Abstract
A β-lactamase gene was cloned from a Nocardia asteroides sensu stricto clinical isolate. A recombinant plasmid, pAST-1, expressed the β-lactamase AST-1 in Escherichia coli JM109. Its pI was 4.8, and its relative molecular mass was 31 kDa. E. coli JM109(pAST-1) was resistant to penicillins and narrow-spectrum cephalosporins. The β-lactamase AST-1 had a restricted hydrolytic activity spectrum. Its activity was partially inhibited by clavulanic acid but not by sulbactam and tazobactam. AST-1 is an Ambler class A β-lactamase sharing 65% amino acid identity with β-lactamase FAR-1, the most closely related enzyme.
The Nocardia genus includes several species that are opportunistic pathogens in immunocompromised patients (3, 13). Species of Nocardia asteroides sensu stricto are the predominant human pathogens and are involved in pulmonary and brain abscesses (13). Since nocardiosis requires a long treatment (6 to 12 months or longer) and may cause a high mortality rate, the choice of the optimal antibiotic treatement is crucial (7).
β-Lactams have been used as first-line treatment with little concern for the β-lactam susceptibility of Nocardia sp. isolates (13). Knowledge of the mechanisms of β-lactam resistance profiles of Nocardia isolates may be critical for assessing the potential clinical efficacy of β-lactams. A study of the antimicrobial susceptibility patterns of 78 clinical isolates belonging to the N. asteroides complex found that 95% of the isolates exhibit one of the four major antibiotic resistance patterns (24). Type I (20% of the isolates) is susceptible to ampicillin and carbenicillin but intermediate in susceptibility to imipenem; type III (18%) is susceptible to ampicillin and erythromycin; type V (17%) is resistant to broad-spectrum cephalosporins; and type VI, the most prevalent group (35%), is resistant to ampicillin but susceptible to extended-spectrum cephalosporins and imipenem. Type II and type IV are extremely rare and not well characterized. Wallace et al. show that drug resistance patterns of type III and type V correlate with taxonomic groups and have been reclassified as Nocardia nova and Nocardia farcinica, respectively (21, 22). Isolates belonging to types I, IV, and VI are grouped into the same subspecies, named N. asteroides sensu stricto.
Although some nocardial β-lactamases have been characterized biochemically in N. asteroides (9, 17), Nocardia brasiliensis (19, 23), and N. farcinica (11, 20), the accurate role of β-lactamase in the β-lactam resistance pattern has scarcely been explored. Sequences of β-lactamase genes are available only for N. farcinica and the nonhuman pathogen Nocardia lactamdurans (5, 11).
We report on the molecular and biochemical characterization of a class A β-lactamase named AST-1 from a clinical isolate belonging to the most prevalent group of N. asteroides sensu stricto species. Hydrolytic activity of β-lactamase AST-1 was compared to that of β-lactamase FAR-1.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The N. asteroides sensu stricto isolate JPL was from a pulmonary abscess of a 30-year-old human immunodeficiency virus-infected man. It was identified by molecular methods based on the restriction analysis of PCR fragments corresponding to the heat shock protein gene, as described previously (12, 18). The recipient strain Escherichia coli JM109 for cloning experiments and phagemid cloning vector pBK-CMV have been reported previously (11).
Antimicrobial agents and MIC determinations.
Antibiotic powders and their sources have been described previously (11). Antibiogram disks were used for routine antibiograms (Sanofi-Diagnostics Pasteur, Marnes-La-Coquette, France). MICs were determined by an agar dilution technique on Mueller-Hinton agar (Sanofi-Diagnostics Pasteur) with an inoculum of 104 CFU per spot as reported previously (11). All plates were incubated at 35°C for 18 h for E. coli and for 72 h for N. asteroides according to NCCLS guidelines (15). MICs of β-lactams were determined alone or in combination with a fixed concentration of clavulanic acid (2 μg/ml), sulbactam (8 μg/ml), and tazobactam (4 μg/ml).
Cloning experiments and genetic analysis.
Genomic DNA from N. asteroides sensu stricto JPL was extracted as previously described (13). Partially digested Sau3AI fragments of genomic DNA of N. asteroides JPL were ligated into BamHI-restricted phagemid pBK-CMV (Stratagene, La Jolla, Calif.). Ligation was performed at a 1:2 vector/insert ratio at a final concentration of 200 ng of DNA in a ligation mixture containing 1 U of T4 DNA ligase at 4°C for 18 h. Recombinant plasmids were transformed by electroporation (Gene Pulser II; Bio-Rad, Ivry-sur-Seine, France) into electrocompetent E. coli JM109 cells. Antibiotic-resistant colonies were selected onto Trypticase soy (TS) agar plates containing amoxicillin (50 μg/ml) and kanamycin (30 μg/ml) that were analyzed as described previously (13). Plasmid DNAs of recombinant strains were obtained using Qiagen columns (Qiagen, Courtaboeuf, France). Plasmid mapping was performed after double restriction analysis. DNA of one recombinant plasmid with the shortest insert was sequenced on both strands by using an ABI 377 sequencer (Applied Biosystems, Foster City, Calif.). The nucleotide and the deduced protein sequences were analyzed with software available over the internet at the National Center of Biotechnology Information webwite (http://www.ncbi.nlm.nih.gov) and at Pedro's Biomolecular Research Tools website (http://www.fmi.chbiology/research_tools.html).
β-Lactamase preparation.
Cultures of E. coli JM109 harboring recombinant plasmid pAST-1 were grown overnight at 37°C in 4 liters of TS broth with amoxicillin (50 μg/ml). The bacteria were harvested for 10 min at 6,000 × g, and the bacterial pellet was resuspended in 30 ml of 20 mM bis-Tris (pH 5.5) [bis(2-hydroxyethylimino)tris(hydroxymethyl)methane] at 4°C. The bacterial cells were disrupted by sonication (two times for 20 s at 20 Hz) (Vibra Cell 75022 Phospholyser; Bioblock, Illkirch, France) and were centrifuged (30 min, 10,000 × g, 4°C). The supernatant containing the enzyme extract was purified by ion-exchange chromatography with AGMP-1 exchanger (Bio-Rad). The exchanger in the chloride form was treated with 0.1 M ammonia in water and was then washed extensively with water. After adsorption of the extracts, elution was performed with 0.1 M NaCl. The active fractions were pooled, dialyzed extensively, and lyophilized.
Kinetic measurements.
Kinetic measurements were performed with the semipurified β-lactamase preparation extracted from E. coli JM109(pAST-1). The kinetic constants were determined by the online computerized microacidimetric method at pH 7.0 and 37°C as described previously (10). Vmax and Vmax/Km were expressed relative to that of benzylpenicillin (Vmax = 100). The 50% inhibitory concentrations (IC50s) were determined for clavulanate, sulbactam, and tazobactam as the concentration that reduced the hydrolysis rate of 100 μM benzylpenicillin by 50% under conditions in which the enzyme was preincubated with various concentrations of inhibitor for 5 min at 30°C before addition of the benzylpenicillin (10). The specific activity of the semipurified enzyme from E. coli JM109 harboring pAST-1 (AST-1) was obtained as described previously (16). One unit of the enzyme was defined as the activity which hydrolyzed 100 μmol of cephalothin per min per mg of protein. The total protein content was determined with bovin serum albumin as the standard (Bio-Rad DC protein assay kit).
IEF and determination of relative molecular mass.
Cultures of N. asteroides JPL were grown in TS broth at 35°C for 72 h in an aerobic atmosphere. β-Lactamase extracts from these cultures were obtained as described previously (11) and were submitted with the β-lactamase preparation from cultures of E. coli JM109 harboring recombinant plasmid pAST-1 to isoelectric focusing (IEF) analysis on an ampholine polyacrylamide gel, as described previously (11). The relative molecular mass of the β-lactamase from E. coli JM109(pAST-1) culture was estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis, as reported previously (16).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper have been assigned to the GenBank nucleotide database under accession no. AF 279904.
RESULTS AND DISCUSSION
Identification and susceptibility testing of N. asteroides sensu stricto isolate.
The N. asteroides isolate JPL was assigned to the type VI group of Steingrube et al. (18), which includes most of the N. asteroides sensu stricto isolates. MICs of β-lactams showed that this isolate was resistant to amino- and ureidopenicillins, narrow-spectrum cephalosporins, ceftazidime, and aztreonam (Table 1). Addition of clavulanate partially decreased the MICs of amino- and ureidopenicillins, while tazobactam and sulbactam did not have any significant effect (Table 1). These results were consistent with those obtained for other N. asteroides isolates (9), for N. farcinica (11, 20), and for Mycobacterium fortuitum (2, 6). Disk susceptibility testing showed that the N. asteroides isolate JPL was also susceptible to aminoglycosides (except kanamycin), tetracycline, and sulfonamides and resistant to fluoroquinolones, macrolides, fosfomycin, and chloramphenicol.
TABLE 1.
MICs of β-lactams for N. asteroides sensu stricto JPL, E. coli JM109 harboring recombinant plasmid pAST-1, and reference strain E. coli JM109
| β-Lactama | MIC (μg/ml)
|
||
|---|---|---|---|
| N. asteroides JPL | E. coli JM109(pAST-1) | E. coli JM109 | |
| Amoxicillin | 64 | >512 | 2 |
| Amoxicillin + CLA | 4 | 32 | 2 |
| Amoxicillin + TZB | 64 | >512 | 2 |
| Amoxicillin + SUL | 64 | >512 | 2 |
| Ticarcillin | 256 | 256 | 2 |
| Ticarcillin + CLA | 8 | 8 | 1 |
| Ticarcillin + TZB | 128 | 256 | 1 |
| Ticarcillin + SUL | 256 | 256 | 2 |
| Piperacillin | 512 | 128 | 1 |
| Piperacillin + CLA | 32 | 4 | 0.5 |
| Piperacillin + TZB | 128 | 128 | 0.5 |
| Piperacillin + SUL | 256 | 128 | 0.5 |
| Cephalothin | 128 | 8 | 4 |
| Cephalothin + CLA | 64 | 4 | 2 |
| Cephalothin + TZB | 128 | 8 | 2 |
| Cephalothin + SUL | 128 | 8 | 2 |
| Cefoxitin | 128 | 4 | 4 |
| Cefoxitin + CLA | 64 | 4 | 4 |
| Cefoxitin + TZB | 64 | 4 | 4 |
| Cefoxitin + SUL | 64 | 4 | 4 |
| Ceftazidime | >512 | 0.25 | 0.25 |
| Ceftazidime + CLA | >512 | 0.25 | 0.25 |
| Ceftazidime + TZB | >512 | 0.25 | 0.25 |
| Ceftazidime + SUL | >512 | 0.25 | 0.25 |
| Cefotaxime | <0.06 | 0.06 | 0.06 |
| Cefotaxime + CLA | <0.06 | 0.06 | 0.06 |
| Cefotaxime + TZB | <0.06 | 0.06 | 0.06 |
| Cefotaxime + SUL | <0.06 | 0.06 | 0.06 |
| Aztreonam | >512 | 0.12 | 0.12 |
| Aztreonam + CLA | >512 | 0.12 | 0.06 |
| Aztreonam + TZB | >512 | 0.12 | 0.06 |
| Aztreonam + SUL | >512 | 0.12 | 0.06 |
| Cefepime | 0.25 | 0.12 | 0.06 |
| Cefepime + CLA | 0.25 | 0.06 | 0.06 |
| Cefepime + TZB | 0.25 | 0.06 | 0.06 |
| Cefepime + SUL | 0.25 | 0.06 | 0.06 |
| Imipenem | 0.5 | 0.06 | 0.06 |
| Imipenem + CLA | 0.5 | 0.06 | 0.06 |
| Imipenem + TZB | 0.5 | 0.06 | 0.06 |
| Imipenem + SUL | 0.5 | 0.06 | 0.06 |
| Meropenem | 0.25 | <0.06 | <0.06 |
| Meropenem + CLA | 0.25 | <0.06 | <0.06 |
| Meropenem + TZB | 0.25 | <0.06 | <0.06 |
| Meropenem + SUL | 0.25 | <0.06 | <0.06 |
CLA, clavulanic acid at a fixed concentration of 2 μg/ml; TZB, tazobactam at a fixed concentration of 4 μg/ml; SUL, sulbactam at a fixed concentration of 8 μg/ml.
Characterization of the blaAST-1 gene and its expression in E. coli.
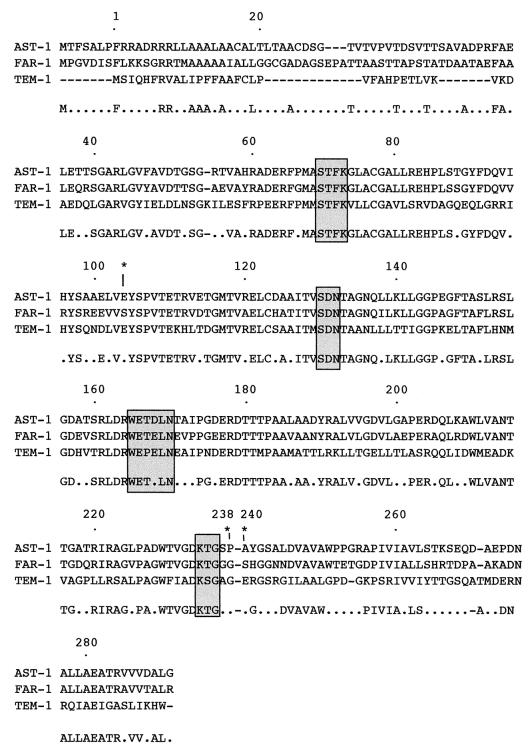
Two recombinant plasmids were obtained harboring the same 1.7-kb insert as a result of cloning experiments. One of them, pAST-1, was further characterized, and its insert was sequenced. It contained a 933-bp open reading frame (ORF), blaAST-1, encoding a 310-amino-acid protein named AST-1 (Fig. 1). The G+C content of blaAST-1 was 71.3%, which lies within the G+C ratios for other chromosomally encoded Nocardia sp. genes as recorded in the EMBL and GenBank sequence databases (64 to 72%). Moreover, 18 bp upstream of this ORF, part of another ORF was identified, the deduced protein of which shared 42% identity with a 561-amino-acid protein of unknown function from Streptomyces coelicolor (GenBank accession no. T35845). Additionally, 244 bp from blaAST-1, another ORF was identified, the protein of which shared 59% amino acid identity (within 89 amino acids) with a probable phosphorylating protein, UreD, from Mycobacterium leprae (GenBank accession no. S72992). These results are consistent with the Actinomycetales origin of blaAST-1. Since no ATG initiation codon was found for blaAST-1, a putative GTG was retained as its initiation codon (data not shown), as in several Streptomyces and Mycobacterium sp. genes (11). Within the deduced protein, structural elements characteristic of serine and Ambler class A β-lactamases were identified (1, 8) (Fig. 1). The comparison of the AST-1 sequence with those of other class A β-lactamases showed that it was distantly related to class A β-lactamases, including those of Streptomyces and Mycobacterium spp. (35 to 50% of amino acid identity). It was related mostly to FAR-1 β-lactamase from N. farcinica VIC, sharing 65% amino acid identity (11).
FIG. 1.
Alignment of the amino acid sequence of AST-1 with those of TEM-1 and FAR-1 β-lactamases. Numbering is performed according to the method described by Ambler et al. (1). The conserved residues among nocardial β-lactamases are reported below the alignment, with dots indicating conserved residues. Four structural elements characteristic of class A β-lactamases are boxed in grey. Some relevant amino acid positions that may correspond to amino acid changes in extended-spectrum TEM derivatives are indicated by asterisks. Dashes indicate gaps within the alignment.
MICs of β-lactams for E. coli JM109(pAST-1) showed mostly resistance to penicillins that was partially antagonized by addition of clavulanate (Table 1). These MICs mirrored those obtained for N. asteroides isolate JPL and for E. coli JM109(pFAR-1) expressing β-lactamase FAR-1, except that in this latter case, a slight increase of the MIC of aztreonam was observed (11).
Biochemical properties of the β-lactamase AST-1.
IEF analysis showed that cultures of N. asteroides isolate JPL and of E. coli JM109(pAST-1) produced an identical β-lactamase with a pI of 4.8 (data not shown). This pI was similar to those observed for β-lactamases of other N. asteroides isolates (4.2 to 4.6) (9) and was different from the pI of 5.8 of an N. asteroides isolate, as reported recently (17). Thus, N. asteroides isolates may possess different β-lactamases of acidic pI values. However, valid comparison of β-lactamase content based on pI values is difficult, since in the previous studies (9, 17), N. asteroides sensu stricto isolates were not differentiated from other N. asteroides spp. by molecular techniques.
The relative molecular mass of the β-lactamase AST-1 expressed in E. coli JM109(pAST-1) was estimated to be 31 kDa (data not shown), close to the value of 32 kDa for the β-lactamase FAR-1 (11).
The β-lactamase AST-1 was very poorly expressed from E. coli JM109(pAST-1) and N. asteroides JPL cultures (data not shown). The specific activity of the semipurified extract of E. coli JM109(pAST-1) was 0.11 mU · mg of protein−1 with 100 μM cephalothin as the substrate. Its purification factor was between 10- and 15-fold. Kinetic parameters of β-lactamase AST-1 revealed its strong activity against penicillins and narrow-spectrum cephalosporins (Table 2). As opposed to β-lactamase FAR-1, hydrolysis of aztreonam was not detected. As assessed by IC50s, the activity of inhibitors was weak, especially for sulbactam and tazobactam (Table 3). Similar results were obtained for β-lactamases extracted from N. asteroides, M. fortuitum, and N. farcinica (FAR-1) isolates (2, 6, 9, 11). Thus, it may be hypothesized that β-lactamases of Nocardia spp. are not susceptible to the β-lactamase inhibitors sulbactam and tazobactam. Susceptibility of N. brasiliensis β-lactamases to clavulanate is, however, greater than that of AST-1 (22). In one report, hydrolytic activity toward cefotaxime was noted for N. asteroides isolates (17). However, comparison with the activity of AST-1 is difficult, again since these N. asteroides isolates have not been grouped by molecular techniques (17).
TABLE 2.
Compared kinetic parameters for AST-1 from N. asteroides sensu stricto and FAR-1 from N. farcinica
| Substrate | Parameter for:
|
|||||
|---|---|---|---|---|---|---|
| AST-1
|
FAR-1
|
|||||
| Vmaxa | Km (μM) | Vmax/Km | Vmax | Km (μM) | Vmax/Km | |
| Benzylpenicillin | 100 | 30 ± 2 | 100 | 100 | 30 ± 2 | 100 |
| Amoxicillin | 53 ± 4 | 50 ± 4 | 32 | 115 ± 12 | 50 ± 3 | 69 |
| Ticarcillin | 8 ± 0.7 | 7 ± 0.5 | 33 | 30 ± 1 | 31 ± 1 | 29 |
| Piperacillin | 90 ± 6 | 330 ± 27 | 8 | 250 ± 26 | 45 ± 2 | 166 |
| Cephalothin | 40 ± 4 | 20 ± 1 | 60 | 85 ± 6 | 104 ± 9 | 2.5 |
| Cephaloridine | 57 ± 3 | >500 | <3.5 | 80 ± 5 | >500 | <5 |
| Cefoperazone | 12 ± 0.7 | >500 | <0.7 | NSb | NS | NS |
| Ceftazidime | <1 | >500 | <6 × 10−2 | <1 | >500 | <6 × 10−2 |
| Cefotaxime | <1 | >500 | <6 × 10−2 | 3 ± 0.2 | >500 | <0.2 |
| Aztreonam | <1 | >500 | <6 × 10−2 | 8 ± 0.5 | 400 ± 36 | 0.6 |
Vmax and Vmax/Km relative to that of benzylpenicillin, which was set at 100. Data are the means and standard deviations from three independent experiments.
NS, not studied.
TABLE 3.
Inhibition profiles of AST-1, FAR-1, and TEM-1 β-lactamases
| β-Lactamase | IC50 (μM) for:
|
||
|---|---|---|---|
| Clavulanic acid | Sulbactam | Tazobactam | |
| AST-1 | 0.7 | 960 | 67 |
| FAR-1 | 0.3 | 600 | 20 |
| TEM-1 | 0.08 | 6.1 | 0.1 |
AST-1, like FAR-1, is tazobactam resistant, like the inhibitor-resistant TEM derivatives occurring as acquired resistance mechanisms. Thus, AST-1 is another example of naturally occurring inhibitor-resistant β-lactamases that mimic molecular mechanisms involved in acquired β-lactam resistance. It would be interesting to investigate whether the N. asteroides isolate JPL produces clavulanate derivatives in a manner similar to that of the β-lactamase-producing N. lactamdurans isolate, which produces cephamycin derivatives (5). Since AST-1 activity is partially or totally resistant to inhibitors, antibiotic combinations containing amoxicillin-clavulanate, piperacillin-tazobactam, and ampicillin-sulbactam should be avoided in treatment of nocardiosis due to N. asteroides sensu stricto.
AST-1, as opposed to β-lactamase FAR-1, did not hydrolyze aztreonam. A few substitutions in TEM-derivative β-lactamases, such as Glu104Lys, Gly238Ser, and Glu240Lys, increase hydrolytic activity toward aztreonam (4, 14). FAR-1 possessed a serine residue in positions 104 and 240, as not found in the AST-1 sequence (Fig. 1). Thus, sequence differences between the two nocardial β-lactamases may account for the observed difference in the hydrolytic activity toward the monobactam aztreonam. Moreover, proline in position 238 in AST-1 sequence may modify the β-3 sheet structure, thus explaining the weak catalytic properties of AST-1.
Conclusion.
β-Lactamase AST-1 is the second class A β-lactamase characterized in a Nocardia sp. clinical isolate. As already mentioned for β-lactamase FAR-1 from N. farcinica, AST-1 expression cannot explain the entire β-lactam resistance profile of the N. asteroides sensu stricto isolate, especially concerning its resistance to aztreonam and ceftazidime. Additionally, other undetected β-lactamases and/or penicillin-binding affinities may account for this naturally occurring β-lactam resistance profile. Since AST-1 and FAR-1 β-lactamases shared significant amino acid identity and similar biochemical properties, they may derive from a common ancestor.
ACKNOWLEDGMENTS
L.P. and F.L. contributed equally to this work.
This work was funded by a grant from the Ministères de l'Education Nationale et de la Recherche (UPRES-JE 2227), Université Paris XI, Faculté de Médecine Paris-Sud, France.
REFERENCES
- 1.Ambler R P, Coulson A F W, Frère J-M, Guysen J M, Joris B, Forsman M, Lévesque R C, Tiraby G, Waley S G. A standard numbering scheme for the class A β-lactamases. Biochem J. 1991;276:269–272. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amicosante G, Franceschini N, Segatore B, Oratore A, Fattorini L, Orefici G, Van Beeumen J, Frère J-M. Characterization of a beta-lactamase produced in Mycobacterium fortuitum D316. Biochem J. 1990;271:729–734. doi: 10.1042/bj2710729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaman B L, Boiron P, Beaman L, Brownell G H, Schaal K, Gombert M E. Nocardia and nocardiosis. J Med Vet Mycol. 1992;30:317–331. [PubMed] [Google Scholar]
- 4.Cantu C, III, Huang W, Palzkill T. Selection and characterization of amino acid substitutions at residues 237–240 of TEM-1 beta-lactamase with altered substrate specificity for aztreonam and ceftazidime. J Biol Chem. 1996;271:22538–22545. doi: 10.1074/jbc.271.37.22538. [DOI] [PubMed] [Google Scholar]
- 5.Coque J J, Liras P, Martin J F. Genes for a β-lactamase, a penicillin-binding protein and a transmembrane protein are clustered with the cephamycin biosynthetic genes in Nocardia lactamdurans. EMBO J. 1993;12:631–639. doi: 10.1002/j.1460-2075.1993.tb05696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fattorini L, Amicosante G, Fiorentino D, Franceschini N, Di Marzio L, Oratore A, Orefici G. Inhibitors and inactivators of beta-lactamase from Mycobacterium fortuitum. J Chemother. 1989;1:293–297. doi: 10.1080/1120009x.1989.11738911. [DOI] [PubMed] [Google Scholar]
- 7.Gombert M E, Berkowitz L B, Aulicino T M, Dubouchet L. Therapy of pulmonary nocardiosis in immunocompromised mice. Antimicrob Agents Chemother. 1990;34:1766–1768. doi: 10.1128/aac.34.9.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joris B, Ledent P, Dideberg O, Fonze E, Lamotte-Brasseur J, Kelly J A, Ghuysen J-M, Frère J-M. Comparison of the sequences of class A β-lactamases and of the secondary structure elements of penicillin-recognizing proteins. Antimicrob Agents Chemother. 1991;35:2294–2301. doi: 10.1128/aac.35.11.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kitzis M D, Gutmann L, Acar J F. In-vitro susceptibility of Nocardia asteroides to 21 beta-lactam antibiotics, in combination with three beta-lactamase inhibitors, and its relationship to the beta-lactamase content. J Antimicrob Chemother. 1985;15:23–30. doi: 10.1093/jac/15.1.23. [DOI] [PubMed] [Google Scholar]
- 10.Labia R, Andrillon J, Legoffic F. Computerized microacidimetric determination of β-lactamase Michaelis-Menten constants. FEBS Lett. 1973;33:42–44. doi: 10.1016/0014-5793(73)80154-1. [DOI] [PubMed] [Google Scholar]
- 11.Laurent F, Poirel L, Naas T, Chaibi E B, Labia R, Boiron P, Nordmann P. Biochemical-genetic analysis and distribution of FAR-1, a class A β-lactamase from Nocardia farcinica. Antimicrob Agents Chemother. 1999;43:1644–1650. doi: 10.1128/aac.43.7.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laurent F J, Provost F, Boiron P. Rapid identification of clinically relevant Nocardia species to genus level by 16S rRNA gene PCR. J Clin Microbiol. 1998;37:99–102. doi: 10.1128/jcm.37.1.99-102.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lerner P I. Nocardiosis. Clin Infect Dis. 1996;22:891–903. doi: 10.1093/clinids/22.6.891. [DOI] [PubMed] [Google Scholar]
- 14.Matagne A, Lamotte-Brasseur J, Frère J-M. Catalytic properties of class A beta-lactamases: efficiency and diversity. Biochem J. 1998;330:581–598. doi: 10.1042/bj3300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7–A3. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 16.Poirel L, Naas T, Guibert M, Chaibi E B, Labia R, Nordmann P. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob Agents Chemother. 1999;43:573–581. doi: 10.1128/aac.43.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scopetti F, Fattorini L, Franceschini N, Amicosante G, Orefici G. Non-inducible, mainly cell-associated β-lactamase from Nocardia asteroides strain 108. J Antimicrob Chemother. 1997;40:5–11. doi: 10.1093/jac/40.1.5. [DOI] [PubMed] [Google Scholar]
- 18.Steingrube V A, Brown B A, Gibson J L, Wilson R W, Brown J, Blacklock Z, Jost K, Locke S, Ulrich R F, Wallace R J., Jr DNA amplification and restriction endonuclease analysis for differentiation of 12 species and taxa of Nocardia, including recognition of four new taxa within the Nocardia asteroides complex. J Clin Microbiol. 1995;33:3096–3101. doi: 10.1128/jcm.33.12.3096-3101.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steingrube V A, Wallace R J, Jr, Brown B A, Pang Y, Zeluff B, Steele L C, Zhang Y. Acquired resistance of Nocardia brasiliensis to clavulanic acid related to a change in β-lactamase following therapy with amoxicillin-clavulanic acid. Antimicrob Agents Chemother. 1991;35:524–528. doi: 10.1128/aac.35.3.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steingrube V A, Wallace R J, Jr, Brown B A, Zhang Y, Steele L C, Young G, Nash D R. Partial characterization of Nocardia farcinica β-lactamases. Antimicrob Agents Chemother. 1993;37:1850–1855. doi: 10.1128/aac.37.9.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallace R J, Jr, Brown B A, Tsukamura M, Brown J M, Onyi G O. Clinical and laboratory features of Nocardia nova. J Clin Microbiol. 1991;29:2407–2411. doi: 10.1128/jcm.29.11.2407-2411.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wallace R J, Jr, Nash D R, Johnson W K, Steele L C, Steingrube V A. Beta-lactam resistance in Nocardia brasiliensis is mediated by beta-lactamase and reversed in the presence of clavulanic acid. J Infect Dis. 1987;156:959–966. doi: 10.1093/infdis/156.6.959. [DOI] [PubMed] [Google Scholar]
- 23.Wallace R J, Jr, Steele L C, Sumter G, Smith J M. Antimicrobial susceptibility patterns of Nocardia asteroides. Antimicrob Agents Chemother. 1988;32:1776–1779. doi: 10.1128/aac.32.12.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallace R J, Jr, Tsukamura M, Brown B A, Brown J, Steingrube V A, Zhang Y S, Nash D R. Cefotaxime-resistant Nocardia asteroides strains are isolates of the controversial species Nocardia farcinica. J Clin Microbiol. 1990;28:2726–2732. doi: 10.1128/jcm.28.12.2726-2732.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]