Abstract
Diplonemids are one of the most abundant groups of heterotrophic planktonic microeukaryotes in the world ocean and, thus, are likely to play an essential role in marine ecosystems. So far, only few species have been introduced into a culture, allowing basic studies of diplonemid genetics, morphology, ultrastructure, metabolism, as well as endosymbionts. However, it remains unclear whether these heterotrophic flagellates are parasitic or free-living and what are their predominant dietary patterns and preferred food items. Here we show that cultured diplonemids, maintained in an organic-rich medium as osmotrophs, can gradually switch to bacterivory as a sole food resource, supporting positive growth of their population, even when fed with a low biovolume of bacteria. We further observed remarkable differences in species-specific feeding patterns, size-selective grazing preferences, and distinct feeding strategies. Diplonemids can discriminate between low-quality food items and inedible particles, such as latex beads, even after their ingestion, by discharging them in the form of large waste vacuoles. We also detected digestion-related endogenous autofluorescence emitted by lysosomes and the activity of a melanin-like material. We present the first evidence that these omnipresent protists possess an opportunistic lifestyle that provides a considerable advantage in the generally food resource-limited marine environments.
Subject terms: Microbiology, Applied microbiology
Introduction
As part of the largest food web on the planet, marine microbial eukaryotes lay a foundation for global biogeochemical cycles, ultimately sustaining life in the ocean and controlling the atmosphere’s composition [1]. Together with prokaryotes, they account for about 70% of the total biomass in the marine environment [2] and, through a multitude of interactions with other microeukaryotes, bacteria and viruses, form highly complex microbial food webs [3]. Consequently, they play a pivotal role in driving primary and secondary production and degradation of organic material and nutrient recycling [4]. Despite their importance, the remarkable diversity of protist-dominated marine microeukaryotes, particularly their most abundant heterotrophic components, has been uncovered only recently [5–7].
Although previously overlooked, recent environmental sequencing studies identified diplonemids (Diplonemea) as one of the major groups of protists populating various habitats of the world ocean [6–10], while they seem to be rarer in freshwater bodies [11]. They inhabit virtually every niche in the water column, including anoxic regions, yet predominate in the mesopelagic and bathypelagic zones [12], as well as in the sediment surfaces [13]. First discovered in 1913 by Griessmann [14], fewer than two dozen diplonemids have been cultivated since then, allowing detailed studies of their morphology and ultrastructure [15–19]. Their uniquely inflated mitochondrial genome [20] and massively edited mitochondrial transcripts have been described to considerable detail [21], although complete nuclear genomes remain unavailable, largely because of their repetitive nature. The genomic complexity of many protists significantly limits our attempts to infer their ecological functions from environmental sequence data alone [5]. To obtain better insights into the biology and ecophysiological traits of protists, environmental sequencing has to be complemented by cultivation efforts, as well as metabolic and biochemical studies, allowing the deciphering of their trophic interactions and modes of behavior.
As heterotrophs, diplonemids depend on the availability of preformed organic compounds in their environment. However, since they dwell preferentially in deep waters and sediments lacking primary photosynthetic production [22], the dominant feeding modes providing a sufficient amount of organic carbon supporting the growth of these relatively large flagellates remain enigmatic. Previously, diplonemids have been associated with an array of organisms, including diatoms, copepods, haptophytes, dinoflagellates, and plants [15, 16, 23], as well as clams, crabs, and lobsters [24, 25]. Some researchers have credited them with a parasitic lifestyle [8, 13], yet an in silico global planktonic interactome [26], as well as genomic features [9] do not support this. Several reports have hinted that diplonemids may possibly ingest bacteria [9, 11, 27, 28], but convincing evidence that these protists might be efficient bacterivores were not provided. A successful cultivation of some diplonemid isolates on carbon-rich artificial media revealed their ability of osmotrophic nutrition [15, 16], however, this also influenced further research. While the use of osmotrophically maintained cultures has greatly contributed to the study of diplonemid cells structure and biology, the understanding of their in situ ecological role as part of marine food webs has been neglected.
One can assume that diplonemids may exploit inherent patchiness of aquatic environments with hotspots of enhanced concentrations of organic particles and slowly sedimenting decaying organisms, which also attract abundant prokaryote populations, forming a rapidly growing biomass of heterotrophic bacteria. These considerations led us to hypothesize that diplonemids can grow on a concentrated bacterial biomass. Here, we explored this hypothesis and aimed to fill the gap in our knowledge on the behavior and feeding modes of these flourishing flagellates.
Materials and methods
Eight diplonemid species (Diplonema aggregatum YPF1606, D. papillatum ATCC 50162, D. japonicum YPF1604, Rhynchopus humris YPF1608, Lacrimia lanifica YPF1601, Sulcionema specki YPF1618, Namystynia karyoxenos YPF1621, and Hemistasia phaeocysticola YPF1303), initially maintained axenically as osmotrophs in the nutrient-rich Hemi medium at 15 °C, were tested for their ability to feed on bacteria. The heat-killed fluorescently labeled bacteria (FLB) were prepared from a mixed bacterial inoculum collected from the Baltic Sea, using the 5-(4,6-dichlorotriazin-2-yl) aminofluorescein [29], and together with unstained stocks of the same bacteria were stored frozen at −20 °C until use in grazing experiments. The bacterial community was characterized via 16S rRNA gene sequences, and its morphometric structure was characterized by sizing of >300 cells in 4’,6-diamidino-2-phenylindole (DAPI)-stained preparations.
Diplonemids were adapted to graze on bacteria through weekly sub-culturing in gradually diluted (50%, 25%, 10%, 5%, 1%, 0%) media by nutrient-free artificial seawater (ASW), with simultaneous additions of heat-killed bacterial mixture (from 5 × 106 progressing to 107 cells/ml). All incubations were performed at 15 °C in 25 cm² tissue culture flasks kept in a horizontal position. For grazing experiments, diplonemids were pre-starved in ASW and subsequently dispensed into ASW containing either 7–9 × 106 cells/ml or 1.5 × 106 cells/ml of FLB that supposedly correspond to the typical densities of prokaryotes in the benthic and pelagic marine habitats, respectively. The growth of diplonemids and the depletion of FLB were assessed by flow cytometry, and the diplonemids’ morphology, ultrastructure, and morphometry were examined using light, fluorescence, and transmission electron microscopies. Transcriptomic analysis of osmotrophic, phagotrophic and starved diplonemids was done on guanidine extracted RNA using NovaSeq 6000 (Illumina) paired-end 150 bp technology.
Grazing rate was calculated as the ratio between the amount of ingested bacteria, based on their decay rate/ml in the culture medium, and the diplonemids’ abundance achieved during this decay phase. The doubling time was calculated using ln-transformed data on diplonemids abundance changing with time. The digestion rates of the bacterial prey were assessed using heat-killed bacteria labeled with amine-reactive dye pHrodo Green. To examine size-dependent species-specific uptake rates as well as the consumption of indigestible particles, diplonemids were fed fluorescent-labeled latex beads (1 µm diameter) resuspended in either ASW or Hemi medium.
The unexpected appearance of red autofluorescence in grazing and starved diplonemids (see Results) was verified by their resuspension into fresh Hemi medium or ASW. The detection of reactive oxygen species in these cultures was performed with dihydrorhodamine 123. To attest the lysosomal origin of autofluorescence, cells were stained with LysoTracker Green, immobilized by low-melting agarose, and observed via confocal laser scanning microscopy. To confirm the presence of melanin, Western blot and immunofluorescence assays were performed, using anti-melanin monoclonal antibody 6D2. The isolation of the melanin-containing fraction was conducted by solubilizing the cells in 1 M NaOH.
Statistical analysis of diplonemid growth and morphometry was performed using SAS software, with α set at 0.05.
For detailed protocols see Supplemental material.
Results
Cell growth
Direct transfer of the osmotrophically grown diplonemids to a particulate bacterial diet resulted in the inability of cells to sustain their growth. Taking into account the high nutritional value of the Hemi medium (total organic carbon - 0.3 mg/ml and nitrogen - 0.08 mg/ml), we applied gradual pre-adaptation of diplonemid cultures by exposing them to a cross-gradient of increasing bacterial densities (mean cell volume 0.37 ± 0.30 µm3, corresponding to 54.9 fg carbon per bacterial cell) and decreasing concentrations of the Hemi medium, inducing bacterivory. Species heterogeneity of the bacterial culture was reflected by pairwise identities of obtained 16S rRNA genes, ranging from 91.8% to 93.7%, with most sequences falling into the genus Pseudomonas.
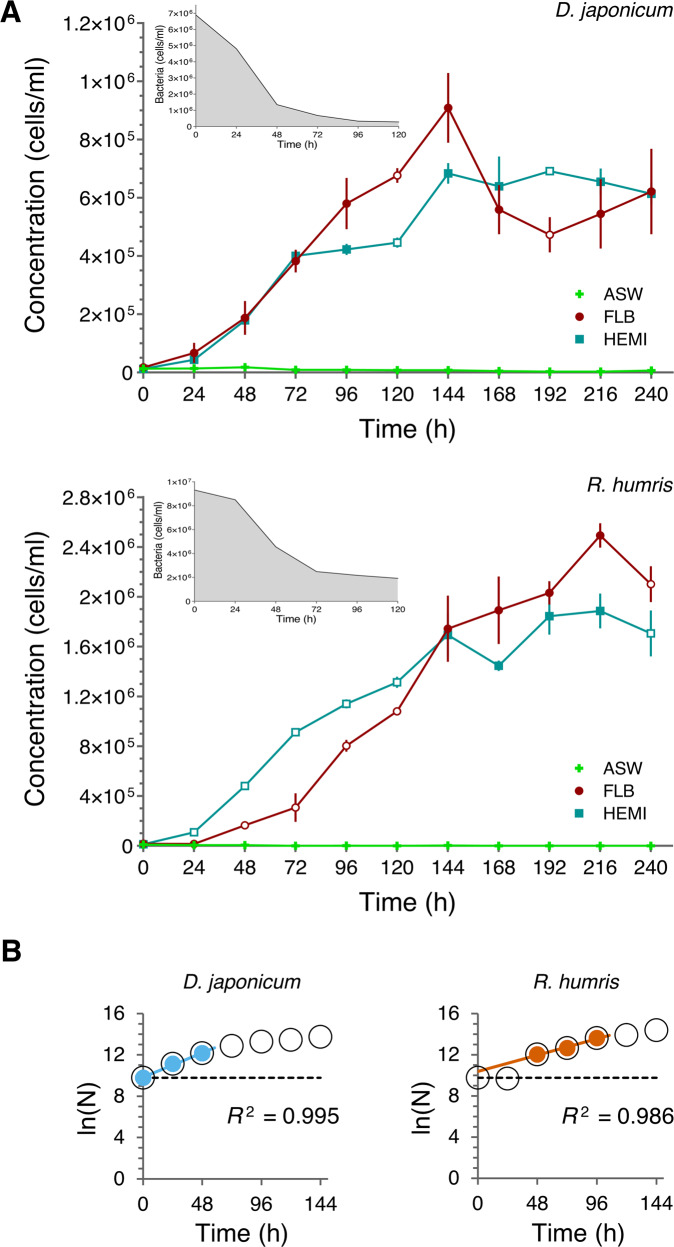
Although most tested species could ingest bacteria, only D. japonicum and R. humris grew well solely on this food source (Fig. 1A). Both species proliferated on the bacterial diet even when fed 5 to 6 times lower concentrations of FLB (Supplementary Fig. S1A, B). While S. specki endured the course of adaptation, as well as prolonged cultivation with bacteria, always showing a high bacterial decay rate (Supplementary Fig. S1C, inset), it remained in the permanent stationary phase irrespective of bacterial concentration used (Supplementary Fig. S1C, data not shown). The dynamics of bacterial uptake significantly differed (T-test, P < 0.05) between D. japonicum and R. humris, consuming 2–31 (17.7 ± 8.1; average ± SD) and 1–24 (13.0 ± 8.2; average ± SD) FLB flagellate−1 h−1, respectively, reflecting their growth dynamics (Fig. 1A, B). Indeed, D. japonicum started to proliferate almost immediately (Fig. 1A, B) with a doubling time of ~14 h (T0–T48), while R. humris displayed growth after a 48 h lag phase (Fig. 1B), yielding the doubling time of ~21 h. The growth of both species was exponential for ~2 days (Fig. 1B). After 96 h of active ingestion of bacteria by R. humris, the number of prey cells remained relatively stable (~2 × 106 cells/ml) for another 144 h (Fig. 1A, inset).
Fig. 1. Dynamics of bacterial uptake and growth of diplonemids.
A Growth curves of Diplonema japonicum and Rhynchopus humris cultured in either the nutrient-rich Hemi medium (turquoise), artificial seawater (ASW) supplemented with FLB [7–9 × 106 cells/ml] (maroon), or nutrient-free ASW (bright green). Values are means for triplicates, and the error bars show SDs. Open symbols in FLB and Hemi curves indicate significantly different values (T-test, P < 0.05). The insets show the exponential decay of FLB. B Growth rates of D. japonicum (blue) and R. humris (orange) represented as the best-fit lines.
Except for two time points (120 h and 192 h), the cell counts of D. japonicum fed on bacteria and cultured in the Hemi medium were similar (Fig. 1A), while the density of R. humris reached the level of osmotrophically maintained cells only by 144 h (Fig. 1A).
Kinetics of consumption and digestion of bacteria
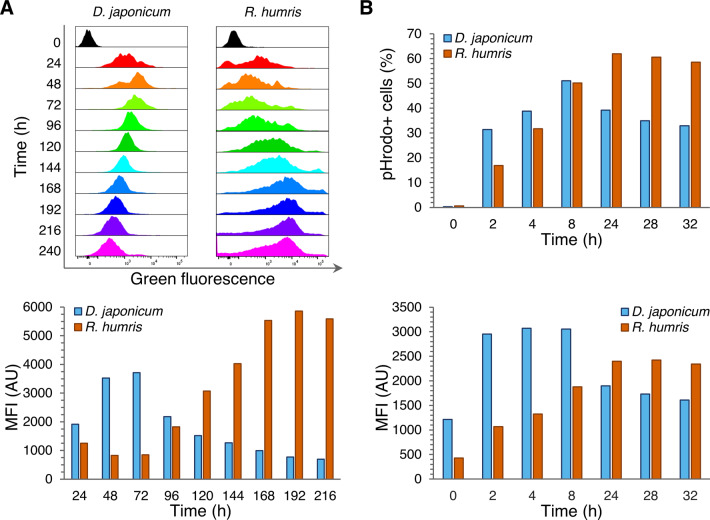
The species-specific uptake rates of FLB were also reflected in variable trends of the green fluorescent signal emitted from food vacuoles (Figs. 2A and 3B, E), suggesting different metabolic and digestion rates. To estimate the bacterial digestion rate, diplonemids were fed a mixture of heat-killed bacteria prelabeled by the pHrodo dye, a fluorescence signal of which gradually increases in the acidic environment. Upon uptake, D. japonicum immediately began to digest the prokaryotes, whereas bacterial consumption and digestion rates were somewhat lower in R. humris (Fig. 2B). Both flagellates rapidly consumed the small amount of bacteria used in this assay, and the emitted fluorescence signal started to decline after active digestion within ~8 h and ~32 h in D. japonicum and R. humris, respectively (Fig. 2B). However, the signal remained relatively stable after 76 h and did not completely disappear even after a week, reflecting the long-term presence of remnants of undigested bacteria in the food vacuoles.
Fig. 2. Kinetics of consumption and digestion of bacteria.
A Time-course changes in green fluorescence signal emitted by FLB in food vacuoles of D. japonicum (blue) and R. humris (orange). Upper panel: each individual histogram represents the intensity of the green fluorescence (corresponding to the amount of ingested FLB) on the x-axis and the number of events on the y-axis. The composite of histograms shows the extent of changes in the relative fluorescence signal over time intervals of 24 h, which are color-coded. Lower panel: column chart showing comparison of mean fluorescence intensity (MFI) of both diplonemids. B Changes in abundance of D. japonicum (blue) and R. humris (orange) with ingested pHrodo Green dye-labeled bacteria over time and MFI emitted by these bacteria inside the food vacuoles of the respective diplonemids.
Fig. 3. Characteristics of diplonemids under different conditions.

General morphology of D. japonicum (A, C) and R. humris (D, F) at 72 h and 120 h time points, respectively, by differential interference contrast (DIC) microscopy. Double arrowheads point to numerous food vacuoles filled with FLB (A, D), and arrowheads indicate storage vacuoles in the Hemi medium-grown cells (C, F). Epifluorescence microscopy of D. japonicum (B) and R. humris (E) showing DAPI-stained nucleus (blue) and food vacuoles (double arrowheads), containing FLB (green) at different stages of digestion. Scale bar: 2.5 µm. G A comparison of major morphometric characteristics (shown as the mean value ± SD and range of cell length, width, and cell volume) of the D. japonicum and R. humris from both culture conditions.
Morphology of diplonemids maintained on bacterial diet
General appearances and morphometrics of both species grown with and without FLB revealed marginal changes in shape but significant differences in cell volume (Fig. 3). While bacteria-fed D. japonicum and R. humris contained several prominent food vacuoles filled with FLB at different degrees of digestion, numerous storage vacuoles of homogeneous consistency were scattered throughout their cytoplasm when maintained on the Hemi medium (Figs. 3A–F and 4A–G). The bacteria-fed D. japonicum were significantly larger than their osmotrophically maintained counterparts at 72 h, while 120-h-old R. humris revealed an opposite trend (Fig. 3).
Fig. 4. Characteristics of FLB-fed diplonemids.
Transmission electron micrographs (TEM) of D. japonicum (A–D) and R. humris (E–G) revealing bacterial prey (double arrowheads) inside the food vacuoles at indicated time points. D Arrows point to the coat enveloping two sessile D. japonicum cells, and the arrowhead shows an excreted vacuole with bacterial residues. H Time-course changes in sizes of D. japonicum and R. humris fed with FLB, represented by forward side scatter (FSC). Time intervals as in Fig. 2A. I DIC micrographs of Sulcionema specki showing successive events of excretion of undigested bacterial residues and released vacuole (J). K TEM shows the content of vacuoles filled with bacteria (double arrowheads) at different stages of digestion. Scale bars: 1 µm (A–G), 5 µm (I–J) and 2 µm (K).
A close inspection of food vacuoles revealed species-specific differences in their content (Fig. 4A–G). While D. japonicum was stuffed with larger bacteria, R. humris ingested smaller prey (Fig. 4A–G). This preference was confirmed by the average volumes 0.36 ± 0.44 µm3 and 0.50 ± 0.61 µm3 of bacteria suspended in the cultures at 72 h and 120 h for D. japonicum and R. humris, respectively. The size-selective variability in grazing rates among the species was further demonstrated using fluorescent 1 µm (0.52 µm3) polystyrene latex beads (Supplementary Fig. S2). While D. japonicum showed a rapid consumption of FLB, R. humris preferentially ingested beads over bacteria, indicating their distinct food selectivity patterns (Supplementary Fig. S2D). Nevertheless, with the proportion of large-sized bacteria gradually increasing in time, R. humris was able to efficiently graze on them as well, as reflected by an increasing proportion of larger bacteria in its food vacuoles (Fig. 4E–G) and cell sizes during the treatment period (Fig. 4H). As for D. japonicum, their food vacuoles content progressively declined over time (Fig. 4A–D), thus reflecting the rapid FLB decay and digestion rates. In one-week-old cultures, almost no ingested prey was found, and only membranous residues of digested bacteria could be distinguished (Fig. 4C). Moreover, as a consequence of the starvation, D. japonicum entered the sessile life stage (Fig. 4D). Notably, in addition to a regular cell coat that D. japonicum produces during this phase for aggregation, the cells become enveloped by another coat, sometimes enclosing several cells (Fig. 4D). By this barrier, sessile D. japonicum creates an electron-transparent void in which no bacteria can be seen. However, a structure reminiscent of a defecation vacuole was occasionally noticed within the layers of the coating (Fig. 4D). Indeed, when most of the cellular body of diplonemids was filled with large vacuoles containing undigested particles, their excretion from the cell’s posterior occurred (Fig. 4I–K). These events lasted ~90 s, during which the cell contracted, and released one to several vacuoles consecutively, usually accompanied by some filaments. It is not yet clear what happens to the plasma membrane and the peripheral microtubular corset at this moment; nevertheless, the cell readily continues its locomotion (Fig. 4I–K). Examination of the excreted vacuoles by transmission electron microscopy revealed that they are filled with aggregates of bacterial residues at different stages of digestion (Fig. 4K).
Autofluorescence of diplonemids
Apart from the green fluorescence signal emitted by the intravacuolar FLB, we have also detected a signal in the red fluorescent channel with an emission maximum of 560 nm. Curiously, this signal appeared only upon feeding (Fig. 5). In D. japonicum, a weak signal can be distinguished after 24 h of feeding on the FLB, with its intensity gradually increasing till 72 h (Fig. 5A). In R. humris, fluorescence at this emission appeared at 48 h, which corresponds to the latency phase, reaching its maximum at 144 h (Fig. 5A). In both cases, the emission maxima correlated with the maxima of cell abundance and ingested bacteria (green fluorescence signal), with the signal progressively declining with the decrease of both parameters (Figs. 2A, 4H, and 5A).
Fig. 5. Characterization of red autofluorescence of diplonemids.
A Panels showing emergence and dynamics of red fluorescence signal emitted by D. japonicum and R. humris. Time intervals as in Fig. 2A. B Time-course changes in mean red autofluorescence intensity of D. japonicum and R. humris freshly subjected to either the Hemi medium (HEMI; solid line) or artificial seawater (ASW; dashed line). C Confocal microscopy images of live D. japonicum showing the presence of red autofluorescence (af) and its colocalization with lysosomes stained with green LysoTracker (lyso); the signal is absent in the ASW-starved cells. Nucleus was stained with NucBlue (nb). Ph, phase contrast. Scale bar: 1 µm. D Time-course changes in abundance of reactive oxygen species produced by D. japonicum grown on bacteria or medium.
The nature of autofluorescence was further evaluated in diplonemids either fed with unlabeled bacteria or grown on the Hemi medium. The red fluorescence signal was emitted in both cases yet ceased upon starvation or transfer into the nutrient-free environment (Fig. 5B). Notably, the autofluorescence did not appear when cells were fed solely with latex beads (Supplementary Fig. S2E). The emergence of the red signal with digestible food uptake implied its association with the lysosomal activity. Indeed, colocalization by confocal microscopy of fluorescently labeled lysosomes and the autofluorescence signal in living cells showed positive spatial overlap (Fig. 5C). Moreover, the appearance of the red signal correlated with the production of reactive oxygen species, suggesting that autofluorescence may be associated with acidification (Fig. 5D).
Transcriptomic analysis
We next sought to identify the molecular mechanisms underlying the response to the different food sources and the red autofluorescence using transcriptomic data. In the absence of a model diplonemid, and due to the large evolutionary distance of diplonemids from well-annotated eukaryotes, we were able to reliably annotate only 28% transcripts in D. japonicum and 40% in R. humris. In both species, most of the highly expressed transcripts (TPM > 400) were related to translation (187 in D. japonicum/175 in R. humris), followed by chromatin structure (25/21), protein modification (17/21), and cytoskeleton (14/13) (Fig. 6). In D. japonicum, the samples cluster according to the respective condition (Fig. 6A). The Hemi medium-fed cells were most distinctive by the increased expression of genes related to the chromatin structure and dynamics, coenzyme transport and metabolism, transcription, and cytoskeleton. Bacteria-fed cultures showed an increased transcription of genes related to RNA processing and modification, while starved D. japonicum invested in translation, energy conversion, metabolism, and transport of lipids. Despite the lack of clustering according to respective cultivation conditions in R. humris (Fig. 6B), in the bacteria-fed cells, we were able to observe some emerging patterns, such as investment in protein synthesis and protection from reactive oxygen species.
Fig. 6. Differentially expressed genes of osmotrophic, phagotrophic, and starved diplonemids.
Overall relative levels of the gene expression for the most expressed transcripts (TPM > 400) in D. japonicum (A) and R. humris (B) maintained on a bacterial diet (Bact) and medium (HEMI) or starved in ASW broken down to the respective functional categories (COG). For each COG category, the sample with the highest expression (TPM) was assigned 100% (red), and the expression of the remaining samples was related to this value. The horizontal bars show the cumulative TPM values of all abundant transcripts belonging to the respective COG category. Numbers count the most expressed transcripts included in a given COG.
Using a conservative filtering approach (passing both edgeR and DESeq2 tests [FDR 0.001], 4-fold change), 3 402 and 2 854 differentially expressed transcripts in D. japonicum and R. humris, respectively, were found. Considerable differences in expression were observed between the Hemi-medium vs. bacteria-fed cells, while only small differences occurred between the medium-fed vs. starved cells. The medium-fed D. japonicum invested more in systemic processes, such as DNA replication, protein assembly, and folding, while the bacteria-fed diplonemids up-regulated genes involved in phagocytosis and oxidation (vacuolar ATP synthase, proton pumps), vesicular transport (serine-threonine kinases), peptidases, cell defense, and synthesis of melanin (Supplementary Fig. S3 and Supplementary Table S1). Starved D. japonicum increasingly invested in ribosome assembly, DNA replication, and translation. The Hemi-medium fed R. humris increased its ribosome assembly, translation, and nutrition metabolism, especially nitrogen. Bacterial prey enhanced vesicular transport, as well as the (endo)membrane system organization and remodeling, whereas starved R. humris up-regulated mainly the expression of genes responsible for cell movement, (endo)membrane organization, and the phosphotransferase system, used in bacteria for import of sugars from the environment (Supplementary Fig. S4 and Supplementary Table S2).
Melanin as possible source of autofluorescence
Based on the upregulation of tyrosine transporters and tyrosinase – a key enzyme of melanin synthesis, the enrichment of melanin synthesis and metabolism (GO:0006582, GO:00482438) as revealed by the Fisher exact test, and also the emission spectrum, we hypothesized that the autofluorescence might be linked to melanin-like pigments. Western blotting with melanin-specific monoclonal antibody 6D2 revealed multiple bands at various molecular weights, consistent with synthetic melanin, which served as a positive control (Supplementary Fig. S5); however, the strongest signal remained in the stacking gel, suggesting that most of the polymer failed to enter the resolving gel (Fig. 7A). As also confirmed by immunofluorescence, melanin was depleted yet still detectable in diplonemids maintained in ASW and did not disappear even after 72 h of starvation (Fig. 7A, B, Supplementary Fig. S5). Instead of being located in the lysosomes, melanin was dispersed throughout the cytoplasm, seemingly in the vicinity of the endoplasmic reticulum (Fig. 7B).
Fig. 7. Characterization of melanin-like pigment in trophic and starved D. japonicum.
A Western blot analysis showing the time course (0–96 h) of melanin expression using anti-melanin antibody 6D2 in the diplonemids freshly inoculated into either the Hemi medium (HEMI) or artificial seawater (ASW). Leishmania mexicana (Lm) and synthetic melanin (SM) served as negative and positive controls, respectively. UM – stain-free membrane. B Confocal microscopy images of melanin detected in 72-h-old cultures by immunofluorescence using 6D2 and Alexa Fluor 647 goat anti-mouse antibody. DAPI stained the nucleus. Ph, phase contrast. Scale bar: 1 µm. Optical absorption spectra (C) and fluorescence (D) emission spectra of pigment-containing fraction extracted from 72-h-old cultures and compared with synthetic melanin (SM).
To further validate that the signal indeed represents melanin, we performed the isolation of pigment-containing fractions from both trophic and starved D. japonicum and characterized their optical properties. The absorption and the emission spectra of the extracted fractions closely resembled the exponential absorption pattern and the profiles, respectively, of synthetic melanin (Fig. 7C, D).
Discussion
Based on extensive data from environmental sequencing [6–10, 12], diplonemids seem to be the most abundant eukaryotic component of the oceanic ecosystem for which any ecological function remains largely speculative. Extensive co-occurrence studies did not reveal any convincing associations between them and other eukaryotes [26], and their size distribution only hinted that some of them might be parasitic [8, 13]. Here we focused on nutritional modes of three marine members of the family Diplonemidae, and demonstrate their capability to switch from osmotrophy to phagotrophy under cultivation preconditioning. In the absence of cultured representatives of Eupelagonemidae, which constitute 98% of diplonemid diversity in the oceans [8], our findings are of ecological relevance. While members of both families inhabit a range of habitats and may or may not (significantly) differ in their biology, they seem to share a lot of similarities in general morphology [30] and feeding behavior [9]. We posit that feeding flexibility of diplonemids is highly advantageous and provides strong clues to the successful life strategy of these understudied yet omnipresent protists.
Oceanic biodiversity is numerically dominated by prokaryotes, recognized to modulate the protistan communities and vice versa. Indeed, protistan bacterivores largely impact the structure and dynamics of bacterial assemblages in the aquatic environment [31, 32]. The ability to grow on bacteria as a sole food source shown here suggests that the abundant diplonemids may considerably contribute to the aggregated predator grazing pressure on prokaryotes. While nothing is known about grazing preferences of diplonemids under natural conditions, our experimental setup indicated substantial differences in their species-specific feeding patterns. Indeed, the growth dynamics of studied diplonemids were impacted by bacterial size, biotic state, composition, and total biomass available. R. humris selectively grazed on smaller prey and only with its depletion switched to larger bacteria, whereas D. japonicum appears to be more flexible in its prey selection and feeds rather erratically, yet this could have been tested only under the constraints inherent to our experimental setup. One possible explanation may be the lineage-specific preferences of flagellates for certain bacterial taxa, although the phenomenon of such grazing specificity is more profound in freshwater habitats [33–37]. As supporting evidence, we have also observed that phagotrophic D. japonicum grazed on living cyanobacteria belonging to the Synechococcus group (Supplementary Fig. S6A), while R. humris ignored this prey. Notably, both species did not grow when the heat-killed bacteria were replaced with their live cultures as prey. Though this may deviate from the general paradigm [34], it is worth noting that some protists have been also reported to prefer dead bacteria to living ones [34]. On the other hand, the well-known virulence factor of Pseudomonas endotoxin, inhibiting the growth of other organisms in the culture, may be an alternative explanation for the limited growth [38]. Another factor leading to moderate consumption of bacteria is a suboptimal prey-size ratio and, as a consequence, the potentially longer food particle processing time and adaptation of a predator to prey [37, 39, 40]. In addition, even closely related prokaryotes may have a distinct nutritional impact on flagellate bacterivores, thereby affecting their community composition and ecophysiological characteristics, such as the growth rate and population biomass [41]. It appears to be the case of S. specki, which exhibits a high rate of total bacterial uptake, but at the same time excretes them almost undigested in numerous waste vacuoles, indicating a low nutritional value of the consumed bacteria. The slower growth and small cell volumes of R. humris fed on FLB may be associated with a deficiency of nutrients in the preferentially ingested small-sized prokaryotes [42], or simply that bacteria alone are not their preferred food items. Apart from the requirement of different types of nutrients, the studied species revealed strikingly distinct feeding strategies. While D. japonicum stuffed itself with the prey and quickly digested it, R. humris accumulated and digested prokaryotes at a much slower rate. These fundamental differences in trophic speciation are likely associated with food preferences and food particle processing. The feeding strategy of R. humris, which may potentially be a lobster parasite [17, 25], may reflect its life strategy accustomed to high food availability not requiring its storage.
The ability to switch between different nutritional modes provides cells with great flexibility and may be critical for their survival in the broad habitat range they occupy. While in our experimental setup diplonemids had to undergo preadaptation, possibly due to their long-term cultivation as osmotrophs, we speculate that the transition would likely be instant under natural conditions. Indeed, the presence of a fully developed cytopharynx [17–19, 43] in the osmotrophic cells indicates their phagotrophic capacity [44]. The species studied here were originally isolated from an aquarium and coastal waters, and utilization of bacterivory is likely beneficial during coastal bacterial blooms or any kind of accumulation of organic matter driven by inherent oceanic processes. The trophic stages of D. japonicum and R. humris are characterized by gliding cells [17, 18], suggesting their colonization of surfaces, which are typically nutrient-rich. At the same time, the starved cells are free swimmers, like most other diplonemids, and while the expression patterns show that they are adapted to utilize bacteria as a food source, bacterivory may not be their main carbon source. Indeed, the generally low C-content of oceanic bacterioplankton compared to cultured bacteria [45] is unlikely to be sufficient to meet metabolic requirements of large flagellates, such as diplonemids. In addition, since in the seawater column the bacterial abundance is decreasing with depth, diplonemids are unlikely to feast on them in the deep sea where any nutritional particle is of great importance [46]. Instead, they likely use other carbon sources, such as sinking marine snow particles, rich in associated bacteria, decaying algae, and other planktonic organisms, representing hotspots of microbial biomass and degradation in the highly diluted, organic particle-poor environments. Near such lavish hotspots (Supplementary Video S1), the opportunistic lifestyle combined with the flexible feeding mode of diplonemids might add a considerable competitive advantage in cropping of food resources compared to suspension-feeding protists [39].
Species-specific feeding responses to a similar prey may indicate a variety of specialized feeding modes of diplonemids, reflecting difference in their thus far neglected ecological roles. The ubiquitous occurrence of diplonemids in different oceanic depths together with the assumed broad array of survival strategies might indicate that this highly diverse group involves many opportunistic strategists with high growth potential once the food resource or decaying biotic particles become abundant. The case of S. specki shows that even when food is depleted, decreased metabolic rates allow survival of dormant stages in adverse conditions, a behavior recently exemplified by flexible metabolic adaptations of diplonemids [47], which suggest an opportunistic lifestyle. Furthermore, the transcriptome-based study revealed an extremely high metabolic versatility of diplonemids [48]. Accordingly, we assume that they are omnivorous or opportunistic generalist predators and scavengers. This assumption is supported by the clearance rates of artificial latex beads as well as grazing on live and dead microalgae (Supplementary Figs. S6B–D). Diplonemids seem to adapt to food dynamics in their habitat, and, at the same time, can discriminate the low-quality or non-digestible food particles, such as latex beads, even after their ingestion, a feature known also for other protists [49]. To a large extent, such selectivity can be attributed to the biotic and physiological state, structural features, particle- or cell-surface properties, or chemical composition of predation-resistant bacteria [33, 40, 50], as well as repulsive chemicals in the environment [49]. In this context, we hypothesize that the appearance of red autofluorescence in cells during food digestion is associated with a self-defense mechanism. It appears that the process of food digestion is accompanied by the production of reactive oxygen species, the excess of which leads to the degradation of the lysosomal membrane [51]. To prevent the damage, diplonemids seem to trigger melanin synthesis, which is known to neutralize free radical toxicity and has other protective functions [52]. While melanin is widely distributed in animals, plants, fungi, and bacteria, it is less common in protists [53], although its presence under specific cultivation conditions was suggested in Leishmania [54], a parasitic relative of diplonemids. We further hypothesize that being produced in the endoplasmic reticulum [55], melanin then enters lysosomes, where its oxidative degradation causes fluorescence [56]. Ultimately, this leads to quenching of the vacuole with the sequestration of waste products and subsequent excretion from the cell. The expulsion of large waste vacuoles, in the absence of any recognizable dedicated cell structures, such as cytopyge or cytoproct, has been observed in cultured cells (this work) as well as freshly captured eupelagonemids (Okamoto N and Keeling PJ, pers. commun.). Since this prominent excretion happens at the posterior end of the cell, it may occur at the convergence of the peripheral microtubular corset or even require disassembly of microtubules, followed by membrane healing [57]. In addition, we revealed another putatively protective extracellular feature of D. japonicum, the electron-transparent coat, which was not observed in cells cultured in the absence of bacteria [18]. These findings imply that in addition to great flexibility in feeding modes, diplonemids also have a broad array of defense and survival strategies as part of their successful lifestyle in permanently changing environments.
Conclusions
We present the first detailed observation of the feeding behavior of diplonemids, one of the understudied yet highly abundant marine protistan groups. We demonstrate that diplonemids possess extensive flexibility in feeding modes, being able to switch from osmotrophy to phagotrophy. Such a broad scale of trophic adaptations can explain their ubiquity in different marine habitats assumed to be largely limited by particulate food sources. In order to unveil all the fascinating adaptations of diplonemids and their fundamental influence on the marine food web, further research on their cell biology, ecology and interaction with other planktonic organisms is needed. In addition, cultured diplonemids can serve as model organisms for studying other possible feeding modes.
Supplementary information
Acknowledgements
We thank Joshua Nosanchuk (Albert Einstein College of Medicine) for the kind gift of 6D2 antibody, Ivan Ivanov (Centrum Algatech) for Porphyridium culture, and Alyson Santoro (University of California), Priscila Peña-Diaz (Charles University), and Ian Butts (Auburn University) for advice and discussions. We also thank Eva Kriegová, Radka Malá, Vojtěch Kasalický, Hana Sehadová and Petr Porcal (Biology Centre) for technical assistance. This work was supported by grants from the Czech Science Foundation 18-23787S (to AH and GP), the ERD project ERDF/ESF project 16_025/0007417 (to KŠ), and ERC CZ LL1601, the ERD project 16_019/0000759, and the Gordon and Betty Moore Foundation (to JL).
Author contributions
GP, TK, JL, and KŠ designed the study. GP and TK carried out the experiments and analyzed the data. VJ and AH performed the transcriptomic analysis. JM carried out Western blotting. GP, KŠ, and JL wrote the manuscript with input from all authors. All authors discussed the results and commented on the manuscript.
Data availability
The raw RNA-Seq reads and transcriptome assemblies used in this study are available at NCBI under the BioProject PRJNA746563.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Galina Prokopchuk, Tomáš Korytář.
Contributor Information
Galina Prokopchuk, Email: prokopchuk@paru.cas.cz.
Julius Lukeš, Email: jula@paru.cas.cz.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01192-0.
References
- 1.Worden AZ, Follows MJ, Giovannoni SJ, Wilken S, Zimmerman AE, Keeling PJ. Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science. 2015;347:1257594. doi: 10.1126/science.1257594. [DOI] [PubMed] [Google Scholar]
- 2.Bar-On YM, Phillips R, Milo R. The biomass distribution on Earth. Proc Natl Acad Sci USA. 2018;115:6506–11. doi: 10.1073/pnas.1711842115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Poi E, Blason C, Corinaldesi C, Danovaro R, Malisana E, Fonda-Umani S. Structure and interactions within the pelagic microbial food web (from viruses to microplankton) across environmental gradients in the Mediterranean Sea. Glob Biogeochem Cycles. 2013;27:1034–45. doi: 10.1002/2013GB004589. [DOI] [Google Scholar]
- 4.Das S, Mangwani N. Ocean acidification and marine microorganisms: responses and consequences. Oceanologia. 2015;57:349–61. doi: 10.1016/j.oceano.2015.07.003. [DOI] [Google Scholar]
- 5.Keeling PJ, del Campo J. Marine protists are not just big bacteria. Curr Biol. 2017;27:R541–R549. doi: 10.1016/j.cub.2017.03.075. [DOI] [PubMed] [Google Scholar]
- 6.de Vargas C, Audic S, Henry N, Decelle J, Mahé F, Logares R, et al. Eukaryotic plankton diversity in the sunlit ocean. Science. 2015;348:1261605. doi: 10.1126/science.1261605. [DOI] [PubMed] [Google Scholar]
- 7.Obiol A, Giner CR, Sánchez P, Duarte CM, Acinas SG, Massana R. A metagenomic assessment of microbial eukaryotic diversity in the global ocean. Mol Ecol Resour. 2020;20:718–31. doi: 10.1111/1755-0998.13147. [DOI] [PubMed] [Google Scholar]
- 8.Flegontova O, Flegontov P, Malviya S, Audic S, Wincker P, de Vargas C, et al. Extreme diversity of diplonemid eukaryotes in the ocean. Curr Biol. 2016;26:3060–5. doi: 10.1016/j.cub.2016.09.031. [DOI] [PubMed] [Google Scholar]
- 9.Gawryluk RMR, Del Campo J, Okamoto N, Strassert JFH, Lukeš J, Richards TA, et al. Morphological identification and single-cell genomics of marine diplonemids. Curr Biol. 2016;26:3053–9. doi: 10.1016/j.cub.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Boeuf D, Edwards BR, Eppley JM, Hu SK, Poff KE, Romano AE, et al. Biological composition and microbial dynamics of sinking particulate organic matter at abyssal depths in the oligotrophic open ocean. Proc Natl Acad Sci USA. 2019;116:11824–32. doi: 10.1073/pnas.1903080116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukherjee I, Salcher MM, Andrei AŞ, Kavagutti VS, Shabarova T, Grujčić V, et al. A freshwater radiation of diplonemids. Environ Microbiol. 2020;22:4658–68. doi: 10.1111/1462-2920.15209. [DOI] [PubMed] [Google Scholar]
- 12.Flegontova O, Flegontov P, Londoño PAC, Walczowski W, Šantić D, Edgcomb VP, et al. Environmental determinants of the distribution of planktonic diplonemids and kinetoplastids in the oceans. Environ Microbiol. 2020;22:4014–31. doi: 10.1111/1462-2920.15190. [DOI] [PubMed] [Google Scholar]
- 13.Schoenle A, Hohlfeld M, Hermanns K, Mahé F, de Vargas C, Nitsche F, et al. High and specific diversity of protists in the deep-sea basins dominated by diplonemids, kinetoplastids, ciliates and foraminiferans. Commun Biol. 2021;4:501. doi: 10.1038/s42003-021-02012-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griessmann K. Über marine Flagellaten. Arch Protistenkd. 1913;32:1–78. [Google Scholar]
- 15.Porter D. Isonema papillatum sp. n., a new colorless marine flagellate: A light- and electronmicroscopic study. J Protozool. 1973;20:351–6. doi: 10.1111/j.1550-7408.1973.tb00895.x. [DOI] [Google Scholar]
- 16.Triemer RE, Ott DW. Ultrastructure of Diplonema ambulator Larsen & Patterson (Euglenozoa) and its relationship to Isonema. Eur J Protistol. 1990;25:316–20. doi: 10.1016/S0932-4739(11)80123-9. [DOI] [PubMed] [Google Scholar]
- 17.Tashyreva D, Prokopchuk G, Yabuki A, Kaur B, Faktorová D, Votýpka J, et al. Phylogeny and morphology of new diplonemids from Japan. Protist. 2018;169:158–79. doi: 10.1016/j.protis.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 18.Tashyreva D, Prokopchuk G, Votýpka J, Yabuki A, Horák A, Lukeš J. Life cycle, ultrastructure, and phylogeny of new diplonemids and their endosymbiotic bacteria. MBio. 2018;9:e02447–17. doi: 10.1128/mBio.02447-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prokopchuk G, Tashyreva D, Yabuki A, Horák A, Masařová P, Lukeš J. Morphological, ultrastructural, motility and evolutionary characterization of two new Hemistasiidae species. Protist. 2019;170:259–82. doi: 10.1016/j.protis.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 20.Lukeš J, Wheeler R, Jirsová D, David V, Archibald JM. Massive mitochondrial DNA content in diplonemid and kinetoplastid protists. IUBMB Life. 2018;70:1267–74. doi: 10.1002/iub.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaur B, Záhonová K, Valach M, Faktorová D, Prokopchuk G, Burger G, et al. Gene fragmentation and RNA editing without borders: eccentric mitochondrial genomes of diplonemids. Nucleic Acids Res. 2020;48:2694–708. doi: 10.1093/nar/gkz1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jürgens K, Massana R. Protist grazing on marine bacterioplankton. In: Kirchman DL (ed). Microbial ecology of the oceans. Hoboken, NJ, USA; John Wiley & Sons, Inc: 2008, pp 383–441.
- 23.Elbrächter M, Schnepf E, Balzer I. Hemistasia phaeocysticola (Scherffel) comb. nov., redescription of a free-living, marine, phagotrophic kinetoplastid flagellate. Arch für Protistenkd. 1996;147:125–36. doi: 10.1016/S0003-9365(96)80028-5. [DOI] [Google Scholar]
- 24.Kent ML, Elston RA, Nerad TA, Sawyer TK. An Isonema-like flagellate (Protozoa: Mastigophora) infection in larval geoduck clams, Panope abrupta. J Invertebr Pathol. 1987;50:221–9. doi: 10.1016/0022-2011(87)90086-3. [DOI] [PubMed] [Google Scholar]
- 25.von der Heyden S, Chao EE, Vickerman K, Cavalier-Smith T. Ribosomal RNA phylogeny of bodonid and diplonemid flagellates and the evolution of euglenozoa. J Eukaryot Microbiol. 2004;51:402–16. doi: 10.1111/j.1550-7408.2004.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 26.Lima-Mendez G, Faust K, Henry N, Decelle J, Colin S, Carcillo F, et al. Determinants of community structure in the global plankton interactome. Science. 2015;348:1262073. doi: 10.1126/science.1262073. [DOI] [PubMed] [Google Scholar]
- 27.Roy J, Faktorová D, Benada O, Lukeš J, Burger G. Description of Rhynchopus euleeides n. sp. (Diplonemea), a free-living marine euglenozoan. J Eukaryot Microbiol. 2007;54:137–45. doi: 10.1111/j.1550-7408.2007.00244.x. [DOI] [PubMed] [Google Scholar]
- 28.Larsen J, Patterson DJ. Some flagellates (Protista) from tropical marine sediments. J Nat Hist. 1990;24:801–937. doi: 10.1080/00222939000770571. [DOI] [Google Scholar]
- 29.Sherr BF, Sherr EB, Fallon RD. Use of monodispersed, fluorescently labeled bacteria to estimate in situ protozoan bacterivory. Appl Environ Microbiol. 1987;53:958–65. doi: 10.1128/aem.53.5.958-965.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okamoto N, Gawryluk RMR, del Campo J, Strassert JFH, Lukeš J, Richards TA, et al. A revised taxonomy of diplonemids including the Eupelagonemidae n. fam. and a type species, Eupelagonema oceanica n. gen. & sp. J Eukaryot Microbiol. 2019;66:519–24. doi: 10.1111/jeu.12679. [DOI] [PubMed] [Google Scholar]
- 31.Jezbera J, Horňák K, Šimek K. Prey selectivity of bacterivorous protists in different size fractions of reservoir water amended with nutrients. Environ Microbiol. 2006;8:1330–9. doi: 10.1111/j.1462-2920.2006.01026.x. [DOI] [PubMed] [Google Scholar]
- 32.Rocke E, Pachiadaki MG, Cobban A, Kujawinski EB, Edgcomb VP. Protist community grazing on prokaryotic prey in deep ocean water masses. PLoS One. 2015;10:e0124505. doi: 10.1371/journal.pone.0124505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pernthaler J. Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol. 2005;3:537–46. doi: 10.1038/nrmicro1180. [DOI] [PubMed] [Google Scholar]
- 34.Massana R, Unrein F, Rodríguez-Martínez R, Forn I, Lefort T, Pinhassi J, et al. Grazing rates and functional diversity of uncultured heterotrophic flagellates. ISME J. 2009;3:588–96. doi: 10.1038/ismej.2008.130. [DOI] [PubMed] [Google Scholar]
- 35.Šimek K, Nedoma J, Znachor P, Kasalicky V, Jezbera J, Hornak K, et al. A finely tuned symphony of factors modulates the microbial food web of a freshwater reservoir in spring. Limnol Oceanogr. 2014;59:1477–92. doi: 10.4319/lo.2014.59.5.1477. [DOI] [Google Scholar]
- 36.Baltar F, Palovaara J, Unrein F, Catala P, Horňák K, Šimek K, et al. Marine bacterial community structure resilience to changes in protist predation under phytoplankton bloom conditions. ISME J. 2016;10:568–81. doi: 10.1038/ismej.2015.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Corte D, Paredes G, Yokokawa T, Sintes E, Herndl GJ. Differential response of Cafeteria roenbergensis to different bacterial and archaeal prey characteristics. Micro Ecol. 2019;78:1–5. doi: 10.1007/s00248-018-1293-y. [DOI] [PubMed] [Google Scholar]
- 38.Matz C, Bergfeld T, Rice SA, Kjelleberg S. Microcolonies, quorum sensing and cytotoxicity determine the survival of Pseudomonas aeruginosa biofilms exposed to protozoan grazing. Environ Microbiol. 2004;6:218–26. doi: 10.1111/j.1462-2920.2004.00556.x. [DOI] [PubMed] [Google Scholar]
- 39.Fenchel T. The ecology of heterotrophic microflagellates. Adv Micro Ecol. 1986;9:57–97. doi: 10.1007/978-1-4757-0611-6_2. [DOI] [Google Scholar]
- 40.González JM, Iriberri J, Egea L, Barcina I. Differential rates of digestion of bacteria by freshwater and marine phagotrophic protozoa. Appl Environ Microbiol. 1990;56:1851–7. doi: 10.1128/aem.56.6.1851-1857.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Šimek K, Kasalický V, Jezbera J, Horňák K, Nedoma J, Hahn MW, et al. Differential freshwater flagellate community response to bacterial food quality with a focus on Limnohabitans bacteria. ISME J. 2013;7:1519–30. doi: 10.1038/ismej.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du Toit A. Growth capacity and cell size. Nat Rev Microbiol. 2019;17:2. doi: 10.1038/s41579-018-0124-y. [DOI] [PubMed] [Google Scholar]
- 43.Montegut-Felkner AE, Triemer RE. Phylogeny of Diplonema ambulator (Larsen and Patterson) 2. Homologies of the feeding apparatus. Eur J Protistol. 1996;32:64–76. doi: 10.1016/S0932-4739(96)80040-X. [DOI] [Google Scholar]
- 44.Leander BS. Predatory protists. Curr Biol. 2020;30:R510–R516. doi: 10.1016/j.cub.2020.03.052. [DOI] [PubMed] [Google Scholar]
- 45.Fukuda R, Ogawa H, Nagata T, Koike II. Direct determination of carbon and nitrogen contents of natural bacterial assemblages in marine environments. Appl Environ Microbiol. 1998;64:3352–8. doi: 10.1128/AEM.64.9.3352-3358.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mestre M, Ruiz-González C, Logares R, Duarte CM, Gasol JM, Sala MM. Sinking particles promote vertical connectivity in the ocean microbiome. Proc Natl Acad Sci USA. 2018;115:E6799–E6807. doi: 10.1073/pnas.1802470115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Škodová-Sveráková I, Prokopchuk G, Peña-Diaz P, Záhonová K, Moos M, Horváth A, et al. Unique dynamics of paramylon storage in the marine euglenozoan Diplonema papillatum. Protist. 2020;171:125717. doi: 10.1016/j.protis.2020.125717. [DOI] [PubMed] [Google Scholar]
- 48.Butenko A, Opperdoes FR, Flegontova O, Horák A, Hampl V, Keeling P, et al. Evolution of metabolic capabilities and molecular features of diplonemids, kinetoplastids, and euglenids. BMC Biol. 2020;18:23. doi: 10.1186/s12915-020-0754-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roberts EC, Legrand C, Steinke M, Wootton EC. Mechanisms underlying chemical interactions between predatory planktonic protists and their prey. J Plankton Res. 2011;33:833–41. doi: 10.1093/plankt/fbr005. [DOI] [Google Scholar]
- 50.Jürgens K, Matz C. Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie Van Leeuwenhoek. 2002;81:413–34. doi: 10.1023/A:1020505204959. [DOI] [PubMed] [Google Scholar]
- 51.Cheville NF (ed). Ultrastructural pathology: the comparative cellular basis of disease, 2nd edn. Ames, Iowa, USA: Wiley-Blackwell; 2009, p 47.
- 52.Simon JD, Peles DN. The red and the black. Acc Chem Res. 2010;43:1452–60. doi: 10.1021/ar100079y. [DOI] [PubMed] [Google Scholar]
- 53.Plonka PM, Grabacka M. Melanin synthesis in microorganisms - biotechnological and medical aspects. Acta Biochimica Polonica. 2006;53:429–43. doi: 10.18388/abp.2006_3314. [DOI] [PubMed] [Google Scholar]
- 54.Lye LF, Kang SO, Nosanchuk JD, Casadevall A, Beverley SM. Phenylalanine hydroxylase (PAH) from the lower eukaryote Leishmania major. Mol Biochem Parasitol. 2011;175:58–67. doi: 10.1016/j.molbiopara.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zelickson AS, Hirsch HM, Hartmann JF. Localization of melanin synthesis. J Invest Dermatol. 1965;45:458–63. doi: 10.1038/jid.1965.159. [DOI] [PubMed] [Google Scholar]
- 56.Kayatz P, Thumann G, Luther TT, Jordan JF, Bartz-Schmidt KU, Esser PJ, et al. Oxidation causes melanin fluorescence. Invest Ophthalmol Vis Sci. 2001;42:241–6. [PubMed] [Google Scholar]
- 57.Boucher E, Mandato CA. Plasma membrane and cytoskeleton dynamics during single-cell wound healing. Biochim Biophys Acta. 2015;1853:2649–61. doi: 10.1016/j.bbamcr.2015.07.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw RNA-Seq reads and transcriptome assemblies used in this study are available at NCBI under the BioProject PRJNA746563.