Abstract
Background and Aims
With high rates of recurrence post-treatment, hepatocellular carcinoma (HCC) is one of the most common types of cancer worldwide and the major cause of cancer death. To improve the overall survival of HCC patients, identification of a reliable biomarker and precise early diagnosis of HCC remain major unsolved problems.
Methods
We initially screened data from the Cancer Genome Atlas liver cancer cohort to identify potential prognosis-related genes. Then, a meta-analysis of five international HCC cohorts was implemented to validate such genes. Subsequently, artificial intelligence models (random forest and neural network) were trained to predict prognosis accurately, and a log-rank test was performed for validation. Finally, the correlation between the molecular hepatocellular carcinoma prognostic score (mHPS) and the stromal and immune scoring in HCC were explored.
Results
A comprehensive list of 65 prognosis-related genes was obtained, most of which have been not extensively studied thus far. A universal HCC mHPS system depending on the expression pattern of only 23 genes was established. The mHPS system had general applicability to HCC patients (log-rank p<0.05) in a platform-independent manner (RNA sequencing or microarray). The mHPS was also correlated with the stromal and immune scoring in HCC, reflecting the status of the tumor immune microenvironment.
Conclusions
Overall, the mHPS is an easy and cost-effective prognosis predicting system, which can disclose previously uncovered heterogeneity among patient subpopulations. The mHPS system can further stratify patients who are at the same clinical stage and should be valuable for precise treatment. Moreover, the prognosis-related genes recognized in this study have potential in targeted and immune therapy.
Keywords: Hepatocellular carcinoma, Random forest, Neural network, Prognostic scoring system, Personalized medicine
Introduction
With a high postoperative recurrence rate, hepatocellular carcinoma (HCC) is currently the sixth most common tumor worldwide (∼850,000 incidence of cases) and the second leading cause of global cancer-associated mortality (∼840,000 deaths).1,2 To improve the prognosis of HCC and identify reliable biomarkers related to its pathogenesis, early diagnosis and prognosis for HCC have become an urgent focus of research. Elucidating the genes related to the occurrence and development of HCC will be helpful for early detection, the development of prognostic markers, and the determination of therapeutic targets.
Methods to better identify high-risk individuals with HCC have been a focused research priority for decades. Patients have been classified based on the American Joint Committee on Cancer (AJCC)-tumor, node, metastasis (TNM) stage3 and the Barcelona Clinic Liver Cancer stage (BCLC),4 which are the most two widely accepted clinical classification systems for HCC. Although these systems have proved to be valuable in prognosis, the overall outcome can differ markedly, even for patients at the same clinical stage. Thus, there is still an urgent need for a highly efficient predictive algorithm for evaluating HCC prognosis.
Recent technological advances have made great strides in developing molecular prognostic indicators for several types of cancers, including breast, lung and colorectal, some of which are recommended in the American Society of Clinical Oncology guidelines. The metastasis risk classifier predicted well recurrence and survival attributed to metastatic HCC in two independent cohorts with mixed etiologies.5 Other useful tools, including various multiple-gene predictive modeling approaches for HCC prognosis, have been also developed.6–9 However, these tools are neither pervasive nor economical, given that they are confined to certain platforms, or the high cost of test for multiple genes. Additionally, none of the tests developed so far are sufficiently accurate to predict overall survival (OS). These restrictions are attributable, to some extent, to the reality that no comprehensive atlas of prognosis-related genes has been discovered, with only finite genes having been substantially checked in this regard. This status quo highlights the lack of unbiased integrated methods to undrape and formulate all prognosis-related molecules, which can act as molecular profiles through the application of large-scale sequencing or array for tumor genomes and transcriptomes.
Nowadays, immunotherapy has revolutionized cancer treatment in a wide range of tumor types.10 However, despite the durable clinical long-term responses, the majority of patients failed to respond to immunotherapy, demonstrating primary resistance. In addition, parts of those who were initially responsive to therapy eventually suffered relapse attributable to acquired resistance. Dozens of mechanisms of resistance have already been unveiled, and more need to be characterized. Identifying the molecular predictor related to the response to immunotherapy has become a focus of recent experimental and clinical lines of research.11–13
Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) is a newly developed algorithm that takes advantage of the unique properties of the transcriptional profiles of cancer tissues to infer tumor cellularity as well as the different infiltrating normal cells.14 This algorithm could infer clinical outcomes of patients with cancer, and analysis of datasets of different cancer types revealed that ESTIMATE scores are useful indicators of tissue-based patient prognosis.15–17 Yet, it has been reported that HCC patients with low immune, stromal and ESTIMATE scores had better clinical outcomes than those with high scores.17
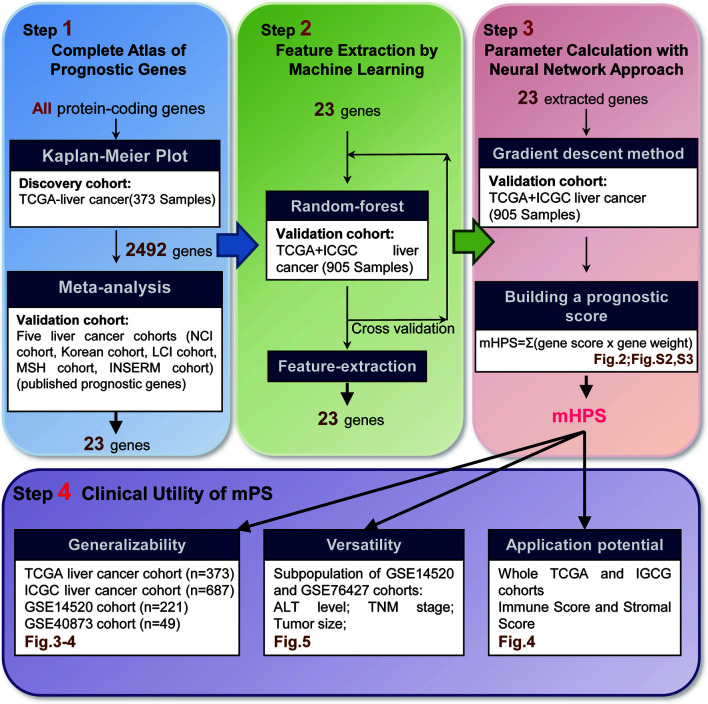
We have now built a new system for the prediction of the prognosis of HCC patients (Fig. 1). To this end, we first inspected all protein-coding genes for their relevance with OS in HCC patients. Then, we identified 23 prognosis-related genes by a meta-analysis of five HCC cohorts assembled. Artificial intelligence-based methods were applied to establish the molecular hepatocellular carcinoma prognostic score (mHPS), a versatile molecular prognostic score system that is capable of precisely stratifying the OS of HCC patients according to the binary expression status of a mere 23 genes.
Fig. 1. Study summary.
All protein-coding genes were examined for their potency as prognosis-related genes with the utility of the data from the TCGA-LIHC cohort and five international multicenter datasets (Step 1). The number of validated genes fell off to 23 with the random forest machine-learning approach (Step 2). A universal prognostic score, appointed as mHPS, was established through a neural network approach (Step 3). Finally, the usage of mHPS was validated in different circumstances (Step 4). LIHC, Liver Cancer Cohort; TCGA, The Cancer Genome Atlas; mHPS, molecular hepatocellular carcinoma prognostic score.
Other than the existing scoring system, the mHPS served well in multiple independent HCC cohorts and reflected the status of the tumor immune microenvironment. We also showed that mHPS can stratify patients even within the same clinical subgroup, emphasizing the importance of the combination of mHPS with conventional staging systems.
Methods
Study design and cohorts
The initial analysis was performed with the TGCA-Liver Cancer Cohort (LIHC; discovery cohort), given that this is the most representative cohort available. The mRNA expression data of the LIHC were downloaded from the website, https://xenabrowser.net/datapages/. The available clinical information of 373 patients was obtained with R package cgdsr, and details are shown in Table 1 and Supplementary Table 1. Then, we conducted a retrospectively comprehensive analysis of five independent HCC cohorts (NCI cohort, Korean cohort, LCI cohort, MSH cohort, INSERM cohort), all published previously.
Table 1. Clinical-pathological features of HCC patients from TCGA.
| Clinical features | Number (%) |
|---|---|
| Age in years | |
| >60 | 195 (52.3) |
| <=60 | 177 (47.5) |
| not available | 1 (0.3) |
| AJCC metastasis pathologic | |
| M0 | 267 (71.6) |
| M1 | 4 (1.1) |
| MX | 102 (27.3) |
| AJCC nodes pathologic | |
| N0 | 253 (67.8) |
| N1 | 4 (1.1) |
| NX | 115 (30.8) |
| not available | 1 (0.3) |
| AJCC tumor pathologic | |
| T1 | 182 (48.8) |
| T2 | 95 (25.2) |
| T3 | 80 (21.4) |
| T4 | 13 (3.5) |
| TX | 1 (0.3) |
| not available | 2 (0.5) |
| AJCC pathologic stage | |
| I | 172 (46.1) |
| II | 87 (23.3) |
| III | 85 (22.8) |
| IV | 5 (1.3) |
| not available | 24 (6.4) |
AJCC, American Joint Committee on Cancer; HCC, hepatocellular carcinoma; TCGA, The Cancer Genome Atlas.
For the establishment of the mHPS, we took in other HCC datasets of the International Cancer Genome Consortium (ICGC). Data of 687 HCC samples with both mRNA expression profile and clinical information (232 cases from ICGC-LIRI-JP, 161 cases from LICA-FR, and 294 cases from LIHC-US, respectively) were downloaded from the ICGC database (https://icgc.org/) (Table 2, Supplementary Table 2).
Table 2. Clinical-pathological features of HCC patients from ICGC.
| Clinical features | Number (%) |
|---|---|
| Sex | |
| female | 194 (28.2) |
| male | 493 (71.8) |
| Status | |
| alive | 413 (60.1) |
| deceased | 119 (17.3) |
| not available | 155 (22.6) |
ICGC, International Cancer Genome Consortium; HCC, hepatocellular carcinoma.
We then validated mHPS with combined TCGA and ICGC cohorts. We also used two independent microarray-based HCC cohorts: dataset GSE145205 with 221 samples and GSE4087318 with 49 samples (including 17 samples from multicentric occurrence and 39 from non-multicentric occurrence) for further validation of the mHPS. The detailed clinicopathological features of the patients are shown in Supplementary Tables 3 and 4.
Gene list
For the integrated analysis of all protein-coding genes, we obtained the intact list of human genes from the HUGO Gene Nomenclature Committee.
Identification of prognosis-related genes
We acquired gene expression profile and survival data of HCC patients from TCGA-LIHC. For each protein-coding gene, the potency as a prognostic marker was interrogated using the TCGA-LIHC discovery cohort, and then we validated the candidate genes in the other five independent HCC cohorts (NCI cohort, Korean cohort, LCI cohort, MSH cohort, and INSERM cohort). We employed a preprocessing pipeline published previously.19 For Affymetrix array data, the MAS5 method20 was applied for normalization before log2 conversion for preprocessing, whereas non-Affymetrix data were downloaded as they were deposited in the public databases. In each cohort, samples were stratified into the high or low expression group according to the median for a particular gene. Kaplan-Meier survival analysis was performed using the survival and survminer R packages,21,22 and the hazard ratios (HRs) and 95% confidence intervals (CIs) were calculated using the Cox regression R package.23
Random forests for classification
The combination of multiple machine learning methods, alleged “ensemble learning”, has been demonstrated to ameliorate the predictive performance. Combining the artificial intelligence-based machine learning algorithm referred to as a random forest with a neural network was regarded as an effective approach among several machine learning tasks.24 Hence, we employed these two methods to build the mHPS system (Supplementary Fig. 1). For the generation and validation of mHPS, we collected and analyzed the TCGA-LIHC and ICGC data.
Introduced by Breiman, random forest is a popular nonparametric tree-ensemble method that combines several randomized decision trees and aggregates their predictive potency by averaging for the analysis of survival data.25,26 Random forest is an algorithm that integrates multiple trees through the idea of integrated learning. Its basic unit is a decision tree, every branch of which depends on a random vector, and all vectors in a random forest are independently, identically distributed. Random forest is used to randomize the column variables and row observations of the dataset to generate multiple numbers of categories. Finally, the classification results will be summarized. In our study, data from the TCGA-LIHC and ICGC cohorts (training dataset, n=905) were first employed to a random forest classifier. Expression levels of the 23 newly identified prognosis-related genes (designated P) and the survival status [designated s, alive (0) or deceased (1)] at 3 years after diagnosis for each patient in the training dataset (n=905) were then put into the random forest classifier. We created the model with the usage of the radomForest R package27 and with default parameters, with the exception of n_trees (500) and max_depth (=10). Based on the importance values (cutoff=1), 23 genes were selected after stratified 10-fold cross-validation for training the neural network.
Calculation of expression status of prognosis-related genes
The 23 genes could be obligated to the OS of HCC patients, with 13 and 10 genes being associated with a poor OS if their expression level is higher or lower than the median, respectively. The expression status (P) of the 23 prognosis-related genes was transformed to “Gene_score Matrix” (M) according to the expression level (above or below the median) and HR for each gene with the steps shown in Table 3.
Table 3. Gene score matrix for the expression status (P) of the 23 prognosis-related genes.
| Gene_score matrix | Gene expression |
||
|---|---|---|---|
| low | high | ||
| HR | <1 | 1 | 0 |
| >1 | 0 | 1 | |
For Cox regression, 2/3 of the samples of ICGC-LIRI-JP were randomly grouped as the training set, with 1/3 as the validation set first. Then, we performed the univariate Cox regression for each clinical feature; the clinical features with p value less than 0.05 were selected for multivariate regression analysis (Surv(time, status) ∼ TNM_STAGE_T + VEIN_INVASION + ALCOHOL + mHPS). At last, the nomograms were constructed, including those variables significant to 0.05 on multivariate analysis. The Cox model of nomogram construction has been described.28 To evaluate the predictive accuracy of the nomogram, we used the receiver operating characteristic curve analysis and Harrell’s concordance index.29 Calibration curves were generated to visualize the discrimination between the actual and predicted 1-, 3- and 5-year OSs. A dense neural network system was then built and trained (Supplementary Fig. 1) using the Python-based Keras library. In each hidden node, the rectified linear unit was utilized as an activation function. In the output layer, two nodes (n1 and n2, for alive and deceased, respectively) were generated and a softmax function was applied to each node, then designated x2 (probability of death; that is, the n2 node) as X. We used categorical_crossentropy as a loss function (F) and optimized each weight with the Adam method (learning rate=0.001; epochs=1,000). After the training, the weights of the nodes (“Gene_Weight”) were utilized to compute mHPS (sum of Gene_Score multiply Gene_Weight for all 23 genes).
Measures of tumor purity and immune cell content
ESTIMATE, which infers the fraction of stromal and immune cells in tumor samples, was applied to predict clinical outcomes of patients with cancer.14,15 For the TCGA-LIHC data, ESTIMATE stromal and immune scores were generated using the estimate R package (https://r-forge.r-project.org/projects/estimate/).30
Results
Limitation of single-gene predicting methods driven by hypothesis
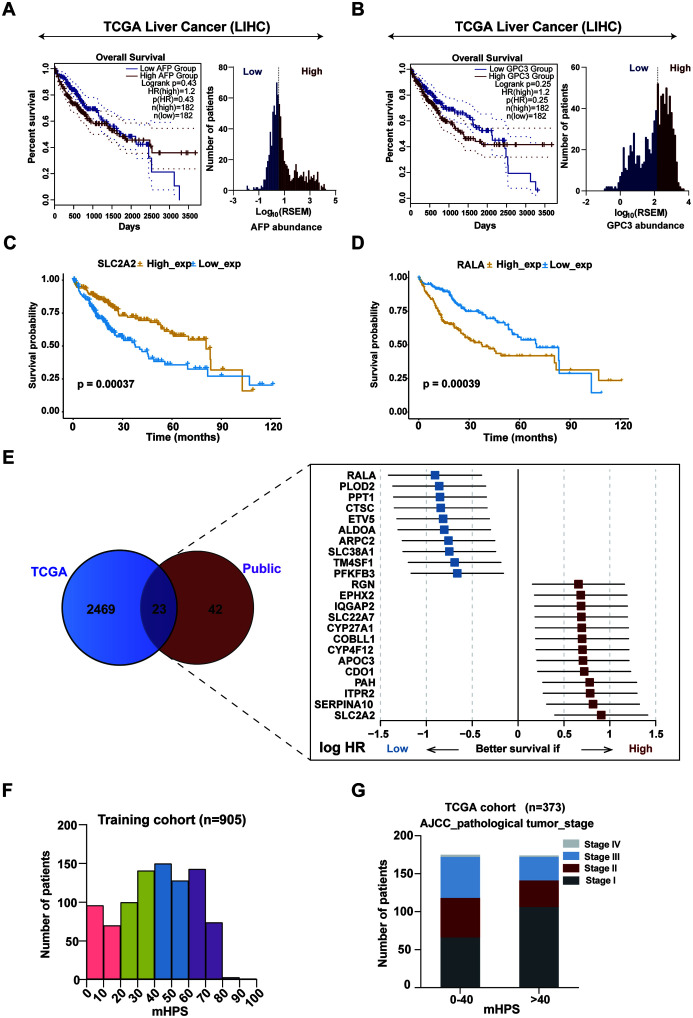
Since the initial discovery of the HCC-predicting genes, such as AFP and GPC3, tremendous advances have been made in the field of oncogenes. Given that these oncogenes play pivotal roles involved in tumor development and progression,31–33 we hypothesized that their expression might be correlated with the OS in HCC patients. HCC patients in the TCGA cohort were divided into two groups (high and low expression level) according to the median mRNA expression of alpha-fetoprotein, and the diversity in survival between the two groups was evaluated. Unexpectedly, there was no significant difference between the two groups (Fig. 2A). Likewise, the mRNA abundance of GPC3, which is often examined as a biomarker for HCC,32 showed no difference in the OS between the two groups of the TCGA-LIHC cohort (Fig. 2B). Moreover, the expression of these two genes also had no significant correlation with the OS in the ICGC-LIRI-JP cohort (Supplementary Fig. 2), which could represent the data of Asian people.
Fig. 2. Identification of all prognosis-related genes in the TCGA-LIHC cohort and validation in five international cohorts.
(A–B) Kaplan-Meier (K-M) curves of OS for the patients in the TCGA-LIHC cohort based on alpha-fetoprotein (AFP) (A) or GPC3 (B) expression levels higher or lower than the median (left), and distribution of AFP expression level (RSEM) among these patients. The log-rank p-value and the HR with corresponding 95% CI are shown, (C and D) K-M curves of OS for the TCGA-LIHC cohort based on SLC2A2 (C) and RALA (D) expression levels, respectively. (E) Logarithm of the integrated HR for all 23 prognosis-related genes in the validation datasets. The complete gene list is provided in Supplementary Table 7. (F) Characteristics of mHPS bins. Distribution of mHPS for all patients in the TCGA+ICGC training cohort (n=905). (G) Numbers of patients classified according to clinical tumor stage in each of the two mHPS bins for the training cohort. See also Supplementary Table 9. LIHC, Liver Cancer Cohort; TCGA, The Cancer Genome Atlas; mHPS, molecular hepatocellular carcinoma prognostic score; ICGC, International Cancer Genome Consortium; HR, hazard ratio; OS, overall survival; K-M curves, Kaplan-Meier curves.
Computational analysis for all prognosis-related genes
According to the median mRNA expression of each protein-coding gene, the relation between mRNA abundance and OS in the TCGA LIHC (as a discovery cohort) dataset were interrogated. Although the expression of most genes was not associated with the clinical outcome, 2,492 protein-coding genes had a significant relationship with OS (OS-related genes). High expression of SLC2A2, for instance, was related to a longer survival, while low expression of RALA was related to a better OS (Fig. 2C, D). The 2,492 OS-related genes in the TCGA-LIHC discovery cohort, with the complete list of these genes and their log-rank p values, has been provided in Supplementary Table 5.
The 2,492 potential prognostic genes identified through the TCGA-LIHC cohort were further validated by meta-analysis composed of five published liver cancer cohorts34 (NCI cohort, Korean cohort, LCI cohort, MSH cohort, and INSERM cohort), and 65 genes were associated with the OS (See Supplementary Table 6). A set of 23 prognosis-related genes were extracted by reduced dimensional analysis (See Fig. 2E, Supplementary Table 7 and Fig. 3). In the gene set obtained by the meta-analysis, SLC2A2 and RALA were the prognosis-related genes with the most potential, having the highest and lowest HRs, respectively (Fig. 2E, Table 4), which suggested that these two genes may be the most promising genes associated with the prognosis of HCC.
Table 4. Twenty-three genes for calculation of the mHPS.
| Gene symbol | Gene ID | Full name | High score | Low score | Weight |
|---|---|---|---|---|---|
| RALA | 5,898 | RAS like proto-oncogene A | 0 | 1 | 3.6386383 |
| PLOD2 | 5,352 | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 | 0 | 1 | 4.571733 |
| PPT1 | 5,538 | Palmitoyl-protein thioesterase 1 | 0 | 1 | 3.6378715 |
| CTSC | 1,075 | Cathepsin C | 0 | 1 | 2.929253 |
| ETV5 | 2,119 | ETS variant transcription factor 5 | 0 | 1 | 3.15026 |
| ALDOA | 226 | Aldolase, fructose-bisphosphate A | 0 | 1 | 3.5781527 |
| ARPC2 | 10,109 | Actin related protein 2/3 complex subunit 2 | 0 | 1 | 3.4750159 |
| SLC38A1 | 81,539 | Solute carrier family 38 member 1 | 0 | 1 | 4.9902163 |
| TM4SF1 | 4,071 | Transmembrane 4 L six family member 1 | 0 | 1 | 3.6512656 |
| PFKFB3 | 5,209 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | 0 | 1 | 4.242654 |
| RGN | 9,104 | regucalcin | 1 | 0 | 3.4342885 |
| EPHX2 | 2,053 | epoxide hydrolase 2 | 1 | 0 | 4.14249 |
| IQGAP2 | 10,788 | IQ motif containing GTPase activating protein 2 | 1 | 0 | 3.2472568 |
| SLC22A7 | 10,864 | Solute carrier family 22 member 7 | 1 | 0 | 3.2640932 |
| CYP27A1 | 1,593 | cytochrome P450 family 27 subfamily A member 1 | 1 | 0 | 3.2004652 |
| COBLL1 | 22,837 | cordon-bleu WH2 repeat protein like 1 | 1 | 0 | 3.4549387 |
| CYP4F12 | 66,002 | cytochrome P450 family 4 subfamily F member 12 | 1 | 0 | 4.133354 |
| APOC3 | 345 | Apolipoprotein C3 | 1 | 0 | 3.7753599 |
| CDO1 | 1,036 | Cysteine dioxygenase type 1 | 1 | 0 | 3.1609652 |
| PAH | 5,053 | Phenylalanine hydroxylase | 1 | 0 | 3.727815 |
| ITPR2 | 3,709 | Inositol 1,4,5-trisphosphate receptor type 2 | 1 | 0 | 3.3972855 |
| SERPINA10 | 51,156 | Serpin family A member 10 | 1 | 0 | 3.569119 |
| SLC2A2 | 6,514 | Solute carrier family 2 member 2 | 1 | 0 | 2.9978447 |
Artificial intelligence-based exploitation of a molecular HCC prognostic score
To testify the application values of the 23 newly identified prognosis-related genes in prediction for the survival rate of HCC patients at the third year, we combined the data from two datasets, TACG and ICGC (Ref.) (training set, n=905), to establish a molecular HCC prognostic score system as described amply in the Methods section and Supplementary Figure 1. The importance values of these 23 genes are shown in Supplementary Table 8.
In short, data from TCGA and ICGC (n=905; 155 cases were eliminated due to the missing prognostic information) were applied to a machine learning algorithm regarded as a random forest classifier, and 23 genes were selected for the further neural network algorithm. Then, the weight for each gene was optimized and a mHPS was computed by summation of “Gene_Score” × “Gene_Weight” for all 23 genes. We took these genes as input layers, and then set thereinto two hidden layers (four and two neurons, respectively). Lastly, we defined two nodes separately for “alive” and “deceased”. With such a four-layer neural network modeling, the accuracy (ACC value) could reach 0.782, while the loss value dropped to 0.488. The weights of each gene were calculated based on the neural network derived from the modeling (Table 4), and subsequently, then a mHPS was constructed according to the integration of gene weights and expression (with the potential value ranging from 0 to 83.37034 (Supplementary Table 9)). Two examples of actual mHPS calculations are presented (Supplementary Fig. 4).
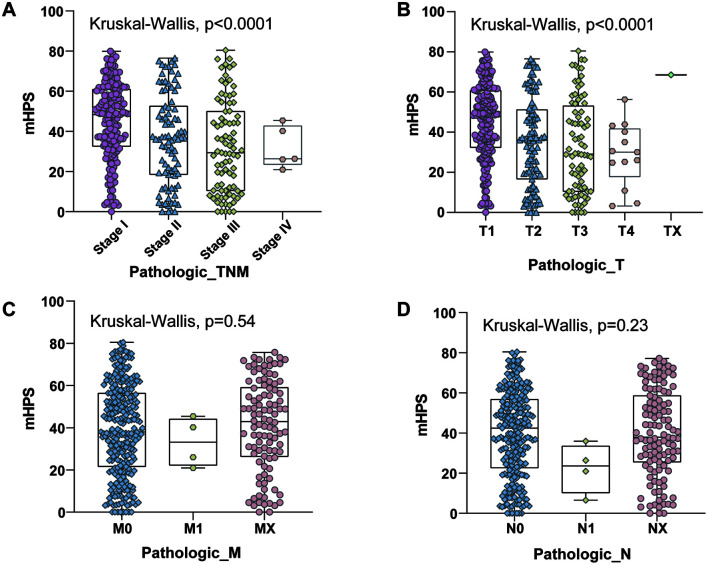
For the training cohort, the mean of mHPS was 41.76 (interquartile range of 26.08–59.01), and its distribution pattern is shown in Figure 2F. The characteristics of mHPS groups based on assignment to two bins (<=mHPS<40 and mHPS>=40) are summarized for the TCGA training cohort in Figure 2G, Supplementary Tables 1 and 9, which highlighted the correlation between mHPS and AJCC pathologic tumor stage. A significant correlation between mHPS and pathologic T and TNM stage was found, although the mHPS was not significantly correlated with pathologic N and M stages (Fig. 3A–D).
Fig. 3. Correlation between clinical characteristics and mHPS.
(A–D) Correlation between mHPS and tumor stage (A), pathologic T (B), pathologic N (C), and pathologic N (D). mHPS, molecular hepatocellular carcinoma prognostic score.
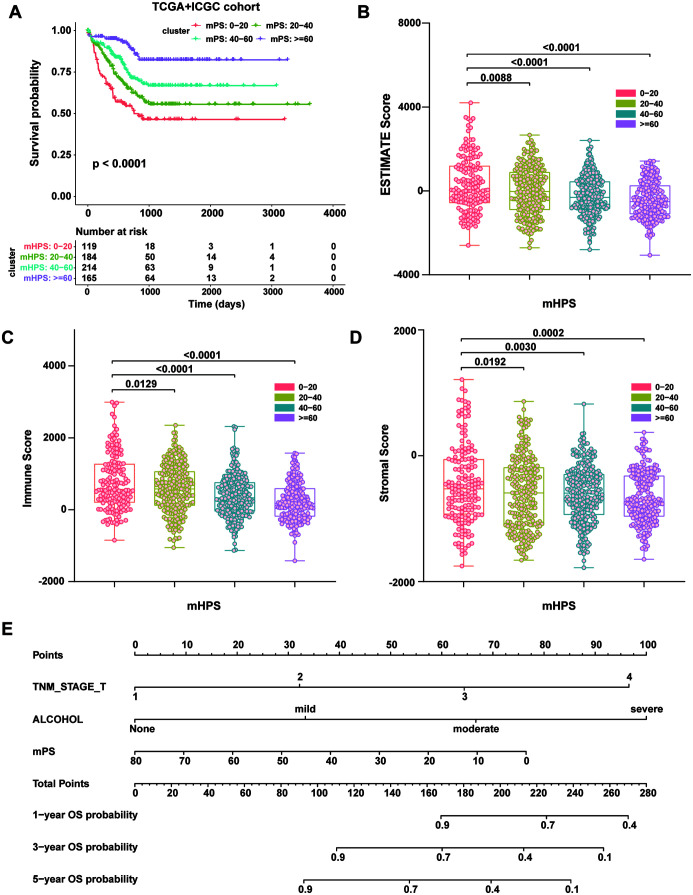
Moreover, we divided the mHPS into four groups: 0<=mHPS<20, 20<=mHPS<40, 40<=mHPS<60, mHPS>=60, and there were significant differences in prognosis between the four groups (Fig. 4A). ESTIMATE is a recently exploited algorithm that is utilized to estimate the immune score and stromal score, which infer the abundance of infiltrating immune cells and tumor purity.14 We further computed the stromal and immune scores with R package estimation using the 905 cases from TCGA+ICGC. As shown in Figure 4B–D, in terms of OS, there was a statistically significant negative association between the ESTIMATE immune/stromal scores and the mHPS of HCC patients (Supplementary Table 10). Based on the univariate and multivariate analyses, we integrated the clinicopathologic risk factors into a nomogram to predict OS of desmoid tumors at 1, 3, and 5 years in HCC patients from ICGC-LIRI-JP. The nomogram is shown in Figure 4E (See also Supplementary Tables 11–14 and had a Harrell’s concordance index of 0.797 (95% CI: 0.701–0.892); the calibration curves demonstrated the agreement between the nomogram predicted and actual survival (Supplementary Fig. 5), indicating its potential clinical utility in predicting OS. Receiver operating characteristic curve analysis showed the areas under the curve of this model at 5-year OS reached 0.83 in the validation cohort (Supplementary Fig. 6).
Fig. 4. The mHPS system accurately stratifies the prognosis and immune status of liver cancer patients.
(A) K-M curves of OS according to the mHPS for the TCGA+ICGC cohort. (B–D) Correlation between the mHPS and ESTIMATE score (B), immune score (C), and stromal score (D). (E) A nomogram estimating the probability of OS at 1, 3, and 5 years in ICGC-LIRI-JP following mHPS scoring, alcohol consumption, and TNM_T stage. K-M curves, Kaplan-Meier curves; ESTIMATE, Estimation of STromal and Immune cells in Malignant Tumor tissues using Expression data; ICGC, International Cancer Genome Consortium; LIHC, Liver Cancer Cohort; mHPS, molecular hepatocellular carcinoma prognostic score; OS, overall survival; TCGA, The Cancer Genome Atlas; TNM, tumor node metastasis classification.
Moreover, we interrogated the potential mechanism by which the 23 genes affected the HCC outcome. Kyoto Encyclopedia of Genes and Genomes enrichment analysis was conducted and none of the adjusted p values was less than 0.05. Even so, we found that the 23 genes might mainly affect the HCC outcome through regulating metabolic pathways, such as the pentose phosphate pathway, fructose and mannose metabolism, cholesterol metabolism, biosynthesis of amino acids, PPAR signaling pathway, taurine and hypotaurine metabolism, and the glucagon signaling pathway (Supplementary Fig. 7 and Table 15).
mHPS predicts prognosis of independent HCC cohorts
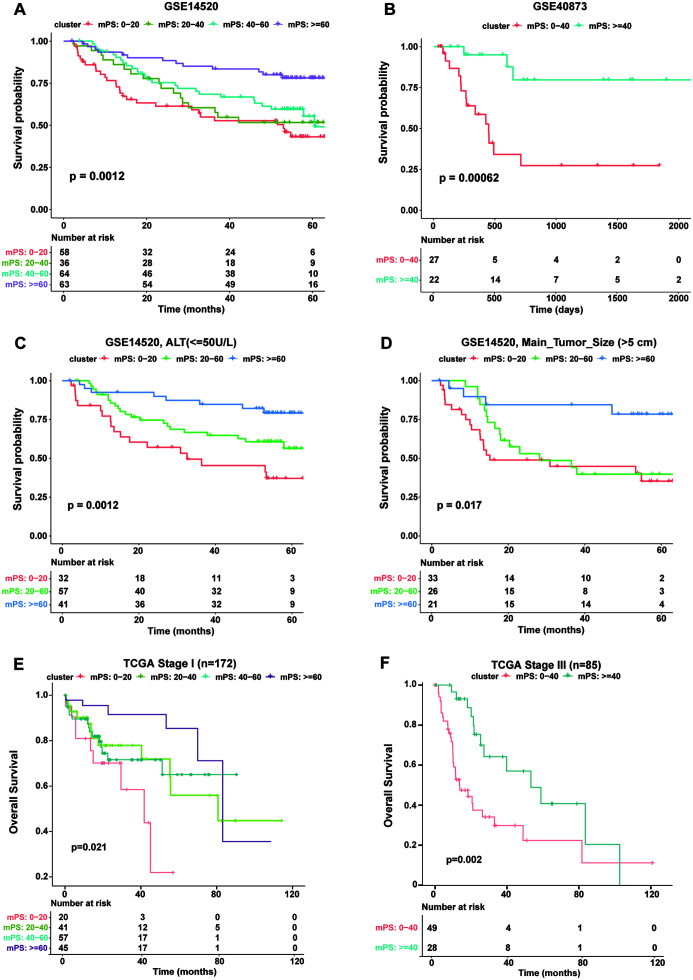
To examine if mHPS can stratify prognosis not only in the TCGA+ICGC cohort but also in other independent HCC cohorts, we inspected the additional two independent datasets of GSE14520 (221 cases; Fig. 5A) and GSE40873 (49 cases; Fig. 5B), calculated the mHPS of each sample (Supplementary Tables 16 and 17), and split them into different groups accordingly. The results showed that the mHPS system could stratify prognosis in both of the test cohorts (Fig. 5A, B), and is generally applicable to HCC in a platform-independent manner (microarray or RNA sequencing). We further explored whether mHPS is also an applicative system for HCC patients with different clinical characteristics, as well as the well-established AJCC tumor stages determined from clinical information. The mHPS showed superiority to the Sixty-Five Gene-Based Risk Score Classifier,7 which was reported to be the prediction for OS in HCC, with regard to the stratification of prognosis (p<0.0001 for mHPS vs. 1.0×10−4 for risk score). It is also of note that mHPS costs lower to apply than risk score, for which 65-gene expression profiling analysis is required.
Fig. 5. The mHPS predicts prognosis of independent HCC cohorts.
(A) K-M curves of OS for the public dataset GSE14520 according to the mHPS. (B) K-M curves of OS according to the mHPS for the public dataset GSE40873. (C) K-M curves according to mHPS for OS of patients in the GSE14520 dataset with serum ALT level lower than 50U/L. (D) K-M curves according to mHPS for OS of patients in the GSE14520 dataset with the main tumor size larger than 5 cm. (E and F) K-M curves according to mHPS for OS of patients in the TCGA cohorts at clinical TNM stage I (E) and stage III (F). K-M curves, Kaplan-Meier curves; ALT, Alanine aminotransferase; BCLC, TNM, tumor node metastasis classification; mHPS, molecular hepatocellular carcinoma prognostic score; OS, overall survival; TCGA, The Cancer Genome Atlas.
mHPS is suitable for various clinical settings
We investigated the utility of mHPS for different subtypes of HCC patients. In the GSE14520 dataset, the mHPS system precisely stratified not only the OS of patients with lower alanine aminotransferase (<50 U/L) level (Fig. 5C) but also of patients got a main tumor diameter greater than 5 cm (Fig. 5D), showing that mHPS is applicable to patients with certain clinical status. Lastly, we examined if mHPS is also suitable for well-established AJCC-TNM tumor stages. The mHPS system revealed that stage I patients in the TCGA test cohort (n=172) are heterogeneous, with only 15% of individuals with a mHPS of <20 surviving for 40 months, whereas ∼40% of patients with a mHPS of >60 survived this long (Fig. 5E). This trend was observed in stage III patients (n=85), with those harboring a higher mHPS showing better prognosis and those with a lower mHPS the worse prognosis (Fig. 5F). Thus, these results suggested that the mHPS system can further stratify patients even at the same clinical stage.
Discussion
Liver cancer is a heterogeneous disease with distinct clinical outcomes. It is crucial to precisely predict the prognosis of patients with HCC for the selection of the appropriate treatment. In the present study, we have depicted a comprehensive atlas of prognosis-related genes for HCC, created a computational framework, and a new prognostic predicting system named mHPS that is applicable in HCC patients. Our algorism is likely to exceed previous scores because mHPS could also stratify patients even at the same clinical stage. The mHPS system is economical and simple to execute and is capable of uncovering previously hidden heterogeneity among patient subpopulations in a platform-independent manner.
There are multiple methods for the storage of tumor samples, extraction of RNA, and analysis of expression levels, which impeded us from building a universal threshold for each of the 23 genes in the present study. We proposed a “platform-independent” scoring system that could be computed from data acquired with any standardly established protocol. Furthermore, the mHPS was related to clinical features. We found that the mHPS was significantly associated with pathologic T and TNM stage. Yet, comparison of the predicting protocols already applied in clinical setting, development of an accurate and robust approach to inspecting the expression status of the 23 genes, and performance of pilot studies therewith to test the distribution patterns are subsistent issues that must be solved before the mHPS will be applicable in clinical practice. Besides, one of the foremost limitations of this study is the retrospective study design. Although cohorts with a sufficient sample size were utilized (n=1,330), including the cohorts of TCGA, ICGC, GSE14520 and GSE40873, prospective studies are needed to validate our conclusions.
The best-characterized gene among the 23 prognosis-related genes identified in our study is regucalcin (RGN). There were 31 papers with regard to a PubMed search for “RGN hepatocellular carcinoma”. Related to the calcium homeostasis, the RGN protein is preferentially expressed in the liver and kidney. RGN acts as a suppressor in cell proliferation that is mediated through various signaling pathways in hepatoma cells.35 Moreover, the RGN gene and protein expression levels have been demonstrated to be specifically reduced in human HCC by studies of gene expression profiles and proteomics.36–39
In contrast, most of the 23 prognosis-related genes (ALDOA, APOC3, CDO1, CTSC, CYP4F12, EPHX2, ETV5, ITPR2, PLOD2, PPT1, RALA, SERPINA10, SLC22A7, SLC38A1, and TM4SF1) have not been deeply studied in relation to HCC, given that PubMed searches for “‘GENE’ hepatocellular carcinoma” yielded less than 10 publications for each gene, with there being no publication at all for two of these genes (ARPC2 and COBLL1). Both basic and clinical research is required for an in-depth illustration of the mechanisms accounting for the biological functions of the 23 mHPS-based genes, and for the development of new drugs to improve the prognosis of HCC patients. Mechanistically, although there was no significant enriched pathway in the Kyoto Encyclopedia of Genes and Genomes analysis, the results still underlined the potential pathway by which the 23 genes of the mHPS scoring system impact the HCC outcome.
The first six pathways involved were all metabolism-related, which indicated that the scoring system we proposed is precise for the HCCs, as the liver is a crucial organ involved in metabolic processes and HCC cells and animal models show commonly metabolic dysregulation. Recently, immune checkpoint inhibitors, especially programmed death receptor 1 (PD-1)/PD-ligand 1 inhibitor, have shown great promise and progress for HCC treatment. However, the CheckMate 040 and the KEYNOTE-224 studies reported that the efficacy of immunotherapy cannot be effectively predicted, suggesting that the predictive biomarkers for PD-1 inhibitors in HCC are still ill-defined.40,41 In that regard, ESTIMATE, an analysis that previously revealed that stromal/immune cell infiltration is associated with the prognosis in patients with various types of tumors,17,42,43 has been applied to investigate the relationship between the mHPS and microenvironment. It has been reported that HCC patients with high immune/stromal scores had a poorer prognosis than those with low scores.17 Consistently, there was a significantly negative association between the mHPS and the ESTIMATE immune/stromal scores, suggesting that mHPS may be a potential predictive biomarker for the OS of HCC patients receiving immunotherapy.
Altogether, the application of the mHPS system defined by the expression pattern of 23 genes can predict the prognosis of HCC patients in a reproducible and reliable manner across independent patient cohorts. Moreover, based on the precise prediction of personal prognosis, the system may not only facilitate the selection of therapeutic approaches but also expands our understanding of the basic biology of HCC and thereby will spur the development of new therapeutic strategies. We also developed a nomogram using Cox regression that assigns predictions for OS based on mHPS score, TNM_T stage, alcohol consumption, and other clinicopathological variables in the ICGC-LIRI-JP cohort. A nomogram was invented by the French engineer Philbert Maurice d’Ocagne and has been extensively applied in the electronic industry for many years. In recent decades, increasing numbers of nomograms have been developed for clinical prognosis of different malignancies, such as non-small cell lung cancer, rectal cancer and HCC.44–46 We propose that the nomogram offers more individualized OS predictions and could be helpful for the decision-making during treatment.47 Besides, the nomogram has potential in estimating risk for clinical trial design, which could be applied to randomization in the studies based on their survival probability. However, this model should also be further evaluated in other independent populations.
Supporting information
Abbreviations
- AJCC
American Joint Committee on Cancer
- BCLC
Barcelona Clinic Liver Cancer
- CI
confidence interval
- ESTIMATE
Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data
- HCC
hepatocellular carcinoma
- HR
hazard ratio
- ICGC
International Cancer Genome Consortium
- LIHC
Liver Cancer Cohort
- mHPS
molecular hepatocellular carcinoma prognostic score
- OS
overall survival
- PD-1
programmed death receptor 1
- RGN
regucalcin
- TCGA
The Cancer Genome Atlas
Data sharing statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- 1.Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69(5):363–385. doi: 10.3322/caac.21565. [DOI] [PubMed] [Google Scholar]
- 2.McGlynn KA, Petrick JL, London WT. Global epidemiology of hepatocellular carcinoma: an emphasis on demographic and regional variability. Clin Liver Dis. 2015;19(2):223–238. doi: 10.1016/j.cld.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 4.Llovet JM, Bru C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis. 1999;19(3):329–338. doi: 10.1055/s-2007-1007122. [DOI] [PubMed] [Google Scholar]
- 5.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70(24):10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iizuka N, Oka M, Yamada-Okabe H, Nishida M, Maeda Y, Mori N, et al. Oligonucleotide microarray for prediction of early intrahepatic recurrence of hepatocellular carcinoma after curative resection. Lancet. 2003;361(9361):923–929. doi: 10.1016/S0140-6736(03)12775-4. [DOI] [PubMed] [Google Scholar]
- 7.Kim SM, Leem SH, Chu IS, Park YY, Kim SC, Kim SB, et al. Sixty-five gene-based risk score classifier predicts overall survival in hepatocellular carcinoma. Hepatology. 2012;55(5):1443–1452. doi: 10.1002/hep.24813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JS, Thorgeirsson SS. Genome-scale profiling of gene expression in hepatocellular carcinoma: classification, survival prediction, and identification of therapeutic targets. Gastroenterology. 2004;127(5 Suppl 1):S51–55. doi: 10.1053/j.gastro.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40(3):667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 10.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36(8):773–779. doi: 10.1200/JCO.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 13.Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19(3):133–150. doi: 10.1038/s41568-019-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu W, Ye H, Liu YF, Xu CQ, Zhong YX, Tian T, et al. Transcriptome-derived stromal and immune scores infer clinical outcomes of patients with cancer. Oncol Lett. 2018;15(4):4351–4357. doi: 10.3892/ol.2018.7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai F, Jin Y, Zhang P, Chen H, Fu Y, Zhang M, et al. Bioinformatic profiling of prognosis-related genes in the breast cancer immune microenvironment. Aging (Albany NY) 2019;11(21):9328–9347. doi: 10.18632/aging.102373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Niu X, Qiu Z. A five-gene signature based on stromal/immune scores in the tumor microenvironment and its clinical implications for liver cancer. DNA Cell Biol. 2020;39(9):1621–1638. doi: 10.1089/dna.2020.5512. [DOI] [PubMed] [Google Scholar]
- 18.Kudo A, Mogushi K, Takayama T, Matsumura S, Ban D, Irie T, et al. Mitochondrial metabolism in the noncancerous liver determine the occurrence of hepatocellular carcinoma: a prospective study. J Gastroenterol. 2014;49(3):502–510. doi: 10.1007/s00535-013-0791-4. [DOI] [PubMed] [Google Scholar]
- 19.Kuhn M. Building predictive models in R using the caret package. J Stat Softw. 2008;28(5):1–26. [Google Scholar]
- 20.Kassambara A. Machine learning essentials: practical guide in R. sthda; 2018.
- 21.Chen Y, Susick L, Davis M, Bensenhaver J, Nathanson SD, Burns J, et al. Evaluation of triple-negative breast cancer early detection via mammography screening and outcomes in African American and White American patients. JAMA Surg. 2020;155(5):440–442. doi: 10.1001/jamasurg.2019.6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Spakowicz D, Burkart J, Patel S, Husain M, He K, et al. Change in neutrophil to lymphocyte ratio during immunotherapy treatment is a non-linear predictor of patient outcomes in advanced cancers. J Cancer Res Clin Oncol. 2019;145(10):2541–2546. doi: 10.1007/s00432-019-02982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang MJ. Cox proportional hazards regression models for survival data in cancer research. Cancer Treat Res. 2002;113:59–70. doi: 10.1007/978-1-4757-3571-0_4. [DOI] [PubMed] [Google Scholar]
- 24.Kong Y, Yu T. A deep neural network model using random forest to extract feature representation for gene expression data classification. Sci Rep. 2018;8(1):16477. doi: 10.1038/s41598-018-34833-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Random Survival Forests. Wiley StatsRef: Statistics Reference Online:1-13.
- 26.Biau G. Analysis of a random forests model. J Mach Learn Res. 2012;13(1):1063–1095. [Google Scholar]
- 27.Liaw A, Wiener M. Classification and regression by randomforest. R News. 2002;2/3:18–22. [Google Scholar]
- 28.Xie D, Marks R, Zhang M, Jiang G, Jatoi A, Garces YI, et al. Nomograms predict overall survival for patients with small-cell lung cancer incorporating pretreatment peripheral blood markers. J Thorac Oncol. 2015;10(8):1213–1220. doi: 10.1097/JTO.0000000000000585. [DOI] [PubMed] [Google Scholar]
- 29.Wolbers M, Koller MT, Witteman JC, Steyerberg EW. Prognostic models with competing risks: methods and application to coronary risk prediction. Epidemiology. 2009;20(4):555–561. doi: 10.1097/EDE.0b013e3181a39056. [DOI] [PubMed] [Google Scholar]
- 30.Block CJ, Dyson G, Campeanu IJ, Watza D, Ratnam M, Wu G. A stroma-corrected ZEB1 transcriptional signature is inversely associated with antitumor immune activity in breast cancer. Sci Rep. 2019;9(1):17807. doi: 10.1038/s41598-019-54282-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu H, Xu Y, Xiang J, Long L, Green S, Yang Z, et al. Targeting alpha-fetoprotein (AFP)-MHC complex with CAR T-cell therapy for liver cancer. Clin Cancer Res. 2017;23(2):478–488. doi: 10.1158/1078-0432.CCR-16-1203. [DOI] [PubMed] [Google Scholar]
- 32.Di Tommaso L, Destro A, Seok JY, Balladore E, Terracciano L, Sangiovanni A, et al. The application of markers (HSP70 GPC3 and GS) in liver biopsies is useful for detection of hepatocellular carcinoma. J Hepatol. 2009;50(4):746–754. doi: 10.1016/j.jhep.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 33.Wu Y, Liu H, Ding H. GPC-3 in hepatocellular carcinoma: current perspectives. J Hepatocell Carcinoma. 2016;3:63–67. doi: 10.2147/JHC.S116513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SM, Leem SH, Chu IS, Park YY, Kim SC, Kim SB, et al. Sixty-five gene-based risk score classifier predicts overall survival in hepatocellular carcinoma. Hepatology. 2012;55(5):1443–1452. doi: 10.1002/hep.24813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamaguchi M. Suppressive role of regucalcin in liver cell proliferation: involvement in carcinogenesis. Cell Prolif. 2013;46(3):243–253. doi: 10.1111/cpr.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernando H, Wiktorowicz JE, Soman KV, Kaphalia BS, Khan MF, Shakeel Ansari GA. Liver proteomics in progressive alcoholic steatosis. Toxicol Appl Pharmacol. 2013;266(3):470–480. doi: 10.1016/j.taap.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graveel CR, Jatkoe T, Madore SJ, Holt AL, Farnham PJ. Expression profiling and identification of novel genes in hepatocellular carcinomas. Oncogene. 2001;20(21):2704–2712. doi: 10.1038/sj.onc.1204391. [DOI] [PubMed] [Google Scholar]
- 38.Schroder PC, Segura V, Riezu JI, Sangro B, Mato JM, Prieto J, et al. A signature of six genes highlights defects on cell growth and specific metabolic pathways in murine and human hepatocellular carcinoma. Funct Integr Genomics. 2011;11(3):419–429. doi: 10.1007/s10142-011-0230-7. [DOI] [PubMed] [Google Scholar]
- 39.Roy L, Laboissiere S, Abdou E, Thibault G, Hamel N, Taheri M, et al. Proteomic analysis of the transitional endoplasmic reticulum in hepatocellular carcinoma: an organelle perspective on cancer. Biochim Biophys Acta. 2010;1804(9):1869–1881. doi: 10.1016/j.bbapap.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 40.Zhu AX, Finn RS, Edeline J, Cattan S, Ogasawara S, Palmer D, et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol. 2018;19(7):940–952. doi: 10.1016/S1470-2045(18)30351-6. [DOI] [PubMed] [Google Scholar]
- 41.He AR, Yau T, Hsu C, Kang YK, Kim TY, Santoro A, et al. Nivolumab (NIVO) plus ipilimumab (IPI) combination therapy in patients (pts) with advanced hepatocellular carcinoma (aHCC): Subgroup analyses from CheckMate 040. J Clin Oncol. 2020;38(4 suppl):512. doi: 10.1200/JCO.2020.38.4_suppl.512. [DOI] [Google Scholar]
- 42.Bai F, Jin Y, Zhang P, Chen H, Fu Y, Zhang M, et al. Bioinformatic profiling of prognosis-related genes in the breast cancer immune microenvironment. Aging (Albany NY) 2019;11(21):9328–9347. doi: 10.18632/aging.102373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu W, Ye H, Liu YF, Xu CQ, Zhong YX, Tian T, et al. Transcriptome-derived stromal and immune scores infer clinical outcomes of patients with cancer. Oncol Lett. 2018;15(4):4351–4357. doi: 10.3892/ol.2018.7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang W, Zhang L, Jiang G, Wang Q, Liu L, Liu D, et al. Development and validation of a nomogram for predicting survival in patients with resected non-small-cell lung cancer. J Clin Oncol. 2015;33(8):861–869. doi: 10.1200/JCO.2014.56.6661. [DOI] [PubMed] [Google Scholar]
- 45.Valentini V, van Stiphout RG, Lammering G, Gambacorta MA, Barba MC, Bebenek M, et al. Nomograms for predicting local recurrence, distant metastases, and overall survival for patients with locally advanced rectal cancer on the basis of European randomized clinical trials. J Clin Oncol. 2011;29(23):3163–3172. doi: 10.1200/JCO.2010.33.1595. [DOI] [PubMed] [Google Scholar]
- 46.Lei Z, Li J, Wu D, Xia Y, Wang Q, Si A, et al. Nomogram for preoperative estimation of microvascular invasion risk in hepatitis B virus-related hepatocellular carcinoma within the Milan criteria. JAMA Surg. 2016;151(4):356–363. doi: 10.1001/jamasurg.2015.4257. [DOI] [PubMed] [Google Scholar]
- 47.Iasonos A, Schrag D, Raj GV, Panageas KS. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008;26(8):1364–1370. doi: 10.1200/JCO.2007.12.9791. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.