Abstract
The phenotypic resistance of selected organisms to ciprofloxacin, levofloxacin, and trovafloxacin was defined as a MIC of ≥4 μg/ml. The dynamics of resistance were studied after single and sequential drug exposures: clinical isolates of methicillin-susceptible and methicillin-resistant Staphylococcus aureus (MSSA and MRSA), Escherichia coli, Klebsiella pneumoniae, Enterobacter cloacae, Serratia marcescens, and Pseudomonas aeruginosa were utilized. After a single 48-h exposure of a large inoculum to four times the initial MIC for the organism, the frequency of selection of resistant mutants of MSSA was greater for trovafloxacin than levofloxacin (P = 0.008); for E. cloacae, the frequency was highest for ciprofloxacin and lowest for levofloxacin and trovafloxacin; for S. marcescens, the frequency was highest for trovafloxacin and lowest for ciprofloxacin (P = 0.003). The results of serial passage experiments were analyzed both by the Kaplan-Meier product-limited method as well as by analysis of variance of mean inhibitory values. By both methods, MSSA and MRSA expressed mutants resistant to ciprofloxacin after fewer passages than were required for either levofloxacin or trovafloxacin. For the aerobic gram-negative bacilli, two general patterns emerged. Mutants resistant to trovafloxacin appeared sooner and reached higher mean MICs than did mutants resistant to levofloxacin or ciprofloxacin. Mutants resistant to ciprofloxacin appeared later and reached mean MICs lower than the MICs of the other two drugs studied. Even though individual strain variation occurred, the mean MICs were reproduced when the serial passage experiment was repeated using an identical panel of E. coli isolates. In summary, the dynamic selection of fluoroquinolone-resistant bacteria can be demonstrated in experiments that employ serial passage of bacteria in vitro.
The clinical use of fluorinated 4-quinolones continues to increase. They now account for roughly 11% of antimicrobial prescriptions worldwide (22). Despite widespread use over 10 or more years, selection of resistant clones has been modest. Clinically significant resistance to ciprofloxacin has occurred for Staphylococcus aureus (especially methicillin-resistant S. aureus [MRSA]), Neisseria gonorrhoeae, Pseudomonas aeruginosa, and Acinetobacter baumanii (1, 3, 12, 19). Surprisingly, resistance of Enterobacteriaceae has been modest. Recent reports of the increasing resistance of Escherichia coli, Campylobacter jejuni, and Salmonella species raise the concern of the potential spread of selected resistant mutants (2, 9, 10, 21, 23).
Microbial resistance to fluoroquinolones results from either mutations in topoisomerase II (DNA gyrase), topoisomerase IV, and/or activation of drug efflux pumps. Selection of resistant organisms evolves from the interplay of gene mutation frequency, the number and type of mutations necessary to express phenotypic resistance, drug potency, concentration of the test drug, and other factors (5, 11). Mutation and selection pressure create the potential for survival advantage and spread of resistant clones.
Assessment of the phenotypic expression of the genotypic changes has largely been based on changes in the MIC. Studies of fluoroquinolone-selected genetic changes indicate that phenotypic change is often the result of multiple sequential changes in the genome of the organism (16). Our intent was to study the dynamics of phenotypic resistance over time.
There is a variable but generally low frequency (<1 × 10−9) of resistant organisms when a test organism is incubated in a concentration of fluoroquinolone that is four times the baseline MIC (6). Earlier studies have demonstrated increasing levels of resistance when test organisms are serially passaged in incrementally increasing concentrations of fluoroquinolone (7, 13; M. E. Evans and W. E. Titlow, Letter, Antimicrob. Agents Chemother. 42:727, 1998). We postulated that differences exist among the fluoroquinolones with respect to the number of sequential passages necessary to develop potentially clinically significant levels of drug resistance.
We studied the propensity of clinical isolates of S. aureus, E. coli, Klebsiella pneumoniae, Enterobacter cloacae, Serratia marcescens, and P. aeruginosa to develop phenotypic resistance to ciprofloxacin, levofloxacin, and trovafloxacin after single and sequential drug exposures.
MATERIALS AND METHODS
Bacterial strains.
Consecutive clinical isolates were collected, including 13 methicillin-susceptible S. aureus (MSSA) and 15 MRSA isolates; 20 isolates each of E. coli, K. pneumoniae, and P. aeruginosa; 16 isolates of S. marcescens; and 19 isolates of E. cloacae.
In vitro susceptibility testing.
Fluoroquinolone in vitro susceptibility was determined by a standard agar dilution method using Mueller-Hinton agar and an inoculum of 104 CFU per spot (15). For testing of S. aureus, 2% sodium chloride was added to the agar. Laboratory standard powders of ciprofloxacin were obtained from the Bayer Company, levofloxacin was obtained from the R. W. Johnson Pharmaceutical Research Institute (Raritan, N.J.), and trovafloxacin was obtained from Pfizer Inc. (Groton, Conn.).
As per the National Committee for Laboratory Standards, in vitro resistance of S. aureus and of enteric gram-negative bacilli to ciprofloxacin is defined as a MIC of ≥4 μg/ml; resistance to levofloxacin is defined as a MIC of ≥8 μg/ml (15). At present, there are no NCCLS-approved breakpoints for trovafloxacin versus staphylococci and enteric bacteria. In evaluating the results of the serial passage experiments, a MIC of ≥4 μg/ml was selected as an end point upon which to base statistical evaluation. The selection of a MIC of ≥4 μg/ml was arbitrary. We believe that a MIC of ≥4 μg/ml, for the quinolones under study, would have potential clinical relevance. The selection of ≥4 μg/ml was based primarily on peak drug concentrations in serum and on recognition that peak trovafloxacin levels and peak levofloxacin levels are usually lower and higher than 4 μg/ml, respectively.
Frequency of selection of resistant strains after single exposure.
Test bacteria were grown overnight in antibiotic-free Mueller-Hinton broth. Inocula of 108 to 1010 CFU were plated on Mueller-Hinton agar containing four times the MIC of the test drugs. The plates were incubated for 48 h. The number of resistant colonies was counted on plates with between 1 and 1,000 visible colonies. Quantitation of each inoculum was done in triplicate. The frequency of resistant isolates selected by each fluoroquinolone was calculated by dividing the number of colonies growing after 48 h by the mean starting inoculum.
One to twenty resistant colonies were selected, and the agar dilution MIC was determined to document the magnitude of resistance.
Selection of resistant strains by serial passage.
Bacteria were grown overnight in antibiotic-free Mueller-Hinton broth to 107 CFU/ml using a McFarland standard. The inoculum was verified by serial plate dilution. An inoculum of 1 μl containing 104 CFU in log-phase growth was plated in a single spot on a series of Mueller-Hinton agar plates containing increasing twofold concentrations of ciprofloxacin, levofloxacin, or trovafloxacin extending from below to above the baseline measured MIC.
After 48 h of incubation at 35°C, bacterial growth on the plates with the highest drug concentration showing growth was collected, regrown in antibiotic-free broth, and reinoculated onto sets of agar plates with increasing twofold-higher concentrations of fluoroquinolone. Each sequential 48-h incubation was repeated for 10 serial passages. The upper limit of quantitation of resistance was 256 μg/ml.
To evaluate reproducibility of results, the serial passage was performed twice using a second collection of 15 isolates of E. coli.
Statistical evaluation.
All statistical analysis was performed using the SPSS statistical package, version 9.0 (Chicago, Ill.).
Differences in mutation frequency among drugs in the single incubation experiments were assessed using two-way analysis of variance on the logarithmic transformation of mutation frequency. Bonferroni's correction was used to adjust for multiple comparisons between drugs.
Data from the serial passage experiments were analyzed by two methods. First, the cumulative risk of an isolate reaching a MIC of ≥4 μg/ml was assessed using Kaplan-Meier (KM) product-limit estimates. This end point was selected as the concentration of most clinical import. The log rank test, with Bonferroni's correction, was used for comparisons. KM end-point-free plots were used for graphical comparison.
KM analysis of the increasing drug concentrations compatible with bacterial growth considers only the first passage through the 4-μg/ml end point and ignores the information beyond that step. As an adjunct to the KM analysis, differences between drugs using all 10 steps (passages) were analyzed by two methods. After logarithmic transformation, the slopes of the inhibitory concentrations between drugs, determined from the interaction term between drug and serial passage step, were compared using analysis of variance. Bonferroni's correction was used for the pairwise comparisons.
RESULTS
Single-incubation mutation frequencies. (i) MIC90.
The baseline and postincubation MIC90s for the seven bacterial strains are shown in Table 1. With the exception of trovafloxacin versus MSSA, all the test organisms exhibited a 16- to 32-fold increase in the MIC90.
TABLE 1.
Susceptibilities of test organisms before and after 48 h of incubation with concentrations of fluoroquinolone four times greater than the baseline MIC
| Microorganism (n) and fluoroquinolone | MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|
| Preincubation
|
Postincubation
|
|||||
| Range | 50%a | 90%b | Ranged | 50% | 90% | |
| MSSA (13) | ||||||
| Ciprofloxacin | 0.12–0.5 | 0.25 | 0.5 | 0.016–4 (7) | 1 | 4 |
| Levofloxacin | 0.12–1 | 0.25 | 0.5 | (0) | —c | — |
| Trovafloxacin | 0.008–0.03 | 0.016 | 0.016 | 0.016–0.06 (6) | 0.06 | 0.06 |
| MRSA (15) | ||||||
| Ciprofloxacin | 0.06–1 | 0.25 | 0.5 | 0.25–32 (7) | 2 | 8 |
| Levofloxacin | 0.12–1 | 0.25 | 0.5 | 1–32 (5) | — | — |
| Trovafloxacin | 0.008–0.12 | 0.03 | 0.12 | 1–4 (2) | — | — |
| E. coli (20) | ||||||
| Ciprofloxacin | 0.004–0.12 | 0.016 | 0.016 | 0.06–1 (15) | 0.12 | 0.25 |
| Levofloxacin | 0.008–0.25 | 0.03 | 0.06 | 0.12–2 (12) | 0.25 | 0.5 |
| Trovafloxacin | 0.002–0.12 | 0.008 | 0.03 | 0.016–2 (17) | 0.12 | 0.25 |
| K. pneumoniae (20) | ||||||
| Ciprofloxacin | 0.004–0.5 | 0.03 | 0.12 | 0.5–8 (17) | 1 | 2.0 |
| Levofloxacin | 0.03–0.5 | 0.06 | 0.12 | 1–2 (16) | 2 | 2 |
| Trovafloxacin | 0.016–0.5 | 0.06 | 0.06 | 0.5–2 (18) | 1 | 2 |
| E. cloacae (19) | ||||||
| Ciprofloxacin | 0.004–0.06 | 0.016 | 0.03 | 0.03–4 (15) | 0.06 | 1 |
| Levofloxacin | 0.03–0.12 | 0.03 | 0.06 | 0.25–2 (11) | 1 | 2 |
| Trovafloxacin | 0.016–0.12 | 0.03 | 0.06 | 0.12–4 (17) | 1 | 2 |
| S. marcescens (16) | ||||||
| Ciprofloxacin | 0.06–1 | 0.25 | 0.25 | (1) | — | — |
| Levofloxacin | 0.06–0.5 | 0.12 | 0.25 | 2–8 (11) | 4 | 8 |
| Trovafloxacin | 0.06–2 | 0.5 | 1 | 1–32 (11) | 8 | 32 |
| P. aeruginosa (20) | ||||||
| Ciprofloxacin | 0.06–0.25 | 0.12 | 0.25 | 2–4 (7) | 4 | 4 |
| Levofloxacin | 0.25–2 | 0.5 | 1 | 1–16 (12) | 8 | 16 |
| Trovafloxacin | 0.12–1 | 0.5 | 1 | 4–32 (9) | 8 | 32 |
MIC50.
MIC90.
—, insufficient number of isolates with growth.
Parentheses, number of resistant isolates.
(ii) Mutation frequencies.
No statistically significant differences in the frequency of detection of resistant mutants to ciprofloxacin, levofloxacin, or trovafloxacin occurred after a single incubation of MRSA, E. coli, K. pneumoniae, or P. aeruginosa in a concentration of test drug that was four times the baseline MIC (data not shown).
Significant differences in the frequency of detection of resistant mutants of MSSA, E. cloacae, and S. marcescens are shown in Table 2. For MSSA, trovafloxacin showed a statistically significant greater frequency of resistant mutants than did levofloxacin. For E. cloacae, the highest frequency of resistant mutants occurred after incubation with ciprofloxacin; differences between ciprofloxacin, levofloxacin, and trovafloxacin were statistically significant. For S. marcescens a higher frequency of resistant mutants occurred for levofloxacin and trovafloxacin than for ciprofloxacin.
TABLE 2.
Mutation frequencies of test bacteria after a 48-h incubation with ciprofloxacin, levofloxacin, or trovafloxacina
| Organism (no. of isolates) | Bacterial mutation frequency after incubation with:
|
||
|---|---|---|---|
| Ciprofloxacin | Levofloxacin | Trovafloxacin | |
| MSSA (13) | 5.9 × 10−7 ± 5.1 × 10−7 | 1.2 × 10−8 ± 8.8 × 10−9b | 6.5 × 10−7 ± 3.2 × 10−7b |
| MRSA (15) | 8.8 × 10−7 ± 8.8 × 10−8 | 3.6 × 10−8 ± 3.1 × 10−8 | 8.5 × 10−9 ± 8.3 × 10−9 |
| E. coli (20) | 1.6 × 10−7 ± 4.9 × 10−8 | 1.6 × 10−7 ± 7.8 × 10−8 | 3.5 × 10−7 ± 2.5 × 10−8 |
| K. pneumoniae (20) | 1.9 × 10−7 ± 1.2 × 10−7 | 6.6 × 10−8 ± 2.0 × 10−8 | 1.2 × 10−7 ± 3.1 × 10−8 |
| E. cloacae (19) | 4.2 × 10−7 ± 1.2 × 10−7c | 2.9 × 10−8 ± 1.1 × 10−8 | 2.0 × 10−8 ± 8.6 × 10−9 |
| S. marcescens (16) | 7.7 × 10−10 ± 7.7 × 10−10d | 1.6 × 10−8 ± 5.2 × 10−9 | 1.8 × 10−7 ± 1.3 × 10−7 |
| P. aeruginosa (20) | 5.4 × 10−8 ± 4.9 × 10−8 | 1.1 × 10−8 ± 3.4 × 10−9 | 4.8 × 10−9 ± 1.7 × 10−9 |
The drug concentrations were four times higher than the baseline MIC. The data represent the mean values of the mutation frequencies expressed as the mean ± standard error.
P = 0.008.
For levofloxacin versus ciprofloxacin, P = 0.003; for trovafloxacin versus ciprofloxacin, P = 0.009.
For ciprofloxacin versus trovafloxacin, P = 0.003; for ciprofloxacin versus trovafloxacin, P = 0.009.
The results were evaluated for a possible relationship between the baseline MIC and the observed mutation frequency. No correlation was found.
Serial passage experiments. (i) Pre- and post-MIC90s.
After 10 serial passages, the MIC90s increased 16- to 1,024-fold for each of the seven bacterial species versus the three test drugs (Table 3). Low postpassage MIC90s observed with trovafloxacin versus MSSA, MRSA, and E. coli still represent 16- to 400-fold increases from baseline values.
TABLE 3.
Susceptibilities of test organisms before and after 10 serial passages with subinhibitory concentrations of ciprofloxacin, levofloxacin, and trovafloxacin
| Microorganism | Initial no. of isolates tested | Fluoroquinolone | MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|---|---|
| Pre-serial passage
|
Post-serial passage
|
|||||||
| Range | 50%a | 90%b | Range | 50% | 90% | |||
| MSSA | 13 | Ciprofloxacin | 0.12–0.5 | 0.25 | 0.5 | 4–256 | 16 | 64 |
| Levofloxacin | 0.12–0.25 | 0.25 | 0.5 | 0.5–4 | 1 | 32 | ||
| Trovafloxacin | 0.004–0.01 | 0.004 | 0.01 | 0.125–32 | 0.25 | 4 | ||
| MRSA | 15 | Ciprofloxacin | 0.12–1 | 0.25 | 0.5 | 1–256 | 8 | 128 |
| Levofloxacin | 0.06–0.5 | 0.25 | 0.5 | 1–32 | 2 | 32 | ||
| Trovafloxacin | 0.008–0.12 | 0.008 | 0.12 | 0.03–2 | 0.25 | 2 | ||
| E. coli | 20 | Ciprofloxacin | 0.004–0.12 | 0.008 | 0.016 | 0.012–128 | 1.0 | 16 |
| Levofloxacin | 0.03–0.25 | 0.03 | 0.06 | 0.5–16 | 2.0 | 8 | ||
| Trovafloxacin | 0.002–0.03 | 0.008 | 0.016 | 0.5–16 | 2.0 | 4 | ||
| K. pneumoniae | 20 | Ciprofloxacin | 0.016–0.25 | 0.06 | 0.12 | 2–64 | 16 | 64 |
| Levofloxacin | 0.03–0.25 | 0.03 | 0.12 | 2–128 | 8 | 32 | ||
| Trovafloxacin | 0.03–0.5 | 0.06 | 0.12 | 2–128 | 16 | 128 | ||
| E. cloacae | 19 | Ciprofloxacin | 0.008–0.06 | 0.03 | 0.06 | 0.5–128 | 8 | 64 |
| Levofloxacin | 0.004–0.12 | 0.06 | 0.12 | 4–64 | 8 | 64 | ||
| Trovafloxacin | 0.03–0.5 | 0.06 | 0.25 | 8–64 | 16 | 64 | ||
| S. marcescens | 16 | Ciprofloxacin | 0.06–0.25 | 0.25 | 0.25 | 2–64 | 16 | 64 |
| Levofloxacin | 0.06–0.5 | 0.25 | 0.5 | 1–256 | 16 | 128 | ||
| Trovafloxacin | 0.06–0.25 | 0.12 | 0.25 | 32–256 | 64 | 256 | ||
| P. aeruginosa | 20 | Ciprofloxacin | 0.03–1 | 0.12 | 0.25 | 2–256 | 8 | 32 |
| Levofloxacin | 0.12–1 | 0.5 | 1 | 16–256 | 128 | 256 | ||
| Trovafloxacin | 0.06–1 | 0.25 | 1 | 32–256 | 256 | 256 | ||
MIC50.
MIC90.
A progressive increase during serial incubations in phenotypic resistance was observed for all organisms and all drugs tested. Differences between drugs and organisms were observed. To demonstrate those differences, the results of the serial incubation experiments are presented in two forms: KM survival curves and regression analysis of mean MICs plotted against number of incubations.
(ii) KM curves.
KM analysis plots of the percentage of isolates remaining susceptible to 4-μg/ml concentrations or less of test fluoroquinolone at each sequential in vitro passage showed that for E. coli, there were no statistically significant differences between the test drugs in the rate of accumulation of resistant mutants. Statistically significant differences between the three test drugs were observed for the other six organisms tested (Fig. 1).
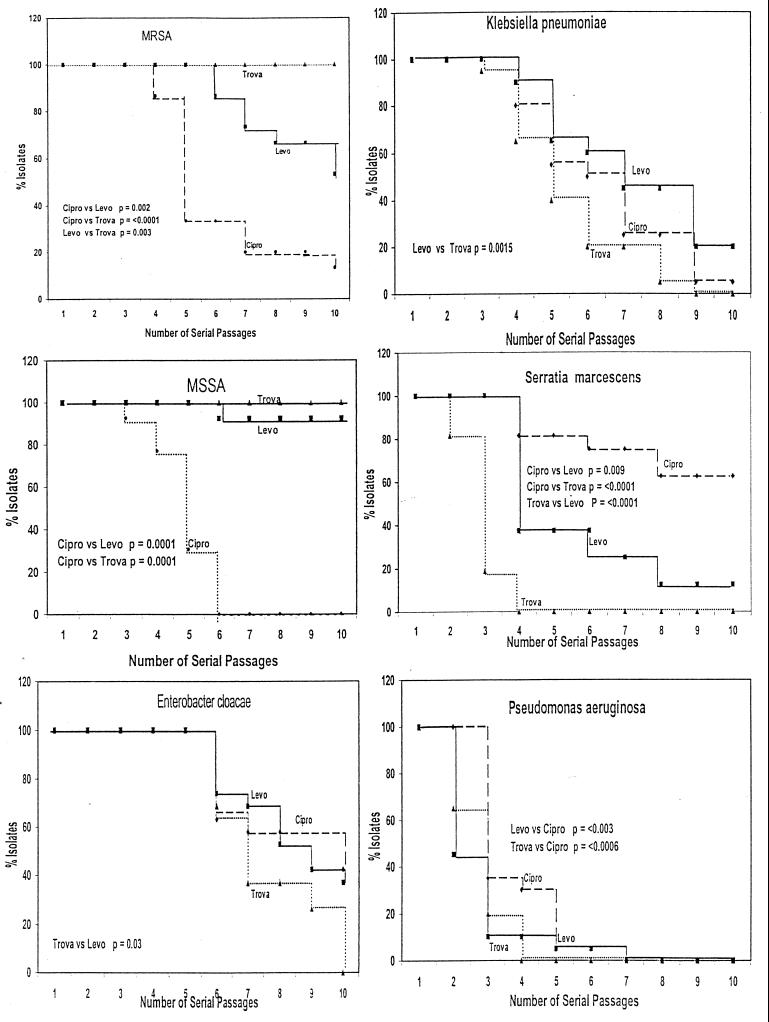
FIG. 1.
The results of the serial passage of clinical isolates of six organisms in increasing concentrations of ciprofloxacin (Cipro)(●), levofloxacin (Levo)(■), or trovafloxacin (Trova)(▴). The data are presented as KM product-limit estimates with a MIC of ≥4 μg/ml arbitrarily selected as indicating an important level of resistance. The vertical axis represents the percentage of isolates with a drug MIC of <4 μg/ml.
Both MSSA and MRSA accumulated mutants with a drug MIC of ≥4 μg/ml significantly faster for ciprofloxacin than for the other two test quinolones. The KM analysis obscures the gradual increase in trovofloxacin phenotypic resistance due to the low baseline trovafloxacin MICs. For the other aerobic gram-negative bacilli tested, two general observations are noteworthy. First, the appearance of resistant phenotypes was faster with trovafloxacin, and second, phenotypes resistant to ciprofloxacin occurred more slowly than phenotypes resistant to the other test drugs. Note that for K. pneumoniae and E. cloacae strains, the rate of accumulation of isolates with a drug MIC of ≥4 μg/ml was slower for levofloxacin and ciprofloxacin than for trovafloxacin. For S. marcescens, the rate of accumulation of mutants with a drug MIC of ≥4 μg/ml was again fastest for trovafloxacin and slowest for ciprofloxacin. For P. aeruginosa, the rate of accumulation of mutants with a drug MIC of ≥4 μg/ml was fastest for levofloxacin and slowest for ciprofloxacin.
Because the KM method of analysis excludes MIC data after test strains reached a MIC of ≥4 μg/ml and failed to consider differences through the 10 incubation cycles, data were also evaluated by regression analysis.
(iii) Regression analysis.
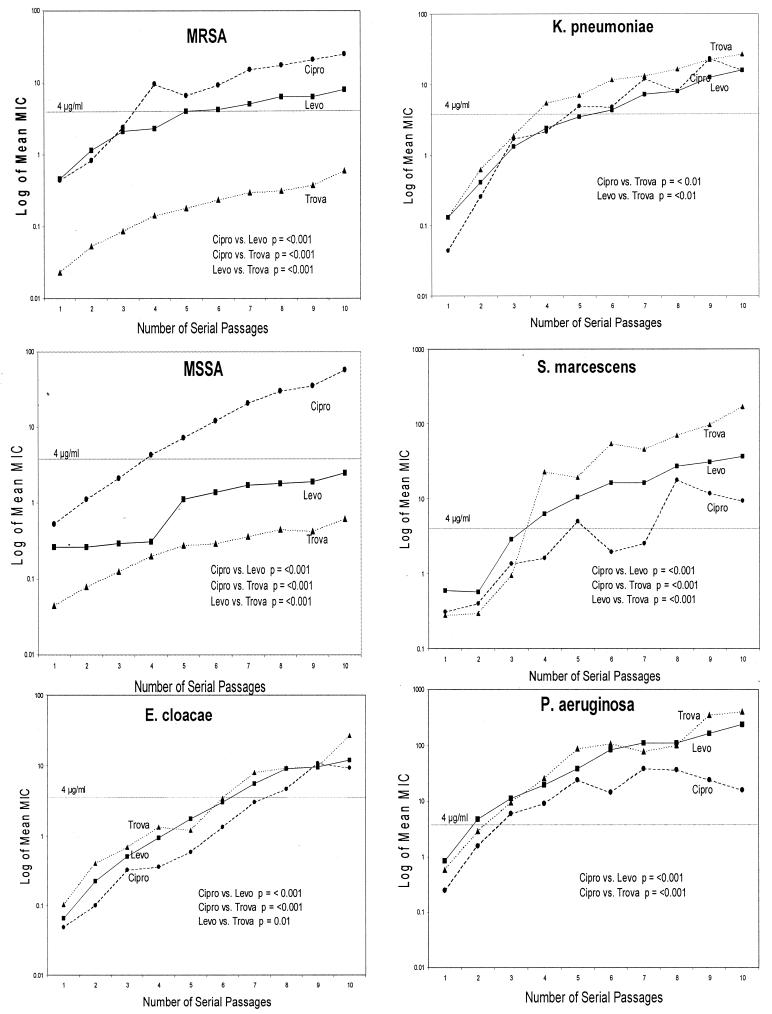
The data were analyzed using two methods. First, the mean log concentration MIC at each incubation step was plotted for each drug and each group of organisms. The results are presented in Fig. 2. As in the KM analysis, rapid identification of mutants of MSSA and MRSA with phenotypic resistance to ciprofloxacin is evident. The difference between levofloxacin and trovafloxacin is statistically significant for MRSA (P < 0.001) but not for MSSA (P = 0.08). The difference between ciprofloxacin and both levofloxacin and trovafloxacin was statistically significant for both MRSA and MSSA (P < 0.001).
FIG. 2.
Differences of log mean MICs through 10 serial passages for ciprofloxacin (Cipro)(●), levofloxacin (Levo)(■), and trovafloxacin (Trova)(▴) versus the number of serial passages.
For E. coli the emergence of mutants resistant to ciprofloxacin is statistically slower than that of mutants resistant to either levofloxacin or trovafloxacin (P < 0.001) (see Fig. 4). For K. pneumoniae, E. cloacae, and S. marcescens, the number of mutants with phenotypic resistance to trovafloxacin emerged faster than the number of mutants resistant to levofloxacin or ciprofloxacin. For all three organisms, the slowest accumulation of resistant phenotypes resulted from incubation with ciprofloxacin. For P. aeruginosa, phenotypic resistance emerged faster to trovafloxacin and levofloxacin than to ciprofloxacin. Also, only when P. aeruginosa was incubated with ciprofloxacin did there appear to be a plateau in mean log MICs. In the second method, the slopes of the regression curves were compared (Fig. 3). Analysis of the slopes results in fewer statistical differences than the differences in mean MICs shown in Fig. 2. The slope for ciprofloxacin was steeper than the levofloxacin or trovafloxacin slope for both MSSA and MRSA. For P. aeruginosa, the slope was steeper for both levofloxacin (P = 0.032) and trovafloxacin (P < 0.001) than for ciprofloxacin.
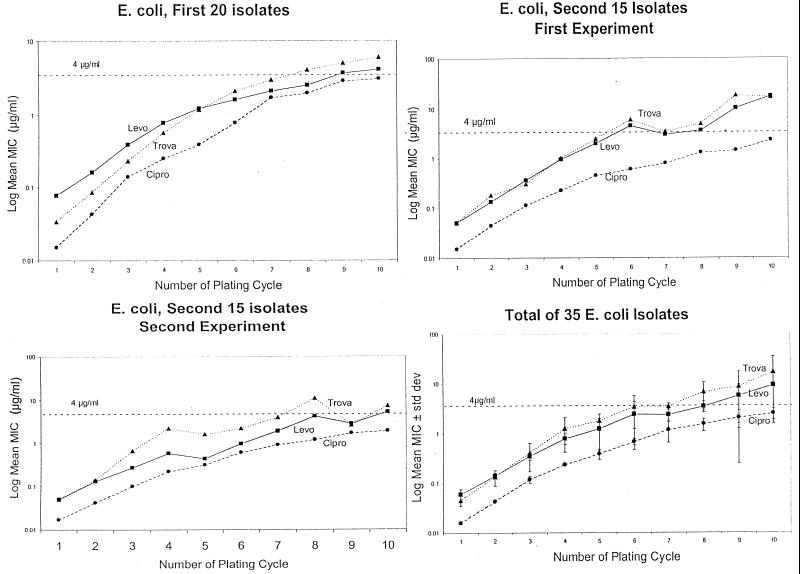
FIG. 4.
The reproducibility of the serial passage experiments was assessed by repeating the experiment sequentially with 15 clinical isolates of E. coli incubated with ciprofloxacin (Cipro)(●), levofloxacin (Levo)(■), and trovafloxacin (Trova)(▴). The 15 isolates were serially passaged in two consecutive experiments, and the results were compared to each other and to those for the earlier initial study with 20 different clinical isolates of E. coli.
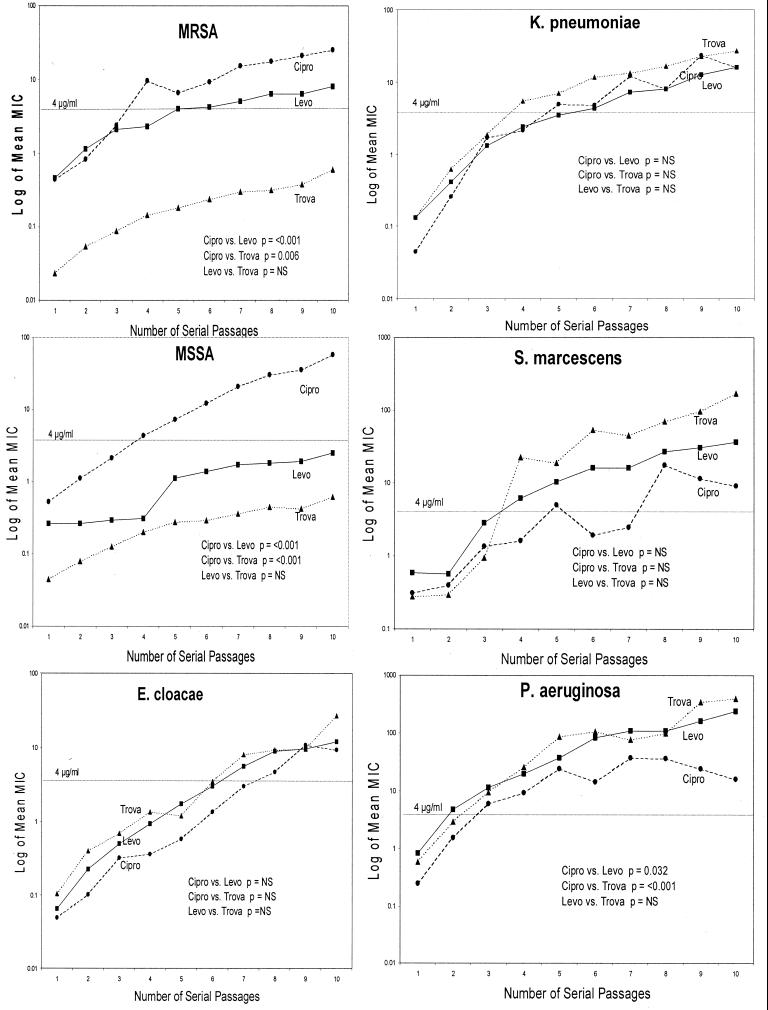
FIG. 3.
Comparison of the slopes of curves derived by plotting the log mean MIC of ciprofloxacin (Cipro)(●), levofloxacin (Levo)(■), and trovafloxacin (Trova)(▴) versus the number of serial passages. The dotted line represents a MIC of 4 μg/ml.
It was postulated that the baseline MICs might predict the subsequent increase in mean MICs. However, data analysis showed no such correlation. As expected, the steeper the slope of the curves, the greater the increase in mean MICs.
(iv) Reproducibility.
Since observed phenotypic changes in resistance might be the result of random genetic mutations, an additional 15 consecutive clinical isolates of E. coli were serially passaged twice and the results were compared with each other, as well as with those for the first 20 E. coli clinical isolates described above. In Fig. 4, the mean MIC is plotted against the result at each serial passage step. In the first 20 clinical isolates, the log mean MIC was higher for trovafloxacin and levofloxacin than for ciprofloxacin (P < 0.0001). Similarly, mean ciprofloxacin MICs were lower than those of levofloxacin or trovafloxacin in the first experiment with the “new” 15 E. coli strains. The second experiment with the 15 E. coli strains yielded a similar relationship of ciprofloxacin to the other two drugs. Although the levofloxacin and trovafloxacin curves appear farther apart on the second experiment, there is no statistically significant difference between trovafloxacin and levofloxacin in either of the two experiments with the 15 clinical isolates or the experiments with the original 20 isolates. The results with all 35 E. coli strains are summarized in Fig. 3D. Standard deviations are included and again indicate the lower mean MICs for ciprofloxacin than for the other two test drugs, as the mean MIC rises with serial passage.
A related question was the behavior of individual E. coli strains, as compared to mean values, when the strains were subjected to serial passage. Individual strain behavior was analyzed using the data from the 15 E. coli strains that were serially passaged twice. At each serial passage, the number of isolates in the second experiment with a MIC that was four times (twofold) higher or lower than in the first experiment was determined. After the fifth passage, as many as 12 of the 15 clinical isolates had a drug MIC that was either fourfold higher or lower than in the initial, identical experiment with the same clinical isolates. Despite the variability of the individual isolate, the mean drug MIC for the entire 15 isolates did not change.
DISCUSSION
The goal of our experiments was to study changes in the MIC over time under the influence of repeated drug exposure, as might happen clinically to the endogenous flora of the skin, nasopharynx, or gastrointestinal tract. The results indicate the potential for creating fluoroquinolone resistance in the organisms and drugs tested. However, there are differences in the ease of selection of resistant mutants between organisms and between drugs for a specific organism. The rapid emergence of phenotypic resistance of both MSSA and MRSA to ciprofloxacin confirms the work of others and also correlates with the rapid loss of the clinical utility of ciprofloxacin for S. aureus infections (4, 14, 17, 18).
Our results document the ease of emergence of resistance of Enterobacteriaceae, especially that of K. pneumoniae, E. cloacae, and S. marcescens to trovafloxacin. Conversely, these organisms showed a reduced propensity to develop mutants with phenotypic resistance to ciprofloxacin. Despite roughly 16 years of clinical use of fluoroquinolones, resistance of Enterobacteriaceae to ciprofloxacin has remained low worldwide (22). However, the recent reports of increasing resistance of E. coli, C. jejuni, and Salmonella enterica serovar Typhi are noteworthy (9, 21, 23).
For some drug-organism combinations, the mutation frequency after a single 48-h incubation did predict the differences observed after serial passages. The differences between drugs observed after a single incubation with S. marcescens and E. cloacae were repeated and magnified during 10 serial passages. On the other hand, only the difference between trovafloxacin and levofloxacin was statistically significant after one incubation with MSSA, while differences between all three drugs appeared during serial passage. Also noteworthy is the absence of any detectable differences in mutation frequency for the other four organisms when incubated with the three test drugs.
KM survival curve analyses have not typically been applied to in vitro studies of susceptibility of bacteria to antimicrobials. The KM analysis is generally applied to data with dichotomous end points: e.g., life versus death. The strength of applying this type of analysis to our data is the immediate visual impact of differences. A potential weakness is the risk of missing subtle differences as a consequence of ignoring data after the “cutoff” of 4 μg/ml is achieved. For the latter reason, additional regression analysis was performed that evaluated both the mean MIC data and the slopes of the resistance curves over the 10 serial passages. Of interest, in no instance were the data in the KM approach in conflict with the data from the regression analysis of the slope. The weakness of the KM analysis is a lack of appreciation of the degree of elevation of MICs after the predetermined “cutoff” value is reached.
The KM curves require a specific value as an end point. The in vitro cutoff for fluoroquinolone resistance varies between 2 and 8 μg/ml, depending on the drug and the organism. Hence we chose a MIC of ≥4 μg/ml as potentially clinically relevant. “Survival” required keeping the drug MIC for the isolate at <4 μg/ml. When the KM approach is used, the results show definite differences between the test drugs for MSSA, MRSA, and S. marcescens. A lower or higher “cutoff” would change the shape of the curves but not the relationship of the drugs to each other.
In general, the three statistical methods employed identified similar differences. The S. marcescens data are the exception. KM curves and plots of the log mean MIC versus serial passage yielded a clear hierarchy of the likelihood of identification of resistant mutants: i.e., more likely with trovafloxacin and least likely with ciprofloxacin. Analysis of the slopes of the resistance curves did not show differences between the three test drugs versus S. marcescens. This is likely due to a greater variability of the drug MICs for S. marcescens than for the other test organisms.
Much of the previously published work in this area has focused on in vitro acquisition of resistance by S. aureus. In investigations of phenotypic resistance that focused on MRSA, Evans and Titlow performed serial incubation studies (7; Evans and Titlow, Letter). They found greater absolute increases in MICs of ciprofloxacin than in MICs of levofloxacin and greater increases in MICs of ciprofloxacin than in MICs of trovafloxacin. Many other studies, focused on the underlying genetic mechanism of resistance, demonstrate that the observed increase in resistance of S. aureus results from stepwise acquisition of at least two genetic mutations (8, 16, 20).
Although the phenotypic resistance data presented cannot compare to the precise delineation of genotypic mechanisms of resistance, analysis of the emergence of resistance of a population of organisms complements genotypic studies by adding the perspective of population dynamics. The serial passage experiments help to delineate differences in emergence of resistance among the organisms and fluoroquinolones tested. When the E. coli serial passage experiment was repeated, the overall results were similar (mean values), but the results for individual isolates were dramatically different. This observation is consistent with an ongoing ever-changing population of genotypes. Nonetheless, the behavior of the population of organisms remains the same with respect to the speed and degree of acquisition of resistant genotypes.
Many questions remain unanswered. How “fit” are the resistant mutants after 10 sequential incubation periods? Is their virulence the same as that of the original susceptible organisms? In the absence of the fluoroquinolone and with continued serial passage in the absence of drug, will the resistant phenotype persist or revert to a more susceptible population of organisms? Lastly, will the changes in in vitro behavior of a population of bacteria repeatedly exposed to a specific fluoroquinolone have any predictive value as to development of clinical resistance?
In summary, the in vitro development of phenotypic resistance was studied in single exposure and serial passage experiments with seven bacterial species and three fluoroquinolones. Determination of the frequency of development of resistant mutants after a single 48-h incubation had a low sensitivity as a predictor of results obtained after serial 48-h incubation periods. Based on the serial passage experiments, MSAA and MRSA phenotypic resistance to ciprofloxacin develops rapidly. For three of the Enterobacteriaceae tested, i.e., K. pneumoniae, E. cloacae, and S. marcescens, phenotypic resistance to trovafloxacin emerged more quickly than resistance to levofloxacin or ciprofloxacin; in contrast, the resistance to ciprofloxacin was slow to emerge. These studies also demonstrate the uniqueness of the interaction between specific bacteria and specific fluoroquinolones; e.g., the results for E. coli were substantively different from those for S. marcescens. For E. coli, repetition of the serial passage experiments yielded similar overall results, even though individual strains displayed marked differences. The results indicate that dynamic studies of acquisition of phenotypic resistance of fluoroquinolones complement studies that determine genetic mechanisms of resistance.
REFERENCES
- 1.Amyes S G B, Thomson C J. Antibiotic resistance in the intensive therapy unit: the eve of destruction. Br J Intensive Care. 1995;5:273–281. [Google Scholar]
- 2.Carratala J, Fernandez-Sevilla A, Tubau F, Dominiguez M A, Gudiol F. Emergence of fluoroquinolone-resistance Escherichia coli in fecal flora of cancer patients receiving norfloxacin prophylaxis. Antimicrob Agents Chemother. 1996;40:503–505. doi: 10.1128/aac.40.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalhoff A. Quinolone resistance in Pseudomonas aeruginosa and Staphylococcus aureus. Development during therapy and clinical significance. Infection. 1994;22(Suppl. 2):S111–S121. doi: 10.1007/BF01793575. [DOI] [PubMed] [Google Scholar]
- 4.Daum T E, Schaberg D R, Terpenning M S, Sottile W S, Kauffman C A. Increasing resistance of Staphylococcus aureus to ciprofloxacin. Antimicrob Agents Chemother. 1990;34:1862–1863. doi: 10.1128/aac.34.9.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eliopoulos G M. In vitro activity of fluoroquinolones against gram-positive bacteria. Drugs. 1995;49(Suppl. 2):48–57. doi: 10.2165/00003495-199500492-00009. [DOI] [PubMed] [Google Scholar]
- 7.Evans M E, Titlow W B. Levofloxacin selects fluoroquinolone-resistant methicillin-resistant Staphylococcus aureus less frequently than ciprofloxacin. J Antimicrob Chemother. 1998;41:285–288. doi: 10.1093/jac/41.2.285. [DOI] [PubMed] [Google Scholar]
- 8.Fukuda H, Hori S, Hiramatasu K. Antibacterial activity of gatifloxacin (AM-1155, CG5501, BMS-206584), a newly developed fluoroquinolone, against sequentially acquired quinolone-resistant mutants and the norA transformant of Staphylococcus aureus. Antimicrob Agents Chemother. 1998;42:1917–1922. doi: 10.1128/aac.42.8.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garau J, Xercavins M, Rodriguez-Carballeiaa M, Gomez-Vera J R, Coll I, Vidal D, Llovet T, Ruiz-Bremon A. Emergence and dissemination of quinolone-resistant Escherichia coli in the community. Antimicrob Agents Chemother. 1999;43:2736–2741. doi: 10.1128/aac.43.11.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoge C W, Gambel J M, Srijan A, Pitarangsi C, Echeverria P. Trends in antibiotic resistance among diarrheal pathogens isolated in Thailand over 15 years. Clin Infect Dis. 1998;26:341–345. doi: 10.1086/516303. [DOI] [PubMed] [Google Scholar]
- 11.Hooper D C. Bacterial topoisomerases, anti-topoisomerases, and anti-topoisomerase resistance. Clin Infect Dis. 1998;27(Suppl. 1):S54–S63. doi: 10.1086/514923. [DOI] [PubMed] [Google Scholar]
- 12.Kam M M, Wong P W, Cheung M M, Ho N K Y, Lo K L. Quinolone-resistant Neisseria gonorrheae in Hong Kong. Sex Transm Dis. 1996;23:103–108. doi: 10.1097/00007435-199603000-00003. [DOI] [PubMed] [Google Scholar]
- 13.Mandell W, Neu H C. In vitro activity of CI-934, a new quinolone, compared with that of other quinolones and other antimicrobial agents. Antimicrob Agents Chemother. 1986;29:852–857. doi: 10.1128/aac.29.5.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulligan M E, Ruane P J, Johnston L, Wong P, Wheelock J P, MacDonald K, Reinhardt J F, Johnson C C, Statner B, Blomquist I. Ciprofloxacin for eradication of methicillin-resistant Staphylococcus aureus colonization. Am J Med. 1987;82:215–219. [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. Antimicrobial susceptibility testing. 5th ed. Wayne, Pa: National Committee for Clinical Laboratory Standards; 2000. [Google Scholar]
- 16.Ng E Y, Trucksis M, Hooper D C. Quinolone resistance mutations in topoisomerase IV: relationship to the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:1881–1888. doi: 10.1128/aac.40.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peterson L R, Quick J N, Jensen B, Homann S, Johnson S, Tenquist J, Stanholtzer C, Petzel R A, Sinn L, Gerding D N. Emergence of ciprofloxacin resistance in nosocomial methicillin-resistant Staphylococcus aureus isolates. Resistance during ciprofloxacin plus rifampin therapy for methicillin-resistant S. aureus colonization. Arch Intern Med. 1990;150:2151–2155. [PubMed] [Google Scholar]
- 18.Pong A, Thomson K S, Moland E S, Chartrand S A, Sanders C S. Activity of moxifloxacin against pathogens with decreased susceptibility to ciprofloxacin. J Antimicrob Chemother. 1999;44:621–627. doi: 10.1093/jac/44.5.621. [DOI] [PubMed] [Google Scholar]
- 19.Shalit I, Berger S A, Gorea A, Frimerman H. Widespread quinolone resistance among methicillin-resistant Staphylococcus aureus isolates in a general hospital. Antimicrob Agents Chemother. 1989;33:593–594. doi: 10.1128/aac.33.4.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sulavik M C, Barg N L. Examination of methicillin-resistant and methicillin-susceptible Staphylococcus aureus mutants with low-level fluoroquinolone resistance. Antimicrob Agents Chemother. 1998;42:3317–3319. doi: 10.1128/aac.42.12.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith K E, Besser J M, Hedberg C W, Leano F T, Bender J B, Wicklund J H, Johnson B P, Moore K A, Osterholm M T. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992–1998. N Engl J Med. 1999;340:1525–1532. doi: 10.1056/NEJM199905203402001. [DOI] [PubMed] [Google Scholar]
- 22.Thomson C J. The global epidemiology of resistance to ciprofloxacin and the changing nature of antibiotic resistance: a 10 year perspective. J Antimicrob Chemother. 1999;43(Suppl. A):31–40. doi: 10.1093/jac/43.suppl_1.31. [DOI] [PubMed] [Google Scholar]
- 23.Threlfall E J, Ward L R, Skinner J A, Smith H R, Lacey S. Ciprofloxacin-resistant Salmonella typhi and treatment failure. Lancet. 1999;353:1590–1591. doi: 10.1016/s0140-6736(99)01001-6. [DOI] [PubMed] [Google Scholar]