Abstract
Many complex disease risk loci map to intergenic regions containing long intergenic noncoding RNAs (lincRNAs). The majority of these is not conserved outside humans, raising the question whether genetically regulated expression of non-conserved and conserved lincRNAs has similar rates of association with complex traits. Here we leveraged data from the Genotype-Tissue Expression (GTEx) project and multiple public genome-wide association study (GWAS) resources. Using an established transcriptome-wide association study (TWAS) tool, FUSION, we interrogated the associations between cis-regulated expression of lincRNAs and multiple cardiometabolic traits. We found that cis-regulated expression of non-conserved lincRNAs had a strikingly similar trend of association with complex cardiometabolic traits as conserved lincRNAs. This finding challenges the conventional notion of conservation that has led to prioritization of conserved loci for functional studies and calls attention to the need to develop comprehensive strategies to study the large number of non-conserved human lincRNAs that may contribute to human disease.
Keywords: Cardiometabolic traits, conservation, expression, GWAS, long intergenic non-coding RNAs (lincRNAs), synteny, TWAS
Numerous complex disease risk loci reside in intergenic regions and map to long intergenic noncoding RNAs (lincRNAs) (1–2). While most human lincRNAs are not conserved across species (3), researchers prioritize research on conserved lincRNAs (4). This is due in large part to the belief that conserved genetic elements are more likely to be functional than non-conserved genetic elements. Because the majority of GWAS findings are in intergenic regions that regulate gene expression (6–7), a question of critical importance is whether cis-regulated expression levels of non-conserved lincRNAs have similar rates of association with complex traits as cis-regulated expression of conserved lincRNAs. Data were leveraged from the Genotype-Tissue Expression (GTEx) project version 7 (8) to identify tissue specific lincRNA expression quantitative trail loci (eQTL) SNPs. Integrating with summary statistics from publicly available large-scale GWAS meta-analyses, we examined the relationships between lincRNAs’ cis-regulated expression and complex disease traits. Our findings challenge the conventional notion of conservation that has led to prioritization of conserved loci for functional studies and calls attention to the need to develop comprehensive strategies to study the large number of non-conserved human lincRNAs that may contribute to human disease.
We used 7089 well-annotated lincRNAs in Human GENCODE v33 (9). The primary definition of lincRNA conservation was based on synteny of lincRNA loci between human and mouse genomes, i.e., positional genomic conservation (10). A secondary definition was based on synteny and lincRNA expression in mouse tissues, as described previously (5). We focused on four complex cardiometabolic traits: waist-to-hip ratio adjusted for body mass index (WHRadjBMI), height, body mass index (BMI) and type 2 diabetes (T2D). Genetic Investigation of Anthropometric Traits (GIANT)/UK Biobank (UKBb) summary data were examined for WHRadjBMI, height and BMI (11–12). DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) data were examined for T2D (13). Using the transcriptome-wide association study (TWAS) tool FUSION (14) to integrate GTEx analysis version 7 summary data on tissue-specific eQTLs (8) and GWAS summary level data, we evaluated the association between cis-regulated expression of lincRNAs and each trait. The FUSION tool produced a TWAS z-score and p-value for each lincRNA under interrogation, and association of each lincRNA’s cis-regulated expression with the trait (termed as having “GWAS signal”) was determined using a Bonferroni corrected p-value threshold of 0.05 (based on the number of lincRNAs being investigated for each trait in each tissue).
To evaluate the association between lincRNA conservation and GWAS signal, Pearson’s chi-square tests were performed for WHRadjBMI, height and BMI, and Fisher’s exact tests were performed for T2D as the number of lincRNAs with GWAS signal was low. Multivariable logistic regression models were fitted, adjusting for the length of the lincRNA. Adjusted odds ratios (ORs) and corresponding 95% confidence intervals were calculated. We examined eQTL SNPs in: 1) visceral adipose tissues (VAT) and subcutaneous adipose tissues (SAT) for WHRadjBMI, 2) skeletal muscle tissues (SMT) and SAT for height; 3) SAT and hypothalamus (Hypo) for BMI; 4) SAT and VAT for T2D. We adjusted for the eight tests (two tissues for each of the four traits) using an additional Bonferroni correction.
Unadjusted analyses using the primary definition of conservation revealed that the proportion of lincRNAs with GWAS signal was consistently higher (though not statistically significant) for non-conserved compared to conserved lincRNAs for all traits and in all tissues, except for height in SAT where the proportion of GWAS signal was slightly higher for conserved lincRNAs but did not reach statistical significance (Table 1). A similar trend was observed using the secondary definition of conservation (Table 2).
Table 1. Unadjusted analysis:
GWAS signal counts for lincRNAs by trait and tissues and definition of conservation. (Conservation defined based on synteny between human and mouse)
| Trait | Tissues | Conservation | Signal | %Signal | Total | p-valuea | Adjusted p-valueb |
|---|---|---|---|---|---|---|---|
| WHRadjBMI | VAT (n=408) | Non-conserved | 11 | 9.4% | 117 | 0.128 | 1.000 |
| Conserved | 14 | 4.8% | 291 | ||||
| SAT (n=506) | Non-conserved | 15 | 10.3% | 146 | 0.150 | 1.000 | |
| Conserved | 22 | 6.1% | 360 | ||||
| Height | SMT (n=392) | Non-conserved | 25 | 22.7% | 110 | 0.799 | 1.000 |
| Conserved | 59 | 20.9% | 282 | ||||
| SAT (n=498) | Non-conserved | 30 | 21.3% | 141 | 1.000 | 1.000 | |
| Conserved | 77 | 21.6% | 357 | ||||
| BMI | SAT (n=506) | Non-conserved | 14 | 9.6% | 146 | 0.859 | 1.000 |
| Conserved | 31 | 8.6% | 360 | ||||
| Hypo (n=200) | Non-conserved | 13 | 19.1% | 68 | 0.071 | 0.568 | |
| Conserved | 12 | 9.1% | 132 | ||||
| T2D | SAT (n=501) | Non-conserved | 5 | 3.5% | 142 | 0.126 | 1.000 |
| Conserved | 4 | 1.1% | 359 | ||||
| VAT (n=400) | Non-conserved | 2 | 1.8% | 114 | 0.626 | 1.000 | |
| Conserved | 3 | 1.0% | 286 |
Corresponds to Pearson’s chi-square test for WHRadjBMI, height and BMI, and Fisher’s exact test for T2D
Bonferroni adjusted p-values accounting for eight tests
Table 2. Unadjusted analysis:
GWAS signal counts for lincRNAs by trait and tissues and definition of conservation. (Conservation defined based on synteny and tissue expression in mouse)
| Trait | Tissues | Conservation | Signal | %Signal | Total | p-valuea | Adjusted p-valueb |
|---|---|---|---|---|---|---|---|
| WHRadjBMI | VAT (n=405) | Non-conserved | 18 | 6.5% | 276 | 0.605 | 1.000 |
| Conserved | 6 | 4.7% | 129 | ||||
| SAT (n=503) | Non-conserved | 26 | 7.8% | 335 | 0.756 | 1.000 | |
| Conserved | 11 | 6.5% | 168 | ||||
| Height | SMT (n=390) | Non-conserved | 59 | 23.0% | 256 | 0.383 | 1.000 |
| Conserved | 25 | 18.7% | 134 | ||||
| SAT (n=495) | Non-conserved | 70 | 21.3% | 328 | 0.926 | 1.000 | |
| Conserved | 37 | 22.2% | 167 | ||||
| BMI | SAT (n=503) | Non-conserved | 30 | 9.0% | 335 | 1.000 | 1.000 |
| Conserved | 15 | 8.9% | 168 | ||||
| Hypo (n=199) | Non-conserved | 18 | 14.2% | 127 | 0.492 | 1.000 | |
| Conserved | 7 | 9.7% | 72 | ||||
| T2D | SAT (n=498) | Non-conserved | 7 | 2.1% | 331 | 0.724 | 1.000 |
| Conserved | 2 | 1.2% | 167 | ||||
| VAT (n=397) | Non-conserved | 5 | 1.9% | 269 | 0.180 | 1.000 | |
| Conserved | 0 | 0.0% | 128 |
Corresponds to Pearson’s chi-square test for WHRadjBMI, height and BMI, and Fisher’s exact test for T2D
Bonferroni adjusted p-values accounting for eight tests
Multivariate modeling results were consistent with unadjusted analyses. Figure 1 illustrates adjusted ORs and corresponding 95% confidence intervals for GWAS signal for non-conserved lincRNAs relative to conserved lincRNAs. Point estimates of ORs were >1.0 for both definitions of conservation and for all 4 traits across all tissues considered, except for height in SAT under primary definition (Figure 1A: OR=0.98, unadjusted p=0.916, adjusted p=1.000) and secondary definition (Figure 1B: OR=0.93, unadjusted p=0.763, adjusted p=1.000) and for BMI in SAT under secondary definition (Figure 1B: OR=0.95, unadjusted p=0.889, adjusted p=1.000) but none of them reached statistical significance.
Figure 1. Multivariable analysis:
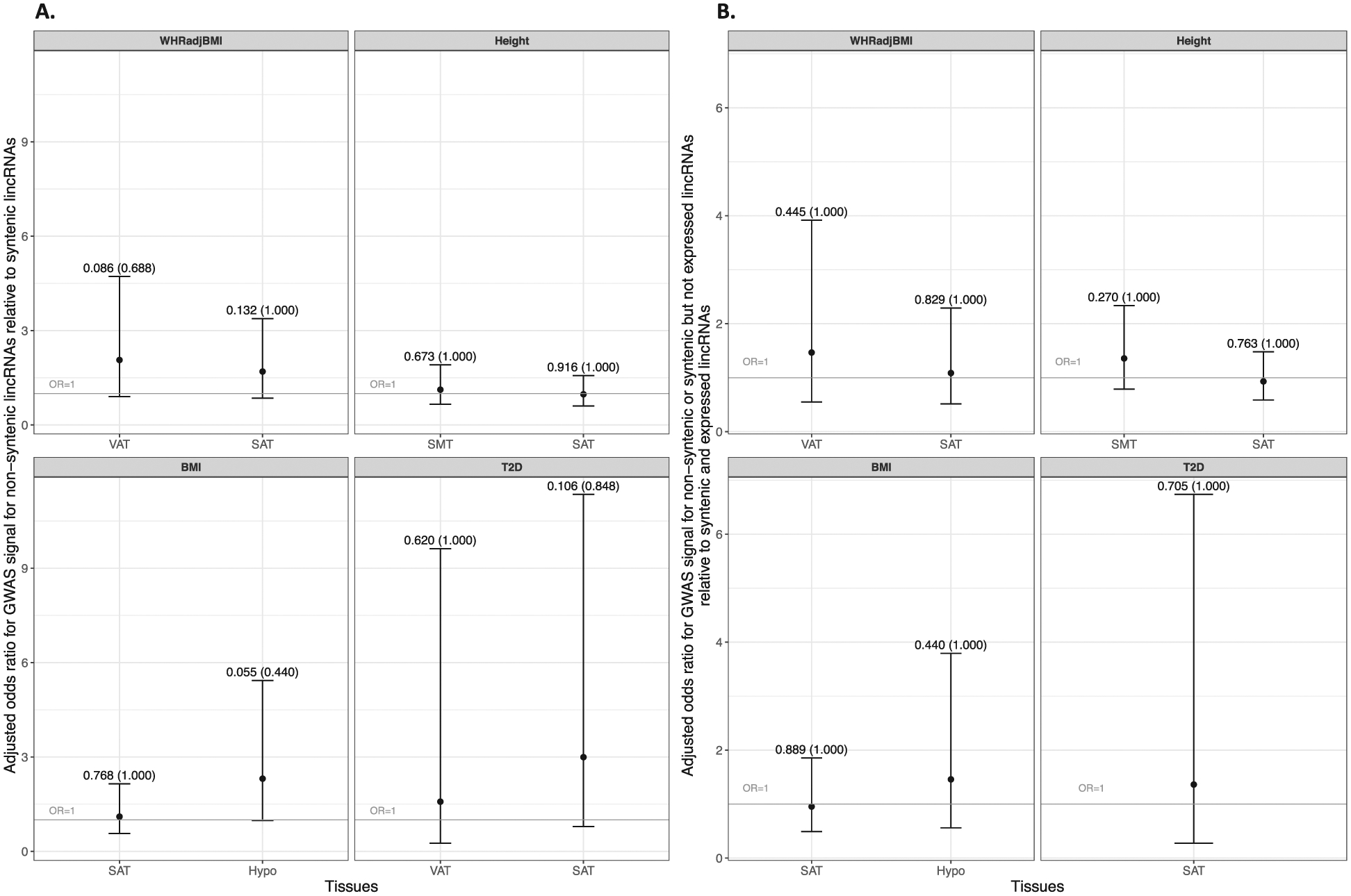
A. Adjusted odds ratio (OR) for GWAS signal for non-syntenic lincRNAs relative to syntenic lincRNAsa, with 95% confidence interval, p-values, and adjusted p-valuesb. B. Adjusted odds ratio (OR) for GWAS signal for non-syntenic or syntenic but not expressed lincRNAs relative to syntenic and expressed lincRNAsa, c, with 95% confidence interval, p-values, and adjusted p-valuesb.
aSeparate multivariable adjusted models are fitted for each trait, adjusting for the length (kb) of lincRNAs. Syntenic lincRNAs (n=2069) tended to be longer (median length: 2624 bps vs. 2471 bps) than non-conserved lincRNAs (n=707). For conservation defined as synteny and expression in mouse, conserved lincRNAs (n=998) were also longer (median length: 3219.5 bps vs. 2359 bps) than non-conserved lincRNAs (n=1761).
bBonferroni adjusted p-values accounting for eight tests
cUnder secondary definition of conservation, there was no conserved lincRNA with GWAS signals for T2D in VAT and thus no multivariate logistic regression model was fitted for T2D in VAT.
The majority of human lincRNAs lacks conservation; yet established examples of functional and disease relevant non-conserved lincRNAs have emerged (15–17). We found that non-conserved lincRNAs’ cis-regulated expression had a strikingly similar trend of association with multiple complex traits as conserved lincRNAs’ expression. Our findings are significant as the majority of human genetic variation that associates with complex traits falls in intergenic regions that overlaps regulatory features. Given the expansive role of regulatory variation, this work provides strong evidence that that lack of conservation does not reduce the probability that a human lincRNA will have genetic variation associated with complex human traits.
Relative to mRNAs, lincRNAs are more abundant in the human transcriptome and are of increasing importance in human diseases (18). Species conservation is an important feature that is often used as triage when determining whether a gene is likely to be functionally important in human disease. However, many cell-specific regulatory elements, including most lincRNAs, are not conserved outside primates (19). Further, established genomic markers of function including tissue-enrichment and binding of tissue-specific transcription factors at lincRNAs do not differ significantly between conserved and non-conserved lincRNAs (17, 19). Of a handful of lincRNAs that overlap loci for human cardiometabolic traits (1, 15), including ANRIL, H19, MALAT1, MEXIS, LOC157273, and LASER, several including LOC157273 and H19 are not conserved in mice (20–21). These results are also consistent with our recent work, in which we found that SNPs physically located within non-conserved lincRNAs have genetic association with complex human cardiometabolic traits at similar rates as SNPs at conserved lincRNAs (5). In conclusion, our work and that of others suggest that strategies considering key regulatory and functional features (13, 15) as well as disease association, rather than an initial triage based on conservation, are required to prioritize important human lincRNAs for translational study.
Acknowledgements.
The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from: dbGaP accession number phs000424.v8.p2 on 05/20/2020.
Sources of Funding.
Support for this research was provided by grants from the National Institutes of Health (NIH) R01 GM127862 to ASF, and R01 HL132561, R01 HL113147 and K24 HL107643 to MPR.
Footnotes
Conflict of Interest Statement
The authors have no financial or non-financial competing interests relevant to this article to disclose.
References:
- 1.Ballantyne RL, Zhang X, Nuñez S, Xue C, Zhao W, Reed E, Salaheen D, Foulkes AS, Li M, Reilly MP. Genome-wide interrogation reveals hundreds of long intergenic noncoding RNAs that associate with cardiometabolic traits. Hum Mol Genet. 2016. Jul 15;25(14):3125–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li MJ, Wang P, Liu X, et al. GWASdb: a database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2012;40(Database issue):D1047–D1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner AW, Wong D, Khan MD, Dreisbach CN, Palmore M, Miller CL. Multi-Omics Approaches to Study Long Non-coding RNA Function in Atherosclerosis. Front Cardiovasc Med. 2019;6:9. Published 2019 Feb 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michalik KM, You X, Manavski Y, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114(9):1389–1397 [DOI] [PubMed] [Google Scholar]
- 5.Foulkes AS, Selvaggi C, Cao T, et al. Nonconserved Long Intergenic Noncoding RNAs Associate With Complex Cardiometabolic Disease Traits. Arterioscler Thromb Vasc Biol. 2021;41(1):501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen CY, Chang IS, Hsiung CA, Wasserman WW. On the identification of potential regulatory variants within genome wide association candidate SNP sets. BMC Med Genomics. 2014. Jun 11;7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020. Sep 11;369(6509):1318–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frankish A, Diekhans M, Ferreira AM, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766–D773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I. Principles of long noncoding rna evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015;11:1110–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pulit SL, Stoneman C, Morris AP, et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum Mol Genet. 2019;28(1):166–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood AR, Esko T, Yang J, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46(11):1173–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, et al. Fine mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50:1505–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gusev A, Ko A, Shi H, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48(3):245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Goede OM, Ferraro NM, Nachun DC, Rao AS, Aguet F, Barbeira AN, et al. Long noncoding RNA gene regulation and trait associations across human tissues. bioRxiv. 2019 [Google Scholar]
- 16.Zhang X, Li DY, Reilly MP. Long intergenic noncoding RNAs in cardiovascular diseases: Challenges and strategies for physiological studies and translation. Atherosclerosis. 2019;281:180–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Xue C, Lin J, Ferguson JF, Weiner A, Liu W, et al. Interrogation of nonconserved human adipose lincRNAs identifies a regulatory role of linc-adal in adipocyte metabolism. Sci Transl Med. 2018;10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiStefano JK. The Emerging Role of Long Noncoding RNAs in Human Disease. Methods Mol Biol. 2018;1706:91–110 [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Xue C, Wang Y, Shi J, Zhang X, Li W, et al. Deep rna sequencing uncovers a repertoire of human macrophage long intergenic noncoding rnas modulated by macrophage activation and associated with cardiometabolic diseases. J Am Heart Assoc. 2017;6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, et al. Long noncoding rna malat1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114:1389–1397 [DOI] [PubMed] [Google Scholar]
- 21.Gao W, Zhu M, Wang H, Zhao S, Zhao D, Yang Y, et al. Association of polymorphisms in long non-coding rna h19 with coronary artery disease risk in a chinese population. Mutation research. 2015;772:15–22 [DOI] [PubMed] [Google Scholar]
- 22.de Goede OM, Ferraro NM, Nachun DC, Rao AS, Aguet F, Barbeira AN, et al. Long non-coding rna gene regulation and trait associations across human tissues. bioRxiv. 2019 [Google Scholar]


