Abstract
The Leptospira interrogans ponA and pbpB genes were isolated and characterized. ponA and pbpB encode the penicillin-binding proteins (PBPs) 1 and 3, respectively. There is little sequence variation between the PBP genes from two L. interrogans strains (serovar icterohaemorrhagiae strain Verdun and serovar pomona strain RZ11). The deduced L. interrogans PBP 1 and PBP 3 protein sequences from the two strains shared over 50% similarity to homologous proteins from Escherichia coli. It was demonstrated for strain Verdun that ponA and pbpB are transcribed individually from their own promoter. The ponA and pbpB genes from both strains are separated by 8 to 10 kb and oriented such that their transcription is convergent. The L. interrogans PBP 1 and PBP 3 proteins were synthesized in E. coli and were modified with ampicillin using a digoxigenin-ampicillin conjugate. These data show that both genes encode functional PBPs.
Leptospirosis is a widespread zoonosis caused by Leptospira interrogans. This bacterial pathogen can infect most mammalian species through either direct or indirect contact with contaminated body fluids from an infected animal (7). Leptospirosis can be fatal in humans. In livestock, Leptospira infection may result in death or a chronic infection may ensue, leading to abortion, stillbirth, infertility, or decreased milk production. Leptospira-infected humans are often treated with β-lactam antibiotics. It has recently been suggested that leptospirosis in livestock can be treated with β-lactam antibiotics (19). In vitro, pathogenic leptospires are very sensitive to β-lactam antibiotics (16). The MIC of ampicillin is between 0.025 and 0.78 μg/ml, and that of penicillin G is between 0.39 and 3.13 μg/ml. The minimal bactericidal concentrations observed for penicillin G are up to 100 μg/ml or more. In contrast, ampicillin exhibits high bactericidal activity, as evidenced by low minimal bactericidal concentrations (<25 μg/ml).
β-Lactams exert their effects by acting as substrate analogs of the peptidoglycan biosynthetic enzymes transpeptidase and d-alanine carboxypeptidase (21). These enzymes are located within the cytoplasmic membrane and play an integral role in the synthesis of peptidoglycan. These proteins are commonly called penicillin-binding proteins (PBPs) because of their ability to covalently bind radiolabeled penicillin (20). There are two distinguishable groups of PBPs: low-molecular-weight PBPs and high-molecular-weight (HMW) PBPs. The low-molecular-weight PBPs are monofunctional enzymes acting as dd-carboxypeptidases involved in the remodeling of peptidoglycan during cell growth. The HMW PBPs have a multidomain structure. These proteins are anchored to the cytoplasmic membrane by an N-terminal pseudo-signal peptide and are essentially composed of two modules localized on the outer face of the cytoplasmic membrane. The N-terminal domain, which is several hundred amino acids long, is fused to the C-terminal penicillin-binding domain. This domain displays the transpeptidase activity that catalyzes cross-linking of the peptidoglycan peptides. Pairwise comparison and multiple alignments of amino acid sequences lead to the conclusion that HMW PBPs fall into two classes, A and B, which differ in their N-terminal domain (8, 13). In Escherichia coli, PBPs 1a and 1b of class A behave as bifunctional proteins exhibiting both transglycosylase (N-terminal module) and transpeptidase (C-terminal module) activities. They catalyze polymerization of the peptidoglycan from undecaprenyl diphosphate-linked disaccharide peptides, probably by producing primers for PBP 2 and PBP 3 to act upon during cell elongation and cell division. PBP 2 and PBP 3 of class B are likewise considered bifunctional proteins, though the role of the N-terminal module is not clearly established. PBP 3 is specifically involved in polymerization of the septal peptidoglycan during cell division (14). Little is known about the PBPs of Leptospira. During analysis of subcellular fractions, five PBPs were identified in Leptospira kirschneri (10). However, neither the proteins nor the genes that encode them have been characterized.
To establish a framework by which leptospiral peptidoglycan structure can be analyzed, we isolated and characterized the L. interrogans ponA and pbpB genes, encoding PBP 1 and PBP 3, respectively, which play an important role in peptidoglycan synthesis. Comparison of these sequences from two strains (serovar icterohaemorrhagiae strain Verdun and serovar pomona strain RZ11) also provides information on genetic drift between distinct serovars of the same species.
(This work represents a portion of a thesis submitted by Audrey Brenot to the University of Paris VII, Paris, France, for the Ph.D. degree.)
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used are detailed in Table 1. E. coli strains were grown at 37°C in Luria-Bertani broth (18). Antibiotics and substrates were used in selective media at the indicated concentrations: isopropyl-β-d-thiogalactopyranoside (IPTG) at 500 μM, 5-bromo-4-chloro-3-indolyl-β-d-galactoside at 80 μg/ml, ampicillin (AMP) at 50 μg/ml, and kanamycin at 30 or 50 μg/ml. L. interrogans serovar icterohaemorrhagiae strain Verdun (National Reference Center for Leptospira, Institut Pasteur, Paris, France) and serovar pomona strain RZ11 (24) were grown in EMJH medium at 28°C (6, 12).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristic | Source or reference |
|---|---|---|
| Strains | ||
| BL21(DE3) | F−ompT hsdSB gal dcm (DE3) | Novagen |
| Novablue (DE3) | endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δlac [F′ proAB+ lacIqZΔM15::Tn10] (DE3) | Novagen |
| TOP10F′ | F′ (lacIq Tn10 [Tetr]) mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| INV F′ | F′ endA1 recA1 hsdR17 supE44 thi-1 gyrA96 relA1 φ80lacZΔM15 (lacZYA-argF)U169 | Invitrogen |
| Verdun | L. interrogans serovar icterohaemorrhagiae strain Verdun (National Reference Center for Leptospira, Institut Pasteur, Paris, France) | |
| RZ11 | L. interrogans serovar pomona strain RZ11 | 24 |
| Plasmids | ||
| pGEM7Zf(+) | Confers resistance to AMP | Promega |
| pET26b(+) | Confers resistance to kanamycin | Novagen |
| pCRII-TOPO | TA vector; confers resistance to AMP and kanamycin | Invitrogen |
| pCR2.1 | TA vector; confers resistance to AMP and kanamycin | Invitrogen |
| pG-PBP1 | ponA gene (XbaI restriction fragment from L. interrogans strain Verdun cosmid library) cloned in XbaI-digested pGEM7Zf(+) | This paper |
| pG-PBP3 | pbpB gene (ClaI restriction fragment from L. interrogans strain Verdun cosmid library) cloned in ClaI-digested pGEM7Zf(+) | This paper |
| pET-PBP3 | Translational fusion of the pelB leader sequence with the predicted periplasmic part of PBP 3 from strain Verdun [PCR product obtained with primers PBP3M and PBP3L cloned in vector pET26b(+)] | This paper |
| p921-1 | Serovar pomona pbpB gene (PCR product using primers 921 and 922) cloned in pCR2.1 vector | This paper |
| p513-3 | Serovar pomona ponA gene (3.1-kb PCR product using primers 155 and 513) cloned in pCR2.1 vector | This paper |
| pKB1 | 5.5-kb BamHI restriction fragment (containing two-thirds of the strain RZ11 ponA gene including the 5′ end) cloned in pBluescript II KS(+) vector | This paper |
| pK127 | 1.3-kb genome walking clone (amplified from RZ11 DNA using the gene-specific primer 173 and linker-specific primer AP1) cloned in pCR2.1 vector | This paper |
Cloning and sequencing of the ponA and pbpB genes.
The ponA gene of L. interrogans serovar pomona strain RZ11 was cloned from a previously described plasmid-based BamHI library (24) and identified during sequence analysis of randomly picked clones (4). The 3′ end of this gene was amplified using a PCR-based genome walking technique (Universal Genome Walker Kit; Clontech, Palo Alto, Calif.) using the conditions described previously (25). Specific amplification of ponA was initiated with primer 173 (Table 2) on BstUI-digested DNA to which adapters had been ligated. The pbpB gene was amplified from strain RZ11 using primers 921 and 922 (Table 2), which were derived from the strain Verdun sequence. Amplicons were ligated with pCR2.1 vector (Invitrogen Corp., Carlsbad, Calif.) and used to transform E. coli INV F′. The resulting plasmid, p921-1, contained the pbpB gene downstream of the T7 promoter.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′) | Location |
|---|---|---|
| 4a | CATTAAAATCCCGATAGC | +651 to +634 downstream of the pbpB start codon (strain Verdun) |
| 8a | GAGTCATCTTTCTCACGT | +88 to +105 downstream of the pbpB start codon (strain Verdun) |
| 11a | TAGTTTTGAGTAGTGATC | −76 to −59 upstream of the pbpB start codon (strain Verdun) |
| 36a | CGGTATTCTCGGGATTAT | −389 to −372 upstream of the pbpB start codon (strain Verdun) |
| 12a | CTCCCGCGACTTCTATGT | −727 to −710 upstream of the pbpB stop codon (strain Verdun) |
| 29a | CCGAAATCCCAAGAGGTC | −92 to −109 upstream of the pbpB stop codon (strain Verdun) |
| 38a | AGGCCGAACTCTGAGAAA | +198 to +181 downstream of the pbpB stop codon (strain Verdun) |
| 39a | TCTAAGTCTACCGGATTC | +520 to +503 downstream of the pbpB stop codon (strain Verdun) |
| 3b | AGCGGCTTCTTGTTTATC | +1091 to +1074 downstream of the ponA start codon (strain Verdun) |
| Z157b | GTCAGTCAAATTCGTCCA | +219 to +236 downstream of the ponA start codon (strain Verdun) |
| 15b | CTTATTCTACATACAAGG | −390 to −373 upstream of the ponA start codon (strain Verdun) |
| 7b | GCCTGTGCAGAAACGATC | −600 to −583 upstream of the ponA stop codon (strain Verdun) |
| Z195b | TCTCCATCTTCTGCTTGA | −53 to −70 upstream of the ponA stop codon (strain Verdun) |
| 12b | TGGCGGAAACAGGGCGCA | +193 to +176 downstream of the ponA stop codon (strain Verdun) |
| 17b | CAAAATCATTCTCATAAG | +498 to +481 downstream of the ponA stop codon (strain Verdun) |
| pbp1begc | GAGGCCGGCTCCAATGGATTTGAA | +38 to +15 downstream of the ponA start codon (strain Verdun) |
| pbp1endc | CCAGAAACTACAGATAATCCTCCA | −27 to −4 upstream of the ponA stop codon (strain Verdun) |
| pbp3begc | TCGTGAGTCTAGTTTTACGGGAAT | +27 to +43 downstream of the pbpB start codon (strain Verdun) |
| pbp3endc | AGCGGGTTTTGTTACGAGCAGGAA | −75 to −52 upstream of the pbpB stop codon (strain Verdun) |
| oligo5.4UPc | GAGTTCAATGGATTCAACACTTGT | Strain Verdun |
| oligo5.4RPc | GAGCGGTCATCTGTTTATGGTTGT | Strain Verdun |
| PBP3Ld | GGGGGGGGATCCGAATGAACGAGAAGTAGTTCTAAAAACTGG | +95 to +124 downstream of the pbpB start codon (BamHI) (strain Verdun) |
| PBP3Md | GGGGGGCTCGAGCTTAAAATATAAATTAAGTCTTCCGTTTTC | −30 to −1 upstream of the pbpB stop codon (XhoI) (strain Verdun) |
| 121b | AGTTCCATCGGAGACAATTC | +1661 to +1642 of ponA (strain RZ11) |
| 173b | GATCAACACAATCGCAGTCA | +1500 to +1519 of ponA for genome walking (strain RZ11) |
| 184c | CTTAAAGCCTGAGTCGTTTC | 84 bp downstream of ponA (strain RZ11) |
| 185c | GTCGTCATCCATTCCTGTAA | +467 to +448 of pbpB (strain RZ11) |
| 186c | GCAGTCAATTGGCTCCTTAT | +681 to +700 of pbpB (strain RZ11) |
| 921 | CCGATCTTGTAAGAAGAGAA | −19 to +1 of pbpB |
| 922 | CCGAACTCTGAGAAATAGAA | 173 bp downstream of the pbpB stop codon |
| 155 | GCGCTTTTGATCCGTTCC | Starts 232 bp upstream of ponA (strain RZ11) |
| 513 | TTAGAATGAGAGGAGACGAGGAGAATA | Starts 389 bp downstream of ponA (strain RZ11) |
Primers used for pbpB RT-PCR.
Primers used for ponA RT-PCR.
Primers used for the LD-PCR.
BamHI and XhoI sites are underlined.
The ponA and pbpB genes from L. interrogans serovar icterohaemorrhagiae strain Verdun were isolated from a cosmid library constructed using methods described previously (2). The strain Verdun cosmid library was screened by colony hybridization (18) using nucleic acid probes labeled with [α-33P]dATP (370 MBq/ml) from NEN (Boston, Mass.) by random priming (Megaprime DNA labeling system; Amersham Life Sciences, Little Chalfont, Buckinghamshire, England). The ponA probe was a 1-kb internal ClaI restriction fragment from pKB1, containing part of the strain RZ11 ponA gene (4). The pbpB probe was a 1.5-kb fragment from L. interrogans strain Verdun identified as part of a pbp gene during sequence analysis of randomly picked clones (4). For strain Verdun, recombinant cosmid DNA identified by hybridization was purified, and inserts were subcloned in pGEM7Zf(+) vector using XbaI for the ponA gene and ClaI for the pbpB gene. The resulting plasmids, pG-PBP1 (ponA) and pG-PBP3 (pbpB), were used as templates for sequence analysis. Plasmid DNA was prepared for subsequent analysis using the QIAprep Spin miniprep kit (Qiagen Inc., Chatsworth, Calif.).
The strain Verdun sequences were determined using the T7 sequencing kit (Pharmacia Biotech, London, United Kingdom) with [α-33P]dATP (370 MBq/ml) from NEN or using an ALFexpress sequencing kit (Pharmacia Biotech). The strain RZ11 genes were sequenced using methods described previously (25).
Sequences were compared to sequences in the GenBank (National Center for Biotechnology Information, Bethesda, Md.; http://www.ncbi.nlm.nih.gov) and EMBL (EMBL Nucleotide Sequence Submissions, Cambridge, United Kingdom; http://www.ebi.ac.uk) databases with the BLASTN, BLASTP, and BLASTX programs (1). Multiple alignments between PBPs were performed using Clustal V software (11).
RT-PCR.
The methods used to extract total genomic L. interrogans strain RZ11 RNA and perform reverse transcriptase PCR (RT-PCR) were described previously (25). Primers used to detect the strain RZ11 ponA transcript were 173 and 121 (Table 2). Total RNA from L. interrogans strain Verdun was prepared from 400 ml of culture at 109 bacteria/ml (3) using Tri reagent (Sigma Chemical Co., St Louis, Mo.). Residual RNA was removed by treatment for 1 h at 37°C with RNase-free DNase (Pharmacia Biotech, 1 U/μg) and extracted using the RNeasy kit (Qiagen, Hilden, Germany). The RT-PCRs were done using the Access RT-PCR System (Promega Corp., Madison, Wis.) according to the manufacturer's recommendations with primers listed in Table 2.
LD-PCR.
PCRs were used to determine the distance and orientation between the ponA and pbpB genes from strains Verdun and RZ11. Long-distance PCR (LD-PCR) products were amplified from strain RZ11 genomic DNA using Tth polymerase (Clontech) using the amplification parameters described previously (25) and primer 184, located downstream of ponA, paired with either primer 185 or 186, oriented in opposite directions within pbpB (Table 2). LD-PCR products were also amplified from a cosmid containing the strain Verdun ponA and pbpB genes with the Advantage 2 PCR kit (Clontech). For strain Verdun, the primers were designed to hybridize at the beginning and the end of both genes and were directed outward of these genes (Table 2). Two additional primers, oligo5.4UP and oligo5.4RP, which anneal to opposite strands of a 5.4-kb XbaI fragment located between ponA and pbpB genes, were also used in LD-PCR analysis of the strain Verdun locus. For amplification of the strain Verdun locus, the cycling parameters were as recommended by the supplier to amplify 10- to 20-kb templates.
Protein expression of pbpB and ponA in E. coli.
The pbpB gene from strain Verdun was amplified by PCR using the PfuTurbo DNA polymerase (Stratagene, La Jolla, Calif.) using primers PBP3M and PBP3L (Table 2). Primers PBP3M and PBP3L (1 μM each) were added to 10 ng of pG-PBP3 plasmid DNA. This allowed amplification of the pbpB sequence from 95 nucleotides after the start codon to the stop codon (which corresponded to the periplasmic predicted part of the protein). The 1,737-bp PCR product was cloned into pCRII-TOPO, and E. coli TOP10F′ cells were screened for lacZ inactivation as described by the supplier (Invitrogen Corp.). A 1,723-bp BamHI-XhoI insert containing the pbpB coding region was inserted into pET26b(+) (Novagen, Inc., Madison, Wis.), resulting in plasmid pET-PBP3. The resulting plasmid created a translational fusion of the pelB leader sequence with the predicted periplasmic part of PBP 3.
The PBP 3 protein from strain Verdun was synthesized from pET-PBP3 in E. coli BL21(DE3) cells as follows. Cells were grown in Luria-Bertani broth at 37°C to a density of 0.6 (A600). IPTG was added to a final concentration of 1 mM to induce expression of pbpB under the control of the T7lac promoter. Cultures were further incubated at 37°C for 3 h before harvesting. Whole-cell lysates were prepared by a sodium dodecyl sulfate (SDS) boiling method (15). Cells collected by centrifugation were resuspended in solubilization buffer and boiled, and the proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). To separate soluble and insoluble fractions from the induced cultures and to purify the protein under denaturing conditions (6 M urea) on His-Bind resin, samples were treated as described by the supplier (Novagen, Inc.). Protein concentrations were determined by the bicinchoninic acid protein assay (Pierce, Rockford, Ill.). Bovine serum albumin was used as a standard.
Complete copies of the strain RZ11 ponA and pbpB genes were amplified and cloned into pCR2.1 vector. The ponA gene from strain RZ11 was located downstream of the lacZ promoter in clone p513-3. This plasmid was used to transform E. coli INV F′ cells. Absence of the lac repressor in this strain allowed constitutive transcription of PBP 1. Copies of the pbpB gene were found only in the opposite orientation, placing it downstream of the vector-encoded T7 RNA polymerase promoter. Thus, p921-1 containing pbpB was used to transform Novablue (DE3) cells, and T7 transcription of the pbpB gene was induced with IPTG as described above.
Identification of PBPs by labeling with DIG-AMP.
Pellets of L. interrogans and E. coli harboring plasmids containing the pbpB and ponA genes and their vector controls were suspended in phosphate-buffered saline and sonicated. Aliquots of the sonicated cells (100 μg of protein) were incubated at 37°C for 10 min with 2.5 μg of AMP per ml conjugated to digoxigenin (DIG) as described by Weigel et al. (22). Of each sample, 12.5 μg was resolved by SDS-PAGE; PBPs were identified by immunoblotting with an anti-DIG–alkaline phosphatase conjugate (Boehringer Mannheim Corp., Indianapolis, Ind.) followed by chemiluminescence from CDP Star (Boehringer Mannheim Corp.). In all competition experiments, samples were incubated for 30 min with a 400-fold excess of free AMP (Sigma Chemical Co.).
Nucleotide sequence accession number.
The pbpB and ponA sequences from strain Verdun have been assigned the EMBL accession no. AJ243720 and AJ278610, respectively. The ponA and pbpB sequences from strain RZ11 have been assigned the EMBL accession no. AF282906 and AF282907, respectively.
RESULTS AND DISCUSSION
Characterization of the L. interrogans pbpB gene.
Partial sequence analysis of a 1.5-kb fragment of L. interrogans serovar icterohaemorrhagiae strain Verdun showed that it contained part of a gene similar to those encoding bacterial HMW PBPs (4). This fragment was used to screen a cosmid library of strain Verdun genomic DNA by colony hybridization to isolate a complete copy of the gene and surrounding DNA. The region surrounding this putative PBP gene was subcloned and sequenced. One open reading frame (ORF) with the potential to encode a 602-amino-acid protein, having an estimated molecular mass of 67.3 kDa according to the compute pI/Mw tool (23), was identified. The protein sequence deduced from this ORF was used to search the GenBank and EMBL databases for homologs using BLASTP. This protein was most similar to several HMW PBPs, including the Bacillus subtilis stage V sporulation protein D; PBP 1 and PBP 3 of Borrelia burgdorferi and Treponema pallidum, respectively; cell division protein FtsI of Streptomyces coelicolor; PBP A2 of Rickettsia prowazekii; and PBP 3 of E. coli. Pairwise comparison revealed that L. interrogans protein shares about 30 and 26% sequence identity with the PBP 3 proteins from T. pallidum and E. coli, respectively. Because of the strong similarity to the gene encoding PBP 3, this gene was designated pbpB.
To determine the level of genetic drift between the genetically similar but distinct serovars icterohaemorrhagiae and pomona, the corresponding pbpB gene of strain RZ11 (serovar pomona) was amplified, cloned, and sequenced. The two L. interrogans pbpB sequences are 99% identical, with 13 base mismatches over an 1,809-bp ORF. Analysis of the derived proteins from both genes revealed that all but two of the sequence changes were silent. The amino acid changes detected were Met435 to Thr435 and Glu468 to Gly468 (changes are written as Verdun to RZ11).
The deduced PBP 3 proteins from both strains had eight sequence motifs that are well conserved among class B PBPs (Table 3) (17). Three motifs found in the C-terminal domain are also common to penicilloyltransferases (8). The active-site serine residue that binds to penicillin is typically part of the motif SXXK (box 6), and this was located at residue 259 in PBP 3. The SXN and KTG motifs present in the active site of every penicillin-binding domain were located at residues 312 and 456 (boxes 7 and 8, respectively). The spacing between these active-site motifs was well conserved, as was the spacing between the other regions of similarity (Table 3).
TABLE 3.
Boxes conserved between the PBPs 3 from both the Verdun and RZ11 strains of L. interrogans and other related PBPsa
| PBP (species) | Box 1
|
Spacing | Box 2
|
Spacing | Box 3
|
Spacing | Box 4
|
Spacing | Box 5
|
Spacing | Box 6
|
Spacing | Box 7
|
Spacing | Box 8
|
Spacing |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RGX3DRNG | RXYPXG | GX2GXEX3(E/D) | GXDX3(T/S)X5Q | TGEX7P(T/S)XDP | SXXK | SXN | K(T/S)G | |||||||||
| PBP 3 (L. interrogans) | 47 | 80 | 136 | 16 | 158 | 13 | 181 | 27 | 221 | 23 | 259 | 48 | 312 | 141 | 456 | 143 |
| RGAIYDRRG | RVYPHG | ALSGLEVQYN | GSNLHLTIDGLIQ | TGKVLASASFPAFDP | STMK | SCN | KTG | |||||||||
| PBP 1 (B. burgdorferi) | 54 | 84 | 147 | 15 | 168 | 18 | 196 | 27 | 236 | 23 | 274 | 55 | 333 | 142 | 478 | 148 |
| RGTIYDRNG | RIYPFR | GLEGIEFSLN | TNNIHLTIDMDIQ | NGEILSMVQFPQYDA | SINK | SSN | KSG | |||||||||
| PBP 3 (T. pallidum) | 79 | 89 | 177 | 15 | 198 | 17 | 225 | 27 | 265 | 15 | 295 | 58 | 353 | 142 | 502 | 169 |
| RGAILDRNG | RIYPES | GLSGIEYTLQ | TVTLFTSIDRTIQ | TGQILSYVSKPSANL | SVFK | SCN | KTG | |||||||||
| PBP 3 (E. coli) | 71 | 87 | 167 | 15 | 188 | 31 | 229 | 27 | 269 | 23 | 307 | 48 | 359 | 132 | 494 | 91 |
| RGMITDRSG | RYYPSG | GIEGVEKSFD | AHNLALSIDERLQ | TGEVLAMANSPSYNP | STVK | SSN | KTG |
PBP 3 sequences from L. interrogans strains Verdun and RZ11 have the same conserved boxes. Numbers above the sequences indicate the positions. Spacing indicates the number of amino acids between boxes. Amino acids conserved between PBPs are indicated in boldface; where they are not conserved, they are underlined.
Characterization of the L. interrogans ponA gene.
A plasmid clone, pKB1, containing a portion of the strain RZ11 ponA gene, encoding PBP 1, was identified during a study using sequence analysis of randomly selected clones to improve resolution of the combined physical and genetic map of L. interrogans (4). Plasmid pKB1 contains about two-thirds of the gene, including the 5′ end. A genomic walking technique was used to amplify the 3′ end of the gene using primer 173. The resulting 1,300-bp amplicon was cloned, generating plasmid pK127, and sequenced. The overlapping sequences of pKB1 and pK127 revealed the presence of a 2,409-bp ORF, with the potential to encode an 802-amino-acid protein with a predicted mass of 89.8 kDa according to the compute pI/Mw tool (23). The deduced protein was used to search the GenBank database using BLASTP. This sequence was most similar to those of HMW PBPs, with 25 and 26% of its amino acids identical to those of Neisseria gonorrhoeae and E. coli PBPs 1 and 1a, respectively. The L. interrogans gene was designated ponA because of its similarity to the E. coli ponA gene. A cosmid containing the strain Verdun ponA gene was identified by colony hybridization using a 1-kb ClaI fragment derived from pKB1 as a probe.
The two L. interrogans ponA sequences are 98% identical with 30 base mismatches over a 2,409-bp ORF. Analysis of the derived proteins from both genes revealed that there are 19 silent mutations, 5 conserved mutations, and 6 nonconserved mutations. Interestingly, most of the mutations occur in the amino-terminal portion of the sequence. There is one nonconservative mutation in the putative transmembrane helix (Thr to Ile).
Further evidence that the L. interrogans ponA gene encoded an HMW PBP was gained by identification of consensus motifs common to class A HMW PBPs. PBPs 1 from both strains, Verdun and RZ11, contain the nine boxes that are conserved in all PBPs of this class (Table 4) Furthermore, each of the three consensus motifs of the active site was identified in the deduced amino acid sequence of ponA, and the intervals between these motifs were consistent with those of other PBPs.
TABLE 4.
Boxes conserved between the PBPs 1 from both the Verdun and RZ11 strains of L. interrogans and other related PBPsa
| PBP (species) | Box 1
|
Spacing | Box 2
|
Spacing | Box 3
|
Spacing | Box 4
|
Spacing | Box 5
|
Spacing | Box 6
|
Spacing | Box 7
|
Spacing | Box 8
|
Spacing | Box 9
|
Spacing |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EDX(R/K)FXEHXG | GAST(I/L)(T/S)QQ | RKX2E | KXEILEXYXN | R(R/Q)X2VL | GX4TTX5Q | SXXK | SXN | K(T/S)G | ||||||||||
| PBP 1 (L. interrogans) | 120 | 20 | 150 | 14 | 172 | 12 | 189 | 55 | 254 | 89 | 349 | 84 | 446 | 50 | 500 | 133 | 636 | 166 |
| EDREFYNHHG | GGSTSTQQ | NKITE | KNTILSMYLN | RQKLVL | GMNVYTTLDYDKQ | STIK | SIN | KTG | ||||||||||
| PBP 1b (Synechocystis sp.) | 113 | 21 | 144 | 14 | 166 | 12 | 183 | 55 | 248 | 60 | 314 | 63 | 390 | 54 | 448 | 119 | 570 | 79 |
| EDSNFYKHQG | GASTLSMQ | RKLAE | KDQILDMYLN | RQEVVL | GLRVQSTVDYDTQ | SAFK | SAN | KTG | ||||||||||
| PBP 1 (N. gonorrhoeae) | 88 | 21 | 119 | 14 | 141 | 12 | 158 | 55 | 223 | 57 | 286 | 162 | 461 | 56 | 521 | 127 | 651 | 146 |
| EDKRFYRHWG | GASTITQQ | RKFNE | KDKILELYFN | RQKYIL | GFKVYTTVRTDHQ | STFK | SKN | KTG | ||||||||||
| PBP 1a (E. coli) | 86 | 21 | 117 | 14 | 139 | 12 | 156 | 55 | 221 | 57 | 284 | 168 | 465 | 55 | 524 | 189 | 716 | 133 |
| EDSRFYEHHG | GASTITQQ | RKIKE | KDEILELYLN | RRNVVL | GYRIYTTITRKVQ | SNIK | SKN | KTG |
PBP 1 sequences from L. interrogans strains Verdun and RZ11 have the same conserved boxes. Numbers above the sequences indicate the positions. Spacing indicates the number of amino acids between boxes. Amino acids conserved between PBPs are indicated in boldface; where they are not conserved, they are underlined.
The pbpB and ponA genes are 8 to 10 kb apart and comprise individual transcription units.
The cloned ponA and pbpB genes were previously localized on the L. interrogans strain RZ11 and Verdun physical maps by hybridization (4). These data showed that the two genes were located in the same region of the genome. However, the methodology used for mapping lacks detailed resolution, and thus, it could not be determined if these two genes were closely linked in the genome. The approximate distance between pbpB and ponA was determined using LD-PCR. The primers used for this analysis were located at the beginning and end of both genes and were directed outward toward flanking sequences (Table 2). Initial LD-PCR results showed that the two genes were about 8 (strain Verdun) and 10 (strain RZ11) kb apart and were in a convergent orientation for both strains. The distances between ponA and pbpB were confirmed for both strains. For strain Verdun, using two additional primers that anneal to a 5.4-kb XbaI fragment found between ponA and pbpB genes, the 8.5-kb distance between these two genes was confirmed. For strain RZ11, the 10.4-kb distance was confirmed, indicating that there may be a small insertion between ponA and pbpB in strain RZ11 compared to strain Verdun.
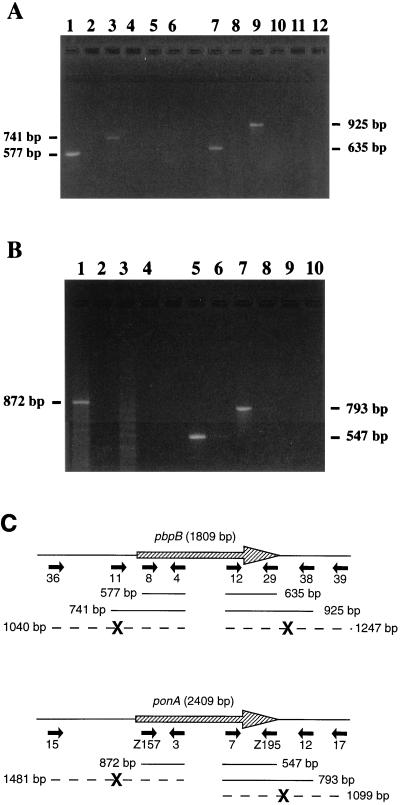
The transcription of both genes from strain Verdun was analyzed by RT-PCR. For the pbpB gene, internal primers allowed reverse transcription of RNA, indicating that pbpB is transcribed (Fig. 1A, lanes 1 and 7). Primers close upstream and downstream of the gene in use with internal primers still allow reverse transcription (Fig. 1A, lanes 3 and 9), while primers located further upstream and downstream of the ORF in use with internal primers do not (Fig. 1A, lanes 5 and 11). The start of transcription can thus be located between 76 and 389 bp upstream of the pbpB gene. Analogously, transcription of the ponA gene was demonstrated (Fig. 1B, lanes 1, 5, and 7). Amplicons were not formed when RT was absent from the reaction mix (Fig. 1A and B, lanes with even numbers). Taken together, the results (Fig. 1C) indicate that both genes are transcribed as single transcription units. Analysis of the strain RZ11 ponA gene confirmed that it was also transcribed (data not shown).
FIG. 1.
Analysis of transcription of pbpB and ponA from L. interrogans strain Verdun by RT-PCR. A sample of each reaction mixture was analyzed on a 1.75% agarose gel with TBE (90 mM Tris, 90 mM borate, 2 mM EDTA [pH 8]) buffer. Lanes with even numbers correspond to negative controls without RT. The size of the amplified products is indicated. (A) RT-PCR for pbpB and adjacent regions. Four reactions were performed with primers internal to the pbpB ORF: primers 4 and 8 (lanes 1 and 2) and primers 12 and 29 (lanes 7 and 8). The other reactions were performed with primers outside the ORF in combination with primers internal to the ORF: primers 4 and 11 (lanes 3 and 4), primers 4 and 36 (lanes 5 and 6), primers 12 and 38 (lanes 9 and 10), and primers 12 and 39 (lanes 11 and 12). (B) RT-PCR for ponA and adjacent regions. Four reactions were performed with primers internal to the ponA ORF: primers 3 and Z157 (lanes 1 and 2) and primers 7 and Z195 (lanes 5 and 6). The other reactions were performed with primers outside the ORF in combination with primers internal to the ORF: primers 3 and 15 (lanes 3 and 4), primers 7 and 12 (lanes 7 and 8), and primers 7 and 17 (lanes 9 and 10). (C) Diagram showing the location of the RT-PCR primers and products of transcription of pbpB and ponA from L. interrogans strain Verdun. The primers (Table 2) are indicated by solid arrows. A thin line shows the presence of a transcript with its length in base pairs. A large boldface X interrupting a broken line indicates the absence of transcript.
PBP 1 and PBP 3 bind AMP.
The L. interrogans PBP 1 and PBP 3 proteins resemble other HMW PBPs, and both proteins retain signature motifs associated with penicillin binding sites (14). Based on these similarities, we predicted that both proteins would covalently bind penicillin. As a first step, we wished to compare the sizes of PBPs in L. interrogans to those of the five PBPs identified for L. kirschneri (10). L. interrogans strain Verdun PBPs were detected by incubation of cell sonicates with DIG-AMP, separated by electrophoresis, and visualized. Several PBPs were detected with estimated masses of 89 (doublet), 64, 41, 32, and 20 kDa (data not shown). Several of the L. interrogans PBPs have estimated masses similar to those previously reported for L. kirschneri serovar grippotyphosa strain RM52 (i.e., 82-kDa doublet and 64, 59, and 33 kDa) (10). Differences were detected with the PBP 1 and 2 proteins (89 kDa for strain Verdun and 82 kDa for strain RM52), PBP 4 proteins (41 kDa for strain Verdun and 59 kDa for strain RM52), and strain Verdun PBP 6 (18 kDa) not reported for L. kirschneri.
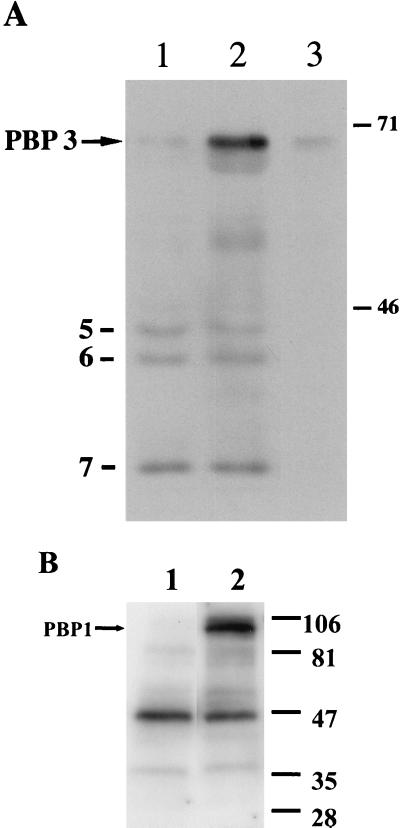
To assay the binding of penicillin to PBP 1 and PBP 3, plasmids containing ponA and pbpB genes downstream of promoters functional in E. coli were constructed. A translational fusion linking the periplasmic portion of the strain Verdun pbpB gene with the vector-encoded pelB leader sequence was constructed. The resulting plasmid, pET-PBP3, contained the pbpB gene downstream of a T7 RNA polymerase promoter. Upon induction with IPTG, a novel protein was observed by SDS-PAGE of the whole-cell lysate samples of E. coli BL21(DE3)/pET-PBP3, indicating that pbpB was efficiently expressed. The protein cofractionated with the insoluble material, indicating that this protein was not correctly folded and aggregated as inclusion bodies. This insoluble protein was partially purified under denaturing conditions with His-Bind resin, but it precipitated after gradual removal of 6 M urea. The DIG-AMP assay confirmed that the pbpB gene product bound AMP. The PBP 3 protein migrated with an apparent molecular mass of 70 kDa (Fig. 2A, lane 2) in agreement with the 69-kDa calculated mass of the fusion product. Preincubation of proteins with free AMP in excess inhibited the binding of DIG-AMP to the polypeptides (Fig. 2A, lane 3).
FIG. 2.
Binding of DIG-AMP to L. interrogans PBP 1 and PBP 3. Lysates of E. coli cells harboring recombinant or vector plasmids were incubated with DIG-AMP, separated by SDS-PAGE, and transferred to a membrane, and the modified proteins were detected as described in Materials and Methods. (A) Production of strain Verdun PBP 3 from pET-PBP3, a recombinant plasmid in E. coli. pET-26b(+) is the expression vector (lane 1). pET-PBP3 is the recombinant plasmid carrying pbpB from strain Verdun (lanes 2 and 3). Binding of DIG-AMP was also assayed in the presence of free AMP (lane 3). The positions of E. coli PBP 5, PBP 6, and PBP 7 and L. interrogans strain Verdun PBP 3 are indicated on the left. The migration of size standards is indicated on the right (in kilodaltons). (B) Production of strain RZ11 PBP 1 from p513-3, a recombinant plasmid in E. coli. pCR2.1 is the expression vector (lane 1). p513-3 is the recombinant plasmid carrying ponA from strain RZ11 (lane 2). The migration of size standards is indicated on the right (in kilodaltons). L. interrogans strain RZ11 PBP 1 is labeled.
Analogously, complete copies of the strain RZ11 ponA and pbpB genes were amplified and cloned into the pCR2.1 vector. Lysates of E. coli that harbored p513-3 (ponA) or p921-1 (pbpB) were incubated with DIG-AMP and compared to lysates of cells harboring the pCR2.1 vector (Fig. 2B, lane 1). These data showed that E. coli synthesized a 90-kDa protein from p513-3, consistent with the predicted full-length PBP 1 (Fig. 2B, lane 2). Binding of DIG-AMP to a 47-kDa protein of unknown origin was also seen with lysates of both vector and p515-3. A 70-kDa protein was synthesized from p921-1, consistent with the predicted mass of PBP 3 (data not shown). Smaller proteins were also detected, suggesting that the proteins may be subjected to proteolysis. AMP competition has been performed, confirming specific binding (data not shown).
Typically, the HMW PBPs 1 and 3 are anchored in the cytoplasmic membrane, using an amino-proximal hydrophobic transmembrane sequence to initiate translation of the protein to the periplasm. Retention of this transmembrane sequence serves to anchor the protein in the cytoplasmic membrane. The L. interrogans PBP 1 and PBP 3 were both predicted to have a single transmembrane segment located near the amino terminus. This may serve as the uncleaved signal sequence and may also anchor the protein in the cytoplasmic membrane. In PBP 1, the putative membrane-spanning region is located between amino acids 33 and 51, and in PBP 3, the potential membrane-spanning segment is between amino acids 11 and 28 (PhdTopology Refinement and Topology Prediction; phd@dodo.cpmc.columbia.edu [PredictProtein]).
Further analysis of the transmembrane-spanning and anchoring functions of these proteins may provide insight into the organization of the spirochete cell wall. The cell envelope organization of spirochetes is unique, having features in common with both gram-positive and gram-negative bacteria. For example, the spirochetal cytoplasmic membrane is intimately associated with the peptidoglycan cell wall, as it is in gram-positive bacteria. Like gram-negative bacteria, spirochetes have an outer membrane, but this membrane is unusual, being the most fluid membrane known to exist in nature (5). These differences may influence the mechanisms of protein secretion. The process of protein secretion is poorly characterized for spirochetes. However, Haake (9) has recently identified a lipoprotein anchor consensus sequence shared by spirochetes that is slightly different from that of other bacteria. Further characterization of the signal sequences for the PBPs should provide insight into the mechanisms by which spirochetal proteins are secreted and the signals used for membrane insertion. These signal sequences may also be useful in targeting leptospiral proteins to the E. coli periplasm to assist in purification.
ACKNOWLEDGMENTS
This work was supported by Institut Pasteur, Paris, France.
A.B. and I.S. thank J. Belfaiza for construction of the cosmid library; C. Boursaux-Eude for the preliminary identification of a PBP; C. Werts for experimental help; D. Margarita for technical help; D. Haake for the detailed DIG-AMP method; G. Baranton for support; and S. Bach, J. Dam, C. Bodenreider, and J.-M. Betton for helpful discussions. R.Z. thanks T. McNunn and A. Olson for excellent technical assistance.
REFERENCES
- 1.Altschul S F, Madden T L, Schaeffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belfaiza J, Martel A, Margarita D, Saint Girons I. Direct sulfhydrylation for methionine biosynthesis in Leptospira meyeri. J Bacteriol. 1998;180:250–255. doi: 10.1128/jb.180.2.250-255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boursaux-Eude C, Margarita D, Gilles A M, Barzu O, Saint Girons I. Borrelia burgdorferi uridine kinase: an enzyme of the pyrimidine salvage pathway for endogenous use of nucleotides. FEMS Microbiol Lett. 1997;151:257–261. doi: 10.1111/j.1574-6968.1997.tb12579.x. [DOI] [PubMed] [Google Scholar]
- 4.Boursaux-Eude C, Saint Girons I, Zuerner R. Leptospira genomics. Electrophoresis. 1998;19:589–592. doi: 10.1002/elps.1150190421. [DOI] [PubMed] [Google Scholar]
- 5.Charon N W, Lawrence C W, O'Brien S. Movement of antibody-coated latex beads attached to the spirochete Leptospira interrogans. Proc Natl Acad Sci USA. 1981;78:7166–7170. doi: 10.1073/pnas.78.11.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellinghausen H C, McCullough W G. Nutrition of Leptospira pomona and growth of 13 other serotypes: fractionation of oleic albumin complex and medium bovine albumin and polysorbate 80. Am J Vet Res. 1965;26:45–51. [PubMed] [Google Scholar]
- 7.Faine S, Adler B, Bolin C, Perolat P. Leptospira and leptospirosis. Melbourne, Australia: MediSci; 1999. [Google Scholar]
- 8.Ghuysen J M. Serine beta-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 9.Haake D A. Spirochetal lipoproteins and pathogenesis. Microbiology. 2000;146:1491–1504. doi: 10.1099/00221287-146-7-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haake D A, Walker E M, Blanco D R, Bolin C A, Miller M N, Lovett M A. Changes in the surface of Leptospira interrogans serovar grippotyphosa during in vitro cultivation. Infect Immun. 1991;59:1131–1140. doi: 10.1128/iai.59.3.1131-1140.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 12.Johnson R C, Harris V G. Differentiation of pathogenic and saprophytic leptospires. I. Growth at low temperatures. J Bacteriol. 1967;94:27–31. doi: 10.1128/jb.94.1.27-31.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massova I, Mobashery S. Kinship and diversification of bacterial penicillin-binding proteins and β-lactamases. Antimicrob Agents Chemother. 1998;42:1–17. doi: 10.1128/aac.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen-Distèche M, Fraipont C, Buddelmeijer N, Nanninga N. The structure and function of Escherichia coli penicillin-binding protein 3. Cell Mol Life Sci. 1998;54:309–316. doi: 10.1007/s000180050157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicas T I, Hancock R E. Outer membrane protein H1 of Pseudomonas aeruginosa: involvement in adaptive and mutational resistance to ethylenediaminetetraacetate, polymyxin B, and gentamicin. J Bacteriol. 1980;143:872–878. doi: 10.1128/jb.143.2.872-878.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oie S, Hironaga K, Koshiro A, Konishi H, Yoshii Z. In vitro susceptibilities of five Leptospira strains to 16 antimicrobial agents. Antimicrob Agents Chemother. 1983;24:905–908. doi: 10.1128/aac.24.6.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piras G, Raze D, El Kharroubi A, Hastir D, Englebert S, Coyette J, Ghuysen J. Cloning and sequencing of the low-affinity penicillin-binding protein 3r-encoding gene of Enterococcus hirae S185: modular design and structural organization of the protein. J Bacteriol. 1993;175:2844–2852. doi: 10.1128/jb.175.10.2844-2852.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Smith C R, Corney B G, McGowan M R, McClintock C S, Ward W, Ketterer P J. Amoxycillin as an alternative to dihydrostreptomycin sulphate for treating cattle infected with Leptospira borgpetersenii serovar hardjo. Aust Vet J. 1997;75:818–821. doi: 10.1111/j.1751-0813.1997.tb15661.x. [DOI] [PubMed] [Google Scholar]
- 20.Spratt B G. Properties of the penicillin-binding proteins of Escherichia coli K12. Eur J Biochem. 1977;72:341–352. doi: 10.1111/j.1432-1033.1977.tb11258.x. [DOI] [PubMed] [Google Scholar]
- 21.Tipper D J, Strominger J L. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D alanyl-D-alanine. Proc Natl Acad Sci USA. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weigel L M, Belisle J T, Radolf J D, Norgard M V. Digoxigenin-ampicillin conjugate for detection of penicillin-binding proteins by chemiluminescence. Antimicrob Agents Chemother. 1994;38:330–336. doi: 10.1128/aac.38.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkins M R, Gasteiger E, Bairoch A, Sanchez J-C, Williams K L, Appel R D, Hochstrasser D F. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 24.Zuerner R L. Physical map of chromosomal and plasmid DNA comprising the genome of Leptospira interrogans. Nucleic Acids Res. 1991;19:4857–4860. doi: 10.1093/nar/19.18.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuerner R L, Hartskeerl R A, Van de Kemp H, Bal A E. Characterization of the Leptospira interrogans S10-spc-alpha operon. FEMS Microbiol Lett. 2000;182:303–308. doi: 10.1111/j.1574-6968.2000.tb08912.x. [DOI] [PubMed] [Google Scholar]