Molecular markers are now unequivocally a requirement for integrative brain tumor diagnostics. The 2021 WHO classification of CNS tumors substantially increases the set of genes required in routine evaluation, and significantly increases the relevance of DNA methylation analysis [12]. The advantages of custom neuropathology NGS panels can only be efficiently exploited when case numbers are sufficient for batchwise processing. Labs with lower specimen submission numbers hence may have to pool samples over multiple weeks. Here we introduce Rapid-CNS2—a custom neurooncology molecular diagnostic workflow using Nanopore “third generation” sequencing for parallel copy-number profiling, mutational and methylation analysis that is highly flexible in target selection, runs efficiently on single samples, and can be initiated immediately upon receipt of frozen sections.
Nanopore sequencing has an advantage over current NGS in read lengths, shorter and easier library preparation, ability to call base modifications, real time analysis, and portability of sequencing devices—all at relatively low cost [2]. However, smaller devices like the MinION and GridION yield low-coverage data when run genome-wide [6]. While nanopore-based approaches to methylation classification, particularly nanoDx, have demonstrated reliable methylation classification with as low as 1000 CpGs, it does not allow for detection of single-nucleotide-variants (SNVs) or MGMT promoter methylation status assessment [4, 7]. Intriguingly, Nanopore provides a “ReadUntil” adaptive sampling toolkit that can reject reads in real-time during sequencing [8]. The tool “ReadFish” harnesses this functionality to enable targeted adaptive sequencing with no additional steps in library preparation, which has been complemented by a built-in adaptive sampling in recent device versions [10]. This considerably increases coverage over “target” regions by real-time enrichment during sequencing.
Rapid-CNS2 leverages adaptive nanopore sequencing through ReadFish and the built-in adaptive sampling and is run here as a proof-of-concept on a portable MinION or GridION device, respectively. We formulated target regions covering our brain tumor NGS panel and CpG sites for methylation classification [4, 13]. Sequenced data was analyzed using an end-to-end bioinformatics pipeline that reports clinically relevant SNVs, copy number alterations, target gene methylation and methylation classification for CNS tumors. Methylation classification was based on nanoDx’s ad-hoc approach but infers CpG importance from the Heidelberg methylation classifier and was trained on the published reference set [3]. MGMT promoter methylation status was estimated by a proposed logistic regression-based prediction model (Supplementary Methods, online resource).
We first sequenced 45 cryoconserved glioma samples for 72 h each to investigate the overall feasibility for detecting relevant alterations (Fig. 1). Targets (Rapid_CNS_A) included the regions of the aforementioned panel and 10,000 CpG sites inferred from the Heidelberg methylation classifier (155 Mb). Molecular alterations were compared to NGS panel sequencing and EPIC array analyses (referred to as “conventional data/analysis”) from corresponding FFPE tissue samples [1, 3, 13]. Incubation time and other parameters were optimized to improve quality, amount of data generated and on-target rate of the libraries (Supplementary Methods, online resource). Samples were selected to cover a variety of clinically relevant pathognomonic alterations (including but not limited to IDH1, 1p/19q codeletion, chr7 gain/chr10 loss, TERT promoter, EGFR amplification, CDKN2A/B deletion, MGMT status) and relevant methylation classes. Second, a smaller panel was developed to compare the effect of target sizes. Rapid_CNS_B comprised regions for mutational analysis only (15 Mb) and was run on 8 glioma samples (Supplementary Fig. 1, online resource). Third, we assessed the reusability of flow cells and the data quality of shorter sequencing times by splitting 5 of the Rapid_CNS_B samples into 24 h and 48 h run intervals.
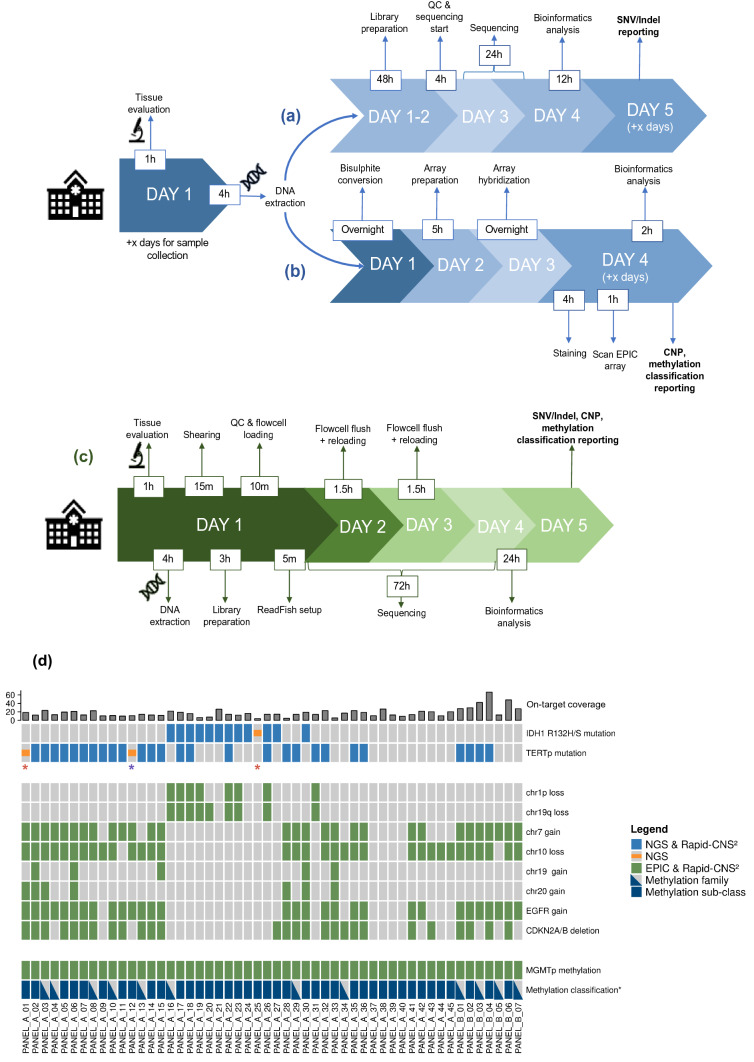
Fig. 1.
Rapid-CNS2 timeline and concordance with standard methods. Timeline for a NGS panel sequencing and analysis pipeline, and b EPIC array analysis pipeline for neuropathology diagnostics (x denotes number of days required to pool sufficient samples). c Timeline for Rapid-CNS2 sequencing and analysis for a single sample on a MinION or up to five samples on a GridION. d Concordance of clinically relevant alterations and classification for diffuse glioma samples with matching NGS panel sequencing and EPIC array analysis. Colored blocks indicate presence of alteration, concordance for detected alterations is denoted in the legend. Triangular denotations for methylation class indicate samples where methylation families were concordant and blocks indicate concordance for sub-classes as well. ‘PANEL_A’ indicates samples sequenced using Rapid_CNS_A, and ‘PANEL_B’ indicates samples sequenced using Rapid_CNS_B (sample by Rapid-CNS2 and 850 k identified as AT/RT excluded, depicted in Supplementary Fig. 4, online resource). Top barplot indicates mean on-target coverage. Red asterisk—different tissue region for Rapid-CNS2 vs conventional analyses, purple asterisk—alteration filtered by PEPPER-Margin-DeepVariant (see main text for details)
The glioma samples run with the Rapid_CNS_A panel as target region were each loaded onto FLO-MIN106 R9.4.1 flow cells and sequencing was controlled in real-time by ReadFish (using a GPU powered consumer notebook). Targeted regions showed an average coverage of 15X. (Supplementary Fig. 1, online resource). IDH1 and TERTp mutation status were correctly identified in 44/45 and 43/45 glioma samples, respectively. Upon re-evaluation of histology and raw data, discrepancy in mutations could be attributed to low tumor cell content in the respective cryopreserved sections in two cases, and variant filtering in another (Supplementary Results, online resource). Copy number alterations displayed complete concordance with EPIC array results and markedly better resolution as compared to NGS panel sequencing (Supplementary Fig. 2, online resource). MGMT promoter status was concordant with both pyrosequencing and EPIC array in all cases (Supplementary Fig. 3, online resource). Methylation families (relevant for WHO-conform diagnosis) were identified in line with the Heidelberg classifier predictions for all samples, with even the methylation subclass correctly identified for 37/45 glioma samples (Fig. 1d).
Reducing the target size with the Rapid_CNS_B panel doubled the mean on-target coverage to 39X with 3.4X average coverage over off-target regions (Supplementary Fig. 1, online resource). IDH1, TERTp, and MGMT status matched Rapid-CNS2 in all eight cases. Methylation family was correctly predicted in all cases, with accurate subclass predictions in 4/8 cases. Despite the reduction in targeted regions, copy number profile resolution was maintained and was concordant with EPIC array for all cases. One sample identified as a glioma by histology was correctly classified as an atypical teratoid/rhabdoid tumor by Rapid-CNS2 (confirmed by EPIC array analysis, Supplementary Fig. 4, online resource).
The histopathological suspicion for diffuse glioma in six additional prospective cases was confirmed by Rapid-CNS2 (Rapid_CNS_A: 2 samples, Rapid_CNS_B: 4 samples) in a real-time clinical setting with respective reports issued (Supplementary Fig. 5, online resource). MGMT promoter status for these samples was in line with pyrosequencing.
Shortening sequencing time to 24 h was sufficient for a comprehensive diagnosis. The flowce ll could also be reliably reused for another sample for the next 48 h (Supplementary Fig. 6, online resource). Detection of clinically relevant fusions and complex genomic rearrangements using targeted Nanopore DNA sequencing still requires considerable manual intervention [5, 9]. As such, automated detection of such alterations is a developing field, and hence beyond the current scope of this study. In its current state, our attempts on FFPE derived DNA showed that Rapid-CNS2 is restricted to use with cryopreserved or fresh tumor samples (Supplementary Fig. 7, online resource). Considering the potential for further advances in Nanopore technology, FFPE sample processing may become feasible in the future.
Targeted regions for Rapid-CNS2 can be easily altered by editing a BED file, in principle allowing even much lower sequencing times than in this study. The recently introduced barcode-aware adaptive sampling by ReadFish proposes an effective way to multiplex samples and further reduce costs [11]. With no additional library preparation steps required, it is possible to modify targeted regions for each individual sample as required. MinION is a portable, handheld device which makes it a rational option for smaller neuropathology labs or in lower-infrastructure locations. Collectively, the Rapid-CNS2 approach can be set-up at low capital expense, is cost-efficient (Supplementary Results, online resource) even in a low throughput setting, and provides a swift and highly flexible alternative to conventional methods for SNV/InDel analysis, MGMT promoter status, methylation classification and detection of copy number alterations.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This study was supported by the Deutsche Forschungsgemeinschaft (DFG) via Collaborative Research Center (SFB) 1389 Unite Glioblastoma and the Brain Tumor Charity UK. PS is a fellow of the Hertie Network of Excellence in Clinical Neuroscience.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was funded by Brain Tumour Charity; Deutsche Forschungsgemeinschaft; Gemeinnützige Hertie-Stiftung.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Areeba Patel, Helin Dogan, Martin Sill, David TW Jones and Matthias Schlesne authors contributed equally as first and senior authors.
References
- 1.Bady P, Sciuscio D, Diserens A-C, Bloch J, van den Bent MJ, Marosi C, et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012;124:547–560. doi: 10.1007/s00401-012-1016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowden R, Davies RW, Heger A, Pagnamenta AT, de Cesare M, Oikkonen LE, et al. Sequencing of human genomes with nanopore technology. Nat Commun. 2019;10:1869. doi: 10.1038/s41467-019-09637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Euskirchen P, Bielle F, Labreche K, Kloosterman WP, Rosenberg S, Daniau M, et al. Same-day genomic and epigenomic diagnosis of brain tumors using real-time nanopore sequencing. Acta Neuropathol. 2017;134:691–703. doi: 10.1007/s00401-017-1743-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorzynski JE, Goenka SD, Shafin K, Jensen TD, Fisk DG, Grove ME, et al. Ultrarapid nanopore genome sequencing in a critical care setting. N Engl J Med. 2022;386:700–702. doi: 10.1056/NEJMc2112090. [DOI] [PubMed] [Google Scholar]
- 6.Jain M, Olsen HE, Paten B, Akeson M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 2016;17:239. doi: 10.1186/s13059-016-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuschel LP, Hench J, Frank S, Hench IB, Girard E, Blanluet M, et al. Robust methylation-based classification of brain tumors using nanopore sequencing. medRxiv. 2021 doi: 10.1101/2021.03.06.21252627. [DOI] [PubMed] [Google Scholar]
- 8.Loose M, Malla S, Stout M. Real-time selective sequencing using nanopore technology. Nat Methods. 2016;13:751–754. doi: 10.1038/nmeth.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller DE, Sulovari A, Wang T, Loucks H, Hoekzema K, Munson KM, et al. Targeted long-read sequencing identifies missing disease-causing variation. The American Journal of Human Genetics. 2021;108:1436–1449. doi: 10.1016/j.ajhg.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Payne A, Holmes N, Clarke T, Munro R, Debebe BJ, Loose M. Readfish enables targeted nanopore sequencing of gigabase-sized genomes. Nat Biotechnol. 2021;39:442–450. doi: 10.1038/s41587-020-00746-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Payne A, Munro R, Holmes N, Moore C, Carlile M, Loose M. Barcode aware adaptive sampling for Oxford Nanopore sequencers. bioRxiv. 2021 doi: 10.1101/2021.12.01.470722. [DOI] [Google Scholar]
- 12.Rushing EJ. WHO classification of tumors of the nervous system: preview of the upcoming 5th edition. MEMO Magaz Eur Med Oncol. 2021 doi: 10.1007/s12254-021-00680-x. [DOI] [Google Scholar]
- 13.Sahm F, Schrimpf D, Jones DTW, Meyer J, Kratz A, Reuss D, et al. Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol. 2016;131:903–910. doi: 10.1007/s00401-015-1519-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.