Abstract
Microtubules deliver essential resources to and from synapses. Three-dimensional reconstructions in rat hippocampus reveal a sampling bias regarding spine density that needs to be controlled for dendrite caliber and resource delivery based on microtubule number. The strength of this relationship varies across dendritic arbors, as illustrated for area CA1 and dentate gyrus. In both regions, proximal dendrites had more microtubules than distal dendrites. For CA1 pyramidal cells, spine density was greater on thicker than thinner dendrites in stratum radiatum, or on the more uniformly thin terminal dendrites in stratum lacunosum moleculare. In contrast, spine density was constant across the cone shaped arbor of tapering dendrites from dentate granule cells. These differences suggest that thicker dendrites supply microtubules to subsequent dendritic branches and local dendritic spines, whereas microtubules in thinner dendrites need only provide resources to local spines. Most microtubules ran parallel to dendrite length and associated with long, presumably stable mitochondria, which occasionally branched into lateral dendritic branches. Short, presumably mobile, mitochondria were tethered to microtubules that bent and appeared to direct them into a thin lateral branch. Prior work showed that dendritic segments with the same number of microtubules had elevated resources in subregions of their dendritic shafts where spine synapses had enlarged, and spine clusters had formed. Thus, additional microtubules were not required for redistribution of resources locally to growing spines or synapses. These results provide new understanding about the potential for microtubules to regulate resource delivery to and from dendritic branches and locally among dendritic spines.
Introduction
Microtubules are cytoskeletal elements inside dendrites and axons that traffic essential components to and from synapses.[1] More than 90% of excitatory synapses occur on dendritic spines throughout the brain. Dendritic spines are highly dynamic structures, and their distribution along dendrites is not uniform. The growth, plasticity, and maintenance of dendritic spines are controlled by the local availability of subcellular resources including mitochondria, polyribosomes, smooth endoplasmic reticulum, and endosomes.[2–8] Transport along microtubules is required to deliver these core structures and proteins throughout the neuron. Hence, our first goal was to learn whether dendritic spine density scales with the distribution of microtubules throughout branching and tapering dendritic arbors.
Recent excitement in the field of nanoconnectomics has produced complete circuit reconstructions for most of C. elegans and parts of a fly brain using serial section electron microscopy.[9, 10] The connectomes have revolutionized understanding of these circuits [11–13], although the dense reconstructions exist for only a couple of individual organisms. Even the most sophisticated technologies have currently produced a few partial dense reconstructions through tiny portions of rat hippocampus and mouse cortex.[14–16]. Thus, this complete approach is not yet mature enough to gain new knowledge about variation and plasticity of synapses during development, learning, memory, and in disease states. Instead, unbiased sampling across multiple conditions and individuals is needed.
We have developed two powerful strategies to obtain unbiased samples using reconstruction from serial section electron microscopy (3DEM).[17] One is the unbiased brick, and the other is the unbiased dendritic segment. Both sampling approaches are subject, however, to nonrandom compositions of small and large caliber dendrites in the neuropil. Thus, sampling must control for intrinsic variation in resource availability in low and high caliber dendrites when comparing across treatment groups and individuals. We demonstrate how sorting dendrites by microtubule number avoids these biases, and provide examples of how it has been used in our prior studies to investigate synaptic plasticity.[3, 4, 18] The findings also provide new understanding about the potential for microtubules to regulate resource availability throughout dendritic arbors and locally to dendritic spines. Preliminary findings were published in abstracts at conferences.[19, 20]
Materials and Methods
Animals
Serial section images were analyzed from series that had been prepared for prior publications as described below. All procedures were carried out in accordance with National Institutes of Health guidelines for the humane care and use of laboratory animals and protocols approved by animal care and use committees at Children’s Hospital (Boston, MA), Medical College of Georgia (Augusta, GA), the University of Texas at Austin, or the University of Otago Animal Ethics Committee. All 3DEM data were collected from adult male Long-Evans rats.
Perfusion-fixation and in vivo electrophysiology
For datasets from area CA1 stratum radiatum, three rats (postnatal days 68–77 and weighing 310–411 g) under heavy pentobarbital anesthesia were transcardially perfusion-fixed with 2% paraformaldehyde, 2.5–6% glutaraldehyde, 2 mM CaCl2, and 0–4 mM MgSO4 in 0.1 M sodium cacodylate buffer (pH 7.35–7.4). The fixative was warmed to 37°C at the tip of the perfusion needle and was delivered with 4 psi pressure.[15, 21, 22]
Twelve datasets from the inner blade of the dorsal dentate gyrus were obtained from 4 rats (121–179 days old; 448–623 g) that underwent in vivo electrophysiology recordings (which induced long-term potentiation in the middle molecular layer of the hippocampal dentate gyrus in one hemisphere), as described previously.[23, 24] Here, we used datasets from the inner, middle, and outer molecular layers of the dorsal dentate gyrus, obtained from the contralateral control hemisphere that received only test pulses and long-term plasticity was not evident. In addition, three datasets were obtained from the CA1 stratum lacunosum-moleculare of two rats. One animal (170 days old) was from the in vivo electrophysiology experiment, obtained from CA1 stratum lacunosum-moleculare in the control hemisphere. The other animal (162 days old) did not undergo any prior experiments and was used to generate two SLM datasets, one from each hemisphere. For the datasets from the CA1 stratum lacunosum-moleculare and the dentate gyrus inner, middle, and outer molecular layers, rats were perfusion-fixed under halothane anesthesia and tracheal supply of oxygen.[25] The perfusion involved brief (~20 s) wash with oxygenated Krebs–Ringer Carbicarb buffer [concentration (in mM): 2.0 CaCl2, 11.0 D-glucose, 4.7 KCl, 4.0 MgSO4, 118 NaCl, 12.5 Na2CO3, 12.5 NaHCO3; pH 7.4; osmolality, 300–330 mmol/kg], followed by 2% formaldehyde and 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) containing 2 mM CaCl2 and 4 mM MgSO4 for ~1 hr (~1,900 mL of fixative was used per animal). The brains were removed from the skull at about 1 h after the end of perfusion.
Acute slices
Additional CA1 datasets were obtained from hippocampal slices. Slices (400 μm thickness) from two animals (postnatal days 50–60 and weighing 236–279 g) were cut at 70° transverse to the long axis from the middle third of the left hippocampus (four to six slices per animal) with a tissue chopper (Stoelting Co., Wood Dale, IL) and placed into ice-cold artificial cerebrospinal fluid (ACSF) containing (in mM) 116.4 NaCl, 5.4 KCl, 3.2 CaCl2, 1.6 MgSO4, 26.2 NaHCO3, 1.0 NaH2PO4, and 10 D-glucose, equilibrated with 95% O2–5% CO2, pH 7.4.[26] Slices from four other animals (postnatal days 56–65 and weighing 263–365 g) were prepared as above, except that they were cut into ACSF at room temperature.[27] All slices were gently transferred directly onto nets over wells filled with ACSF, at the interface of humidified 95% O2–5% CO2 at 32°C in a recording chamber (Stoelting Co.).
Microtubules are sensitive to intracellular calcium and can rapidly depolymerize in response to intense activation, such as that occurring during slice preparation.[2] During the first hour of recovery, when synaptic transmission is silent, microtubules visited some dendritic spines; however, in acute slices recovered for more than three hours, no spines contained microtubules. Furthermore, upon three hours of slice recovery the shaft microtubules regained their full number and length.[2] Hence, all slices were maintained for 4–6.5 hr in vitro prior to fixation. Physiological recordings were done to ensure slice viability. Two concentric bipolar stimulating electrodes were positioned 600–800 μm apart in the middle of stratum radiatum on either side of a single extracellular recording electrode. Slices were judged healthy if the stimulus–response curves were sigmoidal, and the half-maximal responses remained stable for at least 1 hr before experiments were done to compare control test pulses to tetanus or theta-burst stimulation that induced long-term potentiation (LTP). At the end of each experiment, the slices were fixed in mixed aldehydes containing 2% paraformaldehyde, 6% glutaraldehyde, 1 mM CaCl2, and 2 mM MgCl2 in 0.1 M sodium cacodylate buffer at pH 7.4 for 8–20 sec in the microwave oven, which resulted in a measured final temperature that was always < 50°C and usually < 37°C to prevent microtubule destruction that occurs above 60°C.[28] Only the control stimulation datasets were used for these analyses as explained in the results and discussion.
Tissue processing for 3DEM
Perfusion-fixed tissue and fixed slices were either hand- or microwave-processed by procedures as described previously.[15, 21, 22, 26, 27, 29] The fixed tissue was treated with reduced osmium (1.5% K4Fe(CN)6 and 1% OsO4 in 0.1 M sodium cacodylate buffer), followed by 1% OsO4 in 0.1 M sodium cacodylate buffer. The tissue was then stained en bloc with 1% uranyl acetate and dehydrated in a series of ethanol and propylene oxide, or in acetone. The dehydrated tissue was infiltrated with and embedded into LX-112 or Epon/Spurr’s resin. The resin blocks containing the tissue were trimmed to a trapezoid in the regions of interest (middle of stratum radiatum or stratum lacunosum-moleculare of area CA1; inner, middle, or outer molecular layer of dentate gyrus) and serial sections were cut with a diamond knife (DiATOME Ultra35 or Ultra45) on a ultramicrotome and collected on Synaptek slot grids (Ted Pella, Redding, CA) coated with Pioloform or polyetherimide film. Sections were counterstained with saturated ethanolic uranyl acetate, followed by Reynolds lead citrate[30] for 5 min each. A table of the specific processing details from these past studies is located in the Harris lab wiki here: https://wikis.utexas.edu/display/khlab/A+compilation+of+previously+used+tissue+processing+protocols. In addition, when protocols are updated, they can be accessed at the home site here: https://wikis.utexas.edu/display/khlab/KH+Lab+Wiki+Home.
Serial section TEM imaging
Sections were imaged on a JEOL transmission electron microscope (JEM-100B, JEM-1200EX, or JEM-1230) with film (4,000–10,000×) or with a Gatan UltraScan4000 CCD camera (5,000×). Images were acquired from all the SLM serial section series and some of the dentate series using a Zeiss field emission scanning electron microscope operating in the transmission mode using a transmitted electron detector (1.8–2.0 nm/pixel). A diffraction grating replica (Ernest Fullam, Inc., Latham, NY) was imaged along with the serial section series to calibrate pixel size (~2 nm for all datasets).
Reconstructions
The serial section images were aligned using the Reconstruct software ([31], RRID:SCR_002716; https://synapseweb.clm.utexas.edu/software-0) or Fiji with the TrakEM2 plugin. (RRID:SCR_002285; RRID:SCR_008954; http://fiji.sc, http://www.ini.uzh.ch/~acardona/trakem2.html).[32–34] Mean section thickness was calculated for each serial section series using the cylindrical diameters method: dividing the diameters of longitudinally sectioned mitochondria by the number of sections they spanned.[35] Dendrites were reconstructed through serial section electron microscopy using the Reconstruct software. All spines were identified to compute spine density per μm length of the dendritic segment. At the time of initial reconstruction, experimenters were blind as to condition.
Statistical Analysis
Microsoft Excel was used to create graphs and trendlines. The Excel data analysis package and XLSTAT (www.xlstat.com, Addinsoft) were used to compute statistical tests. The specific tests that were used are indicated in each figure legend with outcomes. The nonparametric Spearman’s r was computed on the raw data to determine where the correlations were statistically different from 0, and outcomes are expressed as r and p values in the graphs. Most of the distributions of the data were skewed, and so raw data are plotted on log-log axes with a power analysis used to compute the trendlines and goodness of fit (R2). Estimates for goodness of fit to the trendlines were computed and presented as R2 in power analyses for raw data plotted on the log-log plots. For ratios, the data were plotted on linear axes and R2 was based on a linear distribution. Log-transforms of these data were not used, as none reduced the skew sufficiently to meet parametric criteria. For the non-parametric data, the Kruskal-Wallis test was used, and results are reported as KW (observed, critical values) with p values in the figure legends. Dunn’s post-hoc test was used for multiple pairwise comparisons following a statistically significant Kruskal-Wallis outcome. Box-whisker plots are presented of the range and quartiles around the median. In a couple instances, the distributions of the raw data met parametric criteria and a single or multi-factor analysis of variance and covariance was done, and reported as the F value, degrees of freedom (df), p value, and effect size (η2). The number of dendrites for each analysis is reported either in the graphs or figure legends.
Data availability
Original images, the Reconstruct files, and the Excel spreadsheets will be shared on 3DEM.org via the Texas Data Repository (DOI: - to be obtained upon acceptance).
Results
The first purpose of this study was to identify an important sampling bias encountered when randomly sampling sections through neuropil to assess whether dendritic spines change across conditions. This problem is illustrated first by considering the nonuniform composition of random sections through the neuropil. Solutions to this problem are presented based on selecting dendrites of uniform caliber and resource delivery capacity via microtubules. Then, unbiased outcomes are presented based on comparisons across brain regions and cell types that illustrate how different dendritic morphologies address resource delivery via microtubules to the dendritic arbors in proportion to dendritic branching and synaptic loads.
Nonuniform distribution of processes in hippocampal neuropil
The variety and nonuniform distribution of dendrites, axons, and glia in hippocampal neuropil is illustrated in an example field selected from the middle of a set of serial sections (Figure 1). Even within this relatively small sample area, one can readily distinguish neighboring zones in the same image field which are dominated either by large caliber dendrites, smaller caliber dendrites, or myelinated axons. This lack of uniformity means that random sampling of synapses or spines in single sections may not give representative outcomes; instead, serial reconstruction is required. Thus, when experiments are done to determine if developmental stage, aging, brain region or some experimental or environmental manipulation has altered spine density, it is crucial to control for dendrite caliber and its location in the dendritic arbor. Here, dendrites of varying calibers were reconstructed from control series in hippocampal area CA1 and dentate gyrus. The potential impact of resource delivery was assessed by the strength of the correlation between microtubule number and spine density along dendrites of varying caliber as a reflection of their position in the dendritic arbor.
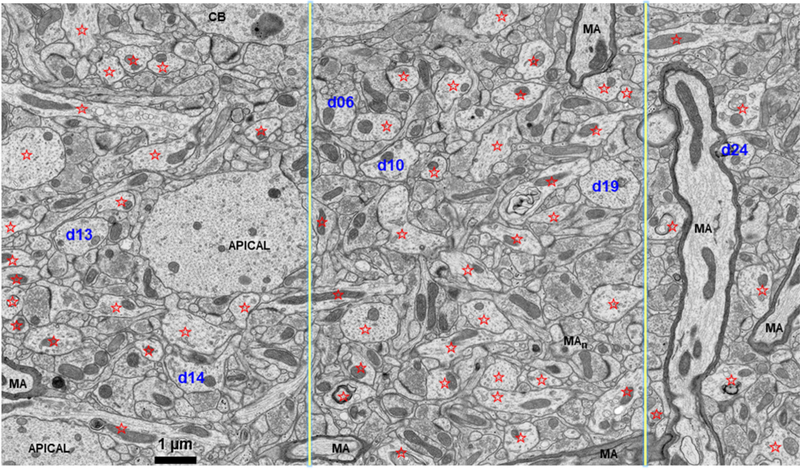
Figure 1:
Image from an adult rat hippocampus (perfusion-fixed in vivo) from the middle of CA1 stratum radiatum illustrating the non-uniform composition of the neuropil. Dendrites are indicated by red stars, and five that were analyzed for this paper are numbered (blue). The two yellow lines divide this image into three parts. The left side is occupied by several longitudinally sectioned thin dendrites and two large caliber apical dendrites that reduce the area where synapses could form. In the middle region, most of the processes are cross-sectioned and constitute smaller dendrites, axons, synapses, and glia. The right side has several longitudinally sectioned myelinated axons (MA, also, the process labelled MAn in the middle region was initially confused with dendrites until viewed through serial sections and then recognized as a node of Ranvier with perinodal folds on later sections in the series).
Quantifying microtubules, dendrite diameters and spine density
Microtubules were quantified at multiple locations along each dendritic segment where they appeared in cross-section, making the count relatively easy to obtain, as illustrated in stratum radiatum of hippocampal area CA1 (Figure 2A, B). Serial sections were viewed to distinguish microtubules from tubules of smooth endoplasmic reticulum (SER) or microfilaments. Microtubule numbers and dendrite diameters were averaged from three locations along each dendrite. The length of the dendrite segment was corrected for the potential bias introduced by over counting spines at the cut ends of the segment.[13] The unbiased dendritic length was determined by starting at the first spine origin and ending at the beginning of the last spine origin, and not including the last spine in the count (Figure 2C). Spine density was computed as the number of spines per micron length of the unbiased dendrite segment. Each branch of a branched spine was counted separately. Mitochondria, SER, and other membrane bound organelles were found in close association with the microtubules, obscuring them in some sections (Figure 2D, E). Usually upon viewing neighboring sections the microtubules were separated from the organelles and became readily distinguished.
Figure 2:
Example cross-sectioned dendrite in the middle of stratum radiatum of area CA1 in adult rat hippocampus illustrating how microtubules and dendrite diameters were quantified in the dendritic shafts and compared to spine density. A-B) Microtubules were readily identified as cylindrical structures (~20 nm diameter) with average inter-microtubule spacing of ~35 nm to accommodate microtubule-associated proteins. A and B are section 110 and 111 of a series, and A’ and B’ illustrate outlines of the same microtubules (blue circles). The dendrite diameter (orange line) was measured across the middle of the narrowest orientation of the cut dendrite. C) Complete three-dimensional reconstruction of the dendritic segment used to compute spine density per unbiased segment length (length, thick black line). Three locations along the length where microtubules were counted are illustrated (blue ellipses, 60 by 10 nm) along with the three dendrite diameter measures (orange lines). Dendrite and spines are a translucent, pale yellow with postsynaptic density areas in red. D) Same dendrite illustrating a section through the origin of a large spine, and three organelles that are associated with microtubules: smooth endoplasmic reticulum (green), a mitochondrion (orange), and a multivesicular body (pink with vesicles outlined in light blue). E) Enlarged reconstruction of this dendrite containing the microtubules (dark blue), mitochondrion (orange), SER (green), and the multivesicular body (pink with black vesicles for illustration in 3D). (Scale bar in A is for A-B and D.) F) Significant correlations (Spearman’s r) between average dendrite diameter and associated number of microtubules in the dendritic shaft, where a power analysis reveals that 52% of the variance can be attributed to this relationship (black values; trendline y=0.2×0.4). However, dendrite diameter was essentially uniform over the range of 9–21 microtubules, where the correlation became non-significant (red values; y=0.3×0.2). G) The significant correlation (Spearman’s r) between spine density and microtubules (black values, y=1.8×0.3) became statistically non-significant when restricted to dendrites with 9–21 microtubules (red; y=2.3×0.2). H) The relationship between spine density and number of microtubules depended on the functional status of the hippocampus. For all three conditions there is a statistically significant correlation between spine density and number of microtubules (Spearman’s r), although the goodness of fit was less in the 30 min condition (perf y = 2×0.3, R2= 0.45; 2 hr y = 1.8×0.3, R2 = 0.70; and 30 min y = 2×0.3, R2=0.29). As expected, an ANCOVA revealed no significant difference in this relationship between the 2 hr and perfusion-fixed hippocampus (green, perfused data points are hidden for clarity). In contrast, ANCOVA comparing the 30-minute data to the combined perf and 2 hr data revealed a significant difference by condition (F= 6 by condition, p<0.02, n=72, df=69), with an effect size of 46% (eta-squared). Overall, ANCOVA results show the relationship between spine density and number of microtubules is strong (F=53 for MT, p<0.0001, n=72, df=69).
There were strong and significant correlations between dendrite diameters (Figure 2F), spine densities (Figure 2G), and the number of microtubules the dendritic segments contained, where more than 50 percent of the variance could be attributed to these relationships. The number of microtubules increased by a factor of 10 while both the diameter of the dendrite and spine density increased by a factor of ~3, close to the square root of 10. These relationships make sense according to the following argument. Suppose it takes 100 microtubules at a certain point on a dendrite to service all the dendritic spines from there out to the distal tips. At that point, the dendrite would need to be large enough to contain 100 microtubules, which means that the cross sectional area of the dendrite must be proportional to 100. However, since the cross sectional area grows as the square of the diameter, the diameter needs only to be proportional to 10. Similarly, if one were to assume that spines have a set density per dendrite diameter (or volume or surface area, all of which are proportional to dendrite diameter), then the local spine density should also grow as the square root of the number microtubules (as observed in Figures 2G, F).
For comparisons across conditions, it is important to determine the range in microtubule number over which dendrite caliber and spine density are essentially constant. Small caliber lateral dendrites are desired because they are highly represented in relatively small EM image volumes, and hence, can be sampled more frequently through a single set of serial sections than large caliber dendrites. Dendrite diameter was uniform over the range of 9–21 microtubules (Figure 2F). Similarly, spine density was also uniform over this same range of 9–21 microtubules (Figure 2G). Thus, restricting analysis to a discrete, relatively small range in microtubule number, produced a sample of dendritic segments of uniform caliber and spine density.
An example from prior experiments shows that changes in spine density for the discrete range of 9–21 microtubules can also reflect changes in spine density across dendrites of larger calibers. Archival data showed that average spine density was initially lower in hippocampal slices from young adult rats but recovered to in vivo (perfusion-fixed) levels under specific conditions in vitro.[18, 36] This spine recovery occurred during a rest period of 3–4 hours after slice preparation that was followed by two hours of control stimulation.[36] However, if the control stimulation was given for just 30 minutes, spine density did not reach the perfusion-fixed values. Here, additional higher caliber dendrites were added and an analysis of covariance showed that spine density was comparable between the perfusion fixed and two hour control stimulation condition across the larger range in dendrite microtubule numbers (Figure 2H). The dendrites that received only 30 minutes of control stimulation had a significantly lower spine density across the larger range in dendritic microtubule numbers (Figure 2H). These results show that the large samples of uniform low caliber dendrites needed for efficient 3DEM sampling, can produce outcomes that reflect a larger range in caliber and microtubule numbers. Whether spine density is altered uniformly across the dendritic arbor under other conditions remains to be determined in future experiments.
Assessing the impact of dendrite branch points on microtubules and spine density
Dendritic segments before and after branch points were evaluated to determine whether the microtubules entered the branches in proportion to spine density (Figure 3). One large dendritic segment was fully reconstructed across 100 serial sections with a branch point, and included the spines, synapses, microtubules, and associated organelles (Figure 3A–D). These structures were also viewed through an additional 100 serial sections. Three mitochondria were located near the branch point (Figure 3E). The pink and dark blue mitochondria were continuous at the beginning and last section of the series, thus spanning more than 10 microns. The dark blue mitochondrion divided at the branchpoint and spanned the entire length of the series containing both the large and small caliber dendritic segments. The small orange mitochondrion was complete within the series, and one end curved from the primary into the lateral branch.
Figure 3:
Conservation of microtubule number and spine density across dendritic branch points. A-C) Three electron microscopic images before, at, and after the branch point. Yellow is the primary dendrite and blue is the lateral branch. D) 3D reconstruction of the primary and lateral dendritic branch with subcellular organelles reconstructed surrounding the branch point (green SER, red synapses). E) Microtubules (MT, gray and blue lines) and three mitochondria (pink, blue, orange) pass by and/or into the lateral branch. F) Microtubules in the primary dendrite before and after the branch point (gray) and crossing into the lateral branch (blue). MT counts and spine densities (Sp/μm) are provided for each location. G-I) Circles were placed on each section just outside the boundaries of the dendrites to measure the lengths of the primary dendrites before (yellow) and after branch points (blue, pink). Unique dendritic spines are illustrated as spheres (20 nm) and color coded to match the dendritic segments or lateral branches (turquoise, orange). The microtubules are illustrated as dark blue 60 nm tall by 10 nm wide ellipses in each branch (black arrows). G) The primary dendrite splits into two nearly equal sized branches that each receive about half of the microtubules. H) A large dendrite gives rise to one thinner lateral branch with a proportionate number of microtubules and spine densities. I) A large dendrite gives rise to two lateral dendrites. J) Range in number of microtubules lost or added across branch points. K) Proportionality of microtubules to spine density across branch points differs depending on the type of branching.
Microtubules before and after the branch points were compared to local changes in spine density along the primary dendrite and branches. Most of the microtubules were oriented along the length of the larger dendritic segment, however several curved and traversed across its diameter to enter the smaller dendritic branch (Figure 3E, F). In this example, some microtubules ended on the small orange mitochondrion, as though they were delivering it into the branch. The 142 microtubules before the branch point were three greater than the sum of the 129 microtubules remaining in the primary branch plus the 10 in the lateral branch. The lost three could be attributed to those ending on the orange mitochondrion, although other microtubules also started and stopped as they traversed the branch point. Nine dendritic branch points were encountered in the CA1 stratum radiatum datasets. Three of the large dendritic segments split into two smaller dendritic branches (Figure 3G), two had a large primary dendrite and one thinner lateral branch (Figure 3H), and two had a large primary dendrite with two thinner lateral branches (4 total branch points, Figure 3I). Across these branch points the differences in microtubule number ranged from 4 less to 7 more microtubules (Figure 3J).
Overall, there was a significant correlation between the ratio of spine densities to the ratio of microtubule numbers across each branch point (r=0.6, p<0.02). Interestingly, this relationship differed substantially when branch type was considered. The Y-branches tended to have 2–3 times as many microtubules before the branch point that distributed roughly equally into the two branches having a ratio of 1–2 spines/micron on each branch (Figure 3K, green). When the branch point involved a larger dendritic segment giving rise to a smaller branch, the ratio of microtubules was proportional to the spine density on each branch (Figure 3K, blue). These relationships were not nearly as strong when multiple branches emerged from a large dendrite in these short series (Figure 3K, pink). Furthermore, one additional dendritic segment had 70 microtubules and gave rise to a branch that itself branched four more times within a few micrometers of the first branch point. These four branches were longitudinally sectioned and partially located at the edge of the series, and so accurate spine and microtubule counts could not be obtained for them (and they are not included in the graphs). This primary dendrite was cross sectioned for 9 microns prior to the complicated branch point, and there were only five dendritic spines for a very low density of 0.55 spines per micron, less than half the lowest density shown over longer stretches without branches in Figure 2H above. Together these findings suggest that the relationship between microtubule number and spine density may depend on the degree of local dendrite branching as well.
Comparing relationships between spine density and microtubules across dendritic arbors
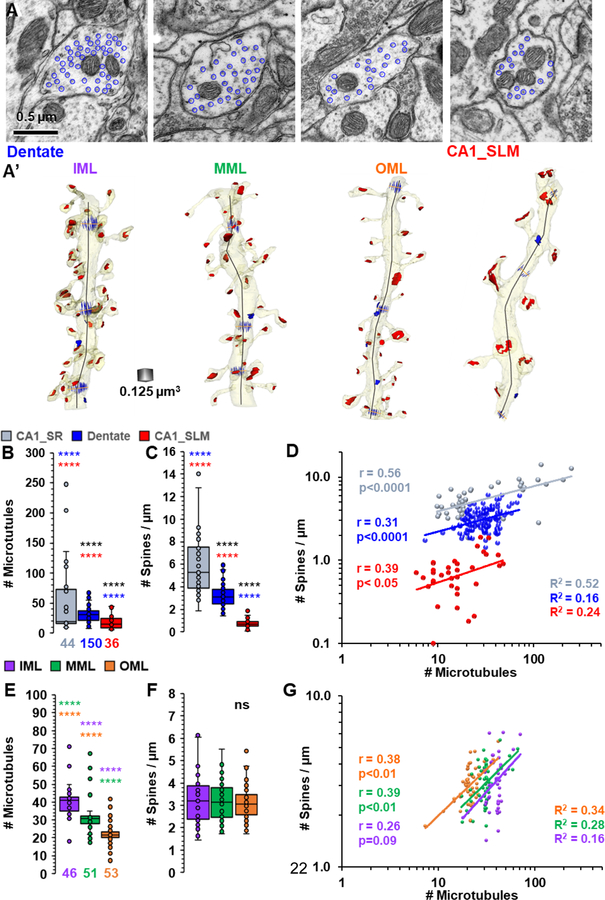
The relationship between spine density and microtubules was determined across axonal input layers in the hippocampal dentate gyrus and compared to those in CA1 stratum radiatum and CA1 stratum lacunosum moleculare. The strategy described above was applied to measure dendrite diameters and count microtubules at all the locations, and to compute spine densities along each dendritic segment (Figure 4A, A’). On average, there were more microtubules in the dendrites from CA1 stratum radiatum than in dentate gyrus or CA1 stratum lacunosum moleculare (Figure 4B). In parallel, spine density was also greater on average in CA1 stratum radiatum than dentate gyrus, which was greater than CA1 stratum lacunosum moleculare (Figure 4C). In all layers, there was a significant correlation between spine density and the number of microtubules in the associated dendrite; however, the goodness of fit was best in CA1 stratum radiatum (Figure 4D). In the dentate gyrus, the microtubule number decreased with distance from the cell body, such that the number was greater in the inner molecular layer than the middle molecular layer, which was greater than in the outer molecular layer (Figure 4E). In contrast, with the dramatic differences between the proximal CA1 stratum radiatum and distal stratum lacunosum moleculare, spine density did not differ significantly across the dendritic arbor of the dentate gyrus (Figure 4F). Nevertheless, there was a small correlation between spine density and microtubule number within the middle and outer molecular layers that did not reach statistical significance in the inner molecular layer (Figure 4G). A uniform spine density was achieved when microtubule number was restricted around the median value in the inner molecular layer to 40–45 microtubules, middle molecular layer to 30–35 microtubules, outer molecular layer to 20–25 microtubules, and CA1 stratum lacunosum moleculare to 11–16 microtubules. Together these data suggest that the absolute values for spine densities and microtubule numbers differ dramatically across brain regions and axonal input types; nevertheless, a correlation between spine density and microtubule number persists.
Figure 4:
Differences in the relationship between spine density and microtubule number across axonal input layers in the hippocampus. A) Representative electron micrographs, and A’) matching 3D reconstructions from dendrites in the inner molecular (IML), middle molecular layer (MML), and outer molecular layer (OML) of the dentate gyrus, and CA1 stratum lacunosum moleculare (CA1_SLM). Scale in A is for all EMs; blue circles surround each microtubule. The reconstructed dendrites are displayed in translucent yellow with red postsynaptic density areas, blue microtubules illustrated at 3 measurement points along their lengths, and the length (black line) measured down the center of the dendrite. B) The average number of microtubules in each dendrite. CA1 stratum radiatum (CA1_SR) dendrites are from the perfusion-fixed and two-hour control dendrites described in Figure 2. C) Spine density along reconstructed dendritic segments. D) Correlations between spine density and the number of microtubules across regions. E) Average microtubule number and F) spine density across input layers in dentate gyrus. G) Correlations between spine density and the number of microtubules for input layers of the dentate gyrus. (B, C; E, F: Total number of dendrites in each location is indicated at the base of each box plot, multiple dendrites overlap each data point; **** indicate p<0.001 in post-hoc Dunn’s test following a significant overall KW(127, 6); p<0.0001; stars above each bar are color coded for differences between locations or layers. D, G: r = Spearman’s correlation coefficient with p values. R2 provides goodness of fit of the power analysis model).
Finally, it was of interest to learn whether the number of spines served by each microtubule differed across the dendritic arbors. CA1 pyramidal cells have a long thick apical dendrite, tapered lateral dendritic branches, and tapered terminal dendritic tufts, and the total arbor extends about 700–900 microns from the soma in adult rats. In contrast, the dentate granule cells have a shorter, conical shaped dendritic arbor with dendritic branches that taper rather more uniformly with distance from the soma, which is about 400–500 microns at the distal tips. Microtubule lengths have been calculated in prior work in area CA1 stratum radiatum and averaged 91.3±19.6 μm.[2] Thus, if this length generalizes across neurons, individual microtubules do not extend from the soma to distal tips, and resources from the soma and elsewhere will be transferred from one microtubule to another during transport. If we assume, that the resources on all the microtubules are available to local dendritic spines, we can then compute the number of spines per microtubule as the spine density (number / micron) being multiplied by average microtubule length (91 μm) and divided by the number of microtubules in each dendritic segment (Figure 5). In area CA1, microtubules in the more proximal dendrites of stratum radiatum on average served more spines than the more distal dendrites of stratum lacunosum moleculare (Figure 5A). Within CA1 stratum radiatum, the microtubules in the thinner lateral dendrites (with lower microtubule numbers) each could serve resources to more spines than individual microtubules in the thicker apical dendrites (Figure 5B). Similarly, within CA1 stratum lacunosum moleculare, microtubules in the thinner dendrites each could serve resources to more spines (Figure 5B). In the dentate gyrus, a somewhat different pattern emerged, where the number of spines served per microtubule increased on average with distance from the soma (Figure 5C). Nevertheless, within each layer of the dentate gyrus, the number of spines served per microtubule was greater along the thinnest dendrites, having fewer microtubules (Figure 5D).
Figure 5:
Number of spines associated with each microtubule. A) The number of spines per microtubule (MT) was greater in CA1_SR than more distally in CA1_SLM (KW(42, 4), p<0.0001, n=number of dendrites beneath the box plots). B) Significant negative correlations between the number of spines per microtubule and number of microtubules in each CA1-SR or CA1_SLM dendrite (Spearman’s r; power analyses R2). C) Number of spines per microtubule increased with distance from the soma in the dentate gyrus (Overall KW(64, 6), p<0.001), and Dunn’s post-hoc tests with Bonferroni correction set to 0.017, ***p=0.005, ****p<0.001 with comparisons color coded above each box plot). D) Significant negative correlations between the number of spines per microtubule and number of microtubules in each MML and OML dendrite, but not in the IML dendrites (Spearman’s r; power analyses R2. For these calculations previously calculated average microtubule lengths were used: 91 μm for perfusion fixed brain and 84 μm for recovered brain slices.)
These observations suggest that microtubules in the proximal thicker dendrites might only serve more distal dendritic branches, which would account for the lower spine number per microtubule on all the thicker relative to the thinner dendrites. In contrast, microtubules in the distal tapering dendrites need serve only the local spines, and hence more spines could be served per microtubule. These observations are interesting because they suggest less resources are trafficked on each microtubule in the distal CA1 dendrites, resulting in lower absolute spine densities (See figure 4 C). In contrast, the shorter conical dendritic arbor of dentate granule cells maintained a constant spine density despite the reduction in microtubule number at the tapered tips. Thus, the absolute capacity for resource distribution by microtubules along dendrites differs not only with distance from the soma, but also across diverse dendritic arbors and brain regions.
Discussion
Microtubules traffic molecules, membrane bound organelles, and other subcellular constituents bidirectionally throughout the neuron.[1, 37] Here we show that dendrite diameters and spine density were correlated with the number of microtubules, suggesting that critical resources are delivered to large and small caliber dendrites in proportion to their synaptic load. However, the strength of this relationship varied dramatically across regions and input layers.
In area CA1, the large apical dendrite contains numerous microtubules that are distributed from the main dendritic shaft into lateral branches in stratum radiatum in proportion to branch caliber and spine density. The large lateral dendrites also branch, and their microtubules are distributed in proportion to spine density. At Y-branch points in stratum radiatum, the ratio of spine density to microtubule density was between 1 and 1.5 as the presumably terminal branches were of nearly equal caliber. In CA1 stratum lacunosum moleculare, there was low variation in absolute microtubule number and spine density, and a weaker correlation between spine density and microtubule number across the terminal tuft dendrites relative to the more proximal dendrites in stratum radiatum. Thus, it appears that the apical dendrite acts like an extension of the soma, distributing microtubules and resources proximally in proportion to dendritic branching and synaptic load, and essentially equally among the branches of the terminal dendritic tufts with their comparable synaptic loads.
Hippocampal dentate granule cells lack a large apical dendrite. Instead, the dendritic arbor is cone-shaped with the proximal thicker dendrites giving rise to thinner branches that taper with distance from the soma. Although microtubule number decreased with distance, spine density remained constant throughout the dentate granule cell’s dendritic arbor. The correlation between microtubule number and spine density increased from inner to middle to outer molecular layers, suggesting that the proximal dendrites distribute microtubules and resources both to and from local dendritic spines, and to and from more distal dendritic branches and spines.
At most, 56% of the variance in spine density was attributed to local variance in microtubule number across thick and thin dendrites. Many other factors could contribute to variance in microtubule number. Two examples were illustrated in this paper. First, near branch points, spine density could be higher, lower, or even absent relative to surrounding dendritic segments. This branch-point variance may reflect competition for resources among local dendritic spines and the emerging dendritic branches, which themselves give rise to numerous dendritic spines past the branch points. A second example relates to mitochondria, as illustrated in our datasets. Most of the microtubules were oriented parallel to the longitudinal axis of the dendritic shaft; however, some microtubules turned to cross the dendritic shaft where they appeared to capture and redirect mitochondria, and possibly other substances, from the main dendritic shaft into branches. These microtubules appeared to end on the mitochondria, and hence the presence or absence of mitochondria and other resources may also contribute to variance in microtubule numbers. Some mitochondria were long and formed branches into multiple dendrites, as noted previously.[38–40] These long branched mitochondria are likely to be quite stable, and would likely require fission to become mobile.[41, 42] Activity levels might help to determine where mitochondrial motors detach from microtubules and allow mitochondria to become tethered by actin filaments to regions of high local protein synthesis [43] It will be interesting to learn more about how microtubules direct resources into dendritic branches, and whether the high variance in spine density around branch points is due to the intrinsic variance in resource distribution or to external hindrance of axonal access to dendrites and spines at branch points.
Microtubules are depolymerized during slice preparation but regain their total length during a 3 hour incubation period.[2] Additional activation is required for spine density to be recovered to in vivo levels.[18, 36] Here we showed that this loss and recovery of dendritic spines is not restricted to the thin dendrites measured in the prior studies but also occurs across dendrites with a broad range in microtubule number and resource availability. In the past, we have also reported a transient visitation of microtubules into about 15% of dendritic spines during the initial silent recovery period (~30 minutes) following the making of hippocampal slice,[2] when activation does not produce a synaptic response.[44] These spine microtubules are not permanent and may be an artifact of the low calcium concentration at inactive synapses, as they are not present in perfusion-fixed brain or following a few hours of slice recovery in vitro.[2] Others have reported microtubule visitation into dendritic spines in developing cultured neurons.[45] No microtubules were found inside dendritic spines in any of the current datasets. Instead, their distribution in the dendritic shafts was proportional to spine density. Our calculations, based on prior estimates of total microtubule length and current spine densities show an average of about 10 spines per microtubule; however, the range could exceed 50 dendritic spines per microtubule in the large CA1 dendrites of stratum radiatum. These findings suggest that microtubules support local delivery of organelles and other important constituents throughout dendritic shafts, but other motility mechanisms, presumably involving actin filaments, are engaged in delivering resources into dendritic spines.[46–48]
Cutting and collecting serial sections across 5–10 microns is routine (i.e., 100–200 serial thin sections each ~35–50 nm thin). Similarly, imaging is routine for 10×10 micron fields with transmission electron microscopes and 50×50 micron fields in transmission mode on a scanning electron microscope.[49] This work is nevertheless exceedingly time consuming, even for these small fields. Thus, it is important to have an unbiased approach to sample dendrites of comparable caliber when assessing spine density. Theoretically, restricting either the ranges of dendrite diameter or microtubule number would obtain unbiased samples. We have chosen microtubule number because it provides insight about the availability to redistribute resources locally, as changes in synaptic efficacy or spine density occur. New tools that automate imaging of larger fields, their reconstruction, and the enhancement of microtubules are needed to improve the efficiency of testing whether microtubule distribution is proportional to synaptic load in other brain regions, and under a variety of experimental and disease states.
Taken together, the present findings suggest that microtubules are distributed in proportion to the total synaptic load of dendritic branches. Our prior studies of synaptic plasticity have restricted analysis to dendrites with comparable microtubule counts to avoid the correlational bias with spine density. In this way, synaptic plasticity could be assessed between control and LTP conditions.[4, 18, 24, 50, 51] For example, local LTP-related synaptic growth and elevated spine density were counterbalanced by reduced spine density in neighboring portions along the same dendritic segment. Where synapses enlarged and spines clustered, there was enhanced local composition of smooth endoplasmic reticulum, polyribosomes, and endosomes in both the dendritic shaft and dendritic spines.[3, 4] Where spine density and synapse sizes were less, subcellular dendritic resources were also less. Microtubule number was the same across the control and LTP dendrites, as well as within individual dendritic segments that showed synapse enlargement and spine clustering, or not. Hence, new microtubules were not needed; instead, existing microtubules provided adequate redistribution of resources locally to growing synapses. Additional work will be needed to determine whether spine density is the best measure of synaptic load. Perhaps the total synaptic input and size, including inhibitory synapses, will be an even stronger predictor of microtubule support of synaptic load and plasticity across dendritic arbors.
Highlights:
3DEM reveals dendrite diameter and spine density correlate with microtubule number.
In thick dendrites, microtubules support both dendritic branches and spines.
In thin unbranched dendrites, microtubules need only support local dendritic spines.
Existing dendritic microtubules are sufficient to supply synapses after plasticity.
Resources are delivered to and from dendrites in proportion to synapse load.
Acknowledgements
National Institutes of Health, Grant/Award Numbers: MH-095980, MH-104319, NS021184, NS033574, NS074644; National Science Foundation, Grant/Award Numbers: 1707356 and 2014862
KMH – designed experiments, did perfusions, curated the data, performed statistical analyses, prepared final figures, and wrote the paper.
DDH – performed substantial final data analyses on the CA1 slice data and the dentate layers; helped curate data and reconstruct dendrites; provided Python programming to aid in data aggregation and curation.
MK – dentate series (tissue processing, imaging, reconstruction); SLM series (perfusion, tissue processing, imaging, reconstruction); helped write the paper and prepare SLM figures, confirmed data sources, and methods.
After MK, co-authors are listed alphabetically by last name for the contributions listed below: WC Abraham – contributed animal experiments for the dentate and SLM analyses, edited final manuscript.
JB – conducted dentate gyrus experiments and analyses and presented an earlier poster on the topic.
JNB – performed slice experiments, performed reconstructions and initial data analyses, and gave poster presentations from preliminary data.
AH – participated in earlier reconstructions, data analysis, and a poster presentation.
PHP – performed reconstructions, curated reconstructed dendrites, and edited the paper.
JMM – helped design prior analyses of branched dendrites, helped prepare dentate series tissue processing and imaging and participated in poster presentations.
BS – performed some prior reconstructions and participated in early analyses.
JS – performed reconstructions and participated in early analyses and poster presentations.
Undergraduate tracers are thanked for help with initial tracing (that was curated by the coauthors): Junho Ahn, Katherine E. Dembny, Ryan Ellis, Corey N. Haines, Dakota Hanka, Alyssa N. Herbort, Matthew Hooper, Zean A. Luna, Jordan N. Mackey, Amy Pohodich, Laura Robertson, Kyle Zatyko. Marcia Lind, Clayton Smith, Libby Perry, and Robert Smith are thanked for EM preparation of some of the older datasets.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Ehlers MD, Dendritic trafficking for neuronal growth and plasticity, Biochem. Soc. Trans, 41 (2013) 1365–1382. [DOI] [PubMed] [Google Scholar]
- [2].Fiala JC, Kirov SA, Feinberg MD, Petrak LJ, George P, Goddard CA, Harris KM, Timing of neuronal and glial ultrastructure disruption during brain slice preparation and recovery in vitro, J Comp Neurol, 465 (2003) 90–103. [DOI] [PubMed] [Google Scholar]
- [3].Chirillo MA, Waters MS, Lindsey LF, Bourne JN, Harris KM, Local resources of polyribosomes and SER promote synapse enlargement and spine clustering after long-term potentiation in adult rat hippocampus, Sci Rep, 9 (2019) 3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kulik YD, Watson DJ, Cao G, Kuwajima M, Harris KM, Structural plasticity of dendritic secretory compartments during LTP-induced synaptogenesis, Elife, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bowen AB, Bourke AM, Hiester BG, Hanus C, Kennedy MJ, Golgi-independent secretory trafficking through recycling endosomes in neuronal dendrites and spines, Elife, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hanus C, Ehlers MD, Specialization of biosynthetic membrane trafficking for neuronal form and function, Curr Opin Neurobiol, 39 (2016) 8–16. [DOI] [PubMed] [Google Scholar]
- [7].Hafner AS, Donlin-Asp PG, Leitch B, Herzog E, Schuman EM, Local protein synthesis is a ubiquitous feature of neuronal pre- and postsynaptic compartments, Science, 364 (2019) 650. [DOI] [PubMed] [Google Scholar]
- [8].Ostroff LE, Watson DJ, Cao G, Parker PH, Smith H, Harris KM, Shifting patterns of polyribosome accumulation at synapses over the course of hippocampal long-term potentiation, Hippocampus, 28 (2018) 416–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jarrell TA, Wang Y, Bloniarz AE, Brittin CA, Xu M, Thomson JN, Albertson DG, Hall DH, Emmons SW, The connectome of a decision-making neural network, Science, 337 (2012) 437–444. [DOI] [PubMed] [Google Scholar]
- [10].Zheng Z, Lauritzen JS, Perlman E, Robinson CG, Nichols M, Milkie D, Torrens O, Price J, Fisher CB, Sharifi N, Calle-Schuler SA, Kmecova L, Ali IJ, Karsh B, Trautman ET, Bogovic JA, Hanslovsky P, Jefferis GSXE, Kazhdan M, Khairy K, Saalfeld S, Fetter RD, Bock DD, A Complete Electron Microscopy Volume of the Brain of Adult Drosophila melanogaster, Cell, 174 (2018) 730–743.e722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sejnowski TJ, Nanoconnectomics, in: Kennedy H, Van Essen DC, Christen Y (Eds.) Micro-, Meso- and Macro-Connectomics of the Brain, Cham (CH), 2016, pp. 1–10. [PubMed] [Google Scholar]
- [12].Motta A, Schurr M, Staffler B, Helmstaedter M, Big data in nanoscale connectomics, and the greed for training labels, Curr Opin Neurobiol, 55 (2019) 180–187. [DOI] [PubMed] [Google Scholar]
- [13].Swanson LW, Lichtman JW, From Cajal to Connectome and Beyond, Annu Rev Neurosci, 39 (2016) 197–216. [DOI] [PubMed] [Google Scholar]
- [14].Motta A, Berning M, Boergens KM, Staffler B, Beining M, Loomba S, Hennig P, Wissler H, Helmstaedter M, Dense connectomic reconstruction in layer 4 of the somatosensory cortex., Science (New York, N.Y.), DOI 10.1126/science.aay3134(2019). [DOI] [PubMed] [Google Scholar]
- [15].Harris KM, Spacek J, Bell ME, Parker PH, Lindsey LF, Baden AD, Vogelstein JT, Burns R, A resource from 3D electron microscopy of hippocampal neuropil for user training and tool development, Sci. Data, 2 (2015) 150046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yin W, Brittain D, Borseth J, Scott ME, Williams D, Perkins J, Own CS, Murfitt M, Torres RM, Kapner D, Mahalingam G, Bleckert A, Castelli D, Reid D, Lee WA, Graham BJ, Takeno M, Bumbarger DJ, Farrell C, Reid RC, da Costa NM, A petascale automated imaging pipeline for mapping neuronal circuits with high-throughput transmission electron microscopy, Nat Commun, 11 (2020) 4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fiala JC, Harris KM, Extending unbiased stereology of brain ultrastructure to three-dimensional volumes, J. Am. Med. Inform. Assoc, 8 (2001) 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bourne JN, Harris KM, Coordination of size and number of excitatory and inhibitory synapses results in a balanced structural plasticity along mature hippocampal CA1 dendrites during LTP, Hippocampus, 21 (2011) 354–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Harris KM, Bourne JN, Mendenhall JM, Spacek J, Hippocampal CA1 dendrites of greater caliber have more spines and contain more microtubules as a subcellular supply route, Society for Neuroscience Abstracts, 147.19 (2007). [Google Scholar]
- [20].Bowden JB, Mendenhall JM, Abraham WC, Harris KM, Microtubule number as a correlate of dendritic spine density in dentate granule cells, Society for Neuroscience Abstracts, 636.20 (2008). [Google Scholar]
- [21].Harris KM, Stevens JK, Dendritic spines of CA1 pyramidal cells in the rat hippocampus: serial electron microscopy with reference to their biophysical characteristics, J. Neurosci, 9 (1989) 2982–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kirov SA, Harris KM, Dendrites are more spiny on mature hippocampal neurons when synapses are inactivated, Nat. Neurosci, 2 (1999) 878–883. [DOI] [PubMed] [Google Scholar]
- [23].Bowden JB, Abraham WC, Harris KM, Differential effects of strain, circadian cycle, and stimulation pattern on LTP and concurrent LTD in the dentate gyrus of freely moving rats, Hippocampus, 22 (2012) 1363–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bromer C, Bartol TM, Bowden JB, Hubbard DD, Hanka DC, Gonzalez PV, Kuwajima M, Mendenhall JM, Parker PH, Abraham WC, Sejnowski TJ, Harris KM, Long-term potentiation expands information content of hippocampal dentate gyrus synapses, Proc Natl Acad Sci U S A, 115 (2018) E2410–E2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kuwajima M, Mendenhall JM, Harris KM, Large-volume reconstruction of brain tissue from high-resolution serial section images acquired by SEM-based scanning transmission electron microscopy, Methods Mol. Biol, 950 (2013) 253–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sorra KE, Harris KM, Stability in synapse number and size at 2 hr after long-term potentiation in hippocampal area CA1, J. Neurosci, 18 (1998) 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bourne JN, Kirov SA, Sorra KE, Harris KM, Warmer preparation of hippocampal slices prevents synapse proliferation that might obscure LTP-related structural plasticity, Neuropharmacology, 52 (2007) 55–59. [DOI] [PubMed] [Google Scholar]
- [28].Jensen FE, Harris KM, Preservation of neuronal ultrastructure in hippocampal slices using rapid microwave-enhanced fixation, J. Neurosci. Methods, 29 (1989) 217–230. [DOI] [PubMed] [Google Scholar]
- [29].Harris KM, Perry E, Bourne J, Feinberg M, Ostroff L, Hurlburt J, Uniform serial sectioning for transmission electron microscopy, J. Neurosci, 26 (2006) 12101–12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Reynolds ES, The use of lead citrate at high pH as an electron-opaque stain in electron microscopy, J Cell Biol, 17 (1963) 208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Fiala JC, Reconstruct: a free editor for serial section microscopy, J. Microsc, 218 (2005) 52–61. [DOI] [PubMed] [Google Scholar]
- [32].Cardona A, Saalfeld S, Schindelin J, Arganda-Carreras I, Preibisch S, Longair M, Tomancak P, Hartenstein V, Douglas RJ, TrakEM2 software for neural circuit reconstruction, PLoS. One, 7 (2012) e38011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Saalfeld S, Fetter R, Cardona A, Tomancak P, Elastic volume reconstruction from series of ultra-thin microscopy sections, Nat. Methods, 9 (2012) 717–720. [DOI] [PubMed] [Google Scholar]
- [34].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Fiji: an open-source platform for biological-image analysis, Nat. Methods, 9 (2012) 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fiala JC, Harris KM, Cylindrical diameters method for calibrating section thickness in serial electron microscopy, J. Microsc, 202 (2001) 468–472. [DOI] [PubMed] [Google Scholar]
- [36].Bell ME, Bourne JN, Chirillo MA, Mendenhall JM, Kuwajima M, Harris KM, Dynamics of nascent and active zone ultrastructure as synapses enlarge during long-term potentiation in mature hippocampus, J Comp Neurol, 522 (2014) 3861–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hanus C, Kochen L, Tom Dieck S, Racine V, Sibarita JB, Schuman EM, Ehlers MD, Synaptic control of secretory trafficking in dendrites, Cell Rep, 7 (2014) 1771–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Popov V, Medvedev NI, Davies HA, Stewart MG, Mitochondria form a filamentous reticular network in hippocampal dendrites but are present as discrete bodies in axons: a three-dimensional ultrastructural study, J. Comp Neurol, 492 (2005) 50–65. [DOI] [PubMed] [Google Scholar]
- [39].Fiala JC, Feinberg M, Peters A, Barbas H, Mitochondrial degeneration in dystrophic neurites of senile plaques may lead to extracellular deposition of fine filaments, Brain Struct. Funct, 212 (2007) 195–207. [DOI] [PubMed] [Google Scholar]
- [40].Faitg J, Lacefield C, Davey T, White K, Laws R, Kosmidis S, Reeve AK, Kandel ER, Vincent AE, Picard M, 3D neuronal mitochondrial morphology in axons, dendrites, and somata of the aging mouse hippocampus, Cell Rep, 36 (2021) 109509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Smith HL, Bourne JN, Cao G, Chirillo MA, Ostroff LE, Watson DJ, Harris KM, Mitochondrial support of persistent presynaptic vesicle mobilization with age-dependent synaptic growth after LTP, Elife, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Li Z, Okamoto K, Hayashi Y, Sheng M, The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses, Cell, 119 (2004) 873–887. [DOI] [PubMed] [Google Scholar]
- [43].Rangaraju V, Lauterbach M, Schuman EM, Spatially Stable Mitochondrial Compartments Fuel Local Translation during Plasticity, Cell, 176 (2019) 73–84 e15. [DOI] [PubMed] [Google Scholar]
- [44].Kirov SA, Sorra KE, Harris KM, Slices have more synapses than perfusion-fixed hippocampus from both young and mature rats, J. Neurosci, 19 (1999) 2876–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Dent EW, Merriam EB, Hu X, The dynamic cytoskeleton: backbone of dendritic spine plasticity, Curr. Opin. Neurobiol, 21 (2011) 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ouyang Y, Wong M, Capani F, Rensing N, Lee CS, Liu Q, Neusch C, Martone ME, Wu JY, Yamada K, Ellisman MH, Choi DW, Transient decrease in F-actin may be necessary for translocation of proteins into dendritic spines, Eur. J Neurosci, 22 (2005) 2995–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Okabe S, Regulation of actin dynamics in dendritic spines: Nanostructure, molecular mobility, and signaling mechanisms, Mol Cell Neurosci, 109 (2020) 103564. [DOI] [PubMed] [Google Scholar]
- [48].Capani F, Martone ME, Deerinck TJ, Ellisman MH, Selective localization of high concentrations of F-actin in subpopulations of dendritic spines in rat central nervous system: a three-dimensional electron microscopic study, J. Comp Neurol, 435 (2001) 156–170. [DOI] [PubMed] [Google Scholar]
- [49].Kuwajima M, Mendenhall JM, Lindsey LF, Harris KM, Automated transmission-mode scanning electron microscopy (tSEM) for large volume analysis at nanoscale resolution, PLoS. One, 8 (2013) e59573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cui-Wang T, Hanus C, Cui T, Helton T, Bourne J, Watson D, Harris KM, Ehlers MD, Local zones of endoplasmic reticulum complexity confine cargo in neuronal dendrites, Cell, 148 (2012) 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Watson DJ, Ostroff L, Cao G, Parker PH, Smith H, Harris KM, LTP enhances synaptogenesis in the developing hippocampus, Hippocampus, 26 (2016) 560–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Original images, the Reconstruct files, and the Excel spreadsheets will be shared on 3DEM.org via the Texas Data Repository (DOI: - to be obtained upon acceptance).