Abstract
Nonmotile microorganisms often enter new habitats by co-transport with motile microorganisms. Here, we report that also lytic phages can co-transport with hyphal-riding bacteria and facilitate bacterial colonization of a new habitat. This is comparable to the concept of biological invasions in macroecology. In analogy to invasion frameworks in plant and animal ecology, we tailored spatially organized, water-unsaturated model microcosms using hyphae of Pythium ultimum as invasion paths and flagellated soil-bacterium Pseudomonas putida KT2440 as carrier for co-transport of Escherichia virus T4. P. putida KT2440 efficiently dispersed along P. ultimum to new habitats and dispatched T4 phages across air gaps transporting ≈0.6 phages bacteria−1. No T4 displacement along hyphae was observed in the absence of carrier bacteria. If E. coli occupied the new habitat, T4 co-transport fueled the fitness of invading P. putida KT2440, while the absence of phage co-transport led to poor colonization followed by extinction. Our data emphasize the importance of hyphal transport of bacteria and associated phages in regulating fitness and composition of microbial populations in water-unsaturated systems. As such co-transport seems analogous to macroecological invasion processes, hyphosphere systems with motile bacteria and co-transported phages could be useful models for testing hypotheses in invasion ecology.
Subject terms: Microbial ecology, Ecosystem ecology, Macroecology, Population dynamics
Introduction
To cope with the heterogeneous and highly changeable soil environment, microorganisms have evolved inter-microbial co-transport strategies to gain motility and colonize new habitats (reviewed by [1]). For instance, bacteria have been found to efficiently disperse along hyphae in soil [2–4]. Fungi embody up to 75% of the subsurface microbial biomass [5]. Their hyphae create fractal-like mycelial networks of 102–104 m g−1 of topsoil and efficiently spread in heterogeneous habitats, penetrate air-water interfaces and cross over air-filled pores [5–7]. They thereby serve as pathways (also called “fungal highways” [2]) for bacteria to efficiently disperse, forage [8] and colonize new habitats [4, 9]. However, hyphae may reduce dispersal of intrinsically nonmotile yet abundant (>108 g−1 soil [10]) soil virus-like particles as they have been shown to retain waterborne phages [11, 12]. Considering the slow diffusion (~0.034 mm/d [13]) and enhanced inactivation at dry conditions [14, 15], transport of phages seems particularly restricted in water-unsaturated habitats. Recent studies have revealed that phages in aquatic environments can adsorb to surfaces [16, 17], mucus [18], flagella [19] of non-host bacteria or even sheath surrounding them [20]. As hyphae are adapted to water-unsaturated habitats we here hypothesize that hyphae allow for phage co-transport with hyphal-riding bacteria and thereby fuel the fitness of invading bacteria in new regions (alien ranges) of water-unsaturated habitats. Co-transported phages, which are not detrimental to the carrier bacteria but specific to resident bacteria in alien ranges thereby may serve as “biological weapons” [21, 22] and increase the competitive ability and fitness of the invading carrier bacteria. Temperate phages integrated in bacterial genome (i.e. as prophages) have been suggested to serve as agents of “bacterial warfare” [22–24]. In aquatic environments, lytic phages adsorbed to bacteria have been shown to facilitate phage infection of biofilm bacteria and to promote biofilm colonization by carrier bacteria [19]. In unsaturated environments like soil, however, little is known about co-transport of phage with motile bacteria and the associated effect on bacterial population dynamics.
In analogy to the previously published MAcroecological Framework of Invasive Aliens (MAFIA [25]) we here explored the possibility to use spatially organized microcosm systems to mimic the stages of the invasion process (i.e. transport, introduction, establishment, and spread) [25–28] of co-transported phages and bacteria in the hyphosphere. Unlike biological invasions in macroecological systems, our model system bears the advantage that all interacting species, their locations and location characteristics as well as invasion events are known and can be manipulated. In our model the “native range” and the “invaded” or “alien range” are two agar patches that are separated by an air gap. The air gap serves as a barrier that only can be overcome by bacterial movement along hyphae as invasion pathways crossing the native and alien ranges. Such water-unsaturated systems allow (i) to evaluate the transport efficiency of flagellated and non-flagellated bacteria as vectors to transport phages into alien ranges and (ii) to quantify possible population effects of co-transported phages in the alien range. Our approach revealed that motile bacteria can help phages to migrate into water-unsaturated habitats and that phage co-transport can fuel the settlement and fitness of hyphal-riding bacteria in host pre-colonized alien ranges.
Materials and methods
Strains, growth conditions, and enumeration methods
GFP-labeled wild type [29] soil-bacterium Pseudomonas putida KT2440 (termed hereafter as WT) and its non-flagellated mutant ΔfilM were used as carriers for phage co-transport. ΔfilM was obtained by allelic exchange with a truncated version of filM [30] and used to test the role of flagella for phage adsorption and phage co-transport. Both strains were kindly provided by Arnaud Dechesne (DTU, Denmark). They were cultivated in LB medium on a gyratory shaker at 30 °C and 150 rpm. For microcosm experiments, an overnight culture (OD600 ≈ 2) was washed once with PBS buffer (100 mM) and adjusted to reach an OD600 ≈ 4 (≈8 × 106 cell µL−1). Hyphae of the oomycete Pythium ultimum [31] were used as model dispersal networks, as P. ultimum is fast growing and provides hydrophilic hyphal surfaces allowing for efficient bacterial dispersal [2]. It was pre-grown on potato dextrose agar (PDA) at room temperature (RT) [32]. Escherichia virus T4 (T4) was selected in co-transport experiments, because it is a commonly used model phage and is known to adsorb to cell surfaces [33]. T4 was propagated on its host E. coli (Migula 1895) using the liquid broth method in DSM544 medium [35]. T4 and E. coli (Fig. 1a) were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Braunschweig, Germany). E. coli was cultivated in DSM544 medium at RT on gyratory shaker at 150 rpm (generation time = 41 ± 0.1 (SD) min in the exponential phase). For microcosm experiments, 10 µL of an overnight E. coli culture were transferred into 20 mL fresh DSM544 medium and cultivated at 30 °C until early exponential growth (OD600 ≈ 0.4).
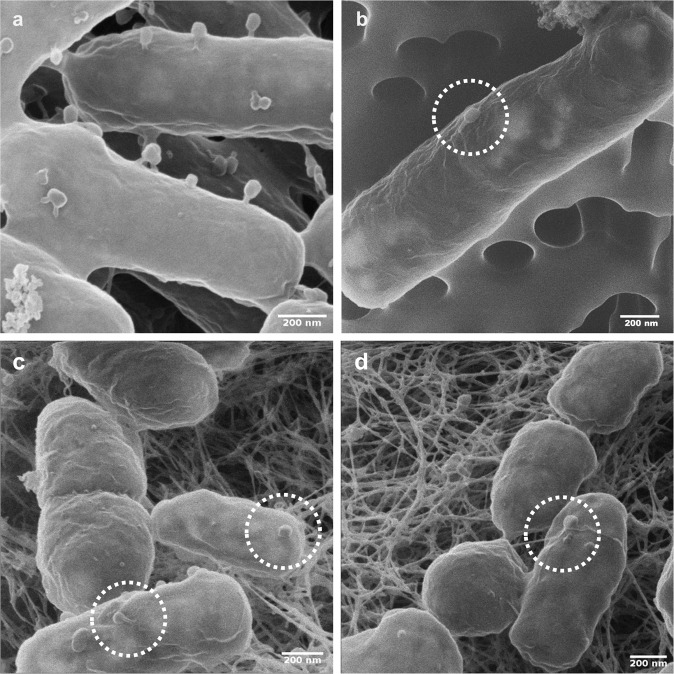
Fig. 1. Helium ion microscopy (HIM) visualization of T4 phage adsorption to E. coli host and non-host P. putida KT2440 cells.
a, b visualize cells from phage adsorption experiments (cf. materials and methods) reflecting tail-mediated adsorption to E. coli host cells (a) and capsid-driven adsorption to non-host P. putida KT2440 (b). c, d visualize tail- (c) and capsid-driven (d) phage adsorption to biofilm cells growing on agar patch B on day 2 in experiments evaluating population effect of T4 co-transport with P. putida KT2440 (cf. Fig. 3).
Enumeration of E. coli and P. putida KT2440 were carried out by counting colony forming units (CFU) on LB agar incubated at 30 °C overnight. When both strains were present in a sample, CFU of GFP-tagged P. putida KT2440 were counted with an epifluorescence microscope equipped with a black-and-white camera (AZ 100 Multizoom; Nikon, Amsterdam, Netherlands) under the GFP channel using NIS Elements software. Plaque forming unit (PFU) enumeration was done using a modified small-drop plaque assay technique as detailed earlier [36] allowing the double-layer counting plates to be incubated overnight at 30 °C. The whole-plate plaque assay (cf. [37]) was also performed to crosscheck PFU counts for samples with zero PFU count by small-drop plaque assay.
Determination of phage adsorption to bacteria
Adsorption efficiencies of T4 to WT, ΔfilM and E. coli were quantified at phage-to-bacteria ratios of 1, 0.1, and 0.01 in 6–8 replicates as described earlier [38]. In brief, suspensions of bacteria (≈108 CFU mL−1) and T4 were incubated in PBS at RT for 1 h (15 min for T4 and E. coli) and centrifuged at 8000 × g at 4 °C to pellet bacteria and adsorbed phages. Amounts of adsorbed phages were estimated by the loss of free phages after centrifugation; i.e. phage adsorption (%) calculated by the ratio of adsorbed phages to total phages prior to centrifugation. A phage-only control was also included to determine the stability and change of infectivity of T4 in the medium and during centrifugation.
Quantification of phage co-transport by hyphal-riding bacterial carriers
T4 co-transport with carrier bacteria was quantified in quintuplicate laboratory microcosms mimicking unsaturated (i.e. vadose) soil zones in the presence and absence of hyphal networks (Fig. 2a). The microcosms consisted of an agar patch A (PDA, 2% agar (w/v), l × w × h = 1 × 1 × 0.6 cm) that was separated from an agar strip (l × w × h = 2 × 1 × 0.6 cm) by a 0.5 cm air gap. P. ultimum was pre-inoculated on agar patch A for 3–4 days to reach >0.5 cm hyphal length prior to finally assembling the microcosms. The agar strip (that was split into equally sized patches B and C before harvesting, Figs. 2a and 3a) however was freshly prepared upon setting up the microcosm. It was made from minimal medium agar (MMA) to avoid bacterial growth and consisted of a top layer (MMA, 0.6% agar (w/v), h = 0.1 cm) and a bottom layer (MMA, 2% agar (w/v), h = 0.5 cm). All agar patches were placed in sterile Petri dishes. To analyze the transport of T4 along hyphae of P. ultimum in presence and absence of carrier bacteria, five different scenarios were used (Fig. 2b): (i) WT, (ii) WT + T4, (iii) T4, (iv) ΔfilM, and (v) ΔfilM + T4. P. ultimum pre-grown agar patches (equal size as agar patch A) with T4 or WT + T4 were used to quantify hyphal effects on T4 infectivity. Inactivation of T4 on agar surfaces was studied on agar patches in the absence of P. ultimum. In scenarios (ii) and (v) bacteria with previously adsorbed phages were added. To do so, T4 (6 × 109 PFU mL−1) was co-incubated in PBS with WT or ΔfilM (≈8 × 109 cells mL−1) at RT for 1 h at 125 rpm and then centrifuged (8000 × g for 10 min at 4 °C) to discard free phages in the supernatant. We used a phage-to-bacteria ratio of ≈1, because phage adsorption to bacteria was observed highest at this ratio (Fig. S1a). The remaining pellet containing bacteria and adsorbed phages was washed once and concentrated by re-suspension in PBS to reach an inoculum density of OD600 (estimation) ≈20. Inocula with either bacterial cells (≈4 × 1010 cells mL−1) or phages (≈6 × 108 PFU mL−1) in PBS served as controls. 1 µL of the respective inoculum (i.e. ≈4 × 107 bacteria or 6 × 105 phages) was placed at 0.25 cm from the left edge of agar patch A. After inoculation the Petri dishes were sealed with Parafilm, placed in a plastic container and incubated at 20 °C in the dark. After 24, 48, and 72 h the microcosms were sacrificed and phage and bacteria numbers were quantified on agar patches A, B, and C by PFU and CFU. Isolation of phage and bacteria from agar was done as described previously [8]; i.e. cut agar pieces were suspended in 3 mL PBS in glass tubes, vortexed at maximal speed for 1 min and then sonicated (2 × 30 s with a break of 1 min). Phage-bacteria suspensions were 1:1 extracted with chloroform in order to inactivate and remove bacterial biomass prior to PFU quantification.
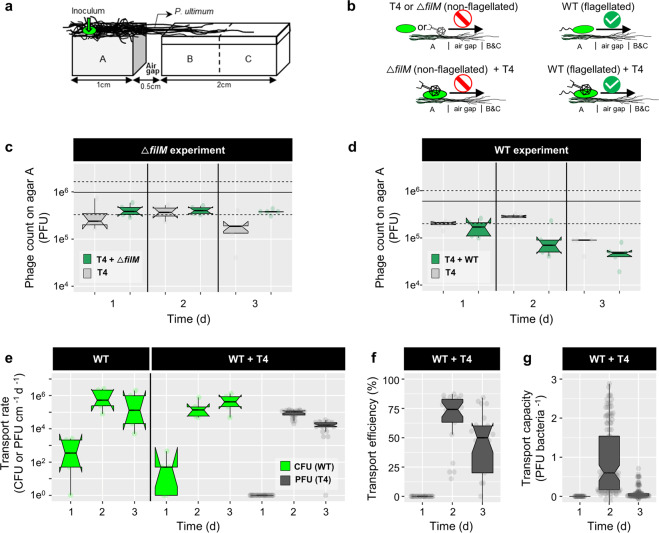
Fig. 2. Co-transport of T4 with P. putida KT2440 (WT or non-flagellated ΔfilM) along hyphae of P. ultimum.
a Scheme of the microcosm setup specifying microbial inoculation points and the spatial arrangement and dimensions of agar patches A, B, and C. b Experimental scenarios and observed results. Upper left panel: T4 or ΔfilM do not disperse along hyphae over the air-gap. Lower left panel: ΔfilM does not transport T4 along hyphae. Upper right panel: WT disperse along hyphae. Lower right panel: WT disperses along hyphae and transports T4 along hyphae over the air-gap. c, d T4 counts on agar patch A after 1 day in the absence and presence of ΔfilM (c) and WT (d). The solid and dashed lines indicate the median of 5 replicates and its 95CI of the initial inocula. e Time-dependent cumulative transport rates of WT (RWT, in green) and phages (RT4, in gray) to agar patches B and C (cf. Eq. 1). f Time-dependent phage transport efficiency in presence of WT (ET4). g Time-dependent phage transport capacity of WT (CT4). Data on T4 transport by ΔfilM are not shown as no transport was observed. Notches of the boxes represent 95CI of 5 replicates. If notches of two conditions do not overlap, it indicates a statistical difference between the two conditions.
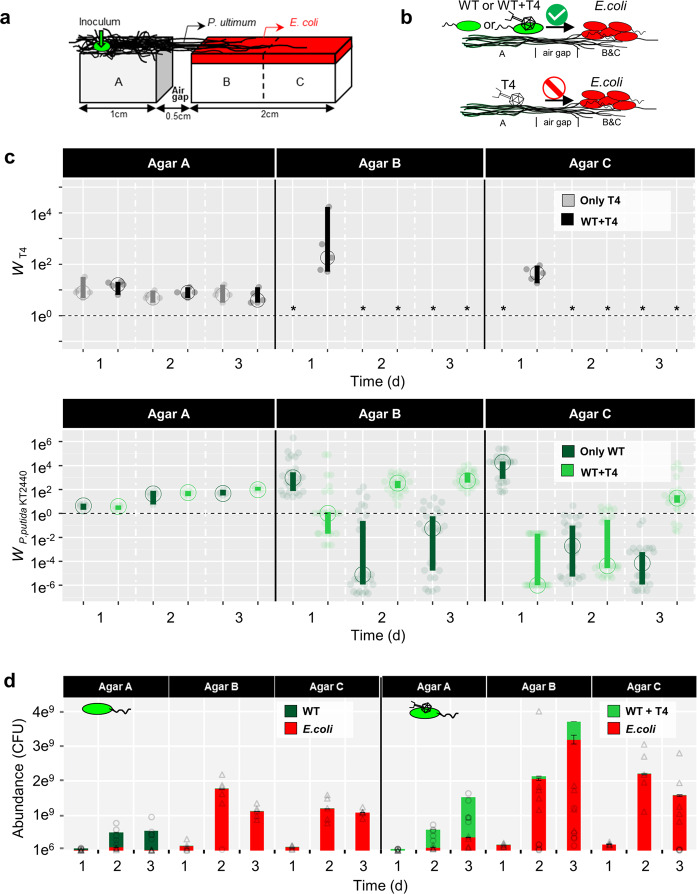
Fig. 3. Effects of T4 co-transport with flagellated P. putida KT2440 (WT) along hyphae of P. ultimum phage, E. coli and WT counts on agar patches A–C.
a Scheme of the microcosm setup specifying microbial inoculation points and pre-colonized E. coli on agar patches A, B, and C. b Overview of the scenarios for evaluating invasion. Upper panel: T4 co-transports with WT along hyphae to an E. coli covered alien range (i.e. agar patches B and C). WT disperses along hyphae to the E. coli pre-colonized alien range. Lower panel: T4 is not able to disperse along hyphae to the alien range. c Time-dependent absolute fitness of T4 (WT4) and of P. putida KT2440 (WWT) on agar patches A, B, and C in presence and absence of phage-bacterial co-transport. Dashed line represents no change of the population size (Wi = 1) and asterisk indicates that no PFU were detected. The vertical bars represent 95CI calculated from 5 biological replicates. If vertical bars of two conditions do not overlap, it indicates a statistical difference between the two conditions. d Abundance of E. coli and WT populations on agar patches A, B, and C with and without T4 co-transport (triangles and circles represented individual data points of E. coli and P. putida KT2440 in 5 replicates). Please note that abundances of WT < 108 CFU (e.g. observed in the case of WT + T4” on 3d) are not visible due to the plot scale.
Evaluation of population effects due to phage co-transport with hyphal-riding bacteria
Effects of phage co-transport on the populations of carrier bacteria invading an alien range occupied by competing bacteria were evaluated in five replicates in similar microcosms as described above. The double-layer of agar patches B and C, however, contained nutrient agar (DSM544) and the upper agar layer was densely populated by E. coli (Fig. 3a). Prior to the experiment, the thin upper agar layer of agar patches B and C was inoculated with E. coli (5 ± 0.5 (SD) × 104 CFU cm−2) and allowed to grow at RT for 3 h (≈4 generations, 2.3 ± 0.1 (SD) × 105 CFU cm−2). Three different scenarios were studied using either (i) T4, (ii) WT, or (iii) WT + T4 as inocula. After 24, 48, and 72 h, all agar patches A, B, and C were harvested and phages and bacteria quantified as described above.
Helium ion microscope imaging
Helium ion microscope (HIM) imaging was performed with suspensions from phage-bacteria adsorption assays and surfaces of agar patches B after invasion of phage-carrying WT P. putida KT2440 into E. coli populations. HIM imaging of surfaces of T4 plaques on a lawn of E. coli served as control. Suspension containing phage-bacteria associations from the adsorption assays were mixed at a ratio of 1:1 with 2% (v/v) paraformaldehyde in 0.2 M sodium‐cacodylate buffer (pH = 7.4) and allowed to stand for 2 h for chemical fixation. The suspension was then transferred onto 0.22-µm pore‐size polycarbonate filter papers (Merck‐Millipore) using a Sartorius hand‐filtration device. The filter papers were rinsed twice for 5 min with Na‐cacodylate buffer to remove salts and debris. The samples were dehydrated in a graded aqueous ethanol series (30, 50, 70, 80, 90, and 100% EtOH) and critical point dried. To observe phage-bacteria associations on agar surfaces, the agar patches were submersed in Petri-dishes in the fixative for 2 h (cf. above). The fixative was then gradually exchanged using a graded aqueous ethanol series (>50% fixative replacement each time to avoid material loss) and the sample finally critical point dried. The dried samples were mounted onto standard stubs for electron microscopy using a conductive silver epoxy glue and imaged by a Zeiss Orion NanoFab (Zeiss, Peabody, MA, USA) scanning HIM using an ion-landing-energy of 25 keV, a 10-µm aperture and an Everhard–Thornley-type secondary electron detector. To achieve both high lateral resolution (≤2 nm) and contrast, the beam current was set between 0.08 pA (high magnification) and 0.25 pA. Charge compensation during imaging was achieved with an electron flood-gun operated in line-flooding mode. In order to avoid beam damage and to allow for efficient charge compensation the dwell time of the beam on a pixel was kept between 0.5 and 1.0 µs.
Data analysis and statistics
The time-dependent transport rate (Ri; shown in Eq.1) of phages (Rp, PFU cm−1 d−1) or bacteria (Rb, CFU cm−1 d−1) was obtained by normalizing the number of phages transported (Np, PFU) or bacteria transported (Nb, CFU) to the dispersal distance (d, cm) and the time (t, d) until harvesting. Because T4 got rapidly inactivated on agar surfaces (99% loss of PFU in <24 h, Fig. S2a) and subsequent low phage numbers were elusive to direct quantification, Np was approximated by the difference of phages counted at the point of inoculation (i.e. agar patch A) in the absence and presence of carrier bacteria. This approach was possible as the presence of either P. ultimum or P. ultimum and P. putida KT2440 co-cultures did not influence T4 infectivity and enumeration (Fig. S2c).
| 1 |
For co-transported phages, time-dependent transport capacity (Cp, PFU bacteria−1) reflect the average number of phages transported by a single bacterium and transport efficiency (Ep, %) the fraction of phages dispatched by carrier bacteria. For calculation details, please refer to extended materials and methods in SI.
To evaluate the effect of phage co-transport on bacterial or phage populations at time t, the absolute fitness (Wi) [39] of a given population i was calculated using Eq. 2. Wi is the time-dependent ratio of the population size (bacteria or phages) on given agar patches in the presence of E. coli (N*i, CFU or PFU) and absence of E. coli (Ni, CFU or PFU). W > 1 and W < 1 indicate an increase and a decrease of the population size, while W = 0 refers to population extinction (c.f. extended materials and methods in SI for calculation details).
| 2 |
Data were plotted as transparent dots and statistics were displayed as median (circle) with 95% confidential interval (95CI, vertical bar) or boxplot notches as modified from [40]. The 95CI was determined by bootstrapping (1000 samples) and derived from the 2.5th and 97.5th percentile [41]. When the 95CIs of two conditions (n ≥ 5) do not overlap, it indicates a statistical difference between these two conditions [42].
Results
Adsorption of T4 phage to WT and ΔfilM
Being a prerequisite for phage co-transport with carrier bacteria we tested the adsorption of T4 phages to the flagellated WT of P. putida KT2440 and its non-flagellated ΔfilM mutant. We found that 13–62% of T4 particles adsorbed to the non-host WT with the highest adsorption (62%) observed at a phage to bacteria ratio of 1 (Fig. S1a). No statistically significant differences between T4 adsorption to flagellated WT and ΔfilM were observed at this phage to bacteria ratio (Fig. S1b). Adsorption to WT was thus consistently lower and more variable than to the host strain E. coli (68–84%; Fig. S1a). T4 adsorption was further evidenced by HIM visualization. It revealed capsid-driven adsorption of T4 (Fig. S1d, e) to the surface of P. putida KT2440 leaving the phages’ tails unattached (Fig. 1b). This is in contrast to T4 adsorption to host E. coli, where perpendicular adsorption with phage tails bound to bacterial surfaces was found (Figs. 1a and S1c).
Effect of hyphal-riding bacteria on phage co-transport
In analogy to the different stages of the plant or animal invasion processes [25, 26] we developed a spatially organized microcosm system to evaluate phage co-transport with hyphal-riding bacteria invading new habitats (i.e. agar patches B and C; Fig. 2a). To challenge the role of bacterial motility for phage co-transport, we used flagellated WT and the non-flagellated ΔfilM mutant in five different scenarios (Fig. 2b): (i) T4, (ii) ΔfilM, (iii) WT, (iv) ΔfilM + T4 and (v) WT + T4. No airborne transport of phages was observed in the microcosms (Fig. S2b). As T4 infectivity got rapidly lost on agar surfaces (>99% loss within 24 h, Fig. S2a) preventing reliable enumeration on agar patches B and C, phage transport rates (Rp; Eq. 1) were calculated by differences of phage counts on agar patch A in all scenarios. Contrary to our observation on sterile agar surfaces (Fig. S2a) T4 maintained its infectivity up to 2 days when placed on agar patches covered by P. ultimum (Fig. 1c). No reduction in T4 counts in the absence of bacteria or in presence of ΔfilM was observed over 2 days pointing at negligible diffusion of viable phages or phage co-transport by ΔfilM. Similarly, inoculation of WT + T4 on isolated P. ultimun agar (i.e. no WT exportation; Fig. S2c) showed no reduction in T4 counts (Fig. S2c). By contrast, inoculation of WT + T4 on interlinked P. ultimum agar (i.e. WT exportation allowed; Fig. 2a) resulted in a significant (>65%) reduction of T4 counts after 2 days and a transport efficiency of Ep ≈ 60% (Fig. 2e). The phage transport rates (Rp ≈ 105 PFU cm−1 d−1, Fig. 1d) thereby coincided with hyphal dispersal rates of the WT (Rb ≈ 1.4 × 105 CFU cm−1 d−1; Fig. 1d) suggesting an apparent transport capacity of Cp ≈ 0.6 PFU bacteria −1 after 2 days (Fig. 1g). After 3 days a 10-fold decreased T4 transport rate (Rp ≈ 1.4 × 104 PFU cm−1 d−1, Fig. 1d) and a 20-fold reduced apparent T4 transport capacity (Cp ≈ 0.03 PFU bacteria−1) were observed. This is likely due to growth of WT bacteria and/or the inability of the WT progeny on agar patch A to get into contact with T4 phages. Transport rates of WT bacteria were similar regardless of the presence of T4 (Fig. 2e).
Effects of phage co-transport on bacterial invasion and invader fitness
To evaluate the community effect of T4 co-transport with hyphal-riding WT, agar patches B and C were covered with 2.3 ± 0.1 × 105 CFU cm−2 of E. coli as local host bacteria of T4 (Fig. 3a). Development of T4, WT, and E. coli counts was quantified over time in four different scenarios (cf. Fig. 3b): (i) PBS only, (ii) WT, (iii) T4 and (iv) T4 + WT. In the absence of WT, no diffusion of infectious T4 along hyphae to agar patches B and C was observed at any time (Fig. 3c). In presence of WT, however, ∼105 and ~104 PFU were recovered from agar patches B and C after 1 day leading to a 45–180-fold increased phage abundance (Fig. 3c; 45 < WT4 < 180 as defined by Eq. 2). The presence of T4 went along with HIM-detectable bacterial lysis of E. coli (Fig. S3e) indicating previous phage lysis on day 1. No T4, however, were detected on day 2 (Fig. 3c) despite HIM-detectable phages adsorbing to bacterial surfaces either in tail-mediated perpendicular (Fig. 1c) or capsid-mediated coaxial positions (Fig. 1d). Tail-mediated adsorption thereby points at an infection of host cells (Fig. 1a) while capsid-mediated adsorption may refer to unspecific interaction with non-host cells (Fig. 1b). In the absence of T4 the carrier WT cells invaded and established on agar patches B and C in the first 24 h (WWT > 930) accounting for 0.08–0.39% of all bacteria. Thereafter WT got strongly inhibited (WWT < 0.06) and comprised less than 0.01% of the population; this, despite a constantly growing WT population on agar patch A (WWT > 1) and a likely on-going WT invasion from agar patch A (Fig. 3c).
By contrast, T4 co-transport with WT promoted invasion and fitness of the carrier bacteria after a delay of 1 day: on agar patch B the absolute fitness of WT increased from WWT ≤ 1 (t = 1 d) to WWT = 556 (≈5 × 108 CFU) at t = 3 d (Fig. 3c). On agar patch C the WT fitness changed from WWT < 1 (t = 1–2 d) to WWT > 1 after 3 days. After 3 days WT accounted for ≈14% and 1% of the total bacterial population on agar patches B and C, resp. (Fig. 3d). Epifluorescence microscopy analysis thereby revealed that gfp-labeled WT established best in the vicinity of the hyphae (Fig. S3b, c), suggesting highest phage-clearance effects on resident E. coli in the hyphosphere. Invasion of WT, however, had no negative impact on the abundances of E. coli cells in agar patches B and C (Fig. 3d). We further checked the effect of phage co-transport on overall phage counts and abundances of host and carrier bacteria in the microcosms (i.e. agar patches A, B, and C) (Fig. S4). While co-transport did not influence T4 abundances (Fig. S4a), it clearly enhanced the abundance of WT (Fig. S4b), yet not influenced the E. coli abundances (Fig. S4c).
Discussion
Phage co-transport with hyphal-riding bacteria
Although described for aquatic environments [19, 43, 44], little is known on viral co-transport with non-host microorganisms in vadose habitats; even though adsorption and subsurface co-transport of nano-colloids with organic or biological materials has been described nearly two decades ago [45]. Viral deposition fluxes though the atmospheric boundary layer of up to 7 × 109 m−2 d−1 [46] or event-driven transport of nano-colloids and viruses of up to 1 × 1010 particles mL−1 seepage water [47] suggest frequent collision of viruses with subsurface biological materials in vadose habitats. Here, we show that Escherichia virus T4 can adsorb to the hyphal-riding soil-bacterium P. putida KT2440 (Fig. S1). It thereby gets efficiently dispersed in water-unsaturated environments (Fig. 2) and invades new habitats (Figs. 2a and 3) even if this involves crossing air-filled spaces (Figs. 2b and 3d). No diffusion of T4 along hyphae of P. ultimum and no T4 co-transport with the non-flagellated ΔfilM was detected. Flagellar mobility hence seems to be a driver for efficient phage transport by bacteria. Although a few studies have reported on preferential adsorption of phages to bacterial flagella [19], we found similar efficiencies of T4 adsorption to flagellated WT and non-flagellated ΔfilM (Fig. S1b). T4 adsorption to the bacterial cell surface rather than to flagella was also confirmed by HIM imaging. Microscopy suggested that capsid-mediated sorption to cells left the phage tails unattached. This allows adsorbed phages to remain infectious, i.e. transferable to thermodynamically favored adsorption to host cells [48]. We chose T4 as model phage as it is known to adsorb to cell surfaces and to be less adsorptive than smaller phages [33, 34]. T4 adsorption to and co-transport by carrier bacteria hence may lead to conservative estimates for phage adsorption and co-transport. For instance, a recent study using Bacillus cereus as a model bacterium for waste water has shown its ability to adsorb phages of different morphologies from 10 viral families [19]. T4, furthermore, was also found to adsorb to the soil-bacterium P. fluorescens LP6a despite of its highly distinct physico-chemical surface properties [49] from P. putida KT2440 (Fig. S1a). As hyphal-riding bacteria are widespread in many unsaturated environments (e.g. bulk soil [3, 50, 51], the vicinity of roots [52, 53] and cheese rinds [54]), we hence speculate that other combinations of phages with hyphal-riding carrier bacteria may also lead to phage co-transport. As phage co-transport with non-host bacteria can take place over days (Fig. 2) one may speculate that such co-transport may be more efficient than recently reported co-transport of phages with their host cells (“virocells”) during the (typically small, e.g. ≥20 min for T4) latent period [55].
Benefits of phage co-transport for bacterial carriers
Our data show that T4 co-transport fosters fitness of hyphal-riding WT cells if invading alien ranges occupied by resident T4 host cells (Figs. 3 and 4). Over 48 h the invasion and colonization of WT occurred in close vicinity (i.e. in the hyphosphere) of colonizing hyphae acting as preferential transport pathways for bacteria carrying adsorbed lytic phages (Fig. 4). Co-transported phages provided significant fitness gain for carrier bacteria in the alien range after 1 day (Fig. 3c). Yet, no phages could be detected at later stages although HIM analyses clearly revealed both phage particles adsorbing to bacterial surfaces (Fig. 1c, d) and lysed E. coli cells (Fig. S3e) in the hyphosphere (Fig. S3b). Such increase of the T4 abundance during initial WT carrier invasion may have been promoted by high accessibility of E. coli cells to T4 introduced by hyphal-riding WT. Lysis of host cells coupled with exponential growth of carrier WT cells in the hyphosphere however may have reduced E. coli cell density, and, hence, T4 accessibility and subsequent host infection. Similar phenomenon has been reported in spatially organized biofilms with two Pseudomonas strains, where the growth of the phage insensitive strain largely reduced the phage abundance by blocking their access to their host [56]. Likewise, on-going T4 transport may have triggered antiphage mechanisms in the E. coli biofilms [57, 58]. Stationary E. coli (i.e. after 1d, Fig. 3d) are well-known to inhibit T4 infection by adsorbing phage particles without producing viral progenies for weeks [59–63]. Such inhibitions has been described for T4, yet not for other phages (e.g. T7 can indefinitely infect stationary E. coli [64]). We thus believe the undetectable T4 (after 1d) is a special case in this study and cannot be generalized for other phages. Our data indicate that initial WT invasion in the absence of T4 was more efficient (WWT > 930) than in its presence (WWT ≈ 1). As transport rates of WT and WT + T4 did not differ (Fig. 2e, P > 0.05), we speculate that phage infection may lead to growth inhibition of WT carrier cells as demonstrated in a mix-culture of Enterococcus faecalis [65]. Hyphal-riding invader WT cells however could not establish in the absence of T4 and—likely due to higher fitness of resident E. coli—got eliminated after initial colonization (Figs. 3c, d and 4). Hyphae thereby served as dispersal pathways allowing for access to and competition of T4-carrying WT with E. coli in the alien range. While hyphae had been described as scaffolds for microbial transport [4] and evolution [66], less is known on their role as drivers for phage transport and bacterial invasion and competition. Diffusion of viral particles is extremely slow (~0.03 mm d−1) [13, 67] and mostly hampered when facing air gaps (e.g. macropores). In such unsaturated environment, hyphae is the indispensable dispersal network, which bridges zones separated by air gaps and enables efficient co-transport of phages and bacteria (e.g. >2.5 mmd−1 in this study). As demonstrated in our model system that fungi can act as corridor for phage-bacterial co-transport, future work needs to challenge the ability of hyphae to transport unknown phage-bacterial pairs in a soil realm or to use hyphae as selective pathway to isolate co-transporting phage-bacterial pairs.
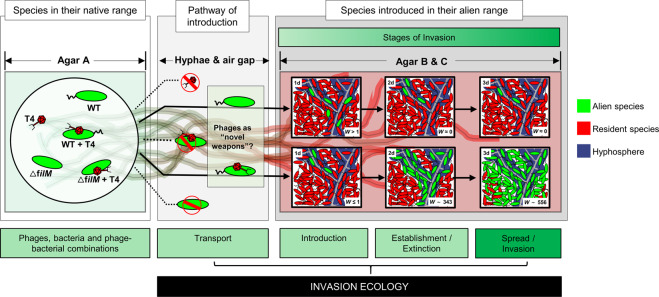
Fig. 4. Equivalence between the MAcroecological Framework of Invasive Aliens (MAFIA) and the microsphere model system used in this study (cf. Fig. 3a).
Based on the background sketch of the microsphere model system, the main findings of the study are summarized and illustrated in the flow chart. Boxes and contents are directly mirrored from the recently published MAcroecological Framework of Invasive Aliens [25] built upon [26–28].
Microbial co-transport as a model for studying biological invasion
Biological invasion experiments are frequently impossible [68] or even unethical, due to the harm they can produce [69]. Hence, conceptual models (e.g. frameworks) exist to provide a better overview to explain and predict biological invasions and to provide a testing bed for hypotheses. Here we tailored a well-controllable model ecosystem in analogy to the MAFIA [25, 26] to study transport, introduction, establishment and spread of hyphal-riding bacteria in presence and absence of co-transported phages (Fig. 4). We see the potential of our model system as an in-vitro testing bed for specific mechanisms proposed in the biological invasion frameworks (Fig. 4). Due to its high degree of controllability and based on extended knowledge on bacterial-fungal interactions [2, 3, 8, 54], phage sorption to bacteria [18, 19, 44], or the role of phages promoters of bacterial colonization [19, 22, 70] it may be manipulated in order to simulate real-world situations at micro-scale. For example, our findings resemble the concept of “novel weapon” in plant ecology (e.g. biochemical possessed by invading species that is fatal to resident species) [21] and are analogous to the spread of infectious disease in animal ecology (e.g. grey squirrel being vectors for squirrel pox infecting European red squirrels [71]). Many elements in our model simulate real-world situations at micro-scale. For example, the biogeographic barriers (e.g. oceans or mountain ranges) [72] can be represented by the air gap (in our model) or other material (e.g. heavy metals) that prevents the bacterial dispersal to agar B (or the alien range, in biological invasions terminology, Fig. 4) from agar A (the native range). To test for the efficiency of different pathways, such as shipping routes [73, 74] in the real world, we can manipulate the volume of the hyphae or their physical properties (e.g. hydrophilic hyphae are more efficient in transporting bacteria than hydrophobic ones) [2]. Similarly, many other parameters can be manipulated in our system (see an overview and references for further examples in Fig. S5). Please note, however, we do not expect that our model system will generalize all aspects for invasion ecology depicted in the MAFIA framework. It is most suitable to reflect similar co-transport scenarios in the real world, which hypothesized that alien species can act in synergy to amplify invasion impacts (e.g. pests or pathogens being transported by other non-native species [75, 76]). As phage interaction with non-host bacteria seems to be a widespread mechanism [19], future exploration of hyphae-associated phage-bacteria communities will not only resolve phage activities in regulating hyphosphere life, but also offer a powerful tool for testing hypotheses of invasion ecology at large scale (e.g. regarding spatial and trait relationships of alien biota) in spatially tailored microbial model systems.
Supplementary information
Acknowledgements
This study is part of the Collaborative Research Centre AquaDiva of the Friedrich Schiller University Jena, funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—SFB 1076—project number 218627073 and the Helmholtz Centre for Environmental Research—UFZ. The authors wish also to thank Maria Fabisch and Anke Hädrich for great coordination of the CRC AquaDiva and iRTG AquaDiva. The authors are thankful for the use of the helium-ion microscope at the Centre for Chemical Microscopy (ProVIS) at UFZ Leipzig, which is supported by European Regional Development Funds (EFRE—Europe funds Saxony) and the Helmholtz Association.
Author contributions
XY and LYW designed the study. XY performed the experiments. XY and MS performed the HIM analysis. IK interpreted the data in the context of MAFIA framework. XY, LYW, and IK wrote the manuscript. LYW, RK, HH, and AC supervised the experiments. All authors read, revised, and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-01155-x.
References
- 1.Muok AR, Briegel A. Intermicrobial hitchhiking: how nonmotile microbes leverage communal motility. Trends Microbiol. 2021;29:542–50. doi: 10.1016/j.tim.2020.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Kohlmeier S, Smits THM, Ford RM, Keel C, Harms H, Wick LY. Taking the fungal highway: mobilization of pollutant-degrading bacteria by fungi. Environ Sci Technol. 2005;39:4640–6. doi: 10.1021/es047979z. [DOI] [PubMed] [Google Scholar]
- 3.Simon A, Bindschedler S, Job D, Wick LY, Filippidou S, Kooli WM, et al. Exploiting the fungal highway: development of a novel tool for the in situ isolation of bacteria migrating along fungal mycelium. FEMS Microbiol Ecol. 2015;91:fiv116. doi: 10.1093/femsec/fiv116. [DOI] [PubMed] [Google Scholar]
- 4.Deveau A, Bonito G, Uehling J, Paoletti M, Becker M, Bindschedler S, et al. Bacterial–fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol Rev. 2018;42:335–52. doi: 10.1093/femsre/fuy008. [DOI] [PubMed] [Google Scholar]
- 5.Harms H, Schlosser D, Wick LY. Untapped potential: exploiting fungi in bioremediation of hazardous chemicals. Nat Rev Microbiol. 2011;9:177. doi: 10.1038/nrmicro2519. [DOI] [PubMed] [Google Scholar]
- 6.Otten W, Hall D, Harris K, Ritz K, Young IM, Gilligan CA. Soil physics, fungal epidemiology and the spread of Rhizoctonia solani. N. Phytol. 2001;151:459–68. doi: 10.1046/j.0028-646x.2001.00190.x. [DOI] [Google Scholar]
- 7.Sun B, Chen X, Zhang X, Liang A, Whalen JK, McLaughlin NB. Greater fungal and bacterial biomass in soil large macropores under no-tillage than mouldboard ploughing. Eur J Soil Biol. 2020;97:103155. doi: 10.1016/j.ejsobi.2020.103155. [DOI] [Google Scholar]
- 8.Otto S, Bruni EP, Harms H, Wick LY. Catch me if you can: dispersal and foraging of Bdellovibrio bacteriovorus 109J along mycelia. ISME J. 2017;11:386–93. doi: 10.1038/ismej.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kjeldgaard B, Listian SA, Ramaswamhi V, Richter A, Kiesewalter HT, Kovács ÁT. Fungal hyphae colonization by Bacillus subtilis relies on biofilm matrix components. Biofilm. 2019;1:100007. doi: 10.1016/j.bioflm.2019.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narr A, Nawaz A, Wick LY, Harms H, Chatzinotas A. Soil viral communities vary temporally and along a land use transect as revealed by virus-like particle counting and a modified community fingerprinting approach (fRAPD) Front Microbiol. 2017;8:1975. doi: 10.3389/fmicb.2017.01975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosner A, Gutstein R. Adsorption of actinophage Pal 6 to developing mycelium of Streptomyces albus. Can J Microbiol. 1981;27:254–7. doi: 10.1139/m81-039. [DOI] [PubMed] [Google Scholar]
- 12.Ghanem N, E. Stanley C, Harms H, Chatzinotas A,Y, Wick L. Mycelial effects on phage retention during transport in a microfluidic platform. Environ Sci Technol. 2019;53:11755–63. doi: 10.1021/acs.est.9b03502. [DOI] [PubMed] [Google Scholar]
- 13.Dennehy JJ. What ecologists can tell virologists. Annu Rev Microbiol. 2014;68:117–35. doi: 10.1146/annurev-micro-091313-103436. [DOI] [PubMed] [Google Scholar]
- 14.Hurst CJ, Gerba CP, Cech I. Effects of environmental variables and soil characteristics on virus survival in soil. Appl Environ Microbiol. 1980;40:1067–79. doi: 10.1128/aem.40.6.1067-1079.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeager JG, Brien RT. Enterovirus inactivation in soil. Appl Environ Microbiol. 1979;38:694–701. doi: 10.1128/aem.38.4.694-701.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz DA, Lindell D. Genetic hurdles limit the arms race between Prochlorococcus and the T7-like podoviruses infecting them. ISME J. 2017;11:1836–51. doi: 10.1038/ismej.2017.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shan J, Ramachandran A, Thanki AM, Vukusic FBI, Barylski J, Clokie MRJ. Bacteriophages are more virulent to bacteria with human cells than they are in bacterial culture; insights from HT-29 cells. Sci Rep. 2018;8:5091. doi: 10.1038/s41598-018-23418-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaudhry W, Lee E, Worthy A, Weiss Z, Grabowicz M, Vega NM, et al. Mucoidy, a general mechanism for maintaining lytic phage in populations of bacteria. FEMS Microbiology Ecology. 2020;96:fiaa162. [DOI] [PMC free article] [PubMed]
- 19.Yu Z, Schwarz C, Zhu L, Chen L, Shen Y, Yu P. Hitchhiking behavior in bacteriophages facilitates phage infection and enhances carrier bacteria colonization. Environ Sci Technol. 2020;55:2462–72. doi: 10.1021/acs.est.0c06969. [DOI] [PubMed] [Google Scholar]
- 20.Tarafder AK, von Kügelgen A, Mellul AJ, Schulze U, Aarts DGAL, Bharat TAM. Phage liquid crystalline droplets form occlusive sheaths that encapsulate and protect infectious rod-shaped bacteria. Proc Natl Acad Sci. 2020;117:4724–31. doi: 10.1073/pnas.1917726117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Callaway RM, Ridenour WM. Novel weapons: invasive success and the evolution of increased competitive ability. Front Ecol Environ. 2004;2:436–43. doi: 10.1890/1540-9295(2004)002[0436:NWISAT]2.0.CO;2. [DOI] [Google Scholar]
- 22.Granato ET, Meiller-Legrand TA, Foster KR. The evolution and ecology of bacterial warfare. Curr Biol. 2019;29:521–37. doi: 10.1016/j.cub.2019.04.024. [DOI] [PubMed] [Google Scholar]
- 23.Gama JA, Reis AM, Domingues I, Mendes-Soares H, Matos AM, Dionisio F. Temperate Bacterial viruses as double-edged swords in bacterial warfare. PLoS One. 2013;8:e59043. doi: 10.1371/journal.pone.0059043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dragoš A, Andersen AJC, Lozano-Andrade CN, Kempen PJ, Kovács ÁT, Strube ML. Phages carry interbacterial weapons encoded by biosynthetic gene clusters. Curr Biol. 2021;31:3479–89. doi: 10.1016/j.cub.2021.05.046. [DOI] [PubMed] [Google Scholar]
- 25.Pyšek P, Bacher S, Kühn I, Novoa A, Catford JA, Hulme PE, et al. Macroecological framework for invasive aliens (MAFIA): disentangling large-scale context dependence in biological invasions. NeoBiota. 2020;62:407–61. doi: 10.3897/neobiota.62.52787. [DOI] [Google Scholar]
- 26.Blackburn TM, Pyšek P, Bacher S, Carlton JT, Duncan RP, Jarošík V, et al. A proposed unified framework for biological invasions. Trends Ecol Evol. 2011;26:333–9. doi: 10.1016/j.tree.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 27.Richardson DM, Pyšek P. Plant invasions: merging the concepts of species invasiveness and community invasibility. Prog Phys Geogr Earth Environ. 2006;30:409–31. doi: 10.1191/0309133306pp490pr. [DOI] [Google Scholar]
- 28.Williamson M. Explaining and predicting the success of invading species at different stages of invasion. Biol Invasions. 2006;8:1561–8. doi: 10.1007/s10530-005-5849-7. [DOI] [Google Scholar]
- 29.Demerec M, Adelberg EA, Clark AJ, Hartman PE. A proposal for a uniform nomenclature in bacterial genetics. Genetics. 1966;54:61–76. doi: 10.1093/genetics/54.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dechesne A, Wang G, Gülez G, Or D, Smets BF. Hydration-controlled bacterial motility and dispersal on surfaces. Proc Natl Acad Sci. 2010;107:14369–72. doi: 10.1073/pnas.1008392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maurhofer M, Keel C, Schnider U, Voisard C, Haas D, Defao G. Influence of enhanced antibiotic production in Pseudomanas fluorescens strain CHA0 on its disease suppressive capacity. Phytopathol. 1992;82:190–5. doi: 10.1094/Phyto-82-190. [DOI] [Google Scholar]
- 32.Schamfuß S, Neu TR, van der Meer JR, Tecon R, Harms H, Wick LY. Impact of mycelia on the accessibility of fluorene to PAH-degrading bacteria. Environ Sci Technol. 2013;47:6908–15. doi: 10.1021/es304378d. [DOI] [PubMed] [Google Scholar]
- 33.Bichet MC, Chin WH, Richards W, Lin Y-W, Avellaneda-Franco L, Hernandez CA, et al. Bacteriophage uptake by mammalian cell layers represents a potential sink that may impact phage therapy. iScience. 2021;24:102287. doi: 10.1016/j.isci.2021.102287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu F, Wu S-H, Hung Y, Mou C-Y. Size effect on cell uptake in well-suspended, uniform mesoporous silica nanoparticles. Small. 2009;5:1408–13. doi: 10.1002/smll.200900005. [DOI] [PubMed] [Google Scholar]
- 35.Fortier L-C, Moineau S Phage production and maintenance of stocks, including expected stock Lifetimes. In: Clokie MRJ, Kropinski AM, editors. Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions. Totowa: Humana Press; 2009. p. 203–19. [DOI] [PubMed]
- 36.Mazzocco A, Waddell TE, Lingohr E, Johnson RP Enumeration of Bacteriophages Using the Small Drop Plaque Assay System In: Clokie MRJ, Kropinski AM, editors. Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions Totowa: Humana Press; 2009. p. 81–85. [DOI] [PubMed]
- 37.Kropinski AM, Mazzocco A, Waddell TE, Lingohr E, Johnson RP Enumeration of Bacteriophages by Double Agar Overlay Plaque Assay In: Clokie MRJ, Kropinski AM, editors. Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions. Totowa: Humana Press; 2009. p. 69–76. [DOI] [PubMed]
- 38.Thanki AM, Taylor-Joyce G, Dowah A, Yakubu Nale J, Malik D, Rebecca Jane Clokie M. Unravelling the Links between Phage Adsorption and Successful Infection in Clostridium difficile. Viruses. 2018;10:441. [DOI] [PMC free article] [PubMed]
- 39.Nair RR, Fiegna F, Velicer GJ. Indirect evolution of social fitness inequalities and facultative social exploitation. Proc R Soc B Biol Sci. 2018;285:20180054. doi: 10.1098/rspb.2018.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Postma M, Goedhart J. PlotsOfData—A web app for visualizing data together with their summaries. PLOS Biol. 2019;17:e3000202. doi: 10.1371/journal.pbio.3000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wood M. Statistical inference using bootstrap confidence intervals. Significance. 2004;1:180–2. doi: 10.1111/j.1740-9713.2004.00067.x. [DOI] [Google Scholar]
- 42.Cumming G, Finch S. Inference by eye: confidence interval and how to read pictures of data. Am Psychol. 2005;60:170–80. doi: 10.1037/0003-066X.60.2.170. [DOI] [PubMed] [Google Scholar]
- 43.Frada MJ, Schatz D, Farstey V, Ossolinski JE, Sabanay H, Ben-Dor S, et al. Zooplankton may serve as transmission vectors for viruses infecting algal blooms in the ocean. Curr Biol. 2014;24:2592–7. doi: 10.1016/j.cub.2014.09.031. [DOI] [PubMed] [Google Scholar]
- 44.Frada MJ, Vardi A. Algal viruses hitchhiking on zooplankton across phytoplankton blooms. Commun Integr Biol. 2015;8:e1029210. doi: 10.1080/19420889.2015.1029210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Totsche KU, Kögel-Knabner I. Mobile organic sorbent affected contaminant transport in soil: numerical case studies for enhanced and reduced mobility. Vadose Zo J. 2004;3:352–67. doi: 10.2136/vzj2004.0352. [DOI] [Google Scholar]
- 46.Reche I, D’Orta G, Mladenov N, Winget DM, Suttle CA. Deposition rates of viruses and bacteria above the atmospheric boundary layer. ISME J. 2018;12:1154–62. doi: 10.1038/s41396-017-0042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehmann K, Lehmann R, Totsche KU. Event-driven dynamics of the total mobile inventory in undisturbed soil account for significant fluxes of particulate organic carbon. Sci Total Environ. 2021;756:143774. doi: 10.1016/j.scitotenv.2020.143774. [DOI] [PubMed] [Google Scholar]
- 48.Storms ZJ, Sauvageau D. Modeling tailed bacteriophage adsorption: insight into mechanisms. Virology. 2015;485:355–62. doi: 10.1016/j.virol.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 49.Shan Y, Harms H, Wick LY. Electric field effects on bacterial deposition and transport in porous media. Environ Sci Technol. 2018;52:14294–301. doi: 10.1021/acs.est.8b03648. [DOI] [PubMed] [Google Scholar]
- 50.Junier P, Cailleau G, Palmieri I, Vallotton C, Trautschold OC, Junier T, et al. Democratization of fungal highway columns as a tool to investigate bacteria associated with soil fungi. FEMS Microbiol Ecol. 2021;97:fiab003. doi: 10.1093/femsec/fiab003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Furuno S, Remer R, Chatzinotas A, Harms H, Wick LY. Use of mycelia as paths for the isolation of contaminant-degrading bacteria from soil. Micro Biotechnol. 2012;5:142–8. doi: 10.1111/j.1751-7915.2011.00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang F, Zhang L, Zhou J, George TS, Feng G. Arbuscular mycorrhizal fungi enhance mineralisation of organic phosphorus by carrying bacteria along their extraradical hyphae. N. Phytol. 2021;230:304–15. doi: 10.1111/nph.17081. [DOI] [PubMed] [Google Scholar]
- 53.Jansa J, Hodge A. Swimming, gliding, or hyphal riding? On microbial migration along the arbuscular mycorrhizal hyphal highway and functional consequences thereof. N. Phytol. 2021;230:14–16. doi: 10.1111/nph.17244. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Kastman EK, Guasto JS, Wolfe BE. Fungal networks shape dynamics of bacterial dispersal and community assembly in cheese rind microbiomes. Nat Commun. 2018;9:336. doi: 10.1038/s41467-017-02522-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ping D, Wang T, Fraebel DT, Maslov S, Sneppen K, Kuehn S. Hitchhiking, collapse, and contingency in phage infections of migrating bacterial populations. ISME J. 2020;14:2007–18. doi: 10.1038/s41396-020-0664-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Testa S, Berger S, Piccardi P, Oechslin F, Resch G, Mitri S. Spatial structure affects phage efficacy in infecting dual-strain biofilms of Pseudomonas aeruginosa. Commun Biol. 2019;2:405. doi: 10.1038/s42003-019-0633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.May T, Tsuruta K, Okabe S. Exposure of conjugative plasmid carrying Escherichia coli biofilms to male-specific bacteriophages. ISME J. 2011;5:771–5. doi: 10.1038/ismej.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abedon ST. Phage “delay” towards enhancing bacterial escape from biofilms: a more comprehensive way of viewing resistance to bacteriophages. AIMS Microbiol. 2017;3:186. doi: 10.3934/microbiol.2017.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams MH Bacteriophages (Interscience Publishers, Inc., New York - London, 1959)
- 60.Schrader HS, Schrader JO, Walker JJ, Bruggeman NB, Vanderloop JM, Shaffer JJ, et al. Effects of host starvation on bacteriophage dynamics. Bact Oligotrophic Environ Starvation-Survival Lifestyle. 1997; 368-85.
- 61.Schrader HS, Schrader JO, Walker JJ, Wolf TA, Nickerson KW, Kokjohn TA. Bacteriophage infection and multiplication occur in Pseudomonas aeruginosa starved for 5 years. Can J Microbiol. 1997;43:1157–63. doi: 10.1139/m97-164. [DOI] [PubMed] [Google Scholar]
- 62.Łoś M, Golec P, Łoś JM, Węglewska-Jurkiewicz A, Czyż A, Węgrzyn A, et al. Effective inhibition of lytic development of bacteriophages λ, P1 and T4 by starvation of their host, Escherichia coli. BMC Biotechnol. 2007;7:13. doi: 10.1186/1472-6750-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bryan D, El-Shibiny A, Hobbs Z, Porter J, Kutter EM. Bacteriophage T4 infection of stationary phase E. coli: life after log from a phage perspective. Front Microbiol. 2016;7:1391. doi: 10.3389/fmicb.2016.01391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yin J. A quantifiable phenotype of viral propagation. Biochem Biophys Res Commun. 1991;174:1009–14. doi: 10.1016/0006-291X(91)91519-I. [DOI] [PubMed] [Google Scholar]
- 65.Chatterjee A, Willett JLE, Dunny GM, Duerkop BA. Phage infection and sub-lethal antibiotic exposure mediate Enterococcus faecalis type VII secretion system dependent inhibition of bystander bacteria. PLOS Genet. 2021;17:e1009204. doi: 10.1371/journal.pgen.1009204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berthold T, Centler F, Hübschmann T, Remer R, Thullner M, Harms H, et al. Mycelia as a focal point for horizontal gene transfer among soil bacteria. Sci Rep. 2016;6:36390. doi: 10.1038/srep36390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee KL, Hubbard LC, Hern S, Yildiz I, Gratzl M, Steinmetz NF. Shape matters: the diffusion rates of TMV rods and CPMV icosahedrons in a spheroid model of extracellular matrix are distinct. Biomater Sci. 2013;1. 10.1039/C3BM00191A. [DOI] [PMC free article] [PubMed]
- 68.Hudson P, Greenman J. Competition mediated by parasites: biological and theoretical progress. Trends Ecol Evol. 1998;13:387–90. doi: 10.1016/S0169-5347(98)01475-X. [DOI] [PubMed] [Google Scholar]
- 69.Sax DF, Stachowicz JJ, Brown JH, Bruno JF, Dawson MN, Gaines SD, et al. Ecological and evolutionary insights from species invasions. Trends Ecol Evol. 2007;22:465–71. doi: 10.1016/j.tree.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 70.Wagner PL, Waldor MK. Bacteriophage control of bacterial virulence. Infect Immun. 2020;70:3985–93. doi: 10.1128/IAI.70.8.3985-3993.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chantrey J, Dale TD, Read JM, White S, Whitfield F, Jones D, et al. European red squirrel population dynamics driven by squirrelpox at a gray squirrel invasion interface. Ecol Evol. 2014;4:3788–99. doi: 10.1002/ece3.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Essl F, Bacher S, Genovesi P, Hulme PE, Jeschke JM, Katsanevakis S, et al. Which taxa are alien? Criteria, applications, and uncertainties. Bioscience. 2018;68:496–509. doi: 10.1093/biosci/biy057. [DOI] [Google Scholar]
- 73.Seebens H, Gastner MT, Blasius B. The risk of marine bioinvasion caused by global shipping. Ecol Lett. 2013;16:782–90. doi: 10.1111/ele.12111. [DOI] [PubMed] [Google Scholar]
- 74.Seebens H, Essl F, Blasius B. The intermediate distance hypothesis of biological invasions. Ecol Lett. 2017;20:158–65. doi: 10.1111/ele.12715. [DOI] [PubMed] [Google Scholar]
- 75.Hulme PE, Bacher S, Kenis M, Klotz S, Kühn I, Minchin D, et al. Grasping at the routes of biological invasions: a framework for integrating pathways into policy. J Appl Ecol. 2008;45:403–14. doi: 10.1111/j.1365-2664.2007.01442.x. [DOI] [Google Scholar]
- 76.Liebhold AM, Brockerhoff EG, Garrett LJ, Parke JL, Britton KO. Live plant imports: the major pathway for forest insect and pathogen invasions of the US. Front Ecol Environ. 2012;10:135–43. doi: 10.1890/110198. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.