Summary
We recently developed a system to create human chimeric antigen receptor (CAR)-T cells using conjugated Cas12a (cCas12a) in which Cas12a is covalently linked to its CRISPR RNA (crRNA). This protocol describes site-specific modification of Cas12a and the preparation of Cas12a-crRNA complex using bio-orthogonal chemistry, followed by CAR-T cell generation through electroporation and AAV infection. This system shows robust editing efficiency in human cells and can be used for precisely targeted, highly efficient integration of CAR genes into T cell genome.
For complete details on the use and execution of this protocol, please refer to Ling et al. (2021).
Subject areas: Cell Biology, Cell culture, Cell isolation, Flow Cytometry/Mass Cytometry, Immunology, Molecular Biology, CRISPR, Protein Biochemistry, Protein expression and purification, Chemistry
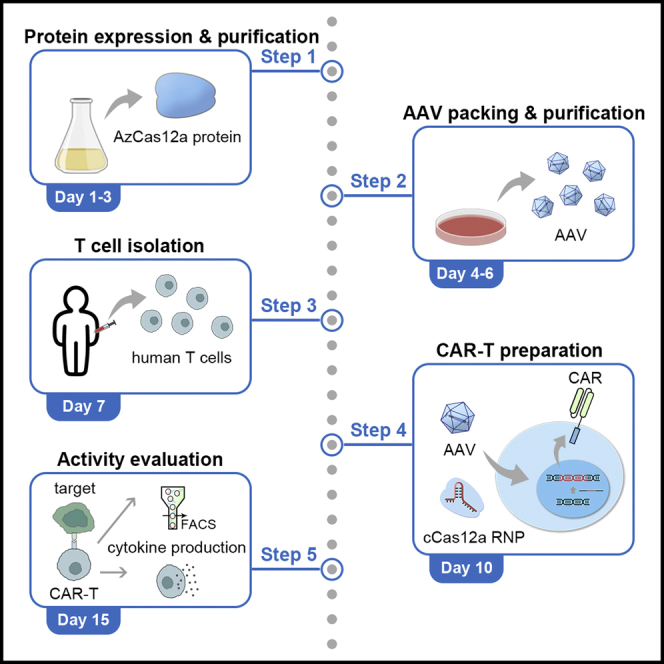
Graphical abstract
Highlights
-
•
Site-specific modification of Cas12a using noncanonical amino acid mutagenesis
-
•
A covalent Cas12a-crRNA complex can be prepared using bio-orthogonal chemistry
-
•
Cas12a-crRNA conjugates afford site-specific CAR-T preparation with high efficiency
We recently developed a system to create human chimeric antigen receptor (CAR)-T cells using conjugated Cas12a (cCas12a) in which Cas12a is covalently linked to its CRISPR RNA (crRNA). This protocol describes site-specific modification of Cas12a and the preparation of Cas12a-crRNA complex using bio-orthogonal chemistry, followed by CAR-T cell generation through electroporation and AAV infection. This system shows robust editing efficiency in human cells and can be used for precisely targeted, highly efficient integration of CAR genes into T cell genome.
Before you begin
The protocol below describes the specific steps for editing Human T cells. This protocol can also be used similarly for editing Jurkat cells and hiPSC cells.
Experimental design considerations
Genome engineering in human primary T cells holds great promise for the development of novel immunotherapeutics (Tebas et al., 2014; Garfall et al., 2015; June and Sadelain, 2018). Currently approved CAR-T cell preparations are based on randomly integrated lentiviral and γ-retroviral vectors and thus carry the risk of insertional mutagenesis-induced carcinogenesis and translational dysregulation (Howe et al., 2008; Wang and Rivière, 2016). To create more consistent and robust CAR-T cells in precise genome locus. Here we used a newly developed cCas12a genome editing system combination with AAV based donor delivery. There are two major considerations to generate locus-specific CAR-T cell at high efficiency. The first is optimizing the crRNA targeting sequence, typically 3–5 different crRNAs should be tested in mammalian cell. Secondly, choose the appropriate length of the homology arm for better precise gene knock-in. For a 2.5 kb gene integration containing promoter and gene of interest, 600 bp homology arm showed high efficiency gene integration.
Institutional approval
This study has received institutional regulatory approval. All recombinant DNA and biosafety work were performed under the guidelines of Peking University Health Science Center. All human PBMC sample are isolated from healthy female donor from Beijing Cord Blood Bank. The work was performed under the guidelines of Peking University Health Science Center Institutional Review Board with an approved protocol (LA2019030).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| c-Myc Monoclonal Antibody (9E10) Dilution Factor: 1:100 |
Invitrogen | Cat# MA1-980; RRID: AB_558470 |
| Anti-mouse IgG (H+L), F(ab')2 Fragment (Alexa Fluor® 647 Conjugate) Dilution Factor: 1:1000 |
Cell Signaling Technology | Cat# 4410 |
| APC anti-human CD3 antibody Dilution Factor: 1:20 |
BioLegend | Cat# 300412; RRID: AB_314066 |
| FITC anti-human beta2-microglobulin antibody Dilution Factor: 1:20 |
BioLegend | Cat# 316304; RRID: AB_492837 |
| PE anti-human CD279 (PD-1) antibody Dilution Factor: 1:20 |
BioLegend | Cat# 329906; RRID: AB_940483 |
| PE/Cyanine7 anti-human CD152 (CTLA-4) antibody Dilution Factor: 1:20 |
BioLegend | Cat# 369613; RRID: AB_2632875 |
| Bacterial and virus strains | ||
| NEB® 5-alpha Competent E. coli (High Efficiency) | New England Biolabs | Cat# C2987I |
| BL21(DE3) Competent E. coli | New England Biolabs | Cat# C2527H |
| Biological samples | ||
| Human PBMC, Female donor ranging from 25-35 years old | N/A | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Isopropyl β-D-1-thiogalactopyranoside | Inalco Pharmaceuticals | Cat# 1758-1400 |
| Tryptone | Oxoid | Cat# LP0042B |
| Yeast extract | Oxoid | Cat# LP0021B |
| Spectinomycin | Inalco Pharmaceuticals | Cat# 1758-9311 |
| Ampicillin | Inalco Pharmaceuticals | Cat# 1758-9314 |
| NaCl | Biodee | Cat# DE0008 |
| Ni-NTA Agarose | QIAGEN | Cat# 30230 |
| Tris | Biodee | Cat# DE0006 |
| KCl | Sigma-Aldrich | Cat# P5405 |
| Imidazole | Sigma-Aldrich | Cat# I5513 |
| Phenylmethylsulfonyl fluoride | Sigma-Aldrich | Cat# 78830 |
| Electroporation buffer | Celetrix LLC, Manassas VA | Cat# 13-0104 |
| 0.4% Trypan Blue Solution | Gibco | Cat# 15250061 |
| Ficoll-Paque PLUS | Cytiva | Cat# 17-1440-02 |
| XYbeads Human CD3/CD28 T Cell Activator | SAILY BIO | Cat# XY-B002-001 |
| X-VIVOTM15 | Lonza | Cat# 04-418Q |
| Fetal Bovine Serum | Gibco | Cat# 10099 |
| Fetal Bovine Serum | PAN-Biotech | Cat# P30-3302 |
| Dulbecco’s Modified Eagle’s Medium | MACGENE | Cat# 15019 |
| Trypsin-EDTA (0.25%) phenol red | MACGENE | Cat# CC017 |
| Penicillin-Streptomycin solution | Hyclone | Cat# SV30010 |
| KOD-One PCR Master Mix | TOYOBO | KMM-101 |
| PBS | MACGENE | Cat# CC008 |
| Opti-MEM™ I Reduced Serum Medium | Thermo Fisher Scientific | Cat# 31985070 |
| Benzonase Nuclease | Smart Lifesciences | Cat# SLP00800 |
| DNase I, RNase-free | Thermo Fisher Scientific | Cat# EN0521 |
| Bestar® Sybr Green qPCR Master Mix | DBI® Bioscience | Cat# DBI-2043 |
| RPMI-1640 | MACGENE | Cat# CM10040 |
| IL-2 Protein, Human, Recombinant | SinoBiological | Cat# 11848-HNAE |
| IL-7 Protein, Human, Recombinant | SinoBiological | Cat# 11821-HNAE |
| IL-15 Protein, Human, Recombinant | SinoBiological | Cat# 10360-HNCE |
| BSA | YEASEN | Cat# 36103ES25 |
| EDTA | Sigma-Aldrich | Cat# E9884 |
| PEI MAX® - Transfection Grade Linear Polyethylenimine Hydrochloride (MW 40,000) | Polysciences | Cat# 24765 |
| Critical commercial assays | ||
| Dynabeads™ Untouched™ Human T Cells Kit | Invitrogen | Cat# 11344D |
| Human IFN gamma Uncoated ELISA kit | Invitrogen | Cat# 88-7316-88 |
| Human TNF alpha Uncoated ELISA kit | Invitrogen | Cat# 88-7346-88 |
| LDH-Glo™ Cytotoxicity Assay | Promega | Cat# J2380 |
| DNA Clean & Concentrator-25 (Capped) | Zymo Research | Cat# D4033 |
| FastPure Cell/Tissue DNA Isolation Mini Kit | Vazyme | Cat# DC102 |
| Experimental models: Cell lines | ||
| NALM-6 Passage numbers: 1–20 |
ATCC | CRL-3273™ |
| AAV293 cells Passage numbers: 1–20 |
Agilent | Cat# 240073 |
| Oligonucleotides | ||
| ITR-FW | Genewiz | 5′- GGAACCCCTAGTGATGGAGTT |
| ITR-RV | Genewiz | 5′-CGGCCTCAGTGAGCGA |
| TRAC-1st-FW | Genewiz | 5′- CCCTTGTCCATCACTGGCAT |
| CART-PCR-FW | Genewiz | 5′- GGAGTACGACGTGCTGGATAAG |
| TRAC-2st-RV | Genewiz | 5′- GCACACCCCTCATCTGACTT |
| DBCO modified crRNA target TRAC | GENERAL BIOL | 5′-AAUAAUUUCUACUCUUGUAGAU GAGUCUCUCAGCUGGUACAC |
| Recombinant DNA | ||
| pUltra-CNF (Data S1) | Gifted by Prof. Peter G. Schultz | Addgene 48215 |
| pET22B-AsCas12a-2C-NLS (Data S1) | This paper | N/A |
| pAAV CD19-CAR (Data S1) | This paper | N/A |
| pHelper (Data S1) | Agilent | Cat# 240071 |
| pAAV-RC6 (Data S1) | Cell Biolabs, Inc. | Cat# VPK-426 |
| Software and algorithms | ||
| Tanon Gis | Tanon | N/A |
| CytExpert 2.3 | Beckman Coulter | https://cytexpert.software.informer.com/2.3/ |
| Gen5 | BioTek | https://www.biotek.com/products/software-robotics-software/gen5-microplate-reader-and-imager-software/ |
| QuantStudio™ Real-Time PCR Software | Thermo Fisher Scientific | https://www.thermofisher.com/jp/ja/home/global/forms/life-science/quantstudio-6-7-flex-software.html |
| ÄKTA pure protein purification system | Cytiva | https://www.cytivalifesciences.com/en/us/shop/chromatography/chromatography-systems/akta-pure-p-05844 |
| GraphPad 6.01 | GraphPad | https://www.graphpad.com/dl/96314/10B92408/ |
| Deposited data | ||
| Plasmid map | This paper | Mendeley Data: https://doi.org/10.17632/x7jmxr2497.1) |
| Other | ||
| Amicon® Ultra-15 Centrifugal Filter Units 50K | Millipore | Cat# UFC905024 |
| AzF (Unnatural amino acid) | Toronto Research Chemicals | A922600 |
| NAEK (Unnatural amino acid) | SIRIUS FINE CHEMICALS | SC-8027 |
| AeF (Unnatural amino acid) | Chemical synthesized | N/A |
| Millipore membrane filter, 0.22 mm pore size | Merck | Cat# SLGVR04NL |
| Superdex® 200 increase 10/300 GL | Cytiva | Cat# 28990944 |
| Electroporator | Celetrix LLC, Manassas VA | CTX-1500A LE |
| Electroporation tube | Celetrix LLC, Manassas VA | Cat# 12-0107 |
| DynaMag™-5 Magnet | Invitrogen | Cat# 12303D |
| CytoFLEX LX Flow Cytometer | Beckman Coulter | N/A |
| Synergy H1 Hybrid Multi-Mode Reader | BioTek | Synergy™ H1 |
| NanoDrop One Microvolume UV-Vis Spectrophotometer | Thermo Fisher Scientific | ND-ONE-W |
Materials and equipment
-
•
T cell complete medium
For 50 mL T cell culture medium, the recipe is listed as follow:
| Reagent | Final concentration | Amount |
|---|---|---|
| X-VIVOTM15 | 1× | 45 mL |
| Inactivated fetal bovine serum | 10% | 5 mL |
| IL-2 (30,000 U/mL) | 30 U/mL | 50 μL |
| IL-7 (5 mg/mL) | 5 ng/mL | 50 μL |
| IL-15 (5 mg/mL) | 5 ng/mL | 50 μL |
| Penicillin-Streptomycin solution | 1× | 500 μL |
| Total | n/a | 50 mL |
Store at 4°C for one week, do not prepare too much because the IL-2 is easy to degrade in the culture medium.
-
•
AAV293 cell complete medium
For 50 mL AAV293 cell culture medium, the recipe is listed as follow:
| Reagent | Final concentration | Amount |
|---|---|---|
| Dulbecco’s Modified Eagle’s Medium | 1× | 45 mL |
| Fetal Bovine Serum | 10% | 5 mL |
| Penicillin-Streptomycin solution | 1× | 500 μL |
| Total | n/a | 50 mL |
Store at 4°C for one month.
-
•
NALM-6 cell complete medium
For 50 mL NALM-6 cell culture medium, the recipe is listed as follow:
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI-1640 | 1× | 45 mL |
| Fetal Bovine Serum | 10% | 5 mL |
| Penicillin-Streptomycin solution | 1× | 500 μL |
| Total | n/a | 50 mL |
Store at 4°C for one month.
-
•
2YT medium
For 1 L 2YT medium, the recipe is listed as follow:
| Reagent | Final concentration | Amount |
|---|---|---|
| Tryptone | 16 g/L | 16 g |
| Yeast extract | 12 g/L | 12 g |
| NaCl | 5 g/L | 5 g |
| Total | n/a | 1 L |
Store at room temperature for one month after autoclaving.
-
•
T cell isolation buffer
For 50 mL T cell isolation buffer, the recipe is listed as follow:
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS without Ca2+ and Mg2+ | 1× | 49 mL |
| BSA (10%, w/v) | 0.1% | 500 μL |
| EDTA (200 mM) | 2 mM | 500 μL |
| Total | n/a | 50 mL |
Store at 4°C for one month.
Step-by-step method details
AzCas12a protein expression and purification
Timing: 3–4 days
At this section, AzCas12a is expressed in 2YT medium and purified with Ni-NTA column and size-exclusion chromatography. The total yield of AzCas12a is about 10 mg/L after purification. The protein can be used for further applications.
-
1.E. coli transformation and culture.
-
a.Day 1. Transform E. coli BL21(DE3) cells with pET22B-Cas12a-mutant and pUltra-CNF plasmids and cultivate the cells on a LB plate containing antibiotics (50 μg/mL spectinomycin and 100 μg/mL ampicillin) for 12–16 h.Detail procedures are listed below:
-
i.Remove E. coli BL21(DE3) competent cells from a −80°C freezer and thaw on ice.
-
ii.Mix pET22B-Cas12a-mutant plasmid (1 μL, ∼100 ng) and pUltra-CNF plasmid (1 μL, ∼100 ng) into the cells and then incubate the Eppendorf tube on ice for 30 min.
-
iii.Heat the tube at 42°C for 45 s in a water bath kettle.
-
iv.Place the tube on ice for 5 min without shaking.
-
v.Add 1 mL of 2YT medium to the tube and incubate at 37°C in a shaking incubator (220 rpm) for 1 h. (2YT medium recipe: 16 g of tryptone, 12 g of yeast extract, and 5 g of NaCl for 1 L of medium; use after autoclaving).
-
vi.Centrifuge at 5,000 × g for 3 min, discard the supernatant, and resuspend the cells with 100 μL of 2YT medium.
-
vii.Plate the suspension on a LB plate containing antibiotics (50 μg/mL spectinomycin and 100 μg/mL ampicillin).
-
viii.Incubate the LB plate at 37°C in a constant-temperature incubator for 12–16 h.
-
i.
-
b.Day 2. Pick a single colony and suspend it in 15 mL of 2YT medium containing antibiotics (50 μg/mL spectinomycin and 100 μg/mL ampicillin) for culture.
-
a.
-
2.Cas12a protein expression.
-
a.Day 3. Take 10 mL of overnight culture and dilute it with 1 L of 2YT expression medium containing antibiotics (50 μg/mL spectinomycin and 100 μg/mL ampicillin), and incubate at 37°C and 220 rpm until OD600 reaches 0.3.
-
b.Add unnatural amino acid AeF to the culture at a final concentration of 1 mM. Grow the culture at 37°C until OD600 reaches 0.6–0.8, then cool at 4°C for 15 min, and add isopropyl β-d-1-thiogalactopyranoside at a final concentration of 0.2 mM.
-
c.Maintain the culture at 18°C and 220 rpm in a shaker incubator for 16 h to express Cas12a protein.
-
a.
-
3.Cas12a protein purification.
-
a.Day 4. Harvest cells by centrifugation at 5,000 × g for 15 min at 4°C.
 CRITICAL: All the buffer for purification should be filtered with 0.22 μM filter before use. Otherwise, the protein may be degraded seriously during purification.
CRITICAL: All the buffer for purification should be filtered with 0.22 μM filter before use. Otherwise, the protein may be degraded seriously during purification. -
b.Discard supernatant, and resuspend cells in 100 mL of lysis buffer (50 mM Tris-HCl [pH 8.0], 1 M KCl, 10 mM imidazole) containing 1 mM phenylmethylsulfonyl fluoride to prevent protein degradation and then sonicate the suspension (30% power 5 s on and 5 s off) on ice for 20 min.
-
c.Centrifuge the lysate at 18,000 × g for 20–30 min at 4°C to pellet the cellular debris.
-
d.Balance a Ni-NTA resin gravity column with lysis buffer, load the supernatant on the column, and allow it to flow under the influence of gravity. Repeat this loading step two additional times.
-
e.Wash the resin with 5–10 column volumes of wash buffer (50 mM Tris-HCl [pH 8.0], 1 M KCl, 20 mM imidazole).
-
f.Elute the protein from the column with 10–20 mL of elution buffer (50 mM Tris-HCl [pH 8.0], 1 M KCl, 250 mM imidazole).
-
g.Transfer the eluted protein to an Amicon Ultra-15 Centrifugal Filter Unit 50K and centrifuge at 4,000 rpm for 20 min to concentrate the eluate to a volume of <1 mL. Then add 14 mL of PBS to the filter unit and centrifuge at 4,000 rpm for 20 min to change the buffer. Repeat this step one more time.
-
h.Purify the concentrated protein by size-exclusion chromatography using a Superdex® 200 Increase 10/300 GL column.
-
i.Balance an ÄKTA pure protein purification system (Cytiva) and the Superdex® 200 Increase 10/300 GL column with PBS at 0.5 mL/min.
-
j.Wash the injection loop with PBS buffer and inject the concentrated sample into the loop.
-
k.Continue to wash the column with PBS and collect the peaks according to the absorption of A280nm. Concentrate the collect fractions using an Amicon® Ultra-15 Centrifugal Filter Unit 50K, as described in step 3g.
-
l.Determine the purity of the Cas12a protein by means of 8% SDS-polyacrylamide gel electrophoresis and determine its concentration by means of UV–vis spectroscopy (NanoDrop One Microvolume UV–vis spectrophotometer).
-
m.Filter the concentrated AzCas12a protein with a 0.22 μm filter, aliquot the filtered protein, and store the aliquots at −80°C.
 CRITICAL: The protein stock solution must aliquot the filtered with a 0.22 μM filter for cell electroporation. Otherwise, it may induce cell bacteria contamination.
CRITICAL: The protein stock solution must aliquot the filtered with a 0.22 μM filter for cell electroporation. Otherwise, it may induce cell bacteria contamination.
-
a.
Production and purification of adeno-associated virus
Timing: 3–4 days
In this section, adeno-associated viruses are produced with AAV293 packaging cells and purified further. Then AAV titration is accomplished by Q-PCR. Using this protocol, the total yield of AAV can be 1013–1014 vg from fifteen 10-cm dishes.
-
4.
Packaging of adeno-associated virus.
The following procedure is used to package a large amount of adeno-associated virus (AAV). If a large amount is not required, the procedure can be scaled down proportionally.-
a.Trypsinize and resuspend AAV293 packaging cells from four 15-cm dishes; the cells should be at ∼80% confluence. Culture the cells at 37°C in complete medium: Dulbecco’s Modified Eagle’s Medium with 4.5 g/L glucose, sodium pyruvate, l-glutamine, 25 mM HEPES, and 10% fetal bovine serum (PAN-Biotech).
-
b.For each 15-cm dish: (1) Aspirate medium and add 6 mL of prewarmed PBS; (2) aspirate PBS, add 2 mL of trypsin-EDTA (0.25%) containing phenol red, and then aspirate trypsin-EDTA within 1 min; (3) hold for 1 min at room temperature (∼25°C); and (4) add 10 mL of prewarmed complete medium to stop digestion.
-
c.The day before transfection, seed fifteen 10-cm tissue culture dishes with the cells in the above-mentioned medium at a density of 1 × 107 cells/dish.
-
d.Twenty-four hours later, when the cell density is about 80%, transfect 5 μg of pAAV CD19-CAR, 5 μg of pHelper plasmid, and 5 μg of AAV-RC6 serotype plasmid into each 10-cm dish together with polyethyleneimine (PEI) at a 3:1 PEI/plasmid mass ratio. Specifically, for each 10-cm dish, mix the plasmids with Opti-MEM to a total volume of 250 μL and mix PEI (1 mg/mL, 45 μL) with 205 μL of Opti-MEM. Incubate the two mixtures separately at room temperature for 5 min, then mix them together, and incubate at room temperature (∼25°C) for 20–30 min. After incubation, add the transfection mixture to the dish.
-
e.Twenty-four hours after transfection, wash the cells once with prewarmed PBS and cultivate them in fresh complete medium.
-
f.Seventy-two hours after transfection, suspend the untrypsinized cells with medium supernatant and transfer them to 50-mL centrifuge tubes. Then centrifuge the tubes at 500 × g for 10 min to collect the transfected cells.
-
a.
-
5.AAV purification.
-
a.Resuspend the cells with 1 mL of PBS per 10-cm dish (Total 15 mL cell resuspension from fifteen 10-cm dishes are collected in 50-mL centrifuge tube). Mix 1.5 mL (1/10 v/v) of chloroform with the 15 mL of cell suspension.
-
b.Incubate the mixture at 37°C for 1 h with shaking.
-
c.Then add NaCl to a final concentration of 1 M and centrifuge at 20,000 × g at 4°C for 15 min.
-
d.Carefully transfer the aqueous layer to another 50-mL centrifuge tube and discard the chloroform layer.
-
e.Add PEG8000 to the aqueous layer at 10% (w/v) and incubate the mixture at 4°C for 1 h.
-
f.Centrifuge at 20,000 × g at 4°C for 15 min to collect the AAV pellet and carefully discard the supernatant.
-
g.Resuspend the pellet in 5 mL of PBS buffer and incubate at room temperature (∼25°C) for 10 min to allow it to fully dissolve.
-
h.Add 1 μL of Benzonase nuclease (Smart Lifesciences) at a final working concentration of 0.006% (∼16 U/mL) to the resulting solution and incubate at 37°C for 30 min.
-
i.Add 5 mL 1:1 (v/v) of chloroform to the solution, shake the tube up and down a few times, and centrifuge at 20,000 × g at 4°C for 20 min.
-
j.Carefully transfer the aqueous layer to an Amicon® Ultra-4 Centrifugal Filter Unit 100K and centrifuge at 4,000 rpm for 10 min to concentrate the AAV to ∼1 mL. Then aliquot the AAV and store the aliquots at −80°C. By means of this method, 1013–1014 vg/mL AAV can be obtained at a final volume of 1 mL.
-
a.
-
6.AAV titration.
-
a.For standard sample preparation, linearize and purify 20 μg of pAAV CD19-CAR with DNA Clean & Concentrator-25. Then dilute the linearized plasmid to 2 × 1010 molecules/μL to generate a standard curve.
-
b.To eliminate any contaminating plasmid DNA, treat the AAV sample with DNase I by mixing 2 μL of the sample with 6 μL of H2O, 1 μL of 10× DNase I buffer, and 1 μL of DNaseI (Thermo Fisher); then incubate the resulting working solution at 37°C for 30 min.
-
c.Mix 10 μL of 5 mM EDTA into the working solution and incubate the mixture at 65°C for 10 min to inactivate the DNase I.
-
d.Make 6 serial dilutions of standard curve plasmid.
Volume of original stock or previous dilution (μL) Volume of nuclease free water (μL) Molecules per μL 10 90 2 × 109 10 of 2 × 109 dilution 90 2 × 108 10 of 2 × 108 dilution 90 2 × 107 10 of 2 × 107 dilution 90 2 × 106 10 of 2 × 106 dilution 90 2 × 105 10 of 2 × 105 dilution 90 2 × 104 -
e.Dilute DNase I-treated AAV samples according to the dilution scheme in the table below:
Dilution series Volume of sample (μL) Volume of nuclease free water (μL) Total dilution factor Dilution 1 5 μL stock 45 10× Dilution 2 5 μL Dilution 1 95 200× Dilution 3 20 μL Dilution 2 80 1,000× Dilution 4 20 μL Dilution 3 80 5,000× Dilution 5 20 μL Dilution 4 80 25,000× Dilution 6 20 μL Dilution 5 80 125,000× -
f.Set up and load a 96-well plate using Bestar SYBR Green qPCR Mastermix (DBI Bioscience); The PCR procedure is performed according to the manufacturer’s instructions (Bestar SYBR Green qPCR Mastermix), which can be downloaded from this link: http://www.xinghanbio.com/ygdlpcrcp.Primers for qPCR:ITR-FW: 5′-GGAACCCCTAGTGATGGAGTT.ITR-RV: 5′-CGGCCTCAGTGAGCGA.PCR reagents
Reagent Volume (μL) Treated AAV or standard sample 2 ITR-FW (10 μM) 0.5 ITR-RV (10 μM) 0.5 Bestar SYBR Green qPCR Mastermix 10 H2O 7 Total 20 PCR amplification procedureTemperature (°C) Time (s) 95 10 55 34 72 30 -
g.Analyze data using QuantStudio software. Determine the physical titer of samples on the basis of the standard curve and the sample dilutions.
-
a.
Peripheral blood mononuclear cell isolation and T cell isolation and activation
Timing: 1 day
Start with peripheral blood from healthy donors. At this section, PBMCs are isolated from peripheral blood and then T cells are isolated from PBMCs. With this protocol, 5 × 106–1.5 × 107 T cells can be obtained from 20 mL peripheral blood. Finally, T cells are activated with CD3/CD28 T cell activator for electroporation.
-
7.PBMC isolation.
-
a.Isolate PBMCs from healthy donors with Ficoll-Paque PLUS (Cytiva) according to the manufacturer’s instructions, which can be downloaded from this link: https://www.cytivalifesciences.com/en/us/shop/cell-therapy/media/ficoll-paque-plus-density-gradient-media-p-05824#related-documents.
-
b.Count isolated cells with a CellDrop Automated Cell Counter (DeNovix) or a blood counting chamber.
-
c.Transfer fewer than 5 × 107 but more than 5 × 106 cells to a 15-mL centrifuge tube, centrifuge the tube at 90 × g for 10 min, and resuspend the cells with T cell isolation buffer (PBS without Ca2+ and Mg2+, containing 0.1% BSA and 2 mM EDTA) for isolation of primary T cells.
-
a.
-
8.T cell isolation.
-
a.Isolate T cells with a Dynabeads™ Untouched™ Human T Cells Kit (Invitrogen) according to the manufacturer’s instructions. Both CD4 and CD8 T cells are isolated by means of this method. The manufacturer’s instructions can be downloaded from this link: https://www.thermofisher.cn/order/catalog/product/11344D?adobe_mc=MCMID%7C62054544961035212653770489779646200236%7CMCAID%3D2F01986B052A2512-6000010A0000C7A5%7CMCORGID%3D5B135A0C5370E6B40A490D44%40AdobeOrg%7CTS=1614293705.
-
b.Count primary T cells with a CellDrop Automated Cell Counter (DeNovix).
-
c.Centrifuge the tube containing collected T cell at 200 × g for 5 min, discard the supernatant, and wash cells once with 4 mL of PBS.
-
d.Centrifuge the cells at 200 × g for 5 min, discard the PBS, and resuspend the cells with complete medium to a density of 0.5–1 × 106 cells/mL.
-
e.5 × 106–1.5 × 107 T cells can be isolated from 20 mL of peripheral blood from healthy donors. Typically, the total T cell isolation yield ranging from 60%-80% with different healthy donors.
-
f.Culture T cells at 37°C with 5% CO2 in T cell complete medium with density of 0.5–1 × 106 cells/mL. For lab tested conditions, T cell culture in 6-well Plate showed better activity than culture in the 10-cm dishes.
-
a.
| Expected isolated T cell | Culture medium volume (mL) | Culture dishes |
|---|---|---|
| 1 × 106 | 1 | 24-well plate |
| 5 × 106 | 5 | 12-well Plate |
| 1 × 107 | 10 | 6-well plate |
-
9.T cell activation.
-
a.Transfer the XYbeads Human CD3/CD28 T Cell Activator (Sailybio) to a 1.5-mL Eppendorf tube and place the tube on a DynaMag-5 Magnet (Invitrogen) for 1 min.
-
b.Carefully aspirate the supernatant without disturbing the beads.
-
c.Remove the tube from the magnet and add 1 mL of T cell complete medium to resuspend the beads. Then place the tube on the magnet for 1 min and aspirate the supernatant again. Repeat step 9c 3 times.
-
d.Activate the T cells with XYbeads Human CD3/CD28 T Cell Activator (Sailybio) at a 1:1 ratio.
-
e.Culture the cells at 37°C under 5% CO2.
-
a.
T cell electroporation and AAV infection
Timing: 1 day
Started with T cells that are activated for 3 days. At this section, cCas12a RNP targeted TRAC locus is prepared in vitro and then electroporated into T cells. AAVs carried CD19-CAR fragment are added into electroporated T cells to produce locus-specific CAR-T cells.
-
10.cCas12a RNP assembly in vitro.
-
a.Mix 30 pmol of AzCas12a with 36 pmol Dibenzocyclooctyne (DBCO) modified crRNA at a 1:1.2 molar ratio in sterile tubes and then add 1× PBS to each tube to bring the total volume to 4 μL.
Reagent Volume (μL) AzCas12a (30 μM) 1 DBCO-crRNA (36 μM) 1 PBS (1×) 2 Total 4 -
b.Incubate the resulting RNP complex at 25°C for 5 min and shake at 4°C for 3 h before electroporation to generate the covalent cCas12a complex in vitro.
 CRITICAL: The amount of Cas12a RNP and the incubation time are important. If the RNP amount or incubation time are reduced, the genome editing efficiency will decrease.
CRITICAL: The amount of Cas12a RNP and the incubation time are important. If the RNP amount or incubation time are reduced, the genome editing efficiency will decrease.
-
a.
-
11.T cell electroporation and AAV delivery.
-
a.Three days after T cell activation, suspend and collect T cells in 15-mL centrifuge tubes and remove the XYbeads Human CD3/CD28 T Cell Activator with a DynaMag™-5 Magnet (Invitrogen).
-
b.Count the cells with a CellDrop Automated Cell Counter (DeNovix).
-
c.Place the 1.5 × 106 cells per reaction into a new 15-mL centrifuge tube.
-
d.Centrifuge at 200 × g for 3 min and then wash cells once with 4 mL of PBS.
-
e.Aspirate as much supernatant as possible with a p200 tip and resuspend cells in the required electroporation buffer (Celetrix 13-0104) (20 μL per reaction).
-
f.Mix 20 μL of cell suspension and RNP complex well with a p200 pipette and transfer the mixture to a 20-μL electroporation tube (Celetrix 12-0107) with a 1–200 μL pipette tip (QSP) for electroporation.
-
g.Electroporate using a Celetrix CTX-1500A LE electroporator. For primary T cells, electroporation is conducted at 420 V for 20 ms at one pulse.
-
h.After electroporation, transfer the cells to a tube containing 100 μL of 37°C prewarmed complete medium.
-
i.Pipette 30 μL of electroporated cells (about 3.5 × 105 cells) into a well of a 48-well plate for culture. For follow-up CAR-T cell analysis, 30 μL of electroporated cells is enough. But for other evaluation or application, more cells can be used according to the need.
-
j.Four hours after electroporation, add AAV to the T cells at multiplicity of infection = 1 × 106 (∼5–6 μL AAV stock solution) and shake well.
-
a.
CRITICAL: The volume of AAV addition is no more than 10% total volume. Adding too much volume can lead to increased cell mortality.
-
12.CAR-T cell culture.
-
a.During culture, count cells every day. When the cell density exceeds 1 × 106 cells/mL (usually 2 days after transduction), passage the cells to a density of 5 × 105 cells/mL. Generally, expand the culture every 2 days from a 48-well plate (300 μL) to a 24-well plate (500 μL) and finally to a 12-well plate (1 mL) with T cell complete medium.
-
b.Specifically, the cell density usually reaches 1 × 106 cells/mL 2 days after electroporation. At this point, transfer the cells from a 48-well plate to a 24-well plate and culture in 500 μL of complete medium. Four days after electroporation, transfer the cells from the 24-well plate to a 12-well plate and culture in 1 mL of complete medium.
-
a.
Evaluation of CAR-T cell preparation efficiency and assessment of immunological characteristics
Timing: 2 days
Five days after electroporation, at this section, CAR-T cells preparation efficiency is evaluated by flow cytometry and semiquantitative in-and-out PCR. For assessment the immunological characteristics of CAR-T cells, LDH release and cytokines secretion are measured by LDH-Glo™ Cytotoxicity Assay and ELISA individually.
-
13.Flow cytometry analysis of CAR-T cell preparation efficiency.
-
a.Collect 1 × 106 CAR-T cells in a 1.5-mL Eppendorf tube and centrifuge at 400 × g for 5 min. Unless stated otherwise, all remaining centrifuge steps in this section are performed at 400 × g for 5 min.
-
b.Discard the supernatant and wash the cells once with 200 μL of PBS.
-
c.Resuspend the cells in 1 mL of blocking buffer (PBS containing 1% fetal bovine serum) and incubate them for 1 h at 4°C for blocking.
-
d.Resuspend the blocked cells in 100 μL of blocking buffer (PBS containing 1% fetal bovine serum) and 1 μL of c-Myc mouse monoclonal antibodies (Invitrogen). Incubate the cells at 4°C for 1 h.
-
e.Centrifuge and wash the cells twice with 200 μL of PBS buffer to remove as much of the primary antibodies as possible.
-
f.Dilute antimouse IgG (H+L), F(ab′)2 Fragment (Alexa Fluor® 647 Conjugate, Cell Signaling Technology) in PBS at a 1:1000 ratio. Resuspend the cells in the dilutions and incubate at 4°C for 30 min.
-
g.Centrifuge and wash the resulting anti-Myc-antibody-stained cells twice with 200 μL of PBS buffer.
-
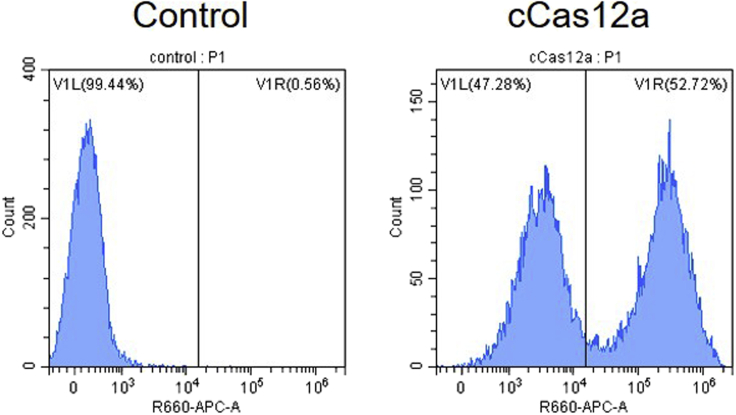
h.Resuspend the stained cells with 200 μL of PBS and analyze the suspension on a CytoFlex flow cytometer (Beckman). Cells are gated on the basis of forward scatter area (FSC-A) and side scatter area (SSC-A), and cells in the gate are analyzed further for percentage of APC660 positive populations using CytExpert software.
-
a.
-
14.Semiquantitative in-and-out PCR.
-
a.Extract genomic DNA from transduced T cells with a FastPure Cell/Tissue DNA Isolation Mini Kit (Vazyme, DC102) according to the manufacturer’s instructions, which can be downloaded from this link: https://www.vazyme.com/product/15.html.
-
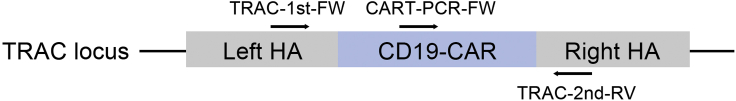
b.Three primers are used for the in-and-out PCR reaction: TRAC-1st-FW (CCCTTGTCCATCACTGGCAT), which binds to the left homologous arm of the TRAC locus; CART-PCR-FW (GGAGTACGACGTGCTGGATAAG), which specifically recognizes a sequence within the anti-CD19 CAR fragment; and TRAC-2st-RV (GCACACCCCTCATCTGACTT), which binds to the right homologous arm of the TRAC locus (Figure 1).
-
c.Analyze the PCR product on a 1% agarose gel with quantification using a Tanon 1600 gel-imaging system and Tanon Gis software.
-
a.
-
15.Co-culture of CAR-T cells with NALM-6 cells.
-
a.Five days after electroporation of T cell, seed 1 × 104 NALM-6 cells in a U-bottom 96-well plate.
-
b.Co-culture CAR-T cells with the NALM-6 cells in fresh RPMI-1640 complete medium (RPMI-1640 containing 10% fetal bovine serum) to a total volume of 200 μL at a 10:1 E/T ratio for 24–48 h. For a control group, co-culture untransduced T cells with NALM-6. All groups are repeated in three more wells.
-
a.
-
16.Evaluation of CAR-T cytotoxicity by LDH-Glo™ Cytotoxicity Assay.
-
a.For cytotoxicity evaluation, use control wells designed for maximum Lactate dehydrogenase (LDH) release. Twenty minutes before analysis, add 2 μL of 10% Triton-X100 per 100 μL to control wells for maximum LDH release.
-
b.Evaluate CAR-T cell cytotoxicity by LDH-Glo™ Cytotoxicity Assay (Promega) following the manufacturer’s instructions, which can be downloaded from this link: https://www.promega.com.cn/products/cell-health-assays/cell-viability-and-cytotoxicity-assays/ldh-glo-cytotoxicity-assay/?catNum=J2380#protocols.
-
c.Centrifuge the cell in the 1.5 mL Eppendorf tube at 500 × g for 10 min to precipitate cells and collect medium supernatant. Dilute 1 μL of medium supernatant at 1:100 with LDH storage buffer for the LDH release assay.
-
d.Transfer 50 μL of diluted sample into a 96-well opaque-walled, nontransparent assay plate.
-
e.Add 50 μL of LDH detection reagent (prepared in advance) to each well.
-
f.Incubate at room temperature (∼25°C) for 60 min in a shaker.
-
g.Measure luminescence with a Synergy™ H1 microplate reader (BioTek).
-
h.Calculate cell cytotoxicity (%) with the following formula: % cytotoxicity = 100(experimental LDH release − medium background)/(maximum LDH release control − medium background).
-
a.
-
17.Immunological characteristics of CAR-T cells.
-
a.Aspirate 90 μL of medium supernatant from step 16c for separate detecting IFN-γ and TNF-α release.
-
b.Perform an enzyme-linked immunosorbent assay to detect cytokine release by CAR-T cells using a Human IFN gamma Uncoated ELISA kit (Invitrogen) and a Human TNF alpha Uncoated ELISA kit (Invitrogen) according to the manufacturer’s instructions, which can be downloaded from these links: https://www.thermofisher.cn/document-connect/document-connect.html?url=https://assets.thermofisher.cn/TFS-Assets%2FLSG%2Fmanuals%2FMAN0017369_88-7316_HumanIFNgamma-ELISA_PI.pdf and https://assets.thermofisher.cn/TFS Assets%2FLSG%2Fmanuals%2FMAN0017366_88-7346_HumanTNFAlphaELISA_PI.pdf.
-
c.Measure absorbances at 450 and 570 nm with a Synergy™ H1 microplate reader (BioTek) and analyze data with A450nm − A570nm. Calculate cytokine concentrations using the standard curves.
-
a.
Figure 1.
Schematic diagram of in-and-out PCR
Expected outcomes
Rapid and precise gene knock-in can be achieved by means of cCas12a-based genome editing. Locus-specifically engineered CAR-T cells can be efficiently prepared by means of multiplex gene editing with the cCas12a system described in this protocol, as shown in Figures 2 and 3. The prepared cells showed robust immunologic characteristics for killing of targeted cells, as shown in Figure 4. Genome-editing details and CAR-T preparation data can be found in our recent Molecular Cell paper (Ling et al., 2021).
Figure 2.
Flow cytometry analysis of CAR-T cell preparation efficiency
T cells were stained with anti-Myc antibody.
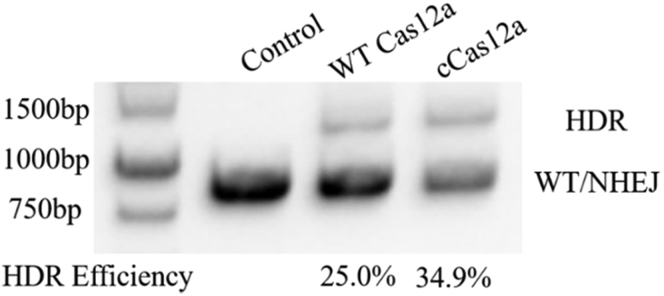
Figure 3.
CAR-T cell genome integration efficiency as determined by semiquantitative in-and-out PCR
The HDR efficiency was analyzed on a 1% agarose gel and quantified with Tanon Gis software.
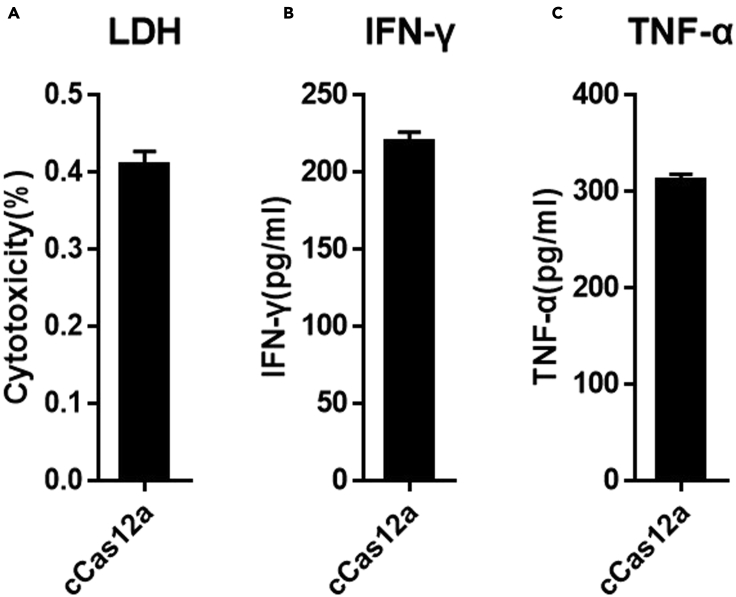
Figure 4.
Evaluation of immunological characteristics of CAR-T cells
(A) LDH-Glo assay of cytotoxicity of CAR-T cells prepared with cCas12a; data are presented as the mean ± SEM derived from triplicate samples (n = 3).
(B) IFN-γ expression levels in CAR-T cells prepared with cCas12a; data are presented as the mean ± SEM derived from triplicate samples (n = 3).
(C) TNF-α expression levels in CAR-T cells prepared with cCas12a; data are presented as the mean ± SEM derived from triplicate samples (n = 3).
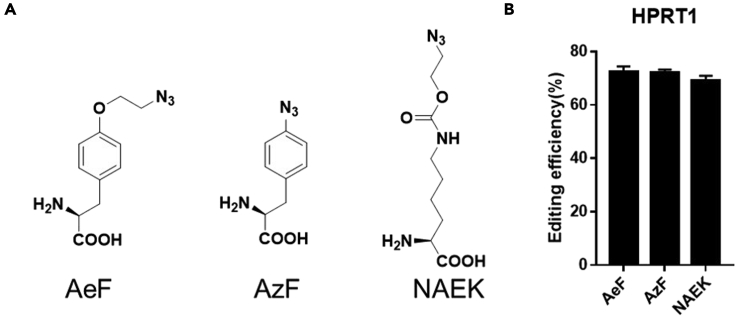
To increase the utility of this protocol, we found that another 2-azido containing commercially available unnatural amino acid, p-azido-phenylalanine (AzF) and N6-((2-azidoethoxy) carbonyl)-l-lysine (NAEK) (Figure 5A), could be site-specifically introduced into the Cas12a protein to form a covalent cCas12a with similar effects. AzF could be purchased at https://www.trc-canada.com/product-detail/?A922600 and N6-((2-Azidoethoxy)carbonyl)-l-lysine can be purchased at https://shop.sichem.de/en/sc-8027.html, Those corresponding aminoacyl-tRNA synthetase/tRNACUA pairs are available from Addgene. As shown if Figure 5B, the AzF and NAEK containing Cas12a mutants show comparable editing efficiency after conjugation to the AeF containing Cas12a mutant.
Figure 5.
Genome editing efficiency with different azido amino acid containing chemically modified Cas12a
(A) Unnatural amino acids used in the study.
(B) The genome editing efficiency were evaluated by Cas12a mutants with its crRNA targeting HPRT1 genome in HEK293 cell; data are presented as the mean ± SEM derived from triplicate samples (n = 3).
Limitations
This protocol provides a covalent-Cas12a-RNP based platform for efficient T cell engineering. One of the important advantages of the Cas12a system is that this enzyme can be used for multiplex genome editing with a single crRNA array (Dai et al., 2019). Unfortunately, our protocol does not have this advantage: each RNP must be prepared individually. However, the stoichiometry of the RNP targeting each locus can be precisely controlled and optimized to achieve a much better overall multiplex genome-editing efficiency. With our protocol, we could achieve locus-specific integration of a CAR gene into the T cell genome with an editing efficiency of >50%, which is one of the highest reported efficiencies for locus-specific CAR integration into T cells (Moço et al., 2020). However, the efficiency is much lower than that of the traditional lentivirus-based CAR-T preparation method, which typically shows an editing efficiency of >90%. Thus, further optimization to increase the overall efficiency of our protocol is necessary. Fortunately, the protocol is theoretically compatible with improved Cas12a mutants, such as PAM-extended and efficiency-enhanced versions of the enzyme. Finally, an electroporator is necessary to perform these experiments, and the electroporation conditions must be optimized.
Troubleshooting
Problem 1
Chemically modified Cas12a protein expression with low yield (steps 1–3: AzCas12a protein expression and purification).
Potential solution
Reexamine the plasmid and colony condition first. The unnatural amino acid should be added into the culture media at least 30 min before protein induction. The final concentration of unnatural amino acid is the key for a successful protein expression. Typically, 1 mM unnatural amino acid is sufficient for protein expression. However, for NAEK containing chemically modified Cas12a, the final concentration of NAEK could be increased to 5 mM for better expression yield and purity.
Problem 2
T cells grow slowly after isolation (step 9: T cell activation).
Potential solution
Using fresh prepared T cell culture medium is the best choice, the IL-2 in the medium would degrade at 4°C in a week.
Problem 3
The conjugation efficiency of AzCas12a and DBCO modified crRNA is not good (step 10: cCas12a RNP assembly in vitro).
Potential solution
The azido containing protein may risk in azide group reduction after long term stock in fridge. The protein should be fresh prepared and should not be used after one-year stock. For conjugation reaction, the final concentration could be increased and the reaction temperature could be increased to room temperature. We have tested that conjugation efficiency is nearly the same at three different reaction conditions: 30 min at 37°C, 1 h at 25°C or 3 h at 4°C.
Problem 4
Low T cell survival rate after electroporation and AAV transduction (step 11: T cell electroporation and AAV delivery).
Potential solution
T cells should not be maintained too long, the best electroporation time should be 3 days after first CD3/CD28 magnetic bead activation, the over activation may reduce the T cell availability after electroporation. For avoiding the toxicity of the virus, the virus volume should not exceed 10% of the final volume.
Problem 5
Low editing efficiency of final CAR-T cell (steps 13–17: Evaluation of CAR-T cell preparation efficiency and assessment of immunological characteristics).
Potential solution
For targeting different sites, the crRNA optimization is needed for a better efficiency. The chemically modified RNA should be stored at −80°C and prevent thaw and freeze. The RNP assembled in vitro should be used immediately, avoiding storage at 4°C for over 24 h. If the CAR-T knock-in efficiency is not high enough, you could optimize the length of homology arms for better knock-in efficiency.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Tao Liu (taoliupku@pku.edu.cn).
Materials availability
Plasmids, primers, recombinant proteins, experimental strains, and any other research reagents generated by the authors will be distributed upon request to other research investigators under a material transfer agreement.
Acknowledgments
This work was financially supported by the National Key Research and Development Program of China (2021YFA0909900), National Natural Science Foundation of China (21922701, 91853111, and 21778005), Beijing Natural Science Foundation (JQ20034), and Peking University and Innovation Fund for Outstanding Doctoral Candidates of Peking University Health Science Center (71006Y2460).
Author contributions
X.L. and L.C. wrote the protocol. H.C. constructed the graph. T.L. edited and revised the protocol.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2022.101321.
Supplemental information
Data and code availability
This study did not generate or analyze datasets or code. The original data containing plasmid map have been deposited to Mendeley Data: https://doi.org/10.17632/x7jmxr2497.1.
References
- Dai X., Park J.J., Du Y., Kim H.R., Wang G., Errami Y., Chen S. One-step generation of modular CAR-T cells with AAV–Cpf1. Nat. Methods. 2019;16:247–254. doi: 10.1038/s41592-019-0329-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfall A.L., Maus M.V., Hwang W.-T., Lacey S.F., Mahnke Y.D., Melenhorst J.J., Zheng Z., Vogl D.T., Cohen A.D., Weiss B.M., et al. Chimeric antigen receptor T cells against CD19 for multiple myeloma. New Engl. J. Med. 2015;373:1040–1047. doi: 10.1056/NEJMoa1504542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe S.J., Mansour M.R., Schwarzwaelder K., Bartholomae C., Hubank M., Kempski H., Brugman M.H., Pike-Overzet K., Chatters S.J., de Ridder D., et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- June C.H., Sadelain M. Chimeric antigen receptor therapy. New Engl. J. Med. 2018;379:64–73. doi: 10.1056/NEJMra1706169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling X., Chang L., Chen H., Gao X., Yin J., Zuo Y., Huang Y., Zhang B., Hu J., Liu T. Improving the efficiency of CRISPR-Cas12a-based genome editing with site-specific covalent Cas12a-crRNA conjugates. Mol. Cell. 2021;81:4747–4756.e7. doi: 10.1016/j.molcel.2021.09.021. [DOI] [PubMed] [Google Scholar]
- Moço P.D., Aharony N., Kamen A. Adeno-associated viral vectors for homology-directed generation of CAR-T cells. Biotechnol. J. 2020;15:1900286. doi: 10.1002/biot.201900286. [DOI] [PubMed] [Google Scholar]
- Tebas P., Stein D., Tang W.W., Frank I., Wang S.Q., Lee G., Spratt S.K., Surosky R.T., Giedlin M.A., Nichol G., et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. New Engl. J. Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Rivière I. Clinical manufacturing of CAR T cells: foundation of a promising therapy. Mol. Ther. Oncolytics. 2016;3:16015. doi: 10.1038/mto.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate or analyze datasets or code. The original data containing plasmid map have been deposited to Mendeley Data: https://doi.org/10.17632/x7jmxr2497.1.