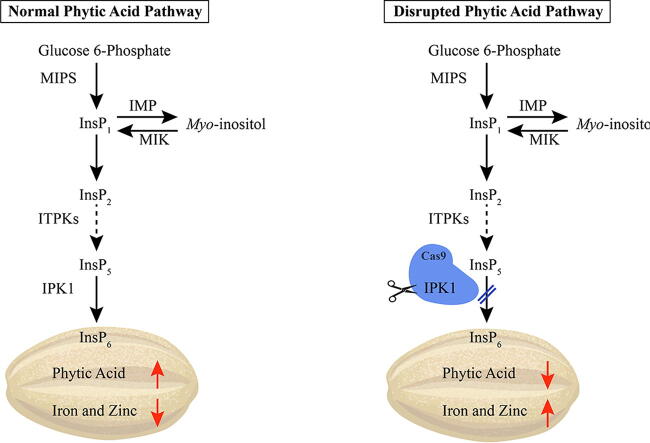
Graphical abstract
Keywords: Biofortification, CRISPR/Cas9, Phytic acid, Iron, Zinc
Highlights
-
•
This study, for the first time, used CRISPR/Cas9 to disrupt TaIPK1 gene in wheat.
-
•
Disruption of TaIPK1 gene reduces phytic acid and enhanced Iron and Zinc in wheat grains.
-
•
This study will help to develop the biofortified wheat and reducing the malnutrition.
-
•
This study provided a great resource for the development of biofortified wheat using CRISPR/Cas9.
Abstract
Introduction
Phytic acid (PA) is an important antinutrient agent present in cereal grains which reduces the bioavailability of iron and zinc in human body, causing malnutrition. Inositol pentakisphosphate 2- kinase 1 (IPK1) gene has been reported to be an important gene for PA biosynthesis.
Objective
A recent genome editing tool CRISPR/Cas9 has been successfully applied to develop biofortified rice by disrupting IPK1 gene, however, it remained a challenge in wheat. The aim of this study was to biofortify wheat using CRISPR/Cas9.
Methods
In this study, we isolated 3 TaIPK1 homeologs in wheat designated as TaIPK1.A, TaIPK1.B and TaIPK1.D and found that the expression abundance of TaIPK1.A was stronger in early stages of grain filling. Using CRISPR/Cas9, we have disrupted TaIPK1.A gene in cv. Borlaug-2016 with two guide RNAs targeting the 1st and 2nd exons.
Results
We got several genome-edited lines in the T0 generation at frequencies of 12.7% and 10.8%. Sequencing analysis revealed deletion of 1–23 nucleotides and even an addition of 1 nucleotide in various lines. Analysis of the genome-edited lines revealed a significant decrease in the PA content and an increase in iron and zinc accumulation in grains compared with control plants.
Conclusion
Our study demonstrates the potential application of CRISPR/Cas9 technique for the rapid generation of biofortified wheat cultivars.
Introduction
Malnutrition or hidden hunger is a serious issue around the globe affecting about two billion people, predominantly in developing and African countries [1]. This is mainly due to the dependency of people on selected grain crops such as maize, wheat, or rice having an inadequate amount of vital micronutrients mainly iron, zinc, and vitamin A [2], [3]. Malnutrition due to the deficiency of these micronutrients often leads to muscular dysfunction, weak immune system, anaemia, body weight loss, diarrhea and pneumonia. Cereals are deficient in micronutrients due to the presence of phytic acid (PA) or phytate (inositol hexakisphosphate/IP6), that acts as a major chelator of important micronutrients [4].
Phytic acid is a member of the inositol phosphates (InsPs) family which comprises soluble signaling molecules [5]. As a second messenger, these molecules play important roles in cell metabolism including mRNA export, DNA repair, chromatin structure regulation and apoptosis [6], [7]. Phytic acid is a major source of phosphorus (P) in wheat and other cereal grains, however, it cannot be efficiently utilized by monogastric animals including humans due to the absence of phytase enzyme [4]. Additionally, it acts as a chelator of metal ions such as Fe+2 and Zn+2 and reduces the bioavailability of these important micronutrients, and thus considered as an antinutrient substance [8]. Considering the antinutritional properties of PA, several approaches were used to reduce the seed PA content in cereal grains such as the development of low PA mutants using induced mutations by ethyl‐methanesulfonate (EMS) or gamma irradiation [9], [10]. For example, low PA mutants, called as Lpa5, Lpa9, and Lpa59, were developed in rice through induced mutations (gamma rays 60Co) which showed 54–63% reduction in phytic acid [10]. Similarly, a low PA mutant was screened from 562 ethyl‐methanesulfonate (EMS) mutagenized M2 lines in wheat with 43% less PA contents [9]. A low phytic acid mutant (lpa241) was also found in maize that showed 90% reduction of phytic acid in seeds [11]. However, these induced mutations are often random and associated with other agronomic defects such as poor germination and low yield, in addition to low PA content [10], [11]. Therefore, a targeted mutagenesis approach should be used that can specifically target the desired gene to avoid other lethal phenotypes [12].
Several genes encoding the enzymes involved in different steps of PA biosynthesis pathway have been reported in different crops [13], [14], [15], [16]. The first step in the PA biosynthesis involves the formation of inositol-3-phosphate (Ins3P) from glucose-6-phosphate which is catalyzed by myo-inositol-3-phosphate synthase (MIPS) [17]. Next steps involve the phosphorylation of the inositol ring through various enzymes at the remaining five positions [16], [18]. Other enzymes involved in catalyzing these phosphorylation reactions include inositol monophosphatase (IMP), inositol tris/tetraphosphate kinase (ITPK), inositol polyphosphate kinase (IPK2), and inositol-pentakisphosphate 2-kinase (IPK1). Attempts were made to disrupt the PA biosynthesis pathway by targeting different genes involved in different steps of this pathway [19]. However, disrupting the pathway at early stages would disrupt the myo-inositol-3-phosphate that will negatively affect the plant inositol metabolism and hence disturb other related pathways [20]. Therefore, the best strategy to develop low PA grains is to disrupt the genes involved in final stages of PA biosynthesis
IPK1 (Inositol 1,3,4,5,6-pentakisphosphate 2-kinase) is a member of inositol phosphate kinase (IPK) superfamily which catalyzes the final step of PA biosynthesis by phosphorylating IP5 to IP6 [4], [21], [22]. Targeting this gene using genome editing approach has great potential to develop low PA crops [23]. Previously, low PA rice and wheat were developed using RNAi- mediated knockdown of the IPK1 gene which caused significant increase in seed Fe and Zn contents [4], [8]. However, RNAi- mediated targeting has several limitations towards crop improvement due to high rate of off-target mutations and completely transgenic nature. Recently, a robust genome editing tool known as Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) (CRISPR/Cas9) system has dominated over RNAi and other genome editing tools for its fast and heritable nature with less chances of off-target mutations. Most importantly, we are now able to eliminate the CAS9 transgene in the next generations using simple segregation or crossing which offers a great potential for its utilization for rapid crop improvement using targeted mutagenesis [12], [24].
Up till now, disruption of IPK1 gene through CRISPR/Cas9 has not been attempted in wheat for biofortification. To address the issue, here we developed CRISPR/Cas9-mediated knockout lines for TaIPK1 gene by targeting 1st and 2nd exons of TaIPK1 gene using specific guide RNAs to produce biofortified wheat grains. We found that expression of TaIPK1.A was higher than other homologs, TaIPK1.B and TaIPK1.D in Pakistani elite wheat cv. Borlaug 2016. Disruption of TaIPK1.A led to increase in the accumulation of iron and zinc contents in genome edited Borlaug 2016 plants compared with non-edited plants. We believe that this study will help for the development of biofortified wheat at commercial level and contribute in reducing the malnutrition from developing countries.
Materials and methods
Plant material and growth conditions
Seeds of the high yielding hexaploid wheat (Triticum aestivum L.) cultivars “Pakistan 2013” and “Borlaug 2016” were obtained from Bio-Resource Conservation Institute (BCI), National Agricultural Research Centre (NARC), Islamabad, Pakistan. Non-transgenic control plants (cv. Pakistan 2013 and Borlaug 2016) and transgenic plants (T0, T1 and T2) were grown in glasshouse at the National Institute for Genomics and Advanced Biotechnology (NIGAB), NARC, Islamabad, Pakistan. The greenhouse conditions were maintained in controlled conditions as day light period 16 h with 25 ± 3 °C temperature and night period 8 h with 15 ± 3 °C temperature. Pots were filled with homogenized soil mixture, containing soil, sand and peat moss (1:1:2). Experiments were conducted according to complete randomized design (CRD). Three biological replicates of each variety/line were maintained for RNA sampling. While, six observations were taken for morphological (plant height, spike length, number of spikelets and, grain related traits) and biochemical (total phytic acid, Fe and Zn content) traits.
TaIPK1 homoeologs isolation
Samples of leaf, shoot and root tissues were taken at 3–4 leaf stage plants and frozen in liquid nitrogen. Total RNA was extracted from leaf, shoot and root tissues using GeneJet plant RNA Purification Kit (Thermofisher, catalog number K0732) according to the manufacturer’s instructions. cDNA synthesis was carried out by using First Strand cDNA Synthesis Kit (Fermantas, catalog number K1612) with 5 µg of total RNA.
Coding sequences of TaIPK1 homeologs of three genomes of wheat i.e. A, B and D were retrieved by a BLAST search of rice (OsIPK1; AK102842) sequence in Ensemble Plants database. Sequence alignments were performed by using BioEdit software. Gene specific primers were manually designed for isolation of IPK1 homeologs(A, B and D genomes separately) from wheat cv. Pakistan 2013 and Borlaug 2016 (Additional file 1). For the amplification of full-length coding sequence of IPK1 gene, PCR was performed with rTaq polymerase (TaKaRa, Tokyo, Japan; Code R001) using cDNA as a template. The PCR products were resolved on 1.0% agarose gel. Target bands were recovered and purified by using GeneJETTM gel extraction kit, and cloned into the pTZ vector by InsTAcloneTM PCR cloning kit (Thermo Scientific InsTA clone #K1214). The TaIPK1 homeologs were then confirmed by sequencing of the pTZ vectors.
BioEdit software was used for sequence analysis of newly isolated IPK1 gene homoeologs from the A, B and D genomes of Borlaug 2016 variety. The IPK1 sequences retrieved from Ensemble plant database served as reference genes. Multiple alignments were generated by ClustalW in BioEdit Sequence Alignment Editor.
Target selection and guide RNAs designing
Samples of developing grains were taken at two stages, i.e. 14 days after anthesis (DAA) and 21 DAA. RNA extraction and cDNA synthesis were performed as described above. StepOne Plus real time RT-PCR system (Applied Biosystems) was employed to observe the expression pattern of TaIPK1.A, TaIPK1.B and TaIPK1.D in cv. Borlaug 2016. SYBR Green was used as fluorescent dye. Total RNA extracted was converted into cDNA using reverse transcriptase (RT) PCR (Fermantas) in 20 µl reaction volume with 5 µg of total RNA. Genome specific primers were designed from 3 homeologs of TaIPK1 gene and TaActin was used as an endogenous control for relative expression [25]. Expression levels were normalized by housekeeping gene (TaActin). The quantification of relative expression level was done by using 2−ΔΔCt method. The primer sequences are available in additional file2. Target homeolog was selected based on expression pattern.
In order to design unique gRNAs, target sequences were identified in the coding region of the TaIPK1 gene. And a BLAST search was done to identify the possible off-target sites. Containing a PAM sequence these manually selected guide RNA seed sequences were close to the start codon with a suitable restriction site. The CRISPR/Cas9 system was provided by Dr. Bing Yang (Iowa State University, Genetics, Development and Cell Biology department, Ames, USA) [26]. The guide RNA expression cassette was constructed according to expression vector pENTR4- gRNA. Two complementary oligonucleotides of 21–25 bp with 4-nucleotide overhang of TGTT at the 5′ end of sense strand and an extra AAAC at the 5′ end of antisense strand for BtgZI restriction site were synthesized commercially. Similarly, another two oligos from the 2nd exon with BsaI restriction site were designed with 4 nucleotide overhang of GTGT at the 5′ end of sense strand and the same AAAC at the 5′ end of antisense strand (Table 1).
Table 1.
Guide RNA sequences used for construction of RNA expression cassettes.
| Name Gene and Oligo | Guide RNA sequence | Size (bp) |
|---|---|---|
| TaIPK-A-1st Oligo TaIPK-A-1st C Oligo |
5′TGTTGGTCTACAAGGGAGAGGGCG3′ 5′AAACCGCCCTCTCCCTTGTAGACC3′ |
24 |
| TaIPK-A-2nd Oligo TaIPK-A-2nd C Oligo |
5′GTGTGTCTGTCAAGCAAGGTTCTG3′ 5′AAACCAGAACCTTGCTTGACAGAC3′ |
24 |
Vector construction for CRISPR/Cas9
In order to unveil the function of TaIPK1 in wheat, gene disruption using CRISPR/Cas9 system was employed. Two single guide RNAs i.e. gRNA1 and gRNA2 with PAM sequences were designed from the first and second exons of TaIPK1. Potential off-target sequences were detected via BLAST searches. These targets were also evaluated for the presence of non-CpG sensitive restriction site sequences predicted to be disrupted by Cas9 induced indels, which also had to be unique within a PCR amplicon. Final target sequences were chosen to be as specific as possible to the intended target sequence (that is, keeping the number of mismatches to off-target sequences high), close to the start codon. The gateway vector system of pENTR4 and pOs-Cas9 was used for genome editing purpose. The constructs were generated according to Bing Yang’s method [26], and wheat cv. Borlaug 16 was used for genome editing. Both the guide RNAs were inserted into pENTR4 vector using BtgZ1 and Bsa1 sites. Finally this cassette was inserted in final pOsCas9 vector through gateway LR-recombination process. The constructed vector of CRISPR/Cas9 carrying both the guide RNAs was introduced into LBA4404 strain of Agrobacterium tumefaciens.
Wheat transformation and screening of transformed plants
For wheat transformation, immature seeds of wheat cv. Borlaug 2016 were surface sterilized with sodium hypochlorite for 10 min followed by washing with distilled water. Immature embryos were isolated from seeds and treated with Agrobacterium tumefaciens carrying CRISPR/Cas9 construct and grown on MS medium for plant regeneration. The regenerated plants were shifted to another medium containing hygromycin for selection of positive transgenic plants. The transgenic seedlings were transferred to glasshouse in pots and allowed to grow under controlled condition (22/16 °C with 16/8h day/night photoperiod and 70% humidity) till maturity.
In order to detect the efficiencies of gRNA-1 and gRNA-2 into wheat plants, PCR based restriction enzyme assay was carried out. Genomic DNA was extracted and PCR was performed with primers flanking the target sites using genomic DNA as template (Additional file 3). PCR products were digested with T7-E1 restriction enzyme and resolved on agarose gel electrophoresis. Finally, undigested PCR products were gel purified and sequenced from commercial company. The presence of homozygous or biallelic indels was detected by the comparison of sequences with wild type sequences at To stage.
Determination of phytic acid (PA) and micronutrients (Fe, Zn) contents
Estimation of total phytic acid in mature seeds of T2 lines and non-transgenic cv. Borlaug 2016 was done by using K-PHYT kit (Megazyme Inc, Bray, Ireland) according to the manufacturer’s protocol. Briefly, the mature seeds were subjected to grinding to make fine powder and extraction was done using 0.66 N NH4Cl with continuous stirring for 10–12 h. For colorimetric procedure, supernatant was used as per manufacturer’s instructions.
For Micronutrients (Fe, Zn) analysis, mature seeds of T2 transgenic lines and non-transgenic cv. Borlaug 2016 plants were subjected to grinding to make fine powder. The samples were transferred to a carbonator for ashing at 600 °C for 16 h. Quantification of micronutrients was done by using Atomic Absorption Spectrometer [27].
Morphological parameters were also measured in three transgenic lines compared to control plant (for each line 6 observations were made). Morphological data measured include plant height, spike length, spikelet number, grain length, grain width, 1000 grain weight.
Statistical analysis
Gene expression, PA accumulation, Fe and Zn content, plant height, spike length, spikelet number, grain length, grain width, 1000 grain weight were statistically analyzed. Bar graphs are made in excel for representation of mean values. T-tests were performed in excel to determine whether the average values were significantly distinct between transgenic (T2) and wild plants (n = 6, *= P < 0.05, **=P < 0.01, ***= P < 0.001).
Results
Isolation of TaIPK1 homeologs
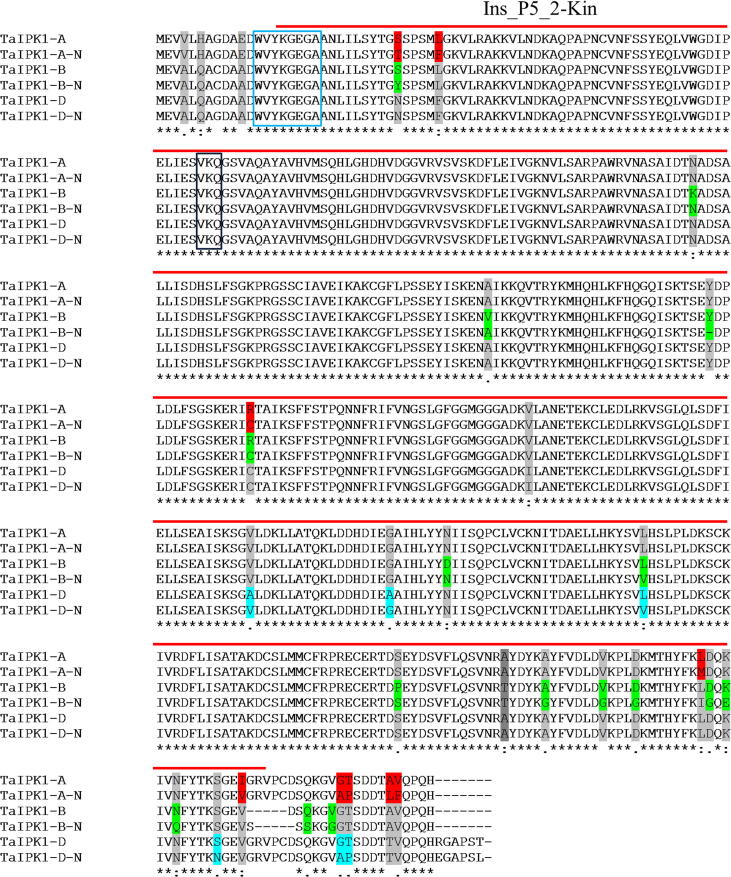
In order to isolate IPK1 homeologs from indigenous wheat varieties “Pakistan 2013” and “Borlaug 2016”, PCR was performed with gene specific primers for the A, B and D genomes using cDNA as template. The amplified sequence contained an open reading frame of 1365 bp which was predicted to encode a protein of 454 amino acid residues. Nucleotide alignment showed that the sequences of new IPK1 homeologs were highly similar to the TaIPK1 sequences retrieved from NCBI database. Protein alignment of newly isolated TaIPK1-A, TaIPK1-B and TaIPK1-D genes and their homeologs retrieved from NCBI showed that the size of the proteins is conserved (Fig. 1). Pairwise alignment of newly isolated genes TaIPK1-A-N revealed 9 amino acids substitutions and conservation of 445 amino acids. Six out of 9 substitutions were synonymous, while 3 were non-synonymous. Interestingly, TaIPK1-B- N showed 16 amino acids substitutions. Eleven out of 16 substitutions were synonymous, while 5 were non-synonymous. In case of TaIPK1-D-N, 6 amino acids substitutions were detected. These mutations were randomly distributed over the entire length of the sequence. These data allow us to infer that newly isolated genes are homoeologs of IPK1 in wheat.
Fig. 1.
Protein sequence alignment showing conserved domain and sequence variation among A, B & D genomes. Variation in amino acids is represented by different colors. Red bar above the amino acid sequences (15–420) represent “Ins_P5_2-Kinase” domain. Light blue rectangular box show 13–20 a.a deletion by gRNA1, and dark blue rectangular box show 76–78 a.a deletion by gRNA2.
Selection of target homeolog of TaIPK1 gene
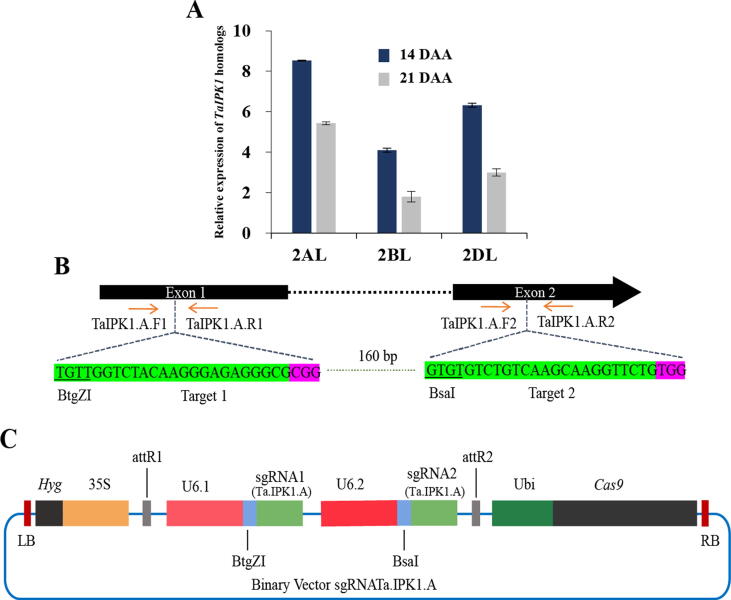
In order to observe gene expression patterns of 3 homeologs in Borlaug 2016, qRT-PCR was performed. Results showed that all the 3 homeologs i.e. TaIPK1.A, TaIPK1.B and TaIPK1.D are expressed in the seeds at 14 days after anthesis (DAA) or in early stages of grain filling (Fig. 2A). However, transcript expression decreases after 21 DAA and a significantly less transcript level was observed at 21 DAA compared to 14 DAA (Fig. 2A). Notably, among the three homeologs, TaIPK1.A (from A genome) exhibits considerable higher expression than other two homeologs i.e. TaIPK1.B and TaIPK1.D at both the stages. TaIPK1.B is the least expressed gene among 3homeologs. We therefore, targeted the TaIPK1.A gene from A genome for CRISPR/Cas9 mediated disruption in Borlaug 2016 cultivar of wheat for the functional characterization of TaIPK1 gene (Fig. 2B).
Fig. 2.
Expression of TaIPK1 homoeologs in seeds and illustration of gRNA expression Cassette. (A) Expression analysis of three TaIPK1 homoeologs at two different developmental stages of seed i.e. 14 and 21 days after anthesis (DAA). 2AL, 2BL, 2DL represent expression of TaIPK1 homoeologs on the long arm of chromosome 2 of the A, B and D genomes. qRT-PCR assays were performed using SYBR green and Ct values were normalized against TaActin as an endogenous control. Blue bars represent expression of TaIPK1 homoeologs at 14 DAA stage and gray bars represent the expression at 21 DAA stage. (B) Wheat Ta.IPK1.A gene model and target sequences are shown. The Ta.IPK1.A sequences for sgRNA1Ta.IPK1.A (Target 1) and sgRNA2Ta.IPK1.A (Target 2) are shown below the target regions with the PAM highlighted in magenta colour. Restriction endonucleases (BtgZ1 andBsa1) recognition sequences are underlined. Primers used for mutation detection are shown in both panels. (C) Schematic of binary plasmid vector delivered to wheat is depicted. The construct sgRNATa.IPK1.A houses a Kanamycin resistant cassette flanked by the Gateway recombination sequence attL1 and attL2 is mobilized to the binary vector pOs-Cas9 through the Gateway recombination. The expression cassette consists of a pair of sgRNAs (sgRNA1Ta.IPK1.A and sgRNA2Ta.IPK1.A) followed by endonuclease Cas9 driven by Ubiquitin promoter from Rice.
CRISPR/Cas9 mediated genome editing unveils mutations in TaIPK1
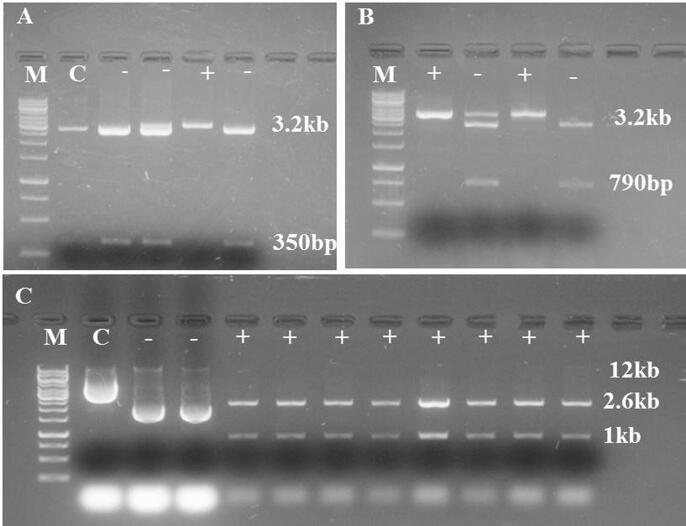
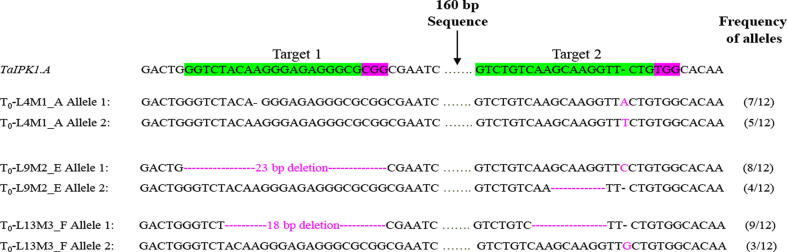
Guide RNAs (gRNA1 and gRNA2) were designed from the 1st and 2nd exons of Ta.IPK1.A gene (Fig. 2B). The construction of binary CRISPR/Cas9 vector sgRNA.TaIPK1 was successfully accomplished using gateway recombination reaction (Fig. 3). A total of 50 independent transgenic lines were produced by Agrobacterium- mediated in planta transformation. Among these, 15 To plantlets were screened for the Cas9 induced mutations through PCR amplification of the targeted regions using target flanking primers. PCR/restriction enzyme assay unveiled the mutation frequency of 12.7% and 10.8% for gRNA1and gRNA2 respectively. In different mutant lines, the Ta.IPK1.A sequences were found to contain various mutant alleles in both target sites (Fig. 4; Additional file 4). PCR amplification followed by sequencing of targeted regions in three To lines; To-L4M1_A, To-L9M2_E and To-L13M3_F, revealed 1 to 23 base pairs deletions and 1 bp insertions as well (Fig. 4). These three transgenic lines with confirmed homozygous edits in T1 generation were selected for T2 embryo rescue and further analysis.
Fig. 3.
Construction of gRNA expression Cassette. (A) Shows the restriction analysis of clones from the ligation of dsOligo at the BtgZ1 site of pENTR4-gRNA1 after digestion with BamH1 restriction enzyme. The clones without the 350 bp fragment are positive clones. (B) Shows the confirmation of insertion of second dsOligo at the Bsa1 site of pENTR4-gRNA1 after digestion with Bsa1 and BamH1 restriction enzymes. The positive clones are without digested fragment of 790 bp. (C) Shows the confirmation of Gateway LR recombination reaction after digestion of clones with HindIII restriction enzyme. The positive clones show the digestion pattern of 12 kb, 2.6 kb and 1 kb. (M, 1 kb DNA marker, C, control plasmid).
Fig. 4.
Confirmation of mutation in TaIPK1.A gene by sequencing. TaIPK1.A mutant alleles detected in To wheat plants. Insertions and deletions are represented by magenta font or magenta hyphens respectively. For each line (L4M1_A to L13M3_F), 12 clones were examined and the frequencies of each mutant allele are represented at the right side of the panel (Additional file 4).
Disruption of TaIPK1 decreases phytic acid but increases Fe and Zn content in wheat
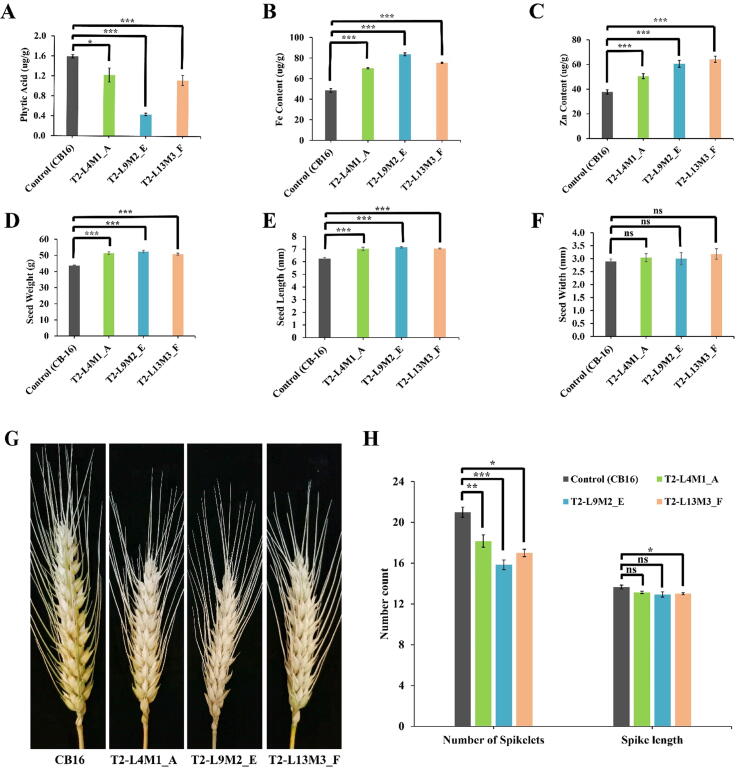
To see if disrupting TaIPK1 gene affects PA, Fe and Zn contents in wheat seeds, we measured PA, Fe and Zn contents in seeds of transgenic genome-edited plants along with CB16. Measurement of PA demonstrated that PA content was significantly reduced in genome edited lines compared with CB16 (Fig. 5A). The maximum reduction in the phytic acid content was observed for T2-L9M2_E plants (0.44ug/g) compared to control plants (1.60ug/g). Remaining two lines i.e. T2-L4M1_A and T2-L13M3_F also showed significant reduction in PA content as 1.22 and 1.12ug/g, respectively.
Fig. 5.
Micronutrient and phenotypic analysis of the mature T2 grains of genome edited mutant lines. (A) Total Phytic acid (PA) in the mature T2 seeds of genome edited (GE) mutant lines. PA was measured in the mature T2 seeds from the primary tiller of each GE mutant line. (B, C) Fe and Zn concentration (μg/g) was estimated by using Atomic absorption spectrometry (AAS). (D) Seed weight of control (CB-16) and GE mutant lines. (E) Seed length of control (CB-16) and GE mutant lines. (F) Seed width of control (CB-16) and GE mutant lines. (G) Representative picture of spikes of wheat GE mutant lines and control (scale bar = 1.5 cm). (H) Spike length (cm) and spikelet count from the primary tiller of GE mutant lines and control CB16 at T2 stage. The data indicates the mean ± SE of six biological replicates. Student’s t-test was used to compare the significant variation between each transgenic line and control. *,** and *** shows statistical significance at P < 0.05, P < 0.01 and P < 0.001, respectively, and ns = non-significant.
Further, micronutrient accumulation of Fe and Zn in grains showed a significant (at P < 0.01) increase in genome edited lines of TaIPK1 compared to control seeds. The results showed approximately 1.5 to 2.1 fold increase in the Fe concentration and 1.6 to 1.9 fold rise in Zn concentration. Maximum Fe content (83.61ug/g) were observed in T2-L9M2_E line plants, while higher Zn content (64.23ug/g) were demonstrated in T2-L13M3_F (Fig. 5B, C). However, no significant difference was observed in the seed germination for the transgenic and non-transgenic control seeds and plant height in transgenic lines compared to CB-16 (Additional file 6). Besides change in micronutrient accumulation, we also observed an altered spike architecture in transgenic plants (Fig. 5G). Spike length and number of spikelets were significantly reduced in genome edited lines compared with CB-16 (Fig. 5H). The maximum reduction in spikelet number was recorded 16 in T2-L9M2_E GE transgenic plants compared to CB-16 as 21, while other transgenic lines, T2-L4M1_A and T2-L13M3_F also demonstrated significant reduction in number of spikelets per spike. (Fig. 5E). Spike length was also reduced in transgenic plants, however, it is significant only for T2-L13M3_F plants (Fig. 5E). It is noteworthy that lowest PA content and highest Fe content were also observed in T2-L9M2_E line which indicates a possible negative correlation among PA and Fe contents. Notably, reduction in spike length and spikelet numbers results in significant increase in grain weight and grain length but not in grain width compared to CB-16 (Fig. 5D, E, F). This increase in grain weight suggested that transgenic lines compensate the reduction in grain yield caused by reduced spike length and number of spikelets. This study reveals that editing of negative regulator of biofortification, results in improvement of Fe and Zn accumulation in grains without compromising significantly on grain yield.
Discussion
Malnutrition is one of the main causes of poor growth and development leading to neurological disorders and poor performance in all spheres of life. The situation is exacerbated in developing countries that harbor most of the world population. The immediate solution to this menace is biofortified food as a cheaper source to mitigate nutritional deficiencies in the diet. Cereal crops are rich sources of micronutrients but their bioavailability is limited due to the presence of certain anti-nutritional factors [28]. In grains of soybean, rice, wheat and other cereal crops, phytic acid (IP6) is a major anti-nutrient that limits the micronutrients bioavailability. At developmental stages of cereal grains it is accumulated as phytate (metal ion chelator) that causes a drastic reduction in the micronutrients bioavailability leading to the micronutrient malnutrition.
Use of functional genomics approaches to disrupt the gene responsible for phytate accumulation thereby leading to bioavailability of micronutrient in the cereal grains especially in wheat can be a cheaper plausible strategy. Inositol 1,3,4,5,6-pentakisphosphate 2- kinase (IPK1) is a suitable candidate gene to reduce phytic acid in wheat [8] as it is a member of inositol phosphate kinase (IPK) superfamily. These enzymes catalyze the phosphorylation of IP5 to IP6. IPK1 gene codes for an enzyme involved with the final step of phytate biosynthesis. Targeting this gene can be one of the most effective methods for developing low phytic acid crops [23]. In the present study, we have successfully isolated and cloned the 3 TaIPK1 homeologs designated as TaIPK1.A, TaIPK1.B and TaIPK1.D and found that the expression of TaIPK1.A gene was stronger in early stages of grain filling. So we disrupted the TaIPK1.A gene in cv. Borlaug-2016 using CRISPR/Cas9 system to develop biofortified wheat grains.
Current advances in genome editing have made it possible to silence or activate multiple genes simultaneously due to availability of the CRISPR/Cas9 system [12]. In maize the IPK1 gene silencing has been done by using ZFNs, however in wheat the genome editing using other genes has been attempted but transiently [24]. We have used this modern but synonymous to natural gene manipulation system to disrupt TaIPK1.A gene in Borlaug 2016 cultivar of wheat. A tremendous reduction of phytic acid in the mature T2 seeds was detected as compared to the control seeds.
Previous studies suggest that disruption of enzymes involved in the early phytic acid synthesis pathway could be injurious for plant development and seed growth [19], [29], [30]. However, CRISPR/Cas9 mediated knock out of three paralogs of BnITPK gene (involved in PA synthesis pathway) in oilseed rape resulted in approximately 35% reduction in PA content in seeds without affecting the oil content and seed growth [31]. Similarly, in rice and Arabidopsis, IPK1 gene silencing “involved in late phase of phytic acid biosynthesis” resulted in 69 and 83% decrease in phytic acid content, respectively, whereas seed physiology was not affected [4], [32]. Compared to previous techniques, genome editing gives a better opportunity to transform specific genes with minimal adverse effects [33]. To avoid pleiotropic effects, one possible approach is to mutate a protein that results in a variant with null or reduced enzymatic activity. The resultant edited protein would show expression and fulfill its function through interaction with other proteins while blocking the concerned reaction. In a metabolic pathway, targeting of upstream reactions would affect all the downstream reactions [34]. Being the most downstream enzyme in the phytic acid pathway, IPK1 is a potential target for attaining decrease in phytate level. Fortunately, IPK1 gene can be mutated in such a way that the protein remains normal for interactions with other proteins. Studies have revealed that in cellular metabolism, inositol pyrophosphates (IP6 and IP7) also perform vital roles [35]. In case of other IPKs null mutants, there is loss of higher pyrophosphates due to abnormality in vesicular morphology, which is not the case with IPK1 mutants. Therefore, reduction in phytic acid would not damage the functions mediated by inositol pyrophosphates (IP7 and IP8). IPK1 enzyme can be slightly altered through disruption of its active site such that the protein remains expressing for important protein–protein interactions but is unable to form phytic acid (IP6) from IP5, without affecting the downstream and upstream processes. Since IPK1 disruption neither compromise plant physiology nor disturb the levels of myo-inositol, thus suggesting the added advantage of IPK1 disruption.
Our results are in corroboration with these findings and significant decrease of phytic acid in the mature T2 seeds was detected. As in developing spikes of mutant lines, awn length and spikelet arrangement was found to be almost similar to the control plants but alterations in spikelet number and spike length were recorded in mutant plants as compared to control. Similarly, OsIPK1 targeting through RNAi technology resulted in low phytic acid without changing the level of amino acids and vital seed storage proteins [4]. Since TaIPK1 gene is highly expressed at the early stages of seed development, it might be a sustainable strategy to operate activity or expression of this gene to decrease phytic acid in wheat grains. Our results demonstrate that the expression of TaIPK1.A transcript was stronger in early stages of grain filling. The TaIPK1.A transcript showed high expression level at both seed developmental stages (14 and 21 DAA) as compared to TaIPK1. B and TaIPK1.D transcripts.
Previously TaIPK1 gene has been downregulated in wheat by using RNAi technology for enhancing the micronutrients (Fe and Zn) bioavailability [8]. In present study, we used highly advanced genome editing tool CRISPR/Cas9 for TaIPK1 gene silencing in order to enhance the micronutrients (Fe and Zn) bioavailability. Overall, silencing results showed approximately 60 to 70% reduction of Ta.IPK1.A transcript in the mature seeds of T2 transgenic lines. Our results showed approximately 1.5 to 2.1 fold increase in the Fe concentration and 1.6 to 1.9 fold rise in Zn concentration due to significant reduction in phytic acid content. Taken together, these results demonstrated that disruption of TaIPK1 gene leads to reduction in PA and increase in Fe and Zn accumulation in wheat grains, suggesting key role of TaIPK1 in wheat biofortification. Thus, our study demonstrates the implications of CRISPR/Cas9 in producing knockouts in wheat for IPK1 gene. These studies have potential application in raising efforts to fortify wheat grains with Fe and Zn for resource poor in developing countries.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
CRediT authorship contribution statement
Saira Ibrahim: Methodology, Investigation, Writing - original draft. Bilal Saleem: Data curation, Writing - original draft. Nazia Rehman: Investigation, Validation. Syed Adeel Zafar: Conceptualization, Validation, Writing - review & editing. Muhammad Kashif Naeem: Validation, Data curation. Muhammad Ramzan Khan: Conceptualization, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank Dr. Bing Yang (Department of Genetics, Development and Cell Biology Iowa State University, Ames, Iowa, USA) for the generous gift of CRISPR/CAS9 vector system for this research work. Sincere appreciation to Amir Mumtaz (FSRI, NARC) for nutrient analysis. This work was supported by research budget from National Centre for Bioinformatics, Quaid-i-Azam University, Islamabad.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2021.07.006.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.McGuire S. FAO, IFAD, and WFP. The state of food insecurity in the world 2015: meeting the 2015 international hunger targets: taking stock of uneven progress. Rome: FAO, 2015. Oxford University Press; 2015. [DOI] [PMC free article] [PubMed]
- 2.Pachón H., Ortiz D.A., Araujo C., Blair M.W., Restrepo J. Iron, zinc, and protein bioavailability proxy measures of meals prepared with nutritionally enhanced beans and maize. J Food Sci. 2009;74(5):H147–H154. doi: 10.1111/j.1750-3841.2009.01181.x. [DOI] [PubMed] [Google Scholar]
- 3.Das J.K., Kumar R., Salam R.A., Bhutta Z.A. Systematic review of zinc fortification trials. Ann Nutr Metab. 2013;62(Suppl. 1):44–56. doi: 10.1159/000348262. [DOI] [PubMed] [Google Scholar]
- 4.Ali N., Paul S., Gayen D., Sarkar S.N., Datta K., Datta S.K. Development of low phytate rice by RNAi mediated seed-specific silencing of inositol 1, 3, 4, 5, 6-pentakisphosphate 2-kinase gene (IPK1) PLoS ONE. 2013;8(7) doi: 10.1371/journal.pone.0068161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michell R.H. Inositol derivatives: evolution and functions. Nat Rev Mol Cell Biol. 2008;9(2):151–161. doi: 10.1038/nrm2334. [DOI] [PubMed] [Google Scholar]
- 6.Agarwal R., Mumtaz H., Ali N. Role of inositol polyphosphates in programmed cell death. Mol Cell Biochem. 2009;328(1–2):155–165. doi: 10.1007/s11010-009-0085-6. [DOI] [PubMed] [Google Scholar]
- 7.Raboy V. myo-Inositol-1, 2, 3, 4, 5, 6-hexakisphosphate. Phytochemistry. 2003;64(6):1033–1043. doi: 10.1016/s0031-9422(03)00446-1. [DOI] [PubMed] [Google Scholar]
- 8.Aggarwal S., Kumar A., Bhati K.K., Kaur G., Shukla V., Tiwari S., et al. RNAi-mediated downregulation of inositol pentakisphosphate kinase (IPK1) in wheat grains decreases phytic acid levels and increases Fe and Zn accumulation. Front Plant Sci. 2018;9:259. doi: 10.3389/fpls.2018.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guttieri M., Bowen D., Dorsch J.A., Raboy V., Souza E. Identification and characterization of a low phytic acid wheat. Crop Sci. 2004;44(2):418–424. [Google Scholar]
- 10.Qamar Z-U, Hameed A, Ashraf M, Rizwan M, Akhtar M. Development and molecular characterization of low phytate basmati rice through induced mutagenesis, hybridization, backcross, and marker assisted breeding. Front Plant Sci 2019; 10: 1525. [DOI] [PMC free article] [PubMed]
- 11.Pilu R., Panzeri D., Gavazzi G., Rasmussen S.K., Consonni G., Nielsen E. Phenotypic, genetic and molecular characterization of a maize low phytic acid mutant (lpa241) Theor Appl Genet. 2003;107(6):980–987. doi: 10.1007/s00122-003-1316-y. [DOI] [PubMed] [Google Scholar]
- 12.Zafar SA, Zaidi SS-e-A, Gaba Y, Singla-Pareek SL, Dhankher OP, Li X, et al. Engineering abiotic stress tolerance via CRISPR/Cas-mediated genome editing. J Exp Botany 2020; 71(2): 470–9. [DOI] [PubMed]
- 13.Stevenson-Paulik J., Odom A.R., York J.D. Molecular and biochemical characterization of two plant inositol polyphosphate 6-/3-/5-kinases. J Biol Chem. 2002;277(45):42711–42718. doi: 10.1074/jbc.M209112200. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki M., Tanaka K., Kuwano M., Yoshida K.T. Expression pattern of inositol phosphate-related enzymes in rice (Oryza sativa L.): implications for the phytic acid biosynthetic pathway. Gene. 2007;405(1–2):55–64. doi: 10.1016/j.gene.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Sweetman D., Stavridou I., Johnson S., Green P., Caddick S.E., Brearley C.A. Arabidopsis thaliana inositol 1, 3, 4-trisphosphate 5/6-kinase 4 (AtITPK4) is an outlier to a family of ATP-grasp fold proteins from Arabidopsis. FEBS Lett. 2007;581(22):4165–4171. doi: 10.1016/j.febslet.2007.07.046. [DOI] [PubMed] [Google Scholar]
- 16.Stiles A.R., Qian X., Shears S.B., Grabau E.A. Metabolic and signaling properties of an Itpk gene family in Glycine max. FEBS Lett. 2008;582(13):1853–1858. doi: 10.1016/j.febslet.2008.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida K.T., Wada T., Koyama H., Mizobuchi-Fukuoka R., Naito S. Temporal and spatial patterns of accumulation of the transcript of myo-inositol-1-phosphate synthase and phytin-containing particles during seed development in rice. Plant Physiol. 1999;119(1):65–72. doi: 10.1104/pp.119.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasmussen S.K., Ingvardsen C.R., Torp A.M. Portland Press Ltd.; 2010. Mutations in genes controlling the biosynthesis and accumulation of inositol phosphates in seeds. [DOI] [PubMed] [Google Scholar]
- 19.Feng X., Yoshida K.T. Molecular approaches for producing low-phytic-acid grains in rice. Plant Biotechnology. 2004;21(3):183–189. [Google Scholar]
- 20.Panzeri D., Cassani E., Doria E., Tagliabue G., Forti L., Campion B., et al. A defective ABC transporter of the MRP family, responsible for the bean lpa1 mutation, affects the regulation of the phytic acid pathway, reduces seed myo-inositol and alters ABA sensitivity. New Phytol. 2011;191(1):70–83. doi: 10.1111/j.1469-8137.2011.03666.x. [DOI] [PubMed] [Google Scholar]
- 21.Stephens L., Irvine R. Stepwise phosphorylation of myo-inositol leading to myo-inositol hexakisphosphate in Dictyostelium. Nature. 1990;346(6284):580–583. doi: 10.1038/346580a0. [DOI] [PubMed] [Google Scholar]
- 22.Brearley C.A., Hanke D.E. Metabolic evidence for the order of addition of individual phosphate esters in the myo-inositol moiety of inositol hexakisphosphate in the duckweed Spirodela polyrhiza L. Biochem J. 1996;314(1):227–233. doi: 10.1042/bj3140227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wakeel A., Arif S., Bashir M.A., Ahmad Z., Rehman H.U., Kiran A., et al. Perspectives of folate biofortification of cereal grains. J Plant Nutr. 2018;41(19):2507–2524. [Google Scholar]
- 24.Zhang Y., Liang Z., Zong Y., Wang Y., Liu J., Chen K., et al. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun. 2016;7(1):1–8. doi: 10.1038/ncomms12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zafar S.A., Hameed A., Ashraf M., Khan A.S., Li X., Siddique K.H. Agronomic, physiological and molecular characterisation of rice mutants revealed the key role of reactive oxygen species and catalase in high-temperature stress tolerance. Funct Plant Biol. 2020;47(5):440–453. doi: 10.1071/FP19246. [DOI] [PubMed] [Google Scholar]
- 26.Char SN, Li R, Yang B. CRISPR/Cas9 for Mutagenesis in Rice. Transgenic Plants: Springer; 2019. p. 279–293.
- 27.Shar G.Q., Kazi T.G., Jakhrani M.A., Sahito S.R. Determination of Iron, Zinc and Manganese in Nine Varieties of Wheat (Triticum aestivum L.) and Wheat Flour by using Atomic Absorption Spectrophotometer. Asian J Plant Sci. 2002;1(2):208–209. [Google Scholar]
- 28.Raboy V. Approaches and challenges to engineering seed phytate and total phosphorus. Plant Sci. 2009;177(4):281–296. [Google Scholar]
- 29.Doria E., Galleschi L., Calucci L., Pinzino C., Pilu R., Cassani E., et al. Phytic acid prevents oxidative stress in seeds: evidence from a maize (Zea mays L.) low phytic acid mutant. J Exp Bot. 2009;60(3):967–978. doi: 10.1093/jxb/ern345. [DOI] [PubMed] [Google Scholar]
- 30.Shi J., Wang H., Schellin K., Li B., Faller M., Stoop J.M., et al. Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat Biotechnol. 2007;25(8):930–937. doi: 10.1038/nbt1322. [DOI] [PubMed] [Google Scholar]
- 31.Sashidhar N., Harloff H.J., Potgieter L., Jung C. Gene editing of three BnITPK genes in tetraploid oilseed rape leads to significant reduction of phytic acid in seeds. Plant Biotechnol J. 2020 doi: 10.1111/pbi.13380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevenson-Paulik J., Bastidas R.J., Chiou S.-T., Frye R.A., York J.D. Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc Natl Acad Sci. 2005;102(35):12612–12617. doi: 10.1073/pnas.0504172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perry M.E., Valdes K.M., Wilder E., Austin C.P., Brooks P.J. Genome editing to're-write'wrongs. Nat Rev Drug Discovery. 2018;17(10):689–690. doi: 10.1038/nrd.2018.91. [DOI] [PubMed] [Google Scholar]
- 34.Livermore T.M., Azevedo C., Kolozsvari B., Wilson M.S.C., Saiardi A. Phosphate, inositol and polyphosphates. Biochem Soc Trans. 2016;44(1):253–259. doi: 10.1042/BST20150215. [DOI] [PubMed] [Google Scholar]
- 35.Shears S.B. Inositol pyrophosphates: why so many phosphates? Adv Biol Regulation. 2015;57:203–216. doi: 10.1016/j.jbior.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.