Abstract
Inter-kingdom belowground carbon (C) transfer is a significant, yet hidden, biological phenomenon, due to the complexity and highly dynamic nature of soil ecology. Among key biotic agents influencing C allocation belowground are ectomycorrhizal fungi (EMF). EMF symbiosis can extend beyond the single tree-fungus partnership to form common mycorrhizal networks (CMNs). Despite the high prevalence of CMNs in forests, little is known about the identity of the EMF transferring the C and how these in turn affect the dynamics of C transfer. Here, Pinus halepensis and Quercus calliprinos saplings growing in forest soil were labeled using a 13CO2 labeling system. Repeated samplings were applied during 36 days to trace how 13C was distributed along the tree-fungus-tree pathway. To identify the fungal species active in the transfer, mycorrhizal fine root tips were used for DNA-stable isotope probing (SIP) with 13CO2 followed by sequencing of labeled DNA. Assimilated 13CO2 reached tree roots within four days and was then transferred to various EMF species. C was transferred across all four tree species combinations. While Tomentella ellisii was the primary fungal mediator between pines and oaks, Terfezia pini, Pustularia spp., and Tuber oligospermum controlled C transfer among pines. We demonstrate at a high temporal, quantitative, and taxonomic resolution, that C from EMF host trees moved into EMF and that C was transferred further to neighboring trees of similar and distinct phylogenies.
Subject terms: Forest ecology, Plant ecology, Microbial ecology, Forest ecology
Introduction
Belowground mutualistic interactions play an essential role in maintaining forest stability around the globe [1]. Ectomycorrhizal fungi (EMF) form extraradical mycelium, a collection of filamentous fungal hyphae emanating from the root, which aid in exploring and exploiting the soil matrix environment [2]. EMF symbiosis is based on the reciprocal exchange of resources [3], and can positively influence the host plant water relations and response to drought [4], and increase its resistance to soil-borne pathogens [5]. Interestingly, EMF symbiosis can extend beyond the single tree-fungus partnership to form common mycorrhizal networks (CMNs) [6]. These networks simultaneously connect multiple plant hosts and mycorrhizal fungi [7], colonizing a large number of plants from the same or different species [8]. For example, CMNs can link hosts belonging to angiosperms and gymnosperms [9], even though these clades diverged during the Jurassic age around 200M years ago [10]. In addition, CMNs have been found to enhance sapling establishment [11–13], transfer water and reduce water stress [14, 15], play a crucial role in C cycling and sequestration [16], and even communicate stress signals among neighboring plants [17].
CMNs have been studied experimentally for many years [18], while their interpretation was continuously criticized [19] and their actual ecological significance for plant fitness has been questioned [20]. Some of the main arguments against these experiments, call for the use of appropriate controls using mesh barriers [21] excluding root-root contact and passive C diffusion through soil. Several studies [22, 23] have shown that C fixed by one plant transferred to the root system, and presumably the hyphae, of the second plant. However, for C to have any eco-physiological importance for the recipient plant, it needs to move out of the roots of the recipient plant. Despite the high prevalence of CMNs in nature, Some of the open questions include the significance of the resources exchange between trees [24], and who are the fungal mediators of the resource exchange [25]. To better understand CMNs role in ecological communities, various labeling methods have been used [26], and particularly 13C has gained popularity among researchers since C is the primary resource traded among the trees and fungi. Labeling techniques have been tested in both artificial [27–30] and natural systems [13, 31, 32]. The results from these studies indicate that the bi-directional C transfer between trees can be dictated via a source-sink relationship [31], the amount of overlap in EMF communities between the various hosts [33], and tree phylogenetic relatedness [34]. However, bidirectional transfer was also found between taxonomically distant mature tree taxa [32].
To unravel the importance of C transfer within CMNs and how it relates to the host species’ identity and function, we need to identify the EMF involved in the process of C transfer among hosts. Most studies on CMNs have presented indirect or circumstantial evidence while ruling out other alternative mechanisms. At the same time, 13C-DNA-Stable Isotope Probing (SIP) has been used to identify microbial partners in several plant-microbe systems [35–37]. DNA-SIP allows identifying which organisms utilized a substrate of interest using a stable isotope tracer. If an organism incorporates 13C into its nucleotide sugar bases, then the 13C-DNA can be separated from the 12C-DNA using density-gradient centrifugation and sequenced. Despite its potential, 13C-SIP has not yet been applied in the study of C transfer between trees.
In this work, we established a simplified network of a tree-fungi-tree system to directly identify the EMF species serving as the mediators within the CMNs using a 13C labeling approach. Further, we tested if species relatedness is important for plant-plant C transfer. We planted the gymnosperm Pinus halepensis (Aleppo Pine) and the angiosperm Quercus calliprinos (Palestine oak) in custom-made containers with mesh barriers, allowing CMNs to develop between the trees while prohibiting direct root-root contact. Plant labeling experiments, mostly employ sterile soil and controlled inoculation of one or two EMF species [38]. In contrast, we used natural forest soil, giving the saplings the possibility to form symbiosis with a variety of fungal species. We used saplings of Pinus and Quercus which belong to the most common tree genera globally, populating vast conifer and broadleaf forests (respectively) across temperate, boreal, and sub-tropical biomes. In the Mediterranean woodland, they colonize similar ecological niches [39, 40], and their mixing in forests seems to mutually improve seedling establishment under xeric conditions [41]. Furthermore, they share EMF species [42], raising the possibility for forming CMNs. We used a 13CO2 labeling system followed by a high-resolution tissue sampling regime and DNA-SIP to explore (i) whether C transfer occurred; (ii) if so, at which species combinations; (iii) at what temporal and quantitative dynamics; and finally, (iv) which EMFs were involved in C transfer between neighboring trees. We hypothesized that C transfer occurs between neighboring trees provided that they share EMF species, regardless of their phylogenetic distance, and that EMF are involved in C transfer via the formation of CMNs.
Materials and methods
Plant and soil material
Soil was collected from the Harel Forest, located ca. 4 km south-west of the town of Beit Shemesh, Israel (31◦ 43′ N, 34◦ 57′ E, 320 m elevation). The vegetation comprises local Mediterranean diversity, such as the gymnosperm tree species Pinus halepensis and Cupressus sempervirens, and local Mediterranean angiosperm woody species, such as Quercus calliprinos, Ceratonia siliqua, and Pistacia lentiscus, accompanied by a rich understory of annual plants that thrive from winter to spring. The soil was taken from the topsoil layer (0–15 cm) and no farther than 10 m of a Pinus or Quercus trunk to obtain the native soil mycobiome. To allow proper aeration in the containers, the soil used to transplant the sapling was mixed with 50% sea sand (v/v), and its final texture was: sand 83 ± 1%, silt 9.5 ± 1.5%, clay 7.3 ± 2% (n = 3; ARO, Gilat, Israel; see Supplementary Table S1). Pinus and Quercus saplings at the age of eight and thirteen months (respectively) were collected from KKL-JNF nursery in Eshtaol, Israel, on 15 December 2019. Following transplantion and immediately prior to labeling the height (cm), diameter (mm), and the number of branches were recorded (Supplementary Table S2).
Experimental design
The saplings were planted in 10 custom-made containers. Each container (10 cm × 50 cm × 30 cm depth) was divided equally into three compartments, hereafter referred to as ‘Control,’ ‘Donor,’ and ‘Recipient,’ each containing one sapling. “Recipient” and “Control” denote unlabeled plants, and “Donor” denotes a labeled plant. We acknowledge that the movement of nutrients is hypothesized to be bi-directional; the compartment names do not indicate the direction the nutrients move but rather the expected transfer direction of the label. The Control sapling was transplanted with a polycarbonate sheet separating the belowground compartment entirely from the rest of the container.
Before inserting soil and saplings into the pots, water was filled in the control compartment and left for 24 h to verify that there were no leaks that would allow a passive transfer between donor and control soil compartments. In the center of the container, the Donor sapling was planted, separated from the Recipient sapling with a 35 µm stainless steel mesh net (Xmd metal mesh, Xinxiang, China), to exclude direct contact of the sapling’s roots [21]. The saplings were planted in four species combinations, Pinus-Pinus-Pinus, Pinus-Quercus-Pinus, Quercus-Quercus-Quercus, Quercus-Pinus-Quercus, alternating the middle Donor sapling and the adjacent Control and Recipient saplings. Saplings grew together for seven months. Ten containers totaling thirty plants of similar size and phenotype were chosen for the labeling experiment, eight containers (n = 24 saplings) which were inserted into the labeling system, and two containers serving as “Unlabeled control”, (n = 6 saplings). Saplings were kept at full sunlight and were irrigated throughout the experiment. We irrigated to field capacity at the end of each sampling day, to ensure adequate soil moisture at all times.
Labeling system
A hermetically sealed labeling system explicitly designed for this experiment was built from two parts (Supplementary Fig. S1). (i) A humidity-controlled glovebox (Coy lab products, Grass Lake, MI, USA) with built-in closed-circuit air condition was used to control and monitor the gas mixture’s humidity and temperature, which was introduced to the saplings. To detect 13CO2/12CO2 concentrations, we attached two sensors: PP systems gas analyzer (PP Systems, Amesbury, MA, USA) and G2131-i Picarro cavity ring-down spectrometer (CRDS; Picarro, CA, USA). (ii) A custom-made enclosure was attached to the glovebox built from plexiglass and polyethylene. The enclosure sealed the crowns of eight Donor plants from the surrounding environment. This design included two replicates from each species combination (n = 2), totaling 24 saplings. All three belowground compartments of the container and the crowns of adjacent Recipient and Control saplings were excluded from the enclosure. Two fans (24 W Europlast; Drautal, Austria) were used in opposite directions to create air circulation. The humidity and temperature were monitored using three data logger sensors (EasyLog EL-USB-2-LCD, Lascar Electronics, Wiltshire, UK), two on opposite sides of the enclosure box and one in the glovebox. An external air conditioner (R-YDH-5500, Feishi, Shanghai, China) was used to control the temperature.
13CO2 labeling
Eight containers, 24 saplings, two of each species combination, were inserted into the labeling system, where only the middle Donor sapling crown was covered (Supplementary Fig. S1). The two remaining containers were kept 150 m away from the labeling apparatus and were not labeled, referred to as Unlabeled control, with the Pinus-Pinus-Pinus, Quercus-Quercus-Quercus species combinationsThe labeling started on 13 July 2020 for three consecutive days, starting each day two hours after sunrise, and finishing at sunset, 19:30, totaling 30 h of labeling. On the first day, 45 g of sodium bicarbonate 99% 13C dissolved in chloric acid (Sigma, Rehovot, Israel) was used. On the second and third days, we used gaseous 13CO2 at equivalent amounts (Sigma, Rehovot, Israel). At the beginning of every labeling day, the CO2 concentration was lowered to 90.6 ± 24.2 ppm by emptying the headspace using a vacuum pump (Vacuubrand, CT, USA) while simultaneously flushing it with an 80% N2, 20% O2 mixture (Maxima, Ashdod, Israel). Light intensity fluctuated around 1,500 µmol m−2 s−1 throughout the day, (Li-250A light meter, Li-cor, NE, USA). Temperature and relative humidity were kept at 33 ± 3 °C and 65 ± 7% inside the enclosure, respectively. Leaf CO2 and H2O gas exchange measurements were done on mature leaves using a photosynthesis system (IRGA; GFS-3000, Walz, Effeltrich, Germany). The conditions inside the IRGA cuvette were set to a CO2 level of 400 ppm; flow rate 750 µmol s−1; no temperature or humidity control; and photosynthetically active radiation (PAR) of 1525 ± 75 µmol m−2 s−1). To estimate isotopic signals in the soil compartment, a hole was drilled at a depth of 5 cm in each of the three compartments at each of the ten containers, and a hard-plastic tube (2 cm diameter) with holes was inserted. The tube was attached to the CRDS unit, which determined δ13C respired from the soil compartment. The high sensitivity of CRDS unit inserted into the soil compartments of the three treatments allowed us to verify that no passive transfer occurred due to leakage from the donor compartment. Further details regarding labeling apparatus appear in previous labeling done in our lab [43].
Plant sampling
Plants were sampled and tissues harvested according to the expected amount of label to avoid isotopic contamination during sample handling, from the lowest (i.e., Unlabeled controls) to the highest (i.e., Donor plants). Eleven sampling days, including baseline samples and post-labeling samples, were carried during 36 days. On each day, first-order lateral roots and leaf tissue were taken for analysis from each plant, and on three sampling days stem samples were also collected. Despite the extensive sampling, all samples were negligible in size, about three orders of magnitude smaller than the biomass of the tissue (e.g., 33 mg root tissue sample from a total 10–25 g root biomass, with an even larger shoot biomass of 30–100 g). In addition, sampling was performed with extra care to minimize disturbance to plant and soil, and was uniform across plants. The roots were thoroughly washed on a 1-mm meshed sieve using DDW, and root tips colonized with mycorrhiza (~33 mg each) were separated using sterilized forceps under a binocular and inserted into a 2 ml Eppendorf tube. The tubes were immersed in liquid nitrogen, lyophilized, and stored at −20 °C until DNA extraction. The remaining root (n = 326), leaf (n = 297), and stem (n = 152) samples were dried for 48 h in a 60 °C oven and then ground for the δ13C analysis. During each sampling day total soil respiration and δ13C were measured as explained above.
Plant harvesting and analysis
On 17 August 2020, following the disassembly of the experiment, extensive sampling was carried out to determine the 13C variation in stem, leaf, and root tissues. Each plant was gently separated from the soil and was divided into its components. The soil and roots were thoroughly checked for the existence of mycelial networks (Supplementary Figs. S2 and S3). Five roots were randomly chosen and treated as described above. Stem and leaf samples were taken at two heights, including two leaves representing mature and young leaves. An additional pooled sample of 30 leaves was ground. Afterward, the remaining biomass was divided to above- and belowground, dried for 48 h in a 60 °C oven, and weighed. Ground tissue samples were weighed to 1.2 g and were measured using a combustion module attached to the Picarro G2131-i unit. After dismantling the experiment and removing the plants and soil, the compartments were examined thoroughly to verify that no leakage occurred due to technical failure of the mesh net or polycarbonate sheet separating the compartments.
DNA extraction
Root tips were collected from four P. halepensis Donors and their respective Recipient partners, two P. halepensis and two Q. calliprinos saplings. Two sampling days were chosen, 9 July 2020 as day 0 and 21 July 2020, 9 days post labeling. Thus, these samples represent the pre-labeling and peak-labeling of the Recipients. Root tips were thawed, and DNA was extracted from them using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol with the following modifications: (1) Pretreatment grinding with a bead beater at 4000 RPM (Restch GmbH, Haan, Germany) for 2 min; (2) Suspension of samples in 700 µl CTAB / PVP buffer and incubation in a water bath (65 °C) for 1 h; (3) 600 µl chloroform and centrifuged 10 min at 13,000 × G. Concentrations of extracted dsDNA were measured fluorometrically using Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, USA). The DNA extracts were used for stable isotope probing (SIP) density gradient ultracentrifugation (below).
DNA stable isotope probing (DNA-SIP)
For DNA-SIP, we used a published protocol [22, 44] with the following modifications: 4 ± 1.6 µg DNA samples were loaded onto gradient buffer (GB) to a total volume of 1.15 ml. The GB + DNA solution was mixed with 5 mL of cesium chloride (CsCl, Thermo Scientific, Waltham, USA). The solution refractive index was measured (AR200 digital Reichert, Depew, NY, USA) to a target value of 1.4030 ± 0.000. The final solution was loaded onto quick-seal polypropylene centrifuge tubes (Beckman Coulter, Brea, CA, USA). The tubes were sealed and centrifuged for 39 h at 170,000 × G at 20 °C (WX model ultracentrifuge Sorvall Thermo Fisher Scientific, Waltham, USA) using an NVT 65.2 rotor (Beckman Coulter). Immediately after centrifugation, 16 fractions containing 350 ± 25 µl each were collected from each gradient, and their refractive index was measured. Next, the DNA was precipitated using PEG solution (Polyethylene glycol 6000, Thermo Scientific [45]; and 2 µl GlycoBlue co-precipitate (Thermo Scientific) and eluted in 30 µl of TE-buffer. In each fraction, DNA concentration was measured with Qubit.
ITS2 region amplification and MiSeq sequencing
Root tip DNA from four Donor pines and the reciprocal Recipient and Control pines and oaks were sequenced to elucidate their fungal community. For this purpose, barcoded amplicon sequencing of the fungal ITS2 region [46] was performed on a MiSeq platform (Illumina, San Diego, CA, USA). From each individual, two sets of root tips harvested on day 0 and day 9 were sampled. From each SIP gradient, 11 out of the 16 fractions (corresponding to densities 1.68–1.77 g ml−1) were sequenced, while the terminal 2–3 fractions from each side were discarded. Two separate sequencing libraries were prepared, the 1st library containing the samples collected from four pine Donors and the 2nd of the corresponding Recipient pairs, two oaks, and two pines. PCR amplification and barcoding was done in a two-step procedure. The first PCR had an initial denaturation at 98 °C for 3 min, followed by 32 cycles of 98 °C for 20 s, 50 °C for 30 s and 72 °C for 30 s, and a final extension at 72 °C for 10 min. Each amplification was carried out in a 50 μl reaction mixture containing 25 μl KAPA HiFi ready mix (Eppendorf-5Prime, Gaithersburg, MD, USA), 0.5 μM forward primer, 0.5 μM reverse primer, 5 μl template DNA, and 15 μl nuclease-free water. Fungus specific primers were used, 5.8-Fun and ITS4-Fun (5’-AAC TTT YRR CAA YGG ATC WCT-3’, 5’ -AGC CTC CGC TTA TTG ATA TGC TTA ART-3’, respectively [46]). PCR products were screened for successful amplification using standard gel electrophoresis and quantified using a Qubit dsDNA HS kit (Life Technologies Inc., Gaithersburg, MD, USA). The PCR products were purified using AMPure magnetic beads (Beckman Coulter Inc., Brea, CA, USA), following the manufacturer’s instructions. Samples were quality checked for amplicon size using the Agilent 2200 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). A second PCR step was done to add an adapter and barcode at the INCPM (Weizmann Institute, Rehovot, Israel). Libraries were prepared using DNA CHIP-seq protocol as described [47]. Briefly, 20 ng from each sample were used for library generation. Each sample went through a process of adapter ligation and PCR with cleanups in between. At the end of the process, each library was quantified by Qubit and was brought to the same molar concentration, then mixed by taking the same volume for each library. The final pool was diluted and loaded into the MiSeq instrument. Sequencing was done on a MiSeq instrument using a V3 600 cycles kit, allocating 0.22 M reads per sample (paired-end sequencing).
Processing of sequence data
We used R (R Core Team, 2018, version 4.0.3) and the RStudio IDE for bioinformatics and statistical analysis. The sequences were processed using the amplicon sequencing DADA2 package v. 1.7.9 in R [48]. Shortly, raw sequences were demultiplexed, and both adapters and barcodes were removed from the samples. Sequences were quality-filtered and trimmed. We only used sequences longer than 50 bases with a mean number of expected errors below 2 (maxN = 0, maxEE = c(2,5) minLen = 50 truncQ = 2). Paired-end sequences were merged using the MergePairs function. We then applied a dereplication procedure on each sample independently, using derepFastq function. Finally, all files were combined in one single Fasta file to obtain a single amplicon sequence variant (ASV) data file. We removed singletons (minuniquesize = 2) and de novo chimera sequences using removeBimeraDenovo function against the reference database (UNITE/UCHIME reference datasets v.7.2). Sequences were then clustered, and taxonomic assignment (id = 0.98) was done against the UNITE database. Non-fungal ASVs were removed. To further validate our results, we used Sequencher software (Sequencher 5.4.6, Gene Codes Corp.) to examine if the recipient and donor ASV from the two libraries cluster together, with a minimum match of 97% and a minimum overlap of 100 bp.
Statistical analysis
The analysis of CRDS data, was implemented on the root, leaf, and stem datasets. In cases where the residuals were not normally distributed, we employed a square root-transformation on the original data. We analyzed the data using a split-plot design (using aov function implemented in the car package), where the identity of the pairs of donors (D) and recipients (R), D-Qc | R-Qc, D-Ph | R-Qc, D-Qc | R-Ph, D-Ph | R-Qc, was considered as the between-subject factor. The two containers of each pair (n = 2) were considered as the experimental units, while the division within each experimental unit into the various treatments (Control, Donor, Recipient, n = 3) was considered as a within plot treatment. The measurement days (n = 11) were considered as an additional, random, within-subject factor. The analysis of DNA-SIP data was done with R package Multiple Window High-Resolution Stable Isotope Probing (MW-HR-SIP) as previously described [49], based on the principles of DESeq2. Briefly, four density windows were set: 1.71–1.72, 1.73–1.74, 1.75–1.76, 1.76–1.77, and for each sapling, the unlabeled gradient from day 0 was compared with the matching labeled gradient from day 9. Day 9 was chosen as it was the peak of labeling that appeared in the recipient treatment. If there was a substantial log fold change (after p-value adjustment and correction for multiple comparisons) per the matching fraction at the set density window, it was recorded and manually examined. MW-HR-SIP is based on DESeq2 gene comparison data, uses t-test and corrections for multiple comparisons, and does not test for interaction or include covariates. Further verification of our results was done using the Corncob R package [50]. Corncob is a beta-binomial regression model for microbial taxon abundances, which allows for an association between the variance of a taxon’s abundance and covariates.
Results
Labeled C was transferred belowground between trees across four tree combinations
Across the four Quercus and Pinus combinations and in six out of eight replicates, 13C was found in the Recipient trees’ roots and stems but not in their leaves (Fig. 1). When Pinus was the Donor, more 13C was found in the Recipient roots than with Quercus as a Donor (Supplementary Fig. S4). On day 0, all saplings’ leaves showed natural δ13C signatures, of −29 – −32‰ (Supplementary Fig. S4). Following the three-day labeling, leaves in the labeled Donor trees showed values of 5000‰ and beyond (5000‰ being the CRDS upper detection limit), and after 36 days averaged 1850 ± 804‰. Notably, in all but three Recipient replicates, Recipient and Control saplings’ leaves did not rise above the δ13C natural variation. These samples originated from three separate Pinus trees on several dates: one sample from day 29 (437‰), the other two from day 5 (18‰ and −2‰). These samples represent three samples which exhibited unexpected values out of the 297 total samples analyzed, and thus was concluded that these outliers were probably the result of human error due to contamination during sample collection and handling, and were removed. The points were handled statistically by replacing their value with the average of the individual sapling the day before and after the removed data point. Comparing all treatments of leaf samples, there was a significant difference (F2,108 = 2844, p < 0.001). However, contrasting only the Recipient and Control treatments, we found no significant difference (F1,92 = 0.0001, p > 0.05), indicating no labeling of the Recipient leaves.
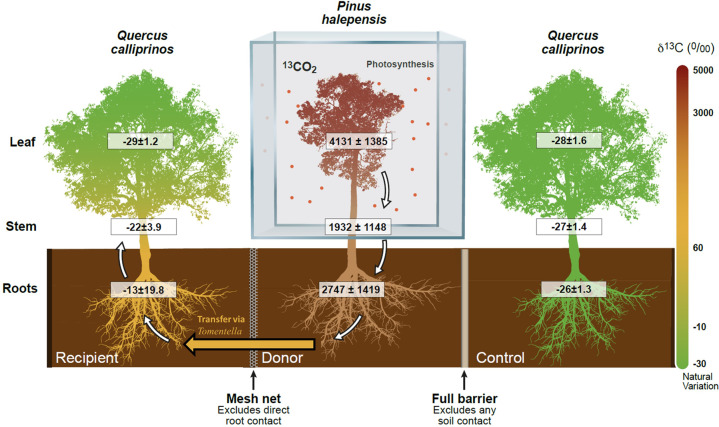
Fig. 1. Evidence for 13C transfer in the Quercus – Pinus – Quercus combination (n = 2).
The orange arrow denotes 13C transfer as it passed across kingdoms, from tree to fungi and on to another tree, facilitated through the EMF specie Tomentella ellisii. White arrows denote 13C allocation within the tree tissues. Values are averages±SE of samples taken along time-points: stem (n = 3), leaves (n = 11) and roots (n = 10).
Labeled C identified in recipient and donor stems and roots but not in control
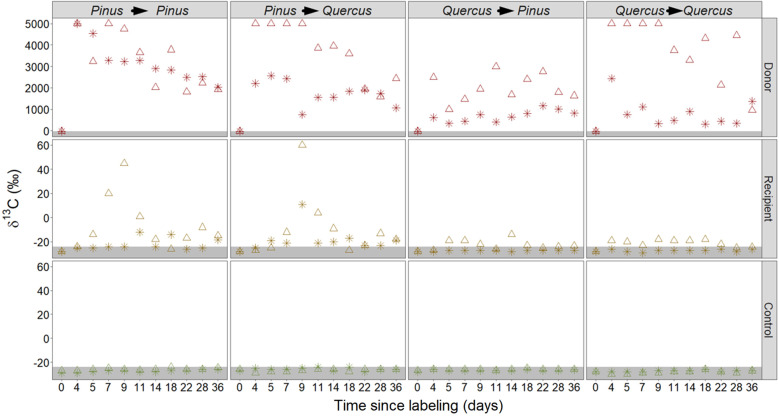
Substantial amounts of 13C were found in the stems of Donors (Fig. 1; Supplementary Fig. S5), 2091 ± 1140‰ averaging all measurement days. Among Recipients, labeling was found on day 18, averaging −21 ± 5.6‰ and decreasing subsequently to −25 ± 2‰ on day 36. The control trees maintained a natural δ13C signature. Comparing all treatments, there was a significant difference (F2,32 = 79, p < 0.001) that prevailed when contrasting the recipient and control treatments only (F1,20 = 7.2, p = 0.015). Donor δ13C increased in roots, 3467 ± 1734‰ at day 4, and 1531 ± 571‰ at day 36 (Fig. 2). In the recipient treatment, a gradual increase of δ13C was observed, peaking at day 9 for both species (7 ± 39‰ in Quercus, −7 ± 34.6‰ in Pinus). Subsequently, there was a decline of δ13C, with day 36 values still above natural variation (−22 ± 4‰ in Quercus, −21 ± 5.3‰ in Pinus). Control treatment showed no increase in δ13C, −27 ± 1.4‰ in Quercus, −26 ± 2.5‰ in Pinus across all measurement days (Table 1). The effects of day and treatment were significant, (Table 1; F10,128 = 3.8, p < 0.0001, F2,128 = 246, p < 0.0001, respectively). The effect of pair combination was not significant (F3,4 = 0.7, p = 0.595). Median tests between Recipient root and corresponding Control roots of the same day found that in 7 out of 10 days (excluding day 0), Recipients and Control trees were significantly different (Supplementary Table S3, see effect sizes in Supplementary Fig. S6). When dismantling the experiment on day 36, the root compartments were inspected thoroughly. In two individual cases, a root breached the neighboring compartment; one Control sapling root was found in the Donor compartment and vice versa. Importantly, corresponding to these breaches, two outliers were found in the data in two Control and Donor roots. The Control that breached the Donor compartment was found to be a single outlier −9‰, and the Donor that breached the Control −12‰. Despite the unintended breach, the actual transfer that was observed reinforces that carbon transfer occurred between the trees. In order to employ a balanced statistical design, the two outlier values (out of n = 326) were removed and an average of the individual sapling results the day before and after were used. The Control saplings, which were adjacent to the labeling apparatus, exhibited, across all tissues and sampling days, δ13C values within natural variation and equivalent to Unlabeled Control treatment, saplings that were separated completely from the labeling apparatus (Supplementary Fig. S7). As root sampling is a destructive measurement that disturbs the soil and mycelium hyphae, we minimized this disturbance by measuring the ratio between 13C and 12C in the gaseous phase of the soil compartment (Supplementary Fig. S8). The three soil compartment denote each of the three treatments, and were measured separately. An increase in the 13C in the soil compartments peaked at the 4th day (18.2 ± 8.7%) for Donors and day 5 (2.66 ± 1.30%) for Recipients, which afterward declined to values similar to unlabeled Control treatment. A linear regression equation was established comparing these respiration proxy values for days where elevated 13C was observed between Donor and Recipient compartments (days 3–7, R2 = 0.64, F(1,80) =147.5, p < 0.001; Supplementary Fig. S9). Additional details are discussed in the Supplementary Information.
Fig. 2. Root δ13C of three treatments: Donor, Recipient, and Control (top to bottom).
Triangles and asterisks denote different biological replicates (n = 2) of each pair combination. Vertical panels represent pair combinations (Donor → Recipient). A gray area marks the δ13C natural variation of −24‰ and below.
Table 1.
The statistical model summarizes the δ13C values in the root tissues (ANOVA on repeated samples).
| Error: Pot ID | Df | Sum Sq | Mean Sq | F value | Pr(>F) |
|---|---|---|---|---|---|
| Pairs | 3 | 12073370 | 4024457 | 0.71 | 0.59 |
| Residuals | 4 | 22689574 | 5672394 |
| Error: Within | Df | Sum Sq | Mean Sq | F value | Pr(>F) |
|---|---|---|---|---|---|
| Days | 10 | 23062398 | 2306240 | 3.87 | 0.0001 *** |
| Treatment | 2 | 293387304 | 146693652 | 246.29 | <2e−16 *** |
| Pairs:Days | 30 | 7193976 | 239799 | 0.40 | 0.99 |
| Pairs:Treatment | 6 | 23684010 | 3947335 | 6.63 | 4.02e−06 *** |
| Days:Treatment | 20 | 45709518 | 2285476 | 3.84 | 1.60e−06 *** |
| Pairs:Days:Treatment | 60 | 14337623 | 238960 | 0.40 | 0.99 |
| Residuals | 128 | 76238157 | 595611 |
Significance codes: <0.001 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05. Df denotes degrees of freedom, Sq denotes square root.
Mycorrhizae amplicon sequence variants were enriched in 13C and colonized both donor and recipient trees
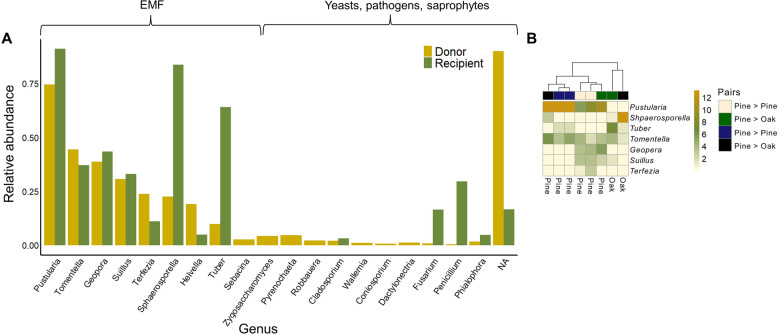
The most abundant species within the ITS2 amplicons of the tree roots were EMF (Fig. 3), reaching 98% of the reads in Donor trees and 90% in the Recipients. Pustularia was the most abundant genus (49% in Donors and 26% in Recipients), and in both libraries, The EMF genera Tomentella, Geopora, Suillus, Terfezia, and Helvella appeared in the top ten. Sphaerosporella and Tuber were notably more common in the Recipient than Donor trees and on Quercus than Pinus roots. After fractionation and sequencing, we compared the relative abundance results of the unlabeled gradient (day 0) and the labeled gradient (day 9) for each sapling (n = 8, four Donors and their four matching Recipients) across all the ASVs (Fig. 4). MW-HR-SIP analysis was performed by defining four density windows for each ASV. MW-HR-SIP compared the normalized abundance of each ASV between the labeled (day 9) and unlabeled (day 0) SIP-gradient of each individual tree, in each density window. The MW-HR-SIP analysis generated 125 Donors’ and 233 Recipients’ significantly differential abundant ASVs. However, despite the significant log fold change found, many of these ASVs had higher relative abundance only in a single SIP fraction, which we assume to be of stochastic origin and to have no biological meaning. In other cases, significant ASVs were detected as such simply because they were all but absent in the control gradients. Hence, these ASVs were ignored. Similar results were obtained after using a prevalence filter.
Fig. 3. The relative abundance of fungal genera on roots of the studied pine and oak trees.
A Rank abundance curve of the relative abundances of fungal genera based on ITS2 amplicon-sequencing. The results are derived from four pine donors and their recipient pairs. The remaining oak donors and their corresponding pairs were not sequenced. B Heat map summarizing the relative abundance of the top seven EMF genera in each of the eight trees that were sequenced. Pairs’ color annotation (top boxes) denotes the donor and recipient tree pairs (Donor → Recipient). Euclidean clustering denotes how the trees clustered together.
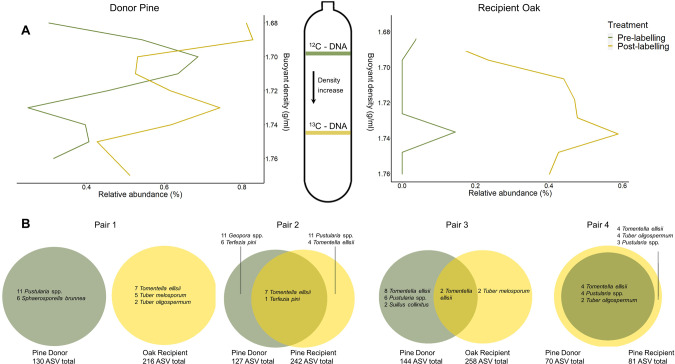
Fig. 4. Labeled carbon in the DNA of mycorrhizal fungi as evidence for their role in belowgruond carbon transfer.
A 13C-DNA-SIP results depicting Tomentella 13C-enriched ASV buoyant density of the fractions compared to the relative abundance; right graph represents DNA from the oak recipient and the left graph from the pine donor; both display an increase of relative abundance in ‘heavy’ (13C) fractions. Pre- and post-labeling are represented by days 0 and 9, respectively. Green and yellow areas highlight the fractions where 13C- and 12C-DNA is expected to be found. B Venn diagrams depicting the number and identity of shared 13C-enriched ASVs of the four pair combinations (donors in gray; recipients in yellow). Total 12C- ASV are shown after prevalence filter and quantitative filters.
To validate our results, further analysis was performed using a different statistical method, the Corncob package. Corncob is a beta-binomial regression model for microbial taxon abundances, which compares an ASV relative abundance with associated covariants of interest. Corncob analysis generated similar significant ASV as MW-HR-SIP with slight variations. The list generated by Corncob of 158 and 168 ASVs for Donor and Recipient trees, respectively, was screened manually to remove false positives, i.e., ASVs that did not show the expected peak shift from unlabeled ‘light’ fractions to labeled ‘heavy’ fractions following labeling. The proportion of false positives was 42% and 56% among donors and recipients, respectively. We note, however, that most of them were either unidentified ASVs, saprophytic, or pathogenic fungal species, and not the dominant EMF species in our system. Tomentella ellisii was identified with labeled carbon in the Donors and Recipients of both the pine-pine and pine-oak pairs (Fig. 4b). Among pines, Pustularia spp., Terfezia pini, and Tuber oligospermum were also identified with 13C. Additional species had labeled carbon either at Recipient or Donor sides (Fig. 4b). We used an additional analysis employing a UPLC-MS/MS protocol on the same fractionated samples of the donor library that were sequenced to ensure the incorporation of 13C atoms within the DNA of the sequenced organisms. In this analysis, each nucleobase (Adenine, Guanine, Cytosine, and Thymine) was examined separately (nucleobase, as illustrated in Supplementary Figs. S10 and S11). An enrichment of +2, +3, and +4 13C atoms was found only in post-labeling samples (Supplementary Table S4). Also, a more significant concentration of all the enrichment atoms was found in the heavier fractions. Further details are provided in the SI.
Discussion
In the present research, we demonstrated an EMF-mediated C transfer between tree saplings, irrespective of phylogenetic relatedness. We used natural forest soil as the inoculum, allowing the formation of diverse CMNs, while a mesh barrier ruled out a direct root-root transfer. The main EMF agent transferring carbon across these CMNs was Tomentella ellisii, the mycorrhizal partner of both pine and oak. Six out of eight tree-pairs and all pair combinations demonstrated transfer of 13C to some extent, indicating that the transfer is not strictly dictated by phylogenetic relatedness.
Leaf (Supplementary Fig. S4), stem (Supplementary Fig. S5), and root (Fig. 2) tissues of the Control treatment did not contain 13C above natural variation, while all the tissues of the Donor trees displayed an increased δ13C signature, a few folds above natural variation. In the Recipient trees, the roots were labeled above natural variation (Fig. 2), and the stem tissues were slightly labeled (Supplementary Figs. S5 and S6), suggesting that the transferred carbon was further distributed in the recipient plant and that the label was not restricted to the EMF mantle that surrounds the root tips, which was one of the main arguments against the idea of plant-plant C transfer (Robinson & Fitter 1999). In contrast, the recipient leaves showed no labeling at all (Supplementary Fig. S4). These results lead us to conclude that 13C is transferred from the roots of the Donor tree to the roots of the Recipient tree, and a small portion of 13C moved against the source-sink gradient to the Recipient stem, as previously shown, albeit without temporal dynamics [32]. Even if some imported C compounds made their way up the plant and into the canopy, they were probably immensely diluted by fresh leaf assimilates, preventing their detection in leaves.
Similar to a review summarizing 47 pulse-labeling studies [26], we found a four-day lag of peak 13CO2 efflux from the soil for pine and oak. In another review [51] focusing on 13C tree labeling, 2.85 days were reported, depending on tree height and phloem structure (the average lag between labeling and efflux from soil was 3.9 ± 0.66 days for gymnosperms, 1.94 ± 0.51 days for angiosperms) placing our results in that range. Labeling done on 2.5-m tall beech trees in field conditions found equivalent temporal 13C dynamics in mycorrhizal roots [52]. Similar 13C labeling done on single Quercus calliprinos and Pinus halepensis saplings in our lab found similar C allocation dynamics, i.e., peak at roots three days post-labeling [43]. The variable that best explained the 13C transfer between trees in our system was the amount of 13C found in the whole root system of the Donor tree (Supplementary Fig. S12). We found no evidence for tree species preference within the CMNs, i.e., carbon moved across species combinations irrespective of the Donor’s or Recipient’s identity. Earlier work found C transfer between phylogenetically distant tree species, Pseudotsuga menziesii and Betula papyrifera, primarily through EMF hyphal pathway and dictated by source-sink relationship [31]. Others showed that closely related sibling pairs exhibited more significant 13C transfer compared with non-sibling pairs [34]. Our results, demonstrating carbon transfer between genetically distant trees, lend further support to the findings of Klein et al. [32] and Rog et al. [33], which demonstrated that the bidirectional transfer occurs between taxonomically distant tree taxa and that the transfer seems to be dictated by EMF that are forming CMNs between mature trees growing in a natural forest.
The pulse labeling coupled with repeated sampling strategy of tissue and respiration allowed us to trace how the 13C was distributed through the Donor tree belowground and onto the Recipient tree. Combining the sequencing and DNA-SIP results, we can create a novel taxonomic list of the main EMF genera involved in C transfer among neighboring trees, pinpointing the exact taxa involved. The EMF genera Pustularia, Terfezia, Tomentella, Tuber, Sphaerosporella, Geopora, and Suillus have been found to directly receive 13C from the pine Donor saplings and integrate the 13C into their DNA. Separate sequencing of the paired Recipients found Pustularia, Terfezia, Tomentella, and Tuber enriched 13C-DNA. Terfezia, Tomentella, and Tuber have been shown to have symbiotic interactions with both pine and oak trees [53–55]. While the host identity of Pustularia is unconfirmed, its function as EMF was demonstrated [56, 57]. Tomentella was found to have 13C enriched DNA in the pine Donor-oak Recipient pair (Fig. 4) and hence is considered the candidate for the formation of CMNs between these distantly related trees. Terfezia, Tomentella, and Tuber were found to have 13C enriched DNA in the pine-Donor pine-Recipient pair and are therefore candidates for forming CMNs between the pines. Intriguingly, two different Tuber EMF species were found in pine and oak trees with 13C enriched DNA, raising the question of whether the C moved across different EMF species forming CMNs (pine → Tuber oligospermum → Tuber X → oak). However, other pathways can explain these results, such as Tuber receiving C from the Recipient tree (tree→fungi→tree→ fungi) or absorbing it through root exudates dispersed in the soil. However, validating such mechanisms requires further observations which are beyond the scope of the current study.
Why were Pustularia, Terfezia, Tomentella, and Tuber species found to transfer C, while other EMF species that were present did not? The DNA-SIP allows us to differentiate between fungal species that were present and metabolically active, and those that came up solely in amplicon sequencing, which only identifies presence or absence. Given the natural soil inoculum that was used and that saplings were well irrigated, a fungal community proliferated, similar in its composition to that of the natural forest (Rog et al. unpublished). In turn, extra-radical mycelium was formed, resulting in ASV sequences that do not necessarily indicate function. Exploring EMF divergences such as different exploration types [58], evolutionary ecology and phylogenetic affinities [59, 60], and generalist vs. specialist strategies [61], we speculate that in our system the generalist, short exploration type fungi dominated and connected dissimilar hosts. Interestingly, the Pezizales order, which was dominant in our findings, originated 150 Ma ago, around the anticipated evolution of EMF in plants. Various Peziza genera form a biotrophic relationship with facultative saprophytic lifestyles are common in arid and semiarid regions, and proliferate in post-fire environments which frequently occur in the Mediterranean. The same EMF species that transfer C in our system inhabited mature trees’ roots in the natural forest ecosystem which the soil was taken from (Rog et al. unpublished), including Terfezia pini, Tomentella ellisii, Suillus collinitus, Tuber melosporum, and Tuber oligospermum. However, these EMF species in the forest did not overlap between the mature pine and oak trees (albeit Inocybe multifolia and Tricholoma terreum did so). We presume this is due to 1. Saplings possibly being more opportunistic, forming symbiotic interactions with a broader range of symbionts to establish fitness, whereas mature trees favor more specific interactions. 2. Our experimental system might have favored short or contact EMF species interactions.
The SIP analysis was performed on root tissues nine days after the labeling, matching the peak of 13C in Recipient roots. This period is long enough for carbon to be transported to root tips [62] and be assimilated into microorganisms in the rhizosphere [63, 64], yet short enough so that the 13C does not substantially leak into saprophytic communities. Long incubation times bear the risk of labeling community members that do not perform the metabolic activity in question: As organisms are linked through trophic interactions, labeled C will eventually spread among multiple trophic levels (cross-feeding). In addition, during sample preparation, we thoroughly washed the roots and only sampled tips that had ectomycorrhizal structures (i.e., mantle, Hartig-net;). The sampling strategy and timing of the SIP at the ninth-day post-labeling helped us avoid a common bias of DNA-SIP, namely, cross-feeding. This is further supported by the lower abundance of saprophytic sequences, compared to EMF, in all our samples (Fig. 3). Still, we cannot completely rule out the option of C transfer through soil (see below). The additional UPLC-MS/MS analysis performed here [65] is an independent analysis of the 13C-DNA-SIP. The results validated that enrichment levels of +2, +3, and +4 13C atoms were found only in post-labeling gradients (Supplementary Figs. S10 and S11). Furthermore, the denser fractions, where the 13C-DNA was expected to drift to after ultracentrifugation, had a larger quantity of enrichment levels than the low-density fractions, where 12C-DNA was found. These results affirm that the DNA-SIP was performed successfully and that the potential bias of DNA-SIP caused by relative GC content of the DNA [66] did not lead to a misinterpretation of the data.
While most of the studies in the field have been dedicated to studying C transfer through CMNs, there are other plausible explanations for how C is being transferred among trees. Other microorganisms might have been involved in the process of 13C transfer, and, in addition, C might move by passive diffusion. These mechanisms are non-mutually exclusive and might co-occur at different spatial and temporal scales. While the current design cannot rule out these other mechanisms, we can inquire about the probability of their role in the temporal timeline 13C appeared in the system. 13CO2 respiration appeared in the recipient compartment as early as three days post-labeling and as early as four days in roots. One possible mechanism of C transfer is direct root grafting among trees. However, this option is ruled out by most CMNs studies, including ours, by using a dense mesh-net control that prevents root to root interaction [21]. Another mechanism of C transfer involves other microorganisms such as bacteria living in the rhizosphere while feeding on root exudates [67]. As elegantly shown by Gorka et al., [28], EMF can receive photosynthetically derived C and further transfer it to bacteria in the soil adjacent to hyphal tips (i.e., hyphosphere). These complex interactions can occur via direct symbiosis [68] or indirectly through C turnover in the soil. For bacteria to be the main C mediators, the bacteria need to absorb 13C exudates secreted by a donor root, finish its life cycle, degrade, and be absorbed by at least a few other microorganisms to bridge over the few millimeters distance between the donor and recipient roots. Bacterial turnover in the soil is a complex interaction involving many factors [69], and separate 16S qPCR or DNA-SIP analysis must be done to understand their role. We speculate that their involvement to be neglectable because the temporal processes that need to occur for the bacteria to transfer C to neighboring trees do not align with the temporal timescale 13C that appeared in the recipient compartment. Lastly, passive C diffusion between Donor and Recipient compartment through the soil matrix is another possible mechanism for C transfer, which requires the uptake of C from the soil by the tree roots. However, evidence for such phenomena in mature forest trees is scarce. We presume passive diffusion of C does not add a significant contribution in the timescale we found 13C in the recipient compartment. Although dissolved inorganic carbon (DIC) processes are instantaneous, they depend on pH, water availability, and temperature. Water infiltration needs to be calculated to convert rates of change in DIC to a function of distance by time. We calculated 72–100 cm year−1 i.e., 0.8–1.1 cm in four days (regression equation; Precipitation (P) = 510 mm year−1; infiltration rate (cm year−1) = 0.4057(P) − 107.13; R² = 0.96 with n = 4. value derived from Beit Shemesh, where the soil was taken from, Qubaja unpublished data). This calculation is adapted from Carmi et al. [70], which calculated the rate in a drier pine forest. In this study, we used similar soil and tree saplings derived from a natural system. Mixing with sand (see Methods) possibly increased the abovementioned rate, since infiltration rate in sand is maximal, and hence the opportunity for C dissolution should be lower. Therefore, it seems that passive diffusion is a few orders of magnitudes slower than when 13C appeared in the recipient compartment. Moreover, all the experimental units contained the same soil mixture and were watered to the same extent, the fact that we did not observe any transfer of C in some of our mesocosms deteriorates passive C diffusion from being the main mechanism for C transfer in our system.
In the broader context of natural forest ecology, questions arise regarding the ecophysiological and ecological significance of the inter-plant C transfer through CMNs. More specifically, what could be the significance of the small amounts of C transferred from donor trees to recipient trees for the carbon balance of the latter? Despite the use of natural soil inoculum in our microcosm design, we tread lightly when wishing to interpret our results in the broader forest ecology context, considering that saplings age and size, irrigation, and the fixed distance between plants are far from representing the forest. Still, we showed that C transfer through mycorrhiza increased the δ13C of recipient saplings from −26‰ to −13‰ in roots, and from −27‰ to −22‰ in the stem (Fig. 1). Using a simple two-end linear mixing model, both of these increases are explained by an import of 0.5% of root C from the donor. This value was maintained across the different species combinations in our experiment, despite the variations among them. Considering that root and stem biomass were 22 and 56 g, respectively, this imported C fraction amounts to 1.1 g and 2.8 g in roots and stem, respectively. These estimates are smaller than those calculated for a 5-year labeling in a forest, where carbon transfer accounted for 4% of tree carbon uptake [33]. Importantly, pulse labeling experiments (as described here) are useful for capturing short-term dynamics [43], yet are limited in their ability to decipher long-term carbon allocation [71]. Therefore, we assume that at the long-term, the rate of imported carbon is higher than 0.5%. Nevertheless, even small amounts of C import might serve to alleviate the EMF partnership C cost of the recipient tree at the local root level [72]. Increased fitness of these roots may play an important role in survival of saplings suffering low C supply due to growth in the shade of older trees [73]. Alternatively, if the transfer is in the form of amino acids or other nitrogen-containing compounds (as would be expected in a mycorrhizal association; [72], C is rather a by-product of nitrogen transfer and hence low in amount. These aspects are yet unresolved and are the topic of follow-up manipulation experiments.
The importance of EMF symbiosis to the balanced functioning of forest ecosystems is well established and unquestionable. However, our understanding stops at the plant-fungi relationship, as data are limited on how these connections distribute further and scale to form networks. For example, Van Der Heijden and Horton [74] elegantly raised the question of “who dictates the symbiotic interaction among plants and their fungal partners?”; is it a “socialist” relationship where both the plant and fungi have equal opportunities and nutrients are evenly distributed, or rather a “capitalist” network where the plant establishes and nourishes the networks, solely controlling the nutrient profit? The first step in shedding light on these essential questions is identifying the key players in this symbiotic relationship, which we successfully achieved in the study. Follow-up studies focus on experimental manipulations to identify the ecological significance of the C being transferred via CMNs. By improving our knowledge of these key players’ identity and ecological role, we will better comprehend the interactions shaping forest biomes.
Supplementary information
Acknowledgements
The authors wish to thank Ido Rog (Weizmann TreeLab) for help with the labeling system; Ishai Sher (WIS) for his help with the graphic design; and Alexander Brandis (WIS) for his help with performing MSMS analysis. RA was supported by MEYS (EF16_013/0001782 - SoWa Ecosystems Research). The study was funded by the European Research Council project RHIZOCARBON, granted to TK.
Author contributions
TK, SLL and RC conceived and designed the experiment. RC and SLL performed the experiment and analyzed the data under the guidance of TK. SIP procedures and analysis were guided by RA. RC wrote the paper with SLL and TK, and all authors contributed substantially to revisions.
Data availability
Sequences were submitted to the National Center for Biotechnology Information Sequence Read Archive with the accession codes: Bioproject PRJNA777920.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Rotem Cahanovitc, Stav Livne-Luzon.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01193-z.
References
- 1.Steidinger BS, Crowther TW, Liang J, Van Nuland ME, Werner GD, Reich PB, et al. Climatic controls of decomposition drive the global biogeography of forest-tree symbioses. Nature. 2019;569:404–8. doi: 10.1038/s41586-019-1128-0. [DOI] [PubMed] [Google Scholar]
- 2.Finlay RD. Ecological aspects of mycorrhizal symbiosis: with special emphasis on the functional diversity of interactions involving the extraradical mycelium. J Exp Bot. 2008;59:1115–26. doi: 10.1093/jxb/ern059. [DOI] [PubMed] [Google Scholar]
- 3.Smith SE, Read D. Nitrogen mobilization and nutrition in ectomycorrhizal plants. Mycorrhizal Symbiosis. 2008;321–48. 10.1016/b978-012370526-6.50011-8.
- 4.Osonubi O, Mulongoy K, Awotoye OO, Atayese MO, Okali DUU. Effects of ectomycorrhizal and vesicular-arbuscular mycorrhizal fungi on drought tolerance of four leguminous woody seedlings. Plant Soil. 1991;136:131–43. doi: 10.1007/BF02465228. [DOI] [Google Scholar]
- 5.Mayerhofer W, Schintlmeister A, Dietrich M, Gorka S, Wiesenbauer J, Martin V, et al. Ectomycorrhizal fungi induce systemic resistance against insects on a nonmycorrhizal plant in a CERK1-dependent manner. N Phytol. 2020;228:728–40. doi: 10.1111/nph.16715. [DOI] [PubMed] [Google Scholar]
- 6.Mayerhofer W, Schintlmeister A, Dietrich M, Gorka S, Wiesenbauer J, Martin V, et al. Mycorrhizal networks: mechanisms, ecology and modelling. Fungal Biol Rev. 2012;26:39–60. doi: 10.1016/j.fbr.2012.01.001. [DOI] [Google Scholar]
- 7.Molina R, Horton TR. Mycorrhiza specificity: its role in the development and function of common mycelial networks. Mycorrhizal Netw. 2015. 10.1007/978-94-017-7395-9_1.
- 8.Van der Heijden MGA, Martin FM, Selosse MA, Sanders IR. Mycorrhizal ecology and evolution: the past, the present, and the future. N Phytol. 2015;205:1406–23. doi: 10.1111/nph.13288. [DOI] [PubMed] [Google Scholar]
- 9.Brundrett MC. Coevolution of roots and mycorrhizas of land plants. N Phytol. 2002;154:275–304. doi: 10.1046/j.1469-8137.2002.00397.x. [DOI] [PubMed] [Google Scholar]
- 10.Linkies A, Graeber K, Knight C, Leubner-Metzger G. The evolution of seeds. N Phytol. 2010;186:817–31. doi: 10.1111/j.1469-8137.2010.03249.x. [DOI] [PubMed] [Google Scholar]
- 11.Nara K. Ectomycorrhizal networks and seedling establishment during early primary succession. N Phytol. 2006;169:169–78. doi: 10.1111/j.1469-8137.2005.01545.x. [DOI] [PubMed] [Google Scholar]
- 12.Horton TR, Molina R, Hood K. Douglas-fir ectomycorrhizae in 40- and 400-year-old stands: mycobiont availability to late successional western hemlock. Mycorrhiza. 2005;15:393–403. doi: 10.1007/s00572-004-0339-9. [DOI] [PubMed] [Google Scholar]
- 13.Teste FP, Simard SW, Durall DM, Guy RD, Jones MD, Schoonmaker AL. Access to mycorrhizal networks and roots of trees: importance for seedling survival and resource transfer. Ecology. 2009;90:2808–22. doi: 10.1890/08-1884.1. [DOI] [PubMed] [Google Scholar]
- 14.Teste FP, Simard SW. Mycorrhizal networks and distance from mature trees alter patterns of competition and facilitation in dry Douglas-fir forests. Oecologia. 2008;158:193–203. doi: 10.1007/s00442-008-1136-5. [DOI] [PubMed] [Google Scholar]
- 15.Egerton-Warburton LM, Querejeta JI, Allen MF. Common mycorrhizal networks provide a potential pathway for the transfer of hydraulically lifted water between plants. J Exp Bot. 2007;58:1473–83. doi: 10.1093/jxb/erm009. [DOI] [PubMed] [Google Scholar]
- 16.Wallander H, Ekblad A. The Importance of ectomycorrhizal networks for nutrient retention and carbon sequestration in forest ecosystems. Mycorrhizal Netw. 2015;69–90. 10.1007/978-94-017-7395-9_3.
- 17.Song YY, Simard SW, Carroll A, Mohn WW, Zeng RS. Defoliation of interior Douglas-fir elicits carbon transfer and stress signalling to ponderosa pine neighbors through ectomycorrhizal networks. Sci Rep. 2015;5:1–9. doi: 10.1038/srep08495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selosse MA, Richard F, He X, Simard SW. Mycorrhizal networks: des liaisons dangereuses? Trends Ecol Evol. 2006;21:621–8. doi: 10.1016/j.tree.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Robinson D, Fitter A. The magnitude and control of carbon transfer between plants linked by a common mycorrhizal network. J Exp Bot. 1999;50:9–13. doi: 10.1093/jxb/50.330.9. [DOI] [Google Scholar]
- 20.Hoeksema JD. Experimentally testing effects of mycorrhizal networks on plant-plant interactions and distinguishing among mechanisms. Mycorrhizal Netw. 2015;255–77. 10.1007/978-94-017-7395-9_9.
- 21.Teste FP, Karst J, Jones MD, Simard SW, Durall DM. Methods to control ectomycorrhizal colonization: effectiveness of chemical and physical barriers. Mycorrhiza. 2006;17:51–65. doi: 10.1007/s00572-006-0083-4. [DOI] [PubMed] [Google Scholar]
- 22.Graves JD, Watkins NK, Fitter AH, Robinson D, Scrimgeour C. Intraspecific transfer of carbon between plants linked by a common mycorrhizal network. Plant Soil. 1997;192:153–9. doi: 10.1023/A:1004257812555. [DOI] [Google Scholar]
- 23.Wu B, Nara K, Hogetsu T. Can 14C-labeled photosynthetic products move between Pinus densiflora seedlings linked by ectomycorrhizal mycelia? N Phytol. 2001;149:137–46. doi: 10.1046/j.1469-8137.2001.00010.x. [DOI] [PubMed] [Google Scholar]
- 24.Bever JD, Dickie IA, Facelli E, Facelli JM, Klironomos J, Moora M, et al. Rooting theories of plant community ecology in microbial interactions. Trends Ecol Evol. 2010;25:468–78. doi: 10.1016/j.tree.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheublin TR, Van Logtestijn RSP, Van Der Heijden MGA. Presence and identity of arbuscular mycorrhizal fungi influence competitive interactions between plant species. J Ecol. 2007;95:631–8. doi: 10.1111/j.1365-2745.2007.01244.x. [DOI] [Google Scholar]
- 26.Epron D, Bahn M, Derrien D, Lattanzi FA, Pumpanen J, Gessler A, et al. Pulse-labelling trees to study carbon allocation dynamics: a review of methods, current knowledge and future prospects. Tree Physiol. 2012;32:776–98. doi: 10.1093/treephys/tps057. [DOI] [PubMed] [Google Scholar]
- 27.Whiteside MD, Werner GD, Caldas VE, Padje A, Dupin SE, Elbers B, et al. Mycorrhizal fungi respond to resource inequality by moving phosphorus from rich to poor patches across networks. Curr Biol. 2019;29:2043–50.e8. doi: 10.1016/j.cub.2019.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorka S, Dietrich M, Mayerhofer W, Gabriel R, Wiesenbauer J, Martin V, et al. Rapid transfer of plant photosynthates to soil bacteria via ectomycorrhizal hyphae and its interaction with nitrogen availability. Front Microbiol. 2019;10:1–20. doi: 10.3389/fmicb.2019.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leake JR, Donnelly DP, Saunders EM, Boddy L, Read DJ. Rates and quantities of carbon flux to ectomycorrhizal mycelium following 14C pulse labeling of Pinus sylvestris seedlings: effects of litter patches and interaction a wood-decomposer fungus. Tree Physiol. 2001;21:71–82. doi: 10.1093/treephys/21.2-3.71. [DOI] [PubMed] [Google Scholar]
- 30.Kiers ET, Duhamel M, Beesetty Y, Mensah JA, Franken O, Verbruggen E, et al. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science. 2011;333:880–2. doi: 10.1126/science.1208473. [DOI] [PubMed] [Google Scholar]
- 31.Simard SW, Perry DA, Jones MD, Myrold DD, Durall DM, Molina R. Net transfer of carbon between ectomycorrhizal tree species in the field. Nature. 1997;388:579–82. doi: 10.1038/41557. [DOI] [Google Scholar]
- 32.Klein T, Siegwolf RTW, Körner C. Belowground carbon trade among tall trees in a temperate forest. Science. 2016;1500:15–8. doi: 10.1126/science.aad6188. [DOI] [PubMed] [Google Scholar]
- 33.Rog I, Rosenstock NP, Körner C, Klein T. Share the wealth: trees with greater ectomycorrhizal species overlap share more carbon. Mol Ecol. 2020;29:2321–33. doi: 10.1111/mec.15351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pickles BJ, Wilhelm R, Asay AK, Hahn AS, Simard SW, Mohn WW. Transfer of 13C between paired Douglas-fir seedlings reveals plant kinship effects and uptake of exudates by ectomycorrhizas. N Phytol. 2017;214:400–11. doi: 10.1111/nph.14325. [DOI] [PubMed] [Google Scholar]
- 35.Lu Y, Conrad R. In situ stable isotope probing of methanogenic Archaea in the rice rhizosphere. Science. 2005;309:1088–90. doi: 10.1126/science.1113435. [DOI] [PubMed] [Google Scholar]
- 36.Haichar Z, Heulin T, Guyonnet JP, Achouak W. Science direct stable isotope probing of carbon flow in the plant holobiont. Curr Opin Biotechnol. 2016;41:9–13. doi: 10.1016/j.copbio.2016.02.023. [DOI] [PubMed] [Google Scholar]
- 37.Sietiö OM, Tuomivirta T, Santalahti M, Kiheri H, Timonen S, Sun H, et al. Ericoid plant species and Pinus sylvestris shape fungal communities in their roots and surrounding soil. N Phytol. 2018;218:738–51. doi: 10.1111/nph.15040. [DOI] [PubMed] [Google Scholar]
- 38.Sapes G, Demaree P, Lekberg Y, Sala A. Plant carbohydrate depletion impairs water relations and spreads via ectomycorrhizal networks. N Phytol. 2021;229:3172–83. doi: 10.1111/nph.17134. [DOI] [PubMed] [Google Scholar]
- 39.Sheffer E. A review of the development of Mediterranean pine-oak ecosystems after land abandonment and afforestation: are they novel ecosystems? Ann Sci. 2012;69:429–43. doi: 10.1007/s13595-011-0181-0. [DOI] [Google Scholar]
- 40.Ajbilou R, Marañón T, Arroyo J. Ecological and biogeographical analyses of Mediterranean forests of northern Morocco. Acta Oecologica. 2006;29:104–13. doi: 10.1016/j.actao.2005.08.006. [DOI] [Google Scholar]
- 41.Loudermilk E, Hiers J, Pokswinski S, O’Brien JJ, Barnett A, Mitchell RJ. The path back: Oaks (Quercus spp.) facilitate longleaf pine (Pinus palustris) seedling establishment in xeric sites. Ecosphere. 2016;7:1–14. [Google Scholar]
- 42.Hynes MM, Smith ME, Zasoski RJ, Bledsoe CS. A molecular survey of ectomycorrhizal hyphae in a California Quercus-Pinus woodland. Mycorrhiza. 2010;20:265–74. doi: 10.1007/s00572-009-0281-y. [DOI] [PubMed] [Google Scholar]
- 43.Rog I, Jakoby G, Klein T. Forest ecology and management carbon allocation dynamics in conifers and broadleaved tree species revealed by pulse labeling and mass balance. Ecol Manag. 2021;493:119258. doi: 10.1016/j.foreco.2021.119258. [DOI] [Google Scholar]
- 44.Jia Z, Cao W, Herna M. DNA-Based stable isotope probing. Springer. 2019;2046:17–29. doi: 10.1007/978-1-4939-9721-3_2. [DOI] [PubMed] [Google Scholar]
- 45.Neufeld JD, Vohra J, Dumont MG, Lueders T, Manefield M, Friedrich MW, et al. DNA stable-isotope probing. Nat Protoc. 2007;2:860–6. doi: 10.1038/nprot.2007.109. [DOI] [PubMed] [Google Scholar]
- 46.Taylor DL, Walters WA, Lennon NJ, Bochicchio J, Krohn A, Caporaso JG, et al. Accurate estimation of fungal diversity and abundance through improved lineage-specific primers optimized for Illumina amplicon sequencing. Appl Environ Microbiol. 2016;82:7217–26. doi: 10.1128/AEM.02576-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blecher-Gonen R, Barnett-Itzhaki Z, Jaitin D, Amann-Zalcenstein D, Lara-Astiaso D, Amit I. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat Protoc. 2013;8:539–54. doi: 10.1038/nprot.2013.023. [DOI] [PubMed] [Google Scholar]
- 48.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buckley DH, Barnett SE, Youngblut ND. Data analysis for DNA stable isotope probing experiments using multiple window high-resolution SIP Chapter 9. Springer. 2019;2046:44–5. doi: 10.1007/978-1-4939-9721-3_9. [DOI] [PubMed] [Google Scholar]
- 50.Martin BD, Witten D, Willis AD. Modeling microbial abundances and dysbiosis with beta-binomial regression. Ann Appl Stat. 2020;14:94–115. doi: 10.1214/19-AOAS1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuzyakov Y, Gavrichkova O. time lag between photosynthesis and carbon dioxide efflux from soil: a review of mechanisms and controls. Glob Chang Biol. 2010;16:3386–406. doi: 10.1111/j.1365-2486.2010.02179.x. [DOI] [Google Scholar]
- 52.Hagedorn F, Joseph J, Peter M, Luster J, Pritsch K, Geppert U, et al. Recovery of trees from drought depends on belowground sink control. Nat Plants. 2016;2:1–5. doi: 10.1038/nplants.2016.111. [DOI] [PubMed] [Google Scholar]
- 53.Moreno-Arroyo B, Infante F, Pulido E, Gómez J. The biogeography and taxonomy of Tuber oligospermum (Tul. and C. Tul.) Trappe (Ascomycota) Cryptogam Mycol. 2000;21:147–52. doi: 10.1016/S0181-1584(00)00115-9. [DOI] [Google Scholar]
- 54.Buscardo E, Rodríguez-Echeverría S, Martín MP, De Angelis P, Pereira JS, Freitas H. Impact of wildfire return interval on the ectomycorrhizal resistant propagules communities of a Mediterranean open forest. Fungal Biol. 2010;114:628–36. doi: 10.1016/j.funbio.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 55.Louro R, Santos-Silva C, Nobre T. What is in a name? Terfezia classification revisited. Fungal Biol. 2019;123:267–73. doi: 10.1016/j.funbio.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 56.Tedersoo L, Arnold AE, Hansen K. Novel aspects in the life cycle and biotrophic interactions in Pezizomycetes (Ascomycota, Fungi) Mol Ecol. 2013;22:1488–93. doi: 10.1111/mec.12224. [DOI] [PubMed] [Google Scholar]
- 57.Tedersoo L, Smith ME. Lineages of ectomycorrhizal fungi revisited: Foraging strategies and novel lineages revealed by sequences from belowground. Fungal Biol Rev. 2013;27:83–99. doi: 10.1016/j.fbr.2013.09.001. [DOI] [Google Scholar]
- 58.Agerer R. Fungal relationships and structural identity of their ectomycorrhizae. Mycol Prog. 2006;5:67–107. doi: 10.1007/s11557-006-0505-x. [DOI] [Google Scholar]
- 59.Tedersoo L, May TW, Smith ME. Ectomycorrhizal lifestyle in fungi: global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza. 2010;20:217–63. doi: 10.1007/s00572-009-0274-x. [DOI] [PubMed] [Google Scholar]
- 60.Miyauchi S, Kiss E, Kuo A, Drula E, Kohler A, Sánchez-García M, et al. Large-scale genome sequencing of mycorrhizal fungi provides insights into the early evolution of symbiotic traits. Nat Commun. 2020;11:1–17. doi: 10.1038/s41467-020-18795-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bruns TD, Bidartondo MI, Taylor DL. Host specificity in ectomycorrhizal communities: what do the exceptions tell us? Integr Comp Biol. 2002;42:352–9. doi: 10.1093/icb/42.2.352. [DOI] [PubMed] [Google Scholar]
- 62.Pumpanen JS, Heinonsalo J, Rasilo T, Hurme KR, Ilvesniemi H. Carbon balance and allocation of assimilated CO2 in Scots pine, Norway spruce, and Silver birch seedlings determined with gas exchange measurements and 14C pulse labelling. Trees Struct Funct. 2009;23:611–21. doi: 10.1007/s00468-008-0306-8. [DOI] [Google Scholar]
- 63.Heinonsalo J, Pumpanen J, Rasilo T, Hurme KR, Ilvesniemi H. Carbon partitioning in ectomycorrhizal Scots pine seedlings. Soil Biol Biochem. 2010;42:1614–23. doi: 10.1016/j.soilbio.2010.06.003. [DOI] [Google Scholar]
- 64.Wallander H, Göransson H, Rosengren U. Production, standing biomass and natural abundance of 15N and 13C in ectomycorrhizal mycelia collected at different soil depths in two forest types. Oecologia. 2004;139:89–97. doi: 10.1007/s00442-003-1477-z. [DOI] [PubMed] [Google Scholar]
- 65.Wilhelm R, Szeitz A, Klassen TL, Mohn WW. Sensitive, efficient quantitation of 13C-enriched nucleic acids via ultrahigh-performance liquid chromatography-tandem mass spectrometry for applications in stable isotope probing. Appl Environ Microbiol. 2014;80:7206–11. doi: 10.1128/AEM.02223-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schildkraut CL, Marmur J, Doty P. Determination of the base composition of deoxyribonucleic acid from its buoyant density in CsCl. J Mol Biol. 1962;4:430–43. doi: 10.1016/S0022-2836(62)80100-4. [DOI] [PubMed] [Google Scholar]
- 67.Jakoby G, Rog I, Megidish S, Klein T. Enhanced root exudation of mature broadleaf and conifer trees in a Mediterranean forest during the dry season. Tree Physiol. 2020;40:1595–605. doi: 10.1093/treephys/tpaa092. [DOI] [PubMed] [Google Scholar]
- 68.Meier IC, Pritchard SG, Brzostek ER, Mccormack ML, Phillips RP. The rhizosphere and hyphosphere differ in their impacts on carbon and nitrogen cycling in forests exposed to elevated CO2. N Phytol. 2015;205:1164–74. doi: 10.1111/nph.13122. [DOI] [PubMed] [Google Scholar]
- 69.Ranjard L, Dequiedt S, Prévost-Bouré NC, Thioulouse J, Saby NPA, Lelievre M, et al. Turnover of soil bacterial diversity driven by wide-scale environmental heterogeneity. Nat. Commun. 2013;4:1–10. doi: 10.1038/ncomms2431. [DOI] [PubMed] [Google Scholar]
- 70.Carmi I, Yakir D, Yechieli Y, Kronfield J, Stiller M. Variations in the isotopic composition of dissolved inorganic carbon in the unsaturated zone of a semi-arid region. Radiocarbon. 2015;57:397–406. doi: 10.2458/azu_rc.57.18356. [DOI] [Google Scholar]
- 71.Klein T, Hoch G. Tree carbon allocation dynamics determined using a carbon mass balance approach. N Phytol. 2015;205:147–59. doi: 10.1111/nph.12993. [DOI] [PubMed] [Google Scholar]
- 72.Mayerhofer W, Schintlmeister A, Dietrich M, Gorka S, Wiesenbauer J, Martin V, et al. Recently photoassimilated carbon and fungus-delivered nitrogen are spatially correlated in the ectomycorrhizal tissue of Fagus sylvatica. N Phytol. 2021;232:2457–74. doi: 10.1111/nph.17591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fraser EC, Lieffers VJ, Landhäusser SM. Carbohydrate transfer through root grafts to support shaded trees. Tree Physiol. 2006;26:1019–23. doi: 10.1093/treephys/26.8.1019. [DOI] [PubMed] [Google Scholar]
- 74.Van Der Heijden MGA, Horton TR. Socialism in soil? the importance of mycorrhizal fungal networks for facilitation in natural ecosystems. J Ecol. 2009;97:1139–50. doi: 10.1111/j.1365-2745.2009.01570.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences were submitted to the National Center for Biotechnology Information Sequence Read Archive with the accession codes: Bioproject PRJNA777920.