Abstract
Severe malarial anemia (SMA) is a leading cause of childhood morbidity and mortality in holoendemic Plasmodium falciparum transmission regions. To gain enhanced understanding of predisposing factors for SMA, we explored the relationship between complement component 3 (C3) missense mutations [rs2230199 (2307C>G, Arg>Gly102) and rs11569534 (34420G>A, Gly>Asp1224)], malaria, and SMA in a cohort of children (n = 1617 children) over 36 months of follow-up. Variants were selected based on their ability to impart amino acid substitutions that can alter the structure and function of C3. The 2307C>G mutation results in a basic to a polar residue change (Arg to Gly) at position 102 (β-chain) in the macroglobulin-1 (MG1) domain, while 34420G>A elicits a polar to acidic residue change (Gly to Asp) at position 1224 (α-chain) in the thioester-containing domain. After adjusting for multiple comparisons, longitudinal analyses revealed that inheritance of the homozygous mutant (GG) at 2307 enhanced the risk of SMA (RR = 2.142, 95%CI: 1.229–3.735, P = 0.007). The haplotype containing both wild-type alleles (CG) decreased the incident risk ratio of both malaria (RR = 0.897, 95%CI: 0.828–0.972, P = 0.008) and SMA (RR = 0.617, 95%CI: 0.448–0.848, P = 0.003). Malaria incident risk ratio was also reduced in carriers of the GG (Gly102Gly1224) haplotype (RR = 0.941, 95%CI: 0.888–0.997, P = 0.040). Collectively, inheritance of the missense mutations in MG1 and thioester-containing domain influence the longitudinal risk of malaria and SMA in children exposed to intense Plasmodium falciparum transmission.
Keywords: C3 missense mutations, complement system, malaria, severe malaria anemia, incident risk ratio, P. falciparum
Impact statement
Malaria remains a major public health problem globally. Recent data show that 229 million cases resulted in 409,000 malaria-related deaths globally in 2019. Genetic variation in immune genes may impact on malaria disease outcomes. However, little is known about the variation on innate immunity genes such as complement 3, which is the focus of this study. As such this piece of work advances the already existing knowledge on relationship between genetic variation and malaria outcomes. For the first time we demonstrate that inheritance of the missense mutations in MG1 and thioester-containing domain influence the longitudinal risk of malaria and SMA in children exposed to intense P. falciparum transmission. These findings will be integral in the development of immunotherapy for malaria.
Introduction
Malaria remains a major global public health challenge. Recent data show that 229 million cases resulted in 409,000 malaria-related deaths globally in 2019. 1 The African region had 215 million cases and 384,000 deaths. These accounted for approximately 94% of the cases and deaths globally. The majority of cases in the African region are due to Plasmodium falciparum (P. falciparum) infections. 1 Severe malaria in children under the age of three years in holoendemic regions presents primarily as severe malarial anemia [SMA, hemoglobin (Hb) <5.0 g/dL] with rare occurrences of cerebral malaria. 2 Previous studies from our group have shown that polymorphic variability, particularly in immune response genes, influences the risk of malaria and SMA at enrollment and over the follow-up period.3–5
The complement system (or complement cascade) of the innate immune surveillance system plays a key role in enhancing phagocytosis and killing invading pathogens. The system is comprised of more than 50 proteins (including complement C3) that work together as a cascade to clear immune complexes and maintain physiological homeostasis through clearance of apoptotic and necrotic cells. 6 There are three distinct complement activation pathways: classical, alternative, and mannose-binding lectin, all of which lead to cleavage of C3 into functional C3a and C3b subunits. 7 C3a, C4a, and C5a are anaphylatoxins that directly stimulate the activation of neutrophils and macrophages, whereas C3b is an opsonin that enhances phagocytosis. 8 The culmination of complement activation is cell lysis via the membrane attack complex (MAC).8,9 Previous investigations have revealed that the complement cascade is activated during acute malaria.10,11 Additionally, complement-fixing antibodies to P. falciparum merozoites show a stronger correlation with protective immunity than parasite growth-inhibiting antibodies. 12
Studies in Kenyan children with SMA suggest that the complement system is an important source of erythrocyte destruction during P. falciparum infections.13,14 Kenyan children with SMA had elevated plasma levels of C3a, C4a, and C5a, high levels of immune complexes, reduced complement hemolytic activity, and diminished activity of the three major complement activation pathways. 15 These results suggest that complement consumption in SMA exceeds the regenerative capacity. In addition, children with SMA have elevated surface levels of IgG deposition on erythrocytes, along with acquired deficiencies of complement receptor 1 (CR1/CD35) and decay-accelerating factor (DAF/CD55).13,14,16 These events result in impaired immune complex (IC) binding capacity and increased C3b deposition on erythrocytes. 17 Furthermore, in vitro studies show that hematin, the oxidized product of heme, activates the complement alternative pathway (AP) and enhances deposition of C3b on erythrocytes, 18 suggesting that breakdown products of intravascular hemolysis, such as hematin, may be an important source of complement activation in malaria.
Although previous investigations have explored the impact of polymorphic variability in the complement mannose-binding lectin (MBL) pathway on severe malaria,19–22 to the best of our knowledge, no studies have focused on the effect of genetic variants in C3 on susceptibility to malaria and its severe disease outcomes. Since C3 has been shown to influence SMA development, a better understanding of the genetic aspects of this protein could reveal important insight into the role of complement in malaria pathogenesis.
The human C3 gene (C3) is located on the short arm of chromosome 19 (19p13.3), contains 41 exons, and is highly polymorphic, suggesting that selection of C3 alleles within a population may confer differences in immune responses to various pathogens.8,23 A common non-synonymous single nucleotide polymorphism (SNP) in the β-chain of C3 (rs2230199, 2307C>G) creates an amino acid replacement at position 102 (Arg/Gly102). In hemolysis assays, the minor mutant allele [2307G (Gly102)] activates the AP more efficiently than the wild-type (WT) major allele [2307C (Arg102)]. It favors AP amplification by having a lower affinity for factor H (FH). 24 A recent meta-analysis illustrates that carriage of the minor allele 2307G (Gly102) increases the risk of advanced age-related macular degeneration (AMD), most strongly pronounced in Middle Eastern populations. 25 Carriage of the minor allele [2307G (Gly102)] is also associated with worse cognitive performance, lower brain parenchymal fraction, and higher lesion burden in patients with multiple sclerosis. 26 Investigations in bacterial meningitis revealed that homozygosity of the major allele 2307C [i.e., genotype CC (Arg/Arg102)] was associated with elevated C3 levels, and lower C5a and terminal complement complex (C5b-9) concentrations in cerebrospinal fluid. 27
Since rs2230199 induces an amino acid replacement in the β-chain of C3 that has been associated with altered risk profiles for numerous diseases, we investigated the impact of this SNP on susceptibility to malaria and SMA across 36 months. In addition, we also examined a SNP (i.e., rs11569534, 34420G>A) in the α-chain that causes a substitution of Gly to Asp at codon 1224 (Gly/Asp1224). The effect of this SNP on disease outcomes in malaria and other diseases has not been reported. Here, we present findings on the impact of the selected C3 variants (genotypes/haplotypes) on susceptibility to malaria and SMA over a 36 months of follow-up in a cohort of Kenyan children (n = 1617) residing in a holoendemic transmission region for P. falciparum.
Materials and methods
Study site and participants
Study participants [children aged 2–48 months (n = 1617) with confirmed P. falciparum malaria] were enrolled at the Siaya County Referral Hospital in Siaya, Kenya in two cohorts between April 2004 and September 2008 (cohort 1) and from February 2009 to September 2015 (cohort 2). Siaya County is a rural region in western Kenya comprised primarily of individuals from the Luo ethnic group (>96%) where P. falciparum malaria transmission is holoendemic. 28 A comprehensive evaluation was performed to collect clinical and demographic information as well as the history of present illness. Exclusion criteria included: testing positive for non-P. falciparum species, any previous hospitalization (for any reason), recent blood transfusion (≤1 month), and diagnosis with cerebral malaria. Children (aged 2–48 months) presenting for routine childhood vaccinations with a negative P. falciparum blood smear were enrolled as aparasitemic controls and also followed quarterly for 36 months. Study participants who presented with P. falciparum malaria (any density) were stratified based on hemoglobin concentrations into uncomplicated malaria (UM, Hb ≥ 5.0g/dL) and SMA (Hb < 5.0g/dL). Additionally, since we have previously shown that HIV and bacteremia influence the severity of malarial anemia, these infections were characterized in all study participants.29,30 Parents/legal guardians of the children were provided with pre- and post-test HIV counselling. All treatment interventions in the study participants were carried out according to the Ministry of Health (MOH)-Kenya guidelines. This study was approved by the Maseno University Ethics Research Committee (MUERC; MSU/DRPI/MUERC/00510/18) and the University of New Mexico Institutional Review Board (16–284). For enrollment into the study, written informed consent was obtained from the parent/legal guardian of every study participant in the language of choice (English, Kiswahili, or Dholuo).
Longitudinal follow-up
Upon enrollment of children into the study (n = 1617, Day 0), parents/guardians were asked to return with their child quarterly. If parents/guardians failed to return to the hospital on their scheduled quarterly visits, study staff would visit children’s residences to check on their health status (including mortality) using our GIS/GPS surveillance system which captured participant locations at enrollment. To document all the malaria episodes, and/or other pediatric infectious diseases during the study period, we asked the parents/guardians to bring the child to the hospital whenever their child had any febrile episode(s) or illnesses. At each acute and quarterly visit, the participants underwent all laboratory tests required for proper clinical management of the patients including complete blood count, malaria parasitemia determination, and evaluation of bacterial infections (if clinically indicated). Additionally, throughout the 36-month follow-up period, all-cause mortality data were collected from the hospital or by verbal autopsy in cases where deaths occurred outside the hospital. The association between the two C3 genetic variations (rs2230199 and rs11569534) and longitudinal outcomes was based on clinical and laboratory measures measured at each visit.
Laboratory procedures
Venipuncture blood samples (≤3.0 mL) were collected into EDTA-containing vacutainer tubes for diagnostic and experimental measures. Quantification of P. falciparum density (parasites/µL) and the reticulocyte production index (RPI) were performed per our previously published methods. 31 Complete hematological profiles were determined using a Beckman Coulter® AcT diff2™ hematology analyzer (Beckman–Coulter Inc., Brea, CA, USA). Hemoglobin variants were determined using cellulose acetate electrophoresis per the manufacturer’s protocol (Helena Bio-Sciences, Oxford, United Kingdom). HIV exposure was assessed by serological testing (DetermineTM HIV-1/2, Abbott, Woodmead, South Africa and Uni-GoldTM HIV-1/2, Trinity Biotech Plc., Wicklow, Ireland). All children with positive results, on either or both of the serological tests, were examined for HIV infection by proviral DNA PCR testing according to our published methods. 29 The presence of bacteremia was determined by microbial cultivation according to standard methods per our previous description. 30
Selection of C3 variants
Selection of C3 SNPs for genotyping was based on variants that resulted in amino acid replacements (i.e., missense mutations) that had the potential to impart changes in protein function/structure (i.e., non-conservative replacements). Visual Molecular Dynamics (VMD) software (v1.9.3) was used to display and analyze the molecular assemblies 32 (see below). The first SNP, rs2230199 (2307C>G, Arg/Gly102) was selected since it is a missense mutation in C3 that has been shown to influence susceptibility to several diseases (non-malarial).25–27 The minor allele frequency (MAF) of this SNP was considered when selecting the second SNP so that the two alleles (potentially) had similar distributions within the cohort. Data from the International HapMap Project and 1000 Genomes Project were used to determine allelic distributions of potential C3 variants for the following populations: Luhya (LWK), Yoruba (YRI; Nigeria), African (AFR), and globally. Additional selection criteria were alleles that did not display strong linkage disequilibrium (LD), as determined by Multiallelic Interallelic Disequilibrium Analysis (MIDAS) software version 1.0. 33 Based on the selection criteria listed above and knowledge known about rs2230199, a second SNP was selected in C3. The second missense mutation, rs11569543, imparts a G to A change at position 34420, resulting in a non-synonymous amino acid change from Gly to Asp at residue 1224 in the protein sequence. The effect of this missence mutation on susceptibility to malaria and other diseases has not been previously reported.
Modeling the effects of the C3 variants on three-dimensional (3D) protein structures
The human C3 protein sequence was retrieved from Genbank at the National Center for Biotechnology Information (NCBI) with the accession number AAA85332.1. To determine the potential impact of the selected C3 SNPs on the overall protein structure, we modeled the mutations in protein sequence with Modeller, 34 and then visualized changes in the 3D protein structures of the two wild-type alleles using the VMD (v1.9.3) software. 32 The 3D protein structure for native (i.e., wild-type) C3 was originally determined by Janssen et al., at 3.3 Å resolution using X-ray diffraction, 35 and is deposited as code 2a73 in the Protein Data Bank (pdb). Amino acid sequence changes for the two mutant C3 alleles were generated using the software Modeller (9.22, r11413). Thereafter, the 3D protein structures of the wild-type and mutant alleles were visualized and analyzed using VMD.
C3 genotyping
Genomic DNA was isolated from cheek cells collected on buccal swabs using the BuccalAmp™ DNA extraction kit (Epicentre Biotechnologies, Madison, WI, USA) followed by amplification using GenomiPhi™ (GE Healthcare Life Sciences, Amersham, UK). C3 variants were genotyped by TaqMan 5’ allelic discrimination Assay-By-Design methods according to the manufacturer’s instructions [Assay ID: C__26330755_10 for rs2230199 (2307C>G, Arg/Gly102) and C__31045915_10 for rs11569543 (34420G>A, Gly/Asp1224), Thermo Fisher Scientific, Carlsbad, CA, USA]. PCR was performed in a total reaction volume of 10 µL with the following amplification cycles: initial incubation at 60 °C for 30 s and 95 °C for 10 min, followed by 45 cycles of 95 °C for 15 s and 60 °C for 1 min with a final extension at 60 °C for 30 s. Allelic discrimination was determined using allele-specific fluorescence on the StepOnePlusTM Real-Time PCR System (Thermo Fisher Scientific) using the manufacturer’s software (Version 2.3, Thermo Fisher Scientific).
Statistical analyses
Data collected at enrollment were analyzed using SPSS, version 23.0 (SPSS Inc., Chicago, IL, USA). Comparisons of demographic, clinical, and laboratory characteristics across the groups were determined using the Kruskal-Wallis tests. Whenever there were significant differences, pairwise comparisons were performed using the Mann-Whitney U test. Fisher’s exact test (two-sided except otherwise noted) for homogeneity was used to compare proportions and performed in R (version 3.1.3). Exact test for Hardy-Weinberg equilibrium (HWE) were performed as described elsewhere. 36 P-values ≤0.05 were considered statistically significant.
Haplotypes of the C3 missense mutants were constructed using HPlus (Version 2.5). 37 The impact of C3 genotypes/haplotypes on longitudinal clinical outcomes was analyzed using R (version 3.1.3). A Poisson rate regression (R glm function, family=Poisson) was used to explore the relationship between C3 genotypes/haplotypes and the number of malaria and SMA episodes over the three-year follow-up period, with the (logarithm of) age at the patients last visits being the offset variable (rate regression). The Poisson regression was performed with a forward-backward model selection minimizing the AIC. Either C3 genotypes or haplotypes entered the model as covariates. Additional covariates were age at enrollment, sex, HIV, cohort, and sickle cell status (sickle trait and sickle cell disease). Lastly, we investigated the relationship between C3 genotypes or haplotypes and all-cause mortality using Cox regression/survival analysis (R survival package version 2.38.2, coxph function). Model selection was performed analogously to the Poisson rate regression with forward-backward selection. Covariates were included analogously to the Poisson rate regression. After model selection in the Poisson rate regression and the Cox model parameter estimates were tested to be significantly different from zero. More precisely, for the Poisson rate regression or Cox models we tested whether the incidence risk ratios or the hazard ratios of the retained covariates were significantly different from 1. Separately for each model Bonferroni-Holm corrections were used to adjust for multiple comparisons.
Results
Demographic, clinical, and laboratory characteristics at enrollment
Study participants (n = 1617) were grouped into three categories: aparasitemic (n = 302), uncomplicated malaria (UM, Hb ≥ 5.0 g/dL, n = 1009) and SMA (Hb < 5.0 g/dL, n = 306). Demographic, clinical, and laboratory characteristics of the study participants upon entry into the study are presented in Table 1 . The distribution of males and females was comparable across the groups (P = 0.822). However, age differed across the three groups (P = 0.001). Children with SMA were younger than those with UM (P < 0.001).
Table 1.
Demographic, clinical, and laboratory characteristics of study participants at enrollment.
| Characteristics | Aparasitemic | UM (Hb ≥ 5.0 g/dL) | SMA (Hb < 5.0 g/dL) | P |
|---|---|---|---|---|
| Demographic parameters | ||||
| Sample size (n) | 302 | 1009 | 306 | |
| Sex, n (%) | ||||
| Female | 153 (50.66) | 497 (49.26) | 156 (50.98) | 0.822a |
| Male | 149 (49.34) | 512 (50.74) | 150 (49.02) | |
| Age (months) | 11.04 (13.20) | 12.67 (10.50) | 9.88 (10.60)** | <0.001b |
| Hematological indices and parasitemia | ||||
| Hematocrit (Hct. %) | 32.90 (7.80) | 25.30 (8.90) | 14.30 (3.80)** | <0.001b |
| Hemoglobin (g/dL) | 10.35 (2.50) | 7.70 (2.90) | 4.30 (1.20)** | <0.001b |
| RBCs (×1012/µL) | 4.65 (1.11) | 3.78 (1.41) | 1.91 (0.69)** | <0.001b |
| MCV | 69.90 (10.25) | 68.80 (11.20) | 73.35 (13.77)** | <0.001b |
| MCH | 22.30 (3.80) | 21.30 (4.00) | 22.05 (4.30)** | <0.001b |
| MCHC | 31.70 (2.70) | 30.70 (2.60) | 30.20 (4.30)* | <0.001b |
| RDW | 18.50 (5.50) | 20.30 (4.60) | 22.75 (5.90)** | <0.001b |
| WBCs (×103/µL) | 11.00 (7.30) | 11.60 (6.50) | 14.40 (9.80)** | <0.001b |
| Monocytes (×103/µL) | 7.60 (4.2) | 7.90 (5.30) | 9.15 (6.80)** | <0.001b |
| Granulocytes (×103/µL) | 36.10 (20.50) | 44.90 (25.00) | 39.50 (21.00)** | <0.001b |
| Lymphocytes (×103/µL) | 55.80 (18080) | 46.35 (21.20) | 49.95 (17.10)* | <0.001b |
| Platelet counts (×103/µL) | 343.00 (224.50) | 152.00 (123.00) | 141.00 (93.50) | 0.001b |
| Parasite density (µL) | 0.00 (0.00) | 29,583.70 (79,084) | 23,242.35 (65,934)* | 0.008c |
| Genetic variants | ||||
| Sickle cell trait, n (%) | ||||
| HbAA | 230 (78.23) | 830 (83.17) | 271 (91.25)* | |
| HbAS | 55 (18.71) | 164 (16.43) | 20 (6.73)* | <0.001a |
| HbSS | 9 (3.06) | 4 (0.40) | 6 (2.21)* | |
| Co-infections | ||||
| Bacteremia, n (%) | 27 (9.03) | 61 (6.45) | 28 (10.11)* | 0.071a |
| HIV-1, n (%) | 12 (4.01) | 29 (2.88) | 26 (8.55)* | <0.001a |
Data presented are medians (interquartile range, IQR), unless otherwise stated.
aStatistical significance determined by Fisher’s exact test.
bDifferences were determined using Kruskal-Wallis tests, and where significant differences were observed, pairwise comparisons between UM and SMA group were performed using Mann-Whitney U tests. Boldface indicates a P value of ≤0.050. *Represents significant pairwise comparisons between UM and SMA P-value < 0.050, and ** represents P-value < 0.001.
cDifferences between UM and SMA group were performed using Mann-Whitney U tests and Fisher’s exact test (one-sided for Bacteremia and HIV-1, two-sided else).RBCs: red blood cells; MCV: Mean corpuscular volume; MCH: mean corpuscular hemoglobin; MCHCH: mean corpuscular hemoglobin concentration; RDW: red blood cell distribution width; WBCs: white blood cells; Hb AA: hemoglobin AA; Hb AS: hemoglobin AS; Hb SS: hemoglobin SS.
Based on a priori assignment, hematological indices, hematocrit, Hb concentrations, and red blood cells (RBCs) counts progressively declined across the three groups and were lowest in children with SMA (P < 0.001, respectively). Mean corpuscular volume (MCV) was elevated in the SMA group compared to UM and aparasitemic groups (P < 0.001). In contrast, both the mean corpuscular hemoglobin (MCH) and mean corpuscular hemoglobin concentration (MCHC) were elevated in the aparasitemic group relative to children with UM and SMA (P < 0.001, respectively). The red cell distribution width (RDW), white blood cells (WBCs), monocyte, and granulocyte counts differed across the groups [(P < 0.001, respectively), with highest values in the SMA group and lowest in the aparasitemic group], while lymphocytes were lowest in the UM group and highest in children with SMA (P < 0.001). Platelet counts progressively decreased across the groups and were lowest in SMA (P = 0.001). Peripheral blood parasite density was lower in children with SMA compared to those with UM (P = 0.008).
Inheritance of genetic variants that can impact the clinical outcomes in children with malaria was also examined. The frequency of HbAS was highest in the UM group and lowest in children who presented with SMA (P < 0.001). Since our previous investigations have revealed that bacteremia and HIV exacerbate the development of SMA,29,30 absence/presence of these co-infections was determined in the study population. The frequency of bacteremia was comparable between the groups (P = 0.071); however, it was significantly higher in the SMA compared with the UM group (P = 0.030, one-sided Fisher’s exact test). The proportion of children with HIV differed across the groups (P = 0.001) and was highest in the SMA group.
Collectively, the demographic, clinical, and laboratory results support our previous findings showing that children with SMA have distinct clinical and hematological features, and elevated co-infections.2,30,38
Potential effects of selected C3 variants on protein structure and function
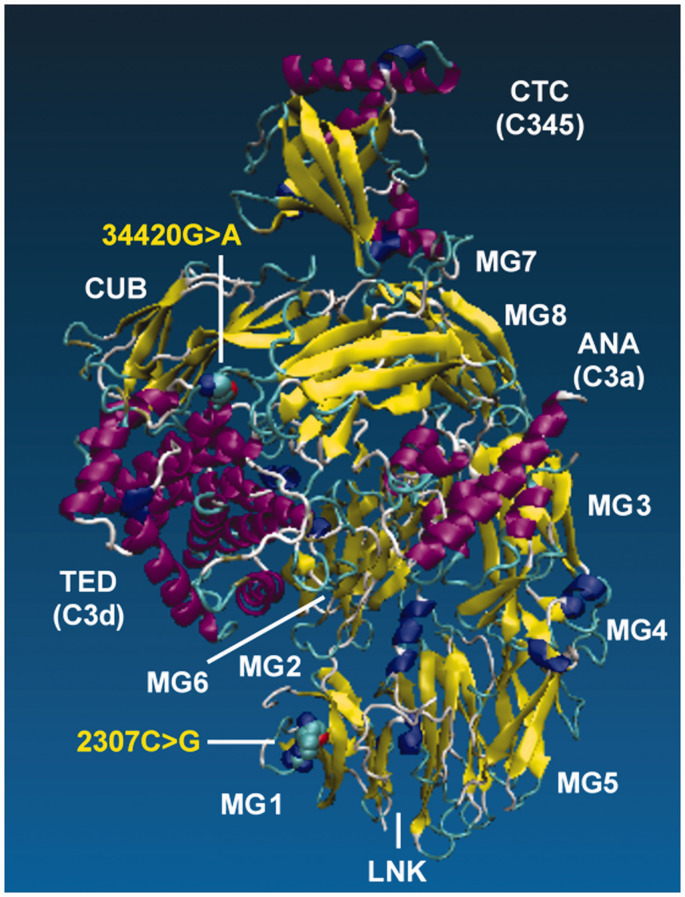
Prior to genotyping the two missense mutations in the cohorts, we first determined whether the selected variants could impart amino acid substitutions that had the ability to alter the structure and function of C3. Examination of the protein structure with VMD revealed that rs2230199 (2307C>G, Arg/Gly102) is located in the macroglobulin 1 (MG1) domain of C3 within the β-chain (shown in Figure 1 ), while rs11569543 (34420G>A, Gly/Asp1224) is located in the thioester-containing domain (TED) region of C3 within the α-chain (shown in Figure 1 ). The Arg to Gly transition at position 102 results in the change from a basic residue (Arg) to that of a polar residue (Gly), while the Gly to Asp transition at position 1224 imparts a change from a polar residue to an acidic residue.
Figure 1.
The human C3 protein sequence was retrieved from Genbank at the National Center for Biotechnology Information (NCBI) with the accession number AAA85332.1. To determine the potential impact of the selected C3 SNPs on the overall protein structure, we modeled the mutations in protein sequence with Modeller, and then visualized changes in the 3D protein structures of the two wild-type alleles using the VMD (v1.9.3) software. The color coding is as follows: a-helix=purple, 3_10 helix=blue, extended b=yellow, bridge b=turn, turn, cyan, and coil=white. Relative to the published C3 structure,8,35 we observed that Arg>Gly102 mutation is located in the macroglobulin MG1domain, while the Gly>Asp1224 is located on the TED.
While (2307C>G, Arg/Gly102) is on the surface of the un-activated C3 structure as shown in Figure 1 , inspection of the C3b structure (pdb code 2i07) 39 shows the positively charged Arg in the WT structure is interacting with a pair of negatively charged Glu on the TED domain, perhaps orienting the TED domain in an active configuration of the C3b molecule. The (34420G>A, Gly/Asp1224) mutation adds a negatively charged sidechain to the TED domain. Homology modeling this residue into the C3 structure using Modeller 40 shows a close interaction with a positively charged lysine residue on the macroglobulin domain. No clear interaction partner of (34420G>A, Gly/Asp1224) is evident in the C3b structure.
Inspection of C3b in complex with Factor B (pdb code 2xwj), 41 or Factors H and I (pdb code 5o32), 42 and C3d with the C3 receptor (pdb code 4m76) 43 shows both mutations considered here are not directly interacting with these binding partners of C3b. Although allosteric and indirect impacts on binding are always possible, the structural evidence suggest both of these mutations impact the C3 → C3b conversion process. Taken together, these results suggest that the amino acid substitutions from both missense mutations have the potential to impact C3 function, most directly impacting the activation of C3 to C3b.
Distribution of C3 genotypic and haplotypic variants
The allelic distributions of rs2230199 and rs11569543 were then explored using available data from the International HapMap Project and 1000 Genomes Project. The minor allelic distributions for rs2230199 (2307C>G, Arg/Gly102) were 0.07 for Luhya (LWK), 0.01 for Yoruba (YRI; Nigeria), 0.03 for African (AFR), and 0.09 globally. The minor allelic distributions for rs11569543 (34420G>A, Gly/Asp1224) were 0.02 for LWK, 0.04 for YRI, 0.03 for AFR, and 0.01 globally. Although the minor allele frequencies (MAF) for the two variants were relatively low in African populations, we proceeded with genotyping the two missense mutations since they had the potential to impart functional consequences and may be reduced due to selective pressure from malaria. Allele frequencies in the overall cohort for rs2230199 were C = 0.890 and G = 0.110, while those for rs11569543 were G = 0.920 and A = 0.080, indicating that the MAFs for both variants were higher than previously cataloged. Analysis of linkage disequilibrium indicated that the two SNPs were not strongly associated (D’=0.079, LOD = 0.04, r2<0.001).
The distribution of genotypes and haplotypes for rs2230199 (2307C>G, Arg/Gly102) and rs11569543 (34420G>A, Gly/Asp1224) is presented in Table 2 . Allele frequencies for rs2230199 were C = 0.930 and G = 0.070 in the aparasitemic group, C = 0.930 and G = 0.070 in UM, and C = 0.880 and G = 0.120 in SMA. Allele frequencies for rs11569543 were G = 0.932 and A = 0.068 in aparasitemic children, G = 0.917 and A = 0.083 in the UM group, and G = 0.900 and A = 0.100 in children with SMA. The proportion of rs2230199 genotypes differed across the study groups (Fisher’s exact test, P = 0.027, Table 2 ) with the overall cohort in HWE (exact test for HWE, P = 0.072). Genotypic distributions in all three groups were also consistent with HWE: aparasitemic (P = 0.342), UM (P = 0.587), and SMA (P = 0.095).
Table 2.
Distribution of C3 genotypic and haplotypic variants.
| Genotype/haplotype | Amino acid | Aparasitemic | UM (Hb ≥ 5.0 g/dL) | SMA (Hb < 5.0 g/dL) | Total | P |
|---|---|---|---|---|---|---|
| 2307C>G (rs2230199) | Arg>Gly102 | n = 300 | n = 1008 | n = 303 | n = 1611 | |
| CC, n (%) | Arg/Arg102 | 223 (74.33) | 824 (81.75) | 240 (79.21) | 1287 (79.89) | |
| CG, n (%) | Arg/Gly102 | 69 (23.00) | 173 (17.16) | 56 (18.48) | 298 (18.50) | 0.027 |
| GG, n (%) | Gly/Gly102 | 8 (2.67) | 11 (1.09) | 7 (2.31) | 26 (1.61) | |
| 34420G>A (rs11569534) | Gly>Asp1224 | n = 299 | n = 1002 | n = 302 | n = 1603 | |
| GG, n (%) | Gly/Gly1224 | 268 (89.63) | 893 (89.12) | 259 (85.80) | 1420 (88.58) | |
| GA, n (%) | Gly/Asp1224 | 23 (7.70) | 56 (5.59) | 26 (8.60) | 105 (6.55) | 0.078 |
| AA, n (%) | Asp/Asp1224 | 8 (2.67) | 53 (5.29) | 17 (5.60) | 78 (4.87) | |
| 2307C>G/34420G>A | Arg>Gly102/Gly>Asp1224 | n = 302 | n = 1009 | n = 306 | n = 1617 | |
| Non-CG, n (%) | Non-Arg102Non-Gly1224 | 19 (6.29) | 73 (7.23) | 33 (10.78) | 125 (7.73) | 0.084 |
| CG, n (%) | Arg102Gly1224 | 283 (93.71) | 936 (92.77) | 273 (89.22) | 1492 (92.27) | |
| Non-CA, n (%) | Non-Arg102Asp1224 | 271 (89.74) | 902 (89.40) | 263 (85.95) | 1436 (88.81) | 0.216 |
| CA, n (%) | Arg102Asp1224 | 31 (10.26) | 107 (10.60) | 43 (14.05) | 181 (11.19) | |
| Non-GG, n (%) | Non-Gly102Gly1224 | 225 (74.50) | 839 (83.15) | 244 (79.74) | 1308 (80.89) | 0.004 |
| GG, n (%) | Gly102Gly1224 | 77 (25.50) | 170 (16.85) | 62 (20.26) | 309 (19.11) | |
| Non-GA, n (%) | Non-Gly102Asp1224 | 302 (100.00) | 995 (98.61) | 305 (99.67) | 1602 (99.07) | 0.048 |
| GA, n (%) | Gly102Asp1224 | 0 (0.00) | 14 (1.39) | 1 (0.33) | 15 (0.093) |
Data are presented as proportions [n (%)] of genetic variants within the study groups. Study participants were categorized into three groups, aparasitemic, SMA (i.e., Hb < 5.0 g/dL with any density parasitemia) or UM (Hb ≥ 5.0 g/dL with any density parasitemia). Statistical significance determined by Fisher’s exact test. Boldface indicates a values of P < 0.05.
Although the frequency of genotypes for rs11569543 was comparable between the groups (Fisher’s exact test, P = 0.078, Table 2 ), there was a significant departure from HWE in the overall population (exact test for HWE,P < 0.001) and within each of the three study groups (exact test for HWE: aparasitemic, P < 0.001; UM, P=<0.001; and SMA, P < 0.001).
Frequency distributions of the haplotypes constructed from rs2230199 and rs2230199 were similar for the CG (Arg102Gly1224, P = 0.073) and CA (Arg102Asp1224, P = 0.209) haplotypes, whereas the GG (Gly102Gly1224, P = 0.003) and GA (Gly102Asp1224, P = 0.042) differed across the groups.
Impact of C3 genotypes/haplotypes on malaria and SMA over 36-months
Children in holoendemic P. falciparum transmission regions encounter multiple malaria episodes prior to either developing naturally acquired immunity or suffering mortality. As such, the impact of inheritance on susceptibility to malaria and SMA can be more rigorously investigated using longitudinal outcome measures. We therefore investigated the impact of the C3 variants on malaria and SMA episodes and all-cause mortality across the 36-month follow-up period. The number of malaria and SMA episodes, along with mortality for each of the genotypes and haplotypes are listed in Table 3 . Poisson rate regression analyses (using the logarithm of age at the last hospital visit as offset variable) were performed in models that included genotype, haplotype, age at enrollment, sex, co-infections (HIV and bacteremia), and sickle cell trait status to account for co-factors that influence the development of SMA.4,29–31 Model selection identified relevant co-variates. All parameter estimates of the final model were tested for being significantly different from zero using Bonferroni-Holm adjustment for multiple testing.
Table 3.
Malaria, SMA, and mortality stratified by genotype/haplotype across the 36-month follow-up period.
| Genotype/haplotype | Amino acid | Malaria episodes | SMA episodes | Mortality |
|---|---|---|---|---|
| 2307C>G (rs2230199) | Arg>Gly102 | Total = 7717 | Total = 444 | Total = 99 |
| CC | Arg/Arg102 | 6127 | 349 | 75 |
| CG | Arg/Gly102 | 1465 | 81 | 21 |
| GG | Gly/Gly102 | 125 | 14 | 3 |
| 34420G>A (rs11569534) | Gly>Asp1224 | Total = 7695 | Total = 442 | Total = 95 |
| GG | Gly/Gly1224 | 6763 | 385 | 82 |
| GA | Gly/Asp1224 | 494 | 36 | 7 |
| AA | Asp/Asp1224 | 438 | 21 | 6 |
| 2307C>G/34420G>A | Arg>Gly102/Gly>Asp1224 | Total = 7755 | Total = 447 | Total = 100 |
| Non-CG | Non-Arg102Non-Gly1224 | 682 | 46 | 9 |
| CG | Arg102Gly1224 | 7073 | 401 | 91 |
| Non-CA | Non-Arg102Asp1224 | 6838 | 390 | 87 |
| CA | Arg102Asp1224 | 917 | 57 | 13 |
| Non-GG | Non-Gly102Gly1224 | 6279 | 355 | 77 |
| GG | Gly102Gly1224 | 1476 | 92 | 23 |
| Non-GA | Non-Gly102Asp1224 | 7639 | 444 | 99 |
| GA | Gly102Asp1224 | 116 | 3 | 1 |
Data are presented as the number of malaria and SMA episodes, and all-cause mortality stratified according genotypes and haplotypes during the 36-month follow-up period.
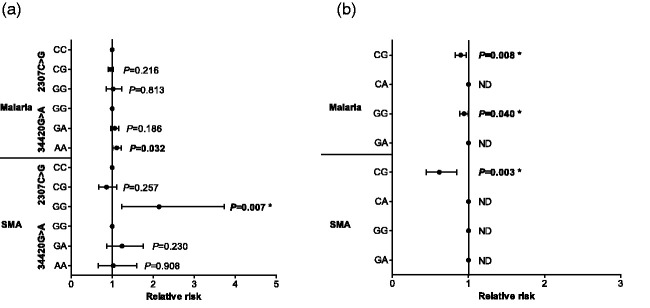
Although none of the 2307C>G genotypes altered the risk for the number of malaria episodes encountered during the longitudinal follow-up period, carriers of 34420AA (Asp/Asp1224) had increased susceptibility to malaria (RR = 1.112, 95%CI: 1.009–1.225, P = 0.032, shown in Figure 2(a) ). However, this finding was not significant after correction for multiple comparisons. Next, we determined the influence of the genotypes on susceptibility to SMA. Children who inherited the 2307GG (Arg/Gly102) genotype were more likely to develop SMA [RR = 2.142, 95%CI: 1.229–3.735, P = 0.007 (significant after Bonferroni-Holm correction), shown in Figure 2(a) ], while none of the other genotypes for either 2307C>G or 34420G>A influenced susceptibility to SMA.
Figure 2.
A Poisson rate regression (R glm function, family=Poisson) was used to explore the relationship between C3 genotypes/haplotypes and the number of malaria and SMA episodes over the three-year follow-up period, with the (logarithm of) age at the patients last visits being the offset variable (rate regression). The Poisson regression was performed with a forward-backward model selection minimizing the AIC. Either C3 genotypes or haplotypes entered the model as covariates. Additional covariates were age at enrollment, sex, HIV, cohort, and sickle cell status (sickle trait and sickle cell disease). Longitudinal relationship between C3 genotypes (2307C>G and 34420G>A), haplotypes (2307C>G/34420G>A), malaria, and SMA. Data presented as relative risk (midline dot) and 95% confidence intervals determined using log-linear regression analyses, adjusting for age at enrollment, sex, HIV, cohort, and sickle cell status (sickle trait and sickle cell disease). Longitudinal follow-up for the cohort was 36 months from the time of enrollment. Correction for multiple comparisons were performed by using the Bonferroni-Holm correction. *significant after Bonferroni-Holm correction. Estimates were not determined (ND) if a covariate was dropped by model selection. (a) Results for models using genotypes as covariates (with the wild types being references absorbed in the models intercept). (b) Results for models with haplotypes as covariates.
Modelling of the relationship between haplotypes and longitudinal outcomes revealed that children with the CG (Arg102Gly1224) haplotype had a reduced risk of malaria during the follow-up [RR = 0.897, 95%CI: 0.828–0.972, P = 0.008 (Bonferroni-Holm significant), shown in Figure 2(b) ]. Malaria incident risk ratio was also reduced in carriers of the GG (Gly102Gly1224) haplotype [RR = 0.941, 95%CI: 0.888–0.997, P = 0.040 (Bonferroni-Holm significant), shown in Figure 2(b) ]. Based on the selection criteria, the CA and GA haplotypes were dropped from the model, indicating that they did not have a significant impact on the number of malaria episodes.
Examination of the longitudinal risk of developing SMA revealed that inheritance of the CG (Arg102Gly1224) haplotype provided a protective effect across the 36-month follow-up [RR = 0.617, 95%CI: 0.448–0.848, P = 0.003 (Bonferroni-Holm significant), shown in Figure 2(b) ]. Model selection dropped the CA and GG haplotypes as covariates, revealing that they did not influence the incident risk ratio of SMA. The GA haplotype was also dropped from the model, but in this case, due to the small number of children with the GA haplotype who had SMA across the follow-up period (n = 3).
The impact of the genotypes/haplotypes on mortality was examined using Cox regression survival analysis with identical covariates as the other models. Although none of the variants significantly influenced all-cause mortality, carriage of the 2307GG (Arg/Gly102) genotype that significantly enhanced susceptibility to SMA was also associated with a 73% higher chance of morality across the 36-month follow-up (HR = 2.750, 95%CI: 0.859–8.780, P = 0.089). Collectively, these results demonstrate that the C3 variants selected for investigation alter the risk of developing both malaria and SMA during the time at which children develop naturally acquired immunity, and may potentially influence all-cause mortality.
Discussion
SMA is one of the leading causes of childhood morbidity and mortality in western Kenya. C3 plays a central role in innate immunity by activating both the classical and alternative complement pathways. 9 Earlier studies showed that complement consumption is excessive in children with SMA. 15 Results presented here extend previous findings by showing that missense mutations in C3 alter the longitudinal susceptibility to both malaria and SMA. Importantly, the variants selected for investigation have the potential to alter the structure and function of C3.
The central step in the activation of complement is the proteolytic cleavage of C3 into C3b, a process that exposes a reactive thioester group that promotes C3b binding to pathogens.8,35 The transition from C3 to C3b repositions the TED for contact with the MG1 domain, thereby generating surfaces that interact with other complement proteins that mediate complement activation and regulation.44–47 Interactions between TED and the MG1 domain are therefore important factors for complement regulation. Examination of C3 and C3b structures shows that the (2307C>G, Arg/Gly102) mutation participates in a stable salt bridge from the MG1 domain to the TED domain in the C3b structure, while homology modeling of the (34420G>A, Gly/Asp1224) mutation creates a salt bridge to another MG domain in the C3 structure. Direct interactions of these two mutation sites with other C3 or C3b binding partners were not identified.
Activation of C3 to C3b also leads to an extension of the CUB-TED interface that contains binding sites for members of the regulators of complement activation (RCA) family, such as CR1. 8 The binding of C3b to CR1 on the surface of RBCs facilitates antibody-mediated phagocytosis in the liver and spleen.48,49 Given the importance of the spleen in malaria’s pathophysiology and the finding that complement activation increases the rate of RBC destruction in children with SMA, the Gly/Asp1224 mutation in TED certainly has the potential to influence the development of SMA. 50
The present study focused on the missense mutations in MG1 and TED given their importance in complement activation, and their potential influence on malarial disease outcomes. After correcting for multiple comparisons, the inheritance of homozygous minor alleles (GG) at 2307 increased the incident risk ratio of SMA, whereas none of the other genotypes influenced susceptibility to either malaria or SMA.
In addition to examining the impact of the C3 genotypes on malarial disease outcomes, we also determined the impact of haplotypes to better understand how combinations of the selected missense mutations influence malaria and SMA. The most prominent signals that emerged from the genetic analyses were protection against both malaria and SMA over 36-months in carriers of both major alleles (CG haplotype). Although the protection against malaria was modest (i.e., ∼9%), we postulate that such a reduction may be important since there are ∼3.5 million clinical malaria cases annually in Kenya of which 16.2% are estimated to present with moderate to severe anemia.1,51 While it is expected that carriage of the minor alleles for both missense mutations would enhance the risk of SMA, the results could not be determined due to the small sample size for GA carriers. The only other haplotype to emerge from the modeling was the longitudinal protection against malaria in GG haplotype carriers.
The factors driving the maintenance of genetic polymorphism in human populations, particularly those linked to a complex phenotype, are hard to elucidate because they depend on multiple factors. Thus, while it would be expected that the MAF for the two variants examined would be lower in the holoendemic region where the study was conducted than in malaria-free regions, this was not the case. The reason for this finding is unknown but may reflect the nature of a high-burden infectious disease region. For example, in addition to malaria, the region suffers from a high rate of HIV, tuberculosis, and bacteremia.29,30 Given the complex nature of the interactions between immune response genes that afford protection against multiple pathogens, there are certainly undefined gene networks that could explain this finding. Consistent with this premise, the alleles studied here, particularly 34420G>A (Gly/Asp1224), are not in HWE, indicating that their frequencies are changing, potentially due to selective pressure. Nevertheless, our results suggest that inheritance of the wild-type alleles for the two missense mutations would be enhanced. In contrast, we expect that the mutant alleles would be selected against due to malaria and SMA pressure in the region. Although we did not find any significant associations between the genotypes/haplotypes and all-cause mortality, there was a 73% higher chance of mortality across the follow-up in individuals who inherited the 2307 mutant (GG) alleles. The lack of a significant association between the C3 variants and mortality is not unexpected given the low number of deaths in our cohorts compared to those in the region, due to our enhanced laboratory diagnostics, improved clinical management with second-line antimicrobial agents, and robust follow-up to quickly identify and treat children with acute disease(s). In any event, the low rate of mortality reduced power to find robust associations.
Conclusions
Although the precise mechanisms and/or the impact of the mutations on the structure and function of C3 remain to be determined, the variants selected for investigation substantially impact on susceptibility to both malaria and SMA. It will be important in future studies to understand how potential structural and functional changes in the MG1 and TED regions impact on the acquisition of malaria and the subsequent development of SMA as identified in our genetic findings.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the assistance of the University of New Mexico-Kenya team: Nicholas Otieno Ondiek and Enos Osanyo for clinical support; Vincent Odhiambo Otieno, Chrispine Wasonga Ochieng, Joan LA Ochieng, and Duncan Njega for laboratory support; Everlyne A Modi, Joseph Oduor, Moses Ebungure, Moses Lokorkeju, and Rodney B Mongare for field work and study participants follow-up; Vincent Omanje for maintaining the databases; and Anne A Ong’ondo for administrative support. The authors are also grateful to all of the parents, guardians, and children who participated in the study.
AUTHORS’ CONTRIBUTIONS: ER, QC, and DJP conceptualized the studies; SBA, EOM, IH, CO, and CN performed data curation; HCJrTO, NWH, CGL, BHM, and KAS performed data analysis; KAS and DJP acquired funding for the studies; ER, SBA, and QC performed the investigations and methodology; CO performed project administration; provision of resources, software, and supervision; CO, MAP, AAE, PDS, CGL, BHM, and DJP performed the final reviews and edits. All authors reviewed and approved the final version of the manuscript.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ETHICAL APPROVAL: This study protocol was reviewed and approved by the Maseno University Ethics Research Committee (MSU/DRPI/MUERC/00510/18) and University of New Mexico Institutional Review Board (16–284).
FUNDING: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The work reported here was supported by grants from the National Institutes of Health [R01 AI130473-01 (DJP) and D43 TW005884 (DJP)]. The work was also supported by the Harvard University-Boston University-Northwestern University-University of New Mexico (HBNU) Fogarty Global Health Postdoctoral Fellowship awarded to Dr Evans Raballah from the Fogarty International Center and National Institute of Mental Health, of the National Institutes of Health [D43 TW010543 (ER, DJP)]; the German Academic Exchange [DAAD, Project-ID 57417782 (KAS)]; the German Federal Ministry for Research and Education [BMBF-DLR, Project-ID 01DQ20002 (KAS)] and Saxony’s State Ministry of Science, Education, and Arts [SMWK-SAB, Project-ID 100257255 (KAS)]. The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
ORCID iDs: Evans Raballah  https://orcid.org/0000-0002-3304-6836
https://orcid.org/0000-0002-3304-6836
Ivy-Foo Hurwitz  https://orcid.org/0000-0003-1566-1111
https://orcid.org/0000-0003-1566-1111
Douglas J Perkins  https://orcid.org/0000-0001-9390-6255
https://orcid.org/0000-0001-9390-6255
References
- 1.WHO. World malaria report, https://cdn.who.int/media/docs/default-source/malaria/world-malaria-reports/9789240015791-eng.pdf?sfvrsn=d7a8ec53_3&download=true (2020, accessed 7 July 2021).
- 2.Novelli EM, Hittner JB, Davenport GC.Ouma C, Were T, Obaro S, Kaplan S, Ong‘echa JM, Perkins DJ. Clinical predictors of severe malarial anaemia in a holoendemic Plasmodium falciparum transmission area. Br J Haematol 2010; 149:711–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Achieng AO, Hengartner NW, Raballah E.Cheng Q, Anyona SB, Lauve N, Guyah B, Foo-Hurwitz I, Ong‘echa JM, McMahon BH, Ouma C, Lambert CG, Perkins DJ. Integrated OMICS platforms identify LAIR1 genetic variants as novel predictors of cross-sectional and longitudinal susceptibility to severe malaria and all-cause mortality in Kenyan children. EBioMedicine 2019; 45: 290–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anyona SB, Hengartner NW, Raballah E.Ong‘echa JM, Lauve N, Cheng Q, Fenimore PW, Ouma C, Lambert CG, McMahon BH, Perkins DJ. Cyclooxygenase-2 haplotypes influence the longitudinal risk of malaria and severe malarial anemia in Kenyan children from a holoendemic transmission region. J Hum Genet 2020; 65:99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kisia LE, Kempaiah P, Anyona SB.Munde EO, Achieng AO, Ong‘echa JM, Lambert CG, Chelimo K, Ouma C, Perkins DJ, Raballah E. Genetic variation in interleukin-7 is associated with a reduced erythropoietic response in Kenyan children infected with Plasmodium falciparum. BMC Med Genet 2019; 20:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaplin H., Jr. Review: the burgeoning history of the complement system 1888–2005. Immunohematology 2005; 21:85–93 [PubMed] [Google Scholar]
- 7.Merle NS, Church SE, Fremeaux-Bacchi V, Roumenina LT. Complement system part I – molecular mechanisms of activation and regulation. Front Immunol 2015; 6:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ricklin D, Reis ES, Mastellos DC, Gros P, Lambris JD. Complement component C3 – the “swiss army knife” of innate immunity and host defense. Immunol Rev 2016; 274:33–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biryukov S, Stoute JA. Complement activation in malaria: friend or foe? Trends Mol Med 2014; 20:293–301 [DOI] [PubMed] [Google Scholar]
- 10.Larsen MD, Quintana MDP, Ditlev SB.Bayarri-Olmos R, Ofori MF, Hviid L, Garred P. Evasion of classical complement pathway activation on Plasmodium falciparum-infected erythrocytes opsonized by PfEMP1-specific IgG. Front Immunol 2018; 9:3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oyong DA, Kenangalem E, Poespoprodjo JR.Beeson JG, Anstey NM, Price RN, Boyle MJ. Loss of complement regulatory proteins on uninfected erythrocytes in vivax and falciparum malaria anemia. JCI Insight 2018; 3:e124854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reiling L, Boyle MJ, White MT.Wilson DW, Feng G, Weaver R, Opi DH, Persson KEM, Richards JS, Siba PM, Fowkes FJI, Takashima E, Tsuboi T, Mueller I, Beeson JG. Targets of complement-fixing antibodies in protective immunity against malaria in children. Nat Commun 2019; 10:610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoute JA, Odindo AO, Owuor BO, Mibei EK, Opollo MO, Waitumbi JN. Loss of red blood cell-complement regulatory proteins and increased levels of circulating immune complexes are associated with severe malarial anemia. J Infect Dis 2003; 187:522–5 [DOI] [PubMed] [Google Scholar]
- 14.Waitumbi JN, Opollo MO, Muga RO, Misore AO, Stoute JA. Red cell surface changes and erythrophagocytosis in children with severe Plasmodium falciparum anemia. Blood 2000; 95:1481–6 [PubMed] [Google Scholar]
- 15.Nyakoe NK, Taylor RP, Makumi JN, Waitumbi JN. Complement consumption in children with Plasmodium falciparum malaria. Malar J 2009; 8:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odhiambo CO, Otieno W, Adhiambo C, Odera MM, Stoute JA. Increased deposition of C3b on red cells with low CR1 and CD55 in a malaria-endemic region of Western Kenya: implications for the development of severe anemia. BMC Med 2008; 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Owuor BO, Odhiambo CO, Otieno WO, Adhiambo C, Makawiti DW, Stoute JA. Reduced immune complex binding capacity and increased complement susceptibility of red cells from children with severe malaria-associated anemia. Mol Med 2008; 14:89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pawluczkowycz AW, Lindorfer MA, Waitumbi JN, Taylor RP. Hematin promotes complement alternative pathway-mediated deposition of C3 activation fragments on human erythrocytes: potential implications for the pathogenesis of anemia in malaria. J Immunol 2007; 179:5543–52 [DOI] [PubMed] [Google Scholar]
- 19.Das BK, Panda AK. MBL-2 polymorphisms (codon 54 and Y-221X) and low MBL levels are associated with susceptibility to multi organ dysfunction in P. falciparum malaria in Odisha. India. Frontiers in Microbiology 2015; 6:778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmberg V, Schuster F, Dietz E.Sagarriga Visconti JC, Anemana SD, Bienzle U, Mockenhaupt FP. Mannose-binding lectin variant associated with severe malaria in young African children. Microbes Infect 2008; 10:342–8 [DOI] [PubMed] [Google Scholar]
- 21.Jha AN, Sundaravadivel P, Singh VK.Pati SS, Patra PK, Kremsner PG, Velavan TP, Singh L, Thangaraj K. MBL2 variations and malaria susceptibility in Indian populations. Infect Immun 2014; 82:52–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luty AJ, Kun JF, Kremsner PG. Mannose-binding lectin plasma levels and gene polymorphisms in Plasmodium falciparum malaria. J Infect Dis 1998; 178:1221–4 [DOI] [PubMed] [Google Scholar]
- 23.Zarkadis IK, Mastellos D, Lambris JD. Phylogenetic aspects of the complement system. Dev Comp Immunol 2001; 25:745–62 [DOI] [PubMed] [Google Scholar]
- 24.Heurich M, Martinez-Barricarte R, Francis NJ.Roberts DL, Rodriguez de Cordoba S, Morgan BP, Harris CL. Common polymorphisms in C3, factor B, and factor H collaborate to determine systemic complement activity and disease risk. Proc Natl Acad Sci U S A 2011; 108:8761–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Li S, Hu S, Yu J, Xiang Y. Association between genetic variation of complement. C3 and the susceptibility to advanced age-related macular degeneration: a Meta-analysis. BMC Ophthalmol 2018; 18:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roostaei T, Sadaghiani S, Mashhadi R.Falahatian M, Mohamadi E, Javadian N, Nazeri A, Doosti R, Naser Moghadasi A, Owji M, Hashemi Taheri AP, Shakouri Rad A, Azimi A, Voineskos AN, Nazeri A, Sahraian MA. Convergent effects of a functional. C3 variant on brain atrophy, demyelination, and cognitive impairment in multiple sclerosis. Mult Scler 2019; 25:532–40 [DOI] [PubMed] [Google Scholar]
- 27.Adriani KS, Brouwer MC, Geldhoff M.Baas F, Zwinderman AH, Paul Morgan B, Harris CL, van der Ende A, van de Beek D. Common polymorphisms in the complement system and susceptiblity to bacterial meningitis. J Infect 2013; 66:255–62 [DOI] [PubMed] [Google Scholar]
- 28.Githeko AK, Service MW, Mbogo CM, Atieli FK, Juma FO. Origin of blood meals in indoor and outdoor resting malaria vectors in Western Kenya. Acta Trop 1994; 58:307–16 [DOI] [PubMed] [Google Scholar]
- 29.Otieno RO, Ouma C, Ong'echa JM.Keller CC, Were T, Waindi EN, Michaels MG, Day RD, Vulule JM, Perkins DJ. Increased severe anemia in HIV-1-exposed and HIV-1-positive infants and children during acute malaria. AIDS 2006; 20:275–80 [DOI] [PubMed] [Google Scholar]
- 30.Were T, Davenport GC, Hittner JB.Ouma C, Vulule JM, Ong‘echa JM, Perkins DJ. Bacteremia in Kenyan children presenting with malaria. J Clin Microbiol 2011; 49:671–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Were T, Hittner JB, Ouma C.Otieno RO, Orago AS, Ong'echa JM, Vulule JM, Keller CC, Perkins DJ. Suppression of RANTES in children with Plasmodium falciparum malaria. Haematologica 2006; 91:1396–9 [PubMed] [Google Scholar]
- 32.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph 1996; 14:33–8, 27–38 [DOI] [PubMed] [Google Scholar]
- 33.Gaunt TR, Rodriguez S, Zapata C, Day IN. MIDAS: software for analysis and visualisation of interallelic disequilibrium between multiallelic markers. BMC Bioinformatics 2006; 7:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webb B, Sali A. Comparative protein structure modeling using MODELLER. Current Protocols in Bioinformatics 2016; 54:5.6.1–5.6.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janssen BJ, Huizinga EG, Raaijmakers HC.Roos A, Daha MR, Nilsson-Ekdahl K, Nilsson B, Gros P. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature 2005; 437:505–11 [DOI] [PubMed] [Google Scholar]
- 36.Schaid DJ, Batzler AJ, Jenkins GD, Hildebrandt MA. Exact tests of Hardy-Weinberg equilibrium and homogeneity of disequilibrium across strata. Am J Hum Genet 2006; 79:1071–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li SS, Khalid N, Carlson C, Zhao LP. Estimating haplotype frequencies and standard errors for multiple single nucleotide polymorphisms. Biostatistics 2003; 4:513–22 [DOI] [PubMed] [Google Scholar]
- 38.Davenport Ouma C, Hittner JB.Were T, Ouma Y, Ong'echa JM, Perkins DJ. Hematological predictors of increased severe anemia in Kenyan children coinfected with Plasmodium falciparum and HIV-1. Am J Hematol 2010; 85:227–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janssen BJ, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature 2006; 444:213–6 [DOI] [PubMed] [Google Scholar]
- 40.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 1993; 234:779–815 [DOI] [PubMed] [Google Scholar]
- 41.Forneris F, Ricklin D, Wu J.Tzekou A, Wallace RS, Lambris JD, Gros P. Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science 2010; 330:1816–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xue X, Wu J, Ricklin D.Forneris F, Di Crescenzio P, Schmidt CQ, Granneman J, Sharp TH, Lambris JD, Gros P. Regulator-dependent mechanisms of C3b processing by factor I allow differentiation of immune responses. Nat Struct Mol Biol 2017; 24:643–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bajic G, Yatime L, Sim RB, Vorup-Jensen T, Andersen GR. Structural insight on the recognition of surface-bound opsonins by the integrin I domain of complement receptor 3. Proc Natl Acad Sci U S A 2013; 110:16426–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alcorlo M, Martinez-Barricarte R, Fernandez FJ.Rodriguez-Gallego C, Round A, Vega MC, Harris CL, de Cordoba SR, Llorca O. Unique structure of iC3b resolved at a resolution of 24 a by 3D-electron microscopy. Proc Natl Acad Sci Usa U S A 2011; 108:13236–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu J, Wu YQ, Ricklin D, Janssen BJ, Lambris JD, Gros P. Structure of complement fragment. C3b-factor H and implications for host protection by complement regulators. Nat Immunol 2009; 10:728–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abarrategui-Garrido C, Martinez-Barricarte R, Lopez-Trascasa M, de Cordoba SR, Sanchez-Corral P. Characterization of complement factor H-related (CFHR) proteins in plasma reveals novel genetic variations of CFHR1 associated with atypical hemolytic uremic syndrome. Blood 2009; 114:4261–71 [DOI] [PubMed] [Google Scholar]
- 47.de Cordoba SR, de Jorge EG. Translational mini-review series on complement factor H: genetics and disease associations of human complement factor H. Clin Exp Immunol 2008; 151:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merle NS, Noe R, Halbwachs-Mecarelli L, Fremeaux-Bacchi V, Roumenina LT. Complement system part II: Role in immunity. Front Immunol 2015; 6:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verschoor A, Karsten CM, Broadley SP, Laumonnier Y, Kohl J. Old dogs-new tricks: immunoregulatory properties of C3 and C5 cleavage fragments. Immunol Rev 2016; 274:112–26 [DOI] [PubMed] [Google Scholar]
- 50.Buffet PA, Safeukui I, Deplaine G.Brousse V, Prendki V, Thellier M, Turner GD, Mercereau-Puijalon O. The pathogenesis of Plasmodium falciparum malaria in humans: insights from splenic physiology. Blood 2011; 117:381–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.CDC. CDC Activities in Kenya, http://www.cdc.gov/malaria/malaria_worldwide/cdc_activities/kenya.html (2021, accessed 20 September 2021).