Abstract
The Genome Warehouse (GWH) is a public repository housing genome assembly data for a wide range of species and delivering a series of web services for genome data submission, storage, release, and sharing. As one of the core resources in the National Genomics Data Center (NGDC), part of the China National Center for Bioinformation (CNCB; https://ngdc.cncb.ac.cn), GWH accepts both full and partial (chloroplast, mitochondrion, and plasmid) genome sequences with different assembly levels, as well as an update of existing genome assemblies. For each assembly, GWH collects detailed genome-related metadata of biological project, biological sample, and genome assembly, in addition to genome sequence and annotation. To archive high-quality genome sequences and annotations, GWH is equipped with a uniform and standardized procedure for quality control. Besides basic browse and search functionalities, all released genome sequences and annotations can be visualized with JBrowse. By May 21, 2021, GWH has received 19,124 direct submissions covering a diversity of 1108 species and has released 8772 of them. Collectively, GWH serves as an important resource for genome-scale data management and provides free and publicly accessible data to support research activities throughout the world. GWH is publicly accessible at https://ngdc.cncb.ac.cn/gwh.
Keywords: Genome submission, Genome sequence, Genome annotation, Genome Warehouse, Quality control
Introduction
Genome sequences and annotations are fundamental resources for a wide range of genome-related studies, including various omics data analysis such as genome [1], transcriptome [2], epigenome [3], [4], and genome variation [5], [6]. China, as one of the most biodiverse countries in the world, harbors more than 10% of the world’s known species [7]. In the past decades, a large number of genomes of featured and important animals and crops in China have been sequenced and assembled [1], [8], [9], [10], [11], most of which were submitted to International Nucleotide Sequence Database Collaboration (INSDC) members [National Center for Biotechnology Information (NCBI), European Bioinformatics Institute (EBI), and DNA Data Bank of Japan (DDBJ)] [12]. With the rapid growth of genome assembly data, in China, for example, the large genome data size, the slow data transfer rate due to limited international network transfer bandwidth, and the language barrier for communication of technical issues have obstructed researchers from efficiently submitting their data to INSDC members. All these call for an additional centralized genomic data repository to complement the INSDC.
Here, we report the Genome Warehouse (GWH; https://ngdc.cncb.ac.cn/gwh), a centralized resource housing genome assembly data and delivering a series of genome data services. As one of the core resources in the National Genomics Data Center (NGDC), part of the China National Center for Bioinformation (CNCB; https:// ngdc.cncb.ac.cn) [13], GWH aims to accept data submissions worldwide and provide an important resource for genome data quality control, data archive, rapid release, and public sharing (e.g., with INSDC) in support of research activities from all over the world. By May 21, 2021, GWH has received a total of 19,124 genome submissions (including 51 international submissions), demonstrating its increasingly important role in global genome data management and sharing.
Data model
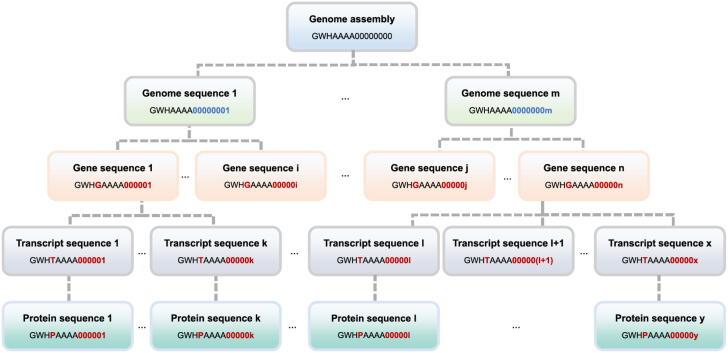
Designed for compatibility with the INSDC data model, each genome assembly in GWH is linked to BioProject (https://ngdc.cncb.ac.cn/bioproject) and BioSample (https://ngdc.cncb.ac.cn/biosample), which are two fundamental resources for metadata description in CNCB-NGDC. Full or partial (chloroplast, mitochondrion, and plasmid) genome assemblies with different assembly levels (complete, draft in chromosome, scaffold, and contig) are all acceptable, and existing genome assemblies are allowed to be updated. Accession numbers are assigned with the following rules (Fig. 1): 1) each genome assembly has an accession number prefixed with “GWH”, followed by four capital letters and eight zeros (e.g., GWHAAAA00000000); 2) genome sequences have the same accession number format as their corresponding genome assembly, with the exception that the eight digits start from 00,000,001 and increase in order (e.g., GWHAAAA00000001); 3) genes have similar accession patterns as those of genome sequences, with the addition of letter “G” between the GWH prefix and the four capital letters, and there are six digits at the end instead of eight (e.g., GWHGAAAA000001); 4) transcripts use the letter “T” to replace “G” in accession numbers for genes (e.g., GWHTAAAA000001); 5) proteins use the letter “P” to replace “G” in accession numbers for genes (e.g., GWHPAAAA000001); 6) if the submission is an update of existing submission in GWH, it will be assigned a dot and an incremental number to represent the version (e.g., GWHAAAA00000000.1).
Fig. 1.
Data model in GWH Genome assembly accession numbers are represented as, for example, “GWHAAAA00000000”, in which the “AAAA” can be replaced by any four other capital English letters representing different genome assemblies. The first genome sequence under the genome assembly is represented as “GWHAAAA00000001”, and other genome sequences under the same genome assembly are represented with the last eight digits increasing in order (“GWHAAAA00000002”, “GWHAAAA00000003”, etc.). For the first gene sequence, transcript sequence, and protein sequence under the genome assembly, the accession numbers are assigned as “GWHGAAAA000001”, “GWHTAAAA000001”, “GWHPAAAA000001”, respectively, and the last six digits are increasing in order for other genes, transcripts, and proteins.
Database components
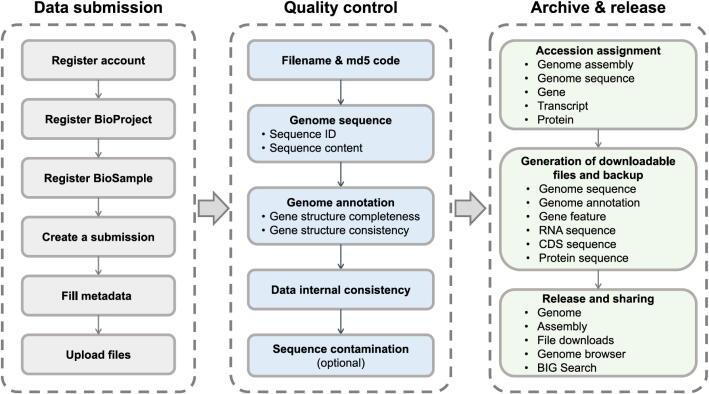
GWH is a centralized resource housing genome-scale data with the purpose to archive high-quality genome sequences and annotation information. GWH is equipped with a series of web services for genome data submission, release, and sharing, accordingly involving three major components, namely, data submission, quality control, and archive and release (Fig. 2).
Fig. 2.
Major components in GWH data processing workflow.
Data submission
GWH not only accepts genome assembly associated data through an online submission system but also allows offline batch submissions. Users need to register first and then provide a complete description of submitted genome sequences. Biological project and sample information should be provided through BioProject and BioSample (two fundamental resources in CNCB-NGDC), respectively, together with genome assembly sequence, annotation, and associated metadata. Metadata mainly consists of a variety of information about submitter, general assembly, file(s), sequence assignment, and publication (if available). After submission, GWH runs an automated quality control pipeline to check the validity and consistency of submitted genome sequence and genome annotation files. Accession numbers are assigned to assemblies and sequences upon the pass of quality control. The updated assembly data can also be submitted to GWH. It should be noted that compatible with the INSDC members (e.g., NCBI GenBank), it is the responsibility of the submitters to ensure the data quality, completeness, and consistency, and GWH does not warrant or assume any legal liability or responsibility for the data accuracy.
Quality control
After metadata and file(s) are received, GWH automatically runs standardized quality control to check 45 different types of errors in submitted genome sequences and annotations, and to scan for contaminated genome sequences (see details at https://ngdc.cncb.ac.cn/gwh/documents) if needed (Fig. 2), which roughly falls into 5 quality control steps. 1) The component will check the consistency of file(s) according to the filename and md5 code. 2) For genome sequences, the component will check the legality of genome sequence ID and sequence content, e.g., unique sequence ID, sequence composition (A/T/C/G or degenerate base), and sequence length (≥200 bp). 3) For genome annotations, the component will check gene structure completeness and consistency, e.g., unique ID, an exon/CDS/UTR coordinate falling within the corresponding gene coordinate, strand consistency for all features (including gene/transcript/exon/CDS/UTR), and codon validity (e.g., valid start/stop codon and no internal stop codon). 4) Finally, it will check the internal consistency of genome sequence and annotation. For example, sequence ID in genome annotation must match genome sequence ID; a feature coordinate must fall within the range of the corresponding genome sequence. 5) Genome sequences will also be scanned to check vectors, adaptors, primers, and indices (collected from UniVec database, ftp://ftp.ncbi.nlm.nih.gov/pub/UniVec) using NCBI’s VecScreen (https://www.ncbi.nlm.nih.gov/tools/vecscreen). If there is an error, a report will be automatically sent to the submitter by email. To finish a successful submission, the submitter needs to fix all errors and resubmit files until they pass the quality control process.
Archive and release
GWH will assign a unique accession number to the submitted genome assembly upon the pass of quality control, allot accession numbers for each genome sequence, gene, transcript, and protein, and generate and backup downloadable files of genome sequence and annotation in FASTA, GFF3, and TSV formats. Data generation is performed with in-house scripts based on submitted genome sequence and annotation files. In order to ensure the security of submitted data, a copy of backup data is stored on a physically separate disk. GWH will release sequence data on a user-specified date, unless a paper citing the sequence or accession number is published prior to the specified release date, in which case the sequence will be released immediately. For the released data, GWH will generate web pages containing two primary tables: genome and assembly. The former shows species taxonomy information and genome assemblies, and the latter contains general information of the assembly (including external links to other related resources), statistics of genome assembly, and its corresponding annotation. All released data are publicly available at GWH FTP site (ftp://download.cncb.ac.cn/gwh). GWH provides data visualization for both genome sequence and genome annotation using JBrowse [14]. It offers statistics and charts in light of total holdings, assembly levels, genome representations, citing articles, submitting organizations, sequencing platforms, assembly methods, and downloads. GWH provides user-friendly web interfaces for data browse and query using BIG Search [13], in order to help users find any released data of interest. For a released genome assembly, GWH also provides machine-readable application programming interfaces (APIs) for publicly sharing and automatically obtaining information on its associated BioProject, BioSample, genome, and assembly metadata and file paths.
Global sharing of SARS-CoV-2 and coronavirus genomes
During the COVID-19 outbreak, GWH, in support of the 2019 Novel Coronavirus Resource (2019nCoVR) [15], [16], has received worldwide submissions of more than a thousand SARS-CoV-2 genome assemblies with standardized genome annotations [17] and has released 298 of them. To expand the international influence of data, 62 of the released sequences have been shared, with the submitters’ permission, in GenBank [18] through a data exchange mechanism established with NCBI. In this model, GWH accessions are represented as secondary accessions in NCBI GenBank records, which are retrievable by the NCBI Entrez system. This model sets a good example for data sharing among different data centers.
In addition, GWH offers sequences of the Coronaviridae family to facilitate researchers to reach the data conveniently and thus to study the relationship between SARS-CoV-2 and other coronaviruses. To promote the data sharing and make all relevant information of the coronaviruses readily available, GWH integrates genomic and proteomic sequences as well as their metadata information from NCBI [19], China National GeneBank Database (CNGBdb) [20], National Microbiology Data Center (NMDC) [21], and CNCB-NGDC. Duplicated records from different sources are identified and removed to gain a non-redundant dataset. As of May 21, 2021, the dataset has 163,637 nucleotide sequences and 1,475,933 protein sequences of the coronaviruses. Filters are implemented to narrow down the required coronavirus sequences using multiple conditions, including country/region, host, isolation source, length, and collection date. Both the metadata and sequences of the filtered results can be selected and downloaded as a separate file. The daily updated sequences and all sequences can also be downloaded from FTP (ftp://download.cncb.ac.cn/Genome/Viruses/Coronaviridae).
Data statistics
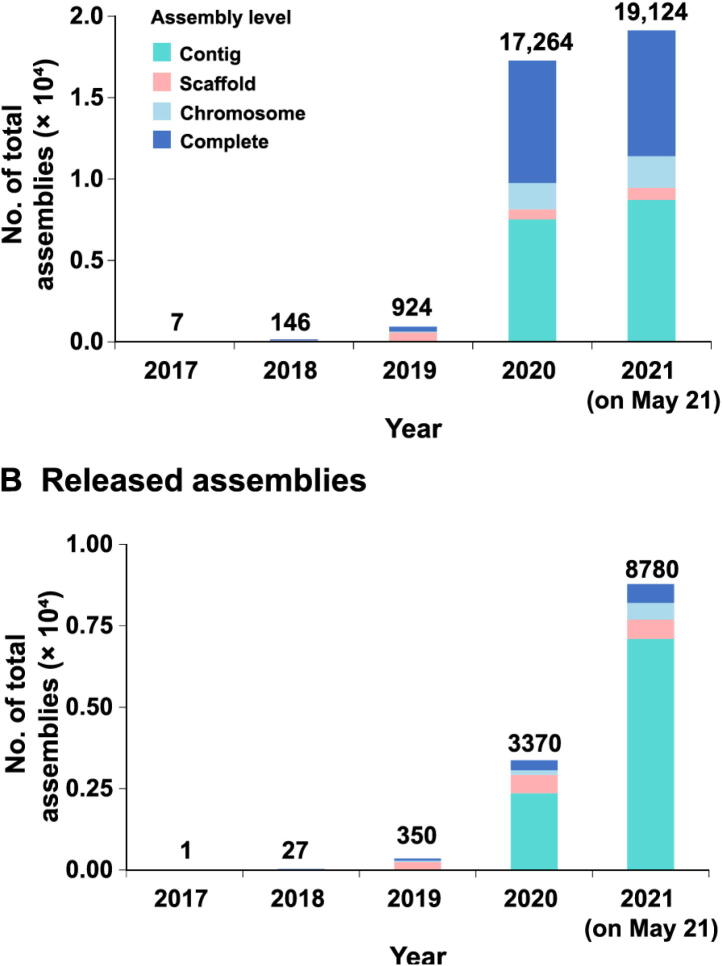
By May 21, 2021, GWH has received 19,124 direct submissions covering a broad diversity of species (Table 1) with different assembly levels (Fig. 3). These genome assemblies link to 367 BioProjects and 17,513 BioSamples, and are submitted by 269 submitters from 61 institutions (including 5 international submitters from 2 countries). There are a total of 8772 released submissions, which were reported in 96 articles from 47 journals. GWH has over 160,000 visits from 157 countries/regions, with ∼ 1,600,000 downloads. The amount of data, visits, and downloads in the GWH has been on a dramatic increase over the past years, clearly showing its great utility in genome-scale data management.
Table 1.
Total data holdings in GWH.
| Status | Type | Animal | Plant | Fungus | Bacterium | Archaea | Virus | Metagenome | Others | Total |
|---|---|---|---|---|---|---|---|---|---|---|
| Released | Assembly | 531 (6.05%) |
251 (2.86%) |
16 (0.18%) |
291 (3.32%) |
103 (1.17%) |
915 (10.43%) |
6651 (75.82%) |
14 (0.16%) |
8772 |
| Species | 90 (21.28%) |
159 (37.59%) |
14 (3.31%) |
109 (25.77%) |
11 (2.60%) |
23 (5.44%) |
5 (1.18%) |
12 (2.84%) |
423 | |
| Unpublic | Assembly | 7490 (72.35%) |
1334 (12.89%) |
104 (1.00%) |
76 (0.73%) |
19 (0.18%) |
858 (8.29%) |
10 (0.10%) |
461 (4.45%) |
10,352 |
| Species | 38 (5.31%) |
642 (89.66%) |
7 (0.98%) |
8 (1.12%) |
5 (0.70%) |
4 (0.56%) |
3 (0.42%) |
9 (1.26%) |
716 | |
| Total | Assembly | 8021 (41.94%) |
1585 (8.29%) |
120 (0.63%) |
367 (1.92%) |
122 (0.64%) |
1773 (9.27%) |
6661 (34.83%) |
475 (2.48%) |
19,124 |
| Species | 125 (11.28%) |
786 (70.94%) |
20 (1.81%) |
113 (10.20%) |
13 (1.17%) |
25 (2.26%) |
7 (0.63%) |
19 (1.71%) |
1108 |
Note: The numbers of genome assemblies and covering species are those directly submitted to GWH, and their percentages (in parentheses) for different organism groups are presented. GWH, Genome Warehouse.
Fig. 3.
Statistics of genome assemblies in GWH (as of May 21, 2021) A. All assemblies. B. Publicly released assemblies. Assemblies at contig, scaffold, chromosome, and complete levels are shown in different colors.
Summary and future directions
Collectively, GWH is a user-friendly portal for genome data submission, release, and sharing associated with a matched series of services. The rapid growth of genome assembly submissions demonstrates the great potential of GWH as an important resource for accelerating worldwide genomic research. With the goal to fully realize the findability, accessibility, interoperability, and reusability (FAIR) of genome data [22], GWH has made ongoing efforts, including but not limited to, improvement of web interfaces for data submission, presentation, and visualization, continuous integration of newly sequenced genomes, and development of useful online tools to help users analyze genome data (such as BLAST [23]). Therefore, we will put in more efforts to provide genome annotation services, especially for bacteria and archaea genomes, with the particular consideration that uniform standardized annotation determines the accuracy of downstream data analysis. Besides, we will expand the coronavirus dataset to other important pathogens to improve the ability of public health emergency response. Finally, we plan to share and exchange all public genome assembly data with the INSDC members to provide comprehensive data for global researchers.
Data availability
Genome Warehouse is freely accessible at https://ngdc.cncb.ac.cn/gwh.
Competing interests
The authors have declared no competing interests.
CRediT authorship contribution statement
Meili Chen: Methodology, Software, Investigation, Data curation, Writing – original draft, Project administration. Yingke Ma: Software, Writing – original draft. Song Wu: Software, Data curation. Xinchang Zheng: Data curation. Hongen Kang: Software. Jian Sang: Investigation, Data curation. Xingjian Xu: Software. Lili Hao: Investigation. Zhaohua Li: Data curation. Zheng Gong: Data curation. Jingfa Xiao: Writing – review & editing. Zhang Zhang: Writing – review & editing. Wenming Zhao: Writing – review & editing. Yiming Bao: Conceptualization, Writing – review & editing, Supervision.
Acknowledgments
We thank Profs. Jingchu Luo and Weimin Zhu for their helpful suggestions and a number of users for reporting bugs and sending comments. We also thank the NCBI GenBank group, especially Ilene Mizrachi, Karen Clark, Mark Cavanaugh, and Linda Yankie, for their valuable advice on sequence contamination scanning and SARS-CoV-2 sequence exchange. This work was supported by the Strategic Priority Research Program of Chinese Academy of Sciences (Grant Nos. XDB38060100 and XDB38030200 to YB; XDB38050300 to WZ; XDB38030400 to JX; XDA19050302 to ZZ), the National Key R&D Program of China (Grant Nos. 2016YFE0206600 to YB; 2020YFC0847000, 2018YFD1000505, 2017YFC1201202, and 2016YFC0901603 to WZ; 2017YFC0907502 to ZZ), the 13th Five-year Informatization Plan of Chinese Academy of Sciences (Grant No. XXH13505-05 to YB), the Genomics Data Center Construction of Chinese Academy of Sciences (Grant No. XXH-13514-0202 to YB), the Open Biodiversity and Health Big Data Programme of International Union of Biological Sciences to YB, the Professional Association of the Alliance of International Science Organizations (Grant No. ANSO-PA-2020-07 to YB), the National Natural Science Foundation of China (Grant Nos. 32030021 and 31871328 to ZZ), and the International Partnership Program of the Chinese Academy of Sciences (Grant No. 153F11KYSB20160008 to ZZ).
Handled by Fangqing Zhao
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences/China National Center for Bioinformation and Genetics Society of China
References
- 1.Liu Y, Du H, Li P, Shen Y, Peng H, Liu S, et al. Pan-genome of wild and cultivated soybeans. Cell 2020;182:162–76.e13. [DOI] [PubMed]
- 2.Guan Y., Chen M., Ma Y., Du Z., Yuan N., Li Y., et al. Whole-genome and time-course dual RNA-Seq analyses reveal chronic pathogenicity-related gene dynamics in the ginseng rusty root rot pathogen Ilyonectria robusta. Sci Rep. 2020;10:1586. doi: 10.1038/s41598-020-58342-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li R., Liang F., Li M., Zou D., Sun S., Zhao Y., et al. MethBank 3.0: a database of DNA methylomes across a variety of species. Nucleic Acids Res. 2018;46:D288–D295. doi: 10.1093/nar/gkx1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong Z., Li M., Yang F., Ma Y., Sang J., Li R., et al. EWAS Data Hub: a resource of DNA methylation array data and metadata. Nucleic Acids Res. 2020;48:D890–D895. doi: 10.1093/nar/gkz840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song S., Tian D., Li C., Tang B., Dong L., Xiao J., et al. Genome Variation Map: a data repository of genome variations in BIG Data Center. Nucleic Acids Res. 2018;46:D944–D949. doi: 10.1093/nar/gkx986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang B., Zhou Q., Dong L., Li W., Zhang X., Lan L., et al. iDog: an integrated resource for domestic dogs and wild canids. Nucleic Acids Res. 2019;47:D793–D800. doi: 10.1093/nar/gky1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McBeath J., McBeath J.H. Biodiversity conservation in China: policies and practice. Journal of International Wildlife Law & Policy. 2006;9:293–317. [Google Scholar]
- 8.Fan H., Wu Q., Wei F., Yang F., Ng B.L., Hu Y. Chromosome-level genome assembly for giant panda provides novel insights into Carnivora chromosome evolution. Genome Biol. 2019;20:267. doi: 10.1186/s13059-019-1889-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia Q., Zhou Z., Lu C., Cheng D., Dai F., Li B., et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- 10.Lin T., Xu X., Ruan J., Liu S., Wu S., Shao X., et al. Genome analysis of Taraxacum kok-saghyz Rodin provides new insights into rubber biosynthesis. Natl Sci Rev. 2018;5:78–87. [Google Scholar]
- 11.Li C., Song W., Luo Y., Gao S., Zhang R., Shi Z., et al. The HuangZaoSi maize genome provides insights into genomic variation and improvement history of maize. Mol Plant. 2019;12:402–409. doi: 10.1016/j.molp.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Arita M., Karsch-Mizrachi I., Cochrane G. The international nucleotide sequence database collaboration. Nucleic Acids Res. 2021;49:D121–D124. doi: 10.1093/nar/gkaa967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.CNCB-NGDC Members and Partners Database resources of the National Genomics Data Center, China National Center for Bioinformation in 2021. Nucleic Acids Res. 2021;49:D18–D28. doi: 10.1093/nar/gkac1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buels R., Yao E., Diesh C.M., Hayes R.D., Munoz-Torres M., Helt G., et al. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016;17:66. doi: 10.1186/s13059-016-0924-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao W.M., Song S.H., Chen M.L., Zou D., Ma L.N., Ma Y.K., et al. The 2019 novel coronavirus resource. Yi Chuan. 2020;42:212–221. doi: 10.16288/j.yczz.20-030. in Chinese with an English abstract. [DOI] [PubMed] [Google Scholar]
- 16.Song S., Ma L., Zou D., Tian D., Li C., Zhu J., et al. The global landscape of SARS-CoV-2 genomes, variants, and haplotypes in 2019nCoVR. Genomics Proteomics Bioinformatics. 2020;18:749–759. doi: 10.1016/j.gpb.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shean R.C., Makhsous N., Stoddard G.D., Lin M.J., Greninger A.L. VAPiD: a lightweight cross-platform viral annotation pipeline and identification tool to facilitate virus genome submissions to NCBI GenBank. BMC Bioinf. 2019;20:48. doi: 10.1186/s12859-019-2606-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sayers E.W., Cavanaugh M., Clark K., Ostell J., Pruitt K.D., Karsch-Mizrachi I. GenBank. Nucleic Acids Res. 2020;48:D84–D86. doi: 10.1093/nar/gkz956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatcher E.L., Zhdanov S.A., Bao Y., Blinkova O., Nawrocki E.P., Ostapchuck Y., et al. Virus Variation Resource – improved response to emergent viral outbreaks. Nucleic Acids Res. 2017;45:D482–D490. doi: 10.1093/nar/gkw1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen F.Z., You L.J., Yang F., Wang L.N., Guo X.Q., Gao F., et al. CNGBdb: China National GeneBank DataBase. Yi Chuan. 2020;42:799–809. doi: 10.16288/j.yczz.20-080. in Chinese with an English abstract. [DOI] [PubMed] [Google Scholar]
- 21.Wu L., Sun Q., Desmeth P., Sugawara H., Xu Z., McCluskey K., et al. World data centre for microorganisms: an information infrastructure to explore and utilize preserved microbial strains worldwide. Nucleic Acids Res. 2017;45:D611–D618. doi: 10.1093/nar/gkw903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z., Song S., Yu J., Zhao W., Xiao J., Bao Y. The elements of data sharing. Genomics Proteomics Bioinformatics. 2020;18:1–4. doi: 10.1016/j.gpb.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Genome Warehouse is freely accessible at https://ngdc.cncb.ac.cn/gwh.