Abstract
Vulvar squamous cell carcinoma (VSCC) pathogenesis is traditionally defined by the presence or absence of human papillomavirus (HPV), but the definition of these groups and their molecular characteristics remains ambiguous across studies. Here, we present a retrospective cohort analysis of 36 patients with invasive VSCC where HPV status was determined using RNA in situ hybridization (ISH) and polymerase chain reaction (PCR). Clinical annotation, p16 immunohistochemistry (IHC), programmed death ligand-1 (PD-L1) IHC, HPV16 circular E7 RNA (circE7) detection, and RNA-sequencing (RNA-seq) of the cases was performed. A combination of ISH and PCR identified 20 cases (55.6%) as HPV-positive. HPV-status did not impact overall survival (HR: 1.36, 95% CI: 0.307 to 6.037, p=0.6857) or progression-free survival (HR: 1.12, 95% CI: 0.388 to 3.22, p=0.8367), and no significant clinical differences were found between the groups. PD-L1 expression did not correlate with HPV status, but increased expression of PD-L1 correlated with worse overall survival. Transcriptomic analyses (n=23) revealed distinct groups, defined by HPV status, with multiple differentially expressed genes previously implicated in HPV-induced cancers. HPV-positive tumors showed higher global expression of endogenous circular RNAs (circRNAs), including several circRNAs that have previously been implicated in the pathogenesis of other cancers.
INTRODUCTION
Vulvar cancer is the fourth most common cancer in women to be caused by high-risk HPV infection after cervical, anal, and vaginal cancer, respectively (Arbyn et al., 2012). Vulvar cancer is a rare disease with only 6,120 women in the United States expected to be diagnosed in 2021, but rates are rising 0.5% per year (2021a, Buchanan et al., 2016). The most common subtype is vulvar squamous cell carcinoma (VSCC), which has poor 5-year progression-free survival and overall survival rates at 48.1% and 61.4%, respectively (Nooij et al., 2016a, Zapardiel et al., 2020).
VSCC has been proposed to develop through one of two independent pathways: HPV-associated (HPV-positive) or HPV-independent (HPV-negative) (Berek and Karam, 2021, Canavan and Cohen, 2002). HPV-positive VSCC is thought to arise from a high-risk HPV-driven precursor lesion. In contrast, factors including chronic inflammation from conditions like vulvar lichen sclerosus (LS) and/or cellular atypia from advanced age have been proposed to contribute to the development of HPV-negative disease (Alkatout et al., 2015, Canavan and Cohen, 2002). In other HPV-related cancers, particularly cervical and head and neck squamous cell carcinomas, HPV positivity is utilized for risk stratification and prognostic outcomes (Blitzer et al., 2014, Li et al., 2017). However, studies of HPV positivity in VSCC cohorts vary widely, from 9–79% of cases (Allo et al., 2020, Gargano et al., 2012, Rasmussen et al., 2018), and the prognostic role of HPV in VSCC is uncertain (Rakislova et al., 2017). We hypothesized that HPV-positive and HPV-negative VSCC are distinct diagnoses with unique biomarkers and clinically distinct behaviors.
RESULTS
Clinical Characteristics and Oncologic Outcomes
Overall Patient Characteristics (Table 1)
Table 1.
Demographic and Tumor Characteristics of Patients with Vulvar Squamous Cell Carcinoma According to Presence or Absence of Human Papillomavirus.
| Variable | Total # (%) | HPV-Positive Total # (%) | HPV-Negative Total # (%) | p-value† |
|---|---|---|---|---|
|
| ||||
| Age | N = 36 | N = 20 | N = 16 | 0.1369 |
| Mean (SD) | 60.3 (14.7) | 57.1 (13.0) | 64.4 (16.2) | |
| Race/Ethnicity | N = 36 | N = 20 | N = 16 | |
| White/Caucasian | 23 (63.9) | 14 (70.0) | 9 (56.3) | 0.4932 |
| Hispanic | 6 (16.7) | 1 (5.00) | 5 (31.3) | 0.0689 |
| African-American | 7 (19.4) | 5 (25.0) | 2 (12.5) | 0.4264 |
| History of Smoking | N = 36 | N = 20 | N = 16 | 0.0910 |
| Yes | 20 (55.6) | 14 (70.0) | 6 (37.5) | |
| No | 16 (44.4) | 6 (30.0) | 10 (62.5) | |
| History of HPV Infection | N=21 | N=12 | N=9 | 0.6605 |
| Yes | 9 (42.9) | 6 (50.0) | 3 (33.3) | |
| No | 12 (57.1) | 6 (50.0) | 6 (66.7) | |
| History of Abnormal Pap | N=33 | N=19 | N=14 | 0.4587 |
| Yes | 11 (33.3) | 5 (26.3) | 6 (42.9) | |
| No | 22 (66.7) | 14 (73.7) | 8 (57.1) | |
| History of Lichen Sclerosus | N=36 | N=20 | N=16 | 0.2853 |
| Yes | 10 (27.8) | 4 (20.0) | 6 (37.5) | |
| No | 26 (72.2) | 16 (80.0) | 10 (62.5) | |
| Initial Symptoms at Presentation | N=36 | N=20 | N=16 | |
| Vulvar Mass | 23 (63.9) | 12 (60.0) | 11 (68.8) | 0.7314 |
| Urinary symptoms | 11 (30.6) | 5 (25.0) | 6 (37.5) | 0.4834 |
| Itching | 9 (25.0) | 5 (25.0) | 4 (25.0) | >0.999 |
| Bleeding/Draining | 18 (50.0) | 12 (60.0) | 6 (37.5) | 0.3145 |
| Surgical Intervention | N=36 | N=20 | N=16 | 0.4921 |
| Wide Local Excision | 2 (5.6) | 2 (10.0) | 0 (0.0) | |
| Vulvectomy | 34 (94.4) | 18 (90.0) | 16 (100.0) | |
| Nodal involvement | N=28 | N=17 | N=11 | 0.4087 |
| Yes | 9 (32.1) | 4 (23.5) | 5 (45.5) | |
| No | 19 (67.9) | 13 (76.5) | 6 (54.5) | |
| Lymphovascular Space Invasion | N=36 | N=20 | N=16 | 0.7225 |
| Yes | 10 (27.8) | 5 (25.0) | 5 (31.3) | |
| No | 26 (72.2) | 15 (75.0) | 11 (68.8) | |
| FIGO Stage (2018) | N=36 | N=20 | N=16 | 0.4834 |
| IB | 25 (69.4) | 15 (75.0) | 10 (62.5) | |
| IIIA/B/C | 11 (30.6) | 5 (25.0) | 6 (37.5) | |
| Histology | N=36 | N=20 | N=16 | 0.6750 |
| Keratinizing | 29 (80.6) | 17 (85.0) | 12 (75.0) | |
| Nonkeratinizing/Basaloid/Mixed | 7 (19.4) | 3 (15.0) | 4 (25.0) | |
| Depth of Invasion (mm) | N=36 | N=20 | N=16 | 0.7410 |
| Mean (SD) | 10.7 (9.3) | 11.1 (11.4) | 10.1 (6.06) | |
| Greatest Dimension of Tumor Size (mm) | N=36 | N=20 | N=16 | 0.5204 |
| Mean (SD) | 37.2 (21.7) | 35.1 (21.5) | 39.8 (22.3) | |
| Tumor Borders | N=22 | N=11 | N=11 | |
| Infiltrating | 17 (77.3) | 7 (63.6) | 10 (90.9) | 0.3108 |
| Pushing | 5 (22.7) | 4 (36.4) | 1 (9.1) | |
| Focality | N=35 | N=19 | N=16 | >0.999 |
| Unifocal | 27 (77.1) | 15 (78.9) | 12 (75.0) | |
| Multifocal | 8 (22.9) | 4 (21.1) | 4 (25.0) | |
| Post-Operative Chemo-radiation | N=30 | N=16 | N=14 | 0.3778 |
| Yes | 6 (20.0) | 2 (12.5) | 4 (28.6) | |
| No | 24 (80.0) | 14 (87.5) | 10 (71.4) | |
| Recurrence | N=28 | N=16 | N=12 | 0.7036 |
| Yes | 14 (50.0) | 7 (43.8) | 7 (58.3) | |
| No | 14 (50.0) | 9 (56.3) | 5 (41.7) | |
| Immunotherapy | N=28 | N=16 | N=12 | 0.1746 |
| Yes | 2 | 0 (0.0) | 2 (16.7) | |
| No | 26 | 16 (100.0) | 10 (83.3) | |
| Death | N=28 | N=16 | N=12 | 0.4319 |
| Yes | 9 (32.1) | 4 (25.0) | 5 (41.7) | |
| No | 19 (67.9) | 12 (75.0) | 7 (58.3) | |
| Length of Follow-Up (months) Mean (SD) | N=36 34.8 (25.3) | N=20 33.1 (24.1) | N=16 36.9 (27.3) | 0.6612 |
Abbreviations: HPV, human papillomavirus; SD, standard deviation; Pap, Papanicolaou; FIGO, International Federation of Gynecology and Obstetrics; mm, millimeters.
Continuous data was analyzed using an unpaired 2-tailed Student’s t-test and categorical data were analyzed using Fisher’s exact test in GraphPad Prism® version 9.0.0. P-values are reported for tests of statistical significance. All comparisons were two-tailed and the alpha error was set at 5%.
A retrospective cohort of patients diagnosed with VSCC was performed with 36 cases that met inclusion criteria and had adequate tissue for analysis. 3 cases of vulvar tissue with no history of VSCC, dysplasia, or HPV infection were obtained as control samples. The mean age of diagnosis was 60 years, 23 (n=36, 63.9%) were Caucasian, 6 (n=36, 16.7%) were Hispanic, and 7 (n=36, 19.4%) were African American; 20 (n=36, 55.6%) had a smoking history. 2 (n=36, 5.6%) patients were immunosuppressed due to autoimmune disease. Relevant medical history included 9 with previous HPV infection (n=21, 42.9%), 11 (n=33, 33.3%) with an abnormal Papanicolaou test results, and 10 (n=36, 27.8%) with a clinical diagnosis of LS.
Treatment and Tumor Characteristics (Table 1)
A radical partial vulvectomy was the most common initial intervention, with 34 (n=36, 94.4%) undergoing this procedure while 2 received a wide local excision (n=36, 5.6%). Most patients, 28 (n=36, 77.8%), underwent a surgical evaluation of regional lymph nodes, which included either a sentinel lymph node mapping or an inguinofemoral lymphadenectomy; of those, 9 (n=28, 32.1%) had nodal involvement. Histologic analyses of the tumors revealed the mean tumor size at greatest dimension was 37.2 mm, and mean depth of invasion was 10.7 mm. Stage was classified based on the International Federation of Gynecology and Obstetrics (FIGO) 2018 staging system (Rogers and Cuello, 2018) and was most commonly stage IB with 25 patients (n=36, 69.4%), followed by 11 patients (n=36, 30.6%) at stage III. For post-operative treatment, 6 received adjuvant chemo-radiation (n=30, 20%). Half of the patients with sufficient follow-up, 14 (n=28, 50%), had either a local (11 patients, n=14, 78.6%), or regional (3 patients, n=14, 21.4%) recurrence. The mean length of follow-up was 34.8 months. Ultimately, 9 patients (n=28, 32.1%) died with 7 (n=9, 77.8%) secondary to VSCC recurrences.
Overall Survival and Progression-Free Survival (Table 2)
Table 2.
Overall Survival and Progression-Free Survival by Clinical Characteristics for Patients with Vulvar Squamous Cell Carcinoma.
| Variable | Overall survival VSCC† | Progression-free survival VSCC† | ||||
|---|---|---|---|---|---|---|
| 95% CI | HR | p-valuea | 95% CI | HR | p-valuea | |
| Recurrence: Y vs. N | 1.529 – 30.36 | 6.814 | 0.0118* | N/A | N/A | N/A |
| Age: ≥60 vs. <60 (REF) | 0.2702 – 5.298 | 1.196 | 0.8132 | 0.3528 – 2.887 | 1.009 | 0.9863 |
| Stage: III vs. I | 0.5904 – 22.74 | 3.664 | 0.1633 | 0.3452 – 3.867 | 1.156 | 0.8146 |
| Margin (mm):<8 vs. ≥8 (REF) | 0.2319 – 10.93 | 1.592 | 0.6360 | 0.3972 – 5.929 | 1.535 | 0.5345 |
| Post-op chemo: N vs. Y | 0.4373 – 24.92 | 3.301 | 0.2468 | 0.5579 – 8.255 | 2.146 | 0.2666 |
| Post-op radiation: N vs. Y | 0.4864 – 23.79 | 3.401 | 0.2173 | 0.6723 – 8.534 | 2.395 | 0.1778 |
| Smoking: Y vs. N | 0.2439 – 4.872 | 1.090 | 0.9100 | 0.2470 – 2.016 | 0.7056 | 0.5151 |
| Focality: Multifocal vs. unifocal | 0.2612 – 10.32 | 1.641 | 0.5972 | 1.312 – 28.38 | 6.103 | 0.0211* |
| LVSI: N vs. Y | 0.3429 – 10.13 | 1.863 | 0.4711 | 0.4019 – 4.393 | 1.329 | 0.6413 |
| LN: pos vs. neg | 1.610 –160.5 | 16.07 | 0.0180* | 0.3149 – 5.818 | 1.353 | 0.6842 |
| Surgery: Wide local excision vs. vulvectomy | 0.03909 – 1820 | 8.435 | 0.0151* | 0.02283 – 4669 | 10.33 | 0.0064* |
| Nodal dissection: N vs. Y | 0.5591 – 142.6 | 8.927 | 0.0003* | 0.6909 –61.38 | 6.512 | 0.0001* |
| Re-excision: N vs. Y | 0.1830 – 9.165 | 1.295 | 0.7956 | 0.04323 –1.553 | 0.2591 | 0.1394 |
| Depth (mm): ≥8 vs. <8 (REF) | 0.2414 – 4.792 | 1.076 | 0.9238 | 0.1453 – 1.306 | 0.4357 | 0.1381 |
| Tumor size (mm):≤30 vs. >30 (REF) | 0.2553 – 5.486 | 1.184 | 0.8294 | 0.5302 – 4.358 | 1.520 | 0.4358 |
| History of Lichen sclerosus: N vs. Y | 0.1743 – 4.965 | 0.9303 | 0.9326 | 0.3492 – 3.418 | 1.092 | 0.8793 |
Abbreviations: VSCC, vulvar squamous cell carcinoma; HR, hazard ratio; CI, confidence interval; Y, yes; N, no; N/A, not applicable; mm, millimeters; post-op, post-operative; chemo, chemotherapy; LVSI, lymphovascular space invasion; LN, lymph nodes.
Calculations were performed using the log-rank test in GraphPad Prism® version 9.0.0.
P-values are reported for tests of statistical significance. All comparisons were two-tailed and the alpha error was set at 5%.
Statistically significant.
Clinical characteristics were analyzed to identify independent predictors of overall survival and progression-free survival. Lower overall survival rate could be predicted by the presence of recurrence (hazard ratio (HR): 6.8, 95% confidence interval (CI): 1.5–30.4, p= 0.0118), lymph node involvement (HR: 16.1, 95% CI: 1.6–160.5, p=0.0180), undergoing a wide local excision rather than a vulvectomy (HR: 8.4, 95% CI: 0.04 to 1820, p=0.0151), and not undergoing a nodal dissection at the time of surgery (HR: 8.9, 95% CI: 0.56 to 142.6, p=0.0003). Reduced progression-free survival could be predicted by the presence of a multifocal tumor (HR: 6.1, 95%, CI: 1.3–28.4, p=0.0211), wide local excision rather than vulvectomy (HR: 10.3, 95% CI: 0.02–4669, p=0.0064), and not undergoing a nodal dissection (HR: 6.5, 95% CI: 0.69–61.4, p=0.0001).
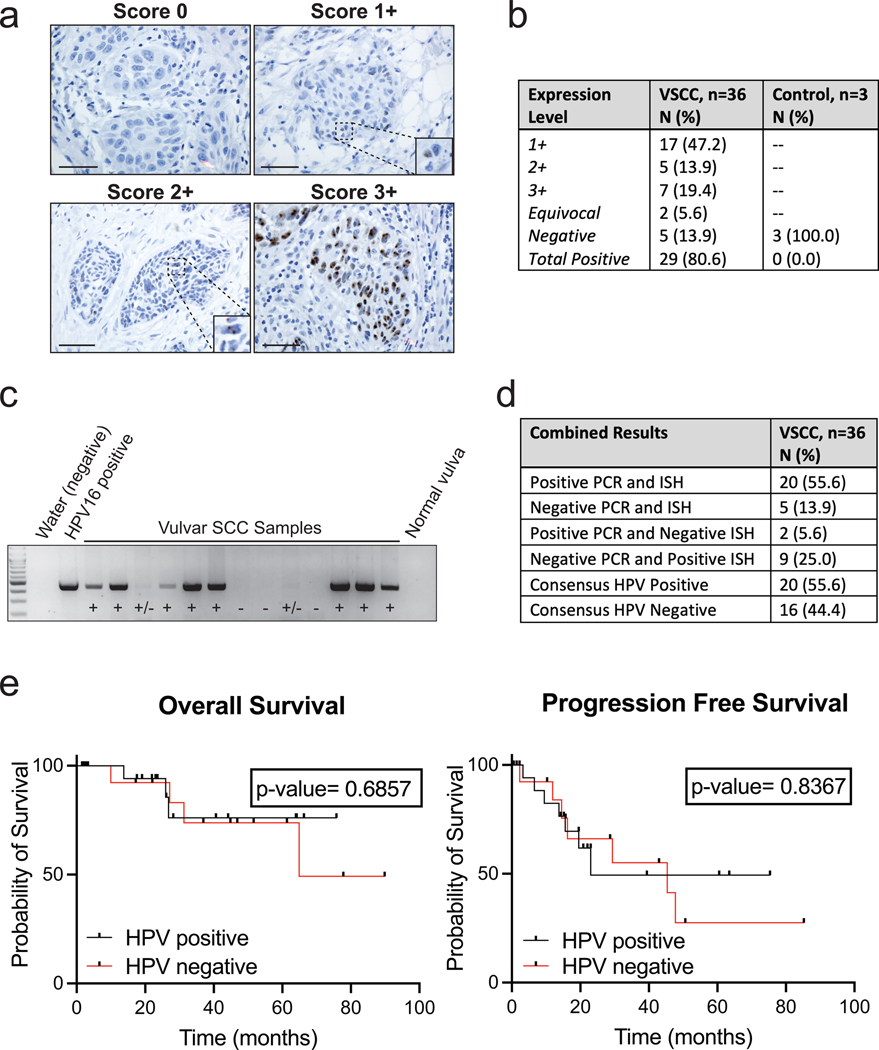
HPV Status Determination and Oncologic Outcomes
HPV status was determined through a combination of HPV RNA ISH and nested PCR for HPV. Expression patterns and scores of HPV RNA ISH were reviewed independently by two pathologists (Figure 1a). A total of 29 out of 36 samples (80.6%) were positive: 7 (19.4%) at 3+ staining, 5 (13.9%) at 2+ staining, and 17 (47.2%) at 1+ staining (Figure 1b). Nested PCR for conserved regions of the HPV genome was also performed to detect HPV (Figure 1c). In this assay, 22 of 36 samples (61.1%) were positive by PCR. There were differences in the detection of HPV by the assays including 2 cases (5.6%) that were negative by ISH but positive by PCR, and 9 cases (25%) that were positive by ISH but negative by PCR. To ensure rigor, only 20 (55.6%) cases that were positive by both ISH and PCR, were considered consensus HPV-positive (Figure 1d).
Figure 1. Determination of Presence or Absence of Human Papillomavirus for Vulvar Squamous Cell Carcinoma.
a. Representative images of RNA HPV-ISH results separated by score. Bar = 50 μm
b. Expression scores for HPV-ISH for VSCC and control samples.
c. Representative image of HPV PCR. Positive control represents 10 pg of the HPV-16 added to the outer PCR reaction. Negative controls used include water and a normal vulvar tissue sample without history of HPV infection or neoplasm.
d. PCR and HPV-ISH comparison results.
e. Kaplan–Meier curves for overall survival and progression-free survival by HPV-final consensus status and compared by log-rank test with a p-value <0.05 considered significant.
Because p16 expression is frequently used as a surrogate marker for HPV, we performed IHC to detect p16. P16 expression significantly correlated with HPV-consensus status (p=0.0172), but there were discrepancies between p16 positive and consensus HPV-positive cases, including 5 cases (13.9%) that were positive for p16 but HPV-negative, and 5 cases (13.9%) that were negative for p16 but HPV-positive.
Finally, we tested for the presence of an HPV16-derived circular RNA containing the E7 oncogene, circE7. The amplification product was confirmed to the expected backsplice junction through Sanger Sequencing (Figure S1). HPV16 circE7 RNA was more frequently detected in HPV-positive cases (N=20, 50%) than HPV-negative cases by either ISH or PCR (N=16, 31.3%). While circE7 was only detected in cases with HPV by either ISH or PCR, the presence of HPV16 circE7 alone was not a reliable marker for consensus HPV status.
Clinical Comparisons by HPV Status
Demographic and clinical characteristics were compared between HPV-positive and HPV-negative exposure variable, and no statistically significant differences were noted (Table 1). Notably, HPV-status did not significantly impact overall or progression-free survival (Figure 1e). Consistent with a lack of impact of HPV status on clinical outcomes, neither positive p16 expression nor the presence of circE7 had a significant impact on overall or progression-free survival (Figure S1).
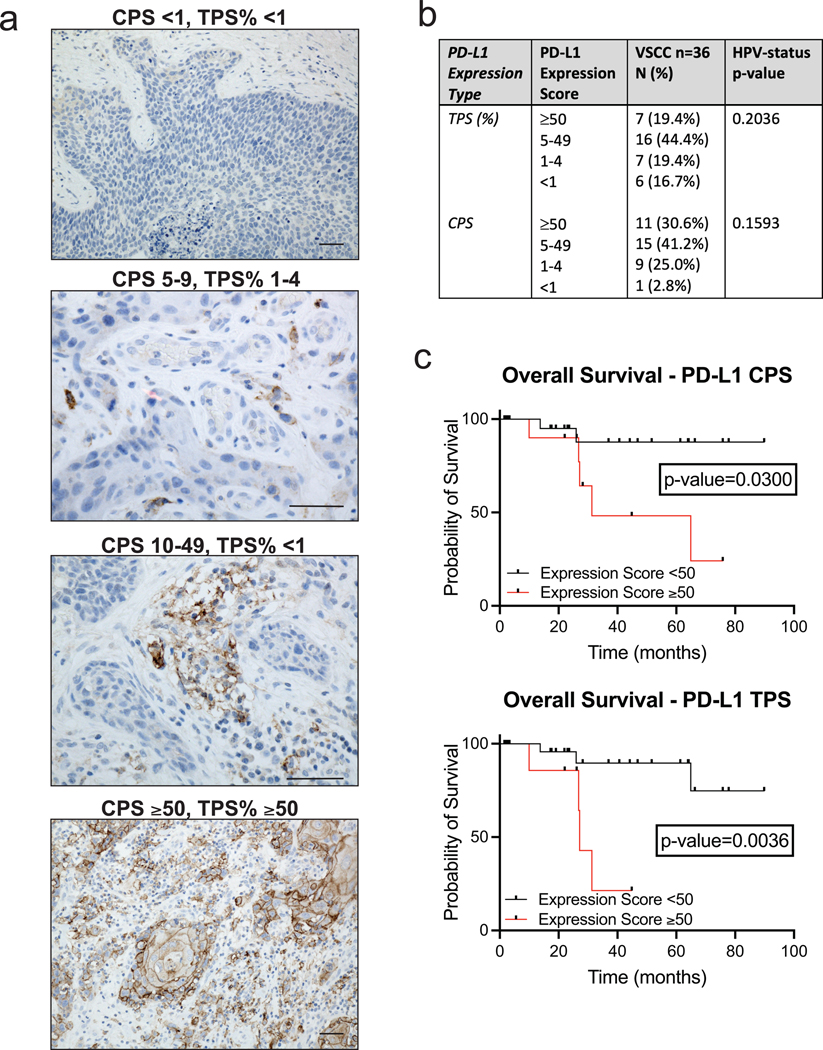
PD-L1, HPV Status, and Oncologic Outcomes
Two pathologists independently assessed PD-L1 expression using combined positive score (CPS) and tumor proportion score (TPS) as depicted by representative images of different expression scores (Figure 2a). Expression scores were reported in ranges (≥50, 5–49, 1–4, and <1) (Lin et al., 2017), numerically for CPS and as percentages for TPS (Figure 2b). The expression of PD-L1 in immune cells was confirmed in 35 cases (n=36, 97.2%). This included widespread expression (≥50) in 11 (30.6%), 15 (41.2%) with scores 5–49, 9 (25.0%) with scores 1–4, and 1 (2.8%) with a score <1, which was considered negative. PD-L1 expression in VSCC tumor cells was observed in 30 patients (n=36, 83.3%). This included widespread expression (≥ 50%) in 7 (19.4%), 16 (44.4%) with scores 5–49%, 7 (19.4%) with scores 1–4%, and 6 (16.7%) with scores <1%. PD-L1 expression (CPS: ≥50 or <50 and TPS: ≥50% or <50%) was compared to HPV status (positive or negative). HPV-negative cases were more likely to have a higher PD-L1 expression score, but this did not reach significance (p=0.2036 and 0.1593) (Figure 2b). We did find PD-L1 expression to be an independent predictor of survival. Patients with a CPS ≥50 had lower overall survival (HR: 5.8, 95% CI: 1.2–28.0, p=0.0300) as did patients with a TPS ≥50% (HR: 6.6, 95% CI: 0.91–47.5, p= 0.0036) compared to patients with lower PD-L1 expression (Figure 2c). Patients with a CPS ≥50 had lower progression-free survival (HR: 1.2, 95% CI: 0.40–3.88, p=0.7074) as did patients with TPS ≥50% (HR: 2.3, 95% CI: 0.56–9.76, p=0.2427), but this did not reach significance.
Figure 2. Programmed Death Ligand-1 Immunohistochemistry Expression Scoring and Prognostic Implications for Vulvar Squamous Cell Carcinoma.
a. Representative images of VSCC PD-L1 IHC expression results separated by scoring. Bar = 50 μm.
b. Table of PD-L1 IHC expression scores for VSCC. Expression ≥50 and <50 expression was compared to HPV-final consensus status using Fisher’s exact test with a p-value <0.05 considered significant.
c. Kaplan–Meier curves for overall survival by CPS and TPS expression levels of PD-L1 and compared by log-rank test with a p-value <0.05 considered significant.
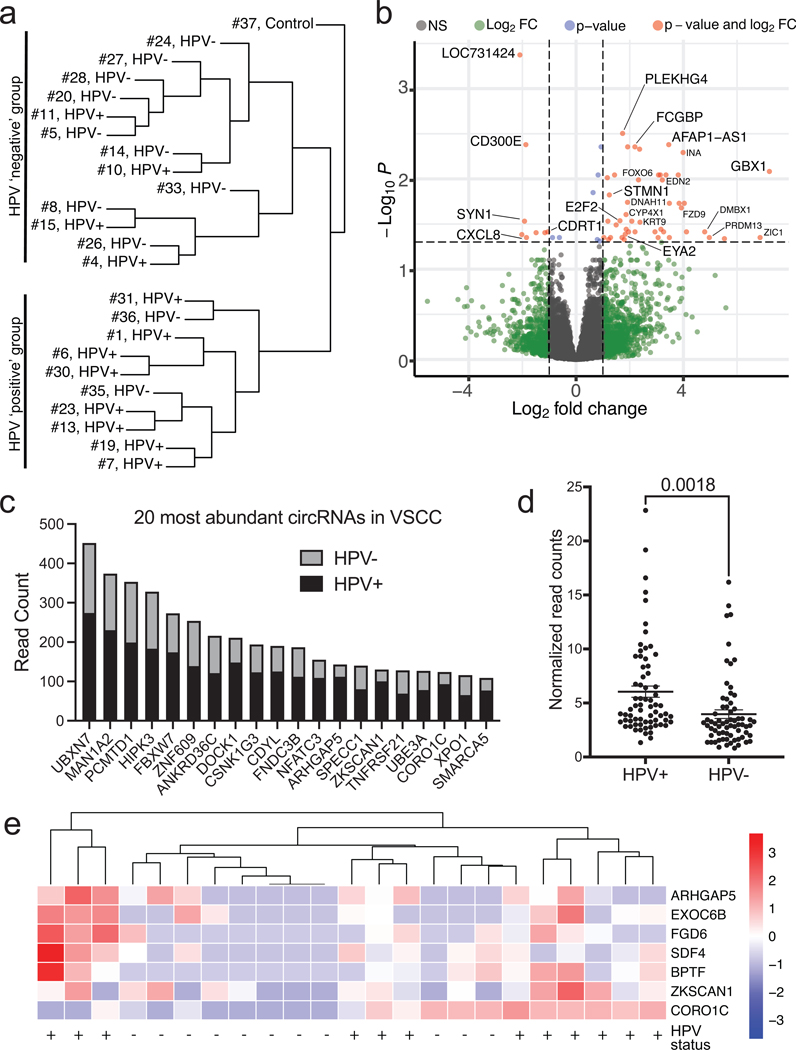
Differential gene expression and circRNA expression in VSCC
RNA sequencing quality was adequate for 23 cases of invasive VSCC and 1 control vulvar tissue. Unsupervised hierarchical clustering confirmed that the VSCC clustered together, distinct from cervical cancer samples that were sequenced in parallel (Figure S2a). Notably, VSCC samples clustered largely, but not exclusively, based on consensus HPV status (Figure 3a, S2b). Differential gene expression (DGE) analysis revealed that a number of linear transcripts were differentially expressed in HPV-negative and HPV-positive tumors (Figure 3b). Notably, several of these genes have been previously reported as potential markers for HPV-positive cancers either in tumor samples (e.g., STMN1, FCGBP) or in infection models (e.g. E2F2, EYA2) (Bierkens et al., 2013, Longworth et al., 2005, Nooij et al., 2016b, Wang et al., 2017). Pathway analyses of the DGE revealed that HPV-positive tumors were enriched for “HSV1 infection”, “viral protein interaction with cytokine receptor”, and infectious and inflammatory signaling pathways (Fig. S3).
Figure 3. RNA-Sequencing Analysis of Vulvar Squamous Cell Carcinoma.
a. Unsupervised hierarchical clustering broken down into HPV-positive and HPV-negative cases. Samples cluster based upon their HPV status.
b. Differential gene expression of HPV-positive vs. HPV-negative illustrated using a volcano plot.
c. Bar graph detailing the 20 most abundant circular RNAs found in VSCC determined by read count. Each bar is broken down into the sum of read counts based from HPV-positive or HPV-negative.
d. Plots of circRNA read counts normalized by sample number. Each dot represents a distinct circRNA with >50 total read counts. HPV-positive samples have significantly higher circRNA read counts.
e. Scaled heatmap demonstrating differential circRNAs in HPV-positive or HPV-negative samples.
The RNA-Seq was further analyzed for genomic variants. Hotspot mutations in tumor suppressors including CDKN2A, FBXW7, and TP53 were identified (Table S4). Mutations in potential oncogenic drivers were also identified in EGFR, FGFR3, HRAS, KRAS, and PIK3CA (Table S4). In contrast to previous exome analyses, a similar distribution of these mutations was found in HPV-positive and negative samples, possibly due to the smaller sample size.
VSCC samples was also assessed for expression of circRNAs. Despite the low levels of HPV read counts that were identified in most samples, three cases with multiple reads corresponding to the expected backsplice junctions for circE7 isoforms were identified (Table S1), with one case showing more than 30% of spliced reads corresponding to the circE7 backsplice junction.
Analysis of the VSCC sequencing revealed the presence of 229 endogenous circRNAs with >50 backsplice reads across all samples. Most of the top 20 circRNA candidates have been identified in previous studies, confirming the accuracy of our circRNA analyses (Figure 3c). Several abundant circRNAs, circular isoforms of HIPK3, FBXW7, and ZNF609, have previously been shown to contribute to the transformed behavior of other tumor types (Legnini et al., 2017, Yang et al., 2018, Zheng et al., 2016).
We next assessed whether HPV status might affect the global circRNA expression profile. Even after normalizing for the slightly higher number of HPV-positive cases, we found that HPV-positive tumors showed a significantly higher expression of circRNAs than HPV-negative tumors (p=0.0018) (Figure 3d). While the 20 most abundant circRNAs showed higher levels in HPV-positive compared to HPV-negative samples (Figure 3c), almost all identified circRNAs were more abundant in HPV-positive samples. Circular isoforms of ARHGAP5, CORO1C, and ZKSCAN1 showed particularly high expression in HPV-positive VSCC (Figure 3e) (Fan et al., 2021, Yao et al., 2017).
DISCUSSION
Outcomes of VSCC by HPV status differ across studies. While some reports demonstrate improved prognosis with HPV-positive cases (Allo et al., 2020, Barlow et al., 2020, Giulia Mantovani et al., 2020, Woelber et al., 2021, Zhang et al., 2018), others suggest that HPV status does not impact survival outcomes (del Pino et al., 2013, Weberpals et al., 2017, Zięba et al., 2018). Because variations in assays utilized for HPV detection may contribute to these differences (del Pino et al., 2013), we used both nested DNA PCR and RNA ISH to determine status prior to testing outcomes. These results were further compared with p16 IHC and circE7 RT-PCR. RNA-sequencing confirmed that HPV transcripts could be detected in all consensus HPV samples (Table S3). However, because RNA-Seq was successful for only a subset of the samples and is not widely available in clinical diagnostic testing, it was not used as the standard for determining HPV status. Even with the use of a very stringent criterion for HPV positivity, we found that HPV status does not act as an independent prognostic factor in VSCC and is not significantly associated with any clinical characteristics. One limitation of this study is the relatively low number of cases that were available for analysis, which may have limited our ability to detect clinical differences between HPV-positive and -negative cases. Also, our stringent definition of HPV-positive cases may underestimate the true contribution of HPV to VSCC. However, even if VSCC cases had been designated HPV-positive by ISH alone, PCR alone, or if either test was positive, there still would have been no significant correlation between HPV status and overall or progression-free survival.
Immunotherapy has emerged as a promising target, particularly for virus-induced cancers due to the expression of non-self, viral proteins (Naumann et al., 2019). The PD-L1 transmembrane protein inhibits T-cell activation and facilitates tumor immune evasion (Reddy et al., 2017). While PD-L1 testing may be beneficial in cases of advanced disease (2021b), response rates to PD-L1 immunotherapy remain poor (How et al., 2021), and the implications of PD-L1 positivity remain unclear (Giulia Mantovani et al., 2020). One study demonstrated high PD-L1 expression to be associated with HPV-negative tumors and worse overall outcomes (Hecking et al., 2017), while others have found no association of PD-L1 with HPV status or outcomes (Choschzick et al., 2018, Thangarajah et al., 2019). While HPV-status was not significantly correlated with PD-L1 expression in our series, PD-L1 expression alone did correlate with the outcomes of patients with VSCC. Notably, our data demonstrate a higher positivity rate of PD-L1 in VSCC than previously reported (83.3% by TPS and 97.2% by CPS) (Choschzick et al., 2018, Giulia Mantovani et al., 2020, Hecking et al., 2017, Thangarajah et al., 2019), and higher expression of PD-L1 in either tumor or immune cells is negatively correlated with outcomes. Our findings support additional studies to investigate PD-L1 expression as a prognostic marker in VSCC.
We sought to confirm the potential utility of circE7 as a biomarker in VSCC, and its utility in distinguishing HPV-positive and HPV-negative VSCC. In previous studies, we could detect a circular RNA generated by HPV16, circE7, in cervical cancer, head and neck SCC, and anal SCC (Zhao et al., 2019). Moreover, higher levels of circE7 predicted improved overall survival in cervical, head and neck, and anal SCC (Chamseddin et al., 2019). We were able to detect HPV16 circE7 in 15 (n=36, 41.7%) VSCC samples, and circE7 was present more often in HPV-positive tumors when compared with HPV-negative tumors. Moreover, the presence of circE7 correlated with improved survival in the cervical cancer cases that were analyzed in parallel (data not shown). However, there was no impact on survival outcomes predicted by circE7 in the VSCC analyzed. Thus, while HPV16-derived circE7 can be detected in VSCC, our studies do not currently support its use as a biomarker in VSCC. Because we limited our analyses to only HPV16 derived circRNAs, our study may underestimate the prevalence of hrHPV derived circE7 sequences. In future studies, it will be interesting to determine how circE7 is regulated and how it may specifically impact the pathogenesis and prognosis of HPV-driven cancers.
While exome sequencing has been completed for VSCC (Dasgupta et al., 2020, Giulia Mantovani et al., 2020, Prieske et al., 2020, Tessier-Cloutier et al., 2020, Weberpals et al., 2017, Zięba et al., 2020a, Zięba et al., 2018, Zięba et al., 2020b), we present the transcriptional profile of VSCC. Our RNA-sequencing results support a biological distinction between HPV-positive and HPV-negative VSCC. We were able to determine two distinct groups roughly defined by HPV status when the cases were subjected to unsupervised hierarchical clustering analysis. Furthermore, upon comparing HPV-positive and HPV-negative VSCC, we were able to identify specific genes likely relevant for disease pathogenesis. Consistent with HPV31 E7’s activation of E2F2 transcription to promote viral replication (Longworth et al., 2005), E2F2 was upregulated by most HPV-positive VSCCs. EYA2, which promotes the viability, growth, and migration of HPV16-positive cervical cancer cell lines, was also upregulated by most HPV-positive VSCCs (Bierkens et al., 2013). Two other HPV-upregulated genes, FCGBP and STMNI, have also been described in other HPV-related tumors—FCGBP in HPV-positive head and neck squamous cell carcinomas (Wang et al., 2017), and STMN1 in HPV-positive vulvar precursor lesions with high grade dysplasia (Nooij et al., 2016b). Thus, analysis of mRNA differential gene expression in VSCC highlights potential biomarkers and therapeutic targets for HPV-related VSCC.
CircRNAs are abundant and have possess biological functions in some cancer (Lu, 2020). Through RNA-seq, we identified hundreds of circRNAs in VSCC. Some, like circ-UBXN7, -PCMTD1, and -HIPK3, promote tumorigenesis by binding and inhibiting specific miRNAs (Yao et al., 2020, Zheng et al., 2016, Zheng et al., 2019), while others, like circFBXW7, effect their biological function after being translated to proteins (Yang et al., 2018). Strikingly, we found significant global increase in circRNA levels in the HPV-positive tumors compared to the HPV-negative ones. Several circRNAs including CORO1C, ZKSCAN1, and BPTF showed exceptionally high expression in HPV-positive tumors, suggesting that both global and allele-specific factors might contribute to the changes in the circRNA expression profile (Bi et al., 2018, Fan et al., 2021, Yao et al., 2017). The regulation of both HPV and endogenous circRNAs in cancer will be an important area for future investigations.
Our study found 80.6% of VSCC cases to be positive by ISH and 61.1% to be positive by PCR. While we used stringent criteria to define HPV-positive cases, our study suggests that rates of HPV-related VSCC may be higher than previously reported due to the poor sensitivity of specific detection methods. Notably, our results revealed a higher rate of HPV positivity in LS-associated VSCC (4/10 cases) than previously reported. Other series have also reported levels of HPV positivity comparable to our study in cases of LS-associated VSCC (Ansink et al., 1994, Hald and Blaakaer, 2018). However, most HPV-positive LS-associated VSCC cases (2/3) still clustered with the HPV-negative cases. Thus, the role of HPV infection in LS-associated VSCC is complex and requires further investigation.
In summary, our retrospective cohort analysis did not detect clinical differences between HPV-positive and HPV-negative cases or an association with biomarkers, PD-L1 and circE7. PD-L1 may have utility as an independent predictor of prognosis. Our transcriptomic analysis of VSCC confirmed the biological distinction between these two groups in VSCC and suggested specific diagnostic and therapeutic targets for the two groups. Finally, we report the circRNA expression profile of VSCC, including the observation that HPV-positive cancers showed a global upregulation of circRNA expression. This comprehensive molecular analysis identifies targets that may be helpful in diagnosing and treating VSCC.
MATERIALS & METHODS
Study Design, Setting, and Cohort
This study was approved by the University of Texas Southwestern (UTSW) Institutional Review Board: STU 072018–067. Because this study utilized archived tissues obtained during standard-of-care treatments, it was granted exemption from the requirement for written, informed consent. Data was collected at Parkland Health and Hospital System and UTSW from January 1, 2010-August 1, 2020. Archived cases of women with invasive VSCC and available FFPE tissue from the initial surgical intervention were collected. Inclusion/exclusion criteria and study design are detailed in Figure S5–S6. Samples were reviewed (E.L.) to confirm adequate tissue was available. Clinical characteristics were assessed through chart review.
Determination of HPV Status
HPV RNA ISH
HPV-ISH was performed as previously described (Chamseddin et al., 2019). Slides were baked, deparaffinized, and treated with hydrogen peroxide reagent (ACD Biosystems) for 10 minutes. The slides were rinsed, incubated in a Citrate Buffer solution (pH 6.0), and incubated in a pressure cooker for 30 minutes. Then, the slides were dehydrated in 100% alcohol. After hydrophobic barrier placement, the slides were incubated with Protease Plus (ACD Biosystems) in a HybEZ Oven (ACD) for 15 minutes at 40°C. Then, slides were incubated with HPV probes (HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68) for 2 hours at 40°C. The slides were washed and recommended amplification steps were performed (RNAScope version 2.5 line by ACD Biosystems). Finally, the slides were placed in 3,3’-diaminobenzidine (DAB) (Vector Labs) for 6 minutes, counterstained, dehydrated, cleared, and covered. ISH was independently assessed by two pathologists according to standards from a previous report (E.L. and G.H.) (Rooper et al., 2016).
HPV DNA Detection
PCR was performed as previously described (Tawe et al., 2018) using a nested primer technique (Table S2). Water was used as a negative control and 10 pg HPV16 genomic plasmid was used as a positive control.
Immunohistochemistry and Interpretation
Tissues were deparaffinized and rehydrated followed by fully submerging the slides in preheated (65°C) EnVision™ FLEX Target Retrieval Solution at high pH (9.0) and incubating at 97°C for 20 minutes. For PD-L1 staining, when the temperature cooled to 70°C, the slides were submerged in 1:20 ratio, laboratory-prepared, wash buffer for 5 minutes. All slides were placed in an Autostainer Link 48 platform (Dako, Agilent) and blocked with FLEX peroxidase block for 5 minutes. For p16, slides were incubated with the rabbit monoclonal anti-human CDKN2A/p16INK4a - C-terminal antibody (clone EPR1473, ABCAM, Cambridge, UK), 1:800 dilution, for 20 minutes, and then incubated with the EnVision™ FLEX HRP visualization reagent for 20 minutes. For PD-L1, slides were incubated with monoclonal mouse anti-human PD-L1 antibody (clone 22C3, Agilent) 1:50 dilution, for 60 minutes, incubated with EnVision™ FLEX+ Mouse LINKER reagent for 30 minutes, and incubated with EnVision™ FLEX/HRP visualization reagent for 30 minutes. After rinsing, the enzymatic conversion of the added DAB chromogen was performed for 10 minutes. For PD-L1, slides were treated with DAB enhancer for 5 minutes. Slides were counterstained with hematoxylin and mounted. Positive and negative controls were included for all stains.
Positive p16 result was defined as strong, diffuse, block-positive nuclear and cytoplasmic staining. Negative result was defined as the absence of both nuclear and cytoplasmic staining or weak, focal, and discontinuous staining. PD-L1 expression was independently evaluated by two pathologists (E.L. and S.H.). TPS was calculated as the percentage of tumor cells with membranous PD-L1 expression. CPS was calculated as the number of PD-L1-staining cells divided by the total number of viable tumor cells, multiplied by 100. Both scores ranged from 0 to 100. A cutoff score ≥1 for CPS and ≥1% for TPS was used to define PD-L1 positivity. For CPS, for scoring of mononuclear immune cells (lymphocytes and macrophages), only intratumoral and peritumoral (within one 20x field from the tumor edge) immune cells were enumerated. IHC analyses of p16 and PD-L1 correlated with CDKN2A and CD274 expression, respectively, confirming consistency between IHC and RNA-Seq results (Fig. S4).
HPV16 circE7 Detection
Total RNA was converted to cDNA with SuperScript IV First-Strand cDNA Synthesis Reaction using random hexamer priming. The cDNA then underwent endpoint PCR for the detection of circE7 using a nested primer technique (Table S2). Positive and negative VSCC samples were used as controls in each reaction.
RNA-Sequencing and Analysis
All FFPE sample sections (36 invasive VSCC and 3 controls) were sent to Admera for RNA quality control, library preparation, and sequencing. Following quality control, 27 VSCC samples and 1 control underwent library preparation using KAPA RNA HyperPrep with RiboErase (HMR) for ribosomal RNA depletion. RNA-Seq was then completed on these samples with a sequencing depth of 60M paired end reads. An additional 4 VSCC samples were excluded from inclusion in downstream analyses due to poor sequencing quality.
Trim Galore was used for quality and adapter trimming. The human reference genome sequence and gene annotation data, hg38, were downloaded from Illumina iGenomes. The qualities of RNA-sequencing libraries were estimated by mapping the reads onto human transcript and ribosomal RNA sequences (Ensembl release 89) using Bowtie (v2.3.4.3) (Langmead and Salzberg, 2012). STAR (v2.7.2b) (Dobin et al., 2013) was employed to align the reads onto the human genome, SAMtools (v1.9) (Li et al., 2009) was employed to sort the alignments, and HTSeq Python package (Anders et al., 2015) was employed to count reads per gene. Picard (2.21.3) was used to assign read group information and remove PCR duplicates. The samples with ribosomal RNA read fractions greater than 10% or PCR duplicate read fractions greater than 95% were excluded from the downstream analyses. DESeq2 R Bioconductor package (Gentleman et al., 2004, McCarthy et al., 2012, Robinson et al., 2010) was used to normalize read counts and identify differentially expressed (DE) genes. Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al., 2017) pathway data was downloaded using KEGG API (https://www.kegg.jp/kegg/rest/keggapi.html). The enrichment of DE genes to pathways were calculated by Fisher’s exact test in R statistical package. HPV16 circular RNAs were identified by mapping the reads onto HPV16 genome sequence (NC_001526.2) downloaded from NCBI nucleotide database using Burrows-Wheeler Aligner (BWA, v0.7.17) (Li et al., 2009) with specific options, “-T 19” to reduce minimum score to output and “-Y” to use soft clipping for supplementary alignments, and vircircRNA ((Zhao et al., 2019); https://github.com/jiwoongbio/vircircRNA). Human circular RNAs were identified by CN-fusion (https://github.com/jiwoongbio/CN-fusion) and selecting back-splice junctions.
Statistical Analysis
Continuous data was analyzed using an unpaired Student’s t-test and categorical data was analyzed using Fisher’s exact test. Three unmatched groups were compared using one-way analysis of variance. Log-rank test analyses were used to compare the overall survival and progression-free survival rate differences. Kaplan–Meier curves were then utilized to construct the visual differences for overall survival and progression-free survival. GraphPad Prism® version 9.0.0 was used for statistical analysis of data.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge the UTSW Tissue Management Shared Resource Center for their assistance in preparation of tissue samples and performance of immunohistochemistry (Grant P30 CA142543). We would also like to thank Admera for the completion of RNA-sequencing. This research received funding from the American Cancer Society, Grant Number: RSG-18–058-0, the National Institute of Arthritis and Musculoskeletal and Skin Diseases, Grant Number: R01AR072655, and Mary Kay Foundation Cancer Research Grant to RCW; Cancer Center Support Grant P30 CA142543, and CPRIT award RP180805 to LX.
Abbreviations:
- VSCC
vulvar squamous cell carcinoma
- HPV
human papillomavirus
- FIGO
International Federation of Gynecology and Obstetrics
- ISH
in situ hybridization
- circRNA,
circular RNA
- PD-L1
programmed death ligand-1
- IHC
immunohistochemistry
- PCR
polymerase chain reaction
- RNA-seq,
RNA-sequencing
- FFPE
formalin fixed paraffin embedded
- CPS
combined positive score
- TPS
tumor proportion score
- LS
lichen sclerosus
- HR
hazard ratio
- CI
confidence interval
Footnotes
CONFLICT OF INTEREST STATEMENT
No conflicts of interest to disclose.
DATA AVAILABILITY STATEMENT
RNA-sequencing available under accession number GSE183454.
REFERENCES
- Key Statistics for Vulvar Cancer. American Cancer Society, https://www.cancer.org/cancer/vulvar-cancer/about/key-statistics.html#:~:text=In%20the%20United%20States%2C%20women,will%20die%20of%20this%20cancer; 2021a. [accessed May 17, 2021].
- NCCN Guidelines Version 3.2021 Vulvar Cancer (Squamous Cell Carcinoma). NCCN, https://www.nccn.org/professionals/physician_gls/pdf/vulvar.pdf; 2021b. [accessed April 28, 2021]. [Google Scholar]
- Alkatout I, Schubert M, Garbrecht N, Weigel MT, Jonat W, Mundhenke C, et al. Vulvar cancer: epidemiology, clinical presentation, and management options. Int J Womens Health 2015;7:305–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allo G, Yap ML, Cuartero J, Milosevic M, Ferguson S, Mackay H, et al. HPV-independent Vulvar Squamous Cell Carcinoma is Associated With Significantly Worse Prognosis Compared With HPV-associated Tumors. Int J Gynecol Pathol 2020;39(4). [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 2015;31(2):166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansink AC, Krul MR, De Weger RA, Kleyne JA, Pijpers H, Van Tinteren H, et al. Human papillomavirus, lichen sclerosus, and squamous cell carcinoma of the vulva: detection and prognostic significance. Gynecol Oncol 1994;52(2):180–4. [DOI] [PubMed] [Google Scholar]
- Arbyn M, de Sanjosé S, Saraiya M, Sideri M, Palefsky J, Lacey C, et al. EUROGIN 2011 roadmap on prevention and treatment of HPV-related disease. Int J Cancer 2012;131(9):1969–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow EL, Lambie N, Donoghoe MW, Naing Z, Hacker NF. The Clinical Relevance of p16 and p53 Status in Patients with Squamous Cell Carcinoma of the Vulva. Journal of Oncology 2020;2020:3739075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berek JS, Karam A. Vulvar Cancer: Epidemiology, diagnosis, histopathology, and treatment, https://www.uptodate.com/contents/vulvar-cancer-epidemiology-diagnosis-histopathology-and-treatment?search=vulvar-cancer&source=search_result&selectedTitle=1~92&usage_type=default&display_rank=1; 2021. [accessed May 2, 2021].
- Bi J, Liu H, Cai Z, Dong W, Jiang N, Yang M, et al. Circ-BPTF promotes bladder cancer progression and recurrence through the miR-31–5p/RAB27A axis. Aging 2018;10(8):1964–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierkens M, Krijgsman O, Wilting SM, Bosch L, Jaspers A, Meijer GA, et al. Focal aberrations indicate EYA2 and hsa-miR-375 as oncogene and tumor suppressor in cervical carcinogenesis. Genes, Chromosomes and Cancer 2013;52(1):56–68. [DOI] [PubMed] [Google Scholar]
- Blitzer GC, Smith MA, Harris SL, Kimple RJ. Review of the clinical and biologic aspects of human papillomavirus-positive squamous cell carcinomas of the head and neck. Int J Radiat Oncol Biol Phys 2014;88(4):761–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan TR, Graybill WS, Pierce JY. Morbidity and mortality of vulvar and vaginal cancers: Impact of 2-, 4-, and 9-valent HPV vaccines. Human Vaccines & Immunotherapeutics 2016;12(6):1352–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canavan TP, Cohen D. Vulvar Cancer. Am Fam Physician 2002;66(7):1269–75. [PubMed] [Google Scholar]
- Chamseddin BH, Lee EE, Kim J, Zhan X, Yang R, Murphy KM, et al. Assessment of circularized E7 RNA, GLUT1, and PD-L1 in anal squamous cell carcinoma. Oncotarget 2019;10(57):5958–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choschzick M, Gut A, Fink D. PD-L1 receptor expression in vulvar carcinomas is HPV-independent. Virchows Arch 2018;473(4):513–6. [DOI] [PubMed] [Google Scholar]
- Dasgupta S, Ewing-Graham PC, Swagemakers SMA, van der Spek PJ, van Doorn HC, Noordhoek Hegt V, et al. Precursor lesions of vulvar squamous cell carcinoma – histology and biomarkers: A systematic review. Crit Rev Oncol Hematol 2020;147:102866. [DOI] [PubMed] [Google Scholar]
- del Pino M, Rodriguez-Carunchio L, Ordi J. Pathways of vulvar intraepithelial neoplasia and squamous cell carcinoma. Histopathology 2013;62(1):161–75. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Liu M, Liu A, Cui N, Chen Z, Yang Q, et al. Depletion of Circular RNA circ_CORO1C Suppresses Gastric Cancer Development by Modulating miR-138–5p/KLF12 Axis. Cancer Manag Res 2021;13:3789–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargano JW, Wilkinson EJ, Unger ER, Steinau M, Watson M, Huang Y, et al. Prevalence of human papillomavirus types in invasive vulvar cancers and vulvar intraepithelial neoplasia 3 in the United States before vaccine introduction. Journal of lower genital tract disease 2012;16(4):471–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004;5(10):R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani Giulia, Fragomeni SM, Inzani F, Fagotti A, Della Corte L, Gentileschi S, et al. Molecular pathways in vulvar squamous cell carcinoma: implications for target therapeutic strategies. Journal of cancer research and clinical oncology 2020;146(7):1647–58. [DOI] [PubMed] [Google Scholar]
- Hald AK, Blaakaer J. The possible role of human papillomavirus infection in the development of lichen sclerosus. Int J Dermatol 2018;57(2):139–46. [DOI] [PubMed] [Google Scholar]
- Hecking T, Thiesler T, Schiller C, Lunkenheimer J-M, Ayub TH, Rohr A, et al. Tumoral PD-L1 expression defines a subgroup of poor-prognosis vulvar carcinomas with non-viral etiology. Oncotarget 2017;8(54):92890–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- How JA, Jazaeri AA, Soliman PT, Fleming ND, Gong J, Piha-Paul SA, et al. Pembrolizumab in vaginal and vulvar squamous cell carcinoma: a case series from a phase II basket trial. Scientific Reports 2021;11(1):3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45(D1):D353–D61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods 2012;9(4):357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell 2017;66(1):22–37 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England) 2009;25(16):2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Tan Y, Zhu L-X, Zhou L-N, Zeng P, Liu Q, et al. Prognostic value of HPV DNA status in cervical cancer before treatment: a systematic review and meta-analysis. Oncotarget 2017;8(39):66352–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Fan X, Zhu W, Huang C, Zhuang W, Xu H, et al. Prognostic significance of PD-L1 expression and tumor infiltrating lymphocyte in surgically resectable non-small cell lung cancer. Oncotarget 2017;8(48):83986–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longworth MS, Wilson R, Laimins LA. HPV31 E7 facilitates replication by activating E2F2 transcription through its interaction with HDACs. EMBO J 2005;24(10):1821–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M Circular RNA: functions, applications and prospects. ExRNA 2020;2(1):1. [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 2012;40(10):4288–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann RW, Hollebecque A, Meyer T, Devlin M-J, Oaknin A, Kerger J, et al. Safety and Efficacy of Nivolumab Monotherapy in Recurrent or Metastatic Cervical, Vaginal, or Vulvar Carcinoma: Results From the Phase I/II CheckMate 358 Trial. J Clin Oncol 2019;37(31):2825–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nooij LS, Brand FAM, Gaarenstroom KN, Creutzberg CL, de Hullu JA, van Poelgeest MIE. Risk factors and treatment for recurrent vulvar squamous cell carcinoma. Crit Rev Oncol Hematol 2016a;106:1–13. [DOI] [PubMed] [Google Scholar]
- Nooij LS, Dreef EJ, Smit VT, van Poelgeest MI, Bosse T. Stathmin is a highly sensitive and specific biomarker for vulvar high-grade squamous intraepithelial lesions. J Clin Pathol 2016b;69(12):1070–5. [DOI] [PubMed] [Google Scholar]
- Prieske K, Alawi M, Oliveira-Ferrer L, Jaeger A, Eylmann K, Burandt E, et al. Genomic characterization of vulvar squamous cell carcinoma. Gynecol Oncol 2020;158(3):547–54. [DOI] [PubMed] [Google Scholar]
- Rakislova N, Saco A, Sierra A, del Pino M, Ordi J. Role of Human Papillomavirus in Vulvar Cancer. Advances in Anatomic Pathology 2017;24(4). [DOI] [PubMed] [Google Scholar]
- Rasmussen CL, Sand FL, Hoffmann Frederiksen M, Kaae Andersen K, Kjaer SK. Does HPV status influence survival after vulvar cancer? Int J Cancer 2018;142(6):1158–65. [DOI] [PubMed] [Google Scholar]
- Reddy OL, Shintaku PI, Moatamed NA. Programmed death-ligand 1 (PD-L1) is expressed in a significant number of the uterine cervical carcinomas. Diagnostic Pathology 2017;12(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26(1):139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers LJ, Cuello MA. Cancer of the vulva. Int J Gynaecol Obstet 2018;143(S2):4–13. [DOI] [PubMed] [Google Scholar]
- Rooper LM, Gandhi M, Bishop JA, Westra WH. RNA in-situ hybridization is a practical and effective method for determining HPV status of oropharyngeal squamous cell carcinoma including discordant cases that are p16 positive by immunohistochemistry but HPV negative by DNA in-situ hybridization. Oral Oncol 2016;55:11–6. [DOI] [PubMed] [Google Scholar]
- Tawe L, Grover S, Narasimhamurthy M, Moyo S, Gaseitsiwe S, Kasvosve I, et al. Molecular detection of human papillomavirus (HPV) in highly fragmented DNA from cervical cancer biopsies using double-nested PCR. MethodsX 2018;5:569–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier-Cloutier B, Pors J, Thompson E, Ho J, Prentice L, McConechy M, et al. Molecular characterization of invasive and in situ squamous neoplasia of the vulva and implications for morphologic diagnosis and outcome. Mod Pathol 2020. [DOI] [PubMed] [Google Scholar]
- Thangarajah F, Morgenstern B, Pahmeyer C, Schiffmann LM, Puppe J, Mallmann P, et al. Clinical impact of PD-L1 and PD-1 expression in squamous cell cancer of the vulva. J Cancer Res Clin Oncol 2019;145(6):1651–60. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu Y, Liu H, Zhang Q, Song H, Tang J, et al. FcGBP was upregulated by HPV infection and correlated to longer survival time of HNSCC patients. Oncotarget 2017;8(49):86503–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weberpals JI, Lo B, Duciaume MM, Spaans JN, Clancy AA, Dimitroulakos J, et al. Vulvar Squamous Cell Carcinoma (VSCC) as Two Diseases: HPV Status Identifies Distinct Mutational Profiles Including Oncogenic Fibroblast Growth Factor Receptor 3. Clinical Cancer Research 2017;23(15):4501. [DOI] [PubMed] [Google Scholar]
- Woelber L, Prieske K, Eulenburg C, Oliveira-Ferrer L, de Gregorio N, Klapdor R, et al. p53 and p16 expression profiles in vulvar cancer: a translational analysis by the Arbeitsgemeinschaft Gynäkologische Onkologie Chemo and Radiotherapy in Epithelial Vulvar Cancer study group. Am J Obstet Gynecol 2021. [DOI] [PubMed] [Google Scholar]
- Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. Journal of the National Cancer Institute 2018;110(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao JN, Zhang XX, Zhang YZ, Li YL, Wang CF, Zhang LF. [Circular RNA-UBXN7 promotes proliferation, migration and suppresses apoptosis in hepatocellular cancer]. Zhonghua gan zang bing za zhi = Zhonghua ganzangbing zazhi = Chinese journal of hepatology 2020;28(5):421–7. [DOI] [PubMed] [Google Scholar]
- Yao Z, Luo J, Hu K, Lin J, Huang H, Wang Q, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Molecular oncology 2017;11(4):422–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapardiel I, Iacoponi S, Coronado PJ, Zalewski K, Chen F, Fotopoulou C, et al. Prognostic factors in patients with vulvar cancer: the VULCAN study. International Journal of Gynecologic Cancer 2020;30(9):1285. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang Y, Zhang Z. Prevalence of human papillomavirus and its prognostic value in vulvar cancer: A systematic review and meta-analysis. PloS one 2018;13(9):e0204162–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Lee EE, Kim J, Yang R, Chamseddin B, Ni C, et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nature communications 2019;10(1):2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nature communications 2016;7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Qi Y, Wu J, Zhou FL, Yu H, Li L, et al. CircPCMTD1 Acts as the Sponge of miR-224–5p to Promote Glioma Progression. Front Oncol 2019;9:398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zięba S, Chechlińska M, Kowalik A, Kowalewska M. Genes, pathways and vulvar carcinoma - New insights from next-generation sequencing studies. Gynecol Oncol 2020a;158(2):498–506. [DOI] [PubMed] [Google Scholar]
- Zięba S, Kowalik A, Zalewski K, Rusetska N, Goryca K, Piaścik A, et al. Somatic mutation profiling of vulvar cancer: Exploring therapeutic targets. Gynecol Oncol 2018;150(3):552–61. [DOI] [PubMed] [Google Scholar]
- Zięba S, Pouwer AW, Kowalik A, Zalewski K, Rusetska N, Bakuła-Zalewska E, et al. Somatic Mutation Profiling in Premalignant Lesions of Vulvar Squamous Cell Carcinoma. Int J Mol Sci 2020b;21(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing available under accession number GSE183454.