Abstract
Climate change-driven ocean warming is increasing the frequency and severity of bleaching events, in which corals appear whitened after losing their dinoflagellate endosymbionts (family Symbiodiniaceae). Viral infections of Symbiodiniaceae may contribute to some bleaching signs, but little empirical evidence exists to support this hypothesis. We present the first temporal analysis of a lineage of Symbiodiniaceae-infecting positive-sense single-stranded RNA viruses (“dinoRNAVs”) in coral colonies, which were exposed to a 5-day heat treatment (+2.1 °C). A total of 124 dinoRNAV major capsid protein gene “aminotypes” (unique amino acid sequences) were detected from five colonies of two closely related Pocillopora-Cladocopium (coral-symbiont) combinations in the experiment; most dinoRNAV aminotypes were shared between the two coral-symbiont combinations (64%) and among multiple colonies (82%). Throughout the experiment, seventeen dinoRNAV aminotypes were found only in heat-treated fragments, and 22 aminotypes were detected at higher relative abundances in heat-treated fragments. DinoRNAVs in fragments of some colonies exhibited higher alpha diversity and dispersion under heat stress. Together, these findings provide the first empirical evidence that exposure to high temperatures triggers some dinoRNAVs to switch from a persistent to a productive infection mode within heat-stressed corals. Over extended time frames, we hypothesize that cumulative dinoRNAV production in the Pocillopora-Cladocopium system could affect colony symbiotic status, for example, by decreasing Symbiodiniaceae densities within corals. This study sets the stage for reef-scale investigations of dinoRNAV dynamics during bleaching events.
Subject terms: Virus-host interactions, Community ecology, Coral reefs
Introduction
Warming seas, driven by climate change, are increasingly causing bleaching events: mass losses of endosymbiotic dinoflagellates (family Symbiodiniaceae) from corals and other invertebrate hosts. Bleaching events often result in coral mortality and are contributing to the degradation of reef ecosystems globally [1, 2]. Viruses, which are diverse and abundant on coral colonies [3–7], are hypothesized to contribute to some coral bleaching signs by lysing Symbiodiniaceae cells (e.g., [8–10]). Alternatively, viral shifts in conjunction with bleaching-associated stressors (e.g., [11–14]) could merely be correlated with bleaching signs or constitute opportunistic secondary infections [15, 16]. Beyond bleaching, viruses may influence colony health by altering the function of resident microbial symbionts or coral tissues (e.g., [17–23]). Although various roles for viruses in coral bleaching, disease, and function have been hypothesized [23, 24], thus far, these roles have been difficult to test empirically.
Symbiodiniaceae are putative target hosts of DNA and RNA viruses (reviewed in [6, 7]), including the “dinoRNAVs”, a group of dinoflagellate-infecting positive-sense single-stranded RNA viruses. Although dinoRNAVs have yet to be isolated, stably propagated, and fully characterized, they have been detected in transcriptomes from Symbiodiniaceae cultures, as well as in viral particles isolated from Atlantic and Pacific corals spanning six genera (Table 1). These associations suggest that dinoRNAVs are prevalent as exogenous, persistent infections in Symbiodiniaceae cells [8–10, 25, 26]. Furthermore, in previous studies, increased dinoRNAV detection [25], and changes in dinoRNAV and host anti-viral transcript expression in a thermosensitive symbiont population only [27], suggest that under stressful conditions, some dinoRNAVs may switch to a more productive replication mode that could culminate in host lysis. This infection strategy has recently been identified in other algal host-virus systems (e.g., coccolithophore Emiliana huxleyi-EhV system, [28]).
Table 1.
Summary of studies in which Symbiodiniaceae-infecting dinoflagellate RNA virus (“dinoRNAV”) genes have been recovered from coral colonies or Symbiodiniaceae cultures.
| Sample type | Symbiodiniaceae ID | Collection location | DinoRNAV detection method | Notes | Reference |
|---|---|---|---|---|---|
| Coral holobiont | |||||
| Montastrea cavernosa (4) | Cladocopiuma | Key West, FL, USA | Viral particle isolation, 454 pyrosequencing | DinoRNAVs detected in heat-treated corals. | Correa et al. 2013 |
| Acropora tenuis | Cladocopiumb | Orpheus Island, Central GBR | Viral particle isolation, RP-SISPA, MiSeq sequencing. | DinoRNAVs ≥99% of ssRNA viruses detected from pooled tissue of 3 colonies. No dinoRNAVs detected in Pocillopora damicornis. | Weynberg et al. 2014 |
| Acropora tenuis | Cladocopiumb | Davies Reef and Orpheus Island, Central GBR | Viral particle isolation, targeted PCR assay, MiSeq sequencing. | DinoRNAVs not detected in Acropora hyacinthus, Acropora millepora or Goniastrea aspera. | Montalvo-Proaño et al. 2017 |
| Fungia fungites | Cladocopiumb | ||||
| Galaxea fascicularis | Cladocopium or Durusdiniumb | ||||
| Pocillopora damicornis | Cladocopiumb | ||||
| Porites cylindrica | Cladocopiumb | ||||
| Porites lutea (6) | Cladocopium C15 | ||||
| Acropora tenuis | Cladocopiumb | Orpheus Island, Central GBR | Viral particle isolation, RP-SISPA, MiSeq sequencing. | Corals collected in triplicate and viral particles pooled prior to sequencing. DinoRNAVs not detected in Goniastrea aspera, Pocillopora acuta, Pocillopora damicornis or Pocillopora verrucosa. | Weynberg et al. 2017 |
| Fungia fungites | Cladocopiumb | ||||
| Galaxea fascicularis | Cladocopium or Durusdiniumb | ||||
| Pocillopora ligulata (2) | Cladocopium latusorum | Mo’orea, French Polynesia | Targeted PCR assay, MiSeq sequencing. Metatranscriptome from rRNA-depleted RNA (P. verrucosa only). | Heat stress increased mcp gene diversity and dispersion. Hits to dinoRNAVs were the tenth-most abundant viral hits in RNA-seq libraries. | This study |
| Pocillopora verrucosa (3) | Cladocopium pacificum | ||||
| Symbiodiniaceae culture | |||||
| Cladocopium C1 (2) | Cladocopium C1 | Magnetic Island and South Molle Island, Central GBR | HiSeq sequencing of poly(A)-purified RNA. | Cultures originally isolated from Acropora tenuis. Higher expression of dinoRNAV major capsid protein genes in a thermo-sensitive Cladocopium strain under ambient temperatures. | Levin et al. 2017 |
Samples sizes are n = 1 unless indicated otherwise in parentheses. Bold Symbiodiniaceae IDs were directly characterized in a given study. All other Symbiodiniaceae IDs indicate the Symbiodiniaceae genus (or genera) typically reported from a given coral species based on published literature [106, 107].
a Serrano et al. 2014.
b Tonk et al. 2013.
Heterocapsa circularisquama RNA virus (“HcRNAV”), which infects free-living dinoflagellates, is among the closest known relatives to Symbiodiniaceae-infecting dinoRNAVs [25, 29]. HcRNAV undergoes a putatively strictly lytic replication cycle following a latent period of 24–48 h in laboratory experiments; during this latent period, the host is infected but not yet lysed and viruses are not yet released [30]. Based on this, we hypothesized that shifts by Symbiodiniaceae-infecting viruses to a more productive replication mode might also be detectable within the first few days of exposure to stress [13, 25, 27]. Examining viral dynamics within individual coral colonies at the onset of thermal stress can help clarify whether viral infections of Symbiodiniaceae contribute to some coral bleaching signs.
With this aim, we quantified, for the first time, the temporal dynamics of dinoRNAVs within coral colonies exposed to control and heat stress conditions. DinoRNAV diversity was then characterized from heat-treated and control fragments of each colony via amplicon sequencing of the major capsid protein (mcp) gene. The mcp gene was targeted to facilitate comparisons with previous dinoRNAV detections (Table 1, [29]) and because the mcp gene resolved ecologically distinct strains in the related H. circularisquama-HcRNAV system [30, 31]. We hypothesized that: (1) dinoRNAVs are present in a majority of Mo’orean pocilloporid colonies; and (2) dinoRNAV richness increases and composition shifts within 48 h of exposure to thermal stress. By analyzing dinoRNAV diversity at the amino acid level, this study partially circumvented methodological challenges arising from the high mutation rates and genetic diversity of single-stranded RNA viruses [32–34], which have previously made it difficult to quantify RNA viral dynamics at ecologically relevant scales [35]. As research on reef-associated dinoRNAVs continues to progress, the aminotypes presented here may eventually merit further collapse into “quasispecies”—heterogeneous mixtures of related genomes [35–38]—a common approach for conceptualizing diversity within RNA virus populations.
Materials and methods
Experimental design
We conducted a replicated aquarium experiment (two treatments; four aquaria per treatment) in which fragments from five colonies of the morphologically cryptic stony coral Pocillopora species complex [39], harboring Cladocopium spp. symbionts, were exposed to control conditions (ambient reef water; 28.2 °C) or a + 2.1 °C heat treatment (summer bleaching temperatures; 30.3 °C) for 5 days (See Fig. S1 and Supplementary Methods, [40, 41]). At the start of the experiment (t(h) = 0), all fragments were photographed with a Coral-Watch Health Monitoring Chart [42] in the frame, and one fragment per colony in the control aquaria (5 fragments in total) were preserved as initial controls. At five time points (t(h) = 4, 12, 24, 72, and 108 h), all fragments were photographed and visually inspected for signs of stress (e.g., excessive mucus production), lesions, and/or paling. To characterize the effect of the heat treatment on Symbiodiniaceae cell densities, we compared brightness values—a proxy for Symbiodiniaceae chlorophyll concentrations, and a metric of coral bleaching status—from standardized photographs of each coral fragment at the start of the experiment and at the time of preservation (see Supplementary Methods, [42, 43]). We tested for changes to brightness values using linear mixed effects models (LMM) by including an interaction between treatment and time, and including colony ID as a random effect.
A control and a heat-stressed fragment per colony were preserved at each time point (generating 10 fragments per time point). DNA and RNA were extracted from each sample (which included coral animal tissue, Symbiodiniaceae cells and viruses) using a ZymoBIOMICS DNA/RNA Miniprep Kit (Zymo Research, Irvine, CA, USA) with an additional enzyme digestion step to improve viral RNA yields.
Coral and symbiont species were delineated from extracted DNA as described in [44]. Briefly, coral species were identified by sequencing the Pocillopora-specific mitochondrial open reading frame (mt-ORF) gene region [44–48]; symbiont species were identified by sequencing the D1/D2 domain of the large ribosomal subunit (LSU rRNA gene) and the non-coding plastid minicircle (psbAncr; [44, 49, 50]). This analysis revealed that colonies 1, 4 and 5 were Pocillopora verrucosa corals harboring Cladocopium pacificum symbionts, and colonies 2 and 3 were Pocillopora ligulata containing Cladocopium latusorum symbionts [44].
Sequencing of dinoRNAV gene amplicons, bioinformatics processing, and phylogenetic analysis
The dinoRNAV major capsid protein (mcp) gene was amplified from cDNA libraries (generated from extracted RNA) using a nested PCR protocol with degenerate primers [29]; cleaned and normalized libraries were sequenced on the Illumina MiSeq platform using PE300 v3 chemistry. To rule out the possibility that the sequenced gene fragments were endogenous viral elements (EVEs) integrated into Symbiodiniaceae genomes, we attempted to amplify the mcp gene from DNA extracted from t(h) = 0 samples. No bands were detectable on agarose gels, indicating that the dinoRNAV mcp sequences detected in this study are most parsimoniously interpreted as exogenous infections, and not EVEs, of Mo’orean C. latusorum or C. pacificum. Processing and analysis of all raw dinoRNAV mcp gene reads were conducted using the program vAMPirus (v1.0; See Supplementary Methods for details; [51]). Briefly, amplicon sequence variants (“ASVs”) were generated via the UNOISE [52] algorithm with vsearch v. 2.14.2 [53], and all ASVs were then translated and collapsed into “aminotypes”—unique amino acid sequences each differentiated by at least one amino acid. Any sequences containing stop codons were removed prior to further analysis.
An alignment was made in MUSCLE (v5) using aminotypes from this study, as well as a dataset from the Great Barrier Reef [29] that was reprocessed using the methods above, and a set of reference best BLASTx hits to the NCBI database [54, 55]. The alignment (Supplementary Data 1) was trimmed to the first column on either side that contained no gaps, and then used to determine the best model for evolution (LG + G4 + I) according to ModelTest-NG [56]. A maximum-likelihood phylogeny was inferred with RAxML-NG (1000 bootstrap iterations) and rooted with HcRNAV as an outgroup.
Statistical analyses of dinoRNAV mcp aminotype composition and diversity
All data processing, visualization, analysis and statistical tests were conducted in R version 4.0.2 and Vegan 2.5-6 [57] on an mcp aminotype counts table (Supplementary Data 2; R code: https://github.com/CorreaLab/Pocillopora-dinoRNAVs). For some analyses, the dataset was rarefied to 59 837 amino acid sequences per sample. First, the overall distribution of dinoRNAV mcp aminotypes among cryptic coral-symbiont species and colonies was explored to determine how subsequent statistical analyses should be conducted. A PERMANOVA based on Bray-Curtis distances from square-root-transformed rarefied data was used to test for differences in overall dinoRNAV composition among species, treatments and colonies, and over time, using adonis() in vegan. We tested for an interaction between treatment and colony, with time and species as additional factors. Although dispersion differed significantly among groups (betadisper test), PERMANOVA is robust to heterogeneous dispersion if the design is balanced [58]. The PERMANOVA revealed that dinoRNAV composition differed between the coral-symbiont species (R2 = 0.21, p < 0.001). However, colony ID was a stronger predictor (R2 = 0.55, p < 0.001), indicating that differences in dinoRNAV composition were mainly structured at the colony level, rather than the coral-symbiont species level.
Venn diagrams were made to identify aminotypes shared among coral-symbiont species, colonies, or treatments based on non-rarified data using the online tool http://bioinformatics.psb.ugent.be/webtools/Venn/ (accessed August 17th, 2020). For comparison, Venn diagrams were also made based on rarified data (results not shown); these Venn diagrams exhibited similar patterns but were less conservative and were therefore not included. Venn diagram analysis indicated that 64% of aminotypes (79 of 124) were shared between coral-symbiont species. Based on these preliminary explorations of the distribution of dinoRNAV mcp aminotypes, in all subsequent statistical tests, we analyzed all five colonies together (i.e., at the Pocillopora-Cladocopium level) but included coral-symbiont species as a separate factor in LMMs.
Shannon index (H) values were calculated based on rarefied data; expected aminotype richness values were calculated using repeated random subsampling of non-rarefied data (sample size = 59 837 amino acid sequences). We tested for differences in Shannon index and aminotype richness values using LMMs with the package LME4 v1.1-23 [59]. We tested for an interaction between treatment and timepoint; by incorporating colony ID as a random effect, this analysis approaches a repeated measures test. Coral-symbiont species were included as an additional factor to test for differences between species. F-tests were used for model selection with car v3.0-8. Assumptions of normality of the residuals were assessed visually with quantile-quantile plots and Shapiro-Wilk tests; the assumption of homogeneity of variance was visually assessed using plots with residuals versus fitted values. We also tested for differences between control and heat-treated fragments at each timepoint (using colonies as replicates), as well as per colony (using time points as replicates), with ANOVAs and controlled for type 1-errors using a Bonferroni correction.
To quantify the dispersion of dinoRNAVs between treatments and over time, a non-metric multidimensional scaling (NMDS) plot was constructed based on Bray-Curtis distances from square-root-transformed rarefied data (k = 2999 iterations), and the distance to centroid for each sample was calculated. Since dinoRNAVs differed among coral colonies, we calculated centroids for each individual colony in the control and heat treatments separately (5 colonies x 2 treatments = 10 centroids) to examine the effect of heat treatment on dinoRNAV dispersion in a given coral colony. For this analysis, different timepoints were used as replicates. We tested for differences in dispersion using the same LMM approach as described above for aminotype alpha diversity, but the values were square root-transformed because they did not follow the assumption of normality of the residuals.
Lastly, we conducted a differential abundance analysis using the non-rarefied amino acid counts table with DESeq2 v1.26.0 [60]. We fitted a negative binomial model and Benjamini-Hochberg FDR-corrected Wald tests (α = 0.05) were used to test for differences in taxon abundance between treatment within each colony and at each timepoint after the start of the experiment (t(h) = 4, 12, 24, 72, 108). We excluded all fragments sampled at timepoint 0, as well as any fragment with < 10,000 reads (and its paired fragment in the other treatment) at a given timepoint (colonies 3 and 5 at timepoint t(h) = 4, colony 3 at t(h) = 72).
Metatranscriptome sequencing and bioinformatics processing
As an additional test of whether the mcp gene amplicons generated in this study represent exogenous dinoRNAVs, we generated 5 metatranscriptomes from random timepoints and treatments for each colony of P. verrucosa-C. pacificum (t(h) = 0 control for colony 1, t(h) = 24 heat for colony 4, t(h) = 108 control and heat for colony 5, t(h) = 72 control for colony 5; see Supplementary Methods for more detail) and queried them for dinoRNAV-derived transcripts. All metatranscriptome reads were trimmed, quality filtered, and merged before alignment to the proteic version of the Reference Viral DataBase (RVDB; v21) using DIAMOND BLASTx [55]. Relative abundances of hits to dinoRNAV reference sequences, and mean alignment metrics (e-values, bitscores and % identity), were calculated. All cleaned forward and reverse reads were then normalized and assembled into contigs using rnaSPAdes v3.13.0 [61]. DinoRNAV contigs were then inferred by DIAMOND BLASTx alignment (minimum bitscore = 50; minimum ORF length = 30 aa; minimum percent identity = 30) to a hybrid database containing the uniprot database and the RVDB [62]. Prodigal (v.2.6.3) was used to predict open reading frames and protein sequences from all dinoRNAV-like contigs, and protein annotation was conducted with another round of DIAMOND BLASTp alignment to the hybrid database. Contigs with best hits to the dinoRNAV mcp gene, as well as mcp gene amplicons, were mapped to a dinoRNAV mcp gene assembly from cultured Cladocopium (species C1) [27].
Results
Coral holobiont traits
Analysis of coral and Symbiodiniaceae gene markers revealed that colonies 1, 4 and 5 were P. verrucosa harboring C. pacificum, and colonies 2 and 3 were P. ligulata containing C. latusorum [44]. Coral fragments in the control and heated aquaria remained apparently healthy throughout the experiment; no signs of stress such as paling, mucus production, or tissue sloughing were observed. Linear mixed effects models of color values did not reveal significant paling in heat-treated coral fragments (treatment F = 0.10, p = 0.75; timepoint F = 0.30, p = 0.59; treatment*timepoint F = 0.09, p = 0.77). Lack of color change was expected; bleaching signs are generally only detectable after weeks of ecologically relevant temperature stress—even though vital molecular processes are affected within several days [63].
DinoRNAV mcp gene sequencing overview
DinoRNAV mcp genes were detected in cDNA libraries from all 5 colonies. Amplicon sequencing of the dinoRNAV mcp gene resulted in 10 222 055 paired raw reads from all samples and one negative control. A total of 7 593 537 reads with a mean length of 423 bases (before trimming to 422 bases) remained after merging and quality control. Denoising resulted in 273 unique amplicon sequence variants (ASVs) across all samples, and translation revealed that 11 ASVs contained stop codons; these ASVs were removed. Translated ASVs collapsed into 124 unique amino acid sequences—“aminotypes”—that were 140 aa in length. Five control samples and the negative control were removed because they contained few (<10,000) amino acid sequences (20 reads were detected in the negative control). All remaining samples had between 50,000 and 210,000 amino acid sequences with a mean read depth of 142,810 ± 36 163 (SD).
DinoRNA viruses in Pocillopora-Cladocopium colonies are similar to dinoRNAVs in other coral-symbiont species
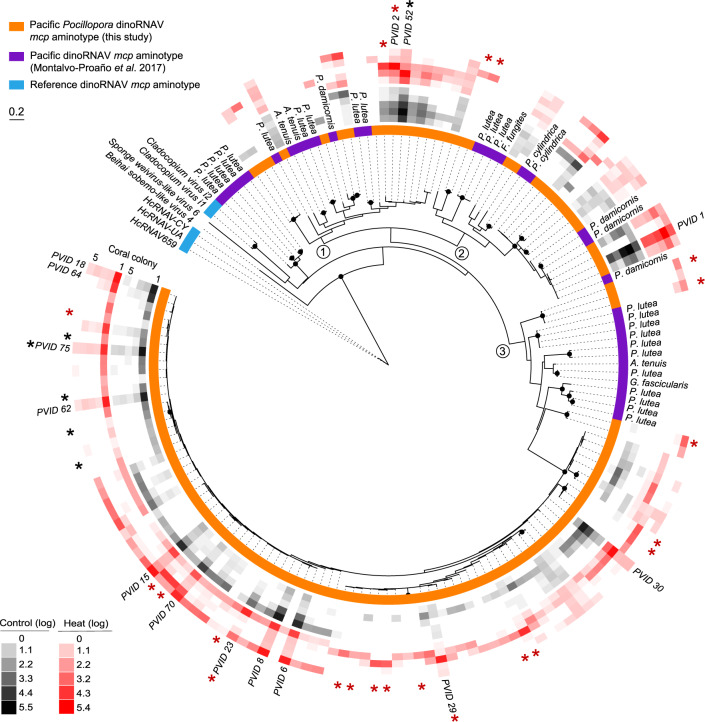
The 124 unique dinoRNAV aminotypes (Fig. 1; Table S1) detected in this study were most similar to 13 amino acid sequences translated from a published library of dinoRNAV mcp nucleotide sequences, generated from exogenous viral particles isolated from six coral-symbiont species via ultracentrifugation of cesium chloride gradients [29]; mean sequence similarities to these mcp sequences ranged from 53.4–98.4% (mean e-values ranged from 1.43·10−71–6.38·10−36). All dinoRNAV mcp aminotypes in this study formed a clade that is more recently derived than reference sequences such as HcRNAV, Beihai sobemo-like virus and sponge weivirus-like virus (Fig. 1). The dinoRNAV aminotypes in this study may form (at least) three quasispecies (e.g., rectangles in Fig. S2, sensu [64]). Strikingly, the majority of dinoRNAV mcp genes identified in our study occupy one clade; on average, sequences in this clade vary 9.8% ± 5.5 (SD) from each other (largest clade in Fig. 1, largest rectangle in Fig. S2; e.g., aminotypes 15, 18, and 30). Ultra-deep sequencing may be necessary to further clarify the drivers of these potential “mutant cloud dynamics” in Symbiodiniaceae hosts [65]. In several branches of the tree, aminotypes from our study are most closely related to aminotypes resolved from Symbiodiniaceae in Acropora tenuis, Favia fungites, Galaxea fascicularis, Pocillopora damicornis, Porites cylindrica, and Porites lutea corals sampled from the Great Barrier Reef (Table S1; [29]). Dominant aminotypes (>1% abundance across total dataset) are present in all major branches of the tree but differ by as much as ~60% in amino acid sequence (three branches in Fig. 1, three rectangles in Fig. S2; e.g., aminotypes 1, 2, and 6). Aminotype detections varied across treatment and timepoint in the experiment (shading of red and black squares in Fig. 1).
Fig. 1. Maximum likelihood tree of major capsid protein (mcp) “aminotypes” (unique amino acid sequences) from Symbiodiniaceae-infecting dinoflagellate RNA viruses (“dinoRNAVs”) isolated from three colonies of Pocillopora verrucosa and two colonies of Pocillopora ligulata containing the Symbiodiniaceae species Cladocopium pacificum and Cladocopium latusorum, respectively.
Aminotypes recovered in this study were similar to those in a previous work that recovered dinoRNAV mcp sequences from viral particles isolated from six coral species via ultracentrifugation of cesium chloride gradients [29]. Three major branches of the tree are indicated with the numbers 1–3. Black dots at nodes represent bootstrap support >75%. Colors adjacent to the tree indicate the study (orange, purple) or NCBI reference (blue) for each mcp aminotype. Reference NCBI accession numbers: Beihai Sobemo-like Virus 4 (YP_009336877), Sponge Weivirus-like Virus 6 (ASM94037), Cladocopium Virus i2 (AOS87317), Cladocopium Virus i1 (AOG17586), HcRNAV-CY (BAE47072), HcRNAV-UA (BAE47070) and HcRNAV-659 (BAU51723). Rings with black or red squares indicate the relative, log(10)-transformed abundances of dinoRNAV aminotypes in fragments from individual coral colonies in the control or heat treatment, respectively. Aminotypes labeled with PVID and a numeral comprise >1% abundance of the total dataset and are included in Fig. 2. Black and red asterisks indicate aminotypes that were significantly associated with control or heat treatments, respectively (see Fig. 4 for details).
DinoRNAV aminotypes differed among coral-symbiont species, colonies, and treatments
Approximately 64% of aminotypes (79 of 124) in this study were shared between the two coral-symbiont species, 19% (24) were unique to the three colonies of P. verrucosa-C. pacificum, and 17% (21) were unique to the two colonies of P. ligulata-C. latusorum. Approximately 34% of aminotypes (42) were present in all colonies, 48% (60) were shared between 2–4 colonies and 18% (22) were unique to individual coral colonies (with individual colonies containing up to 6 unique colony-specific aminotypes, Fig. 2a). All 14 aminotypes that each comprised > 1% reads in the total dataset (listed in Fig. 2b) were detected in all five coral colonies. DinoRNAV aminotype compositions varied among individual colonies (Fig. 2b); colony ID and coral-symbiont species were the most powerful predictors of viral composition (PERMANOVA: R2 = 0.55 and 0.21, respectively; p < 0.001). DinoRNAVs responded to elevated temperatures in colony-specific ways, as indicated by a significant interaction effect between treatment and colony (PERMANOVA: R2 = 0.079, p < 0.001). Treatment was also significant by itself (PERMANOVA: R2 = 0.02, p < 0.001), demonstrating a subtle but consistent response to elevated temperature across all colonies. Time was not a significant predictor (PERMANOVA: R2 = 0.003, p = 0.47; Fig. 2b).
Fig. 2. Overview of Symbiodiniaceae-infecting dinoflagellate RNA virus (“dinoRNAVs”) major capsid protein (mcp) gene “aminotypes” (unique amino acid sequences) associated with the stony corals Pocillopora verrucosa (harboring Cladocopium pacificum, “PV-CP”) and Pocillopora ligulata (harboring Cladocopium latusorum, “PL-CL”).
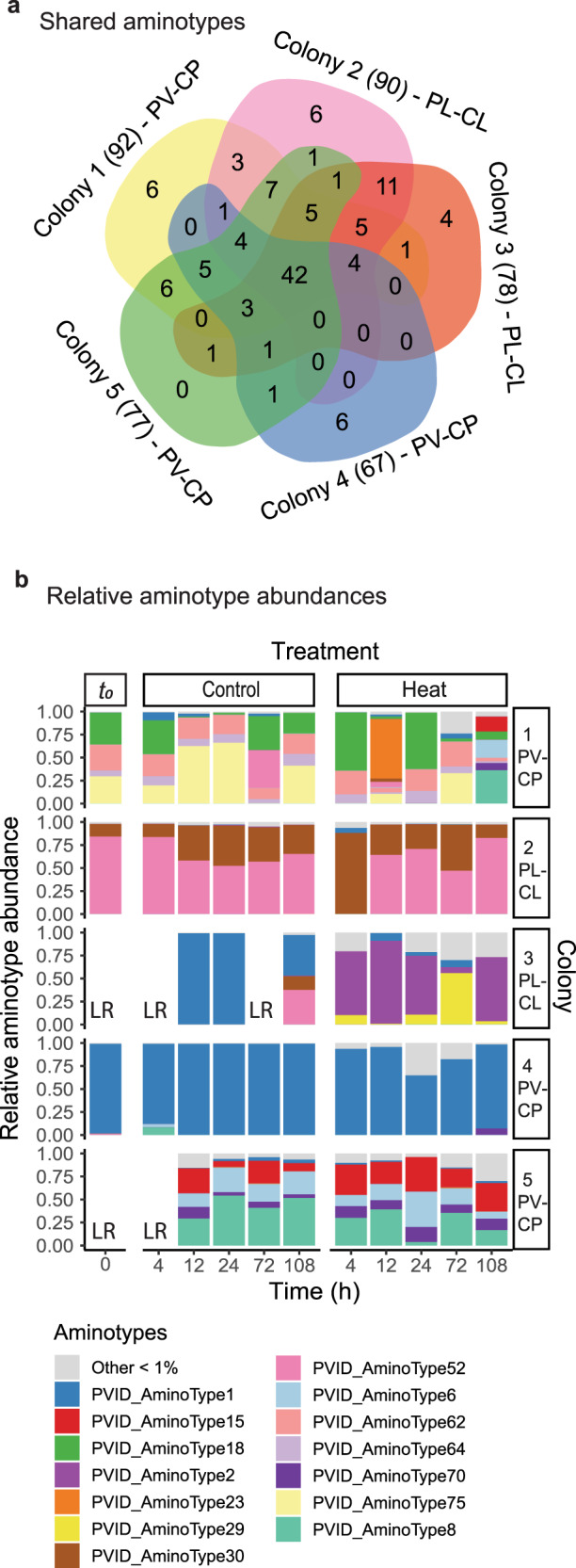
a A Venn diagram based on non-rarefied data indicates that ~82% (102) of 124 unique aminotypes were shared between two or more coral colonies, and 64% (79) were shared between the two coral-symbiont species. Data come from 8–11 fragments per colony sampled from control and heated aquaria. The total number of aminotypes detected per colony is given in parentheses. b The relative abundances of mcp aminotypes differed in fragments exposed to a 2.1 ˚C temperature increase (“Heat” treatment), compared to fragments from the same colonies exposed to ambient reef conditions (“Control”). LR indicates “low reads” and corresponds to samples that had <10 000 reads and were excluded from the analysis. PERMANOVA results: Colony R2 = 0.55, p < 0.001; Coral-symbiont species R2 = 0.21, p < 0.001; Treatment R2 = 0.02, p < 0.001; Treatment*Colony R2 = 0.079, p < 0.001; Time R2 = 0.003, p > 0.05.
Heat treatment rapidly increased the diversity of dinoRNAV aminotypes
A total of 17 aminotypes were unique to the heat treatment; 1 aminotype was unique to the controls (Fig. 3a; Table 2). Heat-specific aminotypes were observed in all colonies and ranged from 1–6 unique aminotypes per colony. Three heat-specific aminotypes were shared among multiple (2–3) coral colonies; one of these was shared between the two coral-symbiont species. The other 14 heat-specific aminotypes were not shared among colonies. A total of nine heat-specific aminotypes were unique to P. ligulata, whereas seven were unique to P. verrucosa (Table 2; Fig. S2). Most heat-specific aminotypes were relatively rare; only two aminotypes comprised >1% of reads in each fragment (aminotype 25: 1.8%; aminotype 114: 1.4%; other aminotypes comprised means of 0.01–0.7% of reads in individual fragments). The single aminotype unique to controls was detected in one fragment (colony 2) after 72 h (0.02% of reads in that fragment).
Fig. 3. Diversity of Symbiodiniaceae-infecting dinoflagellate RNA virus (“dinoRNAV”) major capsid protein (mcp) gene “aminotypes” (unique amino acid sequences) in heat (“Heat”) versus control (“Control”) conditions.
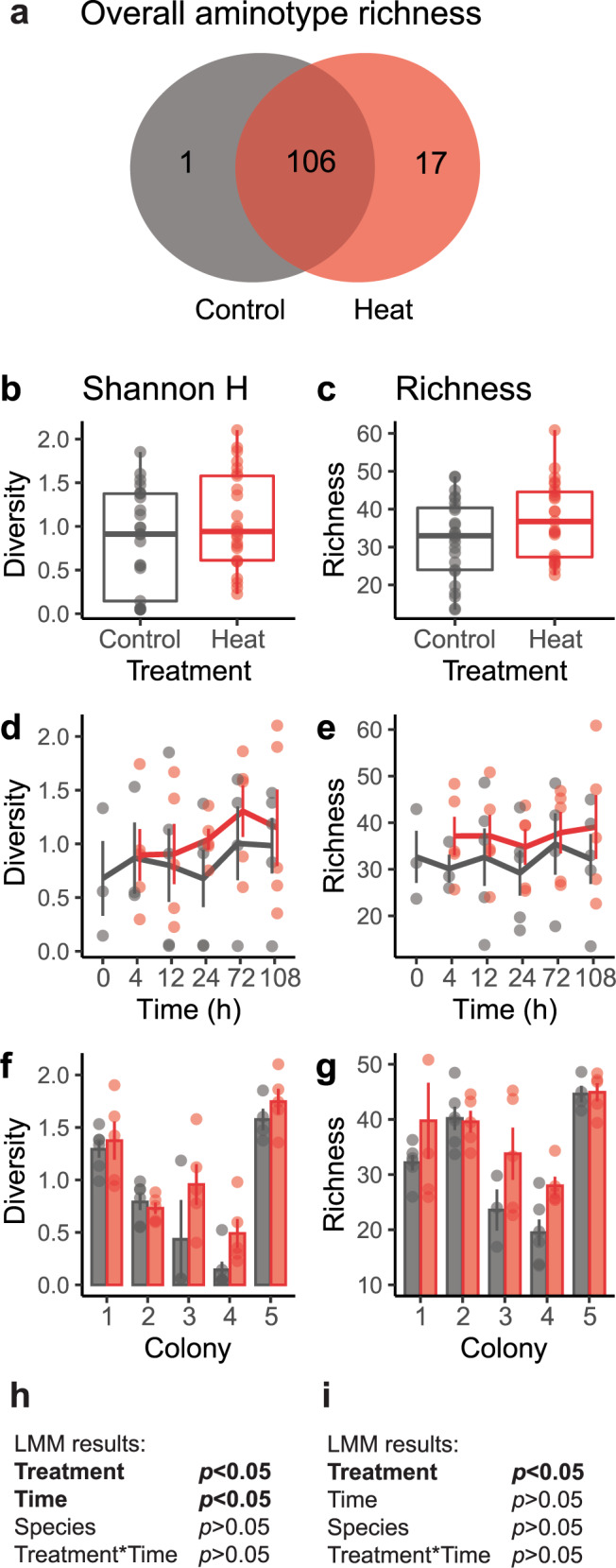
a Venn diagram of non-rarified aminotypes in heat and control treatments. Seventeen unique aminotypes were detected in coral fragments exposed to heat, compared to 1 unique aminotype in control fragments. (b, h) Diversity (Shannon index, H) of aminotypes increased in fragments exposed to heat compared to control fragments, and over time (d, h); variation also appeared to occur among colonies (f). (c, i) Estimated aminotype richness increased in fragments exposed to heat, but not over time (e, i). (g) Mean aminotype richness also appeared to vary among colonies. Linear mixed effects model (LMM) results for Shannon index (h) and estimated richness values (i). Lines and columns in d–g indicate means; error bars indicate SE. Shannon index values were based on sequencing data rarefied to 59,837 amino acid sequences per sample; richness was estimated by repeated random subsampling of unrarefied data. Coral-symbiont combinations were pooled for these analyses. Colonies 1, 4 and 5 are Pocillopora verrucosa (containing Cladocopium pacificum), colonies 2 and 3 are Pocillopora ligulata (containing Cladocopium latusorum).
Table 2.
Summary of Symbiodiniaceae-infecting dinoflagellate RNA virus (“dinoRNAV”) “aminotypes” (unique amino acid sequences) specific to the heat or control treatment based on amplicon sequencing.
| Aminotype ID | N_Observations | Colony | Time observed (h) | Mean relative abundance (%) ± SD | Closest match |
|---|---|---|---|---|---|
| Heat treatment | |||||
| 22 | 3 | 4 | 12, 24, 72 | 0.05 ± 0.06 | Plutea_AminoType17 |
| 25 | 2 | 3 | 24, 72 | 1.70 | Plutea_AminoType21 |
| 37 | 1 | 2 | 4 | 0.10 | Plutea_AminoType21 |
| 44 | 2 | 1 | 4, 12 | 0.01 | Plutea_AminoType13 |
| 68 | 4 | 2,3 | 4 (2,3); 24, 72 (3) | 0.02 ± 0.02 | Plutea_AminoType21 |
| 72 | 1 | 3 | 4 | 0.02 | Pdamicornis_AminoType1 |
| 76 | 5 | 1,2,5 | 4, 24 (2); 12 (1, 2); 72 (5) | 0.03 ± 0.05 | Plutea_AminoType13 |
| 80 | 1 | 2 | 108 | 0.03 | Plutea_AminoType21 |
| 81 | 1 | 4 | 108 | 0.03 | Plutea_AminoType5 |
| 87 | 1 | 4 | 24 | 0.02 | Plutea_AminoType10 |
| 104 | 2 | 3 | 24, 72 | 0.16 | Plutea_AminoType21 |
| 107 | 3 | 4 | 24, 72, 108 | 0.04 ± 0.02 | Gfascicularis_AminoType1 |
| 112 | 4 | 4 | 12, 24, 72, 108 | 0.22 ± 0.03 | Plutea_AminoType17 |
| 114 | 1 | 3 | 72 | 1.38 | Plutea_AminoType4 |
| 115 | 2 | 2,3 | 4 (2); 72 (3) | 0.68 | Plutea_AminoType21 |
| 117 | 1 | 2 | 108 | 0.38 | Plutea_AminoType21 |
| 118 | 1 | 4 | 4 | 0.38 | Plutea_AminoType10 |
| Control treatment | |||||
| 91 | 1 | 2 | 72 | 0.02 | Plutea_AminoType4 |
“Aminotype ID” indicates the name of the aminotype. “N_Observations” lists the total number of fragments in which a given aminotype was observed. “Colony” indicates the coral colony IDs in which a given aminotype was detected. “Time observed (h)” lists the sampling timepoints at which an aminotype was detected; numbers in parentheses indicate in which colonies each aminotype was observed at a given timepoint. “Mean relative abundance (%)” indicates the mean relative abundance of an aminotype across the fragments from which it was detected; standard deviations (SD) are given when aminotypes were identified in three or more samples. “Closest match” lists the strongest similarity of an aminotype in this study to known or putative viruses based on BLASTx against a reference database containing all sequences from the Reference Viral DataBase [63] and aminotypes generated by reprocessing the data in [29]. Colonies 1, 4, and 5 are Pocillopora verrucosa harboring Cladocopium pacificum; colonies 2 and 3 are Pocillopora ligulata harboring Cladocopium latusorum.
Alpha diversity (Shannon index, H) of aminotypes was positively associated with the heat treatment (F = 4.87, p = 0.03) and time (F = 4.91, p = 0.03) in the linear mixed effects model (LMM; Fig. 3b, d, h). There was no significant interaction between heat and time (F = 0.57, p = 0.46), nor was there a significant difference between coral-symbiont species (F = 0.44, p = 0.55). Mean (± SE) Shannon index values were 27% higher in heat-treated fragments (1.1 ± 0.6) than in control fragments (0.8 ± 0.6; Fig. 3b). Within individual timepoints, heated fragments had 4–93% higher mean H values than controls (0.9–1.3 versus 0.7–1.0, respectively; Fig. 3d), but individual comparisons were not significant. Mean Shannon index values per colony ranged between 0.5–1.8 for heat-treated fragments and 0.1–1.6 for control fragments (Fig. 3f).
There was a significant positive association between viral aminotype richness and heat treatment (F = 5.70, p = 0.02), but no effect of time (F = 0.35, p = 0.56) nor coral-symbiont species (F = 0.001, p = 0.99) in the LMM (Fig. 3c, e, i). There was no significant interaction between treatment and time (F = 0.10, p = 0.75). On average, aminotype richness was 16% higher in heat-treated (37.2 ± 2.0) than control fragments (32.0 ± 2.1; Fig. 3c). At individual timepoints, heat-treated fragments had 7–23% higher mean aminotype richness than controls (34.8–39.1 versus 29.2–35.4, respectively; Fig. 3e), but these differences were not significant. Mean aminotype richness for individual colonies (using timepoints as replicates) ranged between 28.0–44.9 for heat-treated fragments and 19.5–44.6 for control fragments (Fig. 3g).
Twenty-two aminotypes had higher relative abundances in heat-treated fragments
DESeq2 analysis revealed 28 aminotypes had significantly altered relative abundances in heat-treated or control fragments (Fig. 4). Twenty-two aminotypes had higher relative abundances in heated fragments, whereas 6 aminotypes had higher relative abundances in controls. Many (16/28) of the differentially abundant aminotypes were identified in both coral-symbiont species (aminotype names without a label, Fig. 4). Ten aminotypes were differentially abundant at multiple (2–4 out of 5) timepoints throughout the experiment; all ten of these were identified in both coral-symbiont species. Aminotypes 29, 38, and 98 were significantly more abundant in the heat treatment in 4 out of 5 sampled timepoints (from 12 to 108 h); aminotype 93 was more abundant in the final 3 timepoints (from 24 to 108 h).
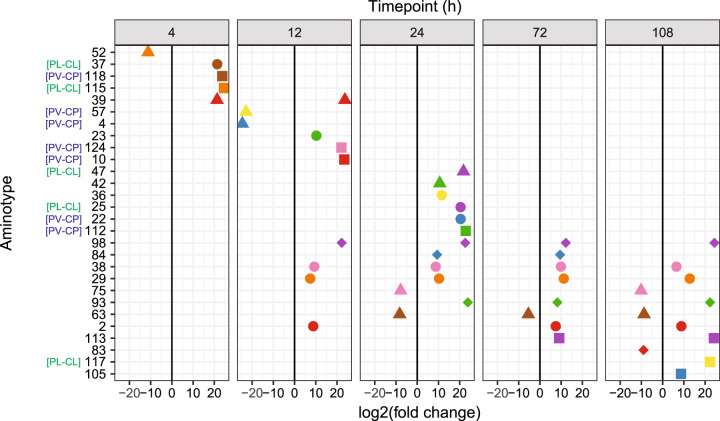
Fig. 4. Differentially abundant Symbiodiniaceae-infecting dinoflagellate RNA virus (“dinoRNAV”) major capsid protein (mcp) “aminotypes” (unique amino acid sequences) across the experiment.
Twenty-eight viral aminotypes (each represented as a point with a unique shape and color) were differentially abundant across control and heated fragments of the coral-symbiont species Pocillopora verrucosa-Cladocopium pacificum (PV-CP) and Pocillopora ligulata-Cladocopium latusorum (PL-CL). Of these, 22 aminotypes occurred at higher relative abundances in heat-treated fragments (right of the 0 lines) and 6 had higher relative abundances in control fragments (left of the 0 lines). Ten aminotypes were differentially abundant at multiple (2–4) timepoints throughout the experiment. This analysis was conducted using DeSeq2 on a non-rarefied dataset including both coral-symbiont species. Aminotypes labeled “[PV-CP]” were unique to PV-CP colonies; aminotypes labeled “[PL-CL]” were unique to PL-CL colonies. DESeq2 analyses were run separately for each time point (t(h) = 4, 12, 24, 72, 108, see Methods for more details).
Dispersion of dinoRNAV mcp amino acid sequences increased in heat-treated fragments
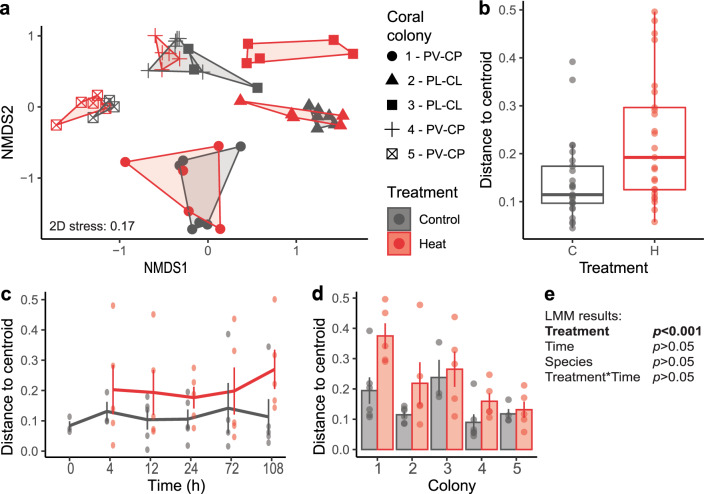
Dispersion (measured as distance to centroid) of dinoRNAVs was positively associated with heat treatment (Fig. 5a, b, e; F = 9.28, p = 0.004) in the LMM. There was no effect of time (F = 0.23, p = 0.95) nor coral-symbiont species (F = 0.20, p = 0.68), and no interaction between treatment and time (F = 0.48, p = 0.75). Overall, mean (±SE) dispersion was 62% higher in the heat treatment when all colonies and timepoints were pooled (heat: 0.23 ± 0.13; control: 0.14 ± 0.09). A trend of increasing dispersion of dinoRNAVs (32–100% higher) was observed in heat-treated samples at individual timepoints (Fig. 5c; heat: 0.2–0.29; controls: 0.12–0.17), but individual comparisons were not significant. Mean dispersion of individual colonies (across timepoints) was 11–93% higher per colony in heat-treated fragments (ranging from 0.13–0.37 in heat-treated fragments versus 0.1–0.24 in controls), but individual comparisons were not significant (Fig. 5d).
Fig. 5. Dispersion of Symbiodiniaceae-infecting dinoflagellate RNA virus (“dinoRNAV”) major capsid protein (mcp) gene “aminotypes” (unique amino acid sequences) associated with coral fragments in heat-treated (“H”) versus control (“C”) conditions.
a A non-metric Multidimensional Scaling (nMDS) plot depicts that dinoRNAVs differ by coral colony ID, coral-symbiont species (Pocillopora verrucosa-Cladocopium pacificum, “PV-CP”; Pocillopora ligulata-Cladocopium latusorum, “PL-CL”), and treatment. b Dispersion of dinoRNAV aminotypes was higher in heat-treated fragments. c Mean (±SE) aminotype dispersion was consistently higher over time in heat-treated fragments and d varied among coral colonies. e Results of a linear mixed effects model testing the effect of treatment and time on dispersion. Centroids were calculated separately for each colony in control and heat-treated conditions based on Bray-Curtis distances from square-root-transformed rarefied data. Dispersion was quantified by measuring the distance of each sample to its centroid.
DinoRNAV mcp and RdRp transcripts were detected in all metatranscriptomes
RNA sequencing resulted in 436 532 reads with similarity to reference viral sequences in the Reference Viral DataBase (RVDB, [62]). A mean of 1.17% ± 1.04 (SD) of viral reads showed homology to the putative RNA-dependent RNA polymerase (RdRp) of dinoRNAVs previously reported in association with Cladocopium C1 (Table S2; [27]). Overall, best hits to the RdRp of Cladocopium C1-infecting dinoRNAVs were identified in all analyzed metatranscriptomes (colonies 1, 4, 5; Table S3). These hits were the tenth-most common viral hits overall, and the fourth most common putative eukaryotic virus hits (Table S2). Other dinoRNAV-like reads matched best to two mcp sequences of Cladocopium C1-infecting dinoRNAV [27], comprising 0.06% ± 0.05 (AOG17586.1) and 0.04% ± 0.03 (AOS87317.1) of the viral transcripts, respectively (Table S3).
RnaSPAdes produced 214,309 contigs, of which 11,804 showed homology to virus reference protein sequences from the RVDB with DIAMOND BLASTx. A total of 128 contigs (lengths 211–2 631 nucleotides) showed homology to reference dinoRNAV ORFs from Levin et al. [27]. Amino acid-based alignment of the 38 recovered mcp contigs (and amplicons) generated in this study to the dinoRNAV mcp ORF from Levin et al. [27] revealed that although the dinoRNAV mcp gene is variable, it also contains five relatively conserved regions (CR I-V in Fig. S3). No significant insertions or deletions were apparent in the mcp contigs or amplicon sequences recovered in this study (Fig. S3).
Discussion
Viruses can have diverse impacts on hosts, ranging from antagonistic to beneficial [24, 66–68]. Efforts to understand how viral infections impact coral colonies have been stymied by the lack of (1) a high-throughput approach to track a viral lineage in colonies across an acute stress event; and (2) established cultures of viruses associated with corals and their symbionts (for use in viral addition experiments). This study tracks a group of Symbiodiniaceae-infecting viruses, the dinoRNAVs, in a controlled experiment to interrogate how infection responds to temperatures associated with bleaching. DinoRNAVs were detected from all five colonies examined, and from every heat-treated coral fragment, but only some controls. These observations, and detections of higher alpha diversity of dinoRNAV mcp aminotypes in heat-treated fragments, along with increased aminotype dispersion, unique aminotypes, and greater relative abundances of specific aminotypes, together strongly indicate that dinoRNAV infections were more active in heat-treated fragments. DinoRNAV responses were detectable within a single day, more quickly than the several weeks over which bleaching signs typically manifest. If viral infection of Symbiodiniaceae cells is enhanced (e.g., increased production, accumulation of viral diversity) during thermal anomalies in situ, then cumulative viral activity, if maintained over weeks, may disrupt coral-symbiont partnerships and potentially modulate some bleaching responses on reefs.
DinoRNAVs as a common, persistent virus of Symbiodiniaceae
DinoRNAV mcp genes were detected in all experimental colonies (N = 5), and in most to all fragments per colony (73–100% of fragments, N = 8/11–11/11 fragments per colony) using the gene amplicon sequencing method. DinoRNAV genes have additionally been reported from colonies of seven other stony coral species across the Atlantic and Pacific Oceans (Table 1, Fig. 1), suggesting that these viruses are commonly associated with coral microbiota. In this experiment, viral aminotype compositions (and potential viral quasispecies) differed among coral-symbiont species and colonies, but were similar within the fragments of a given colony, despite maintenance in separate aquaria (Fig. 2). These results suggest that, under ambient conditions, dinoRNAV populations are driven strongly by within-species and within-colony factors. RNA viruses rely on RNA-dependent RNA polymerase (RdRp) for replication, which is relatively error-prone. Therefore, individual viral progenitors infecting Symbiodiniaceae cells in a given colony may each produce a variety of genetically distinct viral “progeny” during a single replication cycle [35, 69]; hence, the observed pattern of species- and colony-specificity in dinoRNAV aminotypes likely results from diversification within “quasispecies” and (potentially) subsequent purifying selection [35, 70–72]. This production of a “mutant cloud” of dinoRNAV diversity may even help ensure these viruses are successful (at the population level) at infecting Symbiodiniaceae under changing environmental conditions [37]. As a next step, future works can quantify the extent to which dinoRNAV diversity is homogeneously distributed across entire coral-symbiont colonies at a given timepoint using multiple markers (e.g., mcp and RdRp).
Healthy corals contain millions of Symbiodiniaceae cells per cm2 of coral tissue; our results suggest that Symbiodiniaceae in hospite may be infected by one or perhaps several dinoRNAV quasispecies (e.g., aminotypes 30 and 52 in colony 2, Figs. 2 and S2) at any given time. Recent surveys of free-living marine microbial communities reported that viral infections may occur in ~33% [73, 74] to over 60% [75] of marine microorganisms, and many individual cells may be infected by multiple distinct viruses at a given time [73, 76]. Given this, the detection of multiple dinoRNAV mcp genes or quasi-species from a single Symbiodiniaceae cell in hospite might be expected; single cell RNA-Seq should be used to test this possibility.
The dinoRNAV mcp gene was not amplifiable from DNA extractions via PCR [77–79]. This indicates that the dinoRNAV mcp sequences reported here constitute exogenous viral infections, rather than endogenous viral elements (“EVEs”; [80]) within their host’s genome. Considering that the gene amplicons reported here were generated from unfractionated coral tissue, dinoRNAV mcp detections in this study could potentially arise from four sources: RNA genomes within intact dinoRNAV capsids (sensu [29]); mcp genes that are being expressed in host cells during a dinoRNAV replication cycle; free dinoRNAV genomes that may occur as extrachromosomal RNA in a latent [26] or carrier state similar to pseudolysogeny [81, 82]; and /or chimeric RNA viruses with analogous mcp gene sequences [83]. However, the identification of highly abundant transcripts with homology to Symbiodiniaceae-associated dinoRNAV RdRp genes in the metatranscriptomes (Tables S2 and S3) indicates that mcp gene amplicons in this study are most parsimoniously interpreted as derived from dinoRNAVs [77, 78].
The sequencing approaches employed here limit our ability to discern amongst the potential sources of viral mcp gene detections, as does the dearth of information available on dinoRNAV replication cycles in coral symbionts. Approaches such as single cell RNA-seq (e.g., [84]) of Symbiodiniaceae cells, as well as sequencing methods designed to identify and characterize infective viruses (similar to viral tagging [85], adsorption sequencing [86]) represent critical next steps in assessing the dynamics of dinoRNAV infections within individual Symbiodiniaceae cells.
Switching from persistent to more productive infection under stress
Many viruses switch between strategies upon environmental changes [24, 87–91]. RNA virus infections can range from exclusively lytic [30, 92, 93], to infections that are “persistent” or “chronic” and do not immediately kill the host [94, 95]. Persistent RNA viruses of plants, for example, may show altered activity based on seasonality [96, 97], temperature [97, 98], or other environmental factors [99, 100]. Similarly, marine viruses can switch between latent and productive cycles in response to host-related or environmental triggers [8, 101, 102]. For persistent viruses of Symbiodiniaceae, a variety of mechanisms may modulate viral infection strategies; the role of temperature in triggering persistent infections to become more productive has received particular attention due to the link between temperature and coral bleaching [8, 11, 25, 27, 99, 102].
We identified 22 aminotypes that were present at higher relative abundances in heat-treated coral fragments (Fig. 4), and 17 aminotypes were unique to these fragments (Fig. 3a, c). We interpret that these findings indicate a switch from persistent to more productive infections by some dinoRNAV strains (or quasispecies). Of particular interest, aminotypes 29, 38, 93 and 98 exhibited sustained increases in relative abundance in heat-treated corals starting at 12–24 h (Fig. 4). Screening for these dinoRNAV mcp aminotypes in additional Symbiodiniaceae populations (and sequencing libraries) is of interest.
The observation of a rapid increase in dinoRNAV aminotype diversity in some colonies during the onset of thermal stress is consistent with a previous report of increased (but overall low) abundance of dinoRNAV transcripts in the Caribbean coral Montastrea cavernosa following exposure to elevated temperatures for 12 h [25]. Further, a thermosensitive culture of Cladocopium C1 exhibited high expression of transcripts with best hits to dinoRNAV genes under ambient temperatures, while a thermotolerant Cladocopium C1 culture did not, suggesting that dinoRNAVs may modulate resistance to bleaching in some species or populations of Symbiodiniaceae [27]. Similarly, viral metagenomes from bleached pocilloporid colonies in situ contained significantly more eukaryotic virus sequences than unbleached, apparently healthy colonies [11]. Experiments with the cricket paralysis virus—a +ssRNAV with similar genome architecture to dinoRNAVs in Symbiodiniaceae cultures—also showed increased viral replication after two hours when temperatures were raised by 5 °C [100].
Colony-specific dinoRNAV responses to elevated temperatures
Since higher temperatures increase host cell enzymatic activity, increased viral production and accumulation of mutations within quasispecies exposed to heat stress might be expected based on thermodynamics alone [103]. However, dinoRNAV responses to elevated water temperatures differed among individual coral colonies (Figs. 2b, 5a), even in two colonies (3 and 4) that were both dominated by aminotype 1 in control aquaria. While dinoRNAVs in heat-treated fragments from colonies 1 and 3 exhibited strong shifts in putative quasispecies compositions, such changes were less pronounced in the other colonies. These findings suggest that dinoRNAV strains (or quasispecies) in colonies 1 and 3 were more responsive to heat stress. Coral colonies generally exhibit heterogeneous resistance to coral bleaching (e.g., [11, 104, 105]), and bleaching susceptibility in pocilloporids has been correlated to differential communities of eukaryotic viruses [11]. Subsequent experiments that extend multiple weeks to sample both the onset of thermal stress and the onset of bleaching signs are a critical next step in understanding how dinoRNAV dynamics relate to colony health trajectories and relative bleaching resistance.
Conclusions
This is the first study to characterize the dynamics of Symbiodiniaceae-infecting dinoRNAVs in coral colonies exposed to ecologically relevant bleaching temperatures. We identified dinoRNAVs in each sampled coral; temperature stress elicited rapid changes to dinoRNAV diversity and composition, and a subset of viral aminotypes were significantly associated with heat-treated fragments. Multiple lines of evidence suggest that dinoRNAVs are common in pocilloporid corals as persistent, exogenous infections of Symbiodiniaceae. Environmental stress may increase the productivity of these viruses, potentially impacting colony symbiotic status (if stress is prolonged). Overall, these findings add to the growing body of literature demonstrating that viruses of microorganisms affect emergent phenotypes of animal and plant holobionts, and may modulate holobiont responses to changing environmental conditions.
Supplementary information
Acknowledgements
The authors express their sincere appreciation to Kira Turnham and Dr. Todd LaJeunesse for resolving coral and Symbiodiniaceae species in this study, and to Drs. Rebecca L. Vega Thurber, Andrew R Thurber, and Craig E. Nelson for discussions and logistical support related to the experiment. Many thanks to Rebecca L. Maher and J. Grace Klinges for help with sampling, and to Dennis Conetta for assistance with DNA and RNA extractions. We additionally thank Mark Dasenko at Oregon State University’s Center for Genome Research & Biocomputing (Corvallis, OR) for his support in designing the sequencing methods. Lastly, we also thank two reviewers and editors for their suggestions and comments that improved the manuscript. Financial support was provided by a Sigma-Xi Grant-in-aid of Research to C.G., a U.S. National Science Foundation award (OCE #1635798) to AMSC. and an Early-Career Research Fellowship (#2000009651) from the Gulf Research Program of the National Academies of Sciences to AMSC.
Author contributions
CG, LHK and AC conceived of the experiment; CG, LHK, and AJV developed the methods with support from AC; CG, LHK, RB, and AC conducted the experiments and processed samples; CG led data analysis, with contributions by all authors; CG wrote the first draft of the manuscript, with contributions by all authors.
Data availability
All mcp gene amplicon libraries and the 5 metatranscriptomes have been deposited to the Sequence Read Archive under accession PRJNA778019.
Code availability
Code used to generate these results is available at https://github.com/CorreaLab/Pocillopora-dinoRNAVs.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lauren I. Howe-Kerr, Alex J. Veglia.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01194-y.
References
- 1.Hughes TP, Kerry J, Álvarez-Noriega M, Álvarez-Romero J, Anderson K, Baird A, et al. Global warming and recurrent mass bleaching of corals. Nature. 2017;543:373–7. doi: 10.1038/nature21707. [DOI] [PubMed] [Google Scholar]
- 2.Hoegh-Guldberg O. Climate change, coral bleaching and the future of the world’s coral reefs. Mar Freshw Res. 1999;50:839–66. [Google Scholar]
- 3.Patten NL, Harrison PL, Mitchell JG. Prevalence of virus-like particles within a staghorn scleractinian coral (Acropora muricata) from the Great Barrier Reef. Coral Reefs. 2008;27:569–80. doi: 10.1007/s00338-008-0356-9. [DOI] [Google Scholar]
- 4.Leruste A, Bouvier T, Bettarel Y. Enumerating viruses in coral mucus. Appl Environ Microbiol. 2012;78:6377–9. doi: 10.1128/AEM.01141-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen-kim H, Bouvier T, Bouvier C, Bui VN, Le-lan H, Bettarel Y. Viral and bacterial epibionts in thermally-stressed corals. J Mar Sci Eng. 2015;3:1272–86. doi: 10.3390/jmse3041272. [DOI] [Google Scholar]
- 6.Vega Thurber R, Payet JP, Thurber AR, Correa AMS. Virus–host interactions and their roles in coral reef health and disease. Nat Rev Microbiol. 2017;15:205–16. doi: 10.1038/nrmicro.2016.176. [DOI] [PubMed] [Google Scholar]
- 7.Sweet M, Bythell J. The role of viruses in coral health and disease. J Invertebr Pathol. 2017;147:136–44. doi: 10.1016/j.jip.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Wilson WH, Francis I, Ryan K, Davy SK. Temperature induction of viruses in symbiotic dinoflagellates. Aquat Micro Ecol. 2001;25:99–102. doi: 10.3354/ame025099. [DOI] [Google Scholar]
- 9.Lohr J, Munn CB, Wilson WH. Characterization of a latent virus-like infection of symbiotic zooxanthellae. Appl Environ Microbiol. 2007;73:2976–81. doi: 10.1128/AEM.02449-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawrence SA, Wilson WH, Davy JE, Davy SK. Latent virus-like infections are present in a diverse range of Symbiodinium spp. (Dinophyta) J Phycol. 2014;50:984–97. doi: 10.1111/jpy.12242. [DOI] [PubMed] [Google Scholar]
- 11.Messyasz A, Rosales SM, Mueller RS, Sawyer T, Correa AMS, Thurber AR, et al. Coral bleaching phenotypes associated with differential abundances of nucleocytoplasmic large DNA viruses. Front Mar Sci. 2020;7:555474. doi: 10.3389/fmars.2020.555474. [DOI] [Google Scholar]
- 12.Marhaver KL, Edwards RA, Rohwer F. Viral communities associated with healthy and bleaching corals. Environ Microbiol. 2008;10:2277–86. doi: 10.1111/j.1462-2920.2008.01652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Correa AMS, Ainsworth TD, Rosales SM, Thurber AR, Butler CR, Vega Thurber RL. Viral outbreak in corals associated with an in situ bleaching event: atypical herpes-like viruses and a new megavirus infecting Symbiodinium. Front Microbiol. 2016;7:127. doi: 10.3389/fmicb.2016.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bettarel Y, Thuy NT, Huy TQ, Hoang PK, Bouvier T. Observation of virus-like particles in thin sections of the bleaching scleractinian coral Acropora cytherea. J Mar Biol Assoc U K. 2013;93:909–12. doi: 10.1017/S0025315411002062. [DOI] [Google Scholar]
- 15.Lesser MP, Bythell JC, Gates RD, Johnstone RW, Hoegh-Guldberg O. Are infectious diseases really killing corals? Alternative interpretations of the experimental and ecological data. J Exp Mar Biol Ecol. 2007;346:36–44. doi: 10.1016/j.jembe.2007.02.015. [DOI] [Google Scholar]
- 16.Soffer N, Brandt ME, Correa AMS, Smith TB, Thurber RV. Potential role of viruses in white plague coral disease. ISME J. 2014;8:271–83. doi: 10.1038/ismej.2013.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawrence SA, Davy JE, Aeby GS, Wilson WH, Davy SK. Quantification of virus-like particles suggests viral infection in corals affected by Porites tissue loss. Coral Reefs. 2014;33:687–91. doi: 10.1007/s00338-014-1168-8. [DOI] [Google Scholar]
- 18.Lawrence SA, Davy JE, Wilson WH, Hoegh-Guldberg O, Davy SK. Porites white patch syndrome: associated viruses and disease physiology. Coral Reefs. 2015;34:249–57. doi: 10.1007/s00338-014-1218-2. [DOI] [Google Scholar]
- 19.Pollock FJ, M. Wood-Charlson E, Van Oppen MJH, Bourne DG, Willis BL, Weynberg KD. Abundance and morphology of virus-like particles associated with the coral Acropora hyacinthus differ between healthy and white syndrome-infected states. Mar Ecol Prog Ser. 2014;510:39–43. doi: 10.3354/meps10927. [DOI] [Google Scholar]
- 20.Vega Thurber RL, Correa AMS. Viruses of reef-building scleractinian corals. J Exp Mar Biol Ecol. 2011;408:102–13. doi: 10.1016/j.jembe.2011.07.030. [DOI] [Google Scholar]
- 21.Weynberg KD, Voolstra CR, Neave MJ, Buerger P, Van Oppen MJH. From cholera to corals: viruses as drivers of virulence in a major coral bacterial pathogen. Sci Rep. 2015;5:17889. doi: 10.1038/srep17889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quistad SD, Grasis JA, Barr JJ, Rohwer FL. Viruses and the origin of microbiome selection and immunity. ISME J. 2017;11:835–40. doi: 10.1038/ismej.2016.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oppen MJHV, Leong J, Gates RD. Coral-virus interactions: a double-edged sword? SYMBIOSIS. 2009;47:1–8. doi: 10.1007/BF03179964. [DOI] [Google Scholar]
- 24.Correa AMS, Howard-Varona C, Coy SR, Buchan A, Sullvian MB, Weitz JS. The virus-microbe infection continuum: revisiting the viral rules of life. Nat Rev Microbiol. 2021;19:501–13. doi: 10.1038/s41579-021-00530-x. [DOI] [PubMed] [Google Scholar]
- 25.Correa AMS, Welsh RM, Vega Thurber RL. Unique nucleocytoplasmic dsDNA and +ssRNA viruses are associated with the dinoflagellate endosymbionts of corals. ISME J. 2013;7:13–27. doi: 10.1038/ismej.2012.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence SA, Floge SA, Davy JE, Davy SK, Wilson WH. Exploratory analysis of Symbiodinium transcriptomes reveals potential latent infection by large dsDNA viruses. Environ Microbiol. 2017;19:3909–19. doi: 10.1111/1462-2920.13782. [DOI] [PubMed] [Google Scholar]
- 27.Levin RA, Voolstra CR, Weynberg KD, Van Oppen M. Evidence for a role of viruses in the thermal sensitivity of coral photosymbionts. ISME J. 2017;11:808–12. doi: 10.1038/ismej.2016.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knowles B, Bonachela JA, Behrenfeld MJ, Bondoc KG, Cael BB, Carlson CA, et al. Temperate infection in a virus–host system previously known for virulent dynamics. Nat Commun. 2020;11:1–13. doi: 10.1038/s41467-020-18078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montalvo-Proaño J, Buerger P, Weynberg KD, Van Oppen MJH. A PCR-based assay targeting the major capsid protein gene of a dinorna-like ssRNA virus that infects coral photosymbionts. Front Microbiol. 2017;8:1665. doi: 10.3389/fmicb.2017.01665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomaru Y, Katanozaka N, Nishida K, Shirai Y, Tarutani K, Yamaguchi M, et al. Isolation and characterization of two distinct types of HcRNAV, a single-stranded RNA virus infecting the bivalve-killing microalga Heterocapsa circularisquama. Aquat Micro Ecol. 2004;34:207–18. doi: 10.3354/ame034207. [DOI] [Google Scholar]
- 31.Miller JL, Chen S, Nagasaki K, Roseman A, Wepf R, Sewell T, et al. Three-dimensional reconstruction of Heterocapsa circularisquama RNA virus by electron cryo-microscopy. J Gen Virol. 2011;92:1960–70. doi: 10.1099/vir.0.031211-0. [DOI] [PubMed] [Google Scholar]
- 32.Shi M, Lin XD, Tian J-H, Chen L-J, Chen X, Li C-IU, et al. Redefining the invertebrate RNA virosphere. Nature. 2016;540:539–43. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 33.Domingo E, Sheldon J, Perales C. Viral quasispecies evolution. Microbiol Mol Biol Rev. 2012;76:159–216. doi: 10.1128/MMBR.05023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanjuán R, Domingo-Calap P. Mechanisms of viral mutation. Cell Mol Life Sci. 2016;73:4433–48. doi: 10.1007/s00018-016-2299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vlok M, Lang AS, Suttle CA. Marine RNA virus quasispecies are distributed throughout the oceans. mSphere. 2019;4:1–18. doi: 10.1128/mSphereDirect.00157-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Domingo E, Martinez-salas E, Sobrino F, de la Torre JC, Portela A, Ortin J, et al. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance—a review. Gene. 1985;40:1–8. doi: 10.1016/0378-1119(85)90017-4. [DOI] [PubMed] [Google Scholar]
- 37.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–8. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. Rapid evolution of RNA genomes. Science. 1982;215:1577–85. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 39.Gélin P, Postaire B, Fauvelot C, Magalon H. Molecular phylogenetics and evolution reevaluating species number, distribution and endemism of the coral genus Pocillopora Lamarck, 1816 using species delimitation methods and microsatellites. Mol Phylogenet Evol. 2017;109:430–46. doi: 10.1016/j.ympev.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 40.Pratchett MS, McCowan D, Maynard JA, Heron SF. Changes in bleaching susceptibility among corals subject to ocean warming and recurrent bleaching in Moorea, French Polynesia. PLoS ONE. 2013;8:1–10. doi: 10.1371/journal.pone.0070443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donovan MK, Adam TC, Shantz AA, Speare KE, Munsterman KS, Rice MM, et al. Nitrogen pollution interacts with heat stress to increase coral bleaching across the seascape. Proc Natl Acad Sci USA. 2020;117:5351–7. doi: 10.1073/pnas.1915395117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siebeck UE, Marshall NJ, Kluter A, Hoegh-Guldberg O. Monitoring coral bleaching using a colour reference card. Coral Reefs. 2006;25:453–60. doi: 10.1007/s00338-006-0123-8. [DOI] [Google Scholar]
- 43.Winters G, Holzman R, Blekhman A, Beer S, Loya Y. Photographic assessment of coral chlorophyll contents: Implications for ecophysiological studies and coral monitoring. J Exp Mar Biol Ecol. 2009;380:25–35. doi: 10.1016/j.jembe.2009.09.004. [DOI] [Google Scholar]
- 44.Turnham KE, Wham DC, Sampayo E, LaJeunesse TC. Mutualistic microalgae co-diversify with reef corals that acquire symbionts during egg development. ISME J. 2021;15:3271–85. doi: 10.1038/s41396-021-01007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinzón JH, Lajeunesse TC. Species delimitation of common reef corals in the genus Pocillopora using nucleotide sequence phylogenies, population genetics and symbiosis ecology. Mol Ecol. 2011;20:311–25. doi: 10.1111/j.1365-294X.2010.04939.x. [DOI] [PubMed] [Google Scholar]
- 46.Flot JF, Tillier S. The mitochondrial genome of Pocillopora (Cnidaria: Scleractinia) contains two variable regions: the putative D-loop and a novel ORF of unknown function. Gene. 2007;401:80–87. doi: 10.1016/j.gene.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 47.Pinzón JH, Sampayo E, Cox E, Chauka LJ, Chen CA, Voolstra CR, et al. Blind to morphology: genetics identifies several widespread ecologically common species and few endemics among Indo-Pacific cauliflower corals (Pocillopora, Scleractinia) J Biogeogr. 2013;40:1595–608. doi: 10.1111/jbi.12110. [DOI] [Google Scholar]
- 48.Johnston EC, Forsman ZH, Flot JF, Schmidt-Roach S, Pinzón JH, Knapp ISS, et al. A genomic glance through the fog of plasticity and diversification in Pocillopora. Sci Rep. 2017;7:5991. doi: 10.1038/s41598-017-06085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wham DC, Carmichael M, LaJeunesse TC. Microsatellite loci for Symbiodinium goreaui and other Clade C Symbiodinium. Coservation Genet Resour. 2014;6:127–9. doi: 10.1007/s12686-013-0023-5. [DOI] [Google Scholar]
- 50.Bay LK, Ulstrup KE, Nielsen HB, Jarmer H, Goffard N, Willis BL, et al. Microarray analysis reveals transcriptional plasticity in the reef building coral Acropora millepora. Mol Ecol. 2009;18:3062–75. doi: 10.1111/j.1365-294X.2009.04257.x. [DOI] [PubMed] [Google Scholar]
- 51.Veglia AJ, Vicéns RER, Grupstra CGB, Howe-Kerr LI, Correa AMS. vAMPirus: an automated, comprehensive virus amplicon sequencing analysis program. 2021: available at https://zenodo.org/record/4549851 (accessed February 17, 2021).
- 52.Edgar RC. UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv 2016;81257: available at 10.1101/081257.
- 53.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;2016:1–22. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinforma. 2004;5:1–19. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2014;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 56.Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol. 2020;37:291–4. doi: 10.1093/molbev/msz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oksanen J, Blanchet G, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: Community Ecology Package. R package version 2.4-6. R Package Version 25-6 2019.
- 58.Anderson MJ, Walsh DCI. PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: what null hypothesis are you testing? Ecol Monogr. 2013;83:557–74. doi: 10.1890/12-2010.1. [DOI] [Google Scholar]
- 59.Bates DM, Maechler M, Bolker B, Walker S. lme4: Mixed-effects modeling with R. R Package Version 11-7 HttpCRANR-Proj 2014.
- 60.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550–550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goodacre N, Aljanahi A, Nandakumar S, Mikailov M, Khan AS. A reference viral database (RVDB) to enhance bioinformatics analysis of high-throughput sequencing for novel virus detection. mSphere. 2018;3:e00069–18. doi: 10.1128/mSphereDirect.00069-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rädecker N, Pogoreutz C, Gegner HM, Cárdenas A, Roth F. Heat stress destabilizes symbiotic nutrient cycling in corals. Proc Natl Acad Sci USA. 2021;118:e2022653118. doi: 10.1073/pnas.2022653118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Biebricher CK, Eigen M. What Is a Quasispecies? In: Domingo E (ed). Quasispecies: concept and implications for virology. 2006. Springer Berlin Heidelberg, Berlin, Heidelberg, pp 1–31.
- 65.Gelbart M, Harari S, Ben-ari Y, Kustin T, Wolf D, Mandelboim M, et al. Drivers of within-host genetic diversity in acute infections of viruses. PLoS Pathog. 2020;16:e1009029. doi: 10.1371/journal.ppat.1009029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Breitbart M, Bonnain C, Malki K, Sawaya NA. Phage puppet masters of the marine microbial realm. Nat Microbiol. 2018;3:754–66. doi: 10.1038/s41564-018-0166-y. [DOI] [PubMed] [Google Scholar]
- 67.Mann NH, Cook A, Millard A, Bailey S, Clokie M. Bacterial photosynthesis genes in a virus. Nature. 2003;424:741. doi: 10.1038/424741a. [DOI] [PubMed] [Google Scholar]
- 68.Marquez LM, Redman RS, Rodriguez RJ, Roossinck MJ. A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science. 2007;315:513–5. doi: 10.1126/science.1136237. [DOI] [PubMed] [Google Scholar]
- 69.Holmes EC. The RNA virus quasispecies: fact or fiction? J Mol Biol. 2010;400:271–3. doi: 10.1016/j.jmb.2010.05.032. [DOI] [PubMed] [Google Scholar]
- 70.Pybus OG, Rambaut A, Belshaw R, Freckleton RP, Drummond AJ, Holmes EC. Phylogenetic evidence for deleterious mutation load in RNA viruses and its contribution to viral evolution. Mol Biol Evol. 2007;24:845–52. doi: 10.1093/molbev/msm001. [DOI] [PubMed] [Google Scholar]
- 71.Holmes EC. Patterns of intra- and interhost nonsynonymous variation reveal strong purifying selection in dengue virus. J Virol. 2003;77:11296–8. doi: 10.1128/JVI.77.20.11296-11298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edwards CTT, Holmes EC, Pybus OG, Wilson DJ, Viscidi RP, Abrams EJ, et al. Evolution of the human immunodeficiency virus envelope gene is dominated by purifying selection. Genetics. 2006;174:1441–53. doi: 10.1534/genetics.105.052019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roux S, Hawley AK, Beltran MT, Scofield M, Schwientek P, Stepanauskas R, et al. Ecology and evolution of viruses infecting uncultivated SUP05 bacteria as revealed by single-cell- and meta-genomics. eLIFE. 2014;3:e03125. doi: 10.7554/eLife.03125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Labonté JM, Swan BK, Poulos B, Luo H, Koren S, Hallam SJ, et al. Single-cell genomics-based analysis of virus-host interactions in marine surface bacterioplankton. ISME J. 2015;9:2386–99. doi: 10.1038/ismej.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Munson-mcgee JH, Peng S, Dewerff S, Stepanauskas R, Whitaker RJ, Weitz JS, et al. A virus or more in (nearly) every cell: ubiquitous networks of virus–host interactions in extreme environments. ISME J. 2018;12:1706–14. doi: 10.1038/s41396-018-0071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Díaz-Muñoz SL. Viral coinfection is shaped by host ecology and virus-virus interactions across diverse microbial taxa and environments. Virus Evol. 2017;3:1–14. doi: 10.1093/ve/vex011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang L, Wu S, Liu T, Sun J, Chi S, Liu C, et al. Endogenous viral elements in algal genomes. Acta Oceano Sin. 2014;33:102–7. [Google Scholar]
- 78.Moniruzzaman M, Weinheimer AR, Martinez-Gutierrez CA, Aylward FO. Widespread endogenization of giant viruses shapes genomes of green algae. Nature. 2020;588:141–5. doi: 10.1038/s41586-020-2924-2. [DOI] [PubMed] [Google Scholar]
- 79.Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, et al. Global organization and proposed megataxonomy of the virus world. Microbiol Mol Biol Rev. 2020;84:e00061–19. doi: 10.1128/MMBR.00061-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holmes EC. The evolution of endogenous viral elements. Cell Host Microbe. 2011;10:368–77. doi: 10.1016/j.chom.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ripp S, Miller RV. The role of pseudolysogeny in bacteriophage-host interactions in a natural freshwater environment. Microbiology. 1997;143:2065–70. doi: 10.1099/00221287-143-6-2065. [DOI] [PubMed] [Google Scholar]
- 82.Onodera S, Olkkonen VM, Gottlieb P, Strassman J, Qiao XY, Bamford DH, et al. Construction of a transducing virus from double-stranded RNA bacteriophage phi6: establishment of carrier states in host cells. J Virol. 1992;66:190–6. doi: 10.1128/jvi.66.1.190-196.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de la Higuera I, Kasun GW, Torrance EL, Pratt AA, Maluenda A, Colombet J, et al. Unveiling crucivirus diversity by mining metagenomic data. mBio. 2020;11:e01410–20. doi: 10.1128/mBio.01410-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ku C, Sheyn U, Sebé-Pedrós A, Ben-Dor S, Schatz D, Tanay A, et al. A single-cell view on alga-virus interactions reveals sequential transcriptional programs and infection states. Sci Adv. 2020;6:eaba4137. [DOI] [PMC free article] [PubMed]
- 85.Deng L, Ignacio-espinoza JC, Gregory AC, Poulos BT, Weitz JS, Hugenholtz P, et al. Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature. 2014;513:242–6. doi: 10.1038/nature13459. [DOI] [PubMed] [Google Scholar]
- 86.Jonge PAD, Costa AR, Franklin L, Brouns SJJ, Jonge PAD, Meijenfeldt FABV, et al. Adsorption sequencing as a rapid method to link environmental bacteriophages to hosts. ISCIENCE. 2020;23:101439. doi: 10.1016/j.isci.2020.101439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jiang SC, Paul JH. Seasonal and diel abundance of viruses and occurrence of lysogeny/bacteriocinogeny in the marine environment. Mar Ecol Prog Ser. 1994;104:163–72. doi: 10.3354/meps104163. [DOI] [Google Scholar]
- 88.Brum JR, Hurwitz BL, Schofield O, Ducklow HW, Sullivan MB. Seasonal time bombs: dominant temperate viruses affect Southern Ocean microbial dynamics. ISME J. 2016;10:437–49. doi: 10.1038/ismej.2015.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Winter C, Bouvier T, Weinbauer MG, Thingstad TF. Trade-offs between competition and defense specialists among unicellular planktonic organisms: the “killing the winner” hypothesis revisited. Microbiol Mol Biol Rev. 2010;74:42–57. doi: 10.1128/MMBR.00034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thingstad TF, Våge S, Storesund JE, Sandaa R, Giske J. A theoretical analysis of how strain-specific viruses can control microbial species diversity. PNAS. 2014;111:7813–8. doi: 10.1073/pnas.1400909111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thingstad TF. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol Oceanogr. 2000;45:1320–8. doi: 10.4319/lo.2000.45.6.1320. [DOI] [Google Scholar]
- 92.Tomaru Y, Hata N, Masuda T, Tsuji M, Igata K, Masuda Y, et al. Ecological dynamics of the bivalve-killing dinoflagellate Heterocapsa circularisquama and its infectious viruses in different locations of western Japan. Environ Microbiol. 2007;9:1376–83. doi: 10.1111/j.1462-2920.2007.01252.x. [DOI] [PubMed] [Google Scholar]
- 93.Sadeghi M, Tomaru Y, Ahola T. RNA viruses in aquatic unicellular eukaryotes. Viruses 2021. [DOI] [PMC free article] [PubMed]
- 94.Randall RE, Griffin DE. Within host RNA virus persistence: mechanisms and consequences. Curr Opin Virol. 2017;23:35–42. doi: 10.1016/j.coviro.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roossinck MJ. Lifestyles of plant viruses. Philos Trans R Soc B Biol Sci. 2010;365:1899–905. doi: 10.1098/rstb.2010.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Honjo MN, Emura N, Kawagoe T, Sugisaka J, Kamitani M, Nagano AJ, et al. Seasonality of interactions between a plant virus and its host during persistent infection in a natural environment. ISME J. 2020;14:506–18. doi: 10.1038/s41396-019-0519-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim Y, Kim YJ, Paek K-H. Temperature-specific vsiRNA confers RNAi-mediated viral resistance at elevated temperature in Capsicum annuum. J Exp Bot. 2020;72:1432–48. doi: 10.1093/jxb/eraa527. [DOI] [PubMed] [Google Scholar]
- 98.Jones RAC. Chapter three - future scenarios for plant virus pathogens as climate change progresses. In: Kielian M, Maramorosch K, Mettenleiter TC (eds).2016. Academic Press, pp 87–147. [DOI] [PubMed]
- 99.Brüwer JD, Agrawal S, Liew YJ, Aranda M, Voolstra CR. Association of coral algal symbionts with a diverse viral community responsive to heat shock. BMC Microbiol. 2017;17:1–11. doi: 10.1186/s12866-017-1084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cevallos RC, Sarnow P. Temperature protects insect cells from infection by cricket paralysis virus. J Virol. 2010;84:1652–5. doi: 10.1128/JVI.01730-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Edgar RS, Lielausis I. Temperature-sensitive mutants of bacteriophage T4D: their isolation and genetic characterization. Genetics. 1964;49:649–62. doi: 10.1093/genetics/49.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vega Thurber RL, Barott KL, Hall D, Liu H, Rodriguez-Mueller B, Desnues C, et al. Metagenomic analysis indicates that stressors induce production of herpes-like viruses in the coral Porites compressa. Proc Natl Acad Sci USA. 2008;105:18413–8. doi: 10.1073/pnas.0808985105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Seifert M, van Nies P, Papini FS, Arnold JJ, Poranen MM, Cameron CE, et al. Temperature controlled high-throughput magnetic tweezers show striking difference in activation energies of replicating viral RNA-dependent RNA polymerases. Nucleic Acids Res. 2020;48:5591–602. doi: 10.1093/nar/gkaa233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wooldridge SA. Differential thermal bleaching susceptibilities amongst coral taxa: re-posing the role of the host. Coral Reefs. 2014;33:15–27. doi: 10.1007/s00338-013-1111-4. [DOI] [Google Scholar]
- 105.Hédouin L, Rouzé H, Berthe C, Perez-Rosales G, Martinez E, Chancerelle Y, et al. Contrasting patterns of mortality in Polynesian coral reefs following the third global coral bleaching event in 2016. Coral Reefs. 2020;39:939–52. doi: 10.1007/s00338-020-01914-w. [DOI] [Google Scholar]
- 106.Tonk L, Sampayo EM, Weeks S, Magno-Canto M, Hoegh-Guldberg O. Host-Specific interactions with environmental factors shape the distribution of Symbiodinium across the Great Barrier Reef. PLoS ONE. 2013;8:e68533–e68533. doi: 10.1371/journal.pone.0068533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Serrano X, Baums IB, O’Reilly K, Smith TB, Jones RJ, Shearer TL, et al. Geographic differences in vertical connectivity in the Caribbean coral Montastraea cavernosa despite high levels of horizontal connectivity at shallow depths. Mol Ecol. 2014;23:4226–40. doi: 10.1111/mec.12861. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All mcp gene amplicon libraries and the 5 metatranscriptomes have been deposited to the Sequence Read Archive under accession PRJNA778019.
Code used to generate these results is available at https://github.com/CorreaLab/Pocillopora-dinoRNAVs.