-methylated DNA-dependent histone ubiquitylation. Mol Cell. 2018;72:753–765.e6. doi: 10.1016/j.molcel.2018.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gross A.M., Jaeger P.A., Kreisberg J.F., et al. Methylome-wide analysis of chronic HIV infection reveals five-year increase in biological age and epigenetic targeting of HLA article methylome-wide analysis of chronic HIV infection reveals five-year increase in biological age and epigenetic targeting of HLA. Mol Cell. 2016;62:157–168. doi: 10.1016/j.molcel.2016.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen L., Zhang S., Pan X., et al. HIV infection alters the human epigenetic landscape. Gene Ther. 2019;26:29–39. doi: 10.1038/s41434-018-0051-6. [DOI] [PubMed] [Google Scholar]
- 88.Ait-Ammar A., Bellefroid M., Daouad F., et al. Inhibition of HIV-1 gene transcription by KAP1 in myeloid lineage. Sci Rep. 2021;11:1–14. doi: 10.1038/s41598-021-82164-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang X., Wang Y., Lu P., et al. PEBP1 suppresses HIV transcription and induces latency by inactivating MAPK NF-κB signaling. EMBO Rep. 2020;21:1–13. doi: 10.15252/embr.201949305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Adam E., Quivy V., Chariot A., et al. Potentiation of tumor necrosis factor-induced NF-KB activation by deacetylase inhibitors is associated with a delayed cytoplasmic reappearance of IKBa. Mol Cell Biol. 2003;23:6200–6209. doi: 10.1128/MCB.23.17.6200-6209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Quivy V., Adam E., Collette Y., et al. Synergistic activation of human immunodeficiency virus type 1 promoter activity by NF-KB and inhibitors of deacetylases: potential perspectives for the development of therapeutic strategies. J Virol. 2002;76:11091–11103. doi: 10.1128/JVI.76.21.11091-11103.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kuiken C., Leitner T., Foley B., et al. Theoretical Biology and Biophysics; Los Alamos, New Mexico: 2008. HIV Sequence Compendium. 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.