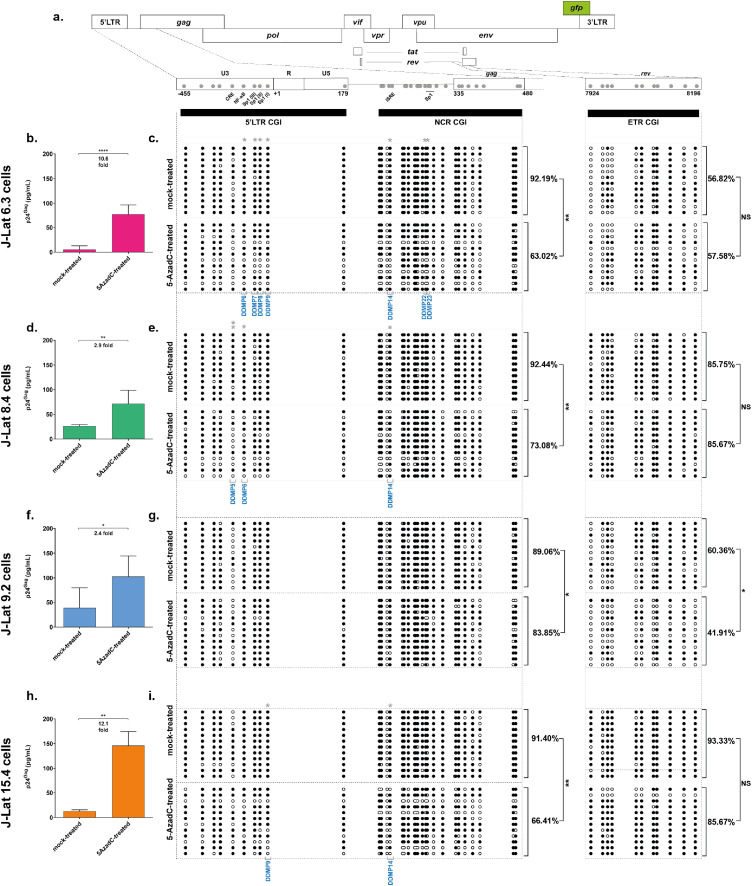
Figure 1.
5-AzadC-induced reactivation of HIV-1 gene expression from latency is associated with 5’LTR CGIs demethylation. (a) Schematic presentation of the three CpG islands studied along the HIV-1 provirus, in the HIV-1 promoter (5’LTR and NCR CGIs) and rev (ETR CGI). The reactivation of HIV-1 production following 72 h treatment with 400 nM of 5-AzadC, quantified by ELISA on p24Gag capsid protein in culture supernatants, and the DNA methylation profile, established by sodium bisulfite sequencing for the three CGIs, are respectively presented for the J-Lat 6.3 cells (b and c), the J-Lat 8.4 cells (d and e), the J-Lat 9.2 cells (f and g) and the J-Lat 15.4 cells (h and i). ELISA results are representative of the means ± SD of three independent 5-AzadC treatments. Reactivation folds are indicated. Unmethylated and methylated CpG dinucleotides are respectively represented with open and closed circles, where each line corresponds to individual sequenced molecules. The global methylation level presented correspond to mean percentages of methylated CpGs for the twelve clones of each condition, either on the promoter CGIs (5’LTR + NCR CGIs considered together) or on the ETR CGI. Statistical significance was determined by [unpaired T-tests].