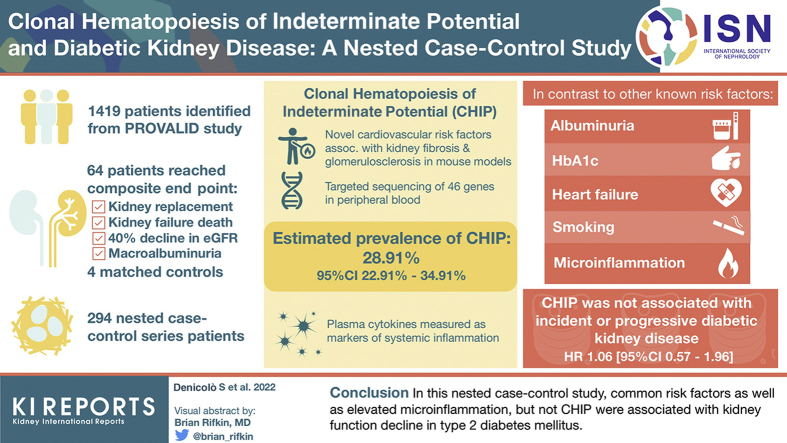
Abstract
Introduction
The disease trajectory of diabetic kidney disease (DKD) shows a high interindividual variability not sufficiently explained by conventional risk factors. Clonal hematopoiesis of indeterminate potential (CHIP) is a proposed novel cardiovascular risk factor. Increased kidney fibrosis and glomerulosclerosis were described in mouse models of CHIP. Here, we aim to analyze whether CHIP affects the incidence or progression of DKD.
Methods
A total of 1419 eligible participants of the PROVALID Study were the basis for a nested case-control (NCC) design. A total of 64 participants who reached a prespecified composite endpoint within the observation period (initiation of kidney replacement therapy, death from kidney failure, sustained 40% decline in estimated glomerular filtration rate or sustained progression to macroalbuminuria) were identified and matched to 4 controls resulting in an NCC sample of 294 individuals. CHIP was assessed via targeted amplicon sequencing of 46 genes in peripheral blood. Furthermore, inflammatory cytokines were analyzed in plasma via a multiplex assay.
Results
The estimated prevalence of CHIP was 28.91% (95% CI 22.91%–34.91%). In contrast to other known risk factors (albuminuria, hemoglobin A1c, heart failure, and smoking) and elevated microinflammation, CHIP was not associated with incident or progressive DKD (hazard ratio [HR] 1.06 [95% CI 0.57–1.96]).
Conclusions
In this NCC study, common risk factors as well as elevated microinflammation but not CHIP were associated with kidney function decline in type 2 diabetes mellitus.
Keywords: chronic kidney disease, clonal hematopoiesis of indeterminate potential, cytokines, diabetic kidney disease, inflammation, type 2 diabetes mellitus
Graphical abstract
Diabetic kidney disease (DKD) develops in 25% to 40% of individuals with type 2 diabetes mellitus (T2DM) and is the leading cause of kidney failure needing replacement therapy in almost all industrialized countries.1, 2, 3, 4 Incidence and progression of DKD have been associated with inadequate glycemic and blood pressure control, dyslipidemia, prolonged diabetes duration, advanced age, genetic predisposition, lifestyle factors, and comorbidities.1 However, the course of the disease shows a considerably high interindividual variability that often cannot be sufficiently explained by conventional risk factors.5,6
Clonal hematopoiesis of indeterminate potential (CHIP) has recently emerged as a novel cardiovascular risk factor linking the innate immune system to specific clonal drivers, aging, and subclinical chronic inflammation (microinflammation).7,8 CHIP is defined as a clonal expansion in the hematopoietic lineages in the absence of a hematological malignancy.9 It is driven by somatic variants providing a proliferative advantage to specific hematopoietic stem cells.9,10 Variants are mainly observed in the epigenetic regulators DNMT3A, TET2, and ASXL1, as well as the cytokine signaling component JAK2 and the tumor suppressor protein TP53.11, 12, 13 As somatic variants accumulate in different cell types throughout life,14 the prevalence of CHIP increases considerably with advancing age.11, 12, 13
In previous studies the presence of CHIP has been associated with increased risk of coronary heart disease, ischemic stroke, and heart failure11,15, 16, 17 as well as adverse outcomes in patients with established heart failure18, 19, 20 or degenerative aortic valve stenosis.21 Experimental data suggest endothelial as well as tissue inflammation and fibrosis via clonally-derived macrophages and monocytes as possible mechanisms behind adverse outcomes.15,22,23
Studies in mouse models simulating myeloid TET2, DNMT3A, and JAK2 variants revealed increased atherogenesis as well as myocardial inflammation and fibrosis.15,22,23 Furthermore, increased glomerulosclerosis15 and greater kidney fibrosis when challenged with angiotensin II22 were observed in these models. CHIP and DKD are both associated with advanced age and microinflammation and, in both conditions, macrophages seem to play a pivotal role.7,24, 25, 26 On the basis of these observations, we hypothesized that CHIP may be associated with inflammatory cytokines and kidney function decline in patients with T2DM.
Methods
Study Population and Design
This study is based on participants of the PROVALID Study, a noninterventional, prospective cohort study including 4000 patients with T2DM in 5 European countries (Austria, Hungary, United Kingdom, Poland, and The Netherlands). Patients with T2DM >18 years were recruited between 2011 and 2014 at the primary level of health care and followed up annually with a tolerance interval of ±3 months. All study visits were integrated into the routine medical care. Clinical and laboratory parameters were obtained at baseline and on every annual follow-up visit. Serum creatinine and urinary albumin and creatinine were measured by local laboratories and entered into a web-based data repository. Estimated glomerular filtration rate was calculated using the Modification of Diet in Renal Disease equation.27 For the diagnosis of albuminuria, successive urinary albumin and creatinine measurements at 3 different time points were used to calculate urine albumin-to-creatinine ratios (mg/g). Albuminuria was then classified on the basis of a two-out-of-three principle (e.g., normoalbuminuria, normoalbuminuria, and microalbuminuria was classified as normoalbuminuria). When fewer than 3 urine collections were available, albuminuria was determined by calculating the arithmetic mean. Further details regarding the study design and baseline characteristics of PROVALID are published elsewhere.28
A nested case-control (NCC) design was used for this study. The study design has been proposed to be highly efficient for analyses requiring high-cost technologies where testing of the whole cohort would not be feasible and event rates are low.29 A total of 1419 participants from PROVALID were eligible for this study (selection cohort). Inclusion and exclusion criteria and the selection process are shown in Supplementary Table S1 and Supplementary Figure S1, respectively. The median follow-up was 5.00 years (25% and 75% quantiles 3.95 and 6.04). Kidney function decline was defined as reaching a prespecified composite end point within the observation period. The composite end point comprised (i) initiation of kidney replacement therapy, (ii) death from kidney failure, (iii) decline in estimated glomerular filtration rate of ≥40% from baseline to <60 ml/min per 1.73 m2, or (iv) progression to macroalbuminuria including a 30% increase in the mean urinary albumin-to-creatinine ratio from baseline. To differentiate incidence or progression of DKD from acute kidney injury or transient increases in albuminuria, both estimated glomerular filtration rate decline and progression to macroalbuminuria had to be sustained over at least 1 year as assessed on 2 consecutive in-study measurements. Initiation of kidney replacement therapy (dialysis or transplantation) and death from kidney failure were investigator-reported. The components of the composite end point were selected on the basis of the proposed definition of major adverse renal events30 and the international consensus definition of clinical trial outcomes for kidney failure.31 Of 1419 eligible PROVALID participants (selection cohort), 64 patients were identified who reached the composite end point within the observation period (cases). Details on the distribution of the different components of the composite end point are shown in Supplementary Table S2. Under the assumption of a CHIP prevalence of 16% (to 33%),11,15,18,21 the study was powered (80%) to detect a clinically meaningful harzard ratio (HR) of 2.6 (to 2.1).32 Thus, to each of the 64 cases 4 controls were matched by age decade and country. As in an NCC design, a subject may act as a control for more than one case, and a case may serve as control before becoming a case,29 this particular stratified random sampling and matching procedure resulted in a study population of 294 individuals (NCC sample).
The PROVALID study protocol was approved in each participating country by the responsible local Institutional Review Boards. Signing an informed consent form was a prerequisite for study participation in all countries. The analysis was further approved by the Ethics Committee of Medical University Innsbruck (EK Nr. 1015/2020). This study is in line with the guidelines laid out by the Declaration of Helsinki.
Sample Preparation
Peripheral blood EDTA and plasma EDTA stored at −80 °C in the PROVALID study biobank was utilized. DNA was isolated with QIAsymphony DNA Midi Kit (Qiagen, Hilden, Germany) on QIAsymphony (Qiagen) from deep-frozen samples of peripheral blood. DNA concentrations were obtained by Qubit dsDNA BR Assay Kit (Life Technologies, Carlsbad, CA) on a Qubit Fluorometer (Life Technologies).
Next-Generation Sequencing
A targeted sequencing approach for the detection of CHIP variants was applied, using the TruSight Myeloid Sequencing Panel (Illumina, San Diego, CA) covering genes mostly affected in myeloid neoplasms and precursors thereof. Genes available on the TruSight Myeloid Sequencing Panel were categorized by being often, seldom, or not analyzed in the context of CHIP. Only genes at least once associated with CHIP were considered for analysis (Supplementary Table S3). Library preparation with 50 ng DNA per sample was performed according to the manufacturer. Paired-End sequencing was performed on MiSeq devices with 151 cycles per direction as specified by the manufacturer.
Variant Calling and Annotation Strategies
For data analysis and variant calling, the SeqNext Module of JSI Medical Systems (Ettenheim, Germany) was utilized to align raw corresponding sequence reads to the hg19 build of the human reference genome. Regions with a minimum coverage of 50 were analyzed. Common single-nucleotide polymorphisms with a minor allele frequency of at least 1% in the non-Finnish European population and synonymous changes were excluded. In addition, variants present in >10% of the cohort as well as in >10% of patients per sequencing run and variants in areas with excessive local noise were considered technical artifacts and excluded from the data set. Only coding variants with a variant allele frequency (VAF) of at least 2%, a quality score of ≥30, and a variant read number of at least 15 were included. Furthermore, variants with a VAF of 0.45 to 0.55 and 0.98 to 1.00 as well as 0.48 to 0.66 for the gene NOTCH1, according to statistical analysis of variant distribution, were not considered in the results to exclude potential germline variants. The remaining variants were evaluated via MLLi:db (Database of Münchner Leukämie Labor, https://mlli.com), Cosmic (Catalogue of Somatic Mutations in Cancer, https://cancer.sanger.ac.uk/cosmic), HGMD (Human Gene Mutation Database, https://ihgseq13.helmholtz-muenchen.de/hgmd/pro/search_gene.php), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar), and LOVD (Leiden Open Variation Database, https://www.lovd.nl/) for exclusion of benign variants.
Cytokine Assay
Multiplex assay for analysis of inflammatory cytokine release was performed using LEGENDplex Human Inflammation Panel 1 (BioLegend, San Diego, CA) following kit instructions on plasma EDTA samples. A list of all analyzed cytokines is provided in Supplementary Table S4. Deviating from kit instructions, samples were incubated with the primary antibody overnight and washed 3 times for binding of low abundant cytokines and background reduction. Data were collected on a Navios flow cytometer (Beckmann Coulter, Brea, CA) and analyzed using BioLegend LEGENDplex Analysis Software Version 2020.11.19 (https://legendplex.qognit.com, San Diego, CA). The geometric mean of each cytokine is reported per patient according to the standard curves and calculations performed by the LEGENDplex Analysis Software. Out of range values were substituted with the lower limit of detection specific for each analyzed cytokine, an approach which, according to Pfister et al.,33 has shown to only marginally bias group difference estimation and thus not enhance further type II errors in subsequent analyses. Information on inter- and intra-assay variability is provided in the Supplementary Material.
Statistical Analysis
Patient characteristics for continuous variables are described with mean and standard deviations (SD) or, when appropriate (e.g., because of skewness), with median and 25% and 75% quantiles. For discrete variables, absolute and relative frequencies are presented. Separate descriptive statistics are shown for cases and controls and for CHIP carriers and noncarriers. Bar plots are presented to describe the distribution of CHIP variants, the frequency of mutations within patients, and the age-specific prevalence of CHIP in our NCC sample. The prevalence of CHIP in the selection cohort (n = 1419) was estimated applying an inverse probability weighting procedure to our sample.34 For each pair of potential controls, the joint probability of being sampled in the NCC sample was derived by simulation. The resulting inclusion matrix was used to perform a Horvitz-Thompson estimation of the population mean with a respective 95% confidence interval (CI).35, 36, 37
The distributions of cytokine concentrations by cases and controls and by CHIP carrier status are described with boxplots. In line with theory, plasma cytokines levels were found to be highly collinear. Partial correlations were calculated from Pearson correlations to account for and briefly describe their complex directed interplay. In situations of high multicollinearity, one has to be wary, as using such data poses additional uncertainty in any subsequent analyses, that is, it depletes power and gives room for discovering spurious relationships. The NCC design does not allow for structural modeling of the cytokines’ relationships alongside with their effect on the outcome. Instead, as a mean to reduce the dimension of the cytokine data to a single variable while retaining a maximum of variation, a principal component (PC) analysis was used.38 The first PC (PC1), standardized as a so-called PC score, turned out to be a solely positively weighted sum of all cytokines (Supplementary Table S5) that captures 55.2% of their variation and mainly consists of interleukin (IL)-18, MCP-1, and IL-8. PC1 was used in the regression analysis. PC1 is interpreted as a proxy variable for the degree of microinflammation.
To not compromise the study design, all analyses are complete case analyses. To gain efficiency in estimation and circumvent bias through confounding at the stage of sampling, we matched cases on each control’s age decade and country. Both variables capture unobserved heterogeneity and very likely affect the outcome as well as potentially the risk factor of primary interest—the CHIP carrier status.39 For each of the 64 cases, stratified incidence density sampling of 4 controls was performed.39, 40, 41 Under this sampling scheme, the conditional logistic likelihood can be exploited to estimate (hazard) rate ratios comparable to those of the underlying cohort as proposed initially in Liddell et al.39,42, 43, 44 We reweight the Breslow estimator with the inverse of the incidence density sampling probabilities to estimate the cumulative incidence for CHIP carriers and noncarriers as also explained by Samuelsen et al.34 To explore crude absolute risk, we present the estimated cumulative incidence of the composite end point in the selection cohort compared with corresponding inverse probability weighting estimates for CHIP carriers and noncarriers from the NCC sample.
We ran unadjusted and adjusted regressions to estimate the effect of CHIP on the outcome. Risk factors at baseline known from the literature were employed in the adjusted models to reduce bias due to the omission of relevant (confounding) variables. Adding these variables to the regressions significantly increased the concordance statistic of the CHIP (DNMT3A/TET2) specification from 50.6% (51.6%) to 83.2% (81.6%), which hints toward their prognostic power.45,46 To measure relative risk, we present HRs with 95% CIs. Continuous variables used to adjust models were standardized. We allow for a type 1 error of 5%, and all hypotheses are two-sided. All calculations were carried out using R 3.6.3.47
Results
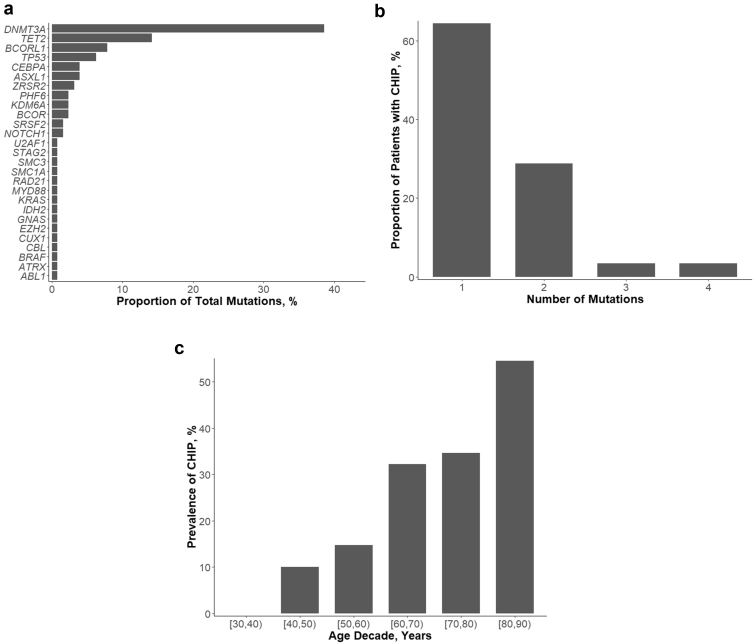
CHIP was identified in 87 of 294 individuals, with a total of 127 aberrations affecting 27 of 46 analyzed CHIP-associated genes (Supplementary Table S6). On the basis of the occurrence of CHIP in the NCC sample (n = 294), the prevalence of CHIP in the selection cohort (n = 1419) was estimated to be 28.91% (95% CI 22.91%–34.91%). The median VAF was 4.60 (25% and 75% quantiles 2.55 and 9.55). Baseline characteristics of the whole cohort as well as grouped by cases and controls and by CHIP status are shown in Table 1. Variants were most commonly found in DNMT3A, TET2, BCORL1, and TP53 (Figure 1a). Most individuals had 1 CHIP-associated variant; however, in some individuals, up to 4 variants were found (Figure 1b). The occurrence of CHP increased with advancing age (Figure 1c).
Table 1.
Baseline characteristics
| Characteristics | Overall | Controls | Cases | CHIP − | CHIP + |
|---|---|---|---|---|---|
| n | 294 | 230 | 64 | 207 | 87 |
| Country, n (%) | |||||
| Austria | 95 (32.3) | 74 (32.2) | 21 (32.8) | 67 (32.4) | 28 (32.2) |
| Hungary | 166 (56.5) | 130 (56.5) | 36 (56.2) | 116 (56.0) | 50 (57.5) |
| UK | 33 (11.2) | 26 (11.3) | 7 (10.9) | 24 (11.6) | 9 (10.3) |
| Sex (male), n (%) | 155 (52.7) | 119 (51.7) | 36 (56.2) | 112 (54.1) | 43 (49.4) |
| Age, yr | 65.26 (9.80) | 65.24 (9.86) | 65.30 (9.63) | 63.68 (9.94) | 69.01 (8.38) |
| Duration of T2DM, yr | 10.00 [5.00–16.00] | 8.00 [4.00–15.00] | 12.50 [8.00–19.00] | 10.00 [5.00–15.00] | 9.00 [4.50–17.50] |
| Duration of hypertension, yr | 12.00 [8.00–19.00] | 12.00 [7.75–17.25] | 15.00 [9.50–22.50] | 12.00 [8.00–18.00] | 13.00 [9.00–20.00] |
| Insulin use, n (%) | 80 (27.2) | 54 (23.5) | 26 (40.6) | 53 (25.6) | 27 (31.0) |
| HbA1c, % | 6.80 [6.20–7.50] | 6.70 [6.20–7.39] | 6.95 [6.50–7.67] | 6.80 [6.23–7.50] | 6.70 [6.20–7.30] |
| Systolic BP, mm Hg | 138.32 (17.59) | 138.05 (16.91) | 139.30 (19.99) | 136.92 (17.78) | 141.64 (16.77) |
| Diastolic BP, mm Hg | 80.00 (9.53) | 80.24 (9.16) | 79.12 (10.79) | 79.90 (9.79) | 80.23 (8.93) |
| Total cholesterol, mg/dl | 183.84 (45.82) | 185.78 (45.30) | 176.86 (47.35) | 185.49 (46.36) | 179.92 (44.52) |
| LDL cholesterol, mg/dl | 104.52 (41.04) | 106.54 (40.17) | 97.26 (43.59) | 105.22 (41.85) | 102.86 (39.24) |
| HDL cholesterol, mg/dl | 48.65 (12.82) | 48.98 (12.84) | 47.46 (12.77) | 48.70 (12.96) | 48.52 (12.54) |
| Triglycerides, mg/dl | 163.29 (80.29) | 162.14 (79.67) | 167.44 (82.98) | 166.42 (80.56) | 155.86 (79.63) |
| eGFR, ml/min per 1.73 m2 | 76.39 (25.24) | 77.54 (21.90) | 72.25 (34.62) | 78.60 (24.97) | 71.14 (25.27) |
| UACR, mg/g creatinine | 11.29 [5.36–36.84] | 8.50 [4.63–20.65] | 52.49 [14.57–294.06] | 10.56 [5.30–31.65] | 14.30 [5.78–47.26] |
| Albuminuria, n (%) | |||||
| Normoalbuminuria | 217 (73.8) | 191 (83.0) | 26 (40.6) | 157 (75.8) | 60 (69.0) |
| Microalbuminuria | 20 (6.8) | 6 (2.6) | 14 (21.9) | 12 (5.8) | 8 (9.2) |
| Macroalbuminuria | 57 (19.4) | 33 (14.3) | 24 (37.5) | 38 (18.4) | 19 (21.8) |
| CRP, mg/dl | 0.72 (1.56) | 0.72 (1.63) | 0.71 (1.26) | 0.64 (1.08) | 0.91 (2.32) |
| Hemoglobin, g/dl | 13.72 (1.55) | 13.90 (1.39) | 13.09 (1.90) | 13.74 (1.57) | 13.66 (1.50) |
| BMI, kg/m2 | 31.34 (5.54) | 31.15 (5.41) | 32.02 (5.98) | 31.49 (5.71) | 30.98 (5.15) |
| Smoking, n (%) | |||||
| Never | 160 (54.4) | 136 (59.1) | 24 (37.5) | 112 (54.1) | 48 (55.2) |
| Current smoker | 31 (10.5) | 22 (9.6) | 9 (14.1) | 24 (11.6) | 7 (8.0) |
| Former smoker | 103 (35.0) | 72 (31.3) | 31 (48.4) | 71 (34.3) | 32 (36.8) |
| Heart failure, n (%) | 14 (4.8) | 6 (2.6) | 8 (12.5) | 9 (4.3) | 5 (5.7) |
| CAD, n (%) | 57 (19.4) | 37 (16.1) | 20 (31.2) | 40 (19.3) | 17 (19.5) |
| PAD, n (%) | 27 (9.2) | 22 (9.6) | 5 (7.8) | 23 (11.1) | 4 (4.6) |
| CD, n (%) | 19 (6.5) | 12 (5.2) | 7 (10.9) | 13 (6.3) | 6 (6.9) |
| Diabetic retinopathy, n (%) | |||||
| No | 223 (75.9) | 182 (79.1) | 41 (64.1) | 164 (79.2) | 59 (67.8) |
| Yes | 44 (15.0) | 32 (13.9) | 12 (18.8) | 27 (13.0) | 17 (19.5) |
| Unknown | 27 (9.2) | 16 (7.0) | 11 (17.2) | 16 (7.7) | 11 (12.6) |
BMI, body mass index; BP, blood pressure; CAD, coronary artery disease; CD, cerebrovascular disease; CHIP, clonal hematopoiesis of indeterminate potential; CRP, c-reactive protein; eGFR, estimated glomerular filtration rate; HbA1c, hemoglobin A1c; HDL, high-density lipoprotein; LDL, low-density lipoprotein; PAD, peripheral artery disease; T2DM, type 2 diabetes mellitus; UACR, urine albumin-to-creatinine ratio; UK, United Kingdom.
Categorical variables are presented as counts (%). For continuous variables, mean (SD) or median [25% and 75% quantile] are presented.
Figure 1.
CHIP in individuals with T2DM; (a) Frequencies of identified variants (n = 127) in CHIP-associated genes, (b) occurrence of 1 and more variants in individuals with CHIP, (c) Prevalence of CHIP by age decade. (a) Frequency distribution of affected genes. A detailed list of all identified variants is provided in Supplementary Table S6. (b) Proportions of CHIP carriers with 1 or more variants in CHIP-associated genes. (c) Prevalence of CHIP by age decade at baseline. CHIP, clonal hematopoiesis of indeterminate potential.
CHIP and Kidney Outcome in T2DM
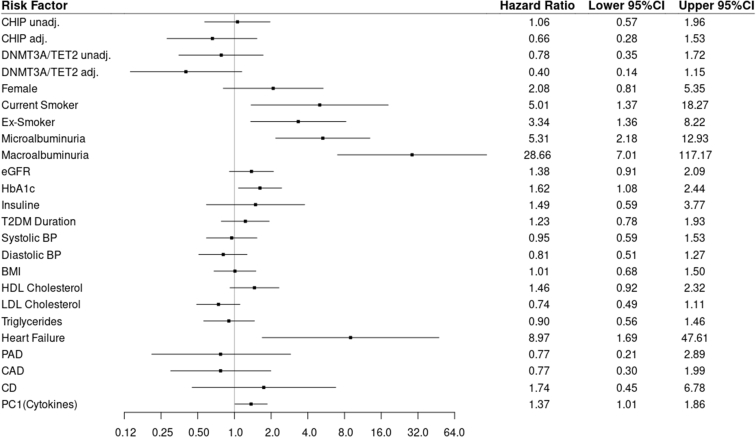
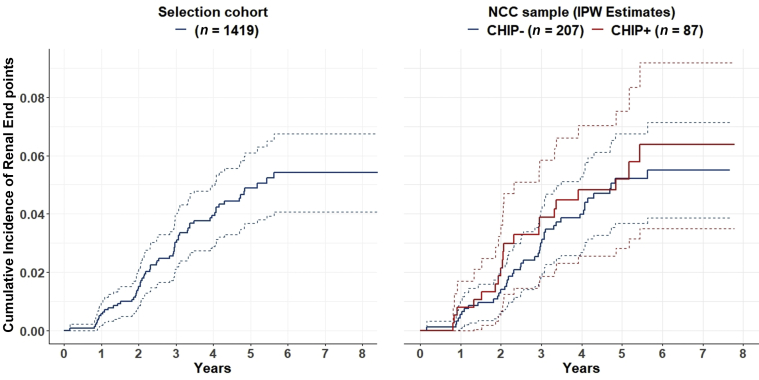
In this NCC study of patients with T2DM, CHIP was not associated with worse kidney outcome (HR 1.06 [95% CI 0.57–1.96]). A subanalysis only considering variants in DNMT3A and TET2 showed similar results (HR 0.78 [95% CI 0.35–1.72]) without change on adjusting the model for different covariates and possible confounders or inflammatory cytokines (HR 0.66 [95% CI 0.28–1.53] and HR 0.40 [95% CI 0.14–1.15]). In addition, on analysis of larger CHIP clones (VAF >0.05, >0.1, or >0.15) no effect on kidney outcome could be observed. While there was no association between CHIP and the composite outcome, known risk factors such as albuminuria, higher hemoglobin A1c, heart failure, and smoking were associated with kidney function decline. Crude and adjusted analyses as well as covariates that were used to adjust the models are shown in Figure 2. In Figure 3, the cumulative incidence rate of the composite end point in the selection cohort is compared with the corresponding inverse probability weighting estimates for CHIP carrier and noncarrier in the NCC sample. The curves show closely resembling patterns for the selection cohort and the NCC sample but no significant differences with regard to the CHIP carrier status.
Figure 2.
Kidney outcome by CHIP carrier status and influence of risk factors. Relative risks are presented as hazard ratios with 95% CIs. Continuous variables used to adjust models were standardized. All listed variables were used to adjust the model for CHIP and DNMT3A/TET2. The hazard ratios of the adjustment variables in the DNMT3A/TET2 model are not shown as they did not differ. The vertical axis is on a logarithmic scale. adj., adjusted; BMI, body mass index; BP, blood pressure; CAD, coronary artery disease; CD, cerebrovascular disease; CHIP, clonal hematopoiesis of indeterminate potential; eGFR, estimated glomerular filtration rate; HbA1c, hemoglobin A1c; HDL, high-density lipoprotein; LDL, low-density lipoprotein; PAD, peripheral artery disease; T2DM, type 2 diabetes mellitus; Unadj., unadjusted.
Figure 3.
Cumulative incidence of the composite end point in the selection cohort and corresponding IPW estimates for CHIP carrier and noncarrier in the NCC sample. The cumulative incidence rate of the composite end point in the selection cohort is shown in the left figure; the corresponding IPW estimates for CHIP carrier and noncarrier in the NCC sample are shown in the right figure. Dashed lines indicate 95% confidence bands. CHIP, clonal hematopoiesis of indeterminate potential; IPW, inverse probability weighting; NCC, nested case-control.
CHIP and Inflammatory Cytokine Profiles in T2DM
A set of 13 inflammatory cytokines was measured in plasma samples of all 294 individuals in the NCC sample. Analyzed cytokines are listed in Supplementary Table S4. Cytokine plasma concentration varied greatly among participants, ranging from being not detected to above physiological levels. Values for IL-1β, IFN-α2, IFN-γ, TNF-α, IL-6, IL-10, IL12p70, IL-17A, IL-23, and IL-33 could not be obtained from all individuals, because of a physiologically low plasma concentration. This was also described by the assay reference plasma samples.
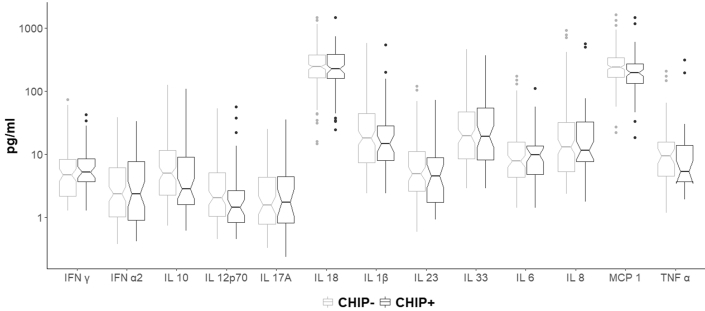
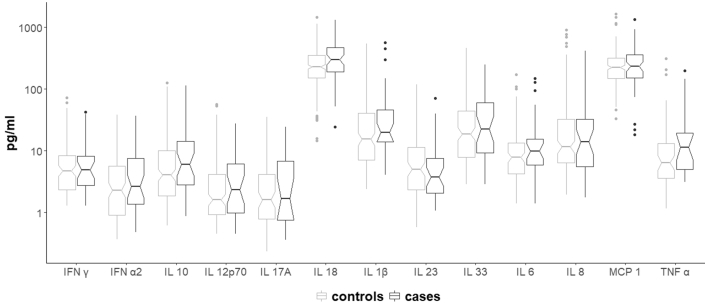
In this study, no difference in plasma cytokine levels between CHIP carriers and noncarriers could be observed (Figure 4). Plasma cytokine levels in cases and controls are shown in Figure 5.
Figure 4.
Plasma cytokine levels in CHIP carriers and noncarriers. Boxplots of plasma cytokine levels by CHIP status. The notch around the median indicates the 95% CI. CHIP, clonal hematopoiesis of indeterminate potential; IL, interleukin.
Figure 5.
Plasma cytokine levels in cases and controls; Boxplots of plasma cytokine levels by cases and control. The notch around the median indicates the 95% CI. IL, interleukin.
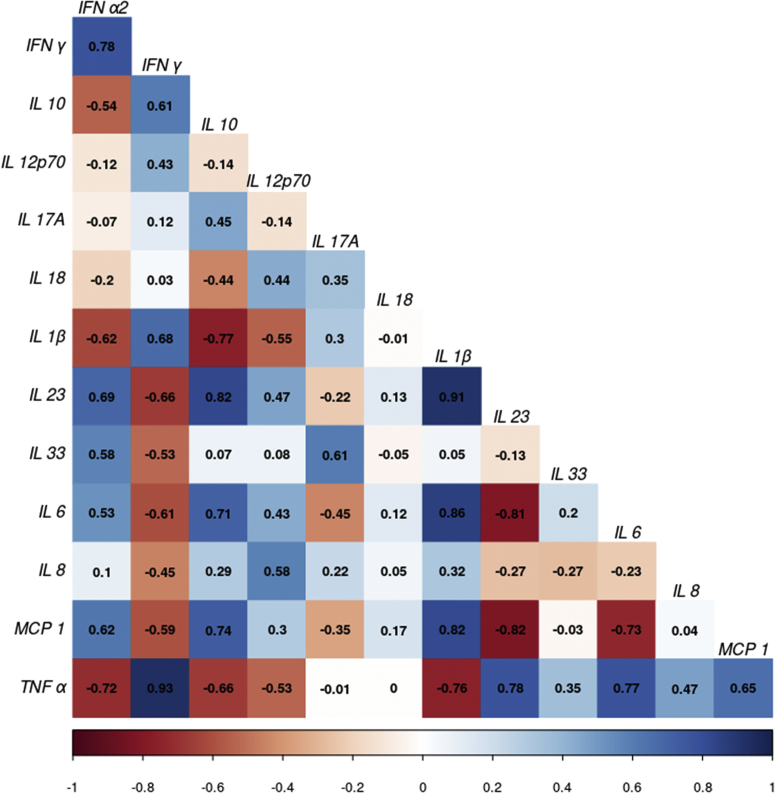
Partial correlations in Figure 6 show that the measured cytokines were highly interdependent. Because of these strong mutual influences, it was not possible to identify direct causal origins of the individual cytokines’ effects on the outcome. Thus, in the regression analysis, a PC score (PC1) derived from the cytokine data was used as a proxy variable for the degree of microinflammation. Elevated microinflammation as represented by PC1 was associated with an increased risk of incidence or progression of DKD (HR 1.37 [95% CI 1.01–1.86]).
Figure 6.
Partial correlations of cytokines. The correlogram displays the partial correlations of the cytokines with blue indicating a positive and red a negative observed relationship between the row and column cytokines. IL, interleukin.
Discussion
The prevalence of CHIP in our study was higher as compared with studies utilizing genome- or exome-wide sequencing approaches, which were among others conducted in 22 cohorts in type 2 diabetes association studies.11,12 This is mostly due to a higher sensitivity of targeted sequencing, as previously discussed by Buscarlet et al.48 Petzer et al.49 could also show a higher prevalence of CHIP using our same sequencing approach as compared to studies using genome- or exome-wide sequencing. However, besides the sensitivity of the sequencing technique, the prevalence of CHIP also depends on how CHIP-associated genes and variants are defined.11 Dorsheimer et al.18 used a similar approach to ours and analyzed 56 genes via targeted amplicon sequencing in bone marrow–derived mononuclear cells of 200 individuals with chronic ischemic heart failure. VAF are highly concordant between blood and bone marrow samples, thus allowing a comparison with our results.50 The mean age was 65 years as it was in our study; however, only 36% had diabetes.18 They reported a prevalence of CHIP of 18.5%.18 There might be an enrichment of CHIP in populations with T2DM, as previously suggested by Jaiswal et al.11 An unhealthy diet quality and a higher body mass index have also been previously associated with a higher prevalence of CHIP.51,52
In our population, most variants were found in DNMT3A and TET2, and most individuals with CHIP carried a single mutation. These findings are in line with previously published data.7,48
Multiple studies indicate an increased cardiovascular risk in CHIP,11,15, 16, 17, 18, 19, 20, 21 presumably via increased microinflammation. Marked glomerulosclerosis and increased kidney fibrosis when challenged with angiotensin II were described in mouse models representing a setting with TET2 and DNMT3A variants.15,22 Thus, it seemed reasonable to hypothesize that CHIP would also increase the risk of kidney function decline in T2DM, a condition where microinflammation plays a major role. However, in this study, we did not find an association between CHIP and a prespecified composite end point in patients with T2DM (HR 1.06 [95% CI 0.57–1.96]). Results did not change when different covariates and possible confounders or inflammatory cytokine levels were added to the model (HR 0.66 [95% CI 0.28–1.53]). In addition, exploratory analyses using alternative definitions of CHIP (DNMT3A/TET2 only, different VAF cut-offs) yielded similar results. These exploratory analyses were based on reports of kidney damage in mouse models having TET2 or DNMT3A variants15,22 and on data suggesting a dose-response association between VAF and cardiovascular outcome.16, 17, 18 Although we could not find an effect of CHIP, strong known risk factors such as albuminuria, higher hemoglobin A1c, heart failure, and smoking1 were associated with kidney function decline in our cohort. It has been suggested that CHIP increases cardiovascular risk via endothelial inflammation that promotes atherogenesis.7,15 It is possible that CHIP, via this pathomechanism, contributes primarily to macrovascular complications and has less impact on the microvasculature of the kidney. As elevated microinflammation was associated with worse kidney outcome but CHIP was not, it is also possible that other mechanism of microinflammation outweigh CHIP in DKD.
Inflammation is a possible trigger for oxidative stress associated DNA damage. An inflammatory environment therefore may enable CHIP emergence.53, 54, 55 On the other hand, CHIP has been proposed to increase inflammation itself.7,53 IL-1β and IL-6 expression was previously reported to be increased in mice simulating TET2-variant CHIP.15,56, 57, 58 Increased levels of IL-8, IL-6, and TNF-α were detected in plasma of individuals with CHIP.15,59 Furthermore, monocytes of patients with DNMT3A- or TET2-variant CHIP showed increased expressions of IL-1β, IL-6R, and the NLRP3 inflammasome complex.60 Bick et al.17 found that, among carriers of large CHIP clones, genetically reduced IL-6 signaling abrogated the increased risk of incident cardiovascular disease. Furthermore, CHIP carriers were reported to have 21% higher hs-CRP levels.61 Here, we analyzed plasma levels of IL-1β, TNF-α, MCP-1, IL-6, IL-8, IFN-α2, IFN-γ, IL-10, IL-12p70, IL-17A, IL-18, IL-23, and IL-33. We could not observe any differences in plasma concentrations between CHIP carriers and noncarriers. Results might be explained by competing inflammatory mechanisms in individuals with T2DM and DKD, as many other processes and stimuli—besides CHIP—can influence cytokine release. It is possible that CHIP does not significantly contribute to the already elevated microinflammation in this patient collective.
Microinflammation is an established contributor to the incidence and progression of kidney disease in T2DM.24,25 Thus, we aimed to analyze whether increased microinflammation, as assessed by a panel of 13 inflammatory cytokines in plasma, is associated with the composite end point regardless of CHIP. Although cytokine concentrations were observed to have strong positive correlations (i.e., when one cytokine was high in a subject, other cytokines tended to be high as well), when looking at partial correlations, a more differentiated picture of mutual influence became apparent. This finding was not surprising, as cytokine interactions in the course of an inflammatory response are known to be complex. Owing to these strong mutual dependencies, direct causal origins of the individual cytokines’ effects could not be identified. Thus, we reduced all cytokine measures to a single score that proxies microinflammation. To a large extent, the score captured the variation in IL-18, MCP-1, and IL-8. Higher circulating IL-18 levels have been previously proposed as a predictor of progression of DKD.62,63 Furthermore, IL-18 seems to contribute more to the progression of DKD than to other diabetic complications.64 MCP-1 has also been reported to promote inflammation, kidney injury, and fibrosis via macrophage accumulation in the kidney,65 and increased plasma levels of MCP-1 were found to be associated with kidney disease progression.66 Serum IL-8 levels were positively correlated with markers of podocyte damage and with albuminuria in T2DM patients.67,68
This study has several limitations. Although we believe that we would have been able to identify a strong effect of CHIP on the composite end point, subtle effects may have been missed in this NCC study. This study is a first report of CHIP in the context of DKD, and we would like to encourage further studies in larger cohorts. As an NCC study only allows the analysis of a single prespecified end point, it was not possible to analyze any other nonsampled outcome. Thus, we were not able to assess whether increased cardiovascular and all-cause mortality risks among CHIP carriers could be reproduced in our cohort. However, an NCC design is a cost-effective design to analyze a clearly defined hypothesis.
The cytokine assay was selected to cover inflammatory cytokines considered most relevant in the context of CHIP and DKD on the basis of existing literature. However, we cannot exclude the involvement of other cytokines that were not part of the assay. The rationale for the cytokine analyses was to add information on microinflammation to the study. These analyses were conceptualized to be exploratory, and the study was not primarily designed to detect differences in cytokine levels between CHIP carriers and noncarriers. Thus, small or complex effects may have been missed.
Although the cytokines were assumed to act as a proinflammatory mediator on the causal pathway from CHIP to the outcome, it has to be pointed out that the regression analysis merely shows an association between the cytokines and the outcome, as the design does not allow for causal inspection of the relationship between CHIP and the cytokine levels.
Cytokines other than IL-18, MCP-1, and IL-8 were less represented in PC1. This may be in part due to lower variation and plasma levels of certain cytokines (especially IL-1β, IFN-γ, IL-10, IL12p70, IL-17A, and IL-33). Although it was not possible to analyze the effect of single cytokines on the composite end point, our results confirm that an increased inflammatory environment is associated with worse kidney outcome in T2DM.
In conclusion, in this NCC study common risk factors as well as markers for elevated microinflammation but not CHIP were associated with kidney function decline in patients with T2DM. To the best of our knowledge, this is the first study to analyze whether CHIP contributes to the incidence or progression of DKD.
Disclosure
GM has received speaker or advisory board honoraria from AbbVie, Astellas, Boehringer Ingelheim, Novo Nordisk, and Sanofi. PBM reports receiving personal fees and nonfinancial support from Vifor, personal fees from AstraZeneca, Astellas, Novartis, and Janssen, grants from Boehringer Ingelheim, personal fees and nonfinancial support from Pharmacosmos, and personal fees and nonfinancial support from Napp, outside the submitted work. PP is also employee at Delta 4 GmbH. AW was on the Steering Committee of the ASCEND Study sponsored by GlaxoSmithKline and received lectures fees by Fresenius, Bayer, Chiesi, ROCHE and Astra-Zeneca. All the other authors declared no competing interests.
Acknowledgments
The authors would like to thank the study personnel of the PROVALID Study and the general practitioners who collaborated with the PROVALID Study. This project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under Grant Agreement number 115974 (BEAt-DKD). This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA. Additional support was provided by the project DC-ren, which has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement number 848011. Furthermore, the project received additional funding from available resources at the Institute of Human Genetics of Medical University Innsbruck.
Footnotes
Figure S1. Selection of the study population.
Table S1. Inclusion and exclusion criteria.
Table S2. Components of the composite endpoint.
Table S3. Next generation sequencing panel.
Table S4. Multiplex cytokine assay.
Table S5. Principal component analysis.
Table S6. List of identified CHIP-associated variants.
Data on intra- and inter-assay variation.
STROBE Statement.
Contributor Information
Gert Mayer, Email: gert.mayer@i-med.ac.at.
Emina Jukic, Email: emina.jukic@i-med.ac.at.
Supplementary Material
Figure S1. Selection of the study population.
Table S1. Inclusion and exclusion criteria.
Table S2. Components of the composite endpoint.
Table S3. Next generation sequencing panel.
Table S4. Multiplex cytokine assay.
Table S5. Principal component analysis.
Table S6. List of identified CHIP-associated variants.
Data on intra- and inter-assay variation.
STROBE Statement .
References
- 1.Macisaac R.J., Ekinci E.I., Jerums G. Markers of and risk factors for the development and progression of diabetic kidney disease. Am J Kidney Dis. 2014;63(suppl 2):S39–S62. doi: 10.1053/j.ajkd.2013.10.048. [DOI] [PubMed] [Google Scholar]
- 2.Kramer A., Boenink R., Noordzij M., et al. The ERA-EDTA Registry Annual Report 2017: a summary. Clin Kidney J. 2020;13:693–709. doi: 10.1093/ckj/sfaa048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ANZDATA Registry, ANZDATA [42nd Annual Report] https://www.anzdata.org.au/report/anzdata-42nd-annual-report-2019/
- 4.Saran R., Robinson B., Abbott K.C., et al. US renal data System 2019 annual data report: epidemiology of kidney disease in the United States. Am J Kidney Dis. 2020;75(suppl 1):A6–A7. doi: 10.1053/j.ajkd.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Spanopoulos D., Okhai H., Zaccardi F., et al. Temporal variation of renal function in people with type 2 diabetes mellitus: a retrospective UK clinical practice research datalink cohort study. Diabetes Obes Metab. 2019;21:1817–1823. doi: 10.1111/dom.13734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heinzel A., Kammer M., Mayer G., et al. Validation of plasma biomarker candidates for the prediction of eGFR decline in patients with type 2 diabetes. Diabetes Care. 2018;41:1947–1954. doi: 10.2337/dc18-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaiswal S., Libby P. Clonal haematopoiesis: connecting ageing and inflammation in cardiovascular disease [published correction appears in Nat Rev Cardiol. 2020;17:828] Nat Rev Cardiol. 2020;17:137–144. doi: 10.1038/s41569-019-0247-5-019-0247-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abplanalp W.T., Cremer S., John D., et al. Clonal hematopoiesis–driver DNMT3A mutations alter immune cells in heart failure. Circ Res. 2021;128:216–228. doi: 10.1161/CIRCRESAHA.120.317104. [DOI] [PubMed] [Google Scholar]
- 9.Steensma D.P., Bejar R., Jaiswal S., et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burns S.S., Kapur R. Putative mechanisms underlying cardiovascular disease associated with clonal hematopoiesis of indeterminate potential. Stem Cell Reports. 2020;15:292–306. doi: 10.1016/j.stemcr.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaiswal S., Fontanillas P., Flannick J., et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genovese G., Kähler A.K., Handsaker R.E., et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie M., Lu C., Wang J., et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martincorena I., Campbell P.J. Somatic mutation in cancer and normal cells [published correction appears in Science. 2016;351:aaf5401] Science. 2015;349:1483–1489. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- 15.Jaiswal S., Natarajan P., Silver A.J., et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377:111–121. doi: 10.1056/NEJMoa1701719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu B., Roberts M.B., Raffield L.M., et al. Supplemental association of clonal hematopoiesis with incident heart failure [published correction appears in J Am Coll Cardiol. 2021;78:762] J Am Coll Cardiol. 2021;78:42–52. doi: 10.1016/j.jacc.2021.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bick A.G., Pirruccello J.P., Griffin G.K., et al. Genetic interleukin 6 signaling deficiency attenuates cardiovascular risk in clonal hematopoiesis. Circulation. 2020;141:124–131. doi: 10.1161/CIRCULATIONAHA.119.044362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorsheimer L., Assmus B., Rasper T., et al. Association of mutations contributing to clonal hematopoiesis with prognosis in chronic ischemic heart failure. JAMA Cardiol. 2019;4:25–33. doi: 10.1001/jamacardio.2018.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pascual-Figal D.A., Bayes-Genis A., Díez-Díez M., et al. Clonal hematopoiesis and risk of progression of heart failure with reduced left ventricular ejection fraction. J Am Coll Cardiol. 2021;77:1747–1759. doi: 10.1016/j.jacc.2021.02.028. [DOI] [PubMed] [Google Scholar]
- 20.Kiefer K.C., Cremer S., Pardali E., et al. Full spectrum of clonal haematopoiesis-driver mutations in chronic heart failure and their associations with mortality. ESC Hear Fail. 2021;8:1873–1884. doi: 10.1002/ehf2.13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mas-Peiro S., Hoffmann J., Fichtlscherer S., et al. Clonal haematopoiesis in patients with degenerative aortic valve stenosis undergoing transcatheter aortic valve implantation. Eur Heart J. 2020;41:93–939. doi: 10.1093/eurheartj/ehz591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sano S., Oshima K., Wang Y., Katanasaka Y., Sano M., Walsh K. CRISPR-mediated gene editing to assess the roles of Tet2 and Dnmt3a in clonal hematopoiesis and cardiovascular disease. Circ Res. 2018;123:335–341. doi: 10.1161/CIRCRESAHA.118.313225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sano S., Wang Y., Yura Y., et al. JAK2 V617F-mediated clonal hematopoiesis accelerates pathological remodeling in murine heart failure. JACC Basic Transl Sci. 2019;4:684–697. doi: 10.1016/j.jacbts.2019.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wada J., Makino H. Innate immunity in diabetes and diabetic nephropathy. Nat Rev Nephrol. 2016;12:13–26. doi: 10.1038/nrneph.2015.175. [DOI] [PubMed] [Google Scholar]
- 25.Sun L., Kanwar Y.S. Relevance of TNF-α in the context of other inflammatory cytokines in the progression of diabetic nephropathy. Kidney Int. 2015;88:662–665. doi: 10.1038/ki.2015.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furuta T., Saito T., Ootaka T., et al. The role of macrophages in diabetic glomerulosclerosis. Am J Kidney Dis. 1993;21:480–485. doi: 10.1016/s0272-6386(12)80393-3. [DOI] [PubMed] [Google Scholar]
- 27.Levey A.S., Coresh J., Greene T., et al. Using standardized serum creatinine values in the modification of diet in renal disease study equation for estimating glomerular filtration rate [published correction appears in Ann Intern Med. 2008;149:519] [published correction appears in Ann Intern Med. 2021;174:584] Ann Intern Med. 2006;145:247–254. doi: 10.7326/0003-4819-145-4-200608150-00004. [DOI] [PubMed] [Google Scholar]
- 28.Eder S., Leierer J., Kerschbaum J., et al. A prospective cohort study in patients with type 2 diabetes mellitus for validation of biomarkers (PROVALID) - study design and baseline characteristics. Kidney Blood Press Res. 2018;43:181–190. doi: 10.1159/000487500. [DOI] [PubMed] [Google Scholar]
- 29.Ohneberg K., Wolkewitz M., Beyersmann J., et al. Analysis of clinical cohort data using nested case-control and case-cohort sampling designs: a powerful and economical tool. Methods Inf Med. 2015;54:505–514. doi: 10.3414/ME14-01-0113. [DOI] [PubMed] [Google Scholar]
- 30.Prischl F.C., Rossing P., Bakris G., Mayer G., Wanner C. Major adverse renal events (MARE): a proposal to unify renal endpoints. Nephrol Dial Transplant. 2021;36:491–497. doi: 10.1093/ndt/gfz212. [DOI] [PubMed] [Google Scholar]
- 31.Levin A., Agarwal R., Herrington W.G., et al. International consensus definitions of clinical trial outcomes for kidney failure: 2020. Kidney Int. 2020;98:849–859. doi: 10.1016/j.kint.2020.07.013. [DOI] [PubMed] [Google Scholar]
- 32.Lachin J.M. Sample size evaluation for a multiply matched case-control study using the score test from a conditional logistic (discrete Cox PH) regression model [published correction appears in Stat Med. 2018;37:1765-1766] Stat Med. 2008;27:2509–2523. doi: 10.1002/sim.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfister I.B., Zandi S., Gerhardt C., Spindler J., Reichen N., Garweg J.G. Risks and challenges in interpreting simultaneous analyses of multiple cytokines. Transl Vis Sci Technol. 2020;9:1–9. doi: 10.1167/tvst.9.7.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samuelsen S.O., Støer N.C. In: Handbook of Statistical Methods for Case-Control Studies. Borgan Ø., Breslow N.E., Chatterjee N., Gail M.H., Scott A., Wild C.J., editors. CRC Press; 2018. Inverse probability weighting in nested case-control studies; pp. 351–371. [Google Scholar]
- 35.Horvitz D.G., Thompson D.J. A generalization of sampling without replacement from a finite universe. J Am Stat Assoc. 1952;47:663–685. doi: 10.2307/2280784. [DOI] [Google Scholar]
- 36.Breslow NE. Case–control study, two-phase. In: Balakrishnan N, Colton T, Everitt B, Piegorsch W, Ruggeri F, Teugels JL, eds. Wiley STATSRef: Statistics Reference Online; 2014.
- 37.Manitz J., Hempelmann M., Kauermann G., et al. samplingbook: survey Sampling Procedures. R package version 1.2.4. https://cran.r-project.org/package=samplingbook Published 2021.
- 38.D’agostino R.B. In: Encyclopedia of Biostatistics. Armitage P., Colton T., editors. Wiley; 2005. Principal components analysis. [Google Scholar]
- 39.Clayton D., Hills M. OUP; 2013. Statistical Models in Epidemiology. [Google Scholar]
- 40.Ury H.K. Efficiency of case-control studies with multiple controls per case: continuous or dichotomous data. Biometrics. 1975;31:643–649. [PubMed] [Google Scholar]
- 41.Carstensen B., Plummer M., Laara E., Hills M. Epi: a package for statistical analysis in epidemiology. R package version 2.40. https://cran.r-project.org/package=Epi Published 2019.
- 42.Langholz B. In: Encyclopedia of Biostatistics. 2nd. Armitage P., Colton T., editors. Vol. 1. Wiley; 2005. Case–Control Study, Nested; pp. 646–655. [Google Scholar]
- 43.Therneau T. A Package for Survival Analysis in S_. version 2.38. https://cran.r-project.org/package=survival Published 2015.
- 44.Liddell F.D.K., McDonald J.C., Thomas D.C., Cunliffe S.V. Methods of cohort analysis: appraisal by application to asbestos mining. J R Statist Soc A. 1977;140:469–491. doi: 10.2307/2345280. [DOI] [Google Scholar]
- 45.Harrell F.E., Jr., Lee K.L., Mark D.B. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 46.Korn E.L., Simon R. Measures of explained variation for survival data. Stat Med. 1990;9:487–503. doi: 10.1002/sim.4780090503. [DOI] [PubMed] [Google Scholar]
- 47.R Core Team R: a language and environment for statistical computing. R Foundation for Statistical Computing. https://www.r-project.org Published 2020.
- 48.Buscarlet M., Provost S., Zada Y.F., et al. DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood. 2017;130:753–762. doi: 10.1182/blood-2017-04-777029. [DOI] [PubMed] [Google Scholar]
- 49.Petzer V., Schwendinger S., Haschka D., et al. Clonal hematopoiesis in patients with Covid-19 is stable and not linked to an aggravated clinical course. Am J Hematol. 2021;96:E331–E333. doi: 10.1002/ajh.26251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guermouche H., Ravalet N., Gallay N., et al. High prevalence of clonal hematopoiesis in the blood and bone marrow of healthy volunteers. Blood Adv. 2020;4:3550–3557. doi: 10.1182/bloodadvances.2020001582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhattacharya R., Zekavat S.M., Uddin M.M., et al. Association of diet quality with prevalence of clonal hematopoiesis and adverse cardiovascular events. JAMA Cardiol. 2021;6:1069–1077. doi: 10.1001/jamacardio.2021.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haring B., Reiner A.P., Liu J., et al. Healthy lifestyle and clonal hematopoiesis of indeterminate potential: results from the women’s health initiative. J Am Heart Assoc. 2021;10 doi: 10.1161/JAHA.120.018789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cook E.K., Luo M., Rauh M.J. Clonal hematopoiesis and inflammation: partners in leukemogenesis and comorbidity. Exp Hematol. 2020;83:85–94. doi: 10.1016/j.exphem.2020.01.011. [DOI] [PubMed] [Google Scholar]
- 54.Kovtonyuk L.V., Fritsch K., Feng X., Manz M.G., Takizawa H. Inflamm-aging of hematopoiesis, hematopoietic stem cells, and the bone marrow microenvironment. Front Immunol. 2016;7:502. doi: 10.3389/fimmu.2016.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Franceschi C., Bonafè M., Valensin S., et al. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- 56.Cull A.H., Snetsinger B., Buckstein R., Wells R.A., Rauh M.J. Tet2 restrains inflammatory gene expression in macrophages. Exp Hematol. 2017;55:56–70.e13. doi: 10.1016/j.exphem.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 57.Fuster J.J., MacLauchlan S., Zuriaga M.A., et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science. 2017;355:842–847. doi: 10.1126/science.aag1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sano S., Oshima K., Wang Y., et al. Tet2-mediated clonal hematopoiesis accelerates heart failure through a mechanism involving the IL-1β/NLRP3 inflammasome. J Am Coll Cardiol. 2018;71:875–886. doi: 10.1016/j.jacc.2017.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cook E.K., Izukawa T., Young S., et al. Comorbid and inflammatory characteristics of genetic subtypes of clonal hematopoiesis. Blood Adv. 2019;3:2482–2486. doi: 10.1182/bloodadvances.2018024729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abplanalp W.T., Mas-Peiro S., Cremer S., John D., Dimmeler S., Zeiher A.M. Association of clonal hematopoiesis of indeterminate potential with inflammatory gene expression in patients with severe degenerative aortic valve stenosis or chronic postischemic heart failure. JAMA Cardiol. 2020;5:1170–1175. doi: 10.1001/jamacardio.2020.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Busque L., Sun M., Buscarlet M., et al. High-sensitivity C-reactive protein is associated with clonal hematopoiesis of indeterminate potential. Blood Adv. 2020;4:2430–2438. doi: 10.1182/bloodadvances.2019000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakamura A., Shikata K., Hiramatsu M., et al. Serum interleukin-18 levels are associated with nephropathy and atherosclerosis in Japanese patients with type 2 diabetes. Diabetes Care. 2005;28:2890–2895. doi: 10.2337/diacare.28.12.2890. [DOI] [PubMed] [Google Scholar]
- 63.Araki S., Haneda M., Koya D., et al. Predictive impact of elevated serum level of IL-18 for early renal dysfunction in type 2 diabetes: an observational follow-up study. Diabetologia. 2007;50:867–873. doi: 10.1007/s00125-006-0586-8. [DOI] [PubMed] [Google Scholar]
- 64.Fujita T., Ogihara N., Kamura Y., et al. Interleukin-18 contributes more closely to the progression of diabetic nephropathy than other diabetic complications. Acta Diabetol. 2012;49:111–117. doi: 10.1007/s00592-010-0178-4. [DOI] [PubMed] [Google Scholar]
- 65.Tesch G.H. Macrophages and diabetic nephropathy. Semin Nephrol. 2010;30:290–301. doi: 10.1016/j.semnephrol.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 66.Srivastava A., Schmidt I.M., Palsson R., et al. The associations of plasma biomarkers of inflammation with histopathologic lesions, kidney disease progression, and mortality—the Boston kidney biopsy cohort study. Kidney Int Rep. 2021;6:685–694. doi: 10.1016/j.ekir.2020.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Milas O., Gadalean F., Vlad A., et al. Pro-inflammatory cytokines are associated with podocyte damage and proximal tubular dysfunction in the early stage of diabetic kidney disease in type 2 diabetes mellitus patients. J Diabetes Complications. 2020;34:107479. doi: 10.1016/j.jdiacomp.2019.107479. [DOI] [PubMed] [Google Scholar]
- 68.Liu S.Y., Chen J., Li Y.F. Clinical significance of serum interleukin-8 and soluble tumor necrosis factor-like weak inducer of apoptosis levels in patients with diabetic nephropathy. J Diabetes Investig. 2018;9:1182–1188. doi: 10.1111/jdi.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.