Abstract
Patients with prurigo nodularis (PN) suffer from intractable itch and dramatic reduction in quality of life. While there is significant clinical heterogeneity in the presentation of PN, disease endotypes remain unknown. We assayed circulating plasma cytokine concentrations in PN patients (n=20) along with matched healthy controls and utilized an unsupervised machine learning algorithm to identify disease endotypes. We found two distinct clusters of PN patients with non-inflammatory (Cluster 1) and inflammatory (Cluster 2) plasma profiles. Cluster 2 had more African-Americans (82%, n=9 vs. 33%, n=3; P=0.028), higher worst-itch numeric rating scale scores (9.5±0.9 vs. 8.3±1.2; P=0.036), and lower quality of life as reflected by higher Dermatology Life Quality Index scores (21.9±6.4 vs. 13.0±4.1; P=0.015). In addition, Cluster 1 had a higher rate of myelopathy (67%, n=6 vs. 18%, n=2; P=0.028). Compared to Cluster 1, Cluster 2 had higher levels of IL-1α, IL-4, IL-5, IL-6, IL-10, IL-17A, IL-22, IL-25, and IFN-α. With population-level analysis, African-American PN patients had higher erythrocyte sedimentation rate, C-reactive protein, ferritin, eosinophils, and lower transferrin than Caucasian PN patients. These findings indicate discrete clusters of PN patients with plasma biomarker profiles corresponding to distinct demographic and clinical characteristics, potentially allowing for precision medicine approaches to treat PN.
INTRODUCTION
Prurigo nodularis (PN) is a chronic inflammatory skin disease featuring intensely pruritic nodules that are often symmetrically distributed on the extensor surfaces of the trunk and limbs (Kwatra, 2020). PN has a tremendous impact on quality of life and is associated with various psychiatric and systemic comorbidities, including cardiovascular disease, diabetes, malignancies, and chronic kidney disease (Boozalis et al., 2018; Huang et al., 2020; Larson et al., 2019; Whang et al., 2020; Sutaria et al., 2021; Whang et al., 2021; Wongvibulsin et al., 2021). The pathogenesis of PN is largely unknown; however, given its strong association with systemic inflammatory comorbidities, it is likely a systemic disease as well. Indeed, PN was recently found to feature a systemic Th22 signature with increased IL-22 expression by circulating CD4+ and CD8+ T-cells and increased γδT cells (Belzberg et al., 2021). This study also suggested trends in immune polarization based on racial endotypes. The heterogeneity of PN is further supported by significant variability in clinical presentation, nodule morphology, and variable responses to immunomodulatory treatments (Huang et al., 2020; Williams et al., 2020a; Williams et al., 2020b). There are currently no U.S. Food and Drug Administration approved therapies for PN, often making treatment challenging (Kwatra 2020; Williams et al., 2020a). The heterogeneity of PN suggests unique disease endotypes, which may facilitate precision medicine approaches for improved treatment.
In the current study, we aimed to better understand systemic disease biomarkers in PN patients by measuring circulating plasma cytokine levels of PN patients and validating these findings in a multicenter cohort of PN patients. Based on the systemic nature and clinical heterogeneity of PN, we hypothesized that discrete clusters of PN patients exist with variable degrees of systemic inflammation. In addition, we employed an unbiased machine learning approach to identify distinct clusters of PN patients based on biomarker profiles.
RESULTS
The demographics of PN (n=20) and control patients (n=15) followed similar age (54.1 ± 14.9 vs. 51.9 ± 14.1 years, P=0.764), sex (75% vs 73% female, P=0.911), and race (40% vs. 33% Caucasian, P=0.686) distributions (Supplementary Table S2). PN patients had a mean Worst Itch Numeric Rating Scale (WI-NRS) of 9.0 ± 1.2 and a mean Investigator’s Global Assessment (IGA) of 3.3 ± 0.7.
Clustering of PN patients
The mean silhouette width was maximized at two, indicating that the optimal number of clusters was two (Figure 1a). The Gower dissimilarity matrix showed two groups of PN patients with different plasma biomarker profiles (Figure 1b). Implementing the Partitioning Around Medoids algorithm on the dissimilarity matrix resulted in two clusters of 9 (Cluster 1) and 11 patients (Cluster 2). A heatmap of biomarker values revealed that Cluster 2 had higher plasma concentrations for most biomarkers (Figure 1c). The Jaccard stability indices of the two clusters were 0.81 and 0.85, where an index of 0.75 to 0.85 indicates a stable and reliable cluster. Representative images of patients from the two clusters can be found in Figure 2.
Figure 1.
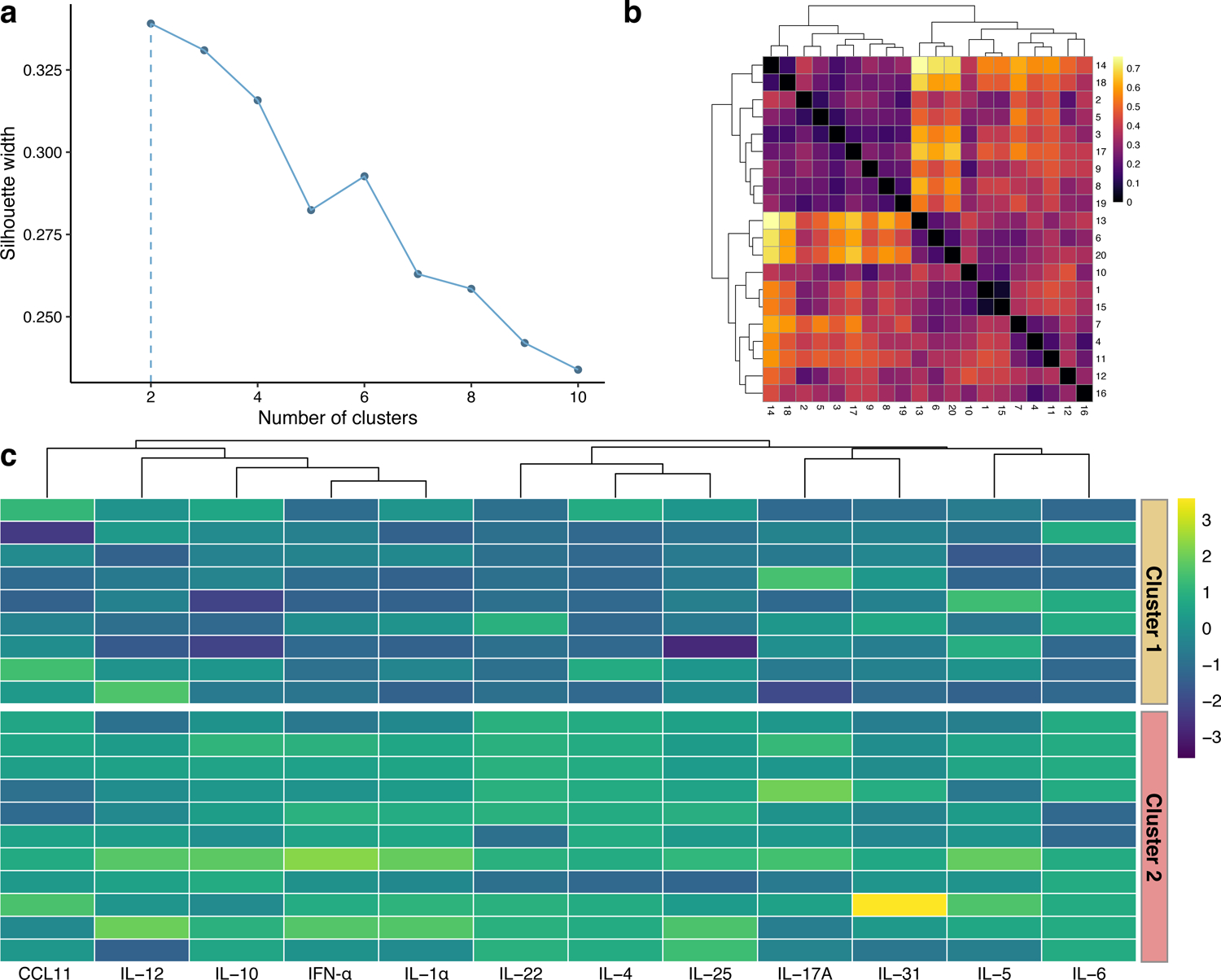
Clustering of prurigo nodularis (PN) patients. (a) Mean silhouette width for different numbers of clusters, with the peak occurring at 2 clusters. (b) Gower dissimilarity matrix of PN patients. (c) Heatmap of z-score transformed biomarker levels for each patient, delineated by cluster. Biomarkers were grouped with agglomerative hierarchical clustering.
Figure 2.
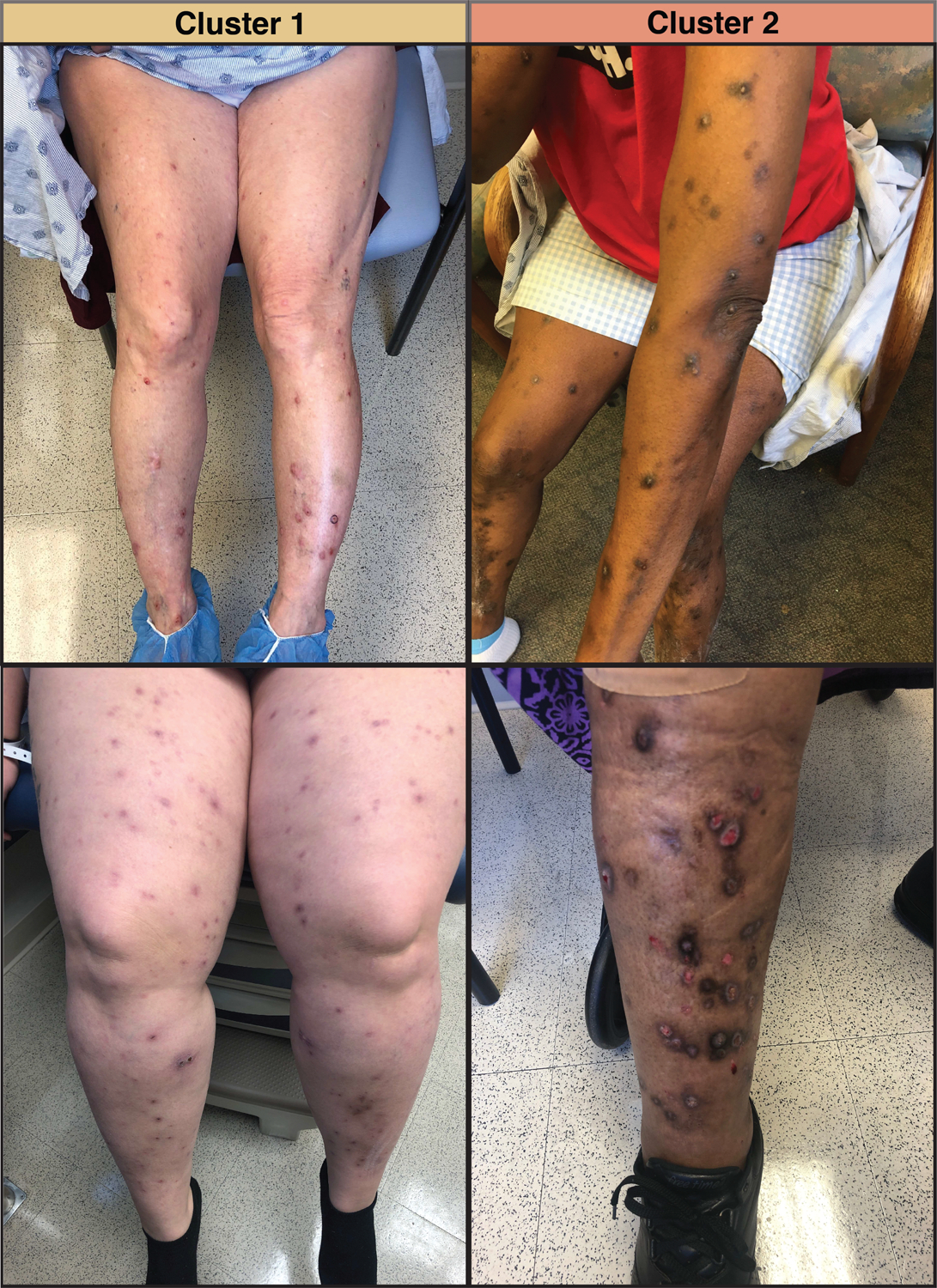
Representative images of prurigo nodularis lesions from two patients in Cluster 1 and two patients in Cluster 2.
Comparing the two PN clusters, Cluster 2 had higher interleukin (IL)-1α, IL-4, IL-5, IL-6, IL-10, IL-17A, IL-22, and IL-25 (Figure 3, Supplementary Table S3). Comparing the PN clusters to control patients, Cluster 2 had higher IL-12, IL-5, IL-17A, interferon (IFN)-α, IL-10, and IL-4 levels than controls, and Cluster 1 had lower IL-22 levels than controls (Figure 3, Supplementary Table S4).
Figure 3.
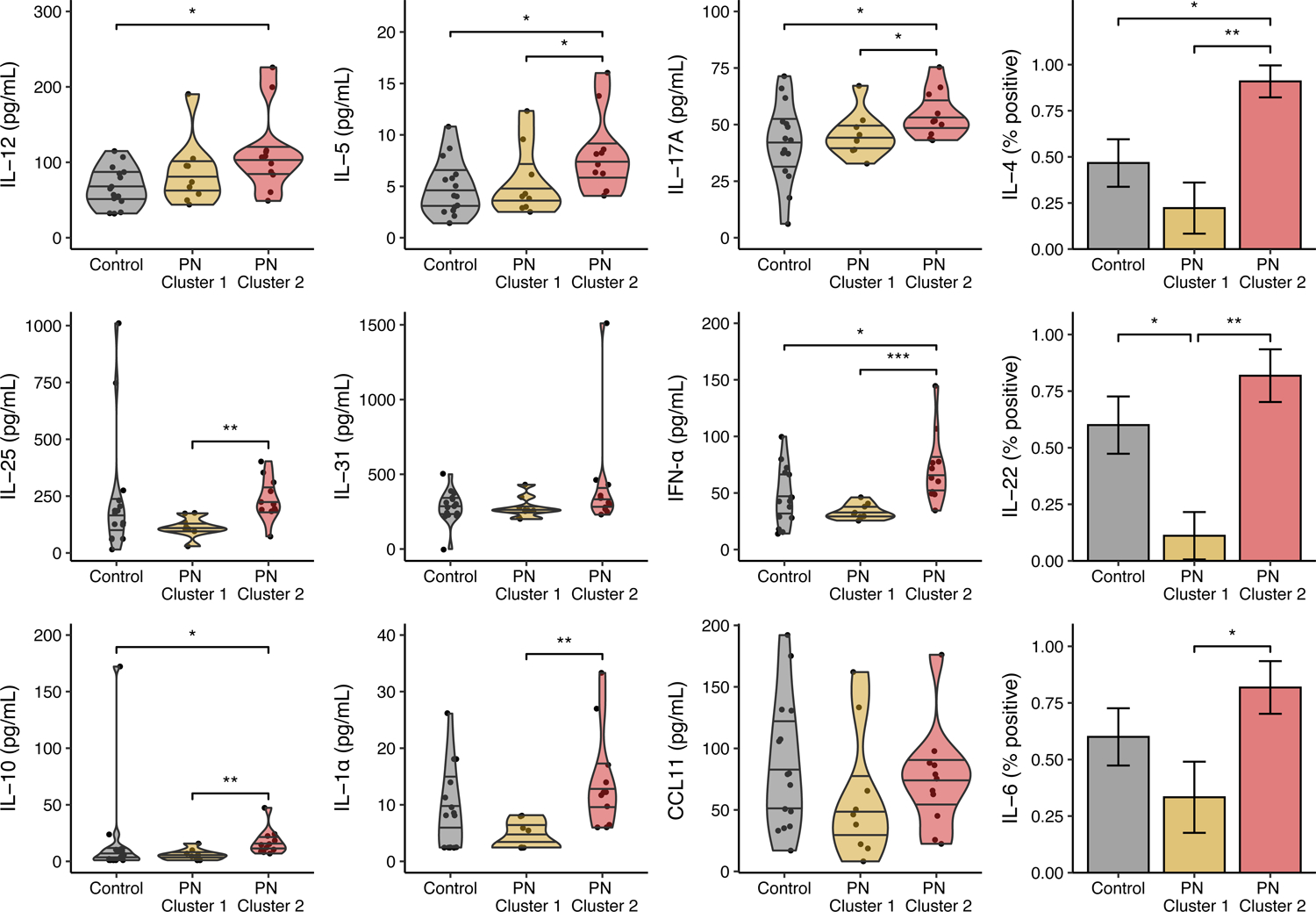
Plasma biomarker levels in each prurigo nodularis (PN) cluster and healthy controls. IL-4, IL-22, and IL-6 were dichotomized to binomial (positive/negative) variables using their detection limits as the cutoff. Biomarkers are expressed as violin plots with median ± interquartile range or bar plots with proportion ± standard error. PN – prurigo nodularis; * – P < 0.05; ** – P < 0.01; *** – P < 0.001.
Cluster 2 had a greater percentage of AA patients compared to Cluster 1 (82% vs. 33%, P=0.0227), with no significant difference in mean age or sex (Supplementary Table S5). Cluster 2 had a higher mean WI-NRS (9.5 ± 0.9 vs. 8.3 ± 1.2, P=0.0359) and Dermatology Life Quality Index (DLQI) (21.9 ± 6.4 vs. 13.0 ± 4.1, P=0.0152). Cluster 1 had a greater percentage of patients with myelopathy (67% vs. 18%, P=0.0277). There were no significant differences in IGA, Beck’s Depression Inventory scores, or history of atopy. Rates of inflammatory comorbidities and ongoing immuno- and neuro-modulating treatments at the time of blood draw were relatively equivalent between the two clusters (Supplementary Tables S5,S6).
Internal validation of PN clusters
For internal validation, principal component analysis (PCA) of plasma biomarkers in PN and healthy patients was performed. The majority of variance (63.3%) was explained by the first two principal components (Figure 4a). There were no major outliers after data cleaning and transformation (Figure 4b). A PCA plot of the first two principal components showed good cluster separation with no overlap (Figure 4c). All biomarkers trended in the same direction with respect to the first principal component (Figure 4d).
Figure 4.
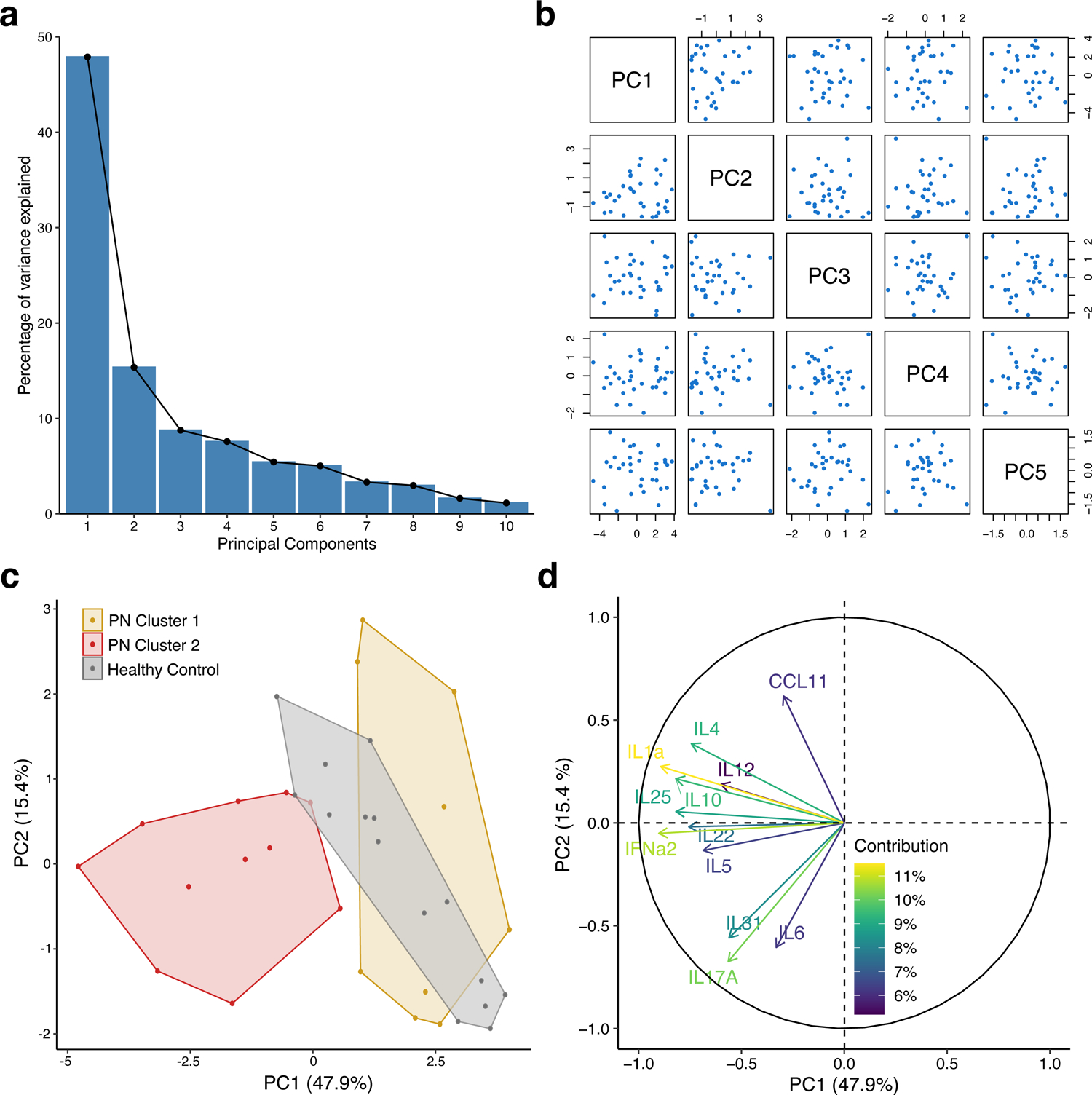
Internal validation of prurigo nodularis (PN) clusters with principal component analysis. (a) Scree plot of principal components. (b) Pairs plot of principal components. (c) Cluster plot PN and healthy patients on the first two principal components. (d) Loadings plot of biomarkers with percent contribution to the first principal component.
Population-level analysis
Based on the findings from our cohort, a population-level analysis of inflammatory markers in a multicenter PN cohort was performed (Figure 5). A total of 5,475 AA PN and 14,117 Caucasian PN patients were identified and matched to 5,475 AA control and 14,117 Caucasian control patients. Clinical lab values within 3 months of the index date were available for only a subset of these patients. Among AAs, PN patients had higher erythrocyte sedimentation rate (ESR) (P<0.0001, n=343), C-reactive protein (CRP) (P=0.0102, n=273), ferritin (P<0.0001, n=399), eosinophils (P<0.0001, n=2003), and alpha-1-antitrypsin (A1A) (P=0.0018, n=23), and lower transferrin (P=0.0031, n=203) compared to control patients. Among Caucasians, PN patients had higher ESR (P<0.001, n=1115), CRP (P=0.0002, n=873), and eosinophils (P<0.0001, n=4803) than control patients. AA PN patients had higher ESR (P<0.0001), CRP (P<0.0001), ferritin (P<0.0001), and eosinophils (P<0.0001), and lower transferrin (P=0.0028) compared to Caucasian PN patients.
Figure 5.
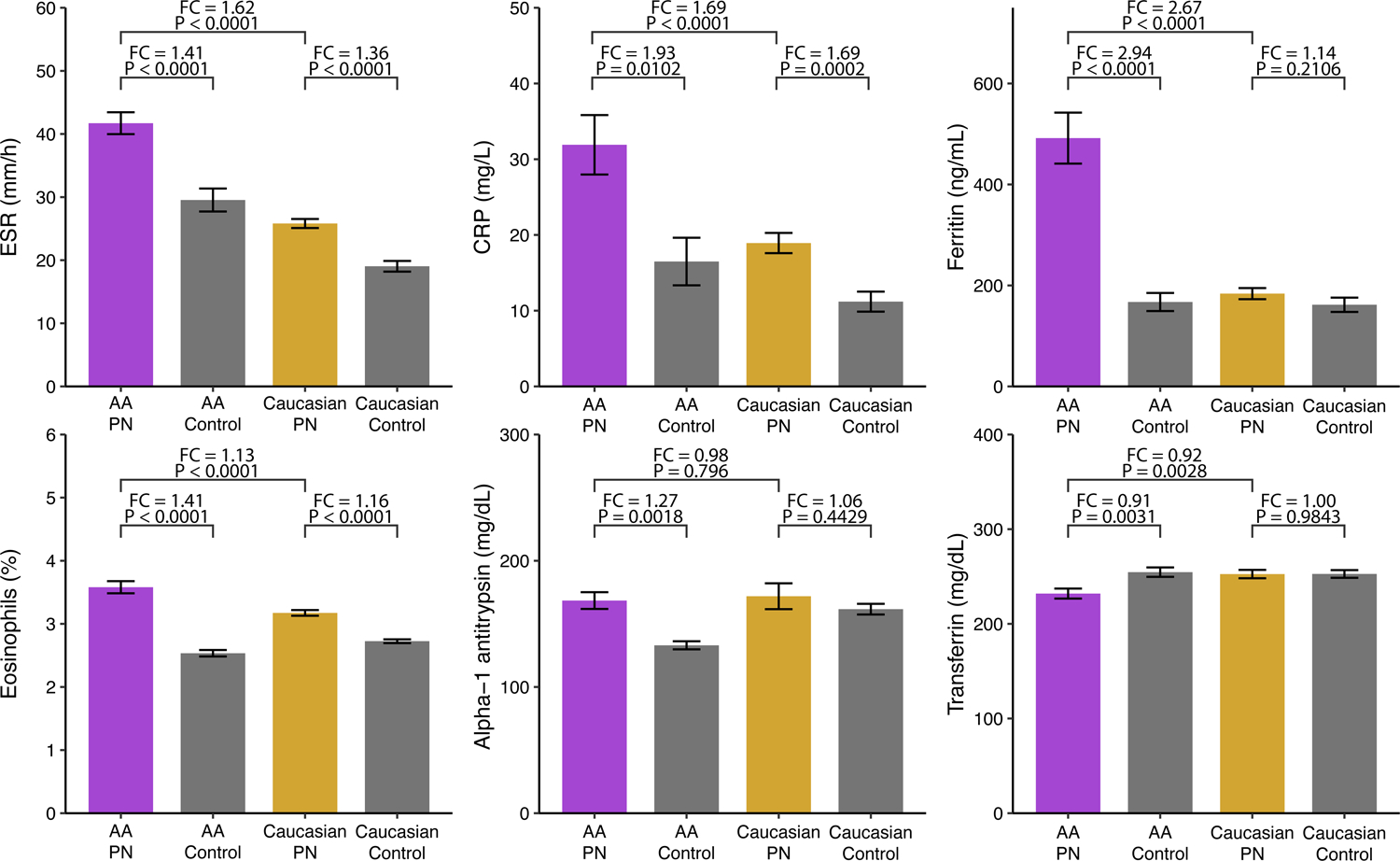
Population-level analysis of prurigo nodularis (PN) clusters using a multicenter PN cohort. Values for erythrocyte sedimentation rate (ESR) (AA: n=343, Caucasian: 1155), C-reactive protein (CRP) (AA: n=273, Caucasian: n=873), ferritin (AA: n=399, Caucasian: n=839), eosinophils (AA: n=2003, Caucasian: 4803), alpha-1-antitrypsin (A1A) (AA: n=23, Caucasian: 31), and transferrin (AA: n=203, Caucasian: 247) were compared between PN and 1:1 matched control patients.
DISCUSSION
PN has been described as a clinically heterogeneous disease, yet little is known about the underlying mechanisms responsible for its diverse phenotypes (Aggarwal et al., 2021; Huang et al., 2020). In this study, we describe heterogeneous plasma biomarker profiles in relation to the clinical presentations in PN patients. We utilized a machine learning approach to assess the levels of 12 biomarkers in PN patients and found two distinct clusters with differing biomarker profiles. In addition, we identified an inflammatory cluster that had more AA patients, greater itch intensity, and a larger effect on quality of life as measured by the DLQI.
Our study of PN patients found that Cluster 2 had a more inflammatory profile than Cluster 1. Nine biomarkers were significantly higher in Cluster 2, including IL-1α, IL-4, IL-5, IL-6, IL-10, IL-17A, IL-22, IL-25, and IFN-α, representing multiple immune axes including Th1, Th2, Th17, and Th22 (Saravia, 2019). In contrast, Cluster 1’s plasma profile was similar to control patients. Inflammatory cytokines, particularly those involved in the Th2, Th17, and Th22 axes, have also been implicated in the pathogenesis of other chronic inflammatory dermatoses, including psoriasis and cutaneous lichen planus (Baliwag et al., 2015; Pietschke et al., 2021). In addition to systemic findings of IL-22 polarization, our findings suggest significant clinical and molecular heterogeneity in PN, with the involvement of multiple immune axes (Belzberg et al., 2021). For example, IL-6 is an inflammatory cytokine that has been previously associated with PN and was also found to be elevated in this study (Konda et al., 2015). Additionally, elevated IL-1α may induce and exacerbate cutaneous inflammation in a variety of inflammatory skin diseases (Hanel et al., 2016; Jensen et al., 2010). Other studies have also identified IFN-α as a potential therapeutic target in inflammatory dermatoses such as AD and dermatomyositis (Klonowska et al., 2018; Kim et al., 2018). Differential levels of these biomarkers among PN patients may therefore highlight the heterogeneity of PN with variable levels of inflammation.
Assessing the demographic and clinical characteristics of the two clusters, Cluster 2 predominantly consisted of AAs, suggesting that this PN population may harbor greater systemic inflammation compared to Caucasian PN patients. Cluster 2 also had a higher mean WI-NRS and DLQI, reflecting a greater reduction in quality of life. Population-level analysis of AA and Caucasian PN patients demonstrated that AAs had greater levels of systemic inflammatory markers such as CRP and ESR and lower levels of anti-inflammatory markers, further reinforcing the hypothesis that AA PN is characterized by greater systemic inflammation. These molecular differences occur in the context of notable clinical differences in the presentation of prurigo nodularis, with lesions often appearing more nodular and firmer in African Americans (McColl et al., 2020).
Notably, Cluster 1 had a higher rate of myelopathy, suggesting a greater role for neural sensitization in these patients. Emerging evidence has suggested PN is neuroimmune mediated, with varying degrees of overexpression of downstream mediators of the Th1, Th2, and Th22 axes, as well as nerve growth factor (Belzberg et al., 2021; Fukushi et al., 2011; Williams et al., 2020a). In murine models, similar neurotrophic factors are elevated following spinal cord injury (Murakami et al., 2002). Therefore, these two clusters may represent distinct endotypes of PN with biases toward either immune or neural dysregulation. The clinical implications of multiple endotypes may guide further research on appropriate initial treatment with neural- or immune-modulating therapies. This strategy has proven effective in treating asthma, in which subgroups of patients with certain inflammatory marker levels respond best to different targeted therapies (Thijs et al., 2017).
Other studies have also demonstrated endotypes of diseases that may be tied to race. Clusters with distinct cytokine profiles correlating with unique clinical characteristics have been described for atopic dermatitis (AD), asthma, chronic obstructive pulmonary disease, and chronic rhinosinusitis (Thijs et al., 2017). Among AD patients, AA individuals are more likely to have increased papular morphology and a treatment-resistant lichenoid phenotype (Czarnowicki et al., 2019). In addition to Th2 polarization, prior molecular profiling studies have shown upregulation of Th17, and Th22-related markers in AA patients (Sanyal et al., 2019), which is linked to keratinocyte and epidermal hyperplasia (Fujita et al., 2013; Wongvibulsin et al., 2021). Although the PN patients in this study did not have concomitant AD, the observed racial patterns are consistent with those in AD. In addition, PN in AA patients frequently features larger, firmer nodules that are often more acanthotic and hyperkeratotic on histology (Huang et al., 2020). As we demonstrate in this study, these unique clinical and histopathological features may be due to greater systemic inflammation in AA patients. This differential systemic inflammation, along with unique molecular characteristics in black skin, may explain the epidemiological and morphological patterns of PN with respect to race (McColl et al., 2020). Regardless of the mechanism, however, the existence of endotypes in PN suggests that patients may benefit from personalized therapies with immunomodulating or neuromodulating treatments. Further neuroimmune phenotyping studies of PN may pave the way for a future precision medicine management approach.
Our findings confirm our original hypothesis that there exist discrete subgroups of PN with variable systemic inflammation. They also confirm our clinical observations of disease heterogeneity, with nodules in AA patients appearing larger, firmer, and often more recalcitrant to treatment. We avoided introducing bias from our clinical observations by utilizing an unsupervised machine model for our analysis. Such models can be advantageous when attempting to delineate disease subgroups without knowing the features that define them. Clinicians must be mindful of clinical heterogeneity in disease presentation and look towards unbiased analytical approaches to identify patterns that may uncover disease endotypes.
Limitations of this study include potential selection bias from recruiting patients from a tertiary medical center. As this was a real-world cohort, some patients were also on immuno-modulating and neuro-modulating treatments at the time of blood draw. The small sample size and observation of only AA and Caucasian patients also limits broad generalizations, particularly about other races. The cross-sectional design precludes inferences about causality. Finally, this biomarker panel may not fully capture the spectrum of circulating biomarkers involved in PN pathogenesis. Despite these limitations, we present two clusters of PN patients based on inflammatory and non-inflammatory plasma profiles that also had differing demographic and clinical characteristics. These findings highlight significant disease heterogeneity, implicating that there are distinct endotypes in PN that may enable precision medicine approaches to the treatment of PN.
MATERIALS AND METHODS
An overview of the study design can be found in Figure 6.
Figure 6.
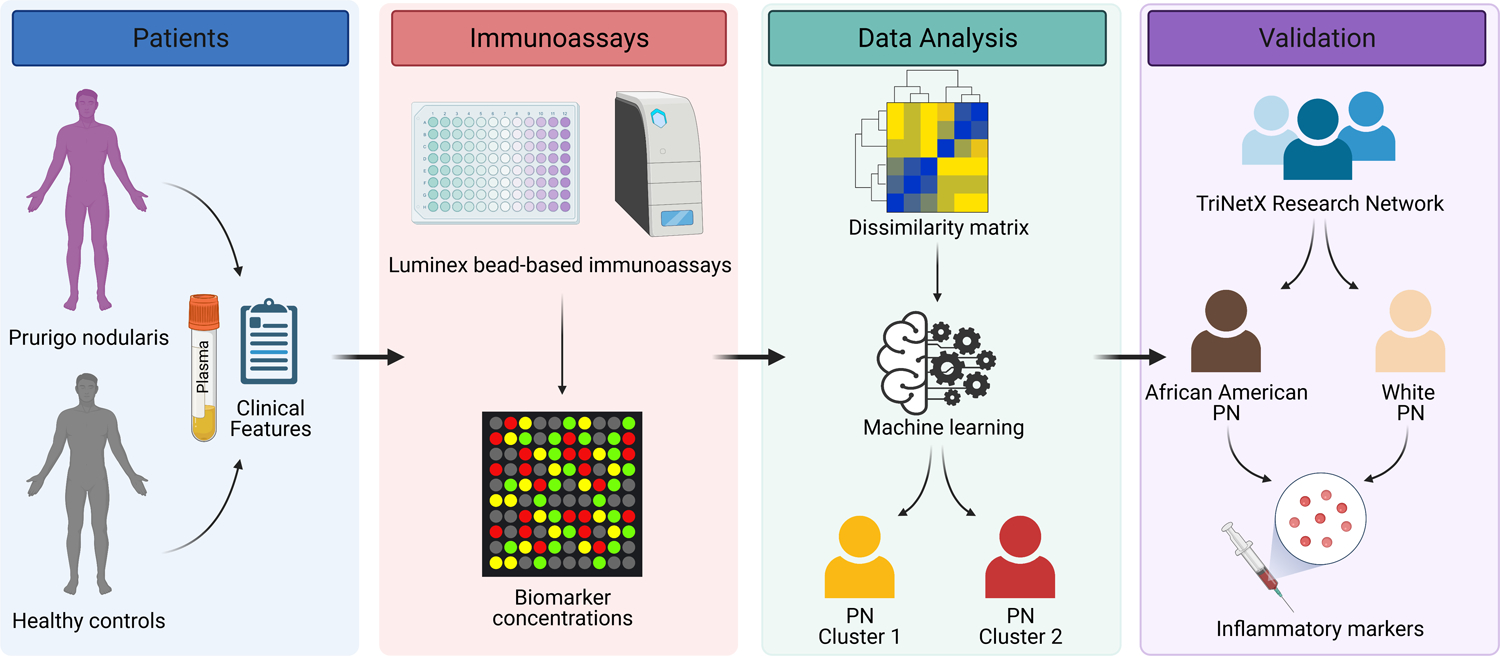
A schematic of the study design. Plasma was collected from prurigo nodularis (PN) and healthy control patients and assayed for a panel of inflammatory biomarkers. An unsupervised machine learning algorithm was used on the biomarker concentrations to identify distinct clusters of PN, which were validated using a large multi-center cohort.
Patient population
This study was approved by the Johns Hopkins Institutional Review Board (IRB00119007). A prospective study was performed comparing circulating plasma biomarkers from adult patients with PN and healthy controls. Inclusion criteria were modeled after previous clinical trials of PN, including diagnosis of PN by a board-certified dermatologist, at least 20 nodules on bilateral regions of the body, and a worst-itch numeric rating scale (WI-NRS) score of at least 7 (Ständer et al., 2020). Patients with clinically healthy skin and no pruritus were included as controls. Demographic and clinical characteristics of patients were collected at the time of blood draw. Severity was assessed with the Investigator’s Global Assessment (IGA) (Zeidler et al., 2021). History of atopy was defined as 2 out of 3 of the following atopic diatheses: asthma, allergic rhinitis, or AD/eczema (however, no patients had concurrent AD or signs of atopy at the time of blood draw). In addition, myelopathy (spinal stenosis, spinal trauma, degenerative disc disease, or disc herniation) was documented due to prior association of PN with a neuropathic etiology (Johansson et al., 2002; Williams et al., 2020a). Written, informed consent was obtained from each study participant after the nature and possible consequences of the studies had been explained.
Plasma biomarker measurement
The Bioplex 200 platform (Biorad, Hercules CA) was used to determine the concentration of multiple target proteins in the extracted specimens. Luminex bead-based immunoassays (Millipore, Billerica NY) were performed following Immune Monitoring Core SOPs, and concentrations were determined using 5 parameter log curve fits (using Bioplex Manager 6.0) with vendor-provided standards and quality controls. A broad range of 12 inflammatory biomarkers was measured, encompassing various roles in helper T-cell axes, pro-inflammatory, and chemotactic capabilities (Supplementary Table S1). The specific biomarkers were chosen based on existing literature demonstrating their dysregulation in PN and other inflammatory dermatoses, such as AD and psoriasis (Takahashi et al., 2010; Vakirlis et al., 2011).
Population-level analysis
We used TriNetX, a global federated health research network of electronic medical records from approximately 69 million patients in 49 healthcare organizations. Patients with PN were identified with the International Classification of Disease, Tenth Revision (ICD-10) code L28.1. The primary years of analysis were from 1995 to 2020, and diagnoses prior to the introduction of ICD-10 were mapped to ICD-10 by TriNetX using SNOMED-CT concepts (Donnelly, 2006). Only patients with ≥2 ICD-10 codes for PN were used for diagnostic accuracy. To further increase accuracy, we restricted the cohort to patients with at least one ICD-10 code after October 2015. Control patients were identified by searching for individuals with no diagnostic codes for PN. Among PN and control patients, labs for inflammatory markers were explored, including ESR, CRP, ferritin, eosinophils, A1A, and transferrin. Labs closest to the initial PN diagnosis date were used and restricted to ± 3 months from the diagnosis date to identify only clinically relevant values. To control for inherent racial differences in inflammatory markers, AA and Caucasian PN patients were compared to AA and Caucasian controls patients, respectively.
Statistical analysis
All statistical analyses were performed in R version 4.0.3. Power calculations were performed to determine the required sample size. Effect sizes were estimated based on prior studies investigating serum cytokines in AD and psoriasis (Takahashi et al., 2010; Vakirlis et al., 2011). With a power of 90%, two-tailed alpha of 0.05, and estimated effect size of 1.5 to 2 for most biomarkers, approximately 10 patients per comparator group would be required. This number was doubled to 20 to allow for comparisons between PN subgroups.
The cleaning/imputation, clustering, and analysis methods were adopted from a previous study investigating inflammatory endotypes of chronic rhinosinusitis (Tomassen et al., 2016). Briefly, biomarkers with >33% of samples below the detection limit (DL) were dichotomized into binomial variables with values below the DL as negative and values above the DL as positive (Supplementary Table S1). For all other biomarkers, values below the DL were imputed with half the DL.
The 12 biomarkers were then used for cluster analysis of PN patients. Continuous biomarkers were first natural log-transformed, and a Gower dissimilarity matrix was calculated, allowing input of both continuous and binomial biomarkers (Gower, 1971). The optimal number of clusters was determined using the silhouette method (Rousseeuw, 1987). The clusters were computed using the unsupervised machine learning algorithm Partitioning Around Medoids, which is more resistant to outliers than the k-means algorithm (Kaufman and Rousseeuw, 1990). Cluster reliability was assessed with the Jaccard stability index and bootstrap resampling (Hennig, 2007). Finally, PCA of PN and healthy patients was performed with log-transformed, scaled, and centered biomarkers.
For the population-level analysis, PN patients were matched to control patients by age, sex, and race using 1:1 propensity score matching. A greedy nearest neighbor matching algorithm with a caliper of 0.1 pooled standard deviations was used. Mean laboratory values were compared using the Student’s t-test. The normality of distributions was assumed under the Central Limit Theorem (Rosenblatt, 1956).
For all analyses, all continuous variables were compared using the Mann-Whitney U test, and categorical variables were compared using Pearson’s χ2 test.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr. Scott Zeger for critically reviewing the manuscript and methodology.
Funding Sources:
SGK is supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number K23AR077073. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Part of this work was also funded by grants to SGK from the Dermatology Foundation and Skin of Color Society.
Abbreviations:
- PN
prurigo nodularis
- AA
African American
- ESR
erythrocyte sedimentation rate
- CRP
C-reactive protein
- IGA
Investigator’s Global Assessment
- AD
atopic dermatitis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
IRB Status: Approved by the Johns Hopkins Institutional Review Board (IRB00119007).
CONFLICTS OF INTEREST
Dr. Kwatra is an advisory board member/consultant for Abbvie, Celldex Therapeutics, Galderma, Incyte Corporation, Pfizer, Regeneron Pharmaceuticals, and Kiniksa Pharmaceuticals and has served as an investigator for Galderma, Kiniksa Pharmaceuticals, Pfizer Inc., and Sanofi.
DATA AVAILABILITY
The datasets generated during the current study are available from the corresponding author on reasonable request.
REFERENCES
- Aggarwal P, Choi J, Sutaria N, Roh YS, Wongvibulsin S, Williams KA, Huang AH, Boozalis E, Le T, Chavda R, Gabriel S, Kwatra SG. Clinical characteristics and disease burden in prurigo nodularis. Clin Exp Dermatol. 2021. May 10. [DOI] [PubMed] [Google Scholar]
- Aleem S, Masood Q, Hassan I. Correlation of C-reactive protein levels with severity of chronic urticaria. Indian J Dermatol. 2014;59(6):636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzberg M, Alphonse MP, Brown I, Williams KA, Khanna R, Ho B, et al. Prurigo Nodularis Is Characterized by Systemic and Cutaneous T Helper 22 Immune Polarization. J Invest Dermatol. 2021; 23:S0022-202X(21)01016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boozalis E, Tang O, Patel S, Semenov YR, Pereira MP, Stander S, Kang S, Kwatra SG. Ethnic differences and comorbidities of 909 prurigo nodularis patients. J Am Acad Dermatol. 2018. Oct;79(4):714–719.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly K SNOMED-CT: The advanced terminology and coding system for eHealth. Stud Health Technol Inform. 2006;121:279–290. [PubMed] [Google Scholar]
- Fukushi S, Yamasaki K, Aiba S. Nuclear localization of activated STAT6 and STAT3 in epidermis of prurigo nodularis. Br J Dermatol. 2011. Nov;165(5):990–6. [DOI] [PubMed] [Google Scholar]
- Gono T, Kawaguchi Y, Hara M, Masuda I, Katsumata Y, Shinozaki M, et al. Increased ferritin predicts development and severity of acute interstitial lung disease as a complication of dermatomyositis. Rheumatology (Oxford). 2010;49(7):1354–60 [DOI] [PubMed] [Google Scholar]
- Gower J A general coefficient of similarity and some of its properties. Biometrics. 1971;27: 857–871 [Google Scholar]
- Hänel KH, Pfaff CM, Cornelissen C, Amann PM, Marquardt Y, Czaja K, et al. Control of the Physical and Antimicrobial Skin Barrier by an IL-31-IL-1 Signaling Network. J Immunol. 2016;196(8):3233–44. [DOI] [PubMed] [Google Scholar]
- Hennig C Cluster-wise assessment of cluster stability. Computational Statistics & Data Analysis. 2007;15;52(1):258–71. [Google Scholar]
- Huang AH, Williams KA, Kwatra SG. Prurigo nodularis: Epidemiology and clinical features. J Am Acad Dermatol. 2020;83(6):1559–1565 [DOI] [PubMed] [Google Scholar]
- Jensen LE. Targeting the IL-1 family members in skin inflammation. Curr Opin Investig Drugs. 2010;11(11):1211–20 [PMC free article] [PubMed] [Google Scholar]
- Johansson O, Liang Y, Emtestam L. Increased nerve growth factor-and tyrosine kinase A-like immunoreactivities in prurigo nodularis skin–an exploration of the cause of neurohyperplasia. Arch Dermatol Res. 2002;293(12), 614–619. [DOI] [PubMed] [Google Scholar]
- Kaufman L, Rousseeuw PJ. Partitioning around medoids (program pam). Finding groups in data: an introduction to cluster analysis. 1990;344:68–125. [Google Scholar]
- Kim HJ, Zeidi M, Bonciani D, Pena SM, Tiao J, Sahu S, et al. Itch in dermatomyositis: the role of increased skin interleukin-31. Br J Dermatol. 2018;179(3):669–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klonowska J, Gleń J, Nowicki RJ, Trzeciak M. New Cytokines in the Pathogenesis of Atopic Dermatitis-New Therapeutic Targets. Int J Mol Sci. 2018;19(10):3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konda D, Chandrashekar L, Rajappa M, Kattimani S, Thappa DM, Ananthanarayanan PH. Serotonin and interleukin-6: Association with pruritus severity, sleep quality and depression severity in Prurigo Nodularis. Asian J Psychiatr. 2015;17:24–8 [DOI] [PubMed] [Google Scholar]
- Kwatra SG. Breaking the Itch–Scratch Cycle in Prurigo Nodularis. N Engl J Med. 2020;382(8):757–758 [DOI] [PubMed] [Google Scholar]
- Larson VA, Tang O, Stander S, Miller LS, Kang S, Kwatra SG. Association between prurigo nodularis and malignancy in middle-aged adults. J Am Acad Dermatol. 2019. Nov;81(5):1198–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl M, Boozalis E, Aguh C, Eseonu AC, Okoye GA, Kwatra SG. Pruritus in Black Skin: Unique Molecular Characteristics and Clinical Features. J Natl Med Assoc. 2021;113(1):30–38 [DOI] [PubMed] [Google Scholar]
- Murakami Y, Furukawa S, Nitta A, & Furukawa Y. Accumulation of nerve growth factor protein at both rostral and caudal stumps in the transected rat spinal cord. J Neurol Sci. 2002;198(1–2), 63–69. [DOI] [PubMed] [Google Scholar]
- Punzi L, Pianon M, Rossini P, Schiavon F, Gambari PF. Clinical and laboratory manifestations of elderly onset psoriatic arthritis: a comparison with younger onset disease. Ann Rheum Dis. 1999;58(4):226–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt M A central limit theorem and a strong mixing condition. Proceedings of the National Academy of Sciences of the United States of America. 1956. Jan;42(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseeuw PJ. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. Computational and Applied Mathematics. 1987;20:53–65. [Google Scholar]
- Thijs JL, Strickland I, Bruijnzeel-Koomen CAFM, Nierkens S, Giovannone B, Csomor E, et al. Moving toward endotypes in atopic dermatitis: Identification of patient clusters based on serum biomarker analysis. J Allergy Clin Immunol. 2017;140(3):730–737 [DOI] [PubMed] [Google Scholar]
- Tomassen P, Vandeplas G, Van Zele T, Cardell LO, Arebro J, Olze H, et al. Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J Allergy Clin Immunol. 2016;137(5):1449–1456.e4. [DOI] [PubMed] [Google Scholar]
- Saravia J, Chapman NM, Chi H. Helper T cell differentiation. Cell Mol Immunol. 2019;16(7):634–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ständer S, Yosipovitch G, Legat FJ, Lacour JP, Paul C, Narbutt J, et al. Trial of Nemolizumab in Moderate-to-Severe Prurigo Nodularis. N Engl J Med. 2020;382(8):706–716. [DOI] [PubMed] [Google Scholar]
- Sutaria N, Choi J, Roh YS, Alphonse MP, Adawi W, Lai J, Pollock JR, Fontecilla Biles N, Gabriel S, Chavda R, Kwatra SG. Association of prurigo nodularis and infectious disease hospitalizations: a national cross-sectional study. Clin Exp Dermatol. 2021. Mar 24. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Tsuji H, Hashimoto Y, Ishida-Yamamoto A, Iizuka H. Serum cytokines and growth factor levels in Japanese patients with psoriasis. Clin Exp Dermatol. 2010;35(6):645–9 [DOI] [PubMed] [Google Scholar]
- Vakirlis E, Lazaridou E, Tzellos TG, Gerou S, Chatzidimitriou D, Ioannides D. Investigation of cytokine levels and their association with SCORAD index in adults with acute atopic dermatitis. J Eur Acad Dermatol Venereol. 2011;25(4):409–16 [DOI] [PubMed] [Google Scholar]
- Vekaria AS, Brunner PM, Aleisa AI, Bonomo L, Lebwohl MG, Israel A, et al. Moderate-to-severe atopic dermatitis patients show increases in serum C-reactive protein levels, correlating with skin disease activity. F1000Res. 2017;6:1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whang KA, Kang S, Kwatra SG. Inpatient Burden of Prurigo Nodularis in the United States. Medicines (Basel). 2019. Aug 11;6(3):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whang KA, Le TK, Khanna R, Williams KA, Roh YS, Sutaria N, Choi J, Gabriel S, Chavda R, Semenov Y, Kwatra SG. Health-related quality of life and economic burden of prurigo nodularis. J Am Acad Dermatol. 2021. May 28:S0190–9622(21)01028–8. [DOI] [PubMed] [Google Scholar]
- Williams KA, Huang AH, Belzberg M, Kwatra SG. Prurigo nodularis: Pathogenesis and management. J Am Acad Dermatol. 2020a;83(6):1567–1575 [DOI] [PubMed] [Google Scholar]
- Williams KA, Roh YS, Brown I, Sutaria N, Bakhshi P, Choi J, et al. Pathophysiology, diagnosis, and pharmacological treatment of prurigo nodularis. Expert Rev Clin Pharmacol. 2020b;14(1):67–77 [DOI] [PubMed] [Google Scholar]
- Wongvibulsin S, Sutaria N, Kannan S et al. Transcriptomic analysis of atopic dermatitis in African Americans is characterized by Th2/Th17-centered cutaneous immune activation. Sci Rep 11, 11175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongvibulsin S, Sutaria N, Williams KA, Huang AH, Choi J, Roh YS, et al. A Nationwide Study of Prurigo Nodularis: Disease Burden and Healthcare Utilization in the United States. J Invest Dermatol. 2021;S0022–202X(21)01130–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler C, Pereira MP, Augustin M, Spellman M, Ständer S. Investigator’s Global Assessment of Chronic Prurigo: A New Instrument for Use in Clinical Trials. Acta Derm Venereol. 2021;101(2):adv00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.


