SUMMARY
Background
Amyotrophic lateral sclerosis (ALS) is known to represent a collection of overlapping syndromes. A better understanding of this heterogeneity and the ability to distinguish ALS subtypes would improve clinical care and enhance our understanding of the disease. Various classification systems have been proposed based on empirical observations, but it is unclear to what extent they reflect ALS population substructure.
Methods
We hypothesized that machine learning techniques could identify the number and nature of ALS subtypes. We applied unsupervised (Uniform Manifold Approximation and Projection, UMAP), semi-supervised (neural network-UMAP), and supervised (ensemble based on LightGBM) modeling to a population-based cohort of 2,858 Italian ALS patients for whom detailed phenotype data were available. We replicated our findings in an independent population-based cohort of 1,097 Italian ALS patients.
Findings
We found that semi-supervised machine learning based on UMAP applied to the output of a multi-layered perceptron neural network produced the optimum clustering of the ALS patients. These clusters roughly corresponded to the six clinical subtypes defined by the Chiò classification system (bulbar, respiratory, flail arm, classical, pyramidal, and flail leg ALS). The same clusters were identified in the replication cohort. In contrast, other ALS classification schema, such as the El Escorial categories, Milano-Torino clinical Staging (MiToS), and King’s clinical stages, did not adequately label the clusters. Ensemble learning identified twelve clinical parameters that predicted ALS clinical subtype with high accuracy (area under the curve = 0·982, 95% confidence interval = 0·980–0·983).
Interpretation
Our data-driven study provides insight into the ALS population’s substructure and demonstrates that the Chiò classification system robustly identifies ALS subtypes. We provide an interactive website (https://share.streamlit.io/anant-dadu/machinelearningforals/main) so that researchers can predict the clinical subtype of an ALS patient based on a small number of clinical parameters.
Funding
National Institute on Aging and the Italian Ministry of Health.
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is one of the most common forms of neurodegeneration in the population, accounting for approximately 6,000 deaths in the United States and 11,000 deaths in Europe annually.1 Characterized by progressive paralysis of limb and bulbar musculature, it typically leads to death within three to five years of symptom onset. Medications only minimally slow the rate of progression, and, as a consequence, treatment focuses on symptomatic management.
Genetic advancements have shown that ALS is not a single entity but consists of a collection of syndromes in which the motor neurons degenerate. Alongside these multiple genetic etiologies, there is a broad variability in the disease’s clinical manifestations in terms of the age of symptom onset, site of onset, rate and pattern of progression, and cognitive involvement. This clinical heterogeneity has hampered efforts to understand the cellular mechanisms underlying this fatal neurodegenerative syndrome and has hindered efforts to find effective therapies.
Given the importance of clinical heterogeneity within ALS, it is not surprising that there has been considerable effort to develop classification systems for patients over the years. Examples include grouping based on family status2, clinical milestones3, neurophysiological measurements4, and diagnostic certainty.5 Though useful, it is unclear if any of these classification systems identify meaningful subgroups within the ALS population or merely represent human constructs based on empirical observations. Determining the correct number and nature of subgroups within the ALS population would be a significant step toward understanding the disease. By extension, a reliable method to predict an individual patient’s subgroup using data collected at the beginning of their illness would be helpful for clinical care and clinical trial design.
Here, we explored the clinical patterns of ALS by applying unsupervised and semi-supervised machine learning to deeply-phenotyped, population-based collections of patients (see Figure 1 for the analysis workflow). Our goal was to determine the disease subtypes existing within this patient population. The advantage of machine learning approaches is their ability to identify complex relationships in a data-driven manner. After successfully identifying the ALS subtypes, we used supervised machine learning to build predictor models that accurately classify individual patients and deployed this as a simple-to-use website that clinicians can access.
Figure 1. Workflow followed in this study.

Unsupervised and semi-supervised machine learning was applied to clinical data collected from two population-based ALS registries (n = 2,858 cases and 1,097 cases) to identify clinical subtypes. Supervised machine learning was used to predict subtypes based on clinical parameters, and a web-based tool was built for clinical researchers to apply to their own data.
METHODS
Study participants
The discovery cohort consisted of 2,858 incident cases who had been diagnosed with ALS and were resident in the Piedmont and Valle d’Aosta regions of Italy between January 1, 1995, and December 31, 2015.6 This population-based registry, known as the Piedmont and Valle d’Aosta Registry for ALS (PARALS), was established in 1995. The registry has near-complete case ascertainment within its catchment population of nearly 4·5 million inhabitants (see appendix p 1).6
The replication cohort consisted of 1,097 incident cases who had been diagnosed with ALS and were resident in the Emilia Romagna region of Italy between January 1, 2009, and March 1, 2018.7 This population-based registry, known as the Emilia Romagna Region registry for ALS (ERRALS), was established in 2008, and its catchment area of 4·4 million includes the cities of Modena and Bologna.7 None of the ALS patients enrolled in ERRALS were enrolled in PARALS, and there were no exclusion criteria for the registries. We used the discovery (PARALS) and the replication (ERRALS) cohorts as the training and testing datasets, respectively, in our machine learning analyses.
A vital feature of these studies is their real-time collection of detailed data on patients throughout their illness by experienced ALS neurologists.6,7 The collection methods were standardized across the registries to facilitate comparisons. Each patient was evaluated according to published classification schema that included: (1) the El Escorial classification system5, (2) family status2, (3) Milano-Torino clinical Staging (MiToS)8, and (4) the King’s clinical stages.3 Patients were given a revised ALS Functional Rating Score (ALSFRS-R)9 and were dichotomized according to their C9orf72 genetic carrier status. The PARALS and ERRALS studies were approved by the local ethics committees (appendix p 2).
Pre-processing of clinical data
The clinical data were filtered before analysis. Features with non-random missingness (e.g., cancer type), high sampling bias (e.g., place of birth), and features that could introduce data leakage (e.g., tracheostomy, initial diagnosis was primary lateral sclerosis) were omitted from the analyses (appendix p 10–11). For unsupervised and semi-supervised subtype identification, samples with missing values in the ALSFRS-R9 feature were also excluded (n = 497 in the discovery cohort, n = 108 in the replication cohort). In contrast, samples with missing ALSFRS-R data were included in the supervised analysis as the ensemble learning methods used in this section can handle missingness. Thus, the prediction modeling used 2,858 cases in the discovery cohort and 1,097 in the replication cohort. Categorical features were encoded to numerical using the one-hot encoding method. Min-max normalization was applied to numeric features to preserve the distribution’s shape and ensure a zero-to-one range.
Data imputation
After filtering and pre-processing, the following features had residual missingness that was distributed randomly across the patients at a rate of 15–20%: (i) forced vital capacity (FVC) percent at diagnosis, (ii) body mass index (BMI) at two years before illness, (iii) rate of decline of BMI per month, (iv) weight two years before illness, (v) BMI at diagnosis, (vi) height, and (vii) weight at diagnosis. We used the k-Nearest Neighbor (kNN) imputation method with k = 5 neighbors to preserve the clusters.10 The discovery and replication cohorts were imputed independently.
Unsupervised machine learning
We hypothesized that machine learning techniques could identify the number and nature of ALS subtypes when applied to a large, well-characterized population cohort. The primary outcome measure of our analyses was a comparison of the subtype clusters defined by the machine learning approaches to the six clinical subtypes (bulbar ALS, respiratory ALS, flail arm ALS, classical ALS, pyramidal ALS, and flail leg ALS) assigned manually by neurologists according to the Chiò classification system.11 The clinical subtypes assigned by the Chiò classification system were not entered into the unsupervised algorithms and were not used to construct the patient clusters.
First, we used an unsupervised clustering approach to identify ALS subtypes by applying Uniform Manifold Approximation and Projection (UMAP) to the processed data. UMAP is used for nonlinear dimension reduction to produce a low dimensional projection of the data with the closest possible equivalent fuzzy topological structure.12 This approach preserves the local and global structures existing within the data, along with reproducible and meaningful clusters. As a comparison, we applied dimension reduction methods such as principal component analysis (PCA), independent component analysis (ICA), and non-negative matrix factorization (NMF) to the data.
Semi-supervised machine learning
To further refine the clusters identified by UMAP alone, we processed the data using a multilayer perceptron neural network consisting of five hidden layers with 200, 100, 50, 25, and 3 neurons (appendix p 3).13 The network was trained with the ‘clinical type at one-year’ outcome labels using a Softmax classifier. After training the network with ten-times cross-validation, we extracted the activations of the last hidden layer and used them as the input for the UMAP algorithm.12 This approach reduced the dataset dimensions from 72 to 3.
Supervised subtype prediction
For supervised machine learning, we used GenoML, an open-source automated machine learning package developed by the authors (https://genoml.com/, accessed 21st July 2021).14 Within this package, ensemble learning was used to develop predictive models forecasting the ALS clinical subtype of a patient based solely on the clinical data obtained at their first neurology visit. The stacking ensembles of three supervised machine learning algorithms (Random forest15, LightGBM16, and XGBoost17) were evaluated, and the best performing ensemble model was selected (see appendix p 4–5 for model selection and hyperparameter tuning). Feature reduction was performed using recursive elimination that did not sacrifice accuracy. Internal and external validation was used to assess performance and determine the best algorithms and parameters to use in the model (appendix p 2).
We used the Shapley additive explanations (SHAP) approach to evaluate each feature’s influence in ensemble learning. This approach is used in game theory and assigns an importance (Shapley) value to each feature to determine a player’s contribution to success.18 Shapley explanations enhance understanding by creating accurate explanations for each observation in a dataset. They bolster trust when the critical variables for specific records conform to human domain knowledge and reasonable expectations. The interactive website (https://share.streamlit.io/anant-dadu/machinelearningforals/main, accessed 5th July 2021) was developed as an open-access and cloud-based platform.
Computational tools and code availability
The data analysis pipeline for this work was performed in Python 3.6 using open-source libraries (numpy, pandas, matplotlib, seaborn, plotly, scikit-learn, umap, xgboost, lightgbm, genoml, and tensorflow). We made our code publicly available at https://github.com/ffaghri1/ALS-ML (accessed 5th July 2021) to facilitate replication and future expansion of our work. Manuscript visualizations were created with tidyverse (version 1.3), ggplot2 (version 3.3.2), and plotly (version 4.9.2.2) implemented in R (version 4.0.3). The reporting guideline checklists are provided in the appendix (p 26–31).
Role of the funding source
The study sponsor had no role in study design, data collection, data analysis, data interpretation, writing of the report, or the decision to submit. The corresponding author had full access to all the data in the study, and all of the authors had final responsibility for the decision to submit for publication.
RESULTS
We aimed to identify the clinical subtypes that exist within the ALS patient population in a data-driven manner. To do this, we applied unsupervised and semi-supervised machine learning approaches to a cohort consisting of 2,858 patients diagnosed with ALS and enrolled in the PARALS registry over twenty-five years. The clinical and demographic details of the discovery cohort are given in the appendix (p 12–15). The sixty-six clinical features collected for each case are listed in the appendix (p 10–11), and an exploratory data analysis describing the content of each feature is provided.
After filtering, data for forty-two clinical features across 2,361 patients were available in the PARALS discovery cohort for analysis. Both the unsupervised and semi-supervised approaches identified multiple clusters of patients, representing distinct subtypes of ALS (see appendix p 6 for the results of the UMAP alone and Figure 2A for the neural network-UMAP). Color-coding the ALS patients according to the clinical subtype assigned by the neurologist showed that the clusters roughly corresponded to the six clinical subtypes previously defined by the Chiò classification system (primary outcome). Visually investigating these three-dimensional projections, the optimum separation of the ALS patients into their clinical subtypes was obtained using the semi-supervised machine learning approach. There was excellent discrimination of the bulbar ALS, respiratory ALS, flail arm ALS, and classical ALS subtypes. In contrast, the pyramidal ALS and flail leg ALS overlapped significantly, though the flail leg ALS variant did form a distinct tail that did not overlap with the other subtypes. Overall, we found that 787 (99·7%) of the bulbar ALS cases, 42 (100%) of the respiratory cases, 150 (92%) of the flail arm cases, and 663 (93%) of the classical cases were assigned to the same subtype by the clinician and the semi-supervised algorithm.
Figure 2. The ALS subtypes identified by machine learning in the discovery and replication cohorts.
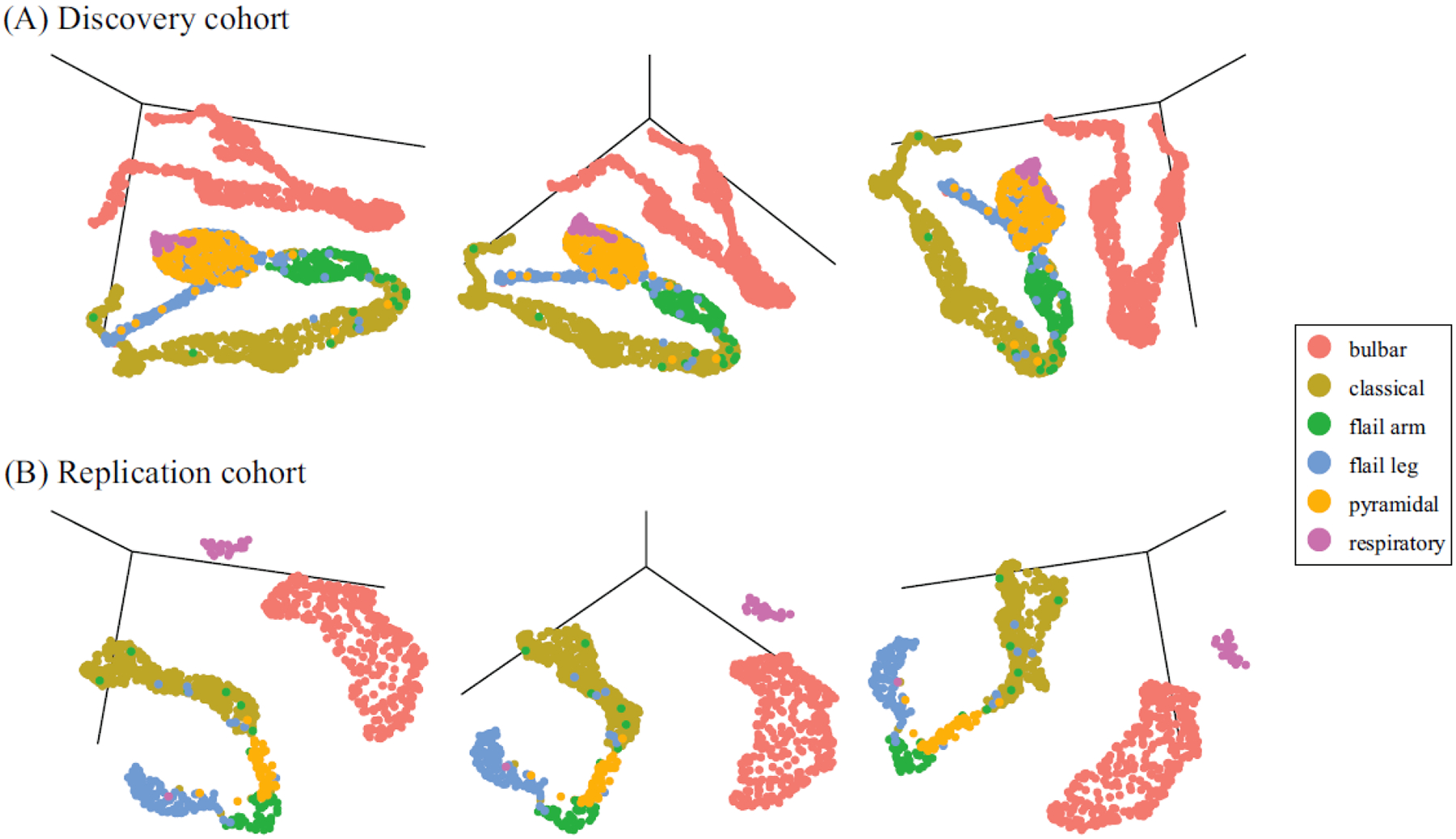
The top row (A) shows the three-dimensional projections of the discovery cohort (n = 2,361) defined by the semi-supervised machine learning algorithm consisting of a UMAP algorithm applied to the output of a five-layer neural network. The same three-dimensional projections (left panel = 100 degrees azimuthal rotation, center panel = 135 degrees, and right panel = 170 degrees) of the replication cohort (n = 989) are shown in the bottom row (B). The projections are symbolic representations of ALS subtypes. Each patient (dot) was color-coded after machine learning cluster generation according to the Chiò classification system. Interactive three-dimensional graphs are available on https://share.streamlit.io/anant-dadu/machinelearningforals/main.
To validate our results, we replicated the ALS subtype identification in an independent cohort consisting of 1,097 incident ALS cases gathered over nine years by a second population-based ALS registry based in the Emilia Romagna Region (ERRALS). After filtering, data for forty-two clinical features across 989 ALS cases were available for analysis. Figure 2B shows the subtypes and clusters identified in the independent replication cohort. The cluster pattern is similar to that observed in the discovery cohort, confirming the reproducibility of our data-driven approach. Interactive three-dimensional graphs are available on https://share.streamlit.io/anant-dadu/machinelearningforals/main (see “Explore the ALS subtype topological space”).
Our semi-supervised machine learning algorithm was more accurate than the other dimension reduction approaches such as principal component analysis (PCA) and independent component analysis (ICA, appendix p 7). Furthermore, other ALS classification schema, such as the El Escorial categories5, family status2, the presence or absence of the pathogenic C9orf72 repeat expansion, Milano-Torino clinical Staging (MiToS)8, ALSFRS-R score9, and King’s clinical stages3, did not label the clusters in a meaningful, clinically-useful manner (Figure 3).
Figure 3. Different classification schema applied to the semi-supervised 3D projection of the ALS discovery cohort (n = 2,361).
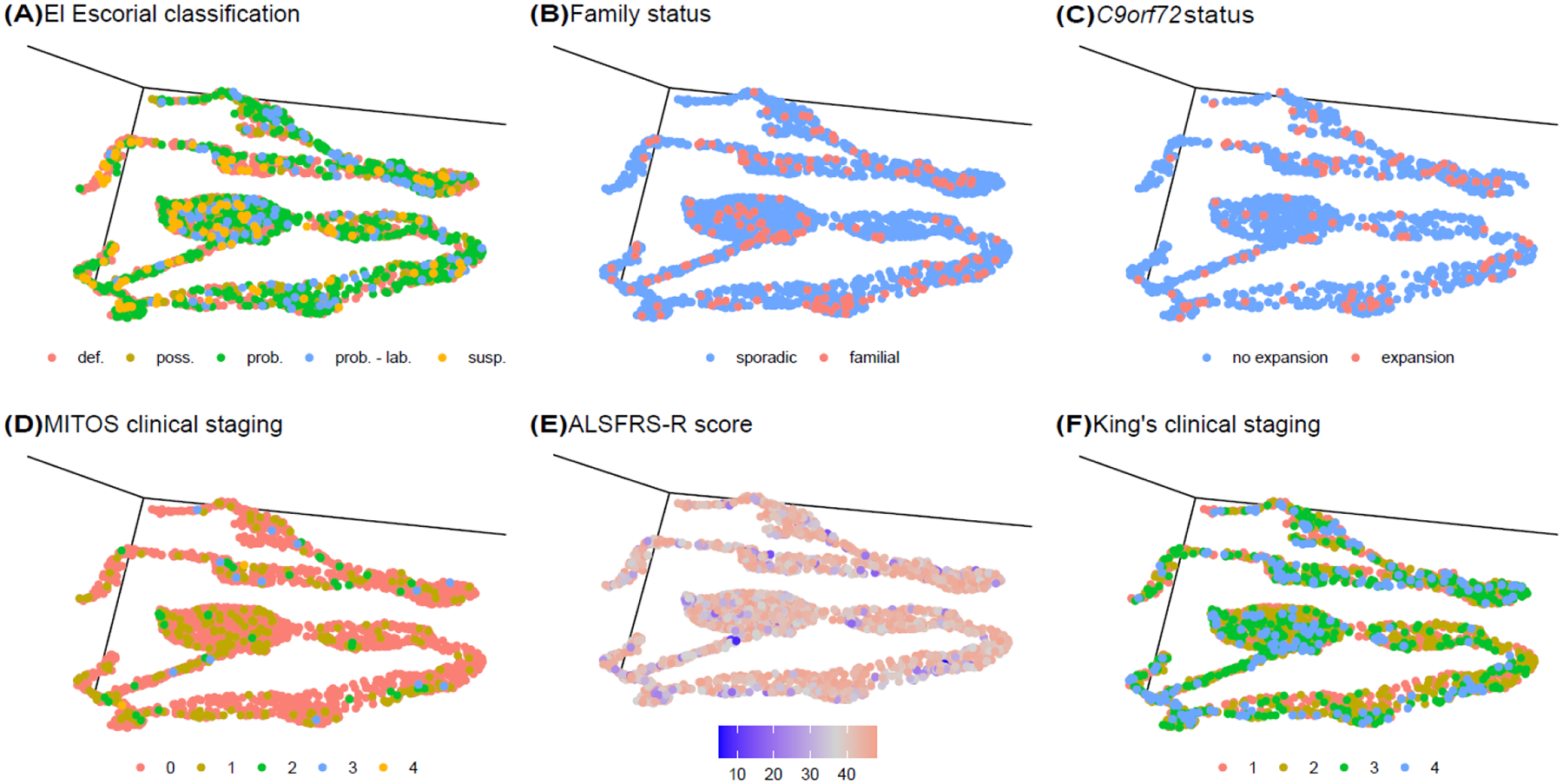
(A) The El Escorial classification system assigns patients to definite (def.), probable (prob.), probable - laboratory supported (prob. - lab.), possible (poss.), and suspected (susp.) categories based on their disability. (B) Patients with a family history of ALS are represented by red dots, and blue dots show patients with sporadic disease. (C) Patients carrying the pathogenic repeat expansion are represented by red dots. (D) The MITOS classification system assigns patients to clinical stages 0 to 4 based on their disability. (E) The ALSFRS-R score rates the severity of disability ranging from 0 to 48 (no disability). (F) The King’s clinical staging system classifies patients into four stages according to their disability level.
Next, we applied a supervised learning approach called ensemble learning to develop predictive models forecasting the ALS clinical subtype of a patient based solely on the clinical data obtained at the first neurology visit. Ensemble learning combines multiple learning algorithms to generate a better predictive model than a single learning algorithm.19 When all available features (n = 66) were included in the model, the clinical subtype of a patient was predicted with high accuracy (internal validation area under the curve (AUC) = 0·982, 95% confidence interval (CI) = 0·979–0·984, and external validation AUC = 0·954, 95% CI = 0·950–0·958, and see appendix p 8, 16–20).
To increase this approach’s clinical utility, we decreased the number of parameters included in the model without sacrificing accuracy. The predictor model built with the top eleven factors was equally robust compared to the all-inclusive model (internal validation AUC = 0·982, 95% CI = 0·980–0·983 and external validation AUC = 0·954, 95% CI = 0·950–0·958, Figure 4 and appendix p 9). Table 1 and Figure 5 lists the eleven parameters selected for the final model and their relative contributions to the model’s precision. Finally, we implemented an interactive website (https://share.streamlit.io/anant-dadu/machinelearningforals/main, see “Predict Patient ALS Subtype”) that allows clinical researchers to determine an ALS patient’s future clinical subtype based on these eleven parameters available in the early stages of the disease. We have also developed a “what-if analysis” functionality to explore how feature changes influence subgroup designation.
Figure 4. Clinical parameters used in the supervised machine learning model to predict ALS clinical subtype.
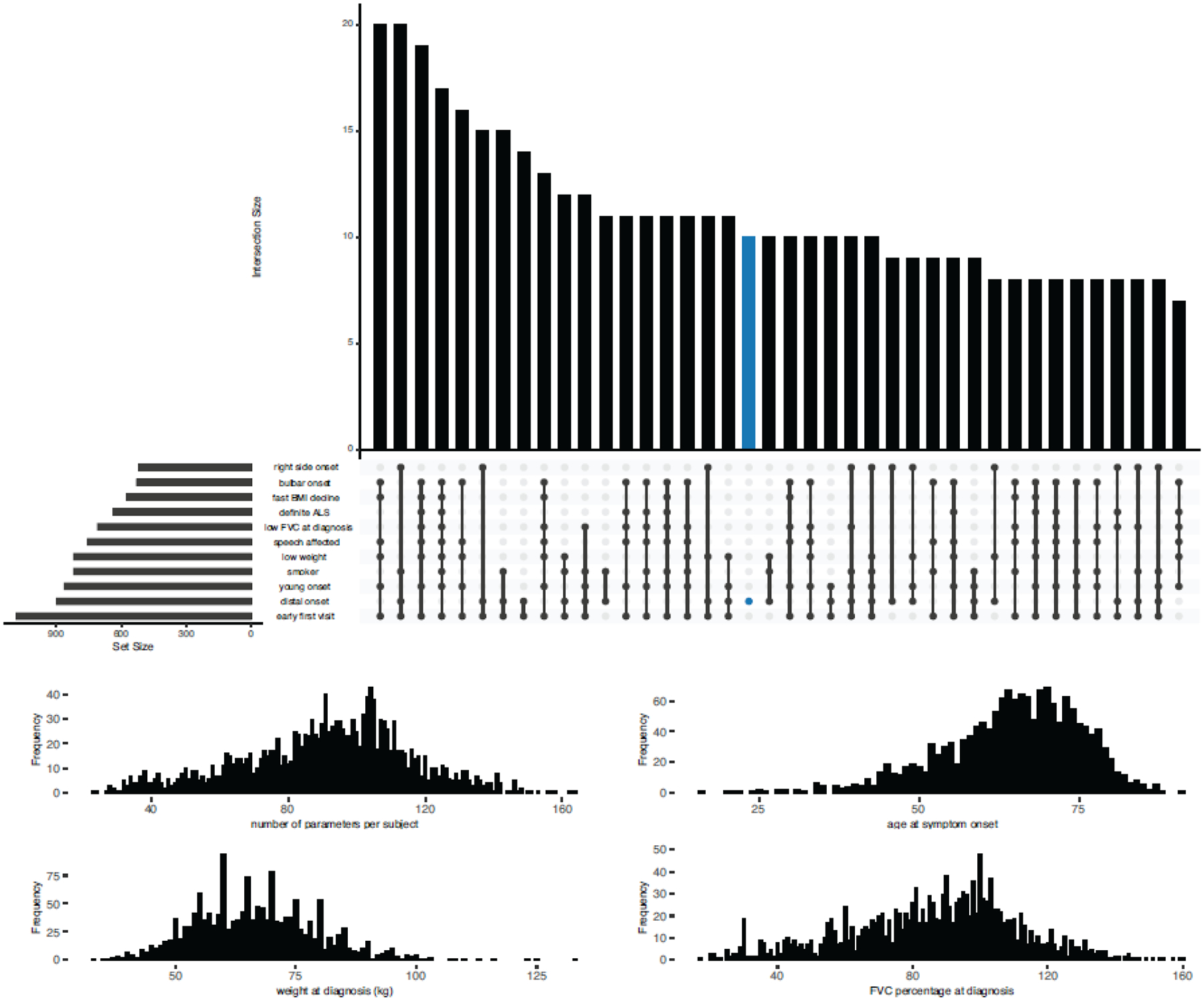
(A) Graphical representation of the overlap between the eleven parameters with the most significant impact on the classification model. The dark circles in the dot plot indicate the parameters that are part of an intersection, and the vertical bar plot reports the number of patients with that parameter combination. The horizontal bar plot reports the set sizes. Analysis was confined to 699 ALS patients with no missing data. (B) Distribution of the parameters in each patient. On average, a patient had five of these clinical features. (C - E) The distribution of the age at onset, weight at diagnosis, and forced vital capacity percent at diagnosis in the analyzed patients.
Table 1.
Clinical features selected for the final model with their relative contributions to the model’s precision.
| Model precision | ||
|---|---|---|
| Relative Importance | Standard Deviation of Relative Importance | |
| Anatomical level at onset | 1·000 | 0·078 |
| Site of symptom onset | 0·460 | 0·021 |
| Onset side | 0·132 | 0·023 |
| Weight at diagnosis (kg) | 0·042 | 4.548×10−4 |
| El Escorial category at diagnosis | 0·033 | 0·003 |
| ALSFRS-R part 1 score | 0·027 | 8.967×10−4 |
| Time of first ALSFRS-R measurement (days from symptom onset) | 0·020 | 0·000 |
| Smoking status | 0·019 | 1.980×10−4 |
| Age at symptom onset | 0·015 | 0·000 |
| Rate of BMI decline (per month) | 0·014 | 0·000 |
| FVC% at diagnosis | 0·013 | 0·000 |
ALSFRS-R part 1 score refers to the first question in the ALS Functional Rating Scale – Revised rating scale concerning speech.
Figure 5. The eleven features used in the supervised machine learning model to predict ALS clinical subtype.
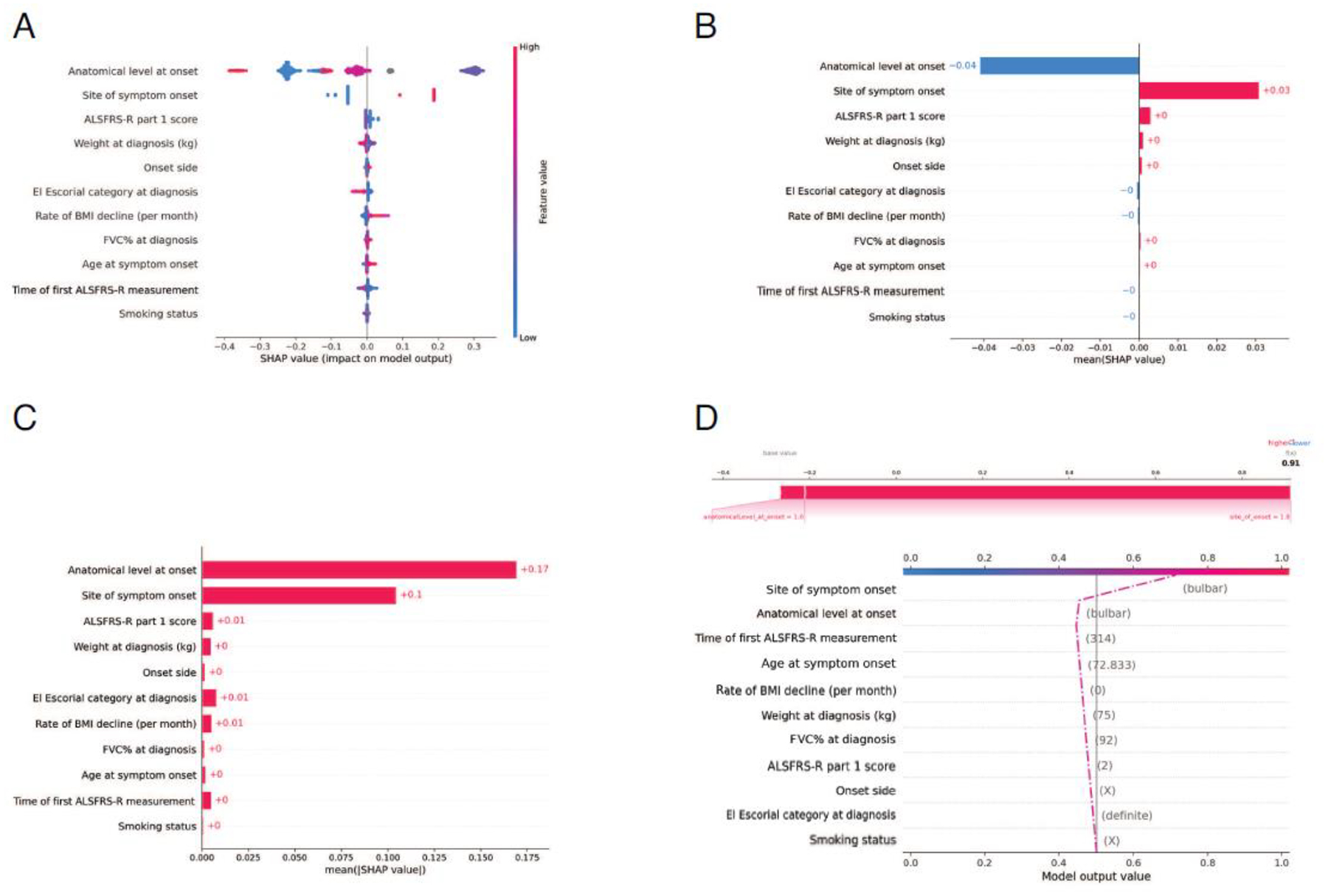
(A) Distribution of the Shap values for the eleven features with the most significant impact on the classification model. Each point represents a subject and may have a positive or negative impact depending on its SHAP value. For instance, high values of the rate of BMI decline in red contribute strongly to the positive class, while low values in blue contribute to a lesser extent to the negative class. (B & C) The aggregate of the Shap values is shown for the top eleven features (ranked from most to least important). (D) Model output trajectory for a single subject with the bulbar subtype of ALS. The predicted probability that the patient had the bulbar subtype of ALS was 0.91, predominantly driven by the patient’s bulbar site of symptom onset and only minorly driven by their smoking status and El Escorial category at diagnosis. See https://share.streamlit.io/anant-dadu/machinelearningforals/main for more examples.
DISCUSSION
Researchers and clinicians have long sought a reliable method to identify the subgroups existing within the ALS population. Knowledge of the ALS substructure would improve our understanding of the clinical heterogeneity associated with this fatal neurodegenerative disease. By extension, it would enhance patient care and provide insights into the underlying pathological mechanisms.20–28 Here, we used a machine learning approach to identify such subtypes within a large cohort of ALS patients and replicated our findings in an independent cohort. This data-driven approach confirmed the existence of subtypes within the ALS disease spectrum. Interestingly, these subtypes roughly corresponded to those previously defined by the Chiò classification system11, demonstrating the schema’s utility. Unlike other subtyping approaches, the Chiò classification system relies on the patient’s clinical data collected during the first year of illness.11 This one-year observation period allows the disease’s symptoms to manifest more clearly and the clinician to assess the progression rate more accurately. Though progression is a fundamental feature of ALS, it is not typically employed in determining the disease subtype.
The primary obstacles to deciphering the clinical heterogeneity observed among ALS patients have been the lack of a sufficiently large dataset and the inability to analyze multi-dimensional relationships. We used data from two large, population-based registries that had enrolled ALS patients over several decades to address these issues. These registries collected data throughout the patient’s illness, and overall, they contained nearly 300,000 pieces of information that we used for our categorization efforts. Our results highlight the value of disease registries that capture deep phenotypes across an entire catchment area. Previous efforts to catalog the various subgroups of ALS hinged on a small number of clinical features, such as family history or site of symptom onset.2–5 Although clinically useful, these univariate or oligovariate classification systems do not capture the complicated clinical patterns existing within the ALS population. In contrast, the machine learning algorithms we applied are adept at deciphering complex and multi-faceted relationships. Indeed, the eleven features selected by the supervised model have not been previously combined to predict ALS subtypes.
Remarkedly, our unsupervised and semi-supervised machine learning algorithms defined the same subgroups outlined by Chiò and colleagues in their 2011 classification system.11 This may not be completely surprising in the context of our semi-supervised approach as the “clinical type at one year” patient labels were used to assist the neural network-UMAP clustering. We do not maintain that our machine learning approach is better at identifying categories than experienced ALS neurologists. Instead, we validated the Chiò classification system using a data-driven approach and provided prima facie evidence that this schema captures the ALS population’s substructure. Classification based on other schemes, such as the El Escorial, MiToS, and King’s systems, did not help assign patients to a disease subtype (see Figure 3).
Nevertheless, our machine learning algorithm provides opportunities to improve and refine the Chiò classification system, especially as the pyramidal and the flail leg subtypes may not be as distinct from each other as other subtypes. This finding was unexpected as these patients are easily distinguished from each other in the clinic, highlighting machine learning’s ability to provide new and essential insights into a complex disease. It also offers a novel starting point for exploring the neurobiology underlying the pyramidal and flail leg ALS variants.
Having established that the six subtypes outlined by the Chiò classification reflect the correct substructure of ALS, we next considered how clinicians and researchers could use this information. The ability to assign patients to subgroups at an early disease stage helps unravel the disease’s clinical heterogeneity and aids in discussions with newly diagnosed individuals about likely disease course and prognosis. For example, patients with the respiratory subtype of ALS had a faster rate of progression. They were more likely to require non-invasive positive pressure ventilation (NIPPV) and gastrostomy feeding at an earlier stage than ALS cases with the upper motor predominant form of the disease. Outcome data from negative clinical trials can be reanalyzed for a therapeutic effect limited to one or two subgroups. A similar approach has been successful in Parkinson’s disease.29 Genetic heterogeneity also handicaps our ability to implicate new loci in the disease’s pathogenesis using genome-wide association analysis. Including the subgroup as a covariate or restricting the search to a single subtype may resolve this issue by focusing gene finding efforts within a more homogeneous patient population.
It has not escaped our attention that the topology representation of the ALS subtypes produced by the machine learning algorithm resembles the central nervous system (CNS). We observed this pattern most clearly in Figure 2. The bulbar subtype delineates the cerebrum, and the spinal cord is represented by a long tail running successively from flail arm, pyramidal, classical, to flail leg subtypes. We speculate that this arrangement hints at a broader anatomical organization within the ALS spectrum, perhaps reflecting subtle differences of the motor neuron subtypes within each segment of the CNS and differing susceptibilities to pathogenic mechanisms of neurodegeneration.
Our study has several limitations. First, machine learning algorithms can identify patterns within a dataset even when no such pattern exists. Such ‘overfitting of the model’ is an inherent problem with this statistical methodology, and the most legitimate remedy is to attempt replication in an independent dataset. To that end, we replicated our findings in an independent, population-based cohort yielded remarkably similar outcomes to the discovery cohort, demonstrating the robustness of our approach. Second, the handling of missing data is increasingly recognized as a critical constraint of machine learning. Our data was remarkably complete, as shown in the exploratory data analysis notebooks. Nonetheless, as with any real-life clinical dataset, information was missing for some parameters, and we aimed to be transparent and cautious in handling these issues.
Third, our modeling may have a bias as we used the same set of patients used by Chiò and colleagues to define their subtypes in their 2011 study.11 However, it is unlikely that the use of this case series led to sampling bias as the clinical information used to create the models is standard across the ALS field. Furthermore, population-based registries decrease the possibility of sampling bias as they capture every case within a catchment area. We also replicated our initial findings in an independent cohort that was not used in the 2011 study, confirming that the clusters identified by the data-driven approach did not arise from spurious within-patient associations between variables in the discovery cohort. Nevertheless, both our discovery and replication data originated from the Northern Italian population. Additional studies in other countries are required to rule out the possibility of population bias and to test our approach’s generalizability. These data will have to be freshly collected, as there is insufficient information to determine the Chiò classification of samples in retrospective data repositories such as the Pooled Resource Open-Access ALS Clinical Trials Database (https://nctu.partners.org/proact, accessed 5th July 2021).30
Like other statistical systems, machine learning algorithms are only practical if they can be applied broadly. To facilitate this, we have established a website where a physician can enter a patient’s characteristics to predict their subtype membership. We have made our programming code publicly available (https://github.com/ffaghri1/ALS-ML) so that other researchers can apply it and modify it as our understanding of ALS and machine learning approaches evolve. Though our current categorization approach is robust, we anticipate that it will improve over time to the point that it will become a valuable tool for clinicians dealing with ALS patients. Here, we provide an early demonstration of machine learning’s ability to unravel highly complex and interrelated disease systems such as ALS.
Supplementary Material
RESEARCH IN CONTEXT.
Evidence before this study
We searched PubMed for articles published in English from database inception until January 5, 2021, about the use of machine learning and the identification of clinical subtypes within the amyotrophic lateral sclerosis (ALS) population, using the search terms “machine learning”, AND “classification ”, AND “amyotrophic lateral sclerosis”. This inquiry identified twenty-nine studies. Most previous studies used machine learning to diagnose ALS (based on gait, imaging, electromyography, gene expression, proteomic, and metabolomic data) or improve brain-computer interfaces. One study used machine learning algorithms to stratify ALS postmortem cortex samples into molecular subtypes based on transcriptome data. Kueffner and colleagues crowdsourced the development of machine learning algorithms to approximately thirty teams to obtain a consensus in an attempt to identify ALS patient subpopulations. In addition to clinical trial information in the PRO-ACT database (www.ALSdatabase.org, accessed July 5, 2021), this effort used data from the Piedmont and Valle d’Aosta Registry for ALS (PARALS). Four ALS patient categories were identified: slow progressing, fast progressing, early stage, and late stage. This approach’s clinical relevance was unclear, as all ALS patients will necessarily pass through an early and late stage of the disease. Furthermore, no attempt was made to discern which of the existing clinical classification systems, such as the El Escorial criteria, the Chiò classification system, and the King’s clinical staging system, can identify ALS subtypes.
ALS subtype identification has previously been explored using t-SNE (Tang, 2019), and UMAP has also been used in the context of ALS patient stratification in two recently published papers (Grollemund, 2020; Westeneng, 2018). Prognosis outcome and patient stratification have been modeled in a classification context by Westeneng and Pfohl using real-life data and by Ong and Beaulieu-Jones using PRO-ACT data. The PARALS data were also previously used for ALS patient stratification by Elamin and colleagues. Our semi-supervised approach based on a neural network and UMAP is similar to work published by Sainburg and colleagues.
We concluded that there remained an unmet need to identify the ALS population’s substructure in a data-driven, non-empirical manner. Building on this, there was a need for a tool that reliably predicts the clinical subtype of an ALS patient. This knowledge would improve our understanding of the clinical heterogeneity associated with this fatal neurodegenerative disease.
Added value of this study
This study developed a machine learning algorithm to detect ALS patients’ clinical subtypes using clinical data collected from the 2,858 Italian ALS patients in PARALS. Ascertainment of these patients within the catchment area was near complete, meaning that the dataset truly represented the ALS population. We replicated our approach using clinical data obtained from an independent cohort of 1,097 Italian ALS patients that had also been collected in a populationbased, longitudinal manner. Semi-supervised learning based on Uniform Manifold Approximation and Projection (UMAP) applied to a multilayer perceptron neural network provided the optimum results based on visual inspection. The observed clusters equated to the six clinical subtypes previously defined by the Chiò classification system (bulbar ALS, respiratory ALS, flail arm ALS, classical ALS, pyramidal ALS, and flail leg ALS). Using a small number of clinical parameters, an ensemble learning approach could predict the ALS clinical subtype with high accuracy (area under the curve = 0·94).
Implications of all available evidence
Additional validation is required to determine these algorithms’ accuracy and clinical utility in assigning clinical subtypes. Nevertheless, our algorithms offer a broad insight into the clinical heterogeneity of ALS and help to determine the actual subtypes of disease that exist within this fatal neurodegenerative syndrome. The systematic identification of ALS subtypes will improve clinical care and clinical trial design.
ACKNOWLEDGEMENTS
This work was supported in part by the Intramural Research Programs of the National Institutes of Health (NIH), National Institute on Aging (Z01-AG000949-02, funding to BJT). This work was in part supported by the Italian Ministry of Health (Ministero della Salute, Ricerca Sanitaria Finalizzata, grant RF-2016-02362405, funding to ACh), the European Commission’s Health Seventh Framework Programme (FP7/2007–2013 under grant agreement 259867, funding to ACh), and the Joint Programme - Neurodegenerative Disease Research (Strength, ALS-Care and Brain-Mend projects funding to ACh), granted by Italian Ministry of Education, University and Research. This study was performed under the Department of Excellence grant of the Italian Ministry of Education, University and Research to ACh and the ‘Rita Levi Montalcini’ Department of Neuroscience, University of Torino, Italy. The Emilia Romagna Registry for ALS (ERRALS) is supported by a grant from the Emilia Romagna Regional Health Authority (funding to JM). FF and MAN participated in this work via a competitive contract between their employer Data Tecnica International and the NIH. This study used the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, Maryland, USA (http://hpc.nih.gov, accessed 5th July 2021). We thank the Laboratory of Neurogenetics (NIH) staff for their collegial support and technical assistance.
DECLARATION OF INTEREST
BJT holds patents on the clinical testing and therapeutic intervention for the hexanucleotide repeat expansion of C9orf72 (patent numbers EP2751284A1, CA2846307A and 20180187262) and has received research grants from The Myasthenia Gravis Foundation, the ALS Association (ALSA), the Center for Disease Control and Prevention (CDC), the United States Department of Veterans Affairs, Merck, and Cerevel. BJT receives funding through the Intramural Research Program at the National Institutes of Health. BJT sits on the scientific advisory committee of the American Neurological Association, is an associate editor of Brain, and sits on the editorial boards of the Journal of Neurology, Neurosurgery, and Psychiatry, Neurobiology of Aging, and eClinical Medicine. JM has received research grants from the Fondazione Italiana di Ricerca per la Sclerosi Laterale Amiotrofica (ARISLA), the Agenzia Italiana del Farmaco (AIFA), the Italian Ministry of Health, the Emilia Romagna Regional Health Authority, and Pfizer. ACh has received research funding and honoraria for lectures from Biogen. ACh also sits on advisory boards for Mitsubishi Tanabe, Roche, Denali Pharma, Cytokinetics, Biogen, Amylyx and Sanofi, and participates in data safety monitoring boards for Lilly and ABScience. RV has received research scholarship funding from the Rotary Club (Global Grant GG2094854 Scholarship).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DATA SHARING
Code for pre-processing and prediction is available at https://github.com/ffaghri1/ALS-ML. The PARALS and ERRALS registry datasets are not publicly available at this moment, since all research or research-related activities that involve an external party might require, at the discretion of the University of Turin or the University Hospital of Modena, a written research agreement to define the obligations and manage the risks.
REFERENCES
- 1.Hirtz D, Thurman DJ, Gwinn-Hardy K, Mohamed M, Chaudhuri AR, Zalutsky R. How common are the “common” neurologic disorders? Neurology 2007; 68: 326–37 [DOI] [PubMed] [Google Scholar]
- 2.Byrne S, Bede P, Elamin M, et al. Proposed criteria for familial amyotrophic lateral sclerosis. Amyotroph Lateral Scler 2011; 12: 157–9 [DOI] [PubMed] [Google Scholar]
- 3.Roche JC, Rojas-Garcia R, Scott KM, et al. A proposed staging system for amyotrophic lateral sclerosis. Brain 2012; 135: 847–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Carvalho M, Dengler R, Eisen A, et al. Electrodiagnostic criteria for diagnosis of ALS. Clin Neurophysiol 2008; 119: 497–503 [DOI] [PubMed] [Google Scholar]
- 5.Brooks BR. El Escorial World Federation of Neurology criteria for the diagnosis of amyotrophic lateral sclerosis. Subcommittee on Motor Neuron Diseases/Amyotrophic Lateral Sclerosis of the World Federation of Neurology Research Group on Neuromuscular Diseases and the El Escorial “Clinical limits of amyotrophic lateral sclerosis” workshop contributors. J Neurol Sci 1994; 124 Suppl: 96–107 [DOI] [PubMed] [Google Scholar]
- 6.Piemonte, Valle d’Aosta Register for Amyotrophic Lateral S. Incidence of ALS in Italy: evidence for a uniform frequency in Western countries. Neurology 2001; 56: 239–44 [DOI] [PubMed] [Google Scholar]
- 7.Mandrioli J, Biguzzi S, Guidi C, et al. Epidemiology of amyotrophic lateral sclerosis in Emilia Romagna Region (Italy): A population based study. Amyotroph Lateral Scler Frontotemporal Degener 2014; 15: 262–8 [DOI] [PubMed] [Google Scholar]
- 8.Chiò A, Hammond ER, Mora G, Bonito V, Filippini G. Development and evaluation of a clinical staging system for amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry 2015; 86: 38–44 [DOI] [PubMed] [Google Scholar]
- 9.Cedarbaum JM, Stambler N, Malta E, et al. The ALSFRS-R: a revised ALS functional rating scale that incorporates assessments of respiratory function. BDNF ALS Study Group (Phase III). J Neurol Sci 1999; 169: 13–21 [DOI] [PubMed] [Google Scholar]
- 10.Beretta L, Santaniello A. Nearest neighbor imputation algorithms: a critical evaluation. BMC Med Inform Decis Mak 2016; 16 Suppl 3: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiò A, Calvo A, Moglia C, Mazzini L, Mora G, group Ps. Phenotypic heterogeneity of amyotrophic lateral sclerosis: a population based study. J Neurol Neurosurg Psychiatry 2011; 82: 740–6 [DOI] [PubMed] [Google Scholar]
- 12.McInnes L, Healy J, Saul N, Großberger L. UMAP: Uniform Manifold Approximation and Projection. Journal of Open Source Software 2018; 3: 861 [Google Scholar]
- 13.Sainburg T, McInnes L, Gentner TQ. Parametric UMAP embeddings for representation and semi-supervised learning. arXiv 2021; arXiv: 2009.12981v3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makarious MB, Leonard HL, Vitale D, et al. GenoML: Automated Machine Learning for Genomics. arXiv 2021; arXiv:2103.03221v1 [Google Scholar]
- 15.Breiman L. Random Forests. Machine Learning 2001; 45: 5–32 [Google Scholar]
- 16.Ke G, Meng Q, Finley T. LightGBM: A Highly Efficient Gradient Boosting Decision Tree. In: G I, L UV, B S, eds. Advances in Neural Information Processing Systems: Curran Associates, Inc., Red Hook; 2017: 3146–54. [Google Scholar]
- 17.Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. New York: ACM; 2016: 785–94. [Google Scholar]
- 18.Lundberg S, Lee S. A unified approach to interpreting model predictions. In: G I, L UV, B S, et al. , editors. 31st Conference on Neural Information Processing Systems (NIPS 2017); Long Beach, CA, USA: Curran Associates, Inc, Red Hook. p. 4765–74. [Google Scholar]
- 19.Rokach L Ensemble-based classifiers. Artificial Intelligence Review 2010; 33: 1–39 [Google Scholar]
- 20.Kueffner R, Zach N, Bronfeld M, et al. Stratification of amyotrophic lateral sclerosis patients: a crowdsourcing approach. Sci Rep 2019; 9: 690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuffner R, Zach N, Norel R, et al. Crowdsourced analysis of clinical trial data to predict amyotrophic lateral sclerosis progression. Nat Biotechnol 2015; 33: 51–7 [DOI] [PubMed] [Google Scholar]
- 22.Tang M, Gao C, Goutman SA, et al. Model-Based and Model-Free Techniques for Amyotrophic Lateral Sclerosis Diagnostic Prediction and Patient Clustering. Neuroinformatics 2019; 17: 407–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grollemund V, Chat GL, Secchi-Buhour MS, et al. Development and validation of a 1-year survival prognosis estimation model for Amyotrophic Lateral Sclerosis using manifold learning algorithm UMAP. Sci Rep 2020; 10: 13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beaulieu-Jones BK, Greene CS, Pooled Resource Open-Access ALSCTC. Semi-supervised learning of the electronic health record for phenotype stratification. J Biomed Inform 2016; 64: 168–78 [DOI] [PubMed] [Google Scholar]
- 25.Elamin M, Bede P, Montuschi A, Pender N, Chio A, Hardiman O. Predicting prognosis in amyotrophic lateral sclerosis: a simple algorithm. J Neurol 2015; 262: 1447–54 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ong ML, Tan PF, Holbrook JD. Predicting functional decline and survival in amyotrophic lateral sclerosis. PLoS One 2017; 12: e0174925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfohl SR, Kim RB, Coan GS, Mitchell CS. Unraveling the Complexity of Amyotrophic Lateral Sclerosis Survival Prediction. Front Neuroinform 2018; 12: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westeneng HJ, Debray TPA, Visser AE, et al. Prognosis for patients with amyotrophic lateral sclerosis: development and validation of a personalised prediction model. Lancet Neurol 2018; 17: 423–33 [DOI] [PubMed] [Google Scholar]
- 29.Leonard H, Blauwendraat C, Krohn L, et al. Genetic variability and potential effects on clinical trial outcomes: perspectives in Parkinson’s disease. J Med Genet 2020; 57: 331–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atassi N, Berry J, Shui A, et al. The PRO-ACT database: design, initial analyses, and predictive features. Neurology 2014; 83: 1719–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


