Abstract
Background:
The regenerative capacity of the heart after myocardial infarction (MI) is limited. Our previous study showed that ectopic introduction of Cdk1/CyclinB1 and Cdk4/CyclinD1 complexes (4F) promotes cardiomyocyte proliferation in 15-20% of infected cardiomyocytes in vitro and in vivo and improves cardiac function after MI in mice.
Methods:
Here, using temporal single-cell RNAseq we aimed to identify the necessary reprogramming stages during the forced cardiomyocyte proliferation with 4F on a single cell basis. Also, using rat and pig models of ischemic heart failure, we aimed to start the first preclinical testing to introduce 4F gene therapy as a candidate for the treatment of ischemia-induced heart failure.
Results:
Temporal bulk and single-cell RNAseq and further biochemical validations of mature hiPS-CMs treated with either LacZ or 4F adenoviruses revealed full cell cycle reprogramming in 15% of the cardiomyocyte population at 48 h post-infection with 4F, which was mainly associated with sarcomere disassembly and metabolic reprogramming (n=3/timepoint/group). Transient overexpression of 4F, specifically in cardiomyocytes, was achieved using a polycistronic non-integrating lentivirus (NIL) encoding the 4F; each is driven by a TNNT2 promoter (TNNT2-4Fpolycistronic-NIL). TNNT2-4Fpolycistronic-NIL or control virus was injected intramyocardially one week after MI in rats (n=10/group) or pigs (n=6-7/group). Four weeks post-injection, TNNT2-4Fpolycistronic-NIL treated animals showed significant improvement in left ventricular ejection fraction and scar size compared with the control virus treated animals. At four months after treatment, rats that received TNNT2-4Fpolycistronic-NIL still showed a sustained improvement in cardiac function and no obvious development of cardiac arrhythmias or systemic tumorigenesis (n=10/group).
Conclusions:
This study provides mechanistic insights into the process of forced cardiomyocyte proliferation and advances the clinical feasibility of this approach by minimizing the oncogenic potential of the cell cycle factors thanks to the use of a novel transient and cardiomyocyte-specific viral construct.
Keywords: Gene therapy, cell cycle, heart failure, cardiomyopathy, metabolism, sarcomere
Introduction
The mammalian cell cycle involves numerous feedback loops that either permit or prevent cell division depending on the cell type1, 2. Although fetal myocytes proliferate to achieve cardiac growth and tissue-specific stem cells undergo cytokinesis postnatally, differentiated cells typically become post-mitotic and permanently exit the cell cycle3. As a result, the regenerative capacity of most organs, including the heart, is limited. The ability to control proliferation in the postnatal heart would represent a powerful approach to promote cardiac repair after infarction.
Recently, we took a combinatorial approach to screen for factors and conditions that could recapitulate the fetal state of cardiomyocyte proliferative activity. We found that ectopic introduction of the Cdk1/CyclinB1 and the Cdk4/CyclinD1 complexes (4F, i.e., four factors) promoted cell proliferation in at least 15% of mouse and human cardiomyocytes in vitro4. Rigorous assessment of cardiomyocyte cytokinesis in vivo using the Cre-recombinase dependent Mosaic Analysis with Double Markers (MADM)5 lineage tracing system revealed similar efficiency in mouse hearts, leading to cardiac regeneration upon delivery of the 4F one week after myocardial infarction4. Moreover, in vitro and in vivo results show that myocytes undergo only one round of division after transduction with these cell cycle factors because the overexpression of the 4F in cardiomyocytes is self-limiting through proteasome-mediated degradation of the protein products4. Interestingly, a recent study showed that AAV-mediated expression of microRNA-199a in infarcted pig hearts could initially stimulate cardiac repair through induction of cardiomyocyte proliferation; however, subsequent persistent and uncontrolled expression of the microRNA resulted in sudden arrhythmic death of most of the treated pigs6. Our approach is currently one of the most robust methods for inducing cardiomyocyte proliferation; however, the timing, dosage, and specificity of this therapy’s expression must be tightly controlled. Therefore, there is a need to transiently and specifically express 4F in cardiomyocytes to induce one cycle of proliferation to avoid any adverse effects in the heart and other tissues if escaped.
Uncertainty regarding the mechanisms underlying the functional improvement observed with cell-based therapies is a significant limitation of these approaches and led to skepticism about their clinical applicability (reviewed in7, 8). Therefore, prior to starting any translational efforts, we first sought to clearly delineate the cell cycle reprogramming events that occur in the cardiomyocytes following 4F cell cycle induction on transcriptomic levels at a single cell level before we moved on to test its clinical applicability in rodent and large animals. Here, we identified the need for sarcomeric disassembly and metabolic reprogramming during the cell cycle reprogramming process, especially at the mitotic phase of the cell cycle. These findings provide an essential foundation that enables one to ascribe subsequent cardiac functional improvements to the generation of new cardiomyocytes. Then, we provided the first proof-of-concept evidence for this approach’s clinical applicability of the cell cycle factors using a novel transient and cardiomyocyte-specific viral construct in rat and pig models of subacute heart failure.
Methods
We made our genomics data available to other researchers as all sequencing data are deposited in GEO. Bulk RNAseq from hiPS-CMs is under accession number: GSE174309 (reviewer access token (wdkfoyoijzsnziv)). Bulk RNAseq from P7 neonatal cardiomyocytes is under accession number: GSE184595 (reviewer access token (cxwbccwsbheptkh)). Single-cell RNAseq is under accession number: GSE162326 (reviewer access token (wnyrgaesbbadrix)).
Animal experiments
All animal procedures were in accordance with the institutional guidelines of the University of Louisville and approved Institutional Animal Care and Use Committee. Due to space limitations, please see Supplemental methods for detailed methods.
Statistical Analyses
For all assays, power analyses were performed to choose the group sizes, which will provide >80% power to detect a 10% absolute change in the parameter with a 5% Type I error rate. These power analyses indicated a minimum of 4 experimental replicates per group; therefore, we used a range of 4 – 15 experimental replicates per group for each assay. Special statistical consideration for RNAseq was detailed under the methods section. The Kolmogorov-Smirnov (K-S) test for normality was conducted; all data sets showed normal distribution. Then, differences between the two groups were examined for statistical significance with unpaired Student t-tests. However, to compare data consisting of more than two groups, we performed one- or Two-way ANOVA tests followed by Bonferroni post hoc multiple comparisons to get the corrected p-value. A value of P<0.05 was regarded as significant, and error bars indicate standard deviation (S.D.). The person who performed the analysis was blinded to the experimental groups. Two independent clinical cardiology fellows who were blinded to group assignments analyzed the echocardiography and MRI data, and the data represented are the average of their analyses. Usually, there were no significant discrepancies between the two readings.
Results
Temporal bulk and single-cell RNAseq of mature hiPS-CMs reveals that cell cycle reprogramming is associated with sarcomere disassembly and metabolic reprogramming during forced cell cycle induction
Previously, we reported that 4F induces at least 15% of mouse and human cardiomyocytes in vitro and in vivo to undergo proliferation within the first 48 h post-infection4. This high percentage of proliferating cells within the bulk population provided confidence that temporal bulk RNAseq could identify the significant transcriptional reprogramming events during the progress of cardiomyocyte through the cell cycle at a transcriptional level. Therefore, to investigate the transcriptional changes during cell cycle progression, we conducted temporal bulk RNAseq on 60-day-old mature hiPS-CMs treated with either LacZ (control) or 4F adenovirus for 24, 48, 72, or 144 h. Overexpression of the 4F induced several transcriptomic changes peaks at 48-72 h post-infection when proliferation is prominent, and they return to normal gene expression 144h post-infection with 4F as shown by the principal component analysis (PCA) (Fig. 1a & Source Data 1). Therefore, we focused our further analysis on the 48-72 h time points when the proliferation is prominent. Comparing the global gene expression at the 48 h time point between the 4F and LacZ control group, there are over 3700 genes that are differentially expressed, as demonstrated by the volcano plot (Fig. 1b & Source Data 1). The gene ontology (GO) analysis of the differentially expressed genes between the 4F-48-h group and the LacZ-48-h group showed significant upregulation of genes involved in cell cycle progression (Fig. 1c & Source Data 2). Furthermore, there is a time-dependent upregulation of cell cycle activation genes following 4F overexpression, which peaks at 48 and 72 h post-infection (Fig. 1d, Supplemental Fig 1a–b & Source Data 1–3). This cell cycle reprogramming was associated with transient downregulation of the sarcomeric and contractile genes at 48 h post-infection and started to decline back at 72 h and completely normalizes to control levels after 144 h (Fig. 2a–b, Supplemental Fig 1c & Source Data 1–3). These data suggest that cardiomyocytes need to withdraw from their primary function of contraction during the cell cycle to enter the cell cycle. Furthermore, bulk RNAseq from P7 neonatal mouse cardiomyocytes treated with either LacZ (control) or 4F adenovirus for 24, 48, or 72 h replicated the cell cycle reprogramming and contractile gene downregulation observed in hiPS-CMs (Supplemental Fig 2a–h & Source data 4–5).
Fig. 1. 4F overexpression induces cell cycle reprogramming in hiPS-CMs.
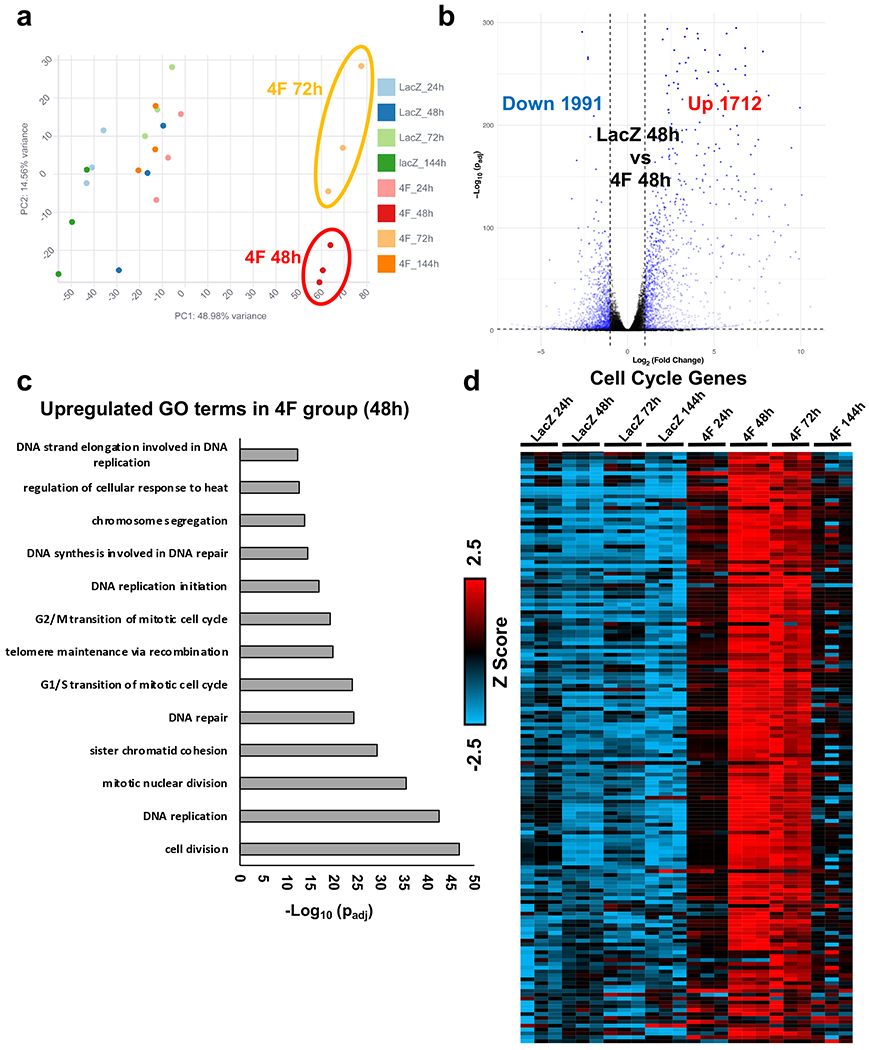
(a) Principal component analysis (PCA) of RNA-seq data from mature hiPS-CMs (60 day old) infected with either LacZ (control) or 4F for 24, 48, 72 or 144 h (n = 3). (b) Volcano plot demonstrating the number of genes that are significantly upregulated or downregulated 48 h post-4F adenovirus overexpression compared to the LacZ group (padj<0.05). (c) The bar graph shows the top GO terms for the significantly upregulated genes from RNA-seq data comparing hiPS-CMs infected with either LacZ or 4F for 48 h (padj<0.05). The GO terms reflect mostly upregulation of cell cycle genes. (d) Row normalized Z score heatmap shows a temporal expression of over 200 genes related to the cell cycle (n=3 in each sample). Source data 1 lists FPKM values for all RNAseq data and for the heart maps. Source data 2 contains the full list of the GO terms and the genes included in each GO term.
Fig. 2. Overexpression of 4F for 48 h induces sarcomere disassembly in hiPS-CMs.
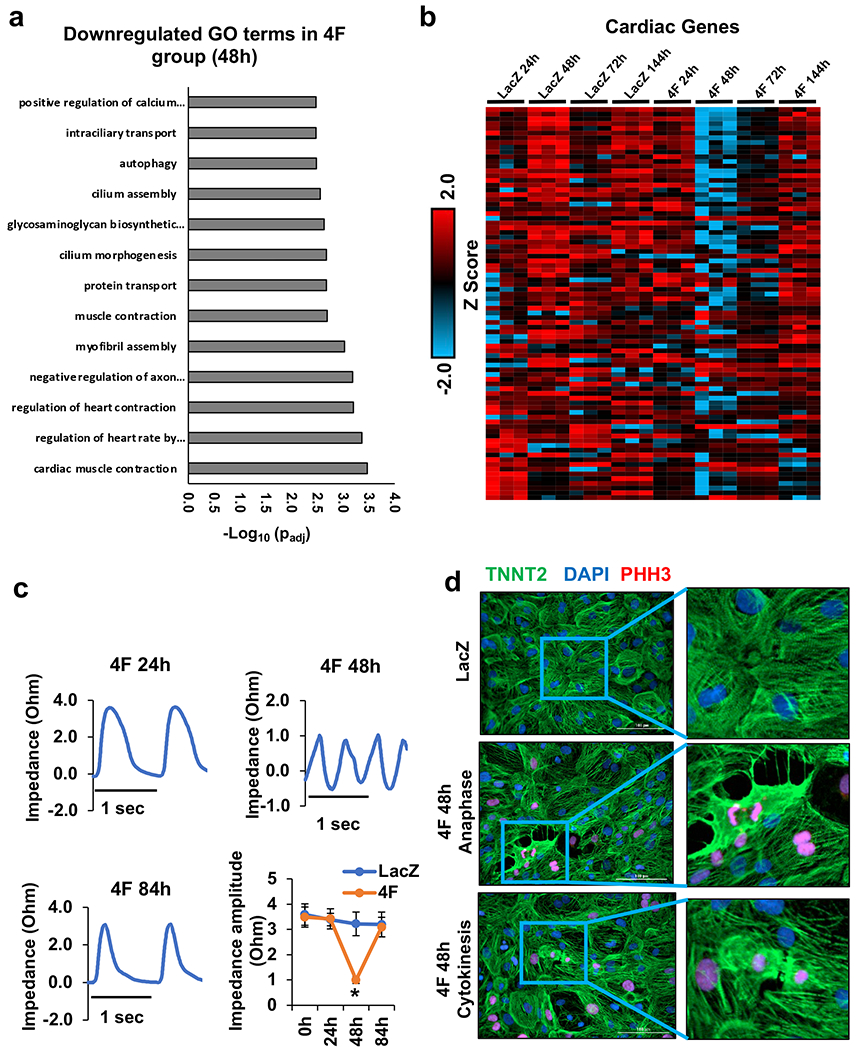
(a) The bar graph shows the top GO terms for the significantly downregulated genes from RNA-seq data comparing gene expression between the hiPS-CMs infected with either LacZ or 4F for 48 h (padj<0.05). The GO terms reflect the downregulation of cardiac contractile and sarcomeric genes. (b) Row normalized Z score heatmap shows the contractile and sarcomeric gene expression in a time-dependent manner following 4F expression (n=3 in each sample). Source data 1 lists FPKM values for all RNAseq data and for the heart maps. Source data 2 contains the full list of the GO terms and the genes included in each GO term. (c) Representative traces of impedance recording for contractile function from hiPS-CMs at 24, 48, and 84 h after infection with 4F. The bottom right panel shows the quantification of the contractile force amplitude comparing LacZ- and 4F-treated hiPS-CMs, 24, 48, and 84 h post-infection (n=3 independent experiments each in triplicate, *p<0.05 compared to LacZ treated cells). (d) representative images of sarcomere disassembly during G2/M phase in hiPS-CMs infected with lacZ (top panel), or 4F adenovirus for 48 h (middle (anaphase example) and bottom (cytokinesis example) panels) and stained with antibodies against the sarcomeric protein (troponin-T) (green), G2/M phase marker (PHH3) (red), and nuclear DAPI (blue).
These transcriptomics changes were functionally confirmed by using time-lapse impedance contractility tracing. We found that cardiomyocyte contractile force declined significantly during proliferation (48 h post-infection), which coincided with the appearance of arrhythmic episodes; normal contractile force and rhythm returned to baseline levels 84 h post-viral infection (Fig. 2c). Furthermore, during the G2/M phase, especially during anaphase or cytokinesis, the sarcomeric structures were disrupted, as shown by troponin-T Immunostaining of the sarcomeres (Fig. 2d). The impedance-based force measurements indicate the contractile force generated by the monolayer sheet of cardiomyocytes, which could have a mixed signal from proliferating and non-proliferating cardiomyocytes. Therefore, we investigated the sarcomeric disassembly implication on the electrophysiological properties and ion current in cardiomyocytes during proliferation. The decrease of the Scn5a transcript suggested a possible change in resulting INa, but the transient nature of the Scn5a decrease, along with the knowledge that transcript levels do not necessarily predict levels of functioning protein, motivated us to assess INa. Mean INa density tended to increase compared to LacZ (Supplemental Fig. 3a), but the difference between lacZ and the 4F 48 or 72 h time points was not significantly different. Cell capacitance was also not different (Supplemental Fig. 3b). We also assessed the voltage-dependence of steady-state inactivation because there is a development shift of the inactivation midpoint (V0.5) whereby V0.5 becomes more negative with embryonic heart maturation9. As with current density, there was no significant difference caused by 4F (Supplemental Fig. 3c), though there is a trend towards a more negative V0.5 (Supplemental Fig. 3d). These data suggest that 4F does not necessarily reduce excitability, and there is an unexpected tendency for larger current with more mature properties in cells treated with 4F for 48 and 72 h compared to control cardiomyocytes.
To further investigate in detail the transcriptional modifications during cardiomyocyte proliferation at a single-cell level, we conducted a temporal single-cell RNAseq of 60-day-old mature hiPS-CMs infected with either LacZ (control) for 48 h or 4F adenovirus for 24, 48, and 72 h. Gene expression data were collected from ~7000 cells/condition as summarized UMAP blots (Fig. 3a). All cells were positive for cardiac markers (TNNT2, TNNC1, and MYH7) (Fig. 3b), which indicates the purity of the hiPS-CMs population as they were selected after differentiation using an α-MHC-Blastocidin selection cassette. Most of the cardiomyocytes demonstrated high expression of the ventricular cardiomyocyte genes and were mostly negative for atrial markers (Supplemental Fig. 4 a–b). The lack of cell cycle activity of control hiPS-CMs was confirmed by the lack of expression of any cell cycle genes in the LacZ infected cells (Fig. 3c).
Fig. 3. Single-cell RNAseq identifies a unique mitotic subpopulation that appears at 48h post-infection with 4F.
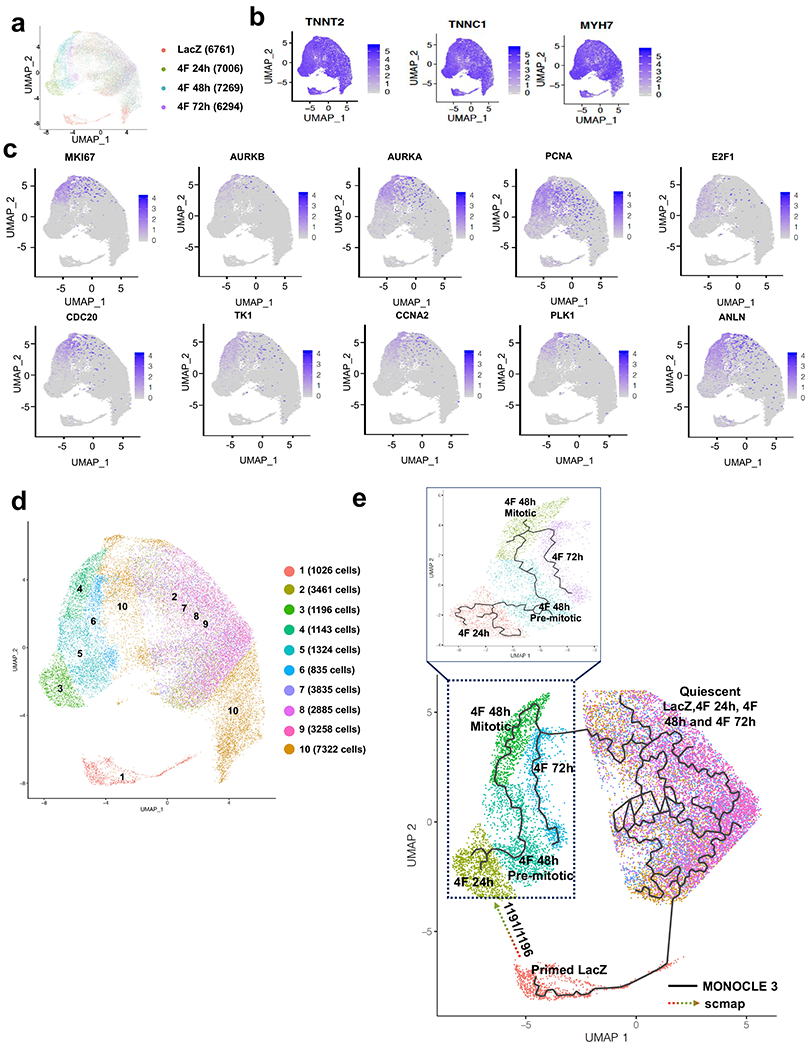
(a) UMAP plots considering global gene expression for all single cardiomyocytes sequenced from LacZ (control group) and 4F-overexpressing cardiomyocytes at 24, 48 h, or 72 h after viral transduction (approximately 7000 cells/sample). (b) All cells show high expression of the cardiac markers, TNNT2, TNNC1, and MYH7, indicating the cardiomyocytes’ purity. (c) A unique cell population appears only at 48 h post-infection with 4F, which express mitotic/cytokinesis genes; this population was identified as the mitotic population that expresses high levels of the G2/M markers (Ki67, Aurora Kinase A and B, PCNA, E2F1, CDC20, TK1, CCNA2, PLK1, and ANLN). (d) re-clustering the cells according to their expression of the mitotic genes and location of the subpopulation of cells associated with a given time-point in the UMAP space. Consistent with our published data, the mitotic population represents ~15% of the total cardiomyocyte population within the 48 h sample. (e) Trajectory analysis using MONOCLE (solid line) and scmap (dotted arrow) algorithms to predict the pseudo time trajectory of each unique population at each time point.
Interestingly, a unique cell population that appears 48 h after infection with the 4F expressed high levels of mitosis/cytokinesis genes (Ki67, Aurora Kinase A and B, E2F1, CDC20, ANLN, TK1, CCNA2, PLK1, and PCNA); this population was identified as the mitotic population (Fig. 3c). Consistent with our published data4, this population represents ~15% of the total cardiomyocyte population expressing 4F (Cluster 4, 1143/7269 cells) (Fig. 3d). Trajectory analysis using the MONOCLE algorithm for pseudo time trajectory suggested that the 24 h population (cluster 3) developed into the pre-mitotic 48 h subpopulation (cluster 5), which progresses to the 48 h mitotic subpopulation (cluster 4). Then cluster 4 progresses to the 72 hours mitotic population (cluster 6), which then projects back to the quiescent state (cluster 2, 7, 8, and 9). Finally, these quiescent cells could progress to cluster 1, which is the unique population in the LacZ group (Fig 3e). To predict the pseudo time projection of cluster 1 and the origin of cluster 3, we used scmap10 (a bioinformatics tool for unsupervised projection of single-cell RNA-seq data). Clusters 1 and 2 were used as the reference atlas, and cluster 3 as input data to be queried. If a cell in cluster 3 was assigned to cluster 1 based on similarity of gene expression, we assumed that it originated from cluster 1. The scmap trajectory analysis indicated that out of the 1195 cells in Cluster 3, 1191 cells have originated from Cluster 1 (Fig 3e). This was in line with the fact that Cluster 1 is the unique population of cardiomyocytes in the LacZ control group [1026 cells out of 6761 cells (~15%)] that disappears after treatment with 4F; and instead, Cluster 3 appears as a unique cluster with similar cell numbers that appears 24 h after 4F transduction. Comparison of the gene expression profiles of this primed population (cluster 1) with the quiescent cells (cluster 2) in the LacZ control sample shows significantly higher levels of 377 genes, many of which are involved in mitochondrial energy generation (Supplemental Fig 5a & Source data 6). These data suggest that cells with higher initial oxidative capacity and energy charge may be primed to proliferate, especially given that biosynthesis is an energy-demanding process11. While investigating the biological significance of these new bioinformatic analyses, we have found that one of the major characteristics of cluster 1 is the expression of CD36 (the fatty acid internalization receptor) (Supplemental Fig 5b) which could be involved in increasing the energetic capacity of the cells by internalizing extra fatty acids. Therefore, we attempted to knock down CD36 in hiPS-CMs; this significantly inhibited the 4F induced cell cycle activation (Supplemental Fig 5c–e). To confirm that CD36 is essential in priming primary cardiomyocytes to proliferate and that these findings are not specific for the hiPS-CMs, we used isolated P7 neonatal cardiomyocytes from CD36 knockout mice. These cardiomyocytes showed significantly less cell cycle activation following 4F cell cycle induction (Supplemental Fig 5f–h). These data suggest that cells with higher CD36 expression can be primed to proliferate.
We performed GO term analysis for the differentially expressed genes between each cluster across the pseudo time trajectory. These GO term analyses showed that cluster 3 (24h post-infection) compared to cluster 1 (LacZ starting population) exhibited differential regulation of the genes involved in ribosomal translational processes, which is likely necessary for translating new proteins to initiate the cell cycle and for building new daughter cells (Source data 7). Compared to cluster 3, cluster 5 (48h pre-mitotic subpopulation) has significantly higher expression of cell cycle genes and lower expression of the cardiac sarcomeric genes, which is evidence of the start of cell cycle engagement (Source data 8). Furthermore, a comparison of gene expression between pre-mitotic (Cluster 5) and mitotic subpopulations (Cluster 4) from the 4F 48 h samples revealed upregulation of the cell cycle genes and downregulation of sarcomeric gene expression in the mitotic subpopulation (Supplemental Fig 6a & Source data 9). Compared with the quiescent population (Cluster 8) from the same sample (48 h post-infection), this unique population of mitotic cardiomyocytes (Cluster 4) shows significant upregulation of the cell cycle program and significant downregulation of the sarcomeric genes as well as the mitochondrial electron transport chain genes (Supplemental Fig 6b & Source data 10). This is an indication of the metabolic shift that needs to occur during mitosis to downregulate catabolic activities and upregulate biosynthetic pathways, a process that was validated biologically in our recent publication12. Furthermore, this mitotic subpopulation of cells at 48 h (Cluster 4) showed higher cell cycle gene expression compared to its 72 h counterpart (Cluster 6) (Source data 11). Finally, there was no significant differential gene expression of genes between the four quiescent populations (clusters 2, 7, 8, and 9).
Transient expression of the 4F using non-integrating lentivirus system for gene therapy
The mechanistic insights gained from the temporal transcriptomics studies of the 4F-mediated induction of complete cell cycle reprogramming in cardiomyocytes encouraged us to standardize this approach to treat heart failure. Our previous studies using adenoviruses provided proof of principle for the efficacy of our approach4; however, the use of adenovirus is not clinically applicable because of the high prevalence of immune response in humans. Furthermore, the induction of long-term cell cycle activity in the heart may become oncogenic and have deleterious effects on the heart6. Therefore, we started to develop and optimize a strategy for 4F delivery and transient expression using a non-integrating lentivirus (NIL). NIL is known for its high infection efficiency and transient expression of the encoded protein, which is limited to 2-3 days, followed by a significant decline in expression13–16. Several ongoing clinical trials are aiming to treat various diseases with the lentivirus gene therapy approach17. This rapid degradation kinetics makes NIL an optimal delivery vehicle for the 4F. Additionally, when using NIL, it is important to target only cardiomyocytes by controlling the 4F expression with the cardiac-specific troponin-T promoter (TNNT2), which we previously used to track direct cardiac reprogramming18.
First, to assess the in vitro kinetics of the gene expression using the proposed viruses, we compared the ability of the CMV early enhancer/chicken beta-actin (CAG) promoter and the TNNT2 promoter to drive the expression of green fluorescent protein (GFP) within the context of integrating lentivirus (IL) and NIL over days in hiPS-CMs. hiPS-CMs infected with CAG-GFP-IL and CAG-GFP-NIL showed high infection efficiency and gene expression as early as 48 h post-infection. In contrast, hiPS-CMs infected with TNNT2-GFP-IL and TNNT2-GFP-NIL did not show high expression until 96 h post-infection, indicating the slower kinetics of the TNNT2 promoter compared to the CAG promoter. However, CAG-GFP-NIL or TNNT2-GFP-NIL kept the expression for only 72 h post-peak, consistent with previous reports13–16 (Supplemental Fig. 7a–c). Then, to investigate the specificity of the TNNT2 promoter to drive the protein expression only in cardiomyocytes, we infected non-cardiomyocyte cells (kidney cells, HEK293), mouse coronary artery endothelial cells (MCAEC), cardiac fibroblasts, and muscle cells (H9C2 and C2C12 myoblasts) with TNNT2-GFP-IL or CAG-GFP-IL (as a control). CAG-GFP-IL resulted in GFP expression over time in all cell types tested at 2, 4, 8 days post-infection; in contrast, TNNT2-GFP-IL did not show any significant GFP expression in any tested cell types (except the cardiomyocytes) up to eight days post-infection. These data suggest that the TNNT2-promoter drives the gene expression specifically in cardiomyocytes (Supplemental Fig. 7d–e).
Polycistronic TNNT2-4F-NIL induced cardiomyocyte proliferation in vitro and in vivo
Each cell cycle factor of the 4F was cloned into a lentivirus backbone under TNNT2 promoter (4F separate lentiviruses); also, all 4F were cloned in one polycistronic lentivirus backbone with each factor is driven by a TNNT2 promoter (4F polycistronic lentivirus) (Supplemental Fig. 8a). First, we assessed each cell cycle factor’s protein expression four days post-infection in hiPS-CMs using western blot. TNNT2-4Fpolycistronic-NIL was significantly more efficient in inducing higher protein expression of all the cell cycle factors compared to the TNNT2-4Fseparate-NIL (Supplemental Fig. 8b–c). Therefore, we only used TNNT2-4Fpolycistronic-NIL for further experiments. TNNT2-4Fpolycistronic-NIL showed 80-100% infection efficiency as assessed by immunofluorescence (Supplemental Fig. 8d–e). Four days post-infection with TNNT2-4Fpolycistronic-NIL, we found that 15-20% of the cardiomyocytes were positive for 5-ethynyl-2´-deoxyuridine (EDU), which marks new DNA synthesis and histone H3 phosphorylation (PHH3), which marks cells in the G2/M phase (Fig. 4a–b). Furthermore, the total cell number was increased by 20-30% (Fig. 4b). Assessments 10 days post-infection showed that cell number increase and EDU labeling for the divided nuclei persisted; however, the PHH3 was abolished, indicating the transient nature of the cell cycle induction in the cardiomyocyte and the likelihood that TNNT2-4Fpolycistronic-NIL induced only one cycle of proliferation.
Fig. 4. Transient expression of the 4F using non-integrating lentivirus (NIL) is efficient in inducing cardiomyocyte proliferation in vitro and in vivo.
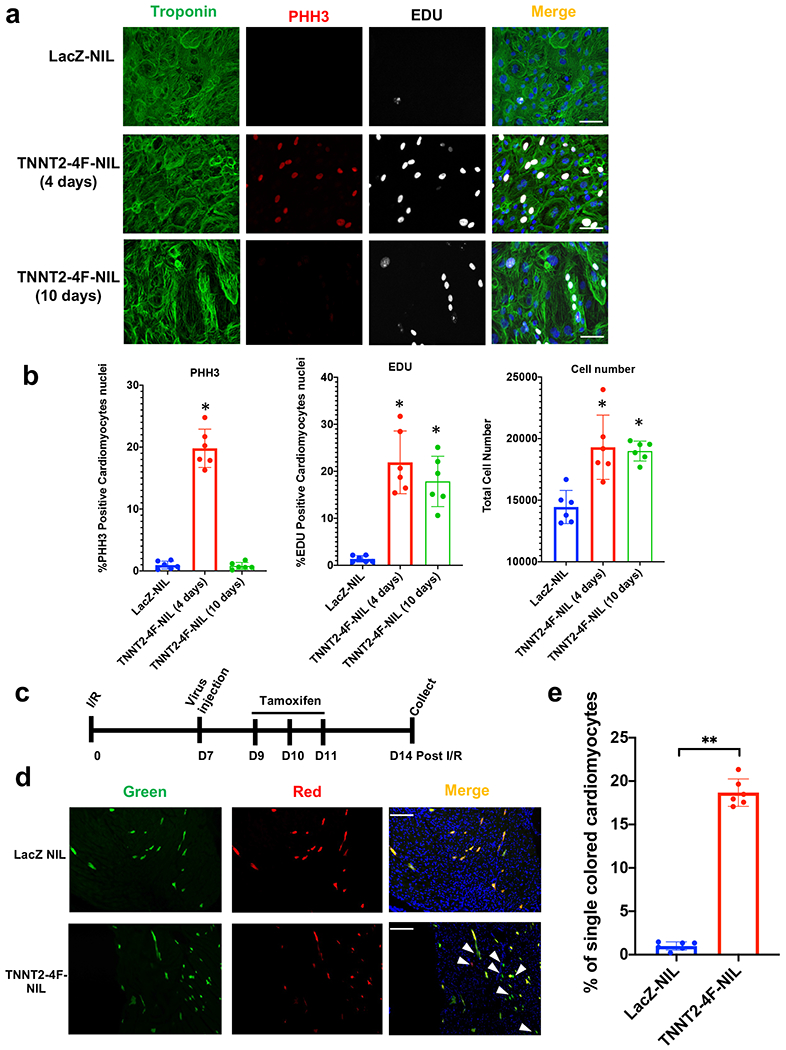
(a) Representative images of hiPS-CMs treated with LacZ-NIL, TNNT2-4Fpolycistronic-NIL for 4 or 10 days, and immuno-stained for troponin-T (green), PHH3 (red), EDU (gray) (scale bar=100μm). (b) Quantification of percentage increase in proliferation markers (PHH3 and EDU) and total cell number/well (n=6 independent experiments conducted in duplicate, *P<0.05 vs. LacZ NIL, error bars indicate S.D.). (c) Schematic diagram of the experimental design for MADM mice injection with the TNNT2-4Fpolycistronic-NIL or LacZ-NIL control. (d) Representative images show single-colored cardiomyocytes of MADM mice hearts treated with LacZ-NIL or TNNT2-4F-polycistronic-NIL (Scale bar=100μm). (e) Quantification of the percentage of the single-colored cells to the total labeled cells (n=6 animals per group, **p<0.01 vs. LacZ NIL, error bars indicate S.D.).
To test the efficacy of the TNNT2-4Fpolycistronic-NIL in inducing cardiomyocyte proliferation in vivo, we used a cardiomyocyte cytokinesis lineage-tracing animal model (inducible α-MHC-Cre::MADM-lineage-tracing)4, 5, 19, 20. In these lineage-tracing mice, cardiomyocytes that undergo cytokinesis produce daughter cells that are either red, green, yellow (red+green), or colorless, based on allelic recombination of fluorescent reporters; if the cardiomyocytes fail to divide, they will remain double-colored (i.e., yellow), or colorless if no recombination occurs. Thus, the presence of single-colored red or green cells definitively indicates cells that have undergone cytokinesis, although dividing cells are underrepresented by single-colored cells because double-colored (yellow) or colorless cells also could have divided. Animals were subjected to a 60-min occlusion of the left anterior descending artery followed by reperfusion, then one week later, TNNT2-4Fpolycistronic-NIL or LacZ-NIL (control) was injected intramyocardially (Fig. 4c). Tamoxifen injection was carried out as described in4, starting 48 h after the virus injection for three days to initiate recombination events. Mice were sacrificed one week after the viral injections, and hearts were sectioned to enumerate the cytokinesis events. All surgeries, imaging, and microscopy analyses were blinded about treatment, and animals were decoded after all data were analyzed. The analysis was done on ten different sections from each heart across the whole myocardium. After intramyocardial injection of TNNT2-4Fpolycistronic-NIL, it showed at least 15% of the recombinant cardiomyocytes were single-colored, compared to <1% in hearts injected with control virus (Fig. 4d–e).
TNNT2-4Fpolycistronic-NIL improves cardiac function in a rat model of subacute heart failure
Before starting in vivo functional studies, we sought to validate that TNNT2-NIL drives cardiomyocyte-specific expression of the 4F in vivo. Therefore, TNNT2-4Fpolycistronic-NIL or GFP-NIL control virus were injected intramyocardially at five different sites, and the rats were sacrificed six days post-injection. Western blotting and Immunostaining confirmed the expression of 4F in the rat hearts six days after injection (Supplemental Fig. 9a–d). Furthermore, RNA expressions of the overexpressed human CDK1, CDK4, CCNB, and CCND in rat hearts were only detected in the heart and not in the other organs six days post-viral injection (Supplemental Fig. 10a). It should be noted that for this experiment, each heart was split longitudinally into one half for immunofluorescence, one quarter for RNA extraction, and another quarter for protein extraction. The immunofluorescence in Supplemental Figure 9c–d shows the protein expression is localized at the injection sites; however, away from the injection sites, there is no expression. However, western blots demonstrate protein expression in a quarter of the heart where the expression of the over-expressed protein in the injection sites is diluted by the other tissue, which is away from the injection site. Despite this dilution, according to the quantification, western blots show at least a 50% increase in the protein expression over the control.
Then we started to test the effects of TNNT2-4Fpolycistronic-NIL on cardiac function following cardiac damage in vivo. Rats were subjected to a 2-h coronary occlusion followed by reperfusion. One week later, TNNT2-4Fpolycistronic-NIL or control TNNT2-GFP-NIL was injected into the peri-infarct region of the heart. Rats were followed for four weeks and then sacrificed, and the cardiac tissue was processed and analyzed (Fig. 5a). The TNNT2-4Fpolycistronic-NIL-treated group exhibited a significantly higher left ventricular ejection fraction four weeks post-viral injection compared to the control group, as assessed by blinded echocardiography, as well as a significant improvement in heart function compared to the starting EF values before injection (Fig. 5b). Comparing the absolute EF values between groups (Fig 5b) could be misleading because not all the animals started from the same EF value one week after I/R. Therefore, we compared the change in EF in each animal before and after the treatment (delta EF); this analysis showed a significant difference in delta EF between the TNNT2-4Fpolycistronic-NIL-treated group and the group that received control TNNT2-GFP-NIL (Fig. 5c).
Fig. 5. Transient expression of the 4F using TNNT2-4Fpolycistronic-NIL improves cardiac function and reduces scar size in rats after I/R.
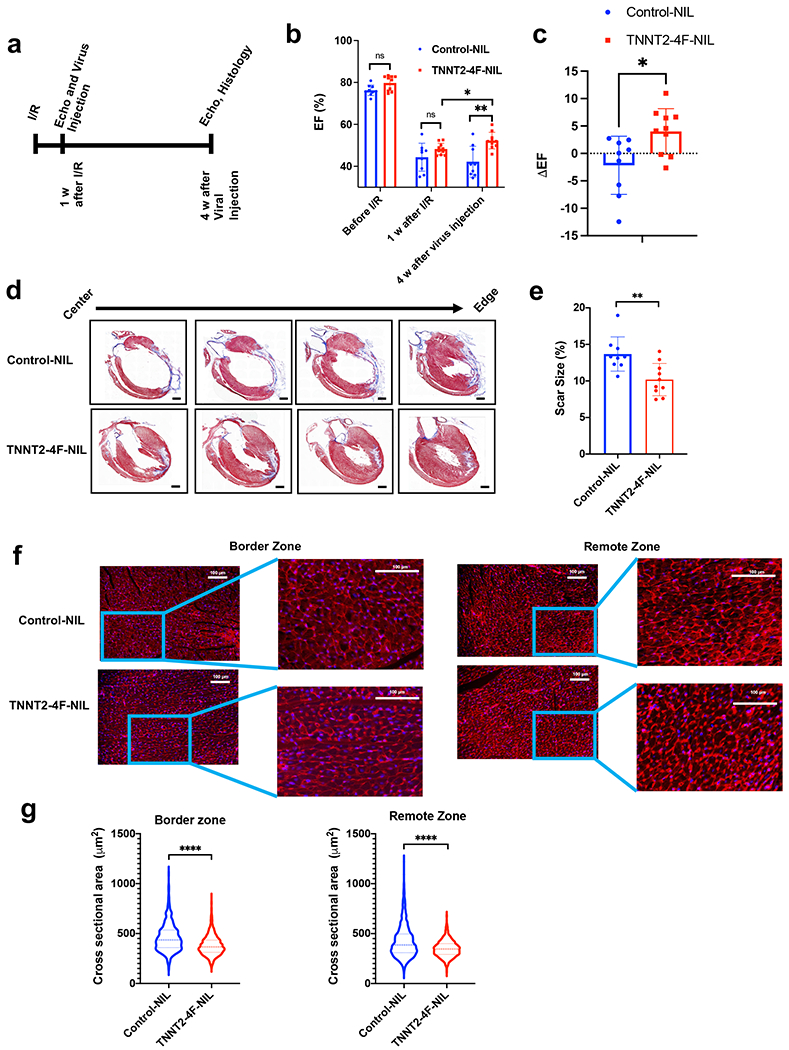
(a) schematic diagram of the experimental design. (b) Ejection fraction (EF), as assessed by echocardiography before Ischemia/reperfusion (I/R), one week after I/R (before viral treatment), and four weeks after viral treatment. (n=9-10 rats per group, *p<0.05, **p<0.01 compared to GFP-NIL control group, error bars indicate S.D.). (c) Quantification of the change in ejection fraction between before and after treatment for each individual rat (n=9-10 rats per group, *p<0.05, compared to GFP-NIL control group, error bars indicate S.D.). (d) Representative images of rat hearts were stained with Masson’s trichrome stain (healthy myocardium stains red and fibrotic tissue stains blue) at the end of the experiment (scale bar=2mm). (e) The scar size quantification as a percentage of total heart tissue (n=9-10 rats per group, 20-25 heart sections per animal, **p<0.01 compared to the control group, error bars indicate S.D.). (f) Representative images of rat heart at the border zone (right panel) or a remote zone (left panel) stained against wheat germ agglutinin, WGA (red), and nuclear DAPI (blue) (scale bar=100 µm). (g) Quantification of the cross-sectional area of cardiomyocytes at border zone (left) or a remote zone (right) (n=1300-1500 cells from each group, 20-25 heart sections per animal ****p<0.0001 compared to control group, error bars indicate S.D.).
Consistent with the improvement in cardiac function, histological analyses of the hearts revealed an approximately 30% reduction in the scar size in hearts treated with TNNT2-4Fpolycistronic-NIL compared to control hearts (Fig. 5d–e & Supplemental Fig 11a). Interestingly, the assessment of cell size at the border and remote zones showed a significant reduction in the cardiomyocyte cross-sectional area (Fig. 5f–g). As the virus was injected in the border zone, the reduction in cardiomyocyte cross-sectional area could be due to the induction of proliferation; however, the reduction in cardiomyocyte cross-sectional area in the remote zone could be due to improvement in the overall function of the heart and the resultant protection from progression towards dilatation after treatment with TNNT2-4Fpolycistronic-NIL. This hypothesis is supported by the findings that TNNT2-4F-NIL treated rats showed a significant decrease in LVEDd, LVEDs compared to Control-NIL treated rats, indicating decreased dilatation (Supplemental Fig 11b–c). This was not accompanied by the development of cardiac hypertrophy, as evident by the significant reduction in the HW/BW compared to control-NIL (Supplemental Fig 11d). Furthermore, there is no significant difference in the thickness of neither the interventricular septum nor the left ventricular free wall, as assessed by echocardiography, between the Control-NIL and TNNT2-4F-NIL treated groups, which again indicates lack of hypertrophy (Supplemental Fig 11e–f). Taken together, these data combined suggest that the TNNT2-4F-NIL treatment mainly alleviates the dilated cardiomyopathy remodeling which resulted from I/R compared to the Control-NIL treatment.
Double reporter to permanently label cardiomyocytes activation of aurora kinase B in vivo in large animals
Please see the expanded results section (Supplemental Materials) for details.
TNNT2-4Fpolyscistronic-NIL induces cardiomyocyte proliferation, preserves cardiac function, and reduces scar size in a porcine model of heart failure
As a proof of concept for our approach’s efficacy in inducing transient cardiomyocyte proliferation in large animals, we injected TNNT2-4Fpolyscistronic-NIL or control LacZ-NIL into the pig myocardium one week after induction of myocardial infarction with a 90-min coronary occlusion followed by reperfusion (Fig. 6a). The double reporter system was co-injected into the border zone with the therapeutic or control virus to assess the extent of cardiomyocyte proliferation induced by the TNNT2-4Fpolyscistronic-NIL.
Fig. 6. Transient expression of the 4F using TNNT2-4Fpolycistronic-NIL improves cardiac function, reduces scar size, and induces cardiomyocyte proliferation in a porcine model of heart failure.
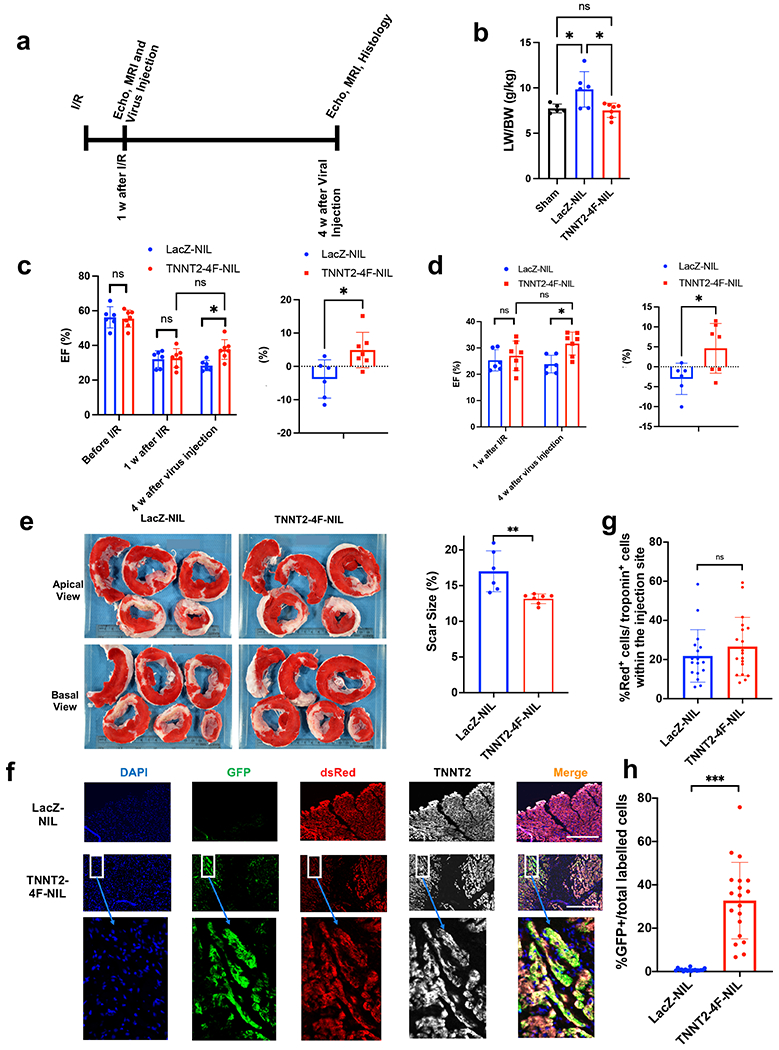
(a) schematic diagram of the experimental design. (b) Quantification of lung weight/body weight ratio (LW/BW). (n=5-7 pigs per group, *p<0.05 vs. control LacZ group, error bars indicate S.D.) (c) Quantification of ejection fraction (EF) as assessed by echocardiography (left panel) before Ischemia/reperfusion (I/R), one week after I/R (before viral treatment), and four weeks after viral treatment. The right panel shows the change in ejection fraction for each pig between before and after the viral injection (n=6-7 pigs per group, *p<0.05 vs. control LacZ group, error bars indicate S.D.). (d) Quantification of ejection fraction (EF) (left panel) and individual changes in EF before and after treatment (right panel) as assessed by MRI 1 week after I/R (before viral treatment) and four weeks after viral treatment (n=6-7 pigs per group, *p<0.05 vs. control LacZ group, error bars indicate S.D.). (e) Left panel, representative images of pig hearts stained with triphenyl tetrazolium chloride (T.T.C.) (healthy myocardium stains red and fibrotic tissue remains white) at the end of the experiment (scale bar in cm). Right panel, quantification of scar size as a percentage of total heart tissue (n=6-7 pigs per group, **p<0.01 vs. control LacZ group, error bars indicate S.D.). (f) Representative images at the site of injection four weeks post-injection in porcine myocardium. LacZ (top panel) or 4F (bottom panel) shows the expression of the proliferation double reporter system GFP and dsRed and their co-localization with cardiomyocytes (TNNT2). (g) Quantification of the percentage of dsRed positive cardiomyocytes at the injection site and (h) percentage of GFP positive cardiomyocytes over the total labeled cardiomyocytes (n=6-7 pigs per group, 3-4 sections from different injection sites ***p<0.001 vs. control LacZ group, error bars indicate S.D.).
Four weeks post-treatment, pigs treated with TNNT2-4Fpolyscistronic-NIL showed significant improvement in gross heart failure measures such as LW/BW compared to the pigs treated with LacZ-NIL (Fig. 6b). Furthermore, the cardiac functional parameter, ejection fraction, assessed by blinded echocardiography (Fig. 6c) and blinded MRI (Fig. 6d, Supplemental Fig. 14 and Supplemental movies 3–6) demonstrated a significantly higher EF and a greater individual change in EF in animals treated with TNNT2-4Fpolyscistronic-NIL compared to control virus-treated pigs four weeks post-treatment. However, compared to the starting EF before injection, at four weeks after injection, the EF in the TNNT2-4Fpolyscistronic-NIL treated pigs tended to increase, but the change did not reach statistical significance (p= 0.12 for Echo and 0.11 for MRI assessment). These findings suggest that the 4F is likely preserved the cardiac function in pigs after I/R. Furthermore, TNNT2-4Fpolyscistronic-NIL treated pigs exhibited a 25% reduction in scar size compared to control pigs (Fig. 6e).
TNNT2-4Fpolyscistronic-NIL-treated animals showed that 30% of the total labeled cardiomyocytes with the double reporter system at the injection site were GFP positive, indicating the cardiomyocyte mitotic activation. Taking into consideration the overestimation nature of this reporter by 30% for the cytokinetic events, it is likely that the accurate quantification would be that 20% of the cells underwent cytokinesis. In contrast, almost no background proliferation was detected in control virus-treated hearts (Fig. 6f–h), supporting the concept that the improvement in function is due to induction of cardiomyocyte proliferation.
Functional improvement following TNNT2-4Fpolyscistronic-NIL treatment is maintained for four months with no obvious systemic toxicity or systemic oncogenicity
To assess the long-term efficacy and toxicity of TNNT2-4Fpolyscistronic-NIL in vivo, rats were subjected to a 2-h coronary occlusion followed by reperfusion. One week later, TNNT2-4Fpolycistronic-NIL or control TNNT2-GFP-NIL was injected into the peri-infarct region of the heart. Rats were followed up for 16 weeks (Fig. 7a). The rats treated with TNNT2-4Fpolyscistronic-NIL demonstrated maintained improvement in cardiac function over the period of 4 months compared to the control group (Fig. 7b). In addition, LW/BW, which is one of the manifestations of heart failure, was significantly improved in animals treated with the TNNT2-4Fpolycistronic-NIL compared to the control group (Fig. 7c). In addition, the significant improvement in the scar size in TNNT2-4Fpolycistronic-NIL treated animals noticed four weeks post-treatment in our earlier experiments was maintained for four months (Fig. 7d). Furthermore, unconscious EKG monitoring for 15 minutes at the end of the experiment did not show any obvious ventricular arrhythmic episodes, nor atrial fibrillation in neither control virus nor TNNT2-4Fpolycistronic-NIL treated rats. However, future studies with conscious telemetry in large animals are needed to assess any arrhythmic potential for this therapeutic approach. We did not find any obvious tumor growth in the visceral cavity nor in tissues collected from the rats after four months of treatment, including liver, kidney, spleen, lung, brain, and skeletal muscle. Furthermore, plasma levels of the major tumor markers AFP and CA-19 showed normal basal levels, which indicates that there is no systemic tumorigenesis developed in these rats four months post-treatment (Supplemental Fig. 15a–b). In addition, the non-specific cell damage marker (LDH) was significantly improved following TNNT2-4Fpolyscistronic-NIL treatment compared to the control group (Supplemental Fig. 15c). Lipid profile (Triglycerides, Total cholesterol, HDL cholesterol, and LDL cholesterol), liver function markers (AST, ALT, and Albumin level), Kidney function marker (creatinine) did not show any significant differences between the control NIL and TNNT2-4Fpolyscistronic-NIL groups even though there is a trend of improvements in the TNNT2-4Fpolyscistronic-NIL treated group especially in the liver and kidney function markers (Supplemental Fig. 15d–k). In addition, we investigated apoptosis in cardiomyocytes at the end of the experiment (16 weeks post-treatment). There was a non-significant trend for decreased apoptosis in the hearts treated with TNNT2-4Fpolyscistronic-NIL compared to the hearts treated with the control-NIL virus (Figure 7E–F).
Fig. 7. TNNT2-4Fpolycistronic-NIL maintained improvement in cardiac function for four months.
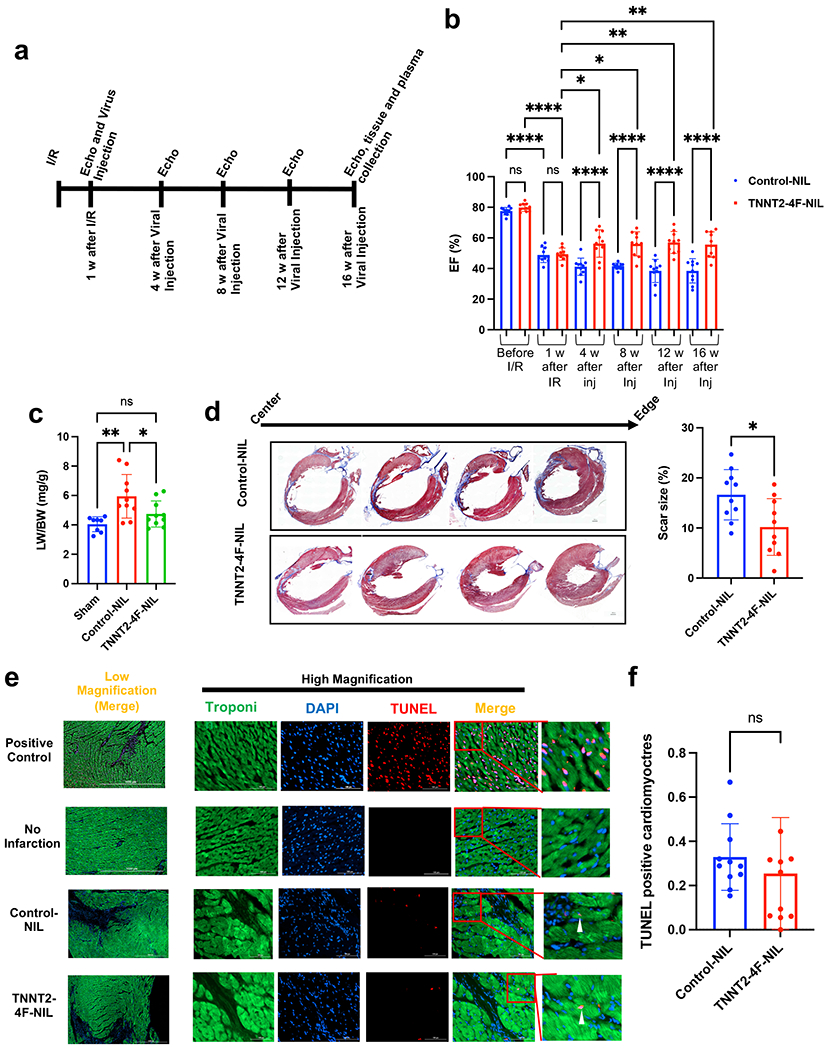
(a) schematic diagram of the experimental design. (b) Ejection fraction (EF), as assessed by echocardiography before ischemia/reperfusion (I/R), one week after I/R (before viral treatment), and every four weeks after viral treatment up to 16 weeks. (n=10 rats per group, *p<0.05, **p<0.01, ****p<0.0001, compared to GFP-NIL control group, error bars indicate the S.D.). (c) Quantification of the lung weight/body weight (LW/BW) (n=8-10 rats per group, *p<0.05, compared to GFP-NIL control group). (d) Representative images of rat hearts were stained with Masson’s trichrome stain (left panel) (healthy myocardium stains red and fibrotic tissue stains blue) at the end of the experiment (scale bar=2mm). The right panel shows scar size quantification as a percentage of total heart tissue (n=10 rats per group, 20-25 heart sections per animal, **p<0.01 compared to the control group, error bars indicate S.D.). (e) Representative images of rat hearts at the border zone stained for apoptotic nuclei using TUNEL assay (red), Troponin-T (green), and nuclear DAPI (blue) (scale bar=1000 µm in the low magnification images and 100 µm in the high magnification images). Positive control was carried out by treating the non-infarct heart section with the DNase-I enzyme. (f) Quantification of the percentage of TUNEL positive cardiomyocytes (n=10 heart sections per animal and 10 animals in each group, error bars indicate S.D.).
Discussion
Direct induction of the cell cycle using 4F is a promising approach for inducing cardiomyocyte proliferation21, 22; however, it is essential to first understand the mechanism of action of this potential heart failure gene therapy and to tightly control its timing, dosage, and specificity of expression in cardiomyocytes. Here, we describe the essential processes associated with forced cardiomyocyte proliferation following direct cell cycle induction, including sarcomeric disassembly and metabolic reprogramming, in a temporal sequence and at a single cell transcriptomic level. Furthermore, we provide the first proof of concept for the efficacy of this approach in improving cardiac function after infarction in a large animal model using a transient and cardiac-specific gene therapy approach.
For comprehensive discussion of the results and the limitations of the study, please see the expanded discussion section (Supplemental Materials).
Supplementary Material
Clinical Perspective.
1). What is new?
Mechanistic demonstration of the process of forced cardiomyocyte proliferation using a combination of four cell cycle factors on a single-cell level.
Development of a transient gene therapy using non-integrating lentivirus to target four cell cycle factors to cardiomyocytes to treat ischemic heart failure.
Demonstrating the efficacy of this transient gene therapy in the treatment of ischemic heart failure in rats and pigs.
2). What are the clinical implications?
Advances the clinical feasibility of the four cell cycle factors in the treatment of ischemic heart failure.
These promising findings in large animals will pave the road for a first-in-human trial to test the efficacy of this gene therapy in patients with ischemic cardiomyopathy.
Further clinical developments for other transient gene therapies, which could include NIL, modRNA, or direct protein delivery, could be targeted for specific tissues or cell subtypes for therapeutic purposes.
Acknowledgments
We acknowledge the Genomics core at the University of Louisville for providing access to the 10X Genomics and the Bioinformatics core at Gladstone Institute for performing all the transcriptomics analyses. We would also like to thank the radiology department at the University of Louisville for performing magnetic resonance imaging.
Funding Sources
T.M.A.M. is supported by NIH grants R01HL147921 and P30GM127607 and American Heart Association grant 16SDG29950012. The authors also acknowledge NIH grants F32HL149140 (R.R.E.A.), P30GM127607 (B.G.H.), R01HL130174 (B.G.H.), R01HL147844 (B.G.H.), R01ES028268 (B.G.H.), P01HL78825 (R.B., B.G.H.), GM127607 (D.J.C.), UM1HL113530 (R.B.), and 2U54HL120163. We also acknowledge the United States of America Department of Defense for the grant W81XWH-20-1-0419 (J.S., T.M.A.M.).
Non-standard Abbreviations and Acronyms
- CDK1
Cyclin Dependent Kinase 1
- CDK4
Cyclin Dependent Kinase 4
- CCNB
Cyclin B1
- CCND
Cyclin D1
- 4F
CDK1, CDK4, CCNB, CCND
- hiPSC-CMs
human induced pluripotent stem cell-derived cardiomyocytes
- MADM
Mosaic Analysis with Double Markers
- EDU
5-ethynyl-2’-deoxyuridine
- PHH3
Phospho-Histone H3
- GO
Gene Ontology
- LacZ
lacZ encodes β-galactosidase enzyme
- GFP
Green Fluorescent Protein
- TNNT2
Cardiac Troponin T isoform 2
- IL
Integrating lentivirus
- NIL
Non-integrating lentivirus
- CAG
CMV early enhancer/chicken beta-actin promoter
- AAV
Adeno-Associated virus
- AFP
Alpha Fetoprotein
- CA-19
Carbohydrate antigen 19
- LDH
Lactate Dehydrogenase
- HDL
high-density lipoprotein
- LDL
low-density lipoproteins
- AST
aspartate aminotransferase
- ALT
alanine aminotransferase
- HEK293
Human Embryonic Kidney cells
- MCAEC
mouse coronary artery endothelial cells
- EF
Ejection Fraction
- LVEDd
Left Ventricular End-Diastolic Diameter at Diastole
- I/R
Ischemia/Reperfusion
- LW/BW
Lung Weight/Body Weight
- EKG
electrocardiogram
- TNNC1
Troponin C1
- MYH7
(β)-myosin heavy chain
- PCNA
Proliferating Cell Nuclear Antigen
- E2F1
E2F Transcription Factor 1
- CDC20
Cell Division Cycle 20
- TK1
Thymidine Kinase 1
- CCNA2
Cyclin A2)
- PLK1
Polo Like Kinase 1
- ANLN
Anillin Actin Binding Protein
- WGA
Wheat Germ Agglutinin
- DAPI
4′,6-diamidino-2-phenylindole
- T.T.C.
triphenyl tetrazolium chloride
- dsRed
constitutively fluorescent red fluorescent protein
- TUNEL
Terminal deoxynucleotidyl transferase dUTP nick end labeling
Footnotes
Conflict of Interest Disclosures
R.R.E.A.: None. A.M.S.: None. Q.O.: None. X-L.T.: None. M.S.: None. K.M.K.: None. Y.G.: None. Y.H.: None. Y.N.: None. L.M.: None. P.K.L.: None. B.G.H.: None. K.C.: None. R.T.: None. B.M.A.: None. J.S.: None. H.R.J.: None. A.S.: None. Z.I.: None. S.H.: None. D.J.C.: None. A.S.E.: None. K.N.I: Holds equities at Tenaya Therapeutics. D.S.: Holds equities at Tenaya Therapeutics. R.B.: None. T.M.A.M.: Holds equities at Tenaya Therapeutics.
References
- 1.Hydbring P, Malumbres M and Sicinski P. Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nat Rev Mol Cell Biol. 2016;17:280–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malumbres M and Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–66. [DOI] [PubMed] [Google Scholar]
- 3.Li VC and Kirschner MW. Molecular ties between the cell cycle and differentiation in embryonic stem cells. Proc Natl Acad Sci U S A. 2014;111:9503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohamed TMA, Ang YS, Radzinsky E, Zhou P, Huang Y, Elfenbein A, Foley A, Magnitsky S and Srivastava D. Regulation of Cell Cycle to Stimulate Adult Cardiomyocyte Proliferation and Cardiac Regeneration. Cell. 2018;173:104–116 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zong H, Espinosa JS, Su HH, Muzumdar MD and Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121:479–92. [DOI] [PubMed] [Google Scholar]
- 6.Gabisonia K, Prosdocimo G, Aquaro GD, Carlucci L, Zentilin L, Secco I, Ali H, Braga L, Gorgodze N, Bernini F, et al. MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature. 2019;569:418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chien KR, Frisen J, Fritsche-Danielson R, Melton DA, Murry CE and Weissman IL. Regenerating the field of cardiovascular cell therapy. Nat Biotechnol. 2019;37:232–237. [DOI] [PubMed] [Google Scholar]
- 8.Eschenhagen T, Bolli R, Braun T, Field LJ, Fleischmann BK, Frisen J, Giacca M, Hare JM, Houser S, Lee RT, et al. Cardiomyocyte Regeneration: A Consensus Statement. Circulation. 2017;136:680–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sada H, Ban T, Fujita T, Ebina Y and Sperelakis N. Developmental change in fast Na channel properties in embryonic chick ventricular heart cells. Can J Physiol Pharmacol. 1995;73:1475–84. [DOI] [PubMed] [Google Scholar]
- 10.Kiselev VY, Yiu A and Hemberg M. scmap: projection of single-cell RNA-seq data across data sets. Nat Methods. 2018;15:359–362. [DOI] [PubMed] [Google Scholar]
- 11.Lane AN and Fan TWM. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Research. 2015;43:2466–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abouleisa RRE, McNally L, Salama ABM, Hammad SK, Ou Q, Wells C, Lorkiewicz PK, Bolli R, Mohamed TMA and Hill BG. Cell cycle induction in human cardiomyocytes is dependent on biosynthetic pathway activation. Redox Biol. 2021;46:102094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Z, Chen F, Zhang L, Lu J, Xu P, Liu G, Xie X, Mu W, Wang Y and Liu D. Non-integrating lentiviral vectors based on the minimal S/MAR sequence retain transgene expression in dividing cells. Sci China Life Sci. 2016;59:1024–1033. [DOI] [PubMed] [Google Scholar]
- 14.Nightingale SJ, Hollis RP, Pepper KA, Petersen D, Yu XJ, Yang C, Bahner I and Kohn DB. Transient gene expression by nonintegrating lentiviral vectors. Mol Ther. 2006;13:1121–32. [DOI] [PubMed] [Google Scholar]
- 15.Shaw A and Cornetta K. Design and Potential of Non-Integrating Lentiviral Vectors. Biomedicines. 2014;2:14–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sultana N, Magadum A, Hadas Y, Kondrat J, Singh N, Youssef E, Calderon D, Chepurko E, Dubois N, Hajjar RJ, et al. Optimizing Cardiac Delivery of Modified mRNA. Mol Ther. 2017;25:1306–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milone MC and O’Doherty U. Clinical use of lentiviral vectors. Leukemia. 2018;32:1529–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohamed TM, Stone NR, Berry EC, Radzinsky E, Huang Y, Pratt K, Ang YS, Yu P, Wang H, Tang S, et al. Chemical Enhancement of In Vitro and In Vivo Direct Cardiac Reprogramming. Circulation. 2017;135:978–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ali SR, Hippenmeyer S, Saadat LV, Luo L, Weissman IL and Ardehali R. Existing cardiomyocytes generate cardiomyocytes at a low rate after birth in mice. Proc Natl Acad Sci U S A. 2014;111:8850–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen NUN, Canseco DC, Xiao F, Nakada Y, Li S, Lam NT, Muralidhar SA, Savla JJ, Hill JA, Le V, et al. A calcineurin–Hoxb13 axis regulates growth mode of mammalian cardiomyocytes. Nature. 2020; 582:271–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadek H and Olson EN. Toward the Goal of Human Heart Regeneration. Cell Stem Cell. 2020;26:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh PCH and Kamp TJ. To Be Young at Heart. Cell Stem Cell. 2018;22:475–476. [DOI] [PubMed] [Google Scholar]
- 23.Ma HT and Poon RY. How protein kinases co-ordinate mitosis in animal cells. Biochem J. 2011;435:17–31. [DOI] [PubMed] [Google Scholar]
- 24.Canseco DC, Kimura W, Garg S, Mukherjee S, Bhattacharya S, Abdisalaam S, Das S, Asaithamby A, Mammen PP and Sadek HA. Human ventricular unloading induces cardiomyocyte proliferation. J Am Coll Cardiol. 2015;65:892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hesse M, Doengi M, Becker A, Kimura K, Voeltz N, Stein V and Fleischmann BK. Midbody Positioning and Distance Between Daughter Nuclei Enable Unequivocal Identification of Cardiomyocyte Cell Division in Mice. Circ Res. 2018;123:1039–1052. [DOI] [PubMed] [Google Scholar]
- 26.Jiang J, Burgon PG, Wakimoto H, Onoue K, Gorham JM, O’Meara CC, Fomovsky G, McConnell BK, Lee RT, Seidman JG, et al. Cardiac myosin binding protein C regulates postnatal myocyte cytokinesis. Proc Natl Acad Sci U S A. 2015;112:9046–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura M, Uchida C, Takano Y, Kitagawa M and Okano Y. Cell cycle-dependent regulation of the human aurora B promoter. Biochem Biophys Res Commun. 2004;316:930–6. [DOI] [PubMed] [Google Scholar]
- 28.Sadek H and Olson EN. Toward the Goal of Human Heart Regeneration. Cell Stem Cell. 2020;26:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cauquil M, Mias C, Guilbeau-Frugier C, Karsenty C, Seguelas M-H, Genet G, Renaud-Gabardos E, Prats A-C, Pons V, Branchereau M, et al. Ephrin-B1 blocks adult cardiomyocyte proliferation and heart regeneration. bioRxiv. Preprint posted online August 15, 2019. 10.1101/735571. [DOI] [Google Scholar]
- 30.Monroe TO, Hill MC, Morikawa Y, Leach JP, Heallen T, Cao S, Krijger PHL, de Laat W, Wehrens XHT, Rodney GG, et al. YAP Partially Reprograms Chromatin Accessibility to Directly Induce Adult Cardiogenesis In Vivo. Dev Cell. 2019;48:765–779 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen NUN, Canseco DC, Xiao F, Nakada Y, Li S, Lam NT, Muralidhar SA, Savla JJ, Hill JA, Le V, et al. A calcineurin-Hoxb13 axis regulates growth mode of mammalian cardiomyocytes. Nature. 2020;582:271–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pettinato AM, Yoo D, VanOudenhove J, Chen YS, Cohn R, Ladha FA, Yang X, Thakar K, Romano R, Legere N, et al. Sarcomere function activates a p53-dependent DNA damage response that promotes polyploidization and limits in vivo cell engraftment. Cell Rep. 2021;35:109088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abouleisa RRE, McNally L, Salama ABM, Hammad SK, Ou Q, Wells C, Lorkiewicz PK, Bolli R, Mohamed TMA and Hill BG. Cell cycle induction in human cardiomyocytes is dependent on biosynthetic pathway activation. Redox Biol. 2021;46:102094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magadum A, Singh N, Kurian AA, Munir I, Mehmood T, Brown K, Sharkar MTK, Chepurko E, Sassi Y, Oh JG, et al. Pkm2 regulates cardiomyocyte cell cycle and promotes cardiac regeneration. Circulation. 2020; 141(15):1249–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honkoop H, de Bakker DE, Aharonov A, Kruse F, Shakked A, Nguyen PD, de Heus C, Garric L, Muraro MJ, Shoffner A, et al. Single-cell analysis uncovers that metabolic reprogramming by ErbB2 signaling is essential for cardiomyocyte proliferation in the regenerating heart. Elife. 2019; 8:e50163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cardoso AC, Lam NT, Savla JJ, Nakada Y, Pereira AHM, Elnwasany A, Menendez-Montes I, Ensley EL, Petric UB, Sharma G, et al. Mitochondrial substrate utilization regulates cardiomyocyte cell-cycle progression. Nature Metabolism. 2020;2:167–178. [PMC free article] [PubMed] [Google Scholar]
- 37.Bae J, Salamon RJ, Brandt EB, Paltzer WG, Zhang Z, Britt EC, Hacker TA, Fan J and Mahmoud AI. Malonate Promotes Adult Cardiomyocyte Proliferation and Heart Regeneration. Circulation. 2021; 143(20):1973–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mills RJ, Parker BL, Quaife-Ryan GA, Voges HK, Needham EJ, Bornot A, Ding M, Andersson H, Polla M, Elliott DA, et al. Drug Screening in Human PSC-Cardiac Organoids Identifies Pro-proliferative Compounds Acting via the Mevalonate Pathway. Cell Stem Cell. 2019;24:895–907 e6. [DOI] [PubMed] [Google Scholar]
- 39.Tran DH and Wang ZV. Glucose Metabolism in Cardiac Hypertrophy and Heart Failure. J Am Heart Assoc. 2019;8:e012673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gabisonia K, Prosdocimo G, Aquaro GD, Carlucci L, Zentilin L, Secco I, Ali H, Braga L, Gorgodze N, Bernini F, et al. MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature. 2019;569:418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohamed TMA, Ang YS, Radzinsky E, Zhou P, Huang Y, Elfenbein A, Foley A, Magnitsky S and Srivastava D. Regulation of Cell Cycle to Stimulate Adult Cardiomyocyte Proliferation and Cardiac Regeneration. Cell. 2018;173:104–116 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ou Q, Jacobson Z, Abouleisa RRE, Tang XL, Hindi SM, Kumar A, Ivey KN, Giridharan G, El-Baz A, Brittian K, et al. Physiological Biomimetic Culture System for Pig and Human Heart Slices. Circ Res. 2019;125:628–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bassat E, Mutlak YE, Genzelinakh A, Shadrin IY, Baruch Umansky K, Yifa O, Kain D, Rajchman D, Leach J, Riabov Bassat D, et al. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature. 2017;547:179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL and Martin JF. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morikawa Y, Zhang M, Heallen T, Leach J, Tao G, Xiao Y, Bai Y, Li W, Willerson JT and Martin JF. Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice. Sci Signal. 2015;8:ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaudhry HW, Dashoush NH, Tang H, Zhang L, Wang X, Wu EX and Wolgemuth DJ. Cyclin A2 mediates cardiomyocyte mitosis in the postmitotic myocardium. J Biol Chem. 2004;279:35858–66. [DOI] [PubMed] [Google Scholar]
- 47.Cheng RK, Asai T, Tang H, Dashoush NH, Kara RJ, Costa KD, Naka Y, Wu EX, Wolgemuth DJ and Chaudhry HW. Cyclin A2 induces cardiac regeneration after myocardial infarction and prevents heart failure. Circ Res. 2007;100:1741–8. [DOI] [PubMed] [Google Scholar]
- 48.Shapiro SD, Ranjan AK, Kawase Y, Cheng RK, Kara RJ, Bhattacharya R, Guzman-Martinez G, Sanz J, Garcia MJ and Chaudhry HW. Cyclin A2 induces cardiac regeneration after myocardial infarction through cytokinesis of adult cardiomyocytes. Sci Transl Med. 2014;6:224ra27. [DOI] [PubMed] [Google Scholar]
- 49.Woo YJ, Panlilio CM, Cheng RK, Liao GP, Atluri P, Hsu VM, Cohen JE and Chaudhry HW. Therapeutic delivery of cyclin A2 induces myocardial regeneration and enhances cardiac function in ischemic heart failure. Circulation. 2006;114:I206–13. [DOI] [PubMed] [Google Scholar]
- 50.Lane AN and Fan TWM. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Research. 2015;43:2466–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cui M, Wang Z, Chen K, Shah AM, Tan W, Duan L, Sanchez-Ortiz E, Li H, Xu L, Liu N, et al. Dynamic Transcriptional Responses to Injury of Regenerative and Non-regenerative Cardiomyocytes Revealed by Single-Nucleus RNA Sequencing. Dev Cell. 2020;53:102–116 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui M, Atmanli A, Morales MG, Tan W, Chen K, Xiao X, Xu L, Liu N, Bassel-Duby R and Olson EN. Nrf1 promotes heart regeneration and repair by regulating proteostasis and redox balance. Nat Commun. 2021;12:5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zangi L, Lui KO, von Gise A, Ma Q, Ebina W, Ptaszek LM, Später D, Xu H, Tabebordbar M, Gorbatov R, et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol. 2013;31:898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaw A and Cornetta K. Design and Potential of Non-Integrating Lentiviral Vectors. Biomedicines. 2014;2:14–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milone MC and O’Doherty U. Clinical use of lentiviral vectors. Leukemia. 2018; 32(7):1529–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao M, Nakada Y, Wei Y, Bian W, Chu Y, Borovjagin AV, Xie M, Zhu W, Nguyen T, Zhou Y, et al. Cyclin D2 Overexpression Enhances the Efficacy of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes for Myocardial Repair in a Swine Model of Myocardial Infarction. Circulation. 2021;144:210–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mohamed TM, Stone NR, Berry EC, Radzinsky E, Huang Y, Pratt K, Ang YS, Yu P, Wang H, Tang S, et al. Chemical Enhancement of In Vitro and In Vivo Direct Cardiac Reprogramming. Circulation. 2017;135:978–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bradley LA, Young A, Li H, Billcheck HO and Wolf MJ. Loss of Endogenously Cycling Adult Cardiomyocytes Worsens Myocardial Function. Circ Res. 2021;128:155–168. [DOI] [PubMed] [Google Scholar]
- 59.Sultana N, Magadum A, Hadas Y, Kondrat J, Singh N, Youssef E, Calderon D, Chepurko E, Dubois N, Hajjar RJ, et al. Optimizing Cardiac Delivery of Modified mRNA. Mol Ther. 2017;25:1306–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mohamed TM, Oceandy D, Prehar S, Alatwi N, Hegab Z, Baudoin FM, Pickard A, Zaki AO, Nadif R, Cartwright EJ, et al. Specific role of neuronal nitric-oxide synthase when tethered to the plasma membrane calcium pump in regulating the beta-adrenergic signal in the myocardium. J Biol Chem. 2009;284:12091–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P and Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kiselev VY, Yiu A and Hemberg M. scmap: projection of single-cell RNA-seq data across data sets. Nat Methods. 2018;15:359–362. [DOI] [PubMed] [Google Scholar]
- 64.Mehra P, Guo Y, Nong Y, Lorkiewicz P, Nasr M, Li Q, Muthusamy S, Bradley JA, Bhatnagar A, Wysoczynski M, et al. Cardiac mesenchymal cells from diabetic mice are ineffective for cell therapy-mediated myocardial repair. Basic Res Cardiol. 2018;113:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Q, Guo Y, Ou Q, Wu WJ, Chen N, Zhu X, Tan W, Yuan F, Dawn B, Luo L, et al. Gene transfer as a strategy to achieve permanent cardioprotection II: rAAV-mediated gene therapy with heme oxygenase-1 limits infarct size 1 year later without adverse functional consequences. Basic Res Cardiol. 2011;106:1367–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang XL, Nakamura S, Li Q, Wysoczynski M, Gumpert AM, Wu WJ, Hunt G, Stowers H, Ou Q and Bolli R. Repeated Administrations of Cardiac Progenitor Cells Are Superior to a Single Administration of an Equivalent Cumulative Dose. J Am Heart Assoc. 2018; 7(4):e007400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tokita Y, Tang XL, Li Q, Wysoczynski M, Hong KU, Nakamura S, Wu WJ, Xie W, Li D, Hunt G, et al. Repeated Administrations of Cardiac Progenitor Cells Are Markedly More Effective Than a Single Administration: A New Paradigm in Cell Therapy. Circ Res. 2016;119:635–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bolli R, Tang XL, Sanganalmath SK, Rimoldi O, Mosna F, Abdel-Latif A, Jneid H, Rota M, Leri A and Kajstura J. Intracoronary delivery of autologous cardiac stem cells improves cardiac function in a porcine model of chronic ischemic cardiomyopathy. Circulation. 2013;128:122–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Swindle MM, Smith AC and Hepburn BJ. Swine as models in experimental surgery. J Invest Surg. 1988;1:65–79. [DOI] [PubMed] [Google Scholar]
- 70.Swindle MM, Horneffer PJ, Gardner TJ, Gott VL, Hall TS, Stuart RS, Baumgartner WA, Borkon AM, Galloway E and Reitz BA. Anatomic and anesthetic considerations in experimental cardiopulmonary surgery in swine. Lab Anim Sci. 1986;36:357–61. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


