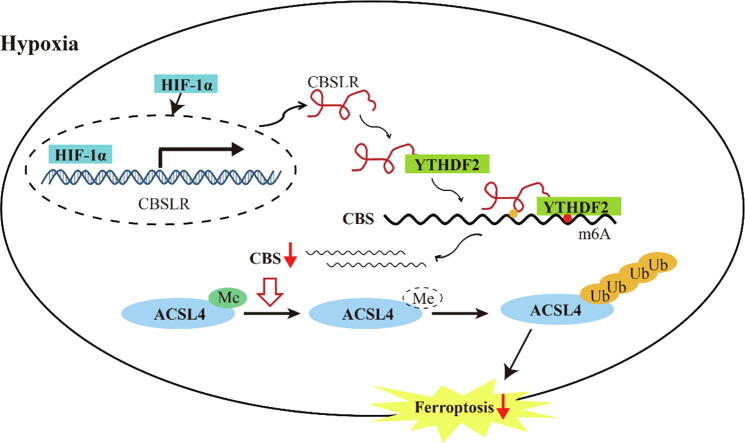
Graphical abstract
Schematic diagram showing that HIF-1α induces lncRNA-CBSLR to recruit YTHDF2 protein and CBS mRNA to form CBSLR/ YTHDF2/CBS complex, which in turn decreases CBS mRNA stability in an m6A dependent manner. The decreased CBS expression reduced methylation of ACSL4 protein, thus, the protein is degraded via the ubiquitination-proteasome pathway. It eventually protects GC from ferroptosis under a hypoxic tumor microenvironment.
Abbreviations: GC, gastric cancer; CDS, coding sequence; lncRNA, long noncoding RNA; HE, Hematoxylin and eosin; IHC, immunohistochemical; YTHDF2, YTH domain family protein 2; AJCC, American Joint Committee on Cancer; TNM, tumor-node-metastasis staging system; FBS, fetal bovine serum; MDA, malondialdehyde; 4-HNE, 4-hydroxynonenal; TCGA, the cancer genome atlas; RIP, RNA immunoprecipitation; qPCR, quantitative real-time PCR; CHIP, chromatin immunoprecipitation; GEO, Gene Expression Omnibus; HREs, hypoxia response elements; DZNeP, 3-deazaneplanocin A
Keywords: Gastric cancer, Chemoresistance, Hypoxia, lncRNA, Ferroptosis
Highlights
-
•
1. The hypoxic microenvironment is a common hallmark of solid tumors and is strongly associated with therapy resistance and poor prognosis.
-
•
2. CBSLR, a long noncoding RNA transactivated by HIF-1α, is upregulated in GC and associated with poor prognosis.
-
•
3. CBSLR inhibition induces ferroptosis under hypoxic conditions and contributes to chemoresistance.
-
•
4. lncRNA-CBSLR recruits YTHDF2 protein and CBS mRNA to form CBSLR/ YTHDF2/CBS complex, which in turn decreases CBS mRNA stability in an m6A dependent manner.
-
•
5. CBSLR/CBS inhibits ferroptosis by modulating ACSL4 methylation to be polyubiquitinated.
Abstract
Introduction
Tumors are usually refractory to anti-cancer therapeutics under hypoxic conditions. However, the underlying molecular mechanism remains to be elucidated.
Objectives
Our study intended to identify hypoxia inducible lncRNAs and their biological function in gastric cancer (GC).
Methods
Differentially expressed lncRNAs were determined by microarray analysis between GC cells exposed to hypoxia (1% O2) and normoxia (21% O2) for 24 h. The expression level of CBSLR was manipulated in several GC cell lines to perform molecular and biological analyses both in vitro and in vivo.
Results
We identified a hypoxia-induced lncRNA-CBSLR that protected GC cells from ferroptosis, leading to chem-resistance. Mechanically, CBSLR interacted with YTHDF2 to form a CBSLR/YTHDF2/CBS signaling axis that decreased the stability of CBS mRNA by enhancing the binding of YTHDF2 with the m6A-modified coding sequence (CDS) of CBS mRNA. Furthermore, under decreased CBS levels, the methylation of the ACSL4 protein was reduced, leading to protein polyubiquitination and degradation of ACSL4. This, in turn, decreased the pro-ferroptosis phosphatidylethanolamine (PE) (18:0/20:4) and PE (18:0/22:4) content and contributed to ferroptosis resistance. Notably, CBSLR is upregulated, whereas CBS is downregulated in GC tissues compared to matched normal tissues; and GC patients with high CBSLR/low CBS levels have a worse clinical outcome and a poorer response to chemotherapy.
Conclusion
Our study reveals a novel mechanism in how HIF1α/CBSLR modulates ferroptosis/chemoresistance in GC, illuminating potential therapeutic targets for refractory hypoxic tumors.
Introduction
Gastric cancer (GC) is the fifth most frequent cancer and the fourth most common cause of cancer death globally [1], [2]. Despite improvements in GC management, the clinical outcome of GC patients at advanced stages remains poor [3]. Chemotherapy is still the first-line adjuvant therapy for these patients. Chemotherapy efficacy has improved modestly in patients with advanced/metastatic GC recently [3]. It is thought that a major obstacle in cancer therapy is chemoresistance. Hence, identifying novel mechanisms of chemoresistance might improve clinical outcomes.
The hypoxic microenvironment is a common hallmark of solid tumors and is strongly associated with an aggressive phenotype and poor prognosis in many cancer types. A growing volume of evidence demonstrates that hypoxia-inducible factor 1/2 alpha (HIF-1/2α) contributes to the treatment resistance, angiogenesis, metastasis and survival of tumor cells under hypoxic conditions [4], [5], [6].
Long noncoding RNAs are a class of long (>200 nucleotides) RNA molecules with limited protein-coding potential that play a key role in a wide array of cell biological processes, including chemoresistance [7], [8]. LncRNAs, such as LUCAT1 [9], KB-1980E6.3 [10], and KB-1980E6.3 [11], participate in hypoxia signaling. However, the role of lncRNAs in hypoxia-related treatment resistance remains to be explored.
Here, in a comprehensive expression profiling of lncRNAs regulated by hypoxia, we identify CBS mRNA-destabilizing lncRNA (CBSLR) as a lncRNA that is significantly induced by hypoxia. We also revealed the upregulation of CBSLR in GC and the clinical relevance of CBSLR. Hypoxic CBSLR could facilitate the survival of GC cells by suppressing ferroptosis. CBSLR interacts with the YTH domain family protein 2 (YTHDF2) to form a CBSLR /YTHDF2/CBS complex that destabilizes CBS mRNA by enhancing the binding of YTHDF2 with the m6A-modified CDS region of CBS mRNA. The decreased CBS expression reduced methylation of ACSL4 protein; thus, the protein was degraded via the ubiquitination-proteasome pathway. High CBSLR/low CBS contributes to resistance to ferroptosis and chemotherapeutics in GC cells. GC patients with high CBSLR/low CBS levels have a worse clinical outcome and a poorer response to chemotherapy.
Materials and Methods
Detailed methods are enclosed in the online supplemental materials and methods.
Patients and samples
This study was sanctioned by the local ethics committee of the Fudan University Shanghai Cancer Center and Yijishan Hospital. Written informed consent was obtained from all participants. Two independent cohorts involving 161 GC patients were enrolled. Two experienced pathologists confirmed the GC tissues. A total of 109 pairs of tumors and matched adjacent normal tissues were obtained from patients who underwent surgery in Fudan university shanghai cancer center between 2008 and 2012 [12]. None of them received preoperative radio- or chemo-therapy. Patients who were lost to follow-up or could not be evaluated were excluded. These patients were in stage IIIA-C based on the American Joint Committee on Cancer (AJCC) tumor-node-metastasis (TNM) staging system (8th edition) according to pathological findings. Clinical information regarding the samples is presented in Supplemental Table S1. Another 52 fresh endoscopic biopsy gastric tissue samples were collected from GC patients before neoadjuvant chemotherapy in Yijishan Hospital between March 2019 and November 2020 (Supplemental Table S2). Upon removal of the surgical specimen, tissues were immediately stored at − 80 °C before further analysis. We did not provide any data that could help identify patients.
Cell culture
All gastric cancer cell lines were purchased from the American Type Culture Collection. The cell lines were routinely characterized by DNA fingerprinting, cell vitality detection, isozyme detection, and mycoplasma detection in our lab12. The last cell characterization was performed in December 2020. The cell culture media were supplemented with 10% fetal bovine serum (FBS, GIBCO), 100 U/mL penicillin, 100 mg/mL streptomycin (Thermo Scientific), and 8 mg/L antibiotic tylosin tartrate against mycoplasma (Sigma-Aldrich, St. Louis, Missouri, USA) at 37 °C in an atmosphere of 5% CO2 with 1% O2 (hypoxic condition) or 21% O2 (normoxic condition)[12]. AGS, MKN-28, and MKN-45 cells were maintained in Roswell Park Memorial Institute 1640 (RPMI-1640, Corning, NY, USA). The other cells (GSE-1, SGC7901, BGC823, MGC803) were cultured in Dulbecco's Modified Eagle Medium (DMEM). The cell lines were passaged in our laboratory less than six months after resuscitation.
Hypoxia-responsive lncRNA Microarray and RNA-sequencing
A detailed description of the hypoxia-responsive lncRNA microarray (GSE114083) and RNA-sequencing (GSE114360) can be found in the Supplemental Materials and Methods.
Subcellular fractionation analysis and 5′ and 3′ rapid amplification of cDNA ends analysis (RACE)
Subcellular fractionation analysis and 5′ and 3′ RACE were performed as described previously [12].
Inhibitors and final concentrations
The following inhibitors were dissolved in DMSO before cell treatment: Necrostatin-1 (11658, Cayman Chemicals, 10 μM), z-VAD-fmk (HY-16658, MedChem Express, 20 μM), Ferrostatin-1 (SML0583, Sigma, 1 μM), and DZNeP (Selleck, S7120, 1 μM), propargylglycine (P7888, Sigma, 2 mM).
Determination of malondialdehyde (MDA), 4-hydroxynonenal (4-HNE), intracellular GSH, thiol, GPX4-specific activity, intracellular iron, lipid ROS, and intracellular metabolites
A detailed description of the determination of various kinds of intracellular metabolites can be found in the Supplemental Materials and Methods.
Cell viability and cell survival assay
Cell viability and cell survival were determined using the CellTiter-Glo luminescent cell viability assay (Promega).
Analysis of the cancer genome atlas (TCGA) data, design of the guide RNAs, RNA isolation, quantitative real-time PCR (qPCR) analysis, m6A-RIP qPCR, western blot analysis, RNA immunoprecipitation (RIP), RNA pull-down and mass spectrometry analysis, chromatin immunoprecipitation (CHIP), detection of ACLY4 polyubiquitination, mRNA stability and polysome profiling
A detailed description of the abovementioned methods can be found in the Supplemental Materials and Methods. The primers are listed in Supplemental Table S3. The antibodies are listed in Supplemental Table S4.
CRISPR/Cas9-mediated KO of CBS, luciferase reporter assay, lentivirus construction, plasmid construction, cell transfections, and xenograft mouse model
A detailed description of the abovementioned methods can be found in the Supplemental Materials and Methods.
Statistical analysis
All statistical analyses were performed using SPSS software (version 17.0, Chicago, IL). The Student's t-test was used to compare the differences between the two groups. Kaplan-Meier analysis and the log-rank test were used to calculate survival and significance. The nonparametric Mann-Whitney-Wilcoxon test was used to determine the relationship between CBSLR/CBS expression levels and tumor regression grade. Spearman's rank correlation test was utilized to determine correlations. Data were presented as means ± S.D. from three independent repeats. A two-sided p-value of<0.05 was treated as statistically significant.
Ethics statement
All experiments involving animals were conducted in accordance with the guidelines for laboratory animals established by the Animal Care and Use Committee of the Fudan University Shanghai Cancer Center.
Results
LncRNA expression profiling of GC cells under hypoxic conditions
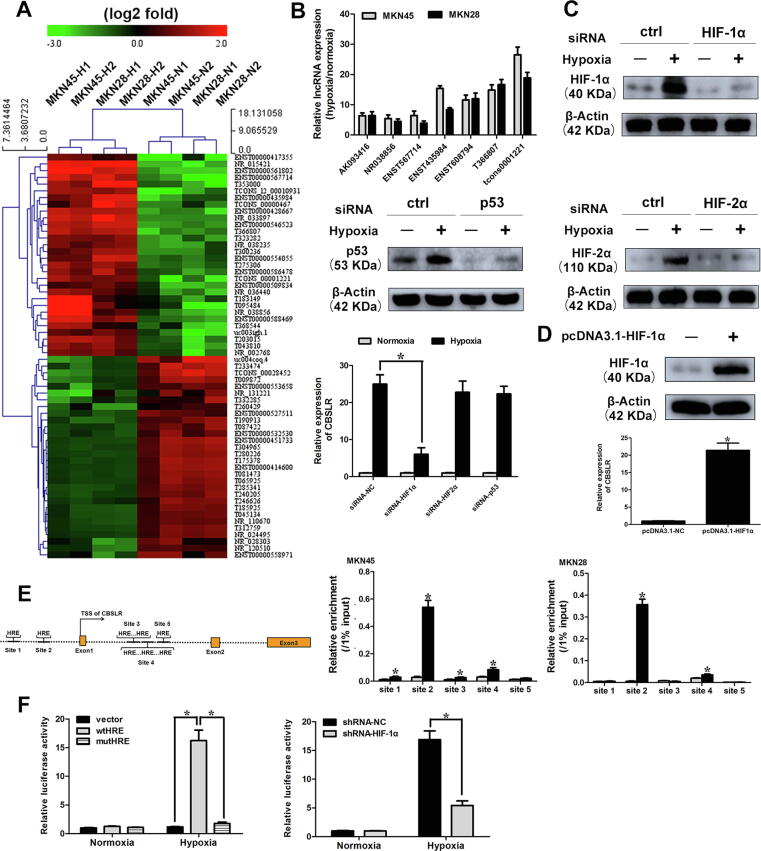
To identify hypoxia-responsive lncRNAs in gastric cancer, lncRNA and protein-coding mRNAs expression profiles in normoxia-treated (21% O2 for 24 h) and hypoxia-treated (1% O2 for 24 h) GC cells (MKN45 and MKN28 cell lines) were examined by microarray analysis. We set a threshold of a fold change > 1.5 and p < 0.05. A hierarchical clustering analysis demonstrated the systematic changes in lncRNA and mRNA expression levels between hypoxia- and normoxia-treated GC cells (Fig. 1A, Supplemental Fig. S1A and 1B). The scatter and volcano plots are demonstrated in Supplemental Fig. S1C-F. The data can be accessed via Gene Expression Omnibus (GEO) GSE114083. It is worth noting that several reported hypoxia-responsive lncRNAs and well-known hypoxia-responsive protein-coding genes (CA9, GLUT1, and BNIP3) were significantly upregulated upon hypoxia [10], [13], [14]. To validate the robustness of the profiling data, dysregulated transcripts upon hypoxia (eight lncRNAs and six mRNAs) were randomly selected and validated with qRT-PCR in hypoxia-treated GC cells (MKN28, MKN45, and HGC27) (Supplemental Fig. S2). Most of their expression levels were consistent with microarray data. Then, we focused on seven lncRNAs upregulated in the microarray data with fold changes > 5. We found that lncRNA-CBSLR (tcons0001221) was the most significantly upregulated lncRNA upon hypoxia (Fig. 1B). The expression level of CBSLR was increased upon hypoxia time-dependently (Supplemental Fig. S3A). CBSLR was also substantially induced in response to cobalt chloride (hypoxia-mimetic agent) treatment in a time- and dose-dependent manner (Supplemental Fig. S3, B and C).
Fig. 1.
tcons1221 expression is induced upon hypoxia. (A) hierarchical clustering analysis of the top 60 lncRNAs (30 upregulated and 30 downregulated) that were differentially expressed between hypoxia-treated (1% O2, 24 h) and normoxia-treated (21% O2) GC cells (MKN45 and MKN28 cell lines). (B) The expression profiling of seven cancer-associated lncRNAs in GC cells under hypoxia (1% O2, 24 h). (C) MKN45 cells were transfected with siRNA-NC, siRNA-HIF-1α, siRNA-HIF-2α, or siRNA-p53. Twenty-four hours after transfection, cells were cultured under normoxic or hypoxic conditions for 24 h. The expression levels of target genes were determined by western blot analysis. The levels of CBSLR expression were determined by qRT-PCR. (D) MKN45 cells were transfected with pcDNA3.1-HIF-1α. Twenty-four hours after transfection, the expression level of HIF-1α was determined by western blot analysis. The levels of CBSLR expression were examined by qRT-PCR. (E) Schematic illustration of HIF-1α responsive element (HRE) in CBSLR locus. MKN45 and MKN28 cells were cultured under normoxia or hypoxic conditions (1% O2) for 24 h, then, a CHIP assay was employed to examine the binding of HIF-1α to each HRE. (F) (Upper) MKN45 cells were cotransfected with Renilla luciferase plasmid and the indicated reporter constructs. (Lower) MKN45 cells expressing shRNA-NC or shRNA-HIF-1a were cotransfected with Renilla luciferase plasmid and reporter constructs containing the sequence around the second HRE site. Twenty-four hours after transfection, cells were cultured under normoxic or hypoxic conditions (1% O2) for 24 h. Luciferase activity was then determined and normalized to Renilla luciferase activity. Results were expressed as mean ± S.D. (n = 3).
Cellular characterization of CBSLR
We found that CBSLR is moderately conserved among species (NONHSAT008421, NONCODE v4 annotation; lnc-SWT1-1, LNCipedia; tcons_0001221, Broad Institute; AL078644.10, Genbank) and is located on human chromosome 1: 185339833–185344123 (Supplemental Fig. S4A). CBSLR is a transcript consisting of three exons with a full length of 794 nt (Supplemental Fig. S4, A and B). We confirmed that CBSLR has no protein-coding capacity with multiple protein-coding potential assessment software (Supplemental Fig. S5, A-C) and an in vitro translation assay (Supplemental Fig. S5D) [12]. The subcellular fraction analysis showed that CBSLR is primarily located in the cytoplasm of GC cells and is polyadenylated (Supplemental Fig. S5E). The prediction of the CBSLR secondary structure suggests that it folds into stem-loop structures, and the middle domain has the highest thermostability (Supplemental Fig. S6A). We determined the exact amount of CBSLR and found that the copy number of CBSLR in GC cells ranged from six to eleven copies per cell and increased significantly in response to hypoxia (Supplemental Fig. S6B). The expression level of CBSLR is comparable to the levels of the known hypoxia-inducible lncRNAs (H19, lincRNA-p21, and MIR31HG) under hypoxic conditions (Supplemental Fig. S6C).
HIF-1α transactivates CBSLR
To explore the molecular mechanism underlying the regulation of CBSLR by hypoxia, we downregulated the noted hypoxia-responsive transcriptional factors (HIF-1α, HIF-2α, and p53) in MKN45 cells (Fig. 1C). We found that silencing of HIF-1α, rather than HIF-2α or p53 rescued the induction of CBSLR by hypoxia (Fig. 1C and Supplemental Fig. S7A). Furthermore, overexpression of HIF-1α upregulated the expression of CBSLR (Fig. 1D). Using JASPAR, we found several hypoxia response elements (HREs) located near the locus of CBSLR (Fig. 1E). ChIP assays illustrated that HIF-1α primarily associates with the second HRE site (Fig. 1E). To confirm whether HRE was responsible for transcriptional activity, the sequence around the second HRE site was inserted into a luciferase reporter construct. The luciferase activity of wild-type, but not the mutant reporter, was induced upon hypoxia and was greatly suppressed by HIF-1α inhibition (Fig. 1F and Supplemental Fig. S7B). CBSLR expression was correlated with that of hypoxia signature genes (p < 0.001, R = 0.342) and HIF-1a (p < 0.001, R = 0.414) in TCGA-STAD (Supplemental Fig. S7C).
CBSLR is upregulated in gastric cancer
We evaluated the expression of CBSLR in TCGA-STAD to explore the expression pattern of CBSLR in GC tissues. CBSLR was upregulated in GC tissues compared with normal tissues (Supplemental Fig. S8A, p < 0.001). Moreover, the expression level of CBSLR was not different between intestinal GC tissues and diffuse GC tissues (Supplemental Fig. S9A). The expression level of CBSLR did not significantly alter with varying degrees of differentiation in GC tissues (Supplemental Fig. S9B). Furthermore, CBSLR expression level was higher in locally advanced tumors (T3 + T4) (Supplemental Fig. S8B and Supplemental Fig. S9C), and in tumors with more advanced TNM stage (Stage III + IV v.s. I + II; Supplemental Fig. S8C and Supplemental Fig. S9D). However, we found no difference in expression levels of CBSLR with or without distant metastasis or lymph node metastasis in GC tissues (Supplemental Fig. S9, E and F). Based on the median ratio of CBSLR expression in GC tissues, high tumoral CBSLR expression positively correlates with shorter overall survival (OS) and disease-free survival (DFS) rates (Supplemental Fig. S8, D and E). Additionally, multivariate analysis revealed that CBSLR expression was an independent prognostic factor that affected the OS (Supplemental table S5) and DFS (Supplemental table S6) of GC patients in the TCGA stomach cancer cohort.
CBSLR inhibition induces ferroptosis under hypoxic conditions
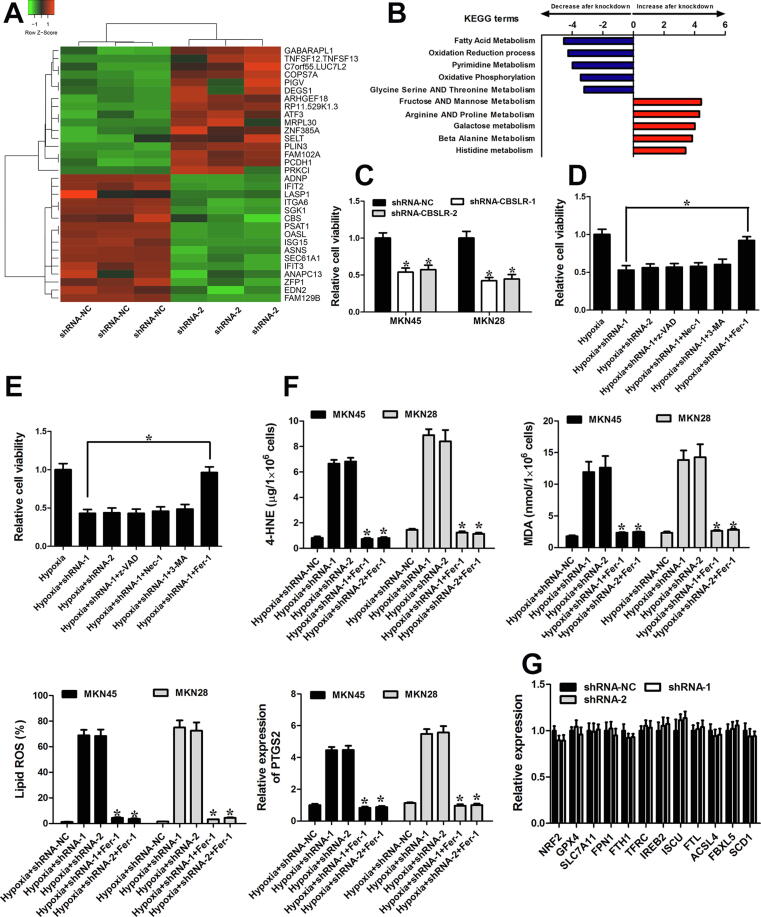
Gain- and loss-of-function studies of CBSLR were conducted in GC cells to explore its biological function. Considering the moderate CBSLR expression levels in different GC cell lines (Supplemental Fig. S10A), we chose MKN45 and MKN28 cell lines for CBSLR expression manipulation (Supplemental Fig. S10, B-E). An RNA transcriptome-sequencing analysis was performed in shRNA-NC or shRNA-CBSLR-1 MKN45 cells cultured under hypoxic conditions for 24 h (Fig. 2A). The top altered cellular pathways included fatty acid metabolism, oxidation reduction process, Fructose AND Mannose Metabolism, Arginine AND Proline Metabolism (Fig. 2B). It suggests that CBSLR takes a part in the cell metabolism of GC. CellTiter Glo assay showed that CBSLR downregulation suppressed cell viability in MKN45 and MKN28 cells under hypoxic conditions (Fig. 2C). To explore the molecular mechanism that is involved in CBSLR-silencing induced cell death, we added inhibitors of apoptosis (z-VAD-FMK, z-VAD), necroptosis (necrostatin-1, Nec-1), autophagy (3-Methyladenine, 3-MA), or ferroptosis (ferrostatin-1, Fer-1) to the GC cells exposed to hypoxia. Only the ferroptosis inhibitor significantly abolished CBSLR silencing-induced tumor cell death. (Fig. 2, D and E) These data suggest that ferroptosis is involved in the CBSLR silencing induced cell death. In the context of ferroptosis, we examined a series of ferroptosis markers. CBSLR silencing induced a significant increase in the intracellular concentrations/expressions of MDA, 4-HNE, PTGS2, and lipid ROS. Such an effect could be attenuated with a ferroptosis inhibitor (Fer-1) (Fig. 2F). We determined the expression of well-characterized regulators in ferroptosis to explore the underlying molecular mechanism by which CBSLR regulates ferroptosis. We observed no significant effects of CBSLR knockdown on the transcript levels of reported ferroptosis-regulating genes (Fig. 2G). As hypoxia and ferroptosis resistance has been linked to therapy resistance, we would like to explore the role of CBSLR in chemoresistance. As hypoxia significantly decreased the chemosensitivity of GC cells to cisplatin (a known ferroptosis inducer) [15], CBSLR silencing attenuated the chemoresistance induced by hypoxia, and this effect could be blocked by ferroptosis inhibitor ferrostatin-1 (Fer-1) (Supplemental Fig. S11, A and B).
Fig. 2.
CBSLR silencing induced ferroptosis and rescued chemoresistance in GC. (A) The heat map demonstrates the top 32 differentially expressed genes (16 upregulated and 16 downregulated) between shRNA-CBSLR and shRNA-NC MKN45 cells cultured under hypoxic conditions for 24 h, with three repeats. (B) Top five GSEA pathways identified by KEGG analysis (false discovery rate of < 0.1) for the differentially expressed genes in (A). The pathways are ranked by normalized enrichment score (NES). (C) Proliferative activity of shRNA-NC and shRNA-CBSLR MKN45/MKN28 cells grown for 24 h of hypoxia and assessed by CellTiter Glo assay. (D, E) Inhibitors of apoptosis (20 μM z-VAD), autophagy (0.5 mM 3-MA), necroptosis (10 μM Necrostatin-1), and ferroptosis (1 μM Fer-1) were added to the shRNA-NC or shRNA-CBSLR MKN45 (D) or MKN28 (E) cells. The cells were cultured under hypoxia (1% O2 for 24 h). Twenty-four hours later, the Celltiter Glo assay was utilized to determine the cell viability. (F) Inhibitors of ferroptosis (1 μM Fer-1) were added to the shRNA-NC or shRNA-CBSLR MKN45 or MKN28 cells. The cells were cultured under hypoxia (1% O2 for 24 h). Twenty-four hours later, MDA and 4-HNE levels were examined in different groups, with three repeats. The levels of lipid peroxidation and PTGS2 were detected using BODIPY staining and qPCR, respectively, with three repeats. *, p < 0.05, significantly different from shRNA-CBSLR transfected GC cells exposed to hypoxia (1% O2 for 24 h). (G) MKN45 cells transfected with shRNA-NC or shRNA-CBSLR-1,2 were cultured under hypoxic conditions (1% O2 for 24 h). Twenty-four hours later, gene levels were detected through qRT-PCR. Results were expressed as mean ± S.D. (n = 3).
Considering the reported cis-regulatory effects of lncRNAs on nearby genes, we attempted to explore the effects of CBSLR on its in cis genes. CBSLR silencing exerted no obvious effects on the mRNA levels of neighboring genes (Supplemental Fig. S12A). This indicates that CBSLR acts in trans. Then, an RNA pull-down assay and subsequent mass spectrometry (MS) analysis were performed to identify CBSLR-associated proteins that might be involved in CBSLR-related biological processes (Fig. 3A and Supplemental table S7). The candidate proteins were filtered based on the criteria: (1) predominantly cytoplasmic localization; (2) reported RNA-binding protein status (RBP); (3) predicted to bind motifs within CBSLR according to RBPmap [16].
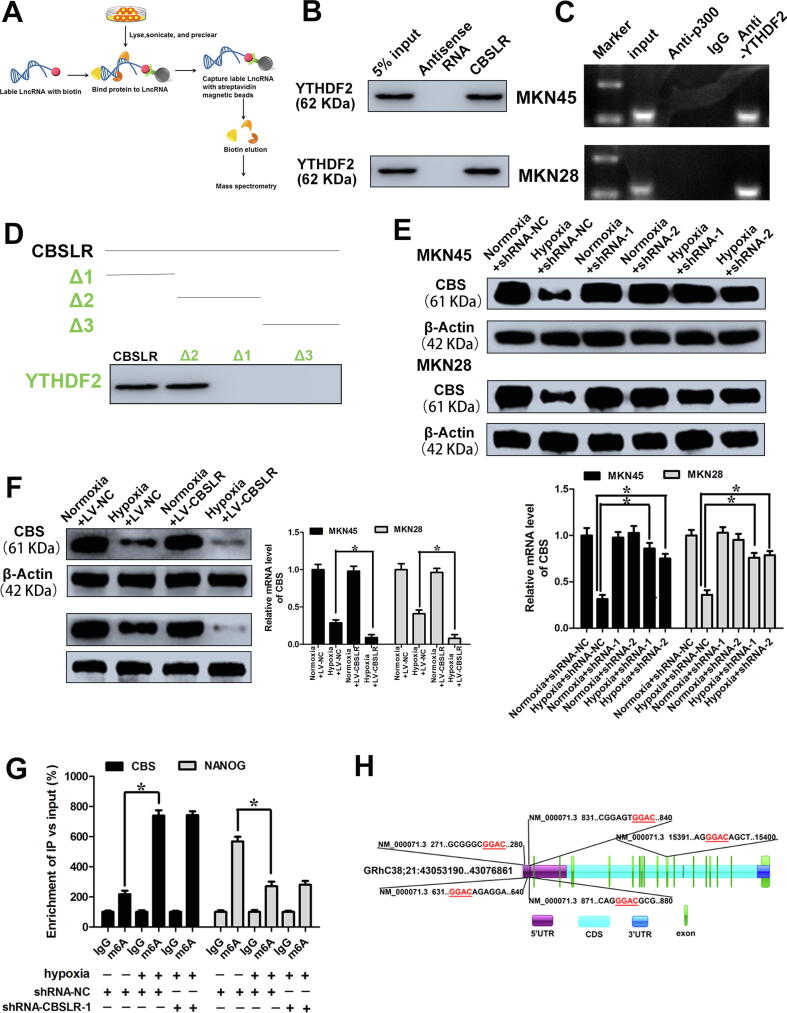
Fig. 3.
CBSLR physically interacts with YTHDF2 and modulates CBS level. (A) Schematic overview of in vitro RNA Antisense Purification and used for identification of proteins interacting with CBSLR. (B) RNA pull-down assay was performed to confirm the association between YTHDF2 and CBSLR. (C) RIP experiments shows the binding of CBSLR with YTHDF2. (D) RNA pull-down using sequentially deleted CBSLR fragments demonstrates the binding segment of CBSLR with YTHDF2. (E) shRNA-NC or shRNA-CBSLR MKN45 or MKN28 cells were cultured under normoxic or hypoxic conditions for 24 h. The mRNA and protein levels of CBS were determined using qRT-PCR or western blot analysis. (F) MKN45, and MKN28 cells stably infected with lentiviruses encoding lncRNA-CBSLR or NC were cultured under normoxic or hypoxic conditions for 24 h. The mRNA and protein levels of CBS were determined using qRT-PCR or western blot analysis. (G) The shRNA-NC and shRNA-CBSLR-1-MKN45 cells were cultured under normal or hypoxic conditions (24 h). m6A RIP-qPCR analysis of CBS or NANOG (positive control) mRNA was performed. (H) Schematic illustrations of m6A motifs positions of CBS mRNA.
RNA pull-down assays confirmed the binding between YTHDF2 and CBSLR in MKN45 and MKN28 cells (Fig. 3B). To further consolidate this finding, we immunoprecipitated YTHDF2 from the cytoplasmic extract of GC cells and analyzed the RNAs that bind to YTHDF2. We found that CBSLR specifically binds to YTHDF2 (Fig. 3C). We further discovered that the middle segment (250– 510 nt) of CBSLR is necessary and sufficient for the association with YTHDF2 with deletion-mapping analyses (Fig. 3D). As the middle domain has the highest thermostability (Supplemental Fig. S6A), it probably provides the required spatial conformation for interaction. Furthermore, CBSLR silencing did not affect the protein level of YTHDF2 (Supplemental Fig. S12B).
As the CBSLR-associated protein YTHDF2 has been reported to promote RNA degradation [17], we screened the dysregulated genes with CBSLR knockdown from RBP CLIP-SEQ data sets, including POSTAR2 [18]. Among the differentially expressed genes after CBSLR silencing, we identified 51 mRNAs bound by YTHDF2 (Supplemental Fig. S12C). Given that CBSLR is involved in cell metabolism, according to the data documented in the literature, metabolism-related genes were further selected (Supplemental Fig. S12C). We found that Cystathionine β-synthase (CBS) was bound by YHTDF2 and is the target of CBSLR, and the transcript level of CBS was negatively correlated with CBSLR, according to TCGA-STAD (Supplemental Fig. S12D). Hypoxic conditions downregulated the transcript and protein levels of CBS compared to normoxic conditions (Fig. 3E,F). CBSLR silencing increased the transcript and protein levels of CBS under hypoxic conditions (Fig. 3E), while exogenous CBSLR suppressed the expression levels of CBS in MKN45 and MKN28 cells under hypoxic conditions (Fig. 3F). We verified that CBS is a target of CBSLR. The CUACUCCUCCG site inside of CBSLR was predicated to be able to directly bind to the CDS of CBS through sequence BLAST analysis (Supplemental Fig. S12E). Therefore, CBS was selected for the subsequent investigation. RNA pull-down assays further confirmed the physical association between CBSLR and CBS (Supplemental Fig. S12F).
CBS, a metabolic enzyme, catalyzes the reaction of homocysteine with either serine or cysteine to form cystathionine and either hydrogen sulfide or water, respectively [19]. The expression level of CBS was higher in colon cancer compared to normal tissue [20]. However, CBS gene expression is downregulated in GC due to promoter hypermethylation [21]. According to data from TCGA, CBS was downregulated in GC compared to normal tissues (Supplemental Fig. S13, A and B). The CBS expression was relatively low in GC compared to other cancer cell lines (Supplemental Fig. S13C).
CBSLR regulates CBS at the post-transcriptional level
We wondered how CBS is regulated by CBSLR. We first determined whether CBSLR modulated CBS gene expression on a transcriptional level. No difference in promoter activities of CBS between shRNA-control and shRNA-CBSLR MKN45 cells was found (Supplemental Fig. S14A). With chromatin immunoprecipitation (ChIP) experiments, we found that the occupancy of DNA polymerase II at the promoter regions of CBS was not affected by CBSLR silencing (Supplemental Fig. S14B). We transfected GC cells with shRNA-CBSLR or shRNA-NC, then exposed them to hypoxia for 18 h, a time course for RNA stability was started with actinomycin D treatment (0.5 mg/mL). CBS induction by CBSLR silencing was not affected by the transcription inhibition (Supplemental Fig. S14C). Furthermore, the stability of precursor mRNA of CBS was not affected by CBSLR downregulation (Supplemental Fig. S14D). This indicates that CBSLR downregulation did not increase CBS expression via transcription mechanisms. Conversely, mature CBS mRNA stability was significantly higher in CBSLR-deletion MKN45 cells than that in the corresponding control cells (Supplemental Fig. S14E). Moreover, neither hypoxic conditions nor CBSLR knockdown affected the subcellular distribution of CBS (Supplemental Fig. S14F). There were no significant differences in CBS mRNA level in the translating pool between the control and CBSLR knockdown MKN45 cells (Supplemental Fig. S15A), suggesting that CBSLR does not regulate CBS mRNA translation. Thus, we speculate that CBSLR decreased CBS expression via post-transcriptional regulation of RNA stability.
m6A regulates mRNA stability of CBS
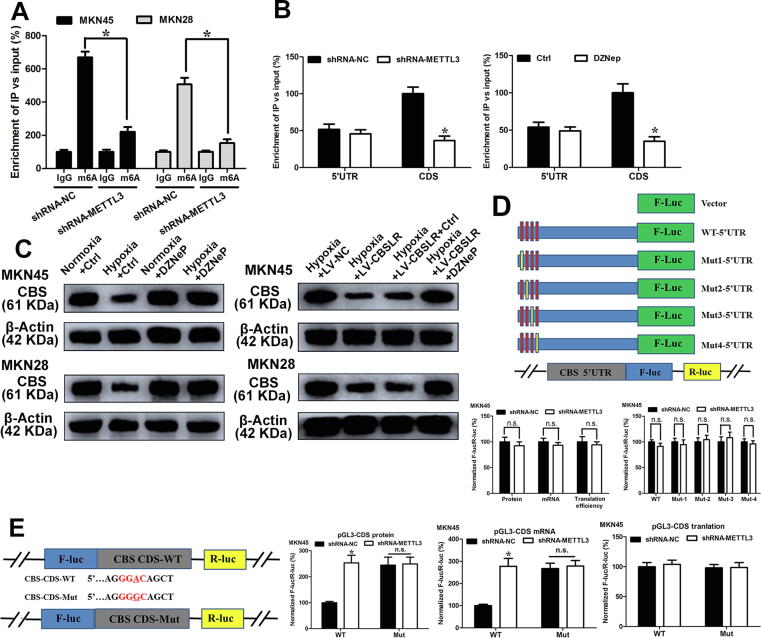
The CBSLR-associated protein YTHDF2 has been reported to promote RNA degradation in an m6A-dependent manner [17]. Previous studies have reported that tumor hypoxia causes m6A epigenetic remodelling [22], [23], [24]. To explore whether CBS is modulated in an m6A-dependent manner, we first determined the m6A abundance in the CBS mRNA. m6A-RIP-PCR showed that the m6A-specific antibody significantly enriched CBS mRNA under hypoxic conditions compared with normoxic conditions, while CBSLR silencing did not affect the m6A-specific antibody enrichment of CBS mRNA (Fig. 3G), NANOG was used as a positive control [24]. According to the results from an online bioinformatics database Whistle [25], we found five RRACU m6A sequence motifs in the exon region (Fig. 3H, at chr21: 44473695, 44488677, 44473896, 44473999, and 44473504). To explore how hypoxia remodels the m6A-mRNA landscape, we examined enzyme expression in hypoxic GC cells. Among methyltransferases, METTL3 was most significantly upregulated upon hypoxia (Supplemental Fig. S15B). m6A-RIP-PCR showed that the m6A enrichment of CBS mRNA was significantly suppressed with METTL3 downregulation (Fig. 4A and Supplemental Fig. S15C). Under hypoxic conditions, CBSLR silencing exerted no effect on the expression level of METTL3 (Supplemental Fig. S15D). We immunoprecipitated fragmented RNA with m6A to further characterize m6A methylation in CBS mRNA. We found that the m6A enrichment of CBS CDS, but not 5′-UTR, was markedly decreased with METTL3 silencing (Fig. 4B). As 3-deazaneplanocin A (DZNeP) has been reported to inhibit RNA methylation [26], GC cells were pretreated with DZNeP (10 μM) before exposure to hypoxia. We found that the m6A modification of CBS decreased dramatically after DZNeP treatment (Fig. 4B). The downregulation of CBS induced by hypoxia was also attenuated with DZNeP treatment. Furthermore, the effect of CBSLR overexpression on the expression CBS was abolished with DZNeP treatment (Fig. 4C).
Fig. 4.
m6A regulates the mRNA stability of CBS. (A) The shRNA-NC and shRNA-METTL3 MKN45 and MKN28 cells were cultured under hypoxic conditions for 24 h. m6A RIP-qPCR analysis of CBS mRNA was performed. (B) (Upper) The shRNA-NC or shRNA-METTL3-MKN45 cells were cultured under hypoxic conditions for 24 h. (Lower) MKN45 cells were pretreated with DZNeP (10 μM) for 1 h and then cultured under hypoxic conditions (24 h). m6A-RIP-qPCR analysis was performed to evaluate the m6A in 5′UTR or CDS of CBS with fragmented RNA. (C) (Left) MKN45 and MKN28 cells pretreated with DZNeP (10 μM) or DMSO for 1 h and then cultured normoxic or hypoxic conditions for 24 h. (Right) LV-NC or LV-CBSLR-MKN45 and MKN28 cells were pretreated with DZNeP (10 μM) or DMSO and then cultured under hypoxic conditions (24 h). The protein level of CBS was determined by western blot analysis. (D) Schematic representation of mutated 5′UTR. shRNA-NC or shRNA-METTL3-MKN45 cells were cotransfected with pGL3-CBS-5′UTR-WT or pGL3-CBS-5′UTR-Mut reporter. Twenty-four hours later, the cells were cultured under hypoxic conditions for 24 h. The protein, mRNA and translation efficiency were determined. (E) Schematic illustrations of mutation in m6A motif in CDS of CBS. shRNA-NC or shRNA-METTL3-MKN45 cells were cotransfected with pmirGLO-CBS-CDS-WT or pmirGLO-CBS-CDS-Mut1 reporter. Twenty-four hours later, the cells were cultured under hypoxic conditions for 24 h. The protein and mRNA were determined.
Luciferase reporters containing wild type CBS 5′UTR, or mutant 1/2/3/4 5′UTR were constructed to clarify the role of m6A methylation in CBS expression (Fig. 4D). The protein expression, mRNA expression, and translation efficiency of CBS-5′UTR in shRNA-METTL3-MKN45 cells was not significantly different from that in shRNA-NC-MKN45 cells, as illustrated by the luciferase assay (Fig. 4D). No significant difference in translational between shRNA-NC and shRNA-METTL3-MKN45 cells in reporter genes from WT-CDS-WT and CDS-Mut was observed as demonstrated by the CBS CDS-reporter luciferase assay (Fig. 4E). These results suggested that m6A methylation in the 5′UTR of CBS is independent of the m6A-regulated expression.
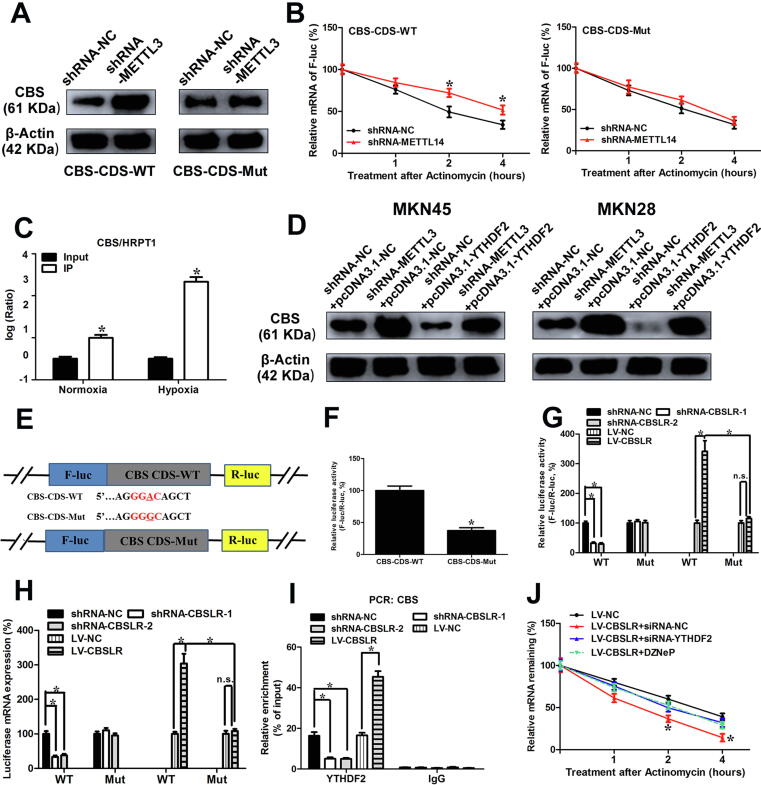
Next, luciferase reporters containing wild type or mutant coding sequence (CDS) region of CBS were constructed (Fig. 4E). The mRNA and protein expression of pmirGLO-CBS-CDS in shRNA-METTL3-MKN45 cells was markedly lower than that in shRNA-NC cells under hypoxic conditions, as demonstrated by the luciferase assay. The m6A motif (CDS-Mut1) mutation in the CDS region induced a decrease in mRNA expression and protein expression of pmirGLO-CDS, but METTL3 knockdown partially attenuated the downregulation (Fig. 4E). shRNA-NC or shRNA-METTL3-MKN45/MKN28 cells were transfected with pcDNA3.1-CBS-CDS-WT or pcDNA3.1-CBS-CDS-Mut, respectively. The upregulation of CBS induced by METTL3 silencing was abolished with pcDNA3.1-CBS-CDS-Mut, compared to that of pcDNA3.1-CBS-CDS-WT as demonstrated by western blot results (Fig. 5A and Supplemental Fig. S16A). Moreover, METTL3 knockdown increased the mRNA stability of pcDNA3.1-CBS-CDS-WT under hypoxic conditions, while pcDNA3.1-CBS-CDS-Mut can improve the difference in mRNA stability between MKN45 cells transfected with shRNA-METTL3 or shRNA-NC (Fig. 5B). Furthermore, the m6A modified GGAC is important for the secondary structure of CBS CDS (Supplemental Fig. S16B). In summary, these data suggest that m6A in CBS CDS is responsible for mRNA stability.
Fig. 5.
CBSLR/YDHDF2/CBS ternary complex promotes degradation of CBS mRNA. (A) pcDNA3.1-CBS-CDS-WT or pcDNA3.1-CBS-CDS-Mut was cotransfected into shRNA-NC or shRNA- METTL3-MKN45 cells. After 24 h, the cells were cultured under hypoxic conditions for 24 h. Cell lysates were subjected to western blot analysis. (B) pcDNA3.1-CBS-CDS-WT or pcDNA3.1-CBS-CDS-Mut was transfected into shRNA-NC or shRNA-METTL3-MKN45 cells. After 24 h, the cells were cultured under hypoxic conditions for 20 h and then further treated with Act-D (0.5 mg/mL) for the indicated times. The mRNA level of CBS was examined by qRT-PCR. (C) The MKN45 cells were cultured under normoxic or hypoxic conditions for 24 h. RIP experiments were performed using the YTHDF2 antibody for immunoprecipitation and qRT-PCR was utilized to detect CBSLR. (D) shRNA-NC or shRNA-METTL3- MKN45 or MKN28 cells were transfected with pcDNA3.1-NC or pcDNA3.1-YTHDF2. Twenty-four hours later, the cells were cultured under hypoxia for 24 h. The protein level of CBS was evaluated by western blot analysis. (E, F) Schematic diagram of CBS WT (CBS-CDS-WT-luc) and CBS mutant (CBS-CDS-MUT-luc) reporters (E). The MKN45 cells were transfected with pmirGLO-CBS-CDS-WT or pmirGLO-CBS-CDS-Mut1 reporter. After 24 h, MKN45 cells were cultured under hypoxia for 24 h. Relative luciferase activity was evaluated. (G, H) shRNA-NC or shRNA-CBSLR-MKN45 cells were cotransfected with pmirGLO-CBS-CDS-WT or pmirGLO-CBS-CDS-Mut1 reporter. MKN45 stably infected with lentiviruses encoding lncRNA-CBSLR or NC were cotransfected with pmirGLO-CBS-CDS-WT or pmirGLO-CBS-CDS-Mut1 reporter. Twenty-four hours later, the cells were cultured under hypoxic conditions for 24 h. Relative luciferase activity and luciferase mRNA expression were determined. (I) RIP assay reveals the relative association of YTHDF2 with CBS mRNA upon CBSLR downregulation or upregulation. (J) shRNA-NC or shRNA-CBSLR-MKN45 cells were transfected with pcDNA3.1-NC or pcDNA3.1-YTHDF2. Twenty-four hours later, the cells were cultured under hypoxic conditions for 20 h in the presence or absence of DZNeP (10 μM). A time course started with treating cells with Act-D (0.5 mg/mL) for the indicated times. The CBS mRNA was examined with qRT-PCR. Data represent mean ± S.D. from three independent experiments.
We further investigated the molecular mechanisms underlying m6A regulated mRNA stability. RIP for YTHDF2 was performed, and a notably higher enrichment of CBS was found in the anti-YTHDF2 immunoprecipitates compared with input, particularly under hypoxic conditions (Fig. 5C). Furthermore, YTHDF2, but not IGF2BP1/2/3, YTHDF1, or YTHDF3, can significantly associate with CBS mRNA in MKN45 cells under hypoxic conditions (Supplemental Fig. S17A). The interaction between YTHDF2 and CBS was reduced by METTL3 silencing (Supplemental Fig. S17B). Moreover, we found that YTHDF2 preferentially interacted with the CBS mRNA's CDS region rather than the 5′-UTR region of CBS mRNA (Supplemental Fig. S17C). Such interaction was suppressed with METTL3 silencing (Supplemental Fig. S17C). We, therefore, manipulated the expression of YTHDF2 as confirmed by western blot analysis (Supplemental Fig. S17D). We then wondered whether YTHDF2 triggers the decay of CBS mRNA. With actinomycin D treatment, the lifetime was prolonged with YTHDF2 knockdown and shortened with exogenous expression of YTHDF2 (Supplemental Fig. S17E); however, DZNeP treatment ameliorated the effect of YTHDF2 on the stability of CBS (Supplemental Fig. S17F). YTHDF2 overexpression inhibited the expression of CBS, while METTL3 silencing abolished the suppressive effect of YTHDF2 on the expression of CBS in GC cells as demonstrated by western blot analysis (Fig. 5D). Furthermore, ectopic expression of YTHDF2 lowered the mRNA lifetime of pcDNA3.1-CBS-CDS-WT, while such an effect was abolished for pcDNA3.1-CBS-CDS-Mut (Supplemental Fig. S18A). This indicates that YTHDF2 plays a role in m6A-mediated CBS mRNA stability.
YTHDF2 has been reported to act as an m6A “reader” relying on the hydrophobic residues W432 and W486 in its carboxy-terminal YTH domain [27]. The catalytically inactive mutant of YTHDF2 was constructed (W432A and W486A) [23] (Supplemental Fig. S18, B and C). Exogenous expression of WT-YTHDF2 rather than the catalytically inactive mutant attenuated the decreased mRNA stability and protein level of CBS induced by YTHDF2 knockdown, while the catalytically inactive mutant had no such effect (Supplemental Fig. S18, D and E). Enforced expression of WT-METTL3, rather than Mut-METTL3 (D395A/W398A) [28] (Supplemental Fig. S18, F and G), restored the increased mRNA stability and protein level of CBS by METTL3 knockdown (Supplemental Fig. S18, H and I). Our data indicate that the m6A reader function of YTHDF2 is necessary for its suppressive role on CBS in GC.
To explore whether CBSLR is required for CBS mRNA destabilization in an m6A-dependent manner, luciferase reporter constructs containing transcript segments harboring m6A motifs were generated (Fig. 5E). The luciferase activity of CBS-CDS-Mut was approximately 40% that of control CBS-CDS-WT under hypoxic conditions (Fig. 5F). Luciferase mRNA expression and luciferase activity of CBS-CDS-WT, but not that of CBS-CDS-Mut was markedly suppressed with CBSLR silencing (Fig. 5, G and H). In contrast, luciferase mRNA expression and luciferase activity of CBS-CDS-WT, but not that of CBS-CDS-Mut was increased with ectopic expression of CBSLR (Fig. 5, G and H).
To determine whether CBSLR/CBS/YTHDF2 formed an RNA-protein ternary complex, we performed RIP assays. RIP assays showed that CBS/YTHDF2 RNA-protein interaction was dramatically reduced by knockdown of CBSLR and was significantly enhanced by exogenous expression of CBSLR (Fig. 5I). These data indicate that CBSLR promotes the interactions between YTHDF2 and CBS and further suggest that CBSLR enhances the mRNA stability of CBS via the formation of a CBSLR/YTHDF2/CBS RNA-protein ternary complex. Furthermore, YTHDF2 did not affect the transcript level of CBSLR (Supplemental Fig. S19A). The effect of CBSLR overexpression on the stability of CBS mRNA was greatly attenuated after METTL3 or YTHDF2 knockdown or DZNeP treatment (Fig. 5J), suggesting that the CBSLR induction of CBS mRNA decay depends on YTHDF2 and METTL3.
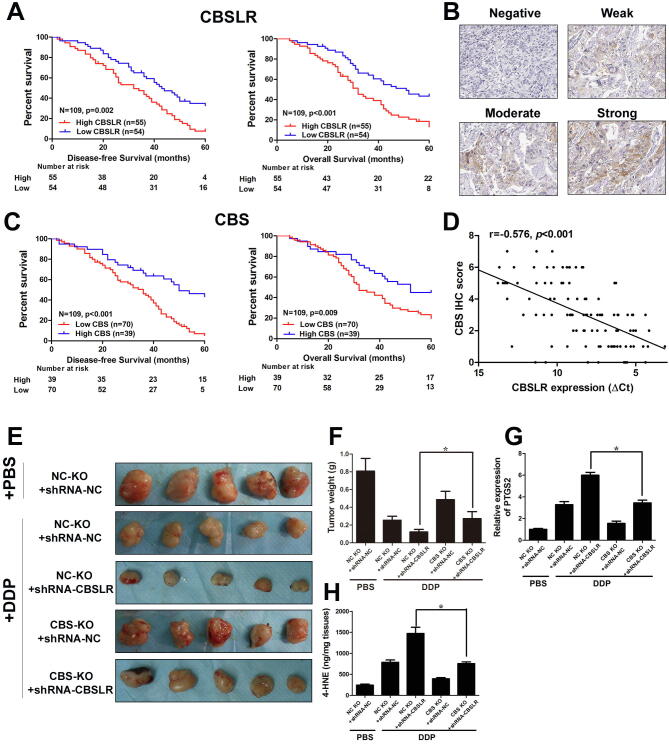
Given that CBS was downregulated in GC tissues, we decided to explore the correlation between CBS levels and the prognosis of GC patients. Surprisingly, CBS high expression correlated with a poor prognosis, based on data from the KMPlot database (Supplemental Fig. S19, B and C). We attempted to unveil the underlying mechanism by which high expression of CBS contributes to a worse clinical outcome and reanalyzed the data. We found that in the cohort of patients that have received 5-Fu based adjuvant (Supplemental Fig. S19, D and E) or other adjuvant therapies (Fig. S20, A and B), low CBS expression was associated with a lower survival rate, but not in patients who received surgery alone (Fig. S20, C and D). Such a phenomenon was observed in an independent cohort of patients that received cisplatin-based adjuvant chemotherapy (Supplemental table S1). High CBSLR expression correlated with more dismal survival rates in this cohort of patients (Fig. 6A). The representative images of negative, weak, moderate and strong CBS expression patterns in GC according to IHC staining was provided in Fig. 6B. Low CBS protein level was associated with a poor prognosis (Fig. 6C). A negative correlation between the expression of CBSLR and CBS was found in this cohort of patients (Fig. 6D). This suggests that CBS might sensitize the GC cells to chemotherapy after surgery, which results in a better clinical outcome. In another cohort of patients that received neoadjuvant chemotherapy (Supplemental table S2), high CBSLR/low CBS expression was associated with worse tumor regression grades (TRG) (Supplemental table S8).
Fig. 6.
CBSLR/CBS correlated with survival rate and response to chemotherapeutics in GC. (A) Kaplan–Meier curves for overall survival and disease-free survival in the CBSLR-high or CBSLR-low groups. (B) The representative images of negative, weak, moderate and strong CBS expression patterns in GC according to IHC staining (×200). (C) Kaplan–Meier curves for overall survival and disease-free survival in the CBS-high or CBS-low groups. (D) Spearman rank-correlation analysis was performed to the correlation between the ΔCt values (normalized to β-actin) of CBSLR and IHC score of CBS in 109 GC tissues. (E) CBSLR knockdown, CBS knockout or control MKN45 cells were grown as xenografts. DDP treatment started after tumors reached approximately 50 mm3 in size. DDP: 7 mg per kg, once a week for three weeks. (F) Xenograft tumor masses were harvested. Tumor weights were determined. (G) The mRNA level of PTGS2 in the indicated tumor was examined with qRT-PCR. (H) The 4-HNE contents in different groups were determined.
CBS has been reported to be involved in ferroptosis as a marker of transsulfuration pathway activity and protects fibrosarcoma and liver cancer cells from ferroptosis [29], [30]. We wondered whether ferroptosis is involved in CBSLR-CBS signaling axis-mediated chemosensitivity. We found that downregulation of CBSLR suppressed cell viability in MKN45 and MKN28 cells under hypoxic conditions, however, such an effect could be abolished by CBS silencing (Supplemental Fig. S21A). Attempting to evaluate the role of the transsulfuration pathway in CBSLR-mediated ferroptosis resistance, we treated shRNA-CBSLR-transfected cells with propargylglycine (PPG), an inhibitor of cystathionine γ-lyase, the enzyme that converts cystathionine to cysteine in the transsulfuration pathway [31] (Supplemental Fig. S21B). Surprisingly, PPG treatment did not affect CBSLR-silencing mediated cell viability suppression (Supplemental Fig. S21A), suggesting that the biological function of CBSLR is mediated by CBS, but not the transsulfuration pathway. The significantly increased intracellular concentrations/expressions of MDA, 4-HNE, PTGS2 and lipid ROS induced by CBSLR silencing could be abolished by CBS silencing (Supplemental Fig. S21C). CBSLR silencing attenuated chemoresistance induced by hypoxia, and this effect could be attenuated by CBS knockdown (Supplemental Fig. S22).
We further validated these findings by xenograft experiments in mice. We next used the CRISPR-Cas9 system to knock out the CBS gene in MKN45 cells. CBSLR knockdown, CBS knockout or control MKN45 cells were grown as xenografts. Cisplatin (DDP) treatment started after tumors reached approximately 50 mm3 in size. DDP treatment significantly retarded tumor growth (NC KO + shRNA-NC + DDP group) compared to PBS treatment (NC KO + shRNA-NC + PBS group) as illustrated with decreased tumor volume (Fig. 7 E) and tumor weight (Fig. 7F), and such an effect was enhanced by CBSLR knockdown (NC KO + shRNA-CBSLR + DDP group); however, CBS knockout (CBS KO + shRNA-CBSLR + DDP group) attenuated the suppressive effect induced by CBSLR knockdown when compared to the single CBSLR knockdown group (NC KO + shRNA-CBSLR + DDP group). Specifically, single CBS knockout (CBS KO + shRNA-NC + DDP group) ameliorated the anti-tumor effect of DDP (NC KO + shRNA-NC + DDP group). The expression/content of ferroptosis markers (PTGS2/4-HNE) was greatly increased with DDP treatment (NC KO + shRNA-NC + DDP group) compared to PBS treatment (NC KO + shRNA-NC + PBS group), and CBSLR silencing further enhanced the increase (NC KO + shRNA-CBSLR + DDP group), with CBS knockout (CBS KO + shRNA-CBSLR + DDP group) greatly attenuated the increase induced by CBSLR knockdown when compared to the single CBSLR knockdown group (NC KO + shRNA-CBSLR + DDP group) (Fig. 7, G and H). Specifically, single CBS knockout (CBS KO + shRNA-NC + DDP group) ameliorated the effect of DDP on expression/content of PTGS2/4-HNE (NC KO + shRNA-NC + DDP group) (Fig. 7, G and H).
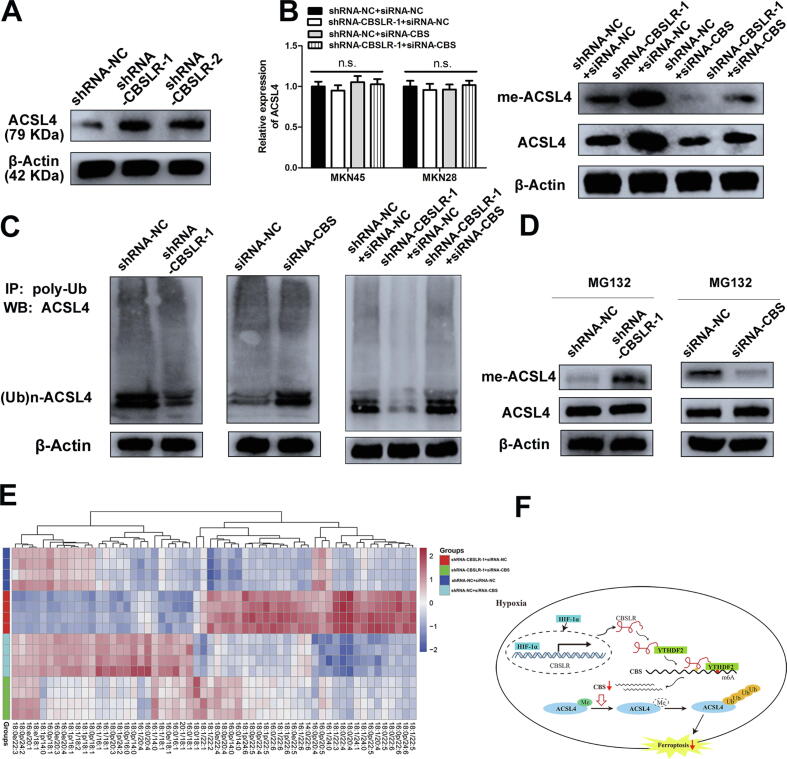
Fig. 7.
CBSLR inhibits ferroptosis by modulating ACSL4 methylation to be polyubiquitinated. (A) shRNA-NC or shRNA-CBSLR-MKN45 cells were cultured under hypoxic conditions for 24 h. Cell lysates were subjected to western blot analysis. (B) siRNA-NC or siRNA-CBS was cotransfected into shRNA-NC or shRNA- CBSLR-MKN45 cells. After 24 h, the cells were cultured under hypoxic conditions (24 h). Anti-dimethyl-ACSL4 and anti-ACSL4 antibodies were used in the western blot analysis. The transcript level of ACSL4 was examined with qRT-PCR. (C) ACSL4 endogenous polyubiquitination ((Ub)n-ACSL4) was observed to be decreased with CBSLR silencing and to be increased with CBS silencing in MKN45 cells under hypoxic conditions (24 h). (D) shRNA-NC, shRNA-CBSLR, siRNA-NC, siRNA-CBS-MKN45 cells were cultured under hypoxic conditions for 18 h. MG132 (10 mM) was treated for 6 h. Cell lysates were analyzed by western blot with anti-dimethyl-ACSL4 and anti-ACSL4 antibodies. (E) Heat map of all major PE species in MKN45 cells with hierarchical clustering of shRNA-NC + siRNA-NC, shRNA-CBSLR + siRNA-NC, shRNA-NC + siRNA-CBS, shRNA-CBSLR + siRNA-CBS. Each PE species was normalized to the corresponding mean value. (F) Schematic diagram showing that HIF-1α induces lncRNA-CBSLR to recruit YTHDF2 protein and CBS mRNA to form CBSLR/ YTHDF2/CBS complex, which in turn decreases CBS mRNA stability in an m6A dependent manner. The decreased CBS expression reduced methylation of ACSL4 protein, thus, the protein is degraded via the ubiquitination-proteasome pathway. It eventually protects GC from ferroptosis under a hypoxic tumor microenvironment.
CBSLR inhibits ferroptosis by modulating ACSL4 methylation to be polyubiquitinated
Then, we explored the mechanism underlying the ferroptosis mediated by CBSLR. However, we demonstrated that CBSLR silencing did not affect the well-recognized regulators of ferroptosis, namely, intracellular GSH and Fe2+ contents, system xc− and GPX4 activity (Supplemental Fig. S23, A-D).
A previous study reported that downregulation of CBS suppressed the methylation of PFKFB3 protein, which shuts glucose metabolism toward the pentose phosphate pathway [32]. It increased cellular NADPH contents to produce reduced glutathione and protected cancer cells from oxidants. We wondered whether the suppressive effect of CBSLR on ferroptosis might be through the modulation of glucose utilization. The difference in glucose biotransformation between shRNA-CBSLR or shRNA-NC-MKN45 cells exposed to hypoxia for 24 h was determined by metabolome analysis utilizing capillary electrophoresis mass spectrometry (CE-MS) as described previously [32]. The data showed that the sum of metabolites belonging to PPP or glycolysis was not significantly different. Furthermore, there were no significant changes in ATP and energy charge values (Supplemental Fig. S24).
It is well recognized that lipid remodelling changes the ferroptosis susceptibility in cancer cells, mainly through the biosynthesis of monounsaturated fatty acids (MUFAs) and polyunsaturated fatty acids (PUFAs). Generally, PUFAs are synthesized by ACSL4, ELOVL2/5, FADS1/2, and LPCAT3 and are more susceptible to ferroptotic cell death, and MUFAs are synthesized by SCD1 and may protect cancer cells from lipid peroxidation by decreasing lipophilic antioxidant coenzyme Q10 [33], [34], [35]. Thus, we assessed the relationship between CBSLR and lipid metabolism regulators. Western blot analysis demonstrated that CBSLR silencing significantly upregulated the protein level of ACSL4, while CBSLR did not affect other regulators (Fig. 7A and Supplemental Fig. S25A). However, there was no difference in mRNA levels of ACSL4 with CBSLR silencing or CBS silencing, supporting the idea that the CBSLR-mediated regulation of ACSL4 might be through a post-transcriptional mechanism (Fig. 7B). Furthermore, the upregulation of ACSL4 induced by CBSLR silencing could be abrogated by CBS knockdown (Fig. 7B). Inspired by the report that CBS could down- and/or upregulate protein arginine methylation [32], [36], we examined whether CBSLR regulated the post-translational arginine methylation of ACSL4. We found that the CBSLR silencing significantly increased methyl-ACSL4 level, and this effect could be abolished with CBS knockdown. Furthermore, increased methylated ACSL4 was concomitant with a noticeable upregulation of total ACSL4 amounts in cells (Fig. 7B and Supplemental Fig. S25B). These data led us to hypothesize that, once methylated, ACSL4 becomes insensitive to polyubiquitination and proteasomal degradation. We immunoprecipitated polyubiquitinated ACSL4 ((Ub)n-ACSL4) by the antipoly-Ub antibody and served as samples for western blot by the anti-ACSL4 antibody, indicating that CBSLR silencing, caused a marked decrease in (Ub)n-ACSL4, such an effect could be abolished with CBS silencing (Fig. 7C and Supplemental Fig. S25C). Finally, after MG132 (a proteasome inhibitor) treatment, CBSLR/CBS silencing upregulated/downregulated amounts of methylated ACSL4, stabilizing ACSL4 (Fig. 7D and Supplemental Fig. S25D). The data indicate that CBSLR/CBS modulates ACSL4 methylation, which affects its polyubiquitination.
Considering the pro-ferroptosis effects of ACSL4-induced PUFA acylation [34], we hypothesized that the CBSLR-inhibition induced upregulation of ACSL4 might mitigate ferroptosis resistance in GC cells. The lipidomic analysis supported this hypothesis (Fig. 7E). The pro-ferroptosis phosphatidylethanolamine (PE)(18:0/20:4) and PE(18:0/22:4) increased with CBSLR silencing and decreased with CBS knockdown (Supplemental Fig. S26A). The increased contents of double- and triple-oxidized arachidonic acid (AA)- and adrenic acid (AdA)-containing PE species induced by the loss of CBSLR could be restored by CBS knockdown (Supplemental Fig. S26B). These data were consistent with the previous report that ACSL4 channels AA and AdA for lipid biosynthesis [34].
Discussion
Hypoxic microenvironments are readily available across almost all solid tumors and profoundly impact tumor progression. The cellular hypoxia signaling is primarily regulated by HIF. HIF can transactive target genes by binding to HRE upon hypoxia. In this study, we characterized CBSLR as a new player in cellular hypoxia signaling. CBSLR is transactivated by HIF-1α under hypoxic conditions and modulates ferroptosis and chemoresistance of GC cells. CBSLR upregulation confers chemoresistance both in vitro and in vivo. Notably, CBSLR is upregulated in GC tissues and high CBSLR expression correlates with a poorer prognosis and a poorer response to chemotherapy. Our data indicate that CBSLR serves as a novel potential indicator for chemotherapy response and helps select individualized treatment in the clinic in GC.
The KEGG pathway enrichment indicated that CBSLR had an impact on cell metabolism and the oxidation–reduction process. Moreover, CBSLR protects GC cells against ferroptosis and does not affect apoptosis, necroptosis, or autophagic cell death upon hypoxia. Previous studies suggest that hypoxia might be involved in ferroptosis. Yang et al. showed that HIF1α prevented ferroptotic cell death [37]. Carbonic anhydrase 9 inhibition leads to higher sensitivity to ferroptosis in malignant mesothelioma under hypoxic conditions by modulating iron metabolism [38].
Thus, CBSLR acts as a novel mediator linking hypoxia and ferroptosis. However, CBSLR exerted no significant effects on the mRNA levels of well-known ferroptosis regulators. The molecular mechanisms of lncRNA's biological function can be diversified, as lncRNAs play a role in multiple levels of gene modulation, namely, transcriptionally via recruiting chromatin-modifying complexes and also post-transcriptionally through interactions with proteins, mRNAs, or miRNAs [8], [9], [10]. Here, we identified that CBSLR associates with YTHDF2. However, CBSLR did not affect the protein level of YTHDF2.
Given that YTHDF2 has been reported to promote RNA degradation in an m6A dependent manner [17], we screened the dysregulated genes with CBSLR knockdown and chose CBS for further investigation. Furthermore, we found that CBSLR had no effect on the subcellular distribution, transcription, and translation of CBS, but markedly decreased the mRNA stability of CBS. Among the identified chemical modifications in RNA, N6-methyladenosine (m6A) (methylated at the N6 position of adenosine) has been regarded as the prominent dynamic mRNA modification [36]. RNA m6A modifications are enriched near stop codon and in the 5′- and 3′-UTR and within long internal exons [23], [24]. m6A modification is dominated by methyltransferase complex (“writers”), demethylases (“erasers”), and RNA-binding proteins (“readers”). m6A has been reported to affect RNA translation, splicing and decay and is implicated in a wide variety of cellular processes, including carcinogenesis [39]. As tumor hypoxia causes m6A epigenetic remodelling [22], [23], [24], we speculated that CBSLR modulated CBS in an m6A-dependent manner. We found significant enrichment of m6A in the CDS regions of CBS in GC cells mediated by METTL3. m6A in CDS, but not 5′UTR, positively regulated the decay rate of CBS mRNA. YTHDF2 was shown to be involved in m6A regulated mRNA stability of CBS. Our results suggested that CBS is involved in CBSLR-mediated ferroptosis- and chemoresistance. A negative correlation between CBSLR and CBS expression was found in GC tissues. High expression of CBSLR and low expression of CBS were associated with a worse prognosis in GC patients that received adjuvant chemotherapy and with a poor response in patients that received neoadjuvant chemotherapy.
The oncogenic or tumor-suppressive role of CBS may depend on the tumor type. CBS promotes colon cancer growth and progression by both autocrine and paracrine mechanisms [20], [40]. However, the decreased expression of CBS has been demonstrated to promote glioma tumorigenesis [41]. Our study was consistent with a study reporting that CBS is downregulated in GC [21], and that CBS expression is further decreased upon hypoxia. However, the biological role of CBS is unknown in GC. Ferroptosis, a ROS-dependent form of cell death, is mainly related to iron accumulation and lipid peroxidation. Dysregulation of ferroptosis is correlated with a diversity of pathological conditions, including the therapeutic response to cancer treatments [42], [43], [44]. CBS has been reported to be involved in ferroptosis as a marker of transsulfuration pathway activity and to protect fibrosarcoma, hepatocellular carcinoma, breast cancer, and lung cancer cells from ferroptosis [29], [30], [45]. Furthermore, CBS was upregulated in breast cancer and lung cancer compared with normal tissues, according to TIMER and Oncomine. When CBS expression is downregulated, the production of endogenous GSH is decreased, which can lead to ferroptosis [45]. However, CBS has also been reported to decrease the intracellular GSH content and antioxidant capacity in lymphoma cell lines, which could contribute to ferroptosis [32]. Thus, the role of CBS in ferroptosis modulation could also depend on the cancer type and be heterogeneous. Therefore, we explored the role of CBS in ferroptosis in GC.
We demonstrated that CBSLR silencing had no effects on the well-recognized ferroptosis regulators, including intracellular GSH and Fe2+ levels, system xc− and GPX4 activity. Considering the function of CBS in catalyzing the reaction of homocysteine with either cysteine or serine to form cystathionine and either hydrogen sulfide or water, respectively, we treated GC cells with propargylglycine (PPG) (an inhibitor of cystathionine γ-lyase). We found that the transsulfuration pathway was not involved in CBSLR-mediated ferroptosis resistance.
However, SLC7A11high results in metabolic reprogramming, causing reliance on GSH and NADPH [46], [47], which increases ferroptosis vulnerabilities. As a study reported that downregulation of CBS increased GSH content and an analysis across human datasets revealed a comparatively high expression of SLC7A11 in GC versus normal tissues [32], [46] (Fig. S27), we wondered whether CBSLR functioned through modulating the glucose metabolism. However, we found that CBSLR had no such effects.
Inspired by the KEGG pathways results that indicated that CBSLR impacted lipid metabolism, we explored whether CBSLR could regulate lipid composition. We found that CBSLR silencing upregulated ACSL4 protein level on a post-transcriptional level and the pro-ferroptosis phosphatidylethanolamine (PE)(18:0/20:4) and PE (18:0/22:4) content. The hypoxia-induced CBSLR/CBS signaling axis modulates ACSL4 methylation to be polyubiquitinated, thus protecting GC cells from ferroptosis.
Conclusions
Overall, we demonstrate that hypoxia-induced CBSLR/CBS/ACSL4 signaling mediates PUFA metabolism and contributes to the ferroptosis resistance in GC cells, which deepens our understanding of the hypoxia-mediated signaling landscape. This study suggests that CBSLR might be a potential therapeutic target for refractory hypoxic tumors.
Ethics approval and consent to participate
This study was sanctioned by the local ethics committee of the Fudan University Shanghai Cancer Center (201736) and Yijishan Hospital (201819). The protocols used in this study were approved by the Ethical Review Committees of Fudan university shanghai cancer center and Yijishan hospital of Wannan medical college, and written informed consent was provided from all the patients. The animal studies were authorized by the Animal Ethic Review Committees of the Yijishan hospital of Wannan medical college. All animal experiments were strictly implemented in compliance with the NIH Guide for the Care and Use of Laboratory Animals.
Consent for publication
All subjects have written informed consent.
Availability of data and materials
All data generated or analyzed during this study are included either in this article or in the supplementary information files. The lncRNA microarray and RNA-seq datasets were deposited in Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), accession numbers: GSE114083 and GSE114360.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Department of Pathology from Yijishan Hospital for technical assistance in pathological evaluation. The authors are thankful to the National Nature Science Foundation of China (Grant Nos. 81972213, 81802503, 81772180, 82072370), the Key University Science Research Project of Anhui Province (KJ2020A0594, KJ2020A0607), Fudan University Shanghai Cancer Center for Outstanding Youth Scholars Foundation (YJYQ201803), Peak Training Program for Scientific Research of Yijishan Hospital, Wannan Medical College (GF2019T01, GF2019G15). The sponsors have no role in study design, in the collection, analysis and interpretation of data, in the writing of the report, and in the decision to submit the article for publication.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2021.10.001.
Contributor Information
Mingzhe Ma, Email: mmz666@163.com.
Yan Zhang, Email: yanyan0921@sina.com.
Hongping Xia, Email: Ncmxhp@nccs.com.sg.
Kun Lv, Email: lvkun315@126.com.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Smyth E.C., Nilsson M., Grabsch H.I., van Grieken N.CT., Lordick F. Smyth, Magnus Nilsson, Heike I Grabsch, Nicole Ct van Grieken Florian Lordick. Gastric cancer. Lancet. 2020;396(10251):635–648. doi: 10.1016/S0140-6736(20)31288-5. [DOI] [PubMed] [Google Scholar]
- 4.de Heer E.C., Jalving M., Harris A.L. HIFs, angiogenesis, and metabolism: elusive enemies in breast cancer. J Clin Invest. 2020;130(10):5074–5087. doi: 10.1172/JCI137552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhandari V., Hoey C., Liu L.Y., Lalonde E., Ray J., Livingstone J., et al. Molecular landmarks of tumor hypoxia across cancer types. Nat Genet. 2019;51(2):308–318. doi: 10.1038/s41588-018-0318-2. [DOI] [PubMed] [Google Scholar]
- 6.Abou Khouzam R., Goutham H.V., Zaarour R.F., Chamseddine A.N., Francis A., Buart S., et al. Integrating tumor hypoxic stress in novel and more adaptable strategies for cancer immunotherapy. Semin Cancer Biol. 2020;65:140–154. doi: 10.1016/j.semcancer.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Statello L., Guo C.-J., Chen L.-L., Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCabe E.M., Rasmussen T.P. lncRNA involvement in cancer stem cell function and epithelial-mesenchymal transitions. Semin Cancer Biol. 2020:30272–30278. doi: 10.1016/j.semcancer.2020.12.012. S1044–579X(20) [DOI] [PubMed] [Google Scholar]
- 9.Huan L., Guo T., Wu Y., Xu L., Huang S., Xu Y.e., et al. Hypoxia induced LUCAT1/PTBP1 axis modulates cancer cell viability and chemotherapy response. Mol Cancer. 2020;19(1) doi: 10.1186/s12943-019-1122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shih J.-W., Chiang W.-F., Wu A.T.H., Wu M.-H., Wang L.-Y., Yu Y.-L., et al. Long noncoding RNA LncHIFCAR/MIR31HG is a HIF-1α co-activator driving oral cancer progression. Nat Commun. 2017;8(1) doi: 10.1038/ncomms15874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu P., He F., Hou Y., Tu G., Li Q., Jin T., et al. A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene. 2021;40(9):1609–1627. doi: 10.1038/s41388-020-01638-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma M., Zhang Y., Weng M., Hu Y.e., Xuan Y.i., Hu YiRen, et al. lncRNA GCAWKR Promotes Gastric Cancer Development by Scaffolding the Chromatin Modification Factors WDR5 and KAT2A. Mol Ther. 2018;26(11):2658–2668. doi: 10.1016/j.ymthe.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi K., Yan I.K., Haga H., Patel T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J Cell Sci. 2014;127(Pt 7):1585–1594. doi: 10.1242/jcs.141069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang F., Zhang H., Mei Y., Wu M. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol Cell. 2014;53(1):88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Su Y., Zhao B., Zhou L., Zhang Z., Shen Y., Lv H., et al. Ferroptosis, a novel pharmacological mechanism of anti-cancer drugs. Cancer Lett. 2020;483:127–136. doi: 10.1016/j.canlet.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 16.Paz I, Kosti I, Ares M Jr, Cline M, Mandel-Gutfreund Y. RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 2014;42(Web Server issue):W361-7. [DOI] [PMC free article] [PubMed]
- 17.Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626. [DOI] [PMC free article] [PubMed]
- 18.Zhu Y, Xu G, Yang YT, Xu Z, Chen X, Shi B, et al. POSTAR2: deciphering the post-transcriptional regulatory logics. Nucleic Acids Res. 2019;47(D1):D203-D211. [DOI] [PMC free article] [PubMed]
- 19.Zhu H, Blake S, Chan KT, Pearson RB, Kang J. Cystathionine β-Synthase in Physiology and Cancer. Biomed Res Int. 2018;2018:3205125. [DOI] [PMC free article] [PubMed]
- 20.Phillips C.M., Zatarain J.R., Nicholls M.E., Porter C., Widen S.G., Thanki K., et al. Upregulation of Cystathionine-β-Synthase in Colonic Epithelia Reprograms Metabolism and Promotes Carcinogenesis. Cancer Res. 2017;77(21):5741–5754. doi: 10.1158/0008-5472.CAN-16-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao H., Li Q., Wang J., Su X., Ng K.M., Qiu T., et al. Frequent Epigenetic Silencing of the Folate-Metabolising Gene Cystathionine-Beta-Synthase in Gastrointestinal Cancer. PLoS ONE. 2012;7(11):e49683. doi: 10.1371/journal.pone.0049683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fry N.J., Law B.A., Ilkayeva O.R., Holley C.L., Mansfield K.D. N6-methyladenosine is required for the hypoxic stabilization of specific mRNAs. RNA. 2017;23(9):1444–1455. doi: 10.1261/rna.061044.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou J., Zhang H.e., Liu J., Zhao Z., Wang J., Lu Z., et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol Cancer. 2019;18(1) doi: 10.1186/s12943-019-1082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang C., Samanta D., Lu H., Bullen J.W., Zhang H., Chen I., et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m⁶A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. 2016;113(14):E2047–E2056. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen K, Wei Z, Zhang Q, Wu X, Rong R, Lu Z, et al. WHISTLE: a high-accuracy map of the human N6-methyladenosine (m6A) epitranscriptome predicted using a machine learning approach. Nucleic Acids Res. 2019;47(7):e41. [DOI] [PMC free article] [PubMed]
- 26.Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155(4):793-806. [DOI] [PubMed]
- 27.Zhu T., Roundtree I.A., Wang P., Wang X., Wang L.i., Sun C., et al. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 2014;24(12):1493–1496. doi: 10.1038/cr.2014.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin H., Zhang X., Yang P., Zhang X., Peng Y., Li D.a., et al. RNA m6A methylation orchestrates cancer growth and metastasis via macrophage reprogramming. Nat Commun. 2021;12(1) doi: 10.1038/s41467-021-21514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayano M., Yang W.S., Corn C.K., Pagano N.C., Stockwell B.R. Loss of cysteinyl-tRNA synthetase (CARS) induces the transsulfuration pathway and inhibits ferroptosis induced by cystine deprivation. Cell Death Differ. 2016;23(2):270–278. doi: 10.1038/cdd.2015.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L.i., Cai H., Hu Y., Liu F., Huang S., Zhou Y., et al. A pharmacological probe identifies cystathionine β-synthase as a new negative regulator for ferroptosis. Cell Death Dis. 2018;9(10) doi: 10.1038/s41419-018-1063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abeles R.H., Walsh C.T. Acetylenic enzyme inactivators. Inactivation of gamma-cystathionase, in vitro and in vivo, by propargylglycine. J Am Chem Soc. 1973;95(18):6124–6125. doi: 10.1021/ja00799a053. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto T., Takano N., Ishiwata K., Ohmura M., Nagahata Y., Matsuura T., et al. Reduced methylation of PFKFB3 in cancer cells shunts glucose towards the pentose phosphate pathway. Nat Commun. 2014;5(1) doi: 10.1038/ncomms4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang X., Stockwell B.R., Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22(4):266–282. doi: 10.1038/s41580-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doll S., Proneth B., Tyurina Y.Y., Panzilius E., Kobayashi S., Ingold I., et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol. 2017;13(1):91–98. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng J., Conrad M. The Metabolic Underpinnings of Ferroptosis. Cell Metab. 2020;32(6):920–937. doi: 10.1016/j.cmet.2020.10.011. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto T., Takano N., Ishiwata K., Suematsu M. Carbon monoxide stimulates global protein methylation via its inhibitory action on cystathionine β-synthase. J Clin Biochem Nutr. 2010;48(1):96–100. doi: 10.3164/jcbn.11-011FR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang M., Chen P., Liu J., Zhu S., Kroemer G., Klionsky D.J., et al. Clockophagy is a novel selective autophagy process favoring ferroptosis. Sci Adv. 2019;5(7):eaaw2238. doi: 10.1126/sciadv.aaw2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z., Jiang L.i., Chew S.H., Hirayama T., Sekido Y., Toyokuni S. Carbonic anhydrase 9 confers resistance to ferroptosis/apoptosis in malignant mesothelioma under hypoxia. Redox Biol. 2019;26:101297. doi: 10.1016/j.redox.2019.101297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lan Q., Liu P.Y., Haase J., Bell J.L., Hüttelmaier S., Liu T. The Critical Role of RNA m6A Methylation in Cancer. Cancer Res. 2019;79(7):1285–1292. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 40.Szabo C., Coletta C., Chao C., Módis K., Szczesny B., Papapetropoulos A., et al. Tumor-derived hydrogen sulfide, produced by cystathionine-β-synthase, stimulates bioenergetics, cell proliferation, and angiogenesis in colon cancer. Proc Natl Acad Sci U S A. 2013;110(30):12474–12479. doi: 10.1073/pnas.1306241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takano N., Sarfraz Y., Gilkes D.M., Chaturvedi P., Xiang L., Suematsu M., et al. Decreased expression of cystathionine β-synthase promotes glioma tumorigenesis. Mol Cancer Res. 2014;12(10):1398–1406. doi: 10.1158/1541-7786.MCR-14-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan H.-f., Zou T., Tuo Q.-Z., Xu S., Li H., Belaidi A.A., et al. Ferroptosis: mechanisms and links with diseases. Signal Transduct Target Ther. 2021;6(1) doi: 10.1038/s41392-020-00428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lei G., Zhang Y., Koppula P., Liu X., Zhang J., Lin S.H., et al. The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell Res. 2020;30(2):146–162. doi: 10.1038/s41422-019-0263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen X, Kang R, Kroemer G, Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol. 2021 Jan 29. doi: 10.1038/s41571-020-00462-0. Epub ahead of print. [DOI] [PubMed]
- 45.Yao X, Yang P, Jin Z, Jiang Q, Guo R, Xie R, et al. Multifunctional nanoplatform for photoacoustic imaging-guided combined therapy enhanced by CO induced ferroptosis. Biomaterials. 2019;197:268-283. [DOI] [PubMed]
- 46.Koppula P, Zhuang L, Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell. 2020 Oct 1. doi: 10.1007/s13238-020-00789-5. Epub ahead of print. PMID: 33000412. [DOI] [PMC free article] [PubMed]
- 47.Liu X, Olszewski K, Zhang Y, Lim EW, Shi J, Zhang X, et al. Cystine transporter regulation of pentose phosphate pathway dependency and disulfide stress exposes a targetable metabolic vulnerability in cancer. Nat Cell Biol. 2020;22(4):476-486. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included either in this article or in the supplementary information files. The lncRNA microarray and RNA-seq datasets were deposited in Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), accession numbers: GSE114083 and GSE114360.