Summary
Pluripotent stem cell (PSC)-derived hepatocyte-like cells (HLCs) have shown great potential as an alternative to primary human hepatocytes (PHHs) for in vitro modeling. Several differentiation protocols have been described to direct PSCs toward the hepatic fate. Here, by leveraging recent knowledge of the signaling pathways involved in liver development, we describe a robust, scalable protocol that allowed us to consistently generate high-quality bipotent human hepatoblasts and HLCs from both embryonic stem cells and induced PSC (iPSCs). Although not yet fully mature, such HLCs were more similar to adult PHHs than were cells obtained with previously described protocols, showing good potential as a physiologically representative alternative to PHHs for in vitro modeling. PSC-derived hepatoblasts effectively generated with this protocol could differentiate into mature hepatocytes and cholangiocytes within syngeneic liver organoids, thus opening the way for representative human 3D in vitro modeling of liver development and pathophysiology.
Keywords: pluripotent stem cells, induced pluripotent stem cells, differentiation, hepatoblasts, hepatocyte-like cells, liver development, liver organoids, human, disease modeling, drug discovery
Highlights
-
•
We generated human hepatoblasts and hepatocyte-like cells (HLCs) from pluripotent stem cells
-
•
Timed action on Wnt/β-catenin and TGFβ pathways improved maturity and yield of HLCs
-
•
Hepatoblasts matured into hepatocytes and bile ducts within complex liver organoids
-
•
The protocol is robust and showed potential for scalability and drug testing
The authors describe a new robust differentiation protocol to generate human hepatoblasts and hepatocyte-like cells (HLCs) from pluripotent stem cells. By leveraging the latest knowledge about the signaling pathways involved in liver development, they managed to improve maturity and yield of the differentiation, thus obtaining cells that show great potential to model and study human liver development, physiology, and disease both in 2D and 3D.
Introduction
For decades, the study of human liver physiology and development, as well as drug discovery, have been limited by the lack of representative models. Animal studies and in vitro models based on tumor cell lines proved to be crucial to build our current knowledge but are inadequate to assess the specificities of the human liver. Primary human hepatocytes (PHHs) suffer from major limitations that have hampered our ability to generate representative data. Although considered the “gold standard” by regulatory agencies, PHHs do not allow for a reliable prediction of a drug’s metabolism and toxicity in patients, which results in a high failure rate at clinical phases and is partially responsible for the high costs and long times of drug development (Kola and Landis, 2004).
Stem-cell-derived hepatocyte-like cells (HLCs) have shown great potential as an alternative to PHHs. Pluripotent stem cells (PSCs), such as embryonic stem cells (ESCs) and induced PSCs (iPSCs), represent an unlimited source from which HLCs can be produced. Various differentiation protocols inspired by liver organogenesis have been described for both ESCs and iPSCs (Mallanna and Duncan, 2013; Shan et al., 2013; Si-Tayeb et al., 2010a; Touboul et al., 2010). Empirical identification of best culture conditions has historically played a central role in establishing such protocols. Our knowledge of liver development has greatly advanced over the last decade, and we now have a reasonable understanding of the signaling pathways involved (Gordillo et al., 2015; Si-Tayeb et al., 2010b). This has helped to improve the quality of PSC-derived HLCs (Fu et al., 2019; Gao et al., 2017; Takayama et al., 2017; Touboul et al., 2016; Xie et al., 2019). Nevertheless, obtained HLCs are still closer to fetal PHHs than to adult PHHs (Baxter et al., 2015). Moreover, the quality of differentiation varies widely among HLCs obtained from different PSC populations, and cell yield is often low with most of described protocols (Kajiwara et al., 2012). Over the last few years, liver organoids that take advantage of cell-to-cell interactions and paracrine signals among the several cellular components of the liver “niche” have been described and are increasingly used for in vitro modeling (Asai et al., 2017; Takebe et al., 2013). Nevertheless, the representativeness of such 3D models relies on the quality and maturation of the liver cells used. Overall, the limitations of currently available protocols constitute a significant barrier to the full implementation of HLCs for in vitro modeling, drug testing, and cell therapy applications (Paganelli, 2019). By leveraging the most recent knowledge about the signaling pathways involved in liver development and their timely acting on Wnt/β-catenin, oncostatin M (OSM), and transforming growth factor beta (TGFβ) pathways during the later stages of differentiation, we developed a new, robust protocol allowing for the high-yield, consistent generation of high-quality bipotent human liver progenitors and HLCs from both ESCs and iPSCs.
Results
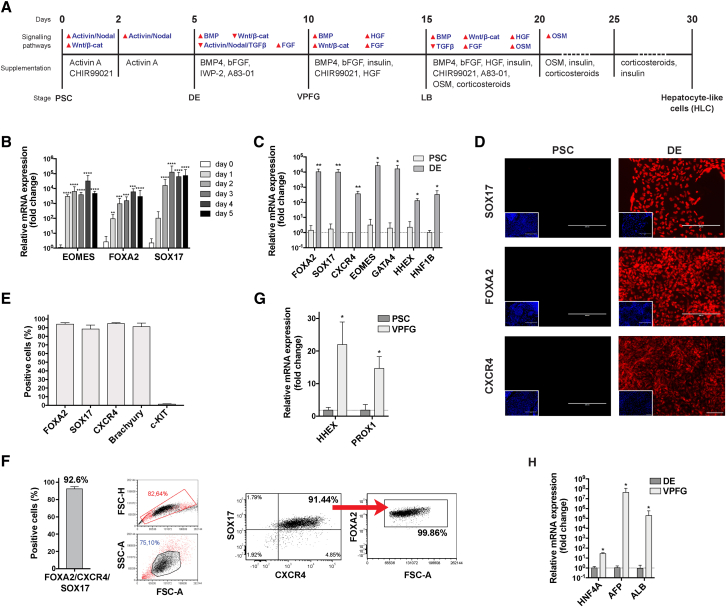
We used 6 different PSC populations (one ESC line and five iPSC lines) and differentiated them through the main developmental stages that lead from the zygote to the fetal liver following an optimized protocol based on an exhaustive review of liver development (Figure 1A).
Figure 1.
Differentiation prototocol and PSC-derived definitive endoderm and posterior foregut
(A) Description of our new differentiation protocol.
(B–F) PSC-derived endoderm (DE).
(B) Expression of EOMES, FOXA2, and SOX17 genes over the first 5 days of differentiation (qRT-PCR, expressed as log10 mean fold change relative to PSCs ± SEM, n = 9, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
(C) Expression of endoderm-enriched genes in PSC-derived cells after 5 days of differentiation compared with undifferentiated PSCs (qRT-PCR, expressed as log10 mean fold change relative to DE ± SEM; n = 3 for PSCs, n = 6 for DEs, ∗p < 0.05, ∗∗p < 0.01; see also Figure S2A).
(D) Expression of DE markers at the end of day 5 of differentiation compared with undifferentiated PSCs (immunofluorescence, representative images, 4′,6-diamidino-2-phenylindole (DAPI) nuclear staining in included images, scale bar, 200 μm).
(E) Expression of DE markers is highly homogeneous in PSC-derived DE cells (flow cytometry, n ≥ 4, mean ± SEM).
(F) FOXA2/CXCR4/SOX17 triple-positive cells constitute >90% of PSC-derived DEs (flow cytometry, n = 4; left: mean ± SEM, center and right: representative experiment).
(G and H) PSC-derived ventral posterior foregut (VPFG): expression of HHEX, PROX1, HNF4A, AFP, and ALB genes in PSC-derived cells (qRT-PCR expressed as log10 mean fold change ± SEM, relative to: G, undifferentiated PSCs and H, PSC-derived DEs; n = 3 for PSCs, n ≥ 3 for DE, n ≥ 5 for VPFG; ∗p < 0.05, ∗∗∗p < 0.001; see also Figures S2E and S2F).
Hepatic specification
After careful characterization of PSCs (Figure S1), we recreated the process of gastrulation in vitro, passing by a primitive streak stage and then pushing Brachyury (T)-positive cells toward the mesendoderm through the epithelial-mesenchymal transition by concurrently activating both the Activin/Nodal signaling (with Activin A) and canonical Wnt/β-catenin pathways (via the selective inhibition of GSK3B with CHIR99021). This led to the timed upregulation of EOMES and FOXA2 (24 h) and SOX17 (48 h; Figure 1B). Stimulation of the Activin/Nodal pathway for 3 additional days allowed the cells to acquire the gene and protein expression profile typical of the definitive endoderm (DE; Figures 1C and 1D). The obtained DE population was highly homogenous, with >90% of FOXA2-, SOX17-, and CXCR4-positive cells (Figures 1E and 1F). The addition of a knockout serum replacement (KOSR) at a low concentration, which, as fibroblast grow factor (FGF) secreted by the cardiac mesoderm, induces hepatic specification through the mitogen-activated protein kinase (MAPK) signaling pathway, significantly reduced cell death without affecting the quality of the obtained DE (Figures S2B and S2C). No difference was seen in the results obtained from applying this protocol to ESC or iPSC populations (Figure S2D).
In the embryo, the DE is then exposed to bone morphogenetic protein (BMP) from the septum transversum mesenchyme (STM) and to FGF from the cardiac mesoderm (Gordillo et al., 2015; Si-Tayeb et al., 2010b). This drives the cells toward the formation of the posterior foregut. Inhibition of the Wnt signaling pathway is needed to relieve the repression of HHEX, which is required for the commitment to the hepatic fate (McLean et al., 2007). TGFβ inhibition allows for the upregulation of albumin (ALB) and PROX1 genes, with the latter playing an essential role in subsequent liver bud formation (Sosa-Pineda et al., 2000). After a 5-day exposure to BMP4, low-dose FGF, A83-01 (TGFβ pathway inhibitor), and IWP2 (Wnt pathway inhibitor), the cells acquired a phenotype suggestive of the commitment of the ventral posterior foregut (VPFG) toward the hepatic fate (corresponding to mouse embryonic day 8.5 [E8.5]), with an overexpression of HHEX, PROX1 (Figure 1G), HNF4A, alpha fetoprotein (AFP), and ALB genes (Figure 1H).
Liver bud formation and expansion
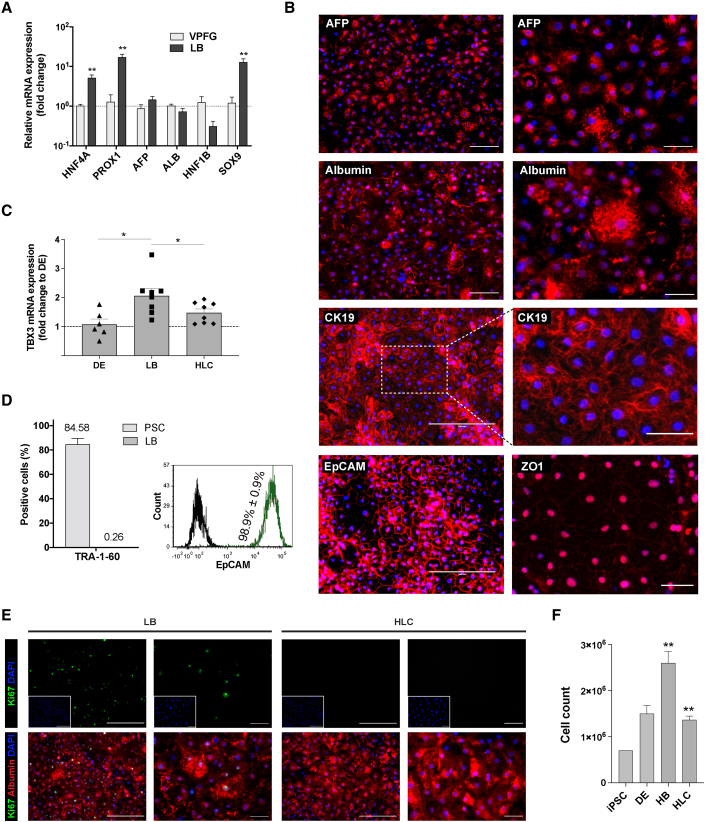
In humans 9 days post-gastrulation, hepatic progenitor cells from the VPFG delaminate and invade the surrounding stroma, forming the liver bud (LB). Hence, liver progenitors (hepatoblasts) enter into contact with the STM, which continues secreting BMP, and are in close proximity with sinusoidal endothelial cells and stellate cells secreting Wnt and hepatocyte growth factor (HGF). Supplementation of hepatoblasts with BMP4, FGF, HGF, and CHIR99021, in the presence of insulin, for 5 days (Figure 1A) triggered the re-establishment of cell-cell contacts and a deep commitment toward the hepatic fate. At the end of this stage, such LB cells further increased the expression of transcription factors HNF4A and of PROX1, which are both required for their proliferation and differentiation (Figure 2A; Parviz et al., 2003; Sosa-Pineda et al., 2000). ALB and AFP continued to be very highly expressed in LB cells. The expression of HNF1B, which at this stage controls cholangiocyte fate, was instead reduced, while the expression of SRY-box 9 (SOX9) was significantly increased in LB cells, which is consistent with the expression of these genes in bipotent hepatic progenitor cells (Figure 2A; Han et al., 2019). Immunostaining showed that 79.1% ± 13.4% of LB cells were AFP-positive and that 80.4% ± 10% were albumin-positive (Figure 2B). The expression of cytokeratin 19 (CK19 or KRT19) and epithelial cell adhesion molecule (EpCAM) was ubiquitous at this stage (Figures 2B and 2D). Zonula occludens-1 (ZO-1) expression at the cell surface confirmed the re-establishment of cell-cell contacts (Figure 2B). Expression of the hepatoblast-enriched TBX3 gene increased in LB cells and subsequently decreased over further maturation into HLCs (Figure 2C). The number of cells still expressing pluripotency markers was negligible at this stage (Figure 2D). The staged activation of HHEX, HNF4A, and PROX1 also led to the proliferation of hepatoblasts (38.6% ± 5.5% of Ki67-positive cells; Figures 2E and 2F), in line with the behavior of the LB at this stage of development (days 24–48 of gestation, corresponding to E10–E14 in mouse; Sosa-Pineda et al., 2000). No difference between ESCs and iPSCs was noted (Figure S2I).
Figure 2.
PSC-derived liver bud (LB)
(A) PSC-derived cells after 15 days of differentiation achieve a gene expression profile consistent with hepatoblasts in the forming LB (qRT-PCR, expressed as log10 mean fold change relative to VPFG ± SEM; n = 3 for VPFG, n = 8 for DE, ∗∗p < 0.01; see also Figure S2G).
(B) Expression of hepatoblast-enriched AFP, albumin, CK19, EpCAM, and ZO-1 in PSC-derived LB cells (immunofluorescence, representative images with DAPI nuclear staining, scale bars, 200 μm, left, and 50 μm, right).
(C) Expression of hepatoblast-enriched TBX3 increases in LB cells and decreases upon further maturation into HLCs (qRT-PCR, expressed as fold change relative to DE ± SEM; n = 8, ∗p < 0.05).
(D) LB cells show high homogeneity for EpCAM expression, while expression of a marker of pluripotency is negligible and comparable to background noise (flow cytometry; TRA-1-60: n = 6 for PSCs, n = 3 for LB, mean ± SEM; EpCAM: n = 5, mean ± SEM, representative experiment).
(E) LB cells replicate actively (left panels), whereas HLCs reach a quiescent state (Ki67-albumin fluorescence co-staining, representative images, DAPI nuclear staining in included images, scale bars, 200 μm, left panels of each group, and 50 μm, right panels).
(F) The differentiation protocol allows for the expansion of LB cells (automated cell count, n = 6 for PSCs and LB, n = 3 for DE, n = 4 for HLCs; mean ± SEM, ∗∗p < 0.01).
Maturation into HLCs
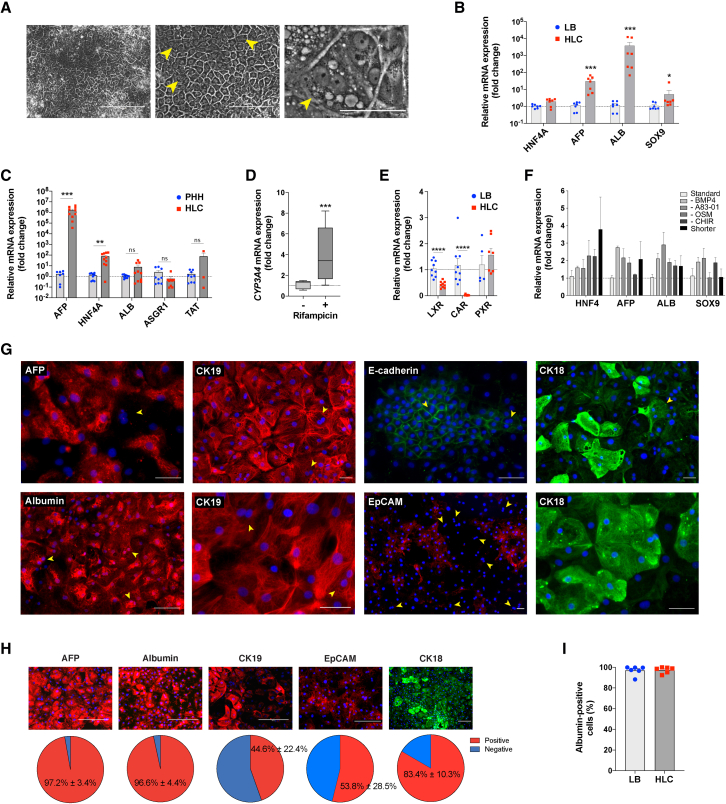
In the developing human embryo, starting at around gestational day 57 (E13.5 in mouse), the close contact between hepatoblasts and hepatic stellate cells (secreting Wnt, HGF, and FGF) and hematopoietic cells (producing OSM) drives their maturation into hepatocytes (Kamiya et al., 1999; Matsumoto et al., 2008; Onitsuka et al., 2010; Schmidt et al., 1995). Mature liver functions are then progressively acquired, with the liver expressing a fully mature phenotype only several weeks after birth. We exposed LB cells to OSM and dexamethasone, in addition to the stimulation of BMP, FGF, HGF, and Wnt signaling pathways and the inhibition of the TGFβ pathway, for 5 days to induce maturation into HLCs (Figure 1A). Subsequently, we maintained only OSM and dexamethasone for another 5 days and then concluded the differentiation processes with 5 more days without OSM in order to mimic the decrease in the stimulus from hematopoietic cells that happens in the later weeks of gestation and post-natal life (Kamiya and Gonzalez, 2004; Khan et al., 2016). At the end of the differentiation process, we obtained a homogeneous monolayer of mono- and binucleated polygonal epithelial cells (Figures 3A and 3G). Unlike LB cells, such PSC-derived HLCs did not actively proliferate (Figure 2E), which was consistent with the achievement of quiescence expected upon hepatocyte maturation (Berasain and Avila, 2015).
Figure 3.
Characterization of PSC-derived HLCs
(A) Representative morphology at the end of the differentiation protocol (phase contrast; arrowheads showing binucleated hepatocytes; scale bars, 200 μm, left, 100 μm, center, and 50 μm, right).
(B) Expression of liver-specific genes compared with LB cells (qRT-PCR, expressed as log10 mean fold change relative to LB ± SEM; n = 7, ∗p < 0.05, ∗∗∗p < 0.001; see also Figure S3C).
(C) Expression of liver-specific genes compared with adult PHHs (qRT-PCR, expressed as log10 mean fold change relative to PHH ± SEM; n = 9 for PHHs, n = 10 for HLCs; ∗∗p < 0.01, ∗∗∗p < 0.001, not significant [ns] p ≥ 0.05; see also Figure S3D).
(D) CYP3A4 expression and induction upon supplementation with 20 μM rifampicin for 72 h (qRT-PCR, mean fold change relative to baseline ± SEM; n = 9, ∗∗∗p < 0.001).
(E) Expression of liver-specific genes LXR, CAR, and PXR in HLCs compared with LB cells (qRT-PCR, log10 mean fold change relative to LB ± SEM; n = 9 for LXR and CAR, n = 5 for PXR, ∗∗∗∗p < 0.0001).
(F) Effect of different maturation conditions between days 16 and 30 on the expression of liver-specific genes by HLCs (qRT-PCR, mean fold change relative to the protocol shown in Figure 1A ± SEM; n = 3; p > 0.05 for all conditions; “shorter” corresponds to 3 days per step instead of 5).
(G) Expression of AFP, albumin, CK19, CK18, E-cadherin, and EpCAM in PSC-derived HLCs (immunofluorescence, representative images, DAPI nuclear staining; yellow arrowheads showing binucleated hepatocytes; scale bar, 50 μm).
(H) Percentage of HLCs expressing AFP, albumin, CK19, EpCAM, and CK18 (immunofluorescence, representative images, DAPI nuclear staining; scale bar, 200 μm; bottom: percentage of positive cells compared with DAPI-positive nuclei, n = 9, mean ± SEM).
(I) HLCs were highly homogeneous for albumin expression (flow cytometry; n = 6, mean ± SEM; p > 0.05 when HLCs were compared with LB cells; see also Figures S3E and S3F).
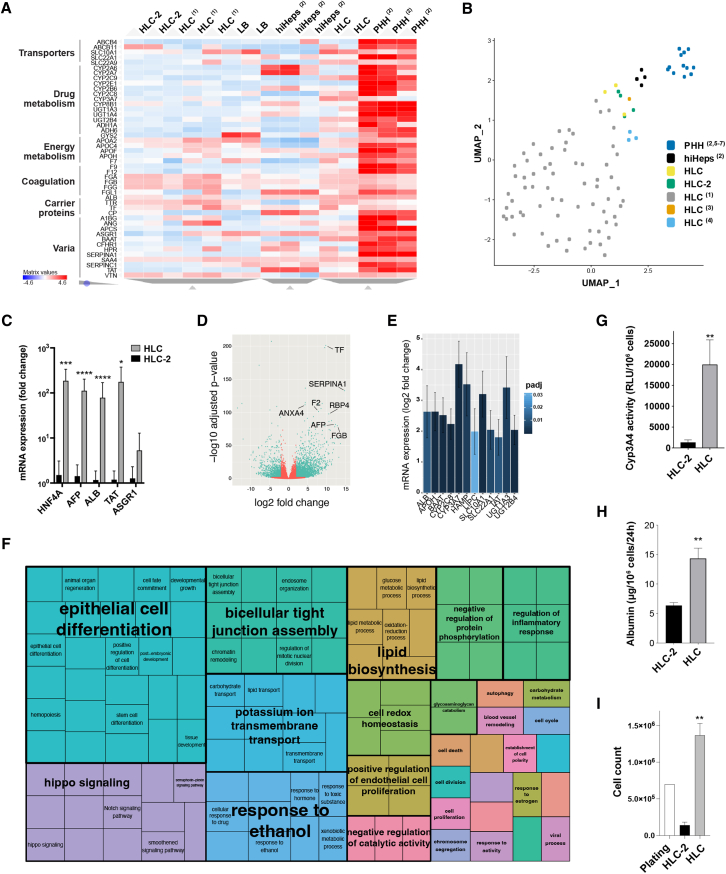
HLCs showed an important increase in the expression of ALB (>1,000-fold change) and AFP (>10-fold change) compared with that in LB hepatoblasts (Figure 3B). The expression of mature liver-specific genes ALB, ASGR1, and TAT in HLCs was comparable to PHHs (Figure 3C), with HNF4A and AFP being more highly expressed. The expression of CYP3A4 was shown to be inducible upon supplementation with rifampicin (Figure 3D). Liver-specific liver X receptor (LXRα or NR1H3), which is upregulated early during liver development and decreases during fetal life, was highly expressed in hepatoblasts and was decreased in HLCs (Figure 3E; Mu et al., 2020). Similarly, the expression of constitutive androstane receptor (CAR or NR1I3), which is known to increase over the first and the beginning of the second gestational trimesters and subsequently decrease, was high in LB cells and decreased in HLCs (but, as in the neonatal liver, it is still expressed; Chen et al., 2013). On the contrary, the expression of pregnane X receptor (PXR or NR1I2) increased in HLCs compared with in hepatoblasts (although it did not reach statistical significance), as observed during later stages of liver development because of the direct binding of HNF4α (Kamiya et al., 2003; Masuyama et al., 2001). Concurrent inhibition of the TGFβ signaling pathway and activation of the Wnt pathway during hepatoblasts maturation, together with BMP4 and OSM supplementation, was superior to alternative protocols (Figure 3F). Shortening this maturation phase (3 days/step instead of 5) resulted in less mature HLCs (Figure 3F), whereas extending it reduced the yield (data not shown). The differentiation protocol proved to be robust, with negligible inter-population and -operator variability (32 differentiations of 6 PSC populations, 3 different operators; Figure S3A). No difference between ESCs and iPSCs was noted (Figure S3B).
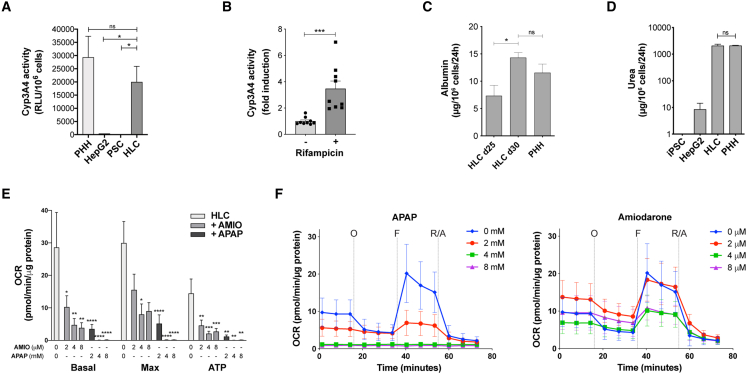
Almost all HLCs expressed albumin, E-cadherin, and AFP, although the latter was not expressed by larger, binucleated cells (Figures 3G–3I). CK18 (KRT18) was expressed by >80% of the cells. Expression of CK19 and EpCAM was still strong but reduced compared with in LB cells (45% and 54% of the HLCs being positive for each protein, respectively) and reduced or absent in larger, binucleated cells. HLCs effectively performed mature liver functions at levels comparable to metabolism-qualified adult PHHs (Figures 4A–4D). The activity of cytochrome P450 3A4 (Cyp3A4) was almost 50 times greater than the commonly used HepG2 cell line (Figure 4A) and was inducible upon supplementation with rifampicin (Figure 4B). Albumin secretion progressively increased in HLCs over maturation, reaching levels comparable to PHHs (Figure 4C). Similarly, HLCs produced urea as effectively as PHH and >100 times more effectively than HepG2 cells (Figure 4D).
Figure 4.
Liver-specific functions performed by HLCs
(A and B) Cyp3A4 activity performed by HLCs in comparison to PHHs and HepG2 cells (A; relative light units per million cells; n = 6 HLCs, n = 7 PHHs, n = 5 PSCs, n = 3 HepG2 cells) and its induction upon supplementation with 20 μM rifampicin for 72 h (B; n = 9; mean ± SEM, ∗p < 0.05, ∗∗∗p < 0.001).
(C) Albumin secretion in HLCs at days 25 and 30 compared with PHHs (ELISA, n = 4 for HLCs and PHHs, mean ± SEM).
(D) Urea production by HLCs compared with PHHs and HepG2 cells (ELISA, n = 3 for HLCs, PSCs, and HepG2 cells, n = 12 for PHHs, mean ± SEM).
(E and F) Hepatotoxicity of acetaminophen (APAP) and amiodarone (AMIO) assessed through the measurement of oxygen consumption rate on HLCs (O, oligomycin; F, FCCP; R/A, Rotenone/Antimycin A; n = 12, mean ± SEM).
As a proof of concept, we used such HLCs to assess hepatotoxicity of two common drugs with a known dose-dependent effect on hepatocytes (Figures 4E and 4F). Mitochondrial dysfunction has emerged as an important mechanism and indicator of drug-induced hepatotoxicity (Ramachandran et al., 2018). By measuring their mitochondrial respiration (oxygen consumption rate [OCR]), we showed that HLCs allow for a representative assessment of the toxic effect of acetaminophen and amiodarone on hepatocytes’ metabolism, with very little variability.
When compared with previously published protocols for which a transcriptome was available (Fu et al., 2019; Gao et al., 2017; Kim et al., 2019; Li et al., 2017; Touboul et al., 2016; Warren et al., 2017; Xie et al., 2019), our new protocol resulted in significantly more mature HLCs (Figures 5 and S4). Our HLCs significantly overexpressed key liver-specific genes compared with other previously described HLCs and resulted in expression profiles more similar to freshly isolated PHH and adult liver samples (Figures 4A, 4B, and S4B). When compared with HLCs obtained with a differentiation protocol not acting on the Wnt/β-catenin and TGFβ pathways beyond the DE stage (Figure S3G), our HLCs showed a 3- to 100-fold higher expression of genes encoding for albumin, HNF4α, AFP, tyrosine aminotransferase, and enzymes involved in xenobiotic metabolism, iron homeostasis, and lipid and bile acid metabolism (Figures 5C–5E). Analysis of Gene Ontology terms (Figure 5F) confirmed the overexpression of superclusters of genes that are essential for the development (epithelial cell differentiation), structural integrity (bicellular tight junction assembly, regulation of endothelial cell proliferation), and function of the human liver (lipid biosynthesis, response to ethanol and drugs, transmembrane transport, cell redox homeostasis, carbohydrate metabolism, etc.). The advantage of the timed regulation of Wnt/β-catenin and TGFβ signaling pathways was even more evident when measuring mature liver functions, with a 10-fold increase in Cyp3A4 activity (Figure 5G) and >2.5-fold increase in albumin secretion (Figure 5H). Moreover, our new protocol allowed us to obtain almost 10 times the HLCs generated by a differentiation protocol not acting on the Wnt/β-catenin and TGFβ pathways beyond the DE stage (Figure 5I), as expected from the relative upregulation of Hippo signaling pathways (Figure 5F), the expression of which is similar to PHHs (Figure S4C; Patel et al., 2017; Valizadeh et al., 2019). Nevertheless, YAP (YAP1) was downregulated in HLCs when compared with LB cells (Figure S4D), which was consistent with their acquired quiescent state and resulted in a relative reduction in the cell number at the end of the differentiation (Figure 2F), all while overall allowing for a 2-fold expansion of seeded cells.
Figure 5.
Comparison of our HLCs with cells obtained with previously described protocols, PHHs, and adult liver samples
(A) Heatmap showing the expression of the top 46 liver-enriched genes in HLCs obtained with our protocol compared with LB cells, HLCs obtained with a protocol not acting on Wnt and TGFβ signaling pathways beyond the DE stage (HLC-2; see Figure S3G), HLCs obtained with other previously described protocols (1: Warren et al., 2017), hepatocytes derived by transdifferentiation (hiHeps; 2: Gao et al., 2017), and PHHs (RNA-seq, unsupervised clustering; 2: Gao et al., 2017; list of genes from the Human Protein Atlas; Uhlén et al., 2015). Transcriptome profile through the differentiation stages and comparison of the HLCs with adult liver samples is shown in Figures S4A and S4B.
(B) Our HLCs are more similar to freshly isolated adult PHHs than most HLCs obtained with previously described protocols (1: Warren et al., 2017; 2: Gao et al., 2017; 3: Li et al., 2017; 4: Touboul et al., 2016; 5: Xie et al., 2019; 6: Fu et al., 2019; 7: Kim et al., 2019). 2D representation of Uniform Manifold Approximation and Projection (UMAP)-based dimensionality reduction of the top 17 principal components obtained analyzing the 3,000 most variable genes across datasets (Dorrity et al., 2020).
(C) Liver-specific genes are overexpressed in HLCs obtained with our new protocol compared with HLC-2 (Figure S3G; qRT-PCR, log10 mean fold change relative to HLC-2 ± SEM, n = 9, ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
(D) Volcano plot showing the most variable genes in our HLCs compared with HLC-2 (n = 2).
(E) Representative sample of liver-enriched genes overexpressed in HLCs obtained with the new protocol compared with HLC-2 (differential gene expression; >3-fold change, p adjusted < 0.05; n = 2).
(F) Semantic similarity-based scatterplot of Gene Ontology terms showing families of genes that are overexpressed in HLCs compared with HLC-2 (REVIGO tree map, n = 2, p adjusted < 0.05).
(G) HLCs obtained with our new protocol show significantly more Cyp3A4 activity than HLC-2 (relative light units per million cells; n = 6 HLCs, n = 4 HLC-2; mean ± SEM, ∗∗p < 0.01).
(H) Our HLCs secrete more albumin per million cells than HLC-2 (ELISA, n = 4 for HLCs, n = 10 for HLC-2; mean ± SEM, ∗∗p < 0.01).
(I) The new differentiation protocol allows for generating HLCs with significantly better efficiency (automated cell count, n = 4; mean ± SEM, ∗p < 0.05).
See also Figures S4C and S4D.
Overall, our new differentiation protocol allowed us to consistently generate more mature HLCs, from both ESCs and iPSCs, with good yield and using reagents that have Good Manufacturing Practice (GMP)-compliant versions available, compared with previously described approaches. Designed for robustness and ease of clinical translation, the protocol has been licensed to a regenerative medicine company, validated by 3 more independent operators, and successfully tech-transferred to an independent contract development and manufacturing organization (CDMO) for scale-up and optimization.
Bipotent hepatic progenitors give rise to hepatocytes and biliary cells in liver organoids
To further prove the bipotency and differentiation potential of iPSC-derived hepatoblasts obtained with our new protocol, as well as to assess the possible applications of such cells to study liver physiology and development and for disease modeling, we generated complex, syngeneic liver organoids derived from iPSCs.
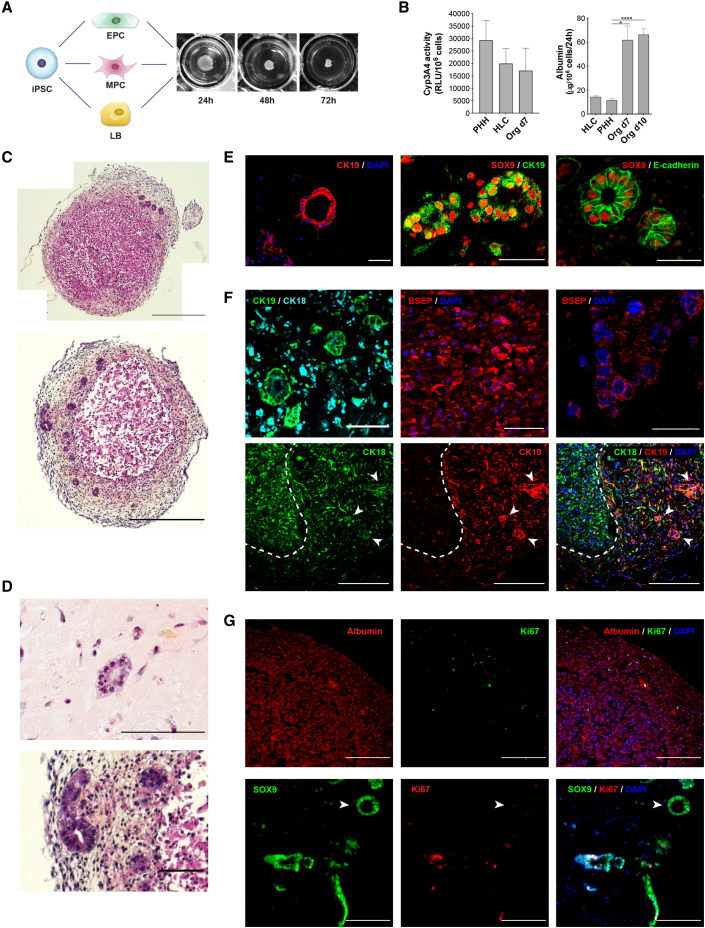
We differentiated each human iPSC population into mesenchymal progenitor cells (MPCs) and endothelial progenitor cells (EPCs). MPCs fulfilled the defining criteria of mesenchymal stromal cells in terms of morphology, surface markers, and multipotency (Figure S5; Dominici et al., 2006). EPCs showed features suggestive of endothelial colony-forming cells (Figure S5). Through the seeding of iPSC-derived LB cells (hepatoblasts at day 16 of the differentiation protocol shown in Figure 1A) with EPCs and MPCs derived from the same iPSC line, in a defined ratio (Takebe et al., 2013), we recreated favorable conditions for self-aggregation and the formation of complex, syngeneic liver organoids (Figure 6A). These organoids formed with dimensions of 1.1 ± 0.2 mm in diameter and allowed for structural and histologic studies after 72 h. Generation of the organoids in suspension, without the use of hydrogels, minimized variability and facilitated processing.
Figure 6.
Complex, syngeneic liver organoids generated with bipotent PSC-derived hepatoblasts
(A) Schematic representation and macroscopic appearance organoids generated with syngeneic, PSC-derived LB cells (hepatoblasts) and mesenchymal and endothelial progenitor cells (MPCs and EPCs, respectively; characterization in Figure S5).
(B) Liver-specific functions performed by the organoids: cyp3A4 activity at day 7 (relative light units per million cells compared with PHHs and HLCs; n = 6 HLCs, n = 7 PHHs, n = 3 organoids; mean ± SEM); albumin secretion at day 7 and 10 compared with PHHs and HLCs (ELISA, n = 4 for HLCs and PHHs, n = 3 organoids day 7, n = 6 organoids day 10; mean ± SEM, ∗p < 0.05, ∗∗∗∗p < 0.0001).
(C) Histological appearance of liver organoids at day 7 showing cords of small hepatocytes at the center, and bile ducts surrounded by extracellular matrix in the outer layer of the structure (H&E staining, representative images, scale bar, 500 μm).
(D and E) Mature bile ducts within the organoids: a single layer of CK19-positive, SOX9-positive cholangiocytes delimiting a lumen surrounded and by the extracellular matrix (D: H&E staining, representative image, scale bar, 100 μm; E, immunofluorescence, scale bar, 50 μm).
(F) Within the organoids, LB cells differentiate into CK18-positive, CK19-negative, BSEP-positive hepatocytes mostly located at the center of the organoids (white dashed line) and CK19- and CK18-positive cholangiocytes forming bile ducts (white arrowheads; immunofluorescence, scale bars, 20 μm, top left and right, 50 μm, top center, and 200 μm, bottom; additional staining in Figure S6).
(G) Within the organoids, hepatocytes reach a state of quiescence (Ki67-negative). Cholangiocytes form bile ducts at different stages of maturation, with immature ducts still showing cell replication and mature ones (arrowhead) being Ki67-negative (immunofluorescence, representative images, scale bar, 200 μm).
The establishment of cell-to-cell contacts and paracrine signals, together with the supplementation of FGF, insulin, OSM, corticosteroids, and vascular endothelial growth factor (VEFG), allowed the rapid maturation of hepatoblasts into hepatocytes performing liver-specific functions (Figure 6B). Seven days after seeding, the organoids were comparable to day-30 HLCs in terms of Cyp3A4 activity, while they secreted significantly more albumin, reaching levels superior to PHHs. Liver functions were maintained until day 10 and then rapidly decreased following the progressive death of liver cells and collagen deposition seen in the central part of the organoids, presumably due to the lack of vascularization and the poor perfusion caused by the size of the aggregates.
Within the organoids, liver cells organized into cords of small hepatocytes surrounded by significant extracellular matrix produced by the mesenchymal cells (Figure 6C). At day 7 post-seeding, bile ducts were clear within the outer layer of the organoids (Figure 6D). These structures were clearly separated from the hepatocytes and surrounded by the mesenchyme. Many such bile ducts showed mature features, such as a lumen and a single layer of cuboidal CK19- and SOX9-positive cholangiocytes (Figure 6E; Vyas et al., 2018). Many bipotent, CK18/CK19 double-positive hepatoblasts quickly matured into CK18- and bile salt export pump (BSEP or ABCB11)-positive, CK19-negative hepatocytes, which mostly localized in the inner layers of the organoids, and CK19/CK18-positive cholangiocytes, forming bile ducts at different stages of maturation in the outer layer (Figure 6F). Hepatocytes homogenously expressed albumin and did not replicate (Ki67-negative; Figure 6G). They also expressed HNF4α and ASGR1 (Figure S6). HNF4α-, BSEP-, and ASGR1-negative bile ducts at different stages were visible within the organoids (Figure S6), with maturing structures composed of Ki67-positive cholangiocytes and more mature ducts showing no replicative activity (Figure 6G). These data confirm the bipotency of hepatoblasts generated with our new protocol and highlight the potential for their use in 3D modeling of complex human liver disease.
Discussion
The study of human liver pathophysiology has been historically hampered by the lack of suitable in vitro models. Our knowledge of liver development has greatly advanced over the last decade, but most of what is known about hepatogenesis derives from studies carried out in rodents. The unavailability of representative in vitro models is partially responsible for the high failure rate of experimental new drugs (Kola and Landis, 2004). Besides being expensive and of limited availability, PHHs suffer from major limitations that hamper their utility for in vitro modeling and drug development. Their quality is extremely variable between donors, which affects in vitro-in vivo extrapolations of obtained data (Braver-Sewradj et al., 2016; Hallifax and Houston, 2009). Freshly isolated PHHs do not replicate in culture, tend to dedifferentiate rapidly with their metabolic activity waning over a few days, and are logistically difficult to procure and use (Elaut et al., 2006). Cryopreserved PHHs are the tool of choice for drug metabolism studies, but they withstand cryopreservation very poorly, with the majority of lots retaining only some of the functions needed for drug development and only for a short time. All these limitations have major consequences on the consistency and costs of conducted experiments and reduce their value to reliably predict the behavior of the drug in patients (Hallifax and Houston, 2009; Kola and Landis, 2004). Several cell lines are available, but, while valuable to answer specific questions, their less mature state makes them less representative of the human liver. Thus, the ability to consistently generate human hepatoblasts and hepatocytes of predictable quality, allowing for representative in vitro modeling, is considered an unmet need for the study of liver physiology and pathophysiology as well as for drug development.
Over the years, differentiation protocols to generate HLCs from either ESCs or iPSCs have been progressively refined by acting on very diverse signaling pathways. Nevertheless, despite the heterogeneity of the growth factors, small molecules, culture media, and coating compounds used, the full maturity of adult hepatocytes has never been achieved in vitro, and a significant variability in the performance of many protocols across various PSC lines was often observed (Baxter et al., 2015; Kajiwara et al., 2012). Here, we described a new protocol that, through the timed activation and inhibition of key signaling pathways playing a role along liver development, allows for the reproducible generation of high-quality hepatoblasts and HLCs from PSCs with no significant variability when applied to various iPSC and ESC populations.
Our HLCs express most of the liver-specific genes responsible for the organ’s synthetic and metabolic activity and perform significantly better at key mature liver functions than do many previously described differentiation protocols. Advanced dimensionality reduction showed that the transcriptome of our HLCs is more similar to freshly isolated adult PHHs than those obtained with previously described protocols, with only hepatocytes generated through the forced expression of specific hepatocyte transcription factors showing a gene expression profile more similar to PHHs (Gao et al., 2017). Our HLCs achieve a state of quiescence and show Cyp3A4 activity and albumin and urea production capabilities comparable to metabolism-qualified cryopreserved adult PHHs and proved suitable for in vitro drug testing. Although cryopreserved PHHs are known to lose functionality compared with freshly isolated PHHs, they are still considered the industry gold standard to assess candidate new drugs despite the high inter-donor variability and costs (Braver-Sewradj et al., 2016). Nevertheless, despite liver-specific gene expression and functions comparable to cultured PHHs, our HLCs are less mature than hepatocytes within the adult liver, as shown by the persistent expression of AFP and CK19 in a significant number of cells. AFP is still expressed at the mRNA level by most adult hepatocytes (MacParland et al., 2018), but the synthesis of the protein decreases over a few weeks after birth. Among our HLCs, only larger/binucleated cells lose the expression of AFP, CK19, and EpCAM, probably as a consequence of a more advanced stage of maturation. Future assessment by single-cell RNA sequencing will allow for the clarification of the different clusters of hepatocytes composing our HLC population. Incomplete HLC maturation was not surprising, as a 3D culture and an interaction with non-parenchymal cells composing the liver niche are indeed required for hepatocytes to achieve full maturity (Berger et al., 2015; Goulart et al., 2019; Takebe et al., 2013). Complete maturation of stem-cell-derived HLCs alone in monolayer culture would depend on the recreation of gradients of paracrine signals that are still unknown to date, which is unlikely to be achievable without the forced expression of specific transcription factors (Du et al., 2014; Gao et al., 2017; Xie et al., 2019). The importance of 3D cell-cell interactions between hepatocytes and non-parenchymal cells and the inadequacy of available culture conditions is also shown by the rapid dedifferentiation of PHHs upon digestion and plating (Elaut et al., 2006). A culture of our hepatoblasts in 3D together with endothelial cells and MPCs recreated such cell-cell interactions and led to a quicker maturation of liver cells, resulting in hepatocytes losing the expression of bipotency marker CK19 and acquiring a functional performance comparable or superior to PHHs over a shorter time span.
The Wnt/β-catenin signaling pathway plays a crucial role in orchestrating liver development. The effect of its activation in synergy with the Activin/Nodal pathway during the early stages of PSC differentiation have previously been used to obtain primitive streak cells and DEs (Nostro et al., 2011; Teo et al., 2014). Although the role of this pathway during the later stages of development has been well shown in rodents and exploited to generate PSC-derived hepatoblasts, the need for its subsequent inhibition and activation in synergy with BMP, FGF, OSM, and TGFβ signaling modulation has not been exploited to generate HLCs from hepatoblasts (Touboul et al., 2016). We stimulate the Wnt pathway during endoderm formation, inhibit it to foster hepatic specification and LB formation through the activation of HHEX, and then activate it once again to mimic the effect of endothelial cells on the expanding LB and developing ductal plate. Sustained Wnt activation during the maturation of hepatoblasts into HLCs and the timed supplementation of OSM to mimic the physiological role of sinusoidal and hematopoietic cells residing in the liver during fetal life allowed for the better maturation of our HLCs compared with the ESC-derived cells previously described by Touboul et al. (2016). We showed above that alternative maturation cocktails not acting on Wnt, TGFβ, OSM, or BMP signaling pathways resulted in less mature HLCs. Not surprisingly, considering that we tried over 4 weeks to reproduce a process that physiologically takes several months, reducing the duration of the maturation step also affected the quality of the HLCs.
It is a widespread experience that most differentiation protocols have an overall low yield with significant cell death, which increases the costs of differentiation and limits scalability. The activation of the Wnt/β-catenin pathway has also been shown to have an important role in hepatocyte proliferation and might contribute to the good yield of our protocol (Reed et al., 2008). Such a better yield compared with protocols not relying as much on the modulation of the TGFβ and Wnt pathways can also be explained by the modulation of Hippo/YAP signaling shown above. Besides promoting cell proliferation at the hepatoblast stage, the activation of this pathway reduces apoptosis and seems to play a key role in liver regeneration (Patel et al., 2017; Valizadeh et al., 2019). The avoidance of NOTCH inhibition resulted in higher SOX9 expression but had no impact on mature liver function, though it did further support cell proliferation and reduced the cost of differentiation (Touboul et al., 2016). Moreover, we avoid the use of serum and use reagents that are easily transitioned to GMP-compliant versions. This will reduce the times and cost of GMP implementation and further ease the protocol’s translation for potential future therapeutic applications. Successfully tech-transferred to a cell therapy company and a CDMO, the protocol confirmed its robustness, although optimization will be needed to allow for the large-scale production required for meaningful in vitro drug testing and cell therapy applications.
We showed above that after 15 days of differentiation, LB cells constitute a very homogenous population of hepatoblasts expressing markers of bipotent progenitor cells capable of generating both hepatocytes and cholangiocytes. Within the liver organoids, such cells differentiate into mature hepatocytes and bile ducts at different stages of maturation. The localization of the bile ducts at the periphery of the organoids is probably secondary to the better oxygenation of the cells on the surface compared with on the deeper layers, which is in accordance with the ducts being found in the portal area of the liver lobule (Carpentier et al., 2011; Eyken et al., 1988). Unfortunately, the absence of vascularization and the poor diffusion of nutrients due to the size of the organoids limits the time window for their use to a few days. Nevertheless, the presence of the main components of the liver niche, all sharing the same genetic background and forming liver structures in a strictly controlled and highly reproducible in vitro environment, opens interesting possibilities to study human liver development as well as complex genetic or multifactorial liver conditions, such as those leading to biliary diseases, fibrosis, or cirrhosis. HLCs are instead a more suitable model to study parenchymal liver diseases or to test the metabolism and toxicity of drugs and compounds because of their maturation and homogeneity.
Overall, we described here a new differentiation protocol to consistently obtain high-quality, homogeneous, and functional hepatoblasts and HLCs with a high yield from both human ESCs and iPSCs. With low variability between starting PSC lines, this protocol might prove useful for 2D and 3D liver development and disease modeling. Combined with genome editing of iPSCs, it might allow for more representative studies of human-specific pathological mechanisms (Pham et al., 2020). Moreover, the protocol design and yield suggest good potential for scalability, which makes it promising as a surrogate to PHHs for in vitro drug testing and development. Once their potential to replace mature liver functions in vivo has been proven, such HLCs might also be considered for cell therapy applications.
Experimental procedures
A detailed description of all experimental procedures is provided as supplemental information.
PSC differentiation into HLCs
PSCs grown in Essential 8 Flex medium (see supplemental information for details on PSC generation, characterization, and maintenance) were dissociated by TrypLE (Life Technologies) to single cells and seeded on human recombinant laminin 521 (BioLamina)-coated plates in Essential 8 Flex medium at a density of 7 × 104 cells/cm2. Differentiation was started (day 0) when the cells reached 70% confluence by changing the medium to RPMI-B27 minus insulin (Life Technologies) supplemented with 1% KOSR (Life Technologies). For the first 2 days, the cells were exposed to 100 ng/mL Activin A (R&D Systems) and 3 μM CHIR-99021 (Stem Cell Technologies) and then for the 3 following days to 100 ng/mL Activin A alone. Subsequently, RPMI-B27 (minus insulin) medium was supplemented with 20 ng/mL BMP4 (Peprotech), 5 ng/mL bFGF (Peprotech), 4 μM IWP-2 (Tocris), and 1 μM A83-01 (Tocris) for 5 days, with daily medium change. At day 10, the medium was changed to RPMI-B27 (Life Technologies) supplemented with 2% KOSR, 20 ng/mL BMP4, 5 ng/mL bFGF, 20 ng/mL HGF (Peprotech), and 3 μM CHIR-99021 for 5 days, with daily medium change. At day 16, the medium was changed to HBM/HCM (minus EGF) medium (Lonza) supplemented with 1% KOSR, 20 ng/mL HGF, 20 ng/mL BMP4, 5 ng/mL bFGF, 3 μM CHIR-99021, 10 μM dexamethasone (Sigma), and 20 ng/ml OSM (R&D System) for 5 days, with daily medium change. From day 20, for 5 days, HBM/HCM medium was supplemented with 1% KOSR, 10 μM dexamethasone, and 20 ng/mL OSM, changing the medium every other day. From day 25, the cells were maintained in HBM/HCM 1% KOSR medium supplemented with 10 μM dexamethasone, with every other day medium change. During all of the differentiation processes, the cells were kept at 37 °C, ambient O2, and 5% CO2. Cells were characterized by real-time qRT-PCR, RNA sequencing (RNA-seq), flow cytometry, and immunofluorescence (see supplemental information for details).
Functional assessment of HLCs
Albumin production was evaluated using the Multigent microalbumin assay on the Architect cSystems. Cyp3A4 activity and urea synthesis were measured using P450-Glo Assay (Promega) and Quantichrom urea assay kit (Gentaur), respectively, according to manufacturers' instructions.
Mitochondrial respiration analysis
Mitochondrial stress testing was carried out using a Seahorse Bioscience XF96 analyzer (Seahorse Bioscience) in 96-well plates at 37°C as per the manufacturer’s instructions, with minor modifications (see supplemental information for details).
Generation of liver organoids
PSCs were differentiated into EPCs and MPCs as previously described, with modifications (see supplemental information for details; Kang et al., 2015; Nguyen et al., 2016). To generate human liver organoids, 2.5 × 105 human PSC-derived hepatoblasts at day 16 of the differentiation protocol, 1.75 × 105 syngeneic EPCs, and 0.5 × 105 syngeneic MPCs were resuspended in a composite medium (50% EBM2-EGM2, 50% HBM-HCM), supplemented with 10 μM dexamethasone, 20 ng/mL OSM, and 20 ng/mL HGF, and seeded in ultra-low attachment 96-well plates (Corning). Culture medium was changed every other day, and supernatant was collected to assess liver functions. After 7–10 days of culture, liver organoids were collected for analysis.
Statistical analysis
Replicates refer to independent experiments on ≥3 different PSC populations. Values are shown as mean ± standard error (SEM). Mann-Whitney U test was used to compare qRT-PCR and functional data. A p value <0.05 was considered significant.
Author contributions
Conception and design, C.R. and M.P.; collection of data, C.R., M.-A.M’C., Q.T.P., P.G., S.S., N.B., and C.L.M.; data analysis and interpretation, C.R., Q.T.P., G.C., J.-S.J., and M.P.; manuscript writing, C.R., J.-S.J., and M.P.; provision of study material, B.R.; financial support and supervision, M.P. All authors approved the final version of the manuscript.
Conflict of interests
C.R. and M.P. filed a patent application to protect this protocol and are co-founders, shareholders, and officers of Morphocell Technologies, Inc. The other authors declare no competing interests.
Acknowledgments
This work was supported by a Stem Cell Network grant FY17/DT8 (M.P.), a Canadian Institutes of Health Research New Investigator in Maternal, Reproductive, Child & Youth Health grant MY6-155373 (M.P.), and a “Fonds de Recherche du Québec – Santé” Junior one Clinician-Scientist grant (M.P.). We thank Kristen Vieira-Lomasney for technical assistance, the Vision Health Research Network for funding the Single-cell Academy platform, and Ines Boufaied (CHU Sainte-Justine’s Flow Cytometry core facility) for her valuable collaboration.
Published: February 3, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.01.003.
Supplemental information
Data and code availability
The RNA-seq data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus under GEO: GSE152390.
References
- Asai A., Aihara E., Watson C., Mourya R., Mizuochi T., Shivakumar P., Phelan K., Mayhew C., Helmrath M., Takebe T., et al. Paracrine signals regulate human liver organoid maturation from iPSC. Development. 2017;144:dev.142794. doi: 10.1242/dev.142794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter M., Withey S., Harrison S., Segeritz C.-P., Zhang F., Atkinson-Dell R., Rowe C., Gerrard D.T., Sison-Young R., Jenkins R., et al. Phenotypic and functional analyses show stem cell-derived hepatocyte-like cells better mimic fetal rather than adult hepatocytes. J. Hepatol. 2015;62:581–589. doi: 10.1016/j.jhep.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berasain C., Avila M.A. Regulation of hepatocyte identity and quiescence. Cell. Mol. Life Sci. 2015;72:3831–3851. doi: 10.1007/s00018-015-1970-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger D.R., Ware B.R., Davidson M.D., Allsup S.R., Khetani S.R. Enhancing the functional maturity of induced pluripotent stem cell-derived human hepatocytes by controlled presentation of cell-cell interactions in vitro. Hepatology. 2015;61:1370–1381. doi: 10.1002/hep.27621. [DOI] [PubMed] [Google Scholar]
- den Braver-Sewradj S.P., den Braver M.W., Vermeulen N.P.E., Commandeur J.N.M., Richert L., Vos J.C. Inter-donor variability of phase I/phase II metabolism of three reference drugs in cryopreserved primary human hepatocytes in suspension and monolayer. Toxicol. Vitro. 2016;33:71–79. doi: 10.1016/j.tiv.2016.02.013. [DOI] [PubMed] [Google Scholar]
- Carpentier R., Suñer R.E., van Hul N., Kopp J.L., Beaudry J.B., Cordi S., Antoniou A., Raynaud P., Lepreux S., Jacquemin P., et al. Embryonic ductal plate cells give rise to cholangiocytes, periportal hepatocytes, and adult liver progenitor cells. Gastroenterology. 2011;141:1432–1438.e4. doi: 10.1053/j.gastro.2011.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Zamule S.M., Coslo D.M., Chen T., Omiecinski C.J. The human constitutive androstane receptor promotes the differentiation and maturation of hepatic-like cells. Dev. Biol. 2013;384:155–165. doi: 10.1016/j.ydbio.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici M., Blanc K.L., Mueller I., Slaper-Cortenbach I., Marini F., Krause D., Deans R., Keating A., Prockop D., Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- Dorrity M.W., Saunders L.M., Queitsch C., Fields S., Trapnell C. Dimensionality reduction by UMAP to visualize physical and genetic interactions. Nat. Commun. 2020;11:1537. doi: 10.1038/s41467-020-15351-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Wang J., Jia J., Song N., Xiang C., Xu J., Hou Z., Su X., Liu B., Jiang T., et al. Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell. Stem Cell. 2014;14:394–403. doi: 10.1016/j.stem.2014.01.008. [DOI] [PubMed] [Google Scholar]
- Elaut G., Henkens T., Papeleu P., Snykers S., Vinken M., Vanhaecke T., Rogiers V. Molecular mechanisms underlying the dedifferentiation process of isolated hepatocytes and their cultures. Curr Drug Metab. 2006;7:629–660. doi: 10.2174/138920006778017759. [DOI] [PubMed] [Google Scholar]
- Eyken P.V., Sciot R., Callea F., Steen K.V.D., Moerman P., Desmet V.J. The development of the intrahepatic bile ducts in man: a keratin-immunohistochemical study. Hepatology. 1988;8:1586–1595. doi: 10.1002/hep.1840080619. [DOI] [PubMed] [Google Scholar]
- Fu G.-B., Huang W.-J., Zeng M., Zhou X., Wu H.-P., Liu C.-C., Wu H., Weng J., Zhang H.-D., Cai Y.-C., et al. Expansion and differentiation of human hepatocyte-derived liver progenitor-like cells and their use for the study of hepatotropic pathogens. Cell. Res. 2019;29:8–22. doi: 10.1038/s41422-018-0103-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Zhang X., Zhang L., Cen J., Ni X., Liao X., Yang C., Li Y., Chen X., Zhang Z., et al. Distinct gene expression and epigenetic signatures in hepatocyte-like cells produced by different strategies from the same donor. Stem Cell. Rep. 2017;9:1813–1824. doi: 10.1016/j.stemcr.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordillo M., Evans T., Gouon-Evans V. Orchestrating liver development. Development. 2015;142:2094–2108. doi: 10.1242/dev.114215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulart E., de Caires-Junior L.C., Telles-Silva K.A., Araujo B.H.S., Kobayashi G.S., Musso C.M., Assoni A.F., Oliveira D., Caldini E., Gerstenhaber J.A., et al. Adult and iPS-derived non-parenchymal cells regulate liver organoid development through differential modulation of Wnt and TGF-β. Stem Cell. Res. Ther. 2019;10:258. doi: 10.1186/s13287-019-1367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallifax D., Houston J.B. Methodological uncertainty in quantitative prediction of human hepatic clearance from in vitro experimental systems. Curr. Drug Metab. 2009;10:307–321. doi: 10.2174/138920009787846341. [DOI] [PubMed] [Google Scholar]
- Han X., Wang Y., Pu W., Huang X., Qiu L., Li Y., Yu W., Zhao H., Liu X., He L., et al. Lineage tracing reveals the bipotency of SOX9+ hepatocytes during liver regeneration. Stem Cell. Rep. 2019;12:624–638. doi: 10.1016/j.stemcr.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara M., Aoi T., Okita K., Takahashi R., Inoue H., Takayama N., Endo H., Eto K., Toguchida J., Uemoto S., et al. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc. Natl. Acad. Sci. U S A. 2012;109:12538–12543. doi: 10.1073/pnas.1209979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya A., Gonzalez F.J. TNF-alpha regulates mouse fetal hepatic maturation induced by oncostatin M and extracellular matrices. Hepatology. 2004;40:527–536. doi: 10.1002/hep.20362. [DOI] [PubMed] [Google Scholar]
- Kamiya A., Kinoshita T., Ito Y., Matsui T., Morikawa Y., Senba E., Nakashima K., Taga T., Yoshida K., Kishimoto T., et al. Fetal liver development requires a paracrine action of oncostatin M through the gp130 signal transducer. The EMBO J. 1999;18:2127–2136. doi: 10.1093/emboj/18.8.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya A., Inoue Y., Gonzalez F.J. Role of the hepatocyte nuclear factor 4α in control of the pregnane X receptor during fetal liver development. Hepatology. 2003;37:1375–1384. doi: 10.1053/jhep.2003.50212. [DOI] [PubMed] [Google Scholar]
- Kang R., Zhou Y., Tan S., Zhou G., Aagaard L., Xie L., Bünger C., Bolund L., Luo Y. Mesenchymal stem cells derived from human induced pluripotent stem cells retain adequate osteogenicity and chondrogenicity but less adipogenicity. Stem Cell. Res. Ther. 2015;6:144. doi: 10.1186/s13287-015-0137-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan J.A., Mendelson A., Kunisaki Y., Birbrair A., Kou Y., Arnal-Estapé A., Pinho S., Ciero P., Nakahara F., Ma’ayan A., et al. Fetal liver hematopoietic stem cell niches associate with portal vessels. Science. 2016;351:176–180. doi: 10.1126/science.aad0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Kang K., Lee S.B., Seo D., Yoon S., Kim S.J., Jang K., Jung Y.K., Lee K.G., Factor V.M., et al. Small molecule-mediated reprogramming of human hepatocytes into bipotent progenitor cells. J Hepatol. 2019;70:97–107. doi: 10.1016/j.jhep.2018.09.007. [DOI] [PubMed] [Google Scholar]
- Kola I., Landis J. Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov. 2004;3:711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- Li Q., Hutchins A.P., Chen Y., Li S., Shan Y., Liao B., Zheng D., Shi X., Li Y., Chan W.-Y., et al. A sequential EMT-MET mechanism drives the differentiation of human embryonic stem cells towards hepatocytes. Nat. Commun. 2017;8:15166. doi: 10.1038/ncomms15166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacParland S.A., Liu J.C., Ma X.-Z., Innes B.T., Bartczak A.M., Gage B.K., Manuel J., Khuu N., Echeverri J., Linares I., et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 2018;9:4383. doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallanna S.K., Duncan S.A. Differentiation of hepatocytes from pluripotent stem cells. Curr. Protoc. Stem Cell Biol. Curr. Protoc. Stem Cell. Biol. 2013;26:1G.4.1–1G.4.13. doi: 10.1002/9780470151808.sc01g04s26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyama H., Hiramatsu Y., Mizutani Y., Inoshita H., Kudo T. The expression of pregnane X receptor and its target gene, cytochrome P450 3A1, in perinatal mouse. Mol. Cell Endocrinol. 2001;172:47–56. doi: 10.1016/s0303-7207(00)00395-6. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Miki R., Nakayama M., Tatsumi N., Yokouchi Y. Wnt9a secreted from the walls of hepatic sinusoids is essential for morphogenesis, proliferation, and glycogen accumulation of chick hepatic epithelium. Developmental Biol. 2008;319:234–247. doi: 10.1016/j.ydbio.2008.04.021. [DOI] [PubMed] [Google Scholar]
- McLean A.B., D’Amour K.A., Jones K.L., Krishnamoorthy M., Kulik M.J., Reynolds D.M., Sheppard A.M., Liu H., Xu Y., Baetge E.E., et al. Activin A efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells. 2007;25:29–38. doi: 10.1634/stemcells.2006-0219. [DOI] [PubMed] [Google Scholar]
- Mu T., Xu L., Zhong Y., Liu X., Zhao Z., Huang C., Lan X., Lufei C., Zhou Y., Su Y., et al. Embryonic liver developmental trajectory revealed by single-cell RNA sequencing in the Foxa2eGFP mouse. Commun. Biol. 2020;3:642. doi: 10.1038/s42003-020-01364-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M.T.X., Okina E., Chai X., Tan K.H., Hovatta O., Ghosh S., Tryggvason K. Differentiation of human embryonic stem cells to endothelial progenitor cells on laminins in defined and xeno-free systems. Stem Cell. Rep. 2016;7:802–816. doi: 10.1016/j.stemcr.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nostro M.C., Sarangi F., Ogawa S., Holtzinger A., Corneo B., Li X., Micallef S.J., Park I.-H., Basford C., Wheeler M.B., et al. Stage-specific signaling through TGFβ family members and WNT regulates patterning and pancreatic specification of human pluripotent stem cells. Development. 2011;138:1445. doi: 10.1242/dev.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onitsuka I., Tanaka M., Miyajima A. Characterization and functional analyses of hepatic mesothelial cells in mouse liver development. Gastroenterology. 2010;138:1525–1535.e6. doi: 10.1053/j.gastro.2009.12.059. [DOI] [PubMed] [Google Scholar]
- Paganelli M. In: Paediatric Hepatology and Liver Transplantation. D’Antiga L., editor. Springer; 2019. Cell therapy in acute and chronic liver disease; pp. 781–797. [Google Scholar]
- Parviz F., Matullo C., Garrison W.D., Savatski L., Adamson J.W., Ning G., Kaestner K.H., Rossi J.M., Zaret K.S., Duncan S.A. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- Patel S.H., Camargo F.D., Yimlamai D. Hippo signaling in the liver regulates organ size, cell fate, and carcinogenesis. Gastroenterology. 2017;152:533–545. doi: 10.1053/j.gastro.2016.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham Q.T., Raad S., Mangahas C.-L., M’Callum M.-A., Raggi C., Paganelli M. High-throughput assessment of mutations generated by genome editing in induced pluripotent stem cells by high-resolution melting analysis. Cytotherapy. 2020;22:536–542. doi: 10.1016/j.jcyt.2020.06.008. [DOI] [PubMed] [Google Scholar]
- Ramachandran A., Duan L., Akakpo J.Y., Jaeschke H. Mitochondrial dysfunction as a mechanism of drug-induced hepatotoxicity: current understanding and future perspectives. J. Clin. Transl. Res. 2018;4:75. doi: 10.18053/jctres.04.201801.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K.R., Athineos D., Meniel V.S., Wilkins J.A., Ridgway R.A., Burke Z.D., Muncan V., Clarke A.R., Sansom O.J. B-catenin deficiency, but not Myc deletion, suppresses the immediate phenotypes of APC loss in the liver. Proc. Natl. Acad. Sci. U S A. 2008;105:18919–18923. doi: 10.1073/pnas.0805778105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C., Bladt F., Goedecke S., Brinkmann V., Zschiesche W., Sharpe M., Gherardi E., Birchmeler C. Scatter factor/hepatocyte growth factor is essential for liver development. Nature. 1995;373:699–702. doi: 10.1038/373699a0. [DOI] [PubMed] [Google Scholar]
- Shan J., Schwartz R.E., Ross N.T., Logan D.J., Thomas D., Duncan S.A., North T.E., Goessling W., Carpenter A.E., Bhatia S.N. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat. Chem. Biol. 2013;9:514–520. doi: 10.1038/nchembio.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K., Noto F.K., Nagaoka M., Li J., Battle M.A., Duris C., North P.E., Dalton S., Duncan S.A. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K., Lemaigre F.P., Duncan S.A. Organogenesis and development of the liver. Developmental Cell. 2010;18:175–189. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Sosa-Pineda B., Wigle J.T., Oliver G. Hepatocyte migration during liver development requires Prox1. Nat. Genet. 2000;25:254–255. doi: 10.1038/76996. [DOI] [PubMed] [Google Scholar]
- Takayama K., Akita N., Mimura N., Akahira R., Taniguchi Y., Ikeda M., Sakurai F., Ohara O., Morio T., Sekiguchi K., et al. Generation of safe and therapeutically effective human induced pluripotent stem cell-derived hepatocyte-like cells for regenerative medicine. Hepatol. Commun. 2017;1:1058–1069. doi: 10.1002/hep4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe T., Sekine K., Enomura M., Koike H., Kimura M., Ogaeri T., Zhang R.-R., Ueno Y., Zheng Y.-W., Koike N., et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- Teo A.K.K., Valdez I.A., Dirice E., Kulkarni R.N. Comparable generation of activin-induced definitive endoderm via additive Wnt or BMP signaling in absence of serum. Stem Cell. Rep. 2014;3:5–14. doi: 10.1016/j.stemcr.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touboul T., Hannan N.R.F., Corbineau S., Martinez A., Martinet C., Branchereau S., Mainot S., Strick-Marchand H., Pedersen R., Santo J.D., et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology. 2010;51:1754–1765. doi: 10.1002/hep.23506. [DOI] [PubMed] [Google Scholar]
- Touboul T., Chen S., To C.C., Mora-Castilla S., Sabatini K., Tukey R.H., Laurent L.C. Stage-specific regulation of the WNT/B-catenin pathway enhances differentiation of hESCs into hepatocytes. J. Hepatol. 2016;64:1315–1326. doi: 10.1016/j.jhep.2016.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Valizadeh A., Majidinia M., Samadi-Kafil H., Yousefi M., Yousefi B. The roles of signaling pathways in liver repair and regeneration. J. Cell. Physiol. 2019;234:14966–14974. doi: 10.1002/jcp.28336. [DOI] [PubMed] [Google Scholar]
- Vyas D., Baptista P.M., Brovold M., Moran E., Gaston B., Booth C., Samuel M., Atala A., Soker S. Self-assembled liver organoids recapitulate hepatobiliary organogenesis in vitro. Hepatology. 2018;67:750–761. doi: 10.1002/hep.29483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren C.R., O’Sullivan J.F., Friesen M., Becker C.E., Zhang X., Liu P., Wakabayashi Y., Morningstar J.E., Shi X., Choi J., et al. Induced pluripotent stem cell differentiation enables functional validation of GWAS variants in metabolic disease. Cell. Stem Cell. 2017;20:547–557.e7. doi: 10.1016/j.stem.2017.01.010. [DOI] [PubMed] [Google Scholar]
- Xie B., Sun D., Du Y., Jia J., Sun S., Xu J., Liu Y., Xiang C., Chen S., Xie H., et al. A two-step lineage reprogramming strategy to generate functionally competent human hepatocytes from fibroblasts. Cell. Res. 2019;29:696–710. doi: 10.1038/s41422-019-0196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus under GEO: GSE152390.