Abstract
The Genome Sequence Archive (GSA) is a data repository for archiving raw sequence data, which provides data storage and sharing services for worldwide scientific communities. Considering explosive data growth with diverse data types, here we present the GSA family by expanding into a set of resources for raw data archive with different purposes, namely, GSA (https://ngdc.cncb.ac.cn/gsa/), GSA for Human (GSA-Human, https://ngdc.cncb.ac.cn/gsa-human/), and Open Archive for Miscellaneous Data (OMIX, https://ngdc.cncb.ac.cn/omix/). Compared with the 2017 version, GSA has been significantly updated in data model, online functionalities, and web interfaces. GSA-Human, as a new partner of GSA, is a data repository specialized in human genetics-related data with controlled access and security. OMIX, as a critical complement to the two resources mentioned above, is an open archive for miscellaneous data. Together, all these resources form a family of resources dedicated to archiving explosive data with diverse types, accepting data submissions from all over the world, and providing free open access to all publicly available data in support of worldwide research activities.
Keywords: Genome Sequence Archive, GSA, GSA-Human, OMIX
Introduction
The Genome Sequence Archive [1] (GSA, https://ngdc.cncb.ac.cn/gsa) is a public archive of raw sequence data in the National Genomics Data Center (NGDC) [2], [3], [4], part of the China National Center for Bioinformation (CNCB). GSA accepts worldwide data submissions, performs data curation and quality control for all submitted data, and provides free open access to all publicly available data without unnecessary restrictions. Since its inception in 2015, GSA has been broadly supported and endorsed by the scientific community, as testified by a total of 359,017 experiments, 395,977 runs, and 8756 terabyte (TB) files submitted by 1574 users from 391 institutions, as well as reported in 711 research articles and 252 scientific journals (as of 30 June 2021). Importantly, GSA serves as one of the core resources in CNCB-NGDC that has stable state funding in biological data management, thus ensuring long-term persistence and preservation of submitted datasets.
Due to the rapid development of sequencing technologies towards higher throughput and lower cost as well as their wider applications in biomedical research, a large number of multi-omics data have been produced at ever-increasing rates and scales, provoking two major challenges for raw data management in GSA. For one thing, several large-scale sequencing projects (such as Earth BioGenome Project [5], Dog 10K Project [6], Protist 10000 Genomes Project [7], Genomic sequencing of SARS-CoV-2 [8], [9]) have been carried out over the past several years, leading to different types of raw sequence data generated around the globe and accordingly requiring a suite of web services for massive data submission and deposition. For another, studies on human population genomics and precision medicine have produced millions of personal genome sequences associated with clinical information, requiring controlled access and security management, which is critically vital in promoting human healthcare and precise medical treatment, and advancing big-data-driven scientific research, while protecting data privacy. These challenges are particularly crucial in China since it not only features the largest population in the world and rich biodiversity resources, but also has a formidable capacity in genome sequencing throughout the country.
To address these challenges, here we provide a family of resources for raw data archive and management, including an updated version of GSA and two newly developed partner resources, namely, GSA for Human (GSA-Human, https://ngdc.cncb.ac.cn/gsa-human) and Open Archive for Miscellaneous Data (OMIX, https://ngdc.cncb.ac.cn/omix). Specially, we updated GSA with significant improvements on data model, online functionalities, and web interfaces. As an important partner to GSA that provides open access to all released data, GSA-Human features controlled-access and security services for human genetics-related data, which is compatible well with the database of Genotypes and Phenotypes (dbGaP) [10] in the National Center for Biotechnology Information (NCBI) [11] and the European Genome-phenome Archive (EGA) [12] in the European Bioinformatics Institute (EBI) [13]. In addition, OMIX, as a critical complement to the two aforementioned resources, is an open archive for miscellaneous data that are unsuitable to store in GSA, GSA-Human, or other databases at CNCB-NGDC. Together, all these resources form a family of resources dedicated to archiving explosive data with diverse types.
Archival resources
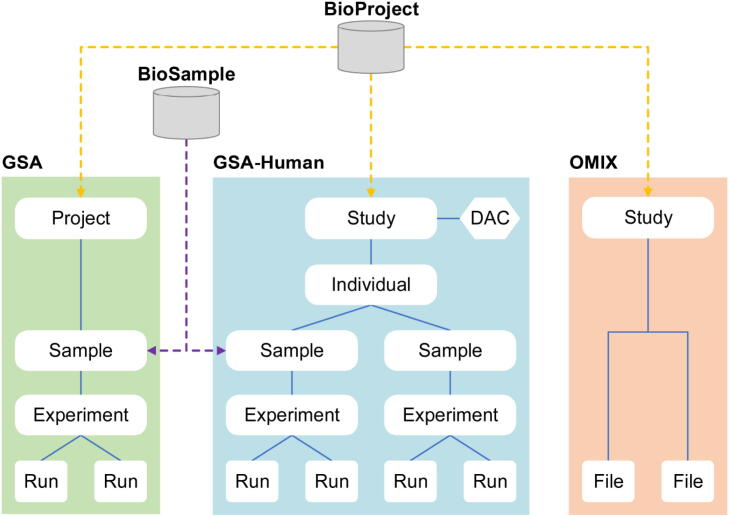
GSA, built based on the International Nucleotide Sequence Database Collaboration (INSDC) [14] data standards and structures, is a public data repository for archiving raw sequence reads. Over the past several years, GSA has been frequently and considerably updated since its establishment in 2015, with significant improvements in data structure, online submission, quality control, and web functionalities (Table 1). First, data structure has been significantly changed (Figure 1); BioProject (https://ngdc.cncb.ac.cn/bioproject/) and BioSample (https://ngdc.cncb.ac.cn/biosample/) have been separated from GSA. They now serve as independent meta-information databases, and act as an organizational framework to provide centralized access to descriptive metadata about research projects and samples, respectively. Second, to help users submit massive data with different types, more sequencing platforms, sample types, and file formats are acceptable, and importantly, batch submission of multiple experiments and runs is enabled in the updated version of GSA. In addition, to provide users with convenient services for uploading raw sequence files, GSA not only provides an FTP server but also equips with Aspera (https://www.ibm.com/products/aspera) to realize high-speed data transmission. Third, GSA was greatly enhanced by improving the expert curation process and integrating an automated quality control pipeline, with the aim to provide value-added services for archiving high-quality data. Fourth, multiple web functionalities for bilingual support (in both English and Chinese), online documentation, data statistics, and visualization charts were updated/newly added. Taken together, the updated version of GSA is more efficient and friendly in big omics data submission, deposition, and management.
Table 1.
Comparison between GSA in 2017 and the GSA family in 2021
| Category | 2017 | 2021 |
|---|---|---|
| Archival resources | GSA | GSA, GSA-Human, OMIX |
| Number of supported sample types* | 7 | 11 |
| Batch submission | NA | Available |
| Data statistics | NA | Available |
| Supported languages | English | English, Chinese |
| Controlled access | NA | Available |
| Data transfer | FTP | FTP, Aspera |
| Number of supported sequencing platforms* | 49 | 66 |
| Number of supported data formats* | 9 | 13 |
| Quality control* | Metadata | Metadata, data |
Note: * More details are available at https://ngdc.cncb.ac.cn/gsa/standards. NA, not available.
Figure 1.
Data model of the GSA family. BioProject and BioSample are two independent meta-information databases, acting as an organizational framework to provide centralized access to descriptive metadata about research projects and samples, respectively. GSA-Human is for archiving human genetic data and OMIX is for various types of data (that are unsuitable for GSA/GSA-Human).
GSA-Human, established in April 2018, is a data repository specialized in the secure management of human genetics-related data. It accepts submissions of various studies, including disease, cohort, cell line, clinical pathogen, and human-associated metagenome. GSA-Human uses the “individual” to organize its metadata and sequence reads, and provides two different data access mechanisms: open access and controlled access. Open access means that all data are public for global researchers, whereas controlled access means that data can be downloadable only after being authorized by the Data Access Committee (DAC) that is responsible for authorizing/declining data access to data requester. Therefore, GSA-Human provides a series of data services including access control, data request, access authorization/declining, and security management. As a specialized database for security management of human genetics-related data, GSA-Human has several major features different from dbGaP. First, data access authorization is controlled by user-defined DAC in GSA-Human, but in dbGaP, it is governed by the National Institutes of Health (NIH) that sponsors each study. Second, GSA-Human is devoted to managing human raw sequencing data, whereas dbGaP is for genotype–phenotype association studies. Third, data in GSA-Human are submitted by users from all over the world, while data in dbGaP are mainly from the NIH sponsored projects.
OMIX, as a new member of the archival resources in CNCB-NGDC, aims to meet users’ needs for submitting various types of data other than sequences. It collects not only raw data from transcriptome, epigenome, and microarray, but also functional data such as lipidome, metabolome, proteome, and even data like clinical information, demographic data, as well as questionnaires. With the concise interface and simplified submission process, OMIX makes data submission and deposition very easy. Of note, similar to GSA-Human, OMIX has a data security management strategy for human genetic data. Any controlled-access dataset in OMIX can be accessed only with the permission of the data submitter.
Data submission and retrieval
Data submission to the GSA family is aided by a series of web services, including BIG Single Sign-On (SSO; https://ngdc.cncb.ac.cn/sso/) that is a user access control system and BIG Submission portal (BIG Sub; https://ngdc.cncb.ac.cn/gsub/) that is a unified one-stop portal providing submission services for a variety of database resources in CNCB-NGDC. To submit data to the GSA family, user needs to register an account and log into any database via SSO, thereby gaining access to multiple independent systems with a single ID and password.
Overall, the GSA family provides a suite of services for data retrieval, download, and access. Public data in these resources can be retrieved via BIG Search (https://ngdc.cncb.ac.cn/search/), a scalable text search engine that performs powerful data retrieval and analytical capabilities. All released data are publicly accessible and downloadable via FTP and HTTP, except the controlled data in GSA-Human and OMIX that require access permission. To access the controlled data, requester needs to create a request and send required documents for data access. Once the request has been reviewed and approved, the requester gains the access to the data.
Data statistics
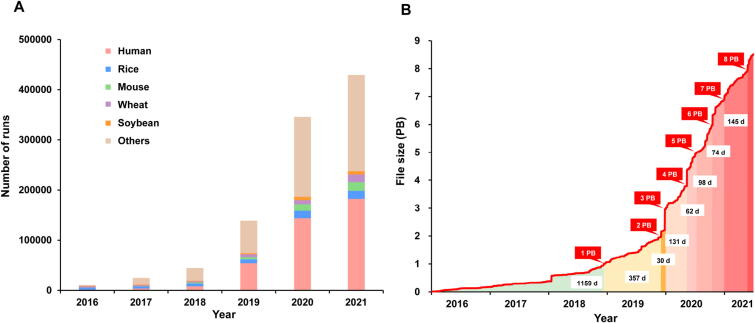
The GSA family has received a large number of data submissions with explosive growth in data and users, thus exhibiting its important roles in raw data management (Figure 2 and Table 2). The volume of archived data has increased by more than 40 times, compared to the 200 TB archived in the previous release of GSA [1]. Till 30 June 2021, GSA and GSA-Human have collected 359,017 experiments, 395,977 runs, and more than 8.5 petabyte (PB) of data submitted from 1574 submitters of 391 organizations (Figure 2A). In particular, GSA-Human has archived 68,241 individuals and housed 5 PB of raw sequence data in one year, clearly showing that human genetic data are growing at an unprecedented rate and scale. More importantly, GSA-Human has received a total of 743 access requests from 501 requesters, with 184 requests approved till 30 June 2021. Regarding the increase in the volume of archived data over time, it took about three years to accumulate the first PB of data and the volume currently reaches 8.5 PB in just over two and a half years after that, with a formidably dramatic decrease in days needed for data accumulation per PB (Figure 2B). Strikingly it took only 30 days for the volume to reach the third PB, primarily attributed to a large-scale sequencing project [15] with 344 TB of data archived. Meanwhile, the number of species involved is also on a rapid increase, from 80 in December 2016 to more than 1000 at present. Also, albeit newly established, OMIX has collected 290 files of 1.614 TB.
Figure 2.
Data statistics of the GSA family. A. Number of runs accumulated from 2016 to 2021, with five major species indicated. B. Increase in the volume of submitted data over time. Time needed to accumulate each PB of data is indicated. All statistics were derived from GSA and GSA-Human as of 30 June 2021. PB, petabyte; d, days.
Table 2.
Data items of the GSA family
| Item | GSA | GSA-Human | OMIX | Total |
|---|---|---|---|---|
| No. of projects | 3157 | 764 | 100 | 3321 |
| No. of individuals | / | 68,241 | / | 68,241 |
| No. of samples | 191,754 | 155,490 | / | 347,244 |
| No. of experiments | 183,441 | 175,576 | / | 359,017 |
| No. of runs | 201,583 | 194,394 | / | 395,977 |
| File size (terabyte) | 3704 | 5052 | 1.614 | 8757 |
| No. of registered users | 2438 | 2563 | 120 | 4365 |
Note: All statistics were derived from the GSA family as of 30 June 2021. / means not applicable.
Currently, the GSA family has more than 4365 registered users and has been visited by 648,274 unique IPs from 111 countries/regions, with a total of 35,010,529 page views and an average of 4 TB of downloads per day. Data housed in these resources have been reported in more than 250 scientific journals (https://ngdc.cncb.ac.cn/gsa/statistics?active=journals), including Cell, Genome Res, Genomics Proteomics Bioinformatics, Nature, Plant Cell, and Proc Natl Acad Sci U S A. More importantly, with frequent updates and improvements in the past several years, GSA has been recognized as one of the certified repositories at FAIRsharing.org and re3data.org, and therefore meets the requirement as a supported repository by Elsevier, Taylor & Francis, Wiley, and Springer Nature. More detailed statistics can be found online at https://ngdc.cncb.ac.cn/gsa/standards.
Future directions
The explosive volume of raw data submitted to the GSA family is still on the increase, posing significant challenges to handle and share such big data [16]. Nowadays, CNCB-NGDC, hosting a suite of database resources including the GSA family, is going to be enhanced by national big data infrastructure, with stable governmental funding investment in upgrading storage, computing, and network resources, thus providing fundamental support in raw data archiving and management of the GSA family. In addition, our future efforts will be made to continuously optimize data models and curation processes in evolution of users’ needs, establishment of cloud infrastructure for big data storage, and development of a variety of tools to facilitate big data submission and high-speed transfer. To make effective use of human genetic data and promote precision healthcare and treatment, efforts will also be devoted to optimizing procedures and mechanisms to enable data sharing with controlled access and security by conforming to applicable regulations and ethical norms. Meanwhile, the security protection measures of the databases will be continuously enhanced, in terms of security mechanisms, hardware facilities, and software technologies, to ensure the long-term secure maintenance and management of data. We also advocate worldwide collaborations in developing data standards, tools, and approaches towards global biodiversity and health big data sharing (Global Biodiversity and Health Big Data Alliance; http://bhbd-alliance.org/).
Data availability
GSA-Human is freely accessible at https://ngdc.cncb.ac.cn/gsa-human/ and OMIX is freely accessible at https://ngdc.cncb.ac.cn/omix/.
Competing interests
The authors have declared no competing interests.
2021 The Authors. Published by Elsevier B.V. and Science Press on behalf of Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Orcid
0000-0003-1296-3093 (Tingting Chen)
0000-0001-6102-1751 (Xu Chen)
0000-0002-3852-4796 (Sisi Zhang)
0000-0003-4689-3513 (Junwei Zhu)
0000-0002-9357-4411 (Bixia Tang)
0000-0002-2565-2334 (Anke Wang)
0000-0003-0953-6306 (Lili Dong)
0000-0002-9422-822X (Zhewen Zhang)
0000-0002-3882-9979 (Caixia Yu)
0000-0002-3175-3625 (Yanling Sun)
0000-0003-4836-0577 (Lianjiang Chi)
0000-0003-1293-4495 (Huanxin Chen)
0000-0002-2084-7132 (Shuang Zhai)
0000-0003-3810-7156 (Yubin Sun)
0000-0002-4761-2245 (Li Lan)
0000-0002-2300-1036 (Xin Zhang)
0000-0002-2835-4340 (Jingfa Xiao)
0000-0002-9922-9723 (Yiming Bao)
0000-0002-7985-7941 (Yanqing Wang)
0000-0001-6603-5060 (Zhang Zhang)
0000-0002-4396-8287 (Wenming Zhao)
CRediT authorship contribution statement
Tingting Chen: Investigation, Methodology, Data curation, Writing – original draft. Xu Chen: Software. Sisi Zhang: Investigation, Methodology, Data curation, Writing – original draft. Junwei Zhu: Software. Bixia Tang: Software. Anke Wang: Writing – original draft, Software. Lili Dong: Data curation. Zhewen Zhang: Data curation. Caixia Yu: Data curation. Yanling Sun: Data curation. Lianjiang Chi: Software. Huanxin Chen: Resources. Shuang Zhai: Resources. Yubin Sun: Resources. Li Lan: Resources. Xin Zhang: Resources. Jingfa Xiao: Writing – review & editing. Yiming Bao: Conceptualization, Writing – review & editing, Funding acquisition. Yanqing Wang: Conceptualization, Investigation, Methodology, Software, Writing – review & editing, Project administration. Zhang Zhang: Conceptualization, Writing – review & editing, Funding acquisition. Wenming Zhao: Conceptualization, Methodology, Writing – review & editing, Supervision, Funding acquisition.
Acknowledgments
This work was supported by grants from National Key R&D Program of China (Grant No. 2017YFC0907502 to ZZ); Strategic Priority Research Program of Chinese Academy of Sciences (Grant Nos. XDB38060100 and XDB38030200 to YB; XDB38050300 to WZ; XDB38030400 to JX; XDA19050302 to ZZ); National Key R&D Program of China (Grant Nos. 2016YFC0901603 to WZ; 2017YFC1201202 to YW; 2020YFC0847000 and 2018YFD1000505 to WZ; 2016YFE0206600 to YB); The 13th Five-year Informatization Plan of Chinese Academy of Sciences (Grant No. XXH13505-05 to YB); Genomics Data Center Construction of Chinese Academy of Sciences (Grant No. XXH-13514-0202 to YB); Open Biodiversity and Health Big Data Programme of the International Union of Biological Sciences to YB; The Professional Association of the Alliance of International Science Organizations (Grant No. ANSO-PA-2020-07 to YB); National Natural Science Foundation of China (Grant Nos. 32030021 and 31871328 to ZZ); International Partnership Program of the Chinese Academy of Sciences (Grant No. 153F11KYSB20160008 to ZZ).
Handled by Ge Gao
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences/China National Center for Bioinformation and Genetics Society of China
Contributor Information
Yanqing Wang, Email: wangyanqing@big.ac.cn.
Zhang Zhang, Email: zhangzhang@big.ac.cn.
Wenming Zhao, Email: zhaowm@big.ac.cn.
References
- 1.Wang Y., Song F., Zhu J., Zhang S., Yang Y., Chen T., et al. GSA: Genome Sequence Archive. Genomics Proteomics Bioinformatics. 2017;15:14–18. doi: 10.1016/j.gpb.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song S., Zhang Z. Database Resources in BIG Data Center: submission, archiving, and integration of big data in plant science. Mol Plant. 2019;12:279–281. doi: 10.1016/j.molp.2019.01.020. [DOI] [PubMed] [Google Scholar]
- 3.National Genomics Data Center Members and Partners Database resources of the National Genomics Data Center in 2020. Nucleic Acids Res. 2020;48:D24–D33. doi: 10.1093/nar/gkz913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CNCB-NGDC Members and Partners Database resources of the National Genomics Data Center, China National Center for Bioinformation in 2021. Nucleic Acids Res. 2021;49:D18–D28. doi: 10.1093/nar/gkac1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewin H.A., Robinson G.E., Kress W.J., Baker W.J., Coddington J., Crandall K.A., et al. Earth BioGenome Project: sequencing life for the future of life. Proc Natl Acad Sci U S A. 2018;115:4325–4333. doi: 10.1073/pnas.1720115115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang B., Zhou Q., Dong L., Li W., Zhang X., Lan L., et al. iDog: an integrated resource for domestic dogs and wild canids. Nucleic Acids Res. 2019;47:D793–D800. doi: 10.1093/nar/gky1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miao W., Song L., Ba S., Zhang L., Guan G., Zhang Z., et al. Protist 10,000 Genomes Project. Innovation (N Y) 2020;1 doi: 10.1016/j.xinn.2020.100058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song S., Ma L., Zou D., Tian D., Li C., Zhu J., et al. The global landscape of SARS-CoV-2 genomes, variants, and haplotypes in 2019nCoVR. Genomics Proteomics Bioinformatics. 2020;18:749–759. doi: 10.1016/j.gpb.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao W., Song S., Chen M., Zou D., Ma L., Ma Y., et al. The 2019 Novel Coronavirus Resource. Hereditas (Beijing) 2020;42:212–221. doi: 10.16288/j.yczz.20-030. (in Chinese with an English abstract) [DOI] [PubMed] [Google Scholar]
- 10.Tryka K.A., Hao L., Sturcke A., Jin Y., Wang Z.Y., Ziyabari L., et al. NCBI’s Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res. 2014;42:D975–D979. doi: 10.1093/nar/gkt1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sayers E.W., Beck J., Bolton E.E., Bourexis D., Brister J.R., Canese K., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021;49:D10–D17. doi: 10.1093/nar/gkaa892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lappalainen I., Almeida-King J., Kumanduri V., Senf A., Spalding J.D., Ur-Rehman S., et al. The European Genome-phenome Archive of human data consented for biomedical research. Nat Genet. 2015;47:692–695. doi: 10.1038/ng.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cantelli G., Cochrane G., Brooksbank C., McDonagh E., Flicek P., McEntyre J., et al. The European Bioinformatics Institute: empowering cooperation in response to a global health crisis. Nucleic Acids Res. 2021;49:D29–D37. doi: 10.1093/nar/gkaa1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cochrane G., Karsch-Mizrachi I., Takagi T. International Nucleotide Sequence Database Collaboration. The international nucleotide sequence database collaboration. Nucleic Acids Res. 2016;44:D48–D50. doi: 10.1093/nar/gkv1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J., Xu C., Lee H.J., Ren S., Zi X., Zhang Z., et al. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature. 2020;580:93–99. doi: 10.1038/s41586-020-2135-x. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z., Song S., Yu J., Zhao W., Xiao J., Bao Y. The elements of data sharing. Genomics Proteomics Bioinformatics. 2020;18:1–4. doi: 10.1016/j.gpb.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
GSA-Human is freely accessible at https://ngdc.cncb.ac.cn/gsa-human/ and OMIX is freely accessible at https://ngdc.cncb.ac.cn/omix/.