Summary
In vitro expansion of human primordial germ cell-like cells (hPGCLCs), a pluripotent stem cell-derived PGC model, has proved challenging due to rapid loss of primordial germ cell (PGC)-like identity and limited cell survival/proliferation. Here, we describe long-term culture hPGCLCs (LTC-hPGCLCs), which actively proliferate in a serum-free, feeder-free condition without apparent limit as highly homogeneous diploid cell populations maintaining transcriptomic and epigenomic characteristics of hPGCLCs. Histone proteomics confirmed reduced H3K9me2 and increased H3K27me3 marks in LTC-hPGCLCs compared with induced pluripotent stem cells (iPSCs). LTC-hPGCLCs established from multiple human iPSC clones of both sexes were telomerase positive, senescence-free cells readily passaged with minimal cell death or deviation from the PGC-like identity. LTC-hPGCLCs are capable of differentiating to DAZL-positive M-spermatogonia-like cells in the xenogeneic reconstituted testis (xrTestis) organ culture milieu as well as efficiently producing fully pluripotent embryonic germ cell-like cells in the presence of stem cell factor and fibroblast growth factor 2. Thus, LTC-hPGCLCs provide convenient access to unlimited amounts of high-quality and homogeneous hPGCLCs.
Key words: primordial germ cell-like cells, embryonic germ cells, induced pluripotent stem cells, xrTestis, human endogenous retroviruses, primordial germ cells, germline reprogramming, serum-free culture, feeder-free culture
Highlights
-
•
A novel system perpetually expands human primordial germ cell-like cells (hPGCLCs).
-
•
HPGCLCs expand as a homogeneous cell population maintaining the germline features
-
•
Long-term culture (LTC) hPGCLCs can be converted to embryonic germ cell-like cells
-
•
The CpG methylation of LTR5_Hs is specifically decreased in LTC-hPGCLCs
In this article, Kobayashi and colleagues describe a novel cell culture system that supports perpetual expansion of telomerase-positive human primordial germ cell-like cells (hPGCLCs) without serum or feeder layers. Large amounts of homogeneous hPGCLCs are readily obtained without repeated re-enrichment by FACS, deviations from the germline cell identity, or accumulation of chromosomal anomalies.
Introduction
Primordial germ cells (PGCs), the common embryonic precursors of gametes, emerge in peri-implantation mammalian embryos (Sasaki et al., 2016). PGC-like cells (PGCLCs) are pluripotent stem cell-derived cell culture models of PGCs (Saitou, 2021). Mouse PGCLCs reflecting post-migration PGCs with advanced global genomic DNA (gDNA) demethylation can generate fertilization-competent sperm and oocytes (Hayashi et al., 2012; Ishikura et al., 2021; Miyoshi et al., 2016). In contrast, human PGCLCs (hPGCLCs) resemble earlier stages of embryonic PGCs with limited global gDNA demethylation (Irie et al., 2015; Mitsunaga et al., 2017; Sasaki et al., 2015; Tang et al., 2015). In organoid culture milieux, hPGCLCs can differentiate to more advanced stages of germline cells mimicking oogonia (Yamashiro et al., 2018) or prospermatogonia (Hwang et al., 2020).
Although the use of PGCLCs as a convenient and reliable surrogate model of embryonic PGCs is becoming increasingly popular (Saitou, 2021), the technical difficulty in cell culture expansion of PGCLCs poses a hurdle to its application for experiments, requiring large amounts of homogeneous cells such as chemical or genetic screening. Ohta et al. reported extended culture of mouse PGCLCs for 9 days on m220 feeders using a medium containing 10 μM each of rolipram and forskolin (FR10), stem cell factor (SCF), and 2.5% FCS (Ohta et al., 2017). Our preceding study maintained hPGCLCs for up to 21 days on STO feeders in a medium containing FR10, SCF, and 2.5% FCS (Gell et al., 2020). Even when hPGCLCs survive and proliferate in vitro, they tend to diverge from the PGC-like identity to other types of cells. By repeatedly eliminating such unwanted cells from the culture by FACS at every passaging step, Murase et al. provided evidence that hPGCLCs can maintain their proliferation potential long term in vitro (Murase et al., 2020). However, it remains a major challenge to expand hPGCLC culture to a large number of homogeneous cells maintaining their PGC-like characteristics.
Here, we describe long-term expansion of hPGCLCs homogenously preserving the PGC-like characteristics in cell culture without serum or feeder layer cells. Even after 150 days of expansion in vitro, the telomerase-positive hPGCLCs maintain their PGC-like morphology and gene expression profile with no signs of senescence or deviation to other types of cells while their global gDNA is modestly demethylated upon increasing passage number. Upon changing cell culture condition, hPGCLCs are effectively converted to pluripotent embryonic germ cell-like cells (EGCLCs), restoring the induced pluripotent stem cell (iPSC)-like transcriptome and gDNA methylome but losing the imprinted allelic restriction of H19 RNA expression. Thus, long-term culture (LTC) hPGCLCs represent a novel type of stably maintained, pluripotency-relevant cell culture resource that permanently retains biological characteristics of hPGCLCs and are easily expanded in vitro as a stable cell line.
Results
Expansion of hPGCLCs on STO feeder layer
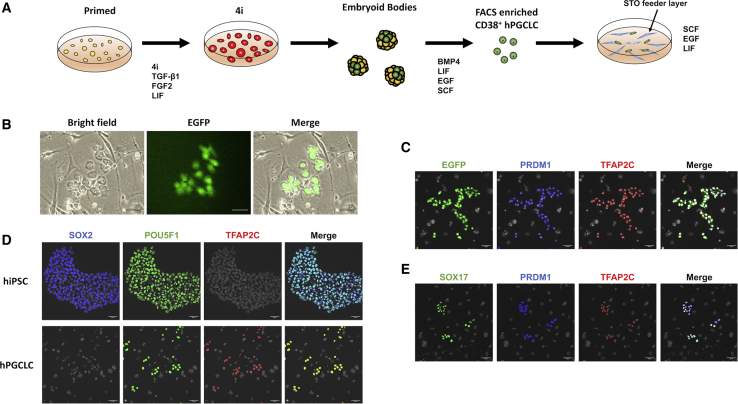
Our preceding study showed an ∼7.6-fold increase in yield of the CD38+ hPGCLCs FACS-isolated from embryoid bodies (EBs) during the last 3 days of the 8-day EB culture in hPGCLC derivation medium containing bone morphogenetic protein 4 (BMP4) (Mitsunaga et al., 2017). However, our subsequent experiments showed that removal of BMP4 from days 5–8 EB culture did not reduce hPGCLC yield (Figure S1), leading us to speculate that other growth factors in hPGCLC derivation medium—namely, stem cell factor (SCF), epidermal growth factor (EGF), and leukemia inhibitory factor (LIF)—may support proliferation of hPGCLCs. To explore this possibility, we inoculated EGFP-labeled CD38+ hPGCLCs freshly isolated from day-8 EBs onto the STO feeder layer in a medium containing SCF, EGF, and LIF (Figure 1A). Seven days later, hPGCLCs formed grape-like loose clusters, reproducing our preceding study (Gell et al., 2020) (Figure 1B). In this condition, hPGCLCs actively proliferated and readily passaged for 4 weeks on STO feeder layer. Immunofluorescence (IF) cell staining showed that these EGFP+ hPGCLCs strongly expressed hPGCLC markers PRDM1, TFAP2C, and SOX17 (Figures 1C–1E) but not the pluripotency-associated marker SOX2 (Figure 1D). The PRDM1+/TFAP2C+ double-positive cells also strongly expressed POU5F1, a marker of both human induced pluripotent stem cells (hiPSCs) and PGCLCs (Figure 1D). These results indicate that hPGCLCs expanded on STO feeder layer in the presence of SCF, EGF, and LIF, preserving expression of hPGCLC markers. Hereafter, we refer to the long-term culture hPGCLCs as LTC-hPGCLCs.
Figure 1.
Initial expansion of hPGCLCs on STO feeder layer
(A) Steps to initiate long-term expansion of hPGCLCs. EGFP-labeled, primed pluripotency hiPSCs were exposed to the 4i/LIF medium for 48 hs and subjected to formation of EBs in microwells. hPGCLCs were induced in a medium containing BMP4, LIF, EGF, and SCF, enriched by FACS as CD38+ cells, and inoculated on STO feeder layer in a medium containing LIF, EGF, and SCF. (B) A small cluster of hPGCLCs formed after 7-day culture. (C–E) IF with nuclei visualized with Hoechst (gray); Scale bars, 50 μm. (C) Expression of PRDM1 and TFAP2C in hPGCLCs maintained in culture for 4 weeks (D) Expression of SOX2, POU5F1, and TFAP2C in hiPSCs and hPGCLCs (maintained for 4 weeks on STO feeder layer). (E) Expression of SOX17, PRDM1, and TFAP2C in hPGCLCs maintained in culture for 4 weeks.
Feeder-free expansion of LTC-hPGCLCs
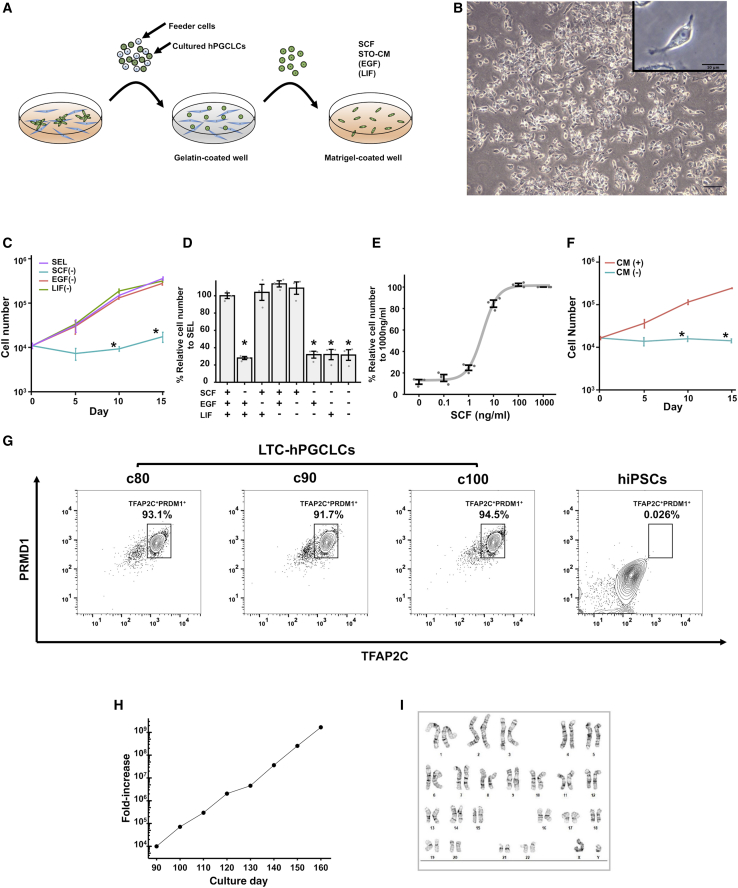
We next attempted to expand LTC-hPGCLCs in a feeder-free condition. When LTC-hPGCLCs reached ∼20% confluence, we removed STO cells by differential adhesion to gelatin-coated dishes, and LTC-hPGCLCs were inoculated on Matrigel-coated dishes in STO-conditioned medium (STO-CM) containing SCF, EGF, and LIF (Figure 2A). In this condition, which is referred to as the SEL condition, LTC-hPGCLCs scattered on the Matrigel-coated surface and randomly migrated (Figure 2B). LTC-hPGCLCs harbored cytoplasmic granules that were not found in hiPSCs (Figure 2B, inset) and still expressed PGC markers PRDM1, TFAP2C, SOX17, CD38, and POU5F1 but not the pluripotency-associated marker SOX2 (Figures S2A–S2C). All LTC-hPGCLCs strongly expressed CD38, and all CD38+ hPGCLCs also expressed both PRDM1 and TFAP2C (Figures S2D–S2F). LTC-hPGCLCs actively proliferated in STO-CM supplemented with SCF, whereas EGF or LIF was dispensable (Figures 2C, 2D). A significant growth-supporting effect of SCF was observed at 10 ng/mL and saturated at 100 ng/mL (Figure 2E). The size of the caspase3/7-activated apoptotic cells in rapidly growing LTC-hPGCLC cell population was minimal (2.28%), whereas cells exposed to staurosporine, a potent apoptosis inducer, increased the apoptotic population size to 10.1% (Figure S2G). The small size of the apoptotic cell population agrees with our microscopic observations supporting the apparent absence of significant cell death during culture and passaging of LTC-hPGCLCs. Whereas LTC-hPGCLCs did not proliferate on non-coated plastic wells, coating with laminin or vitronectin supported short-term proliferation as efficiently as Matrigel for up to 7 days (Figures S3A, S3B). Whether coating with laminin and/or vitronectin supports long-term expansion of a homogeneous population of LTC-hPGCLCs needs to be determined in future studies. The SCF-supported, feeder-free expansion of LTC-hPGCLCs was dependent on STO-CM (Figure 2F). Hereafter, we refer to to the LTC-hPGCLC maintenance medium containing both SCF and STO-CM as S-CM.
Figure 2.
Feeder-free, long-term expansion of hPGCLCs
(A) Steps to initiate feeder-free culture hPGCLCs. After STO feeder cells were removed by differential adhesion to gelatin-coated surface, hPGCLCs were inoculated on Matrigel-coated wells in STO-CM containing SCF, EGF, and/or LIF. (B) Dispersed appearance of LTC-hPGCLC feeder-free culture. Phase contrast images show high-density cells (scale bar, 100 μm) and a magnified single cell (inset; scale bar, 10 μm). (C–E) SCF dependency of LTC-hPGCLC expansion in STO-CM. Data were pooled from three independent experiments. (C) Growth curves of LTC-hPGCLCs in STO-CM containing three growth factors: SCF, EGF, and LIF (SEL; purple line, full condition), or SEL lacking SCF (blue), EGF (orange), or LIF (green). Each data point represents mean ± SEM ∗Statistically significant reduction (p < 0.05) in growth compared with the SEL condition. (D) Relative numbers of LTC-hPGCLCs expanded in a 7-day culture in STO-CM in the presence of indicated growth factors (mean ± SEM). Cell numbers of the condition containing SEL is defined as 100%; ∗ statistically significant reduction from it (p < 0.05). (E) A dose-response curve of SCF for 7-day growth of LTC-hPGCLCs in STO-SCM without EGF or LIF. Each data point represents mean ± SEM (F) Growth curves of LTC-hPGCLCs in the presence of SCF with or without STO-CM (mean ± SEM; ∗p < 0.05). Data were pooled from three independent experiments. (G) Expression of PRDM1 and TFAP2C in hiPSCs and LTC-hPGCLCs expanded for 80–100 days in culture. Fixed and permealized cells were immunostained and analyzed with FACS. Rectangular gates indicate PRDM1+/TFAP2C+ cells. (H) Growth curve of feeder-free LTC-hPGCLCs from 90 to 160 days supported by SCF and STO-CM. (I) Normal diploid karyotype (G-banding) of LTC-hPGCLCs expanded in the SCF-supported STO-CM for 100 days.
Previous studies attempted to maintain PGCLCs in media containing 2.5% fetal calf serum (FCS) (Gell et al., 2020; Murase et al., 2020; Ohta et al., 2017). However, addition of 2.5% hESC-grade FCS to S-CM resulted in rapid emergence of cells morphologically distinct from hPGCLCs and lost expression of both PRDM1 and TFAP2C (Figures S3C and S3D). These PRDM1−/TFAP2C− double-negative cells were morphologically heterogeneous (Figure S3C), with no characteristics of pluripotent stem cells, and many of them rapidly proliferated to dominate the cultures. In contrast, at least 90% of LTC-hPGCLCs expanded in S-CM maintained expression of both PRDM1 and TFAP2C after 80–100 days of culture (Figures 2G, S3C–S3E).
LTC-hPGCLCs actively proliferated for at least 160 days with a 4.5-day population doubling time, expanding an initial population of 10,000 hPGCLCs to more than one billion cells in 70 days (Figure 2H). A G-banding assay (Figure 2I) and digital karyotyping (Figure S4) demonstrated the normal male diploid karyotype of LTC-hPGCLCs after 100 days of feeder-free expansion. The above-mentioned experiments performed with LTC-hPGCLCs derived from the A4 male hiPSCs were reproduced using a female hiPSC clone (ATCC-BXS0115/ACS-1029; referred to to as F2 in this study) as well as two more male hiPSC clones, A5 and 9A13 (Figures S2F and S4). Note that the hiPSC clones A4 and A5 were derived from the same donor, whereas 9A13 was from a different donor.
STO-CM inhibits expression of pluripotency-associated markers in LTC-hPGCLCs
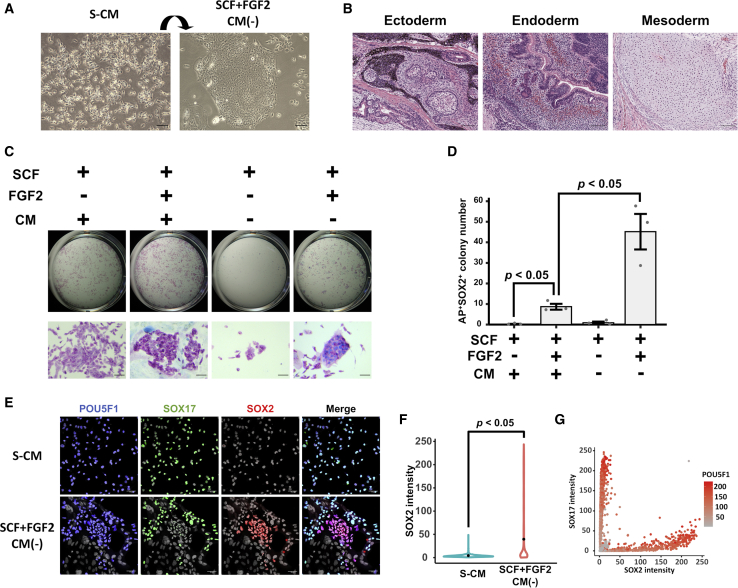
Mammalian PGCs become committed to produce only gametes (i.e., unipotent) after they colonize in the nascent gonads, whereas earlier stages of PGCs are poised to de-differentiate into the pluripotent state embryonic germ cells (Nicholls et al., 2019). To test whether STO-CM prevents hPGCLCs from de-differentiation to pluripotency, we cultured feeder-free LTC-hPGCLCs in the SCF-containing maintenance medium lacking STO-CM but supplemented with fibroblast growth factor 2 (FGF2), which supports the growth of human primed pluripotent stem cells, and we observed that many large colonies morphologically similar to hiPSC colonies emerged in 10 days (Figure 3A). Those hiPSC-like cells formed teratomas when transplanted into NOD scid-γ (NSG) mice (Figure 3B). Differentiation to three germ layers in these teratomas confirmed the pluripotency of the iPSC-like cells. Whereas LTC-hPGCLCs were alkaline phosphatase (AP) positive but SOX2 negative (Figure 3C, leftmost), the hiPSC-like colonies were AP+/SOX2+ double positive (Figure 3C, rightmost). These AP+/SOX2+ cells readily expanded in the mTeSR medium for primed human pluripotent stem cells and were transcriptionally indistinguishable from bona fide primed hiPSCs (Figure 4). On the basis of the above-mentioned observations, we concluded that the AP+/SOX2+ hiPSC-like cells resemble human embryonic germ cells; thus, we refer to to them as human embryonic germ cell-like cells (hEGCLCs). In the absence of both STO-CM and FGF2, hPGCLCs were unable to proliferate, and hEGCLCs did not emerge in the culture either (Figures 3C and 3D). Finally, in the presence of STO-CM, derivation of hEGCLCs was strongly suppressed even in the presence of FGF2 (Figures 3C and 3D). These results support the notion that STO-CM suppresses de-differentiation of hPGCLCs to hEGCLCs. SOX17 and SOX2 were identified as specific markers of LTC-hPGCLCs and hEGCLCs, respectively (Figure 1D; also, see, Figures S2B, and S5). During LTC-hPGCLC conversion to hEGCLC, a small portion of cells lost expression of POU5F1, a common marker of these two cell types (Figure 3E). SOX2 was expressed in LTC-hPGCLCs only when they were exposed to the hEGCLC derivation medium (SCF+/FGF2+/CM−) but not when maintained in the S-CM medium (Figure 3F). In the POU5F1+ LTC-hPGCLCs maintained in the hEGCLC derivation condition, expressions of SOX17 and SOX2 were mutually exclusive, which was confirmed by quantitative image analysis of the IF photographs (Figure 3G). Eventually, all cells maintained in the hEGCLC derivation medium expressed SOX2 (Figure 3C).
Figure 3.
De-differentiation of LTC-hPGCLCs to hEGCLCs
(A) Phase contrast images of hPGCLCs cultured in S-CM (left) and hEGCLCs emerged after 10-day culture containing SCF and FGF2 but lacking STO-CM (right). (B) H&E staining of a teratoma formed from hEGCLCs in NSG mice. (C) hPGCLCs cultured in the indicated conditions were stained for alkaline phosphatase (AP, red) and SOX2 (blue). Top row, global appearance; bottom row, microscopic images (scale bar, 50 μm). (D) Number of colonies including AP+/SOX2+ cells in each well (mean ± SEM). Data were pooled from three independent experiments. (E) IF of POU5F1 (blue), SOX17 (green), and SOX2 (red) in LTC-hPGCLCs maintained for 10 days in S-CM or hEGCLC induction medium containing SCF and FGF2 but lacking CM. Nuclei were visualized by Hoechst (gray). (F) Nuclear expression of SOX2 in LTC-hPGCLCs cultured in S-CM or hEGCLC induction medium for 10 days (violin plots; dots show average intensities). Data were pooled from three independent experiments. (G) Quantitative image analysis on nuclear expression of SOX2, SOX17, and POU5F1 in LTC-hPGCLCs cultured for 10 days in hEGCLC induction medium. Data were pooled from three independent experiments.
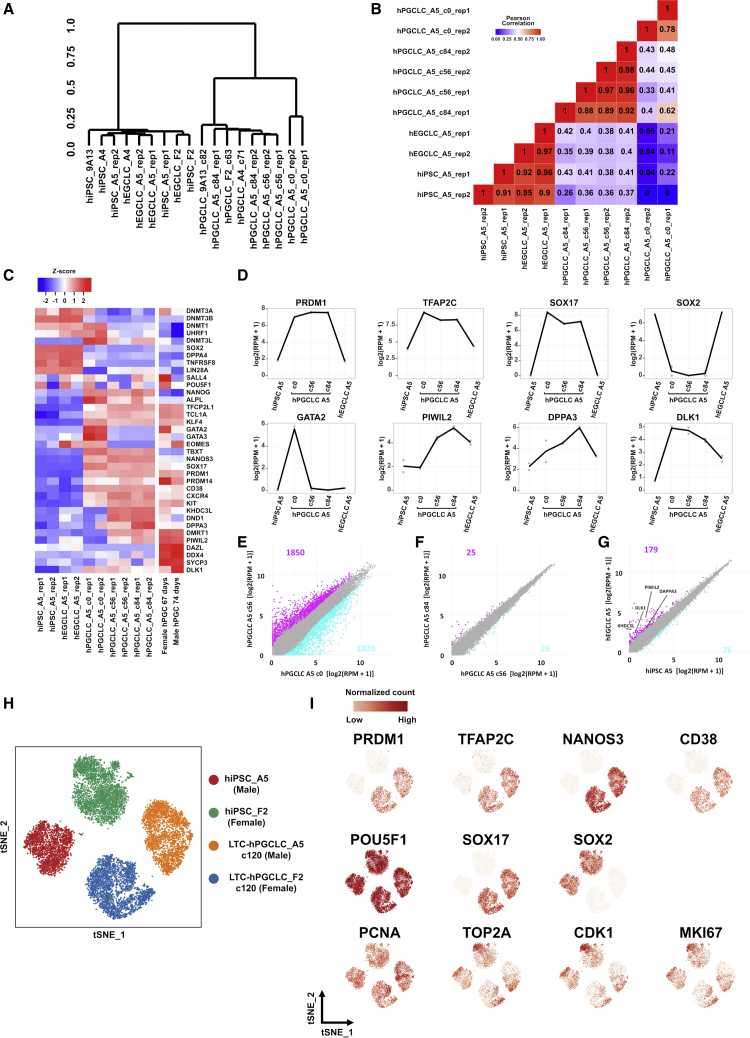
Figure 4.
Transcriptomic profiles of hiPSCs, hPGCLCs, and hEGCLCs
(A–D) Bulk RNA-seq data. (A) Unsupervised hierarchical clustering. hPGCLCs generated from hiPSCs (clones A4, A5, F2, and 9A13) were long-term expanded (culture days are shown as c#). hEGCLCs were derived from hPGCLCs. Rep1 and rep2 are independently performed replicates. (B) Pearson’s correlation coefficients between transcriptomes. (C) Heatmap representation of marker gene expression. hPGC data (gestational days 67–74) are from another study (Gkountela et al., 2015). (D) Gene expression dynamics in hiPSCs, c0-hPGCLCs, LTC-hPGCLCs, and hEGCLCs. (E–G) Changes in gene expression in hiPSCs, hPGCLCs, and hEGCLCs. Scatterplots compare (E) hPGCLCs (c0) versus LTC-hPGCLCs (feeder free, 56 days); (F) LTC-hPGCLCs cultured feeder free for 56 and 84 days; and (G) hiPSCs and hEGCLCs. Axes are in log2 (reads per million + 1), and the numbers and data points of differentially expressed genes (log2 [fold change] ≥ 1) are shown in magenta or cyan. (H and I) Single cell RNA-seq tSNE plot (H) and superimposed expression of marker genes (I).
LTC-hPGCLCs maintain transcriptomic characteristics of early-stage human PGCs
We determined transcriptomic profiles of feeder-free LTC-hPGCLCs by bulk RNA-seq. Transcriptomes of freshly isolated, pre-expansion (c0) hPGCLCs and long-term-expanded (c56–c84) hPGCLCs were similar but distinguishable (Figures 4A, 4B, and 4E), agreeing with the observation made by Murase et al. that transcriptome of the pre-expansion hPGCLCs significantly changed during a short-period expansion culture for 10 days (Murase et al., 2020). Both Murase et al. and we observed that expression of GATA2 and GATA3 strongly diminished from pre- to post-expansion of hPGCLCs (Figure 4C and Murase et al., 2020). Both the pre- and post-expansion hPGCLCs maintained strong expression of representative hPGC markers PRDM1, TFAP2C, SOX17, NANOS3, KIT, CD38, and CXCR4 (Figures 4C, 4D, and S5), supporting their legitimate germline identity. Transcriptomes of LTC-hPGCLCs derived from different iPSC clones and maintained for 56–84 days (c56–c84) showed strong similarities among one another (Figures 4A, 4B, and 4F). Interestingly, LTC-hPGCLCs expressed later-stage hPGC markers DPPA3, PIWIL2, and DND1 more strongly than pre-expansion hPGCLCs (Figures 4C, 4D, and S5); however, expression of germline markers reflecting more advanced stages of development (e.g., DDX4 and DAZL) in LTC-hPGCLCs remained weak (Figures 4C, 4D, and S5). Tyser et al. recently presented single-cell RNA-seq data of in vivo hPGCs in very early stages of development at 16–18 gestational days (Tyser et al., 2021). Their dataset identified eight NANOG+/POU5F1+/NANOS3+ hPGCs, which sporadically expressed hPGC markers TFAP2C, PRDM1, or CD38 and were negative for GATA2, GATA3, DAZL, DDX4, or SYCP3 (Figures S6A and S6B). On the other hand, single-cell RNA-seq data presented by Li et al. for in vivo NANOG+/NANOS3+ hPGCs in a later stage at 4 weeks of gestation (Li et al., 2017) expressed the late-PGC markers DAZL and DDX4 weakly but at unignorable levels (Figures S6C–S6E). In either of these single-cell RNA-seq data, expression of GATA2 or GATA3 was not detected in hPGCs (Figures S6B, S6D, and S6E), placing our c0 hPGCLCs to a unique GATA2+/GATA3+ stage in hPGC development as described by Kojima et al. (2021). Taken together, these results indicate that hPGCLCs experience significant transcriptomic changes during the initial transition from the GATA2/3-positive pre-expansion state to the GATA2/3-negative post-expansion state, whereas both the pre- and post-expansion cells are legitimate hPGCLCs. Once entering the post-expansion state, transcriptomes of LTC-hPGCLCs remain stable.
Homogeneity of LTC-hPGCLCs determined by single-cell RNA-seq
To evaluate cellular heterogeneity in LTC-hPGCLC cultures, we examined their single-cell RNA-seq profiles. The t-distributed stochastic neighbor embedding (t-SNE) plot analysis demonstrated the highly homogeneous nature of LTC-hPGCLC cultures comparable to that of hiPSCs (Figure 4H). Expression of hPGCLC marker genes (PRDM1, TFAP2C, SOX17, NANOS3, and CD38) and a pluripotency-associated marker gene (SOX2) showed strong homogeneity within each cluster of cells, whereas expression of POU5F1 was homogenously ubiquitous in all cells (Figure 4I). The tSNE clusters of LTC-hPGCLCs (as well as those of hiPSCs) were clearly separated by sex, which is consistent with significantly stronger expression of genes located on the X chromosome in female cells than in males’, although expression of autosomal genes is largely unaffected by sex (Figure S5E). Degrees of the female-augmented expression of the X chromosomal genes were largely unaffected by cell types (Figure S5E). The sensitivity of our single-cell RNA-seq analysis was sufficient to detect cellular heterogeneity in expression of cell cycle genes (PCNA for G1-S; TOP2A, CDK1, and MKI67 for G2/M) within each cluster (Figure 4I). PCA analysis separated the four tSNE clusters into two groups by cell types (hiPSC versus hPGCLC), with small degrees of heterogeneity in each group along by sex (Figure S5C), largely agreeing with the tSNE plots (Figure S5D). Thus, the single-cell RNA-seq data confirm that LTC-hPGCLCs maintain highly homogeneous populations of hPGCLCs consisting of actively proliferating cells even after long-term expansion without accumulating other types of cells.
Epigenetic characteristics of LTC-hPGCLCs
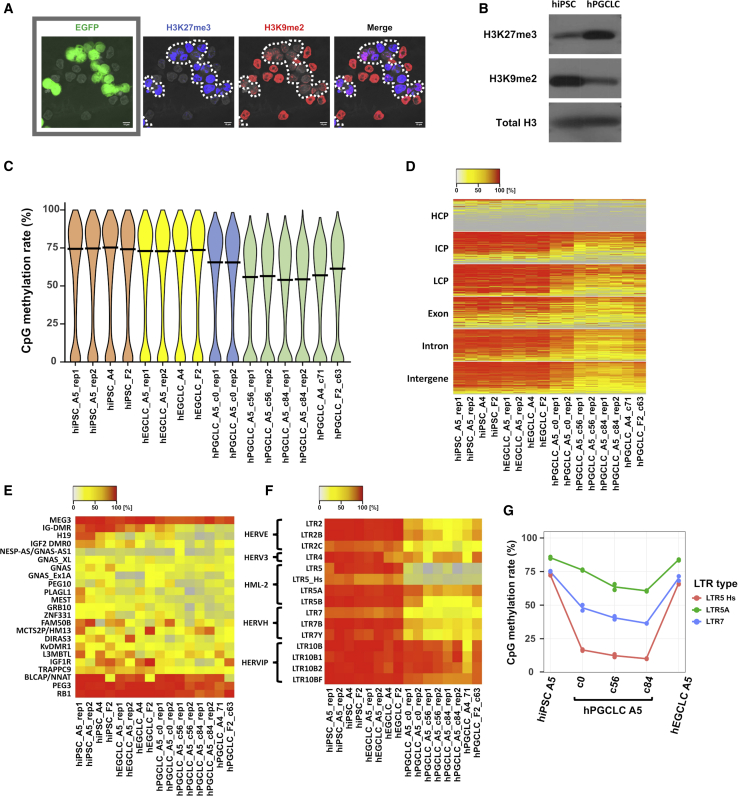
Global deposition of H3K27me3 increases upon differentiation of human pluripotent stem cells to hPGCLCs, whereas deposition of H3K9me2 decreases (Sasaki et al., 2015). IF and western blotting confirmed that these changes are preserved in LTC-hPGCLCs (Figures 5A and 5B), providing further evidence that LTC-hPGCLCs maintain their hPGC-like characteristics.
Figure 5.
Epigenetic profiles of hiPSCs, hPGCLCs, and hEGCLCs
(A) IF comparison of H3K9me2 and H3K27me3 expression between hiPSCs and LTC-hPGCLCs. Mixtures of hiPSCs (EGFP negative) and LTC-hPGCLCs (EGFP positive) were subjected to multiplex staining for H3K27me3 (blue), and H3K9me2 (red). Nuclei were visualized by Hoechst (gray). Dotted lines delineate LTC-hPGCLCs. (B) Western blotting detection of H3K27me3 and H3K9me2 in hiPSCs and LTC-hPGCLCs. (C) Violin plots of DNA methylomes (whole-genome bisulfite sequencing). Horizontal bars indicate global averages. (D–F) Heatmaps of CpG methylation at (D) various genomic features (HCP, ICP, and LCP are gDNA regions with high, intermediate, and low densities of CpG sites, respectively); (E) imprinting-controlling differentially methylated regions (DMRs), and (F) representative human endogenous retroviruses. (G) Line plot of CpG methylation rate of LTR5_Hs, LTR5A, and LTR7 in the indicated cells.
Although proteomics of histone modifications is a powerful approach to quantitative determination of epigenetic characteristics of mammalian cells, it requires large amounts of homogeneous cells that are not readily obtained using previous methods of hPGCLC expansion (Gell et al., 2020; Murase et al., 2020). Taking advantage of our LTC-hPGCLCs, we performed proteomics determination of 84 histone modifications in their genome (Figure S7A) and were able to confirm a reduction in H3K9me2 and an increase in H3K27me3 as differences between LTC-hPGCLCs and hiPSCs, agreeing with IF and western blotting data (Figures 5A and, 5B). Various other changes in histone modifications between LTC-hPGCLCs and hiPSCs (e.g., H4K12Ac) were also evident, although detailed confirmations of such modifications await future studies.
We determined the global gDNA methylome of LTC-hPGCLCs by whole-genome bisulfite sequencing (WGBS; Figure 5C). The global 5-methylcytosine (5mC) level decreased from hiPSCs (mean = 73.3%) to c0-hPGCLCs (64.2%) and then further in LTC-hPGCLCs (54.2%) across all major genomic features (Figure 5D). A small decrease from c56 (56.2%) to c84 (54.2%) LTC-hPGCLCs was statistically insignificant. The global 5meC levels of hiPSCs and hEGCLCs were comparable, indicating that the ∼20% global gDNA demethylation in LTC-hPGCLCs from their pluripotent precursor cells was largely reversible. The differentially methylated regions (DMRs) of the imprinting genes tended to be demethylated in LTC-hPGCLCs more strongly than the global average (Figure 5E). Among the DMRs, the intergenic-DMR (IG-DMR) of the DLK1-DIO3 imprinting cluster and H19 DMR showed relatively robust demethylation in (LTC-)hPGCLCs compared with hiPSCs (IG-DMR: 85.0% in hiPSCs versus 28.1% in LTC-hPGCLCs; H19: 70.3% in hiPSCs versus 18.6% in LTC-hPGCLCs). On the other hand, PEG3-DMR and MEG3-DMR maintained their high levels of CpG methylation (PEG3: 90.0% in hiPSC versus 82.6% in LTC-hPGCLCs; MEG3: 91.0% in hiPSCs versus 82.0% in LTC-hPGCLCs). Thus, DMR demethylation in LTC-hPGCLCs was heterogeneous and partial. The DMRs regulating the imprinted expression of the H19 RNA were almost completed demethylated in LTC-hPGCLCs, whereas these regions are ∼50% methylated in hiPSCs, as confirmed by WGBS (Figure 5E) and methylation-specific multiplexed ligation-dependent probe amplification (MS-MLPA; Figures S7B–S7D). Agreeing with this strong demethylation, H19 RNA expression was bi-allelic in LTC-hPGCLCs of both sexes, whereas it is monoallelic in the precursor hiPSCs (Figures S7E and S7F). Whether other imprinting genes are bi-allelically expressed in LTC-hPGCLCs, and possibly in hEGCLCs as well, needs to be determined in future studies.
The long-terminal repeats (LTRs) of the human endogenous retroviruses (HERVs) generally reduced their 5mC contents in LTR-hPGCLCs, although the degree of reduction was relatively small in the HERV-IP family (Figure 5F). In contrast, LTR5 and LTR5_Hs (Hs, human specific), which are the youngest classes of HERVs, showed the most remarkable LTR demethylation in both c0-hPGCLCs and LTC-hPGCLCs (Figures 5F and 5G).
Transcriptomic and epigenomic characteristics of hEGCLCs
Transcriptomic profiles of hiPSCs and hEGCLCs showed striking similarities (Figures 4A and, 4C) with Pearson’s correlation coefficients greater than 0.90 (Figure 4B). Between them, we identified 179 differentially expressed genes (DEGs; Figure 4G), including KHDC3L, DPPA3, PIWIL2, and DLK1 (Figures 4C, 4D, and 4G). Because these four DEGs were also expressed more strongly in LTC-hPGCLCs than in hiPSCs (Figures 4C and 4D), we speculate that they were incompletely re-suppressed during re-programming of LTC-hPGCLCs to the pluripotent hEGCLCs. However, degrees of CpG methylation in promoter or gene body of these four DEGs determined by WGBS did not show significant differences between hiPSCs and hEGCLCs. Expression of DLK1 protein in LTC-hPGCLCs or hEGCLCs was not detected by IF, apparently due to technical issues. Whereas the imprinting genes differentially expressed between hiPSCs and hEGCLCs may significantly contribute to biological differences between these two pluripotent types of cells, non-imprinting DEGs would certainly affect the distinct characteristics of these cells as well. Future studies are necessary for more detailed comparisons between hiPSCs and hEGCLCs.
Whereas DMRs of several imprinting genes were demethylated in LTR-hPGCLCs compared with hiPSCs, many of them were re-methylated in hEGCLCs but to weaker degrees than in hiPSCs (Figure 5E). For example, the average 5mC level of the IG-DMR, which is involved in regulation of DLK1 expression, was high in hiPSCs (84%), reduced in the c0-hPGCLCs (64%), and decreased further in LTC-hPGCLCs (28%) but partially re-methylated to 70% in hEGCLCs (Figure 5E). These results suggest that de-differentiation of LTC-hPGCLCs to hEGCLCs is accompanied by re-methylation of several imprinting-controlling DMRs although degrees of their re-methylation often do not reach the levels in the precursor hiPSCs. The mechanism of DNA re-methylation during the transition from LTC-hPGCLC to hEGCLC is unknown. During this transition, expression of the de novo DNA methyltransferases DNMT3A and DNMT3B as well as the maintenance methyltransferase DNMT1 (along with its cofactor UHRF1) was upregulated to the level observed in hiPSCs (Figure 4C). The increased expression of these methyltransferases/cofactor may support global DNA re-methylation in hEGCLCs. Although expression of DNMT3L mRNA was rather decreased, DNMT3L protein might have been able to support functions of the de novo methyltransferases during the transition.
LTC-hPGCLCs are telomerase-positive cells whose telomere length is preserved during long-term expansion
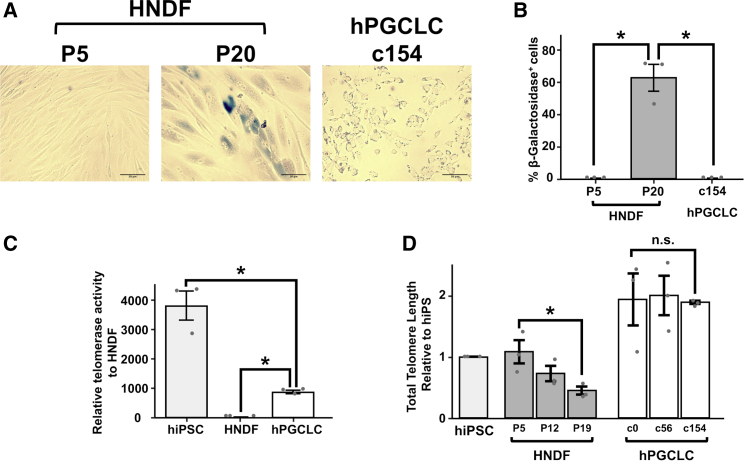
Because the growth curve of LTC-hPGCLCs was linear during their long-term expansion (Figures 2H, S4B, and S4C), we examined whether LTC-hPGCLCs experience cellular senescence. Senescence-associated β-galactosidase activity was detected in human neonatal dermal fibroblasts (HNDFs), from which the A4 and A5 hiPSC clones were generated (Mitsunaga et al., 2017), after 20 passages (Figures 6A, 6B). In contrast, our attempts to detect β-galactosidase activity in c154 LTC-hPGCLCs under the same condition was conclusively negative (Figure 6B). LTC-hPGCLCs had significant telomerase activity that was ∼25% the level of hiPSCs (Figure 6C) and showed no sign of telomere length shortening during long-term expansion in cell culture (Figure 6D). These results indicate that the LTC-hPGCLC is a telomerase-positive cell culture model whose proliferation is not limited by cellular senescence, reminiscent of the ability of human pluripotent stem cells or malignant cells to proliferate without apparent limits of passaging.
Figure 6.
Senescence assessment of LTC-hPGCLCs
(A and B) Enzymatic detection of senescence-associated β-galactosidase (βGal) activity. Human neonatal dermal fibroblasts (HNDF) were maintained for five or 20 passages and subjected to βGal staining. (A) Phase contrast microscopic images (scale bar, 50 μm). (B) Quantification of βGal+ cells (mean ± SEM). Data were pooled from three independent experiments. (C) Telomerase enzymatic activity. Data were normalized to HNDF and shown as mean ± SEM Data were pooled from three independent experiments. (D) Whole-genome telomere length. hiPSCs and LTC-hPGCLCs and HNDF (passages 5–19) were subjected to a telomere length assay, and data are presented as mean ± SEM normalized to hiPSCs. Data were pooled from three independent experiments. ∗Statistical significance (p < 0.05, t test); n.s., not significant.
Differentiation of LTC-hPGCLCs to DAZL-positive cells resembling M-prospermatogonia in the xrTestis organ culture
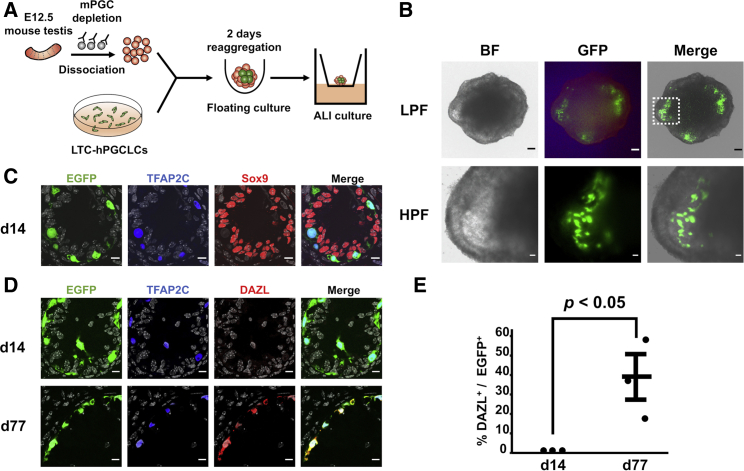
Hwang et al. recently developed the xenogeneic reconstituted testis (xrTestis) organ culture system consisting of mouse fetal testicular somatic cells and hPGCLCs and demonstrated that freshly isolated, DAZL-negative hPGCLCs were capable of differentiating to DAZL-positive cells resembling M-prospermatogonia (Hwang et al., 2020). We constructed the xrTestis organ culture aggregates using EGFP-labeled LTC-hPGCLCs expanded for 120 days in cell culture (Figure 7A). The xrTestis spontaneously formed tubular structures reminiscent of seminiferous tubules, and LTC-hPGCLCs were localized exclusively within these tubules (Figure 7B). IF of the cross sections of the tubules revealed central hollow spaces surrounded by Sox9-positive mouse Sertoli cells, and the EGFP+/TFAP2C+ LTC-hPGCLCs were integrated within the tubules outside the Sertoli cells (Figure 7C). After 77-day of xrTestis culture, approximately 40% of the EGFP+/TFAP2C+ cells expressed DAZL, whereas no DAZL expression was detected at 14 days (Figures 7D, 7E). These data indicate that LTC-hPGCLCs retained their capability of differentiating to “M-prospermatogonia-like cells” (MLCs) in xrTestis after 120 days of expansion in cell culture.
Figure 7.
Differentiation of LTC-hPGCLCs to DAZL-positive gonocyte-like cells in xrTestis
(A) Schematic representation of xrTestis. Single-cell suspensions of E12.5 mouse fetal testes were depleted of mouse PGCs (mPGCs) by MACS and co-cultured with EGFP-labeled LTC-hPGCLCs in a floating culture condition. Resulting cell aggregates were maintained in an air-liquid interphase (ALI) culture condition. (B) Microscopic images of day-77 xrTestis. BF, bright field. The area shown by a dotted rectangle in the low-power field (LPF; scale bar, 100 μm) is enlarged in the high-power field (HPF; scale bar, 20 μm). (C) IF of human TFAP2C and mouse Sox9 in day-14 xrTestis. Scale bars, 10 μm. (D) IF of human TFAP2C and DAZL in day-14 and day-77 xrTestis. Scale bars, 10 μm. (E) Ratio of DAZL+ cells in EGFP+ cells in day-14 and day-77 xrTestis (mean ± SEM of three independently formed xrTestis).
Discussion
Our current study established LTC-hPGCLCs, which can be expanded in cell culture long term in a serum-free and feeder-free condition without losing their homogeneous, PGC-like cellular identity. FACS analyses showed no accumulation of cells that lost expression of the hPGCLC markers PRDM1 or TFAP2C in the LTC-hPGCLC cultures after in vitro expansion for 70–153 days (Figures 2G and S2F). This remarkable stability is in sharp contrast to a preceding study, in which hPGCLCs had to be re-enriched by FACS at every passage since a large portion of them continuously lost their germline identity over time (Murase et al., 2020). Our RNA-seq data demonstrate very strong transcriptomal similarities between LTC-hPGCLCs maintained in culture for 56 and 84 days (Figure 4B) as well as robust and persistent expression of a number of hPGC marker genes (Figure 4C). These results confirmed that our protocol supports FACS-free expansion of LTC-hPGCLCs retaining their hPGC-like identity. Single-cell RNA-seq data demonstrate the highly homogeneous aspects of LTC-hPGCLCs after 120-day expansion in cell culture, whereas heterogeneous expression of cell cycle genes was evident in the homogeneous clusters of cells (Figures 4H, 4I, S5C, and S5D).
During cell culture expansion or passaging, LTC-hPGCLCs show a minimal level of apoptotic cell loss, whereas they are still sensitive to apoptotic induction by staurosporine (Figure S2G). This novel resource would significantly lower a hurdle to using the hPGCLC model for human germline cell research. Successful maintenance of LTC-hPGCLCs in feeder-free cell culture is dependent on four critical factors, namely, the use of SCF, STO-CM, and Matrigel, and the absence of FCS. STO-CM is required to stimulate hPGCLC proliferation (Figures 2F, 3C, and 3D) and to prevent de-differentiation of hPGCLCs to pluripotent hEGCLCs (Figure 3). During the conversion of the POU5F1+/SOX2–/SOX17+ LTC-hPGCLCs to POU5F1+/SOX2+/SOX17– hEGCLCs, expressions of SOX2 and SOX17 were mutually exclusive, with no evidence of SOX2+/SOX17+ double-positive cells (Figures 3E–3G).
In the presence of SCF and FGF2 but the absence of STO-CM, LTC-hPGCLCs were readily converted to hEGCLCs (Figure 3). We observed no difference in the efficiency of hEGCLC generation between freshly isolated hPGCLCs and LTC-hPGCLCs (data not shown). Nicholls et al. showed that early-stage, migrating mouse embryonic PGCs (mPGCs) can generate EGCs but late-stage mPGCs that have settled in the genital ridges rapidly lose this capability (Nicholls et al., 2019). The capability of (LTC-)hPGCLCs to generate hEGCLCs may reflect that hPGCLCs resemble early-stage human embryonic PGCs (Mitsunaga et al., 2017). It is interesting to speculate that late-stage human embryonic PGCs in the genital ridges may be incapable of generating embryonic germ cells and that the xrTestis culture of (LTC-)hPGCLCs might be useful for exploring mechanisms of the retention and/or loss of the reinstatement of the pluripotency; this speculation should be addressed in future studies.
Addition of FCS to our optimized hPGCLC long-term maintenance medium (S-CM; SCF + STO-CM) turned disruptive rather than supportive (Figures S3A–S3C), which may partly explain why preceding efforts of long-term expansion of hPGCLCs in media containing FCS have proved challenging (Gell et al., 2020; Murase et al., 2020). Our attempts to replace the STO-CM with growth factors secreted from STO cells into the medium such as CXCL12, TGFβs, and IGFs (Talbot et al., 2012) have been unsuccessful so far.
Evidence is accumulating that specification of human and monkey PGCs in early-stage embryos require the pioneer transcription factors such as GATA3 and TFAP2A, which bind their target DNA sequences in tightly wrapped heterochromatin regions to induce chromatin remodeling and enhance their access to other transcription factors (Chen et al., 2019; Kojima et al., 2021; Murase et al., 2020; Sasaki et al., 2016). In vivo expression of GATA3 in the PGCs is strong soon after PGC specification but rapidly suppressed during their migration far before they settle in the genital ridges (Sasaki et al., 2016). In vitro expression of GATA3 in hPGCLCs is rapidly induced when precursor cell aggregates are exposed to the hPGCLC induction medium containing BMP4 (Chen et al., 2019; Kojima et al., 2021) but ceased during the course of hPGCLC induction (Chen et al., 2019) or during the initial, short-term (∼10 days) expansion of hPGCLCs in cell culture (Murase et al., 2020). Prolonged expression of GATA3 from an inducible vector in hPGCLCs in the BMP4-containing hPGCLC induction medium resulted in major cell death (Kojima et al., 2021). Thus, our GATA2/3-positive, freshly isolated (c0) hPGCLCs correspond with the GATA2/3-positive hPGCLCs observed by others (Chen et al., 2019; Kojima et al., 2021; Murase et al., 2020), and our GATA2/3-negative LTC-hPGCLCs are equivalent to the post-expansion hPGCLCs described by Murase et al. (2020). Importantly, both the pre- and the post-expansion hPGCLCs obtained by us and others expressed the key hPGC (LC) markers, indicating that the two states of cells are both legitimate hPGCLCs. Thus, our LTC-hPGCLCs stably maintain the GATA2/3-negative, post-expansion state of hPGCLCs, which still resemble the DDX4/DAZL-negative, early-stage hPGCs (Chen et al., 2017). It is unknown whether long-term maintenance of the GATA2/3-positive state of hPGCLCs, which are presumably close to the germline specification stage of very early hPGCs in vivo, is possible or not. Applicability of our method to cell culture expansion of human embryonic PGCs in the early or late stage also remains to be determined by future studies.
Several markers associating late stages of hPGCs such as PIWIL2 or DPPA3 significantly increased their expression during the course of the long-term expansion of LTC-hPGCLCs (Figures 4C and 4D), implying a possibility that LTC-hPGCLCs alone can undergo a certain degree of maturation in vitro. However, the significant degree of global gDNA demethylation in LTC-hPGCLCs observed in our current study (Figure 5C; reaching to 54.2% at c84) was still far weaker than those observed for embryonic hPGCs (∼30% in 59-day male PGCs; Gkountela et al., 2015) or advanced stages of hPGCLC-derived germline cells in xeno-organoid cultures (Hwang et al., 2020; Yamashiro et al., 2018). Therefore, in vitro maturation of LTC-hPGCLCs, if any, seems arrested at a stage mimicking hPGCs before their gonadal settlement. The LTR5 and LTR5_Hs human endogenous retroviruses showed exceptionally strong degrees of demethylation (Figures 5F and 5G) although the reason for the specific strong CpG demethylation at these endogenous retroviruses is unknown. Future studies will be required to elucidate the possible biological significance as well as epigenetic mechanism of the unique and strong CpG demethylation in (LTC)-hPGCLCs.
The global gDNA demethylation involving imprinting genes is a hallmark of epigenetic reprogramming in mammalian PGCs (Seisenberger et al., 2012; Seki et al., 2007). Some of the DMRs controlling imprinted gene expression were significantly demethylated in LTC-hPGCLCs (Figure 5E), and the strong demethylation at the IG-DMR (Figure 5E) may contribute to the increased expression of DLK1 mRNA in (LTC-)hPGCLCs (Figure 4D). Our RNA-seq data provided evidence that the H19 imprinting gene is expressed mono-allelically in the precursor hiPSCs but bi-allelically in LTC-hPGCLCs (Figures S7E and S7F). Prevalence and mechanisms of the apparent loss of imprinting in (LTC-)hPGCLCs need to be characterized further in future studies.
The usefulness of CD38 as a surface marker for FACS enrichment of hPGCLCs was first reported by Irie et al. (2015), and our preceding study showed that the CD38+ hPGCLCs strongly expressed representative hPGC markers such as PRDM1, TFAP2C, SOX17, and NANOS3 (Mitsunaga et al., 2017). We also showed strong expression of CD38 in hPGCLCs generated using a different method, involving intermediate production of the incipient mesoderm-like cells (iMeLCs) and FACS-enriched using other cell surface markers, namely, EpCAM and integrin α6 (Mitsunaga et al., 2017; Sasaki et al., 2015). On the other hand, expression of several key hPGC markers such as NANOS3 or SOX17 in CD38− cells was far weaker than in CD38+ cells (Mitsunaga et al., 2017). Single-cell RNA-seq profiling in our current study presents direct evidence that all individual LTC-hPGCLC cells expressing CD38 simultaneously and strongly express the hPGC markers PRDM1, TFAP2C, NANOS3, and SOX17 (Figures 4H, 4I, S5C, and S5D). Taken together, the CD38-positive hPGCLCs and LTC-hPGCLCs represent the authentic population of cells resembling human embryonic PGCs. On the other hand, there has been no evidence supporting the existence of CD38-negative subpopulations of hPGCs or hPGCLCs.
The protocol described here supports unlimited and rapid expansion of homogeneous hPGCLCs preserving hPGC-like characteristics and normal karyotype in a feeder-free condition. Even after prolonged expansion (120 days), LTC-hPGCLCs retained their capability of differentiating to DAZL-positive MLCs (Figure 7), reproducing a previous observation made with freshly isolated hPGCLCs (Hwang et al., 2020). The emergence of hPGCLC-derived TFAP2C+/DAZL+ double-positive cells in xrTestis was observed in our current study (Figure 7D) and in a preceding study (Hwang et al., 2020), using different rabbit polyclonal antibodies, whose specificity was confirmed by staining cryosections of human adult testes in the present study. Future studies will be necessary to determine whether TFAP2C+/DAZL+ double-positive germline cells also emerge in human embryonic testes. Two male and one female LTC-hPGCLC lines have been established in the current study, and their sex-specific DE-Gs clearly separated their tSNE profiles (Figures 4H, 4I, and S5E). LTC-hPGCLCs can be used for various experiments that require homogeneous populations of live cells, such as high-throughput chemical or genetic screenings, proteomics, or biochemical studies, possibly facilitating pharmacological or toxicological assessments of drugs or environmental toxicants for their germline effects. The histone proteomics experiment presented in our current study (Figure S7A) exemplifies the usefulness of access to large-yield and highly homogeneous populations of LTC-hPGCLCs offered by our method. LTC-hPGCLCs will also be valuable in studying carcinogenic mechanisms of germ cell tumors, which are believed to arise from hPGCs (Oosterhuis and Looijenga, 2019). Since frozen LTC-hPGCLCs are readily stored with high viability, a collection of LTC-hPGCLCs harboring various normal and disease-linked genetic backgrounds (e.g., sex, ethnicities, familial infertility) would be a useful resource that facilitates studies on health of human germline cells.
Experimental procedures
Induction of hPGCLCs
hPGCLCs were generated from hiPSCs as we previously described (Mitsunaga et al., 2017, 2019, 2021) with 5 or 8 days of exposure to BMP4 as shown in Figure S1. Day 8 EBs were dissociated to single cells, using the Embryoid Body Dissociation Kit (Militenyi Biotec, 130-096-348) and stained with an allophycocyanin (APC)-conjugated mouse anti-CD38 antibody (Abcam, Ab134399, dilution 1:20) to collect CD38+ hPGCLCs by using FACSAria Fusion (BD Biosciences).
Preparation of hPGCLC maintenance medium (S-CM medium)
The hPGCLC basal medium contained 13% (v/v) KSR, 1× NEAA, 1 mM sodium pyruvate, and 1× penicillin-streptomycin in Glasgow’s MEM with 2 mM glutamine (ThermoFisher, 11710035). STO-CM was prepared by maintaining 5.0 × 106 mitomycin C-treated STO cells in 12 mL of hPGCLC basal medium for 24 h, removing cells by centrifugation, and storing frozen at −20°C until use. The complete hPGCLC maintenance medium (S-CM for SCF-supplemented CM) was prepared by adding 0.1 mM β-mercaptoethanol, 50 μg/mL L-ascorbic acid, and 100 ng/mL recombinant human SCF to the CM.
hPGCLC expansion culture
FACS-enriched CD38+ hPGCLCs (500–2,000 cells) were inoculated onto STO feeder layers (8-9 × 104 cells/cm2) in a well of six-well plates (Corning, 3506) with SCF-supplemented hPGCLC basal medium containing 10 μM Y27632 dihydrochloride. Medium was changed every other day without Y27632. During the initial phase of hPGCLC expansion, cells were dissociated using Accutase and passaged onto fresh STO feeder layers at every 5–14 days without increasing the number of wells.
When hPGCLCs expanded to ∼20% confluence, cells were dissociated using Accutase and incubated in wells coated with 0.1% gelatin (STEMCELL, 07903) at 37°C for 30 min to remove STO cells, which rapidly adhere to the wells. The feeder-diminished hPGCLCs were transferred onto Matrigel-coated wells in the S-CM and passaged using Accutase. hPGCLCs were also inoculated on wells coated with laminin (Millipore, cc160) or vitronectin (Millipore, cc130).
hEGCLC derivation from hPGCLCs
The hEGCLC derivation medium consisted of the hPGCLC basal medium, 100 ng/mL recombinant human SCF (Peprotech, 300-07), and 20 ng/mL recombinant human FGF2 (R&D Systems, 4114-TC). LTC-hPGCLCs were inoculated into Matrigel-coated six well plates (1,000–2,000 cells/cm2) and maintained in the hEGCLC derivation medium for 10 days with one-half of the medium changed every other day. Thereafter, cells were maintained in the mTeSR plus medium for at least 10 days before use.
Data availability
Bulk RNA-seq, single-cell RNA-seq, WGBS, and digital karyotyping whole genome sequencing data generated in this study are available at NCBI Gene Expression Omnibus and Sequence Read Archive: GSE174485.
Author contributions
Conceptualization, Mu.K. and T.S. Investigation, Mu.K., M. Kobayashi, K. Shioda, J.O., A.R.L., and T.S. Software and formal analysis, Mu.K. and P.C. Writing – original draft, Mu.K. and T.S. Writing – review and editing, Mu.K., Y.S.H., K. Sasaki, J.G., C.L., and T.S. Resources, Mu.K., Y.S.H., K.Sasaki, C.K., R.E.K., H.J., and T.S. Supervision, R.E.K., G.M.C., R.S.W., K.Sasaki, C.L., and T.S. Funding Acquisition, T.S.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
We thank Shino Mitsunaga, Shiomi Yawata, Chie Owa, Maulik Vyas, and the MGH FACS core for technical assistance. We appreciate the Singulomics Corporation for single-cell RNA-seq and the Progressive Assessment of Therapeutics (PATh) PDX Facility at Cornell University College of Veterinary Medicine for PDX support. This work was supported by gifts from The RICBAC Foundation and The Escher Fund for Autism to T.S. The work was also supported in part by NIH grants R21ES024861, R01ES020454, R01ES023316, and R01ES031139 to T.S. and NIH fellowship F30CA247458 to A.R.L.
Published: February 10, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.01.012.
Contributor Information
Mutsumi Kobayashi, Email: m.kobayashi310@gmail.com.
Toshi Shioda, Email: shioda@helix.mgh.harvard.edu.
Supplemental information
References
- Chen D., Gell J.J., Tao Y., Sosa E., Clark A.T. Modeling human infertility with pluripotent stem cells. Stem Cell Res. 2017;21:187–192. doi: 10.1016/j.scr.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Sun N., Hou L., Kim R., Faith J., Aslanyan M., Tao Y., Zheng Y., Fu J., Liu W., et al. Human primordial germ cells are specified from lineage-primed progenitors. Cell Rep. 2019;29:4568–4582 e4565. doi: 10.1016/j.celrep.2019.11.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell J.J., Liu W., Sosa E., Chialastri A., Hancock G., Tao Y., Wamaitha S.E., Bower G., Dey S.S., Clark A.T. An extended culture system that supports human primordial germ cell-like cell survival and initiation of DNA methylation erasure. Stem Cell Rep. 2020;14:433–446. doi: 10.1016/j.stemcr.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountela S., Zhang K.X., Shafiq T.A., Liao W.W., Hargan-Calvopiña J., Chen P.Y., Clark A.T. DNA demethylation dynamics in the human prenatal germline. Cell. 2015;161:1425–1436. doi: 10.1016/j.cell.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Ogushi S., Kurimoto K., Shimamoto S., Ohta H., Saitou M. Offspring from oocytes derived from in vitro primordial germ cell-like cells in mice. Science (New York, N.Y.) 2012;338:971–975. doi: 10.1126/science.1226889. [DOI] [PubMed] [Google Scholar]
- Hwang Y.S., Suzuki S., Seita Y., Ito J., Sakata Y., Aso H., Sato K., Hermann B.P., Sasaki K. Reconstitution of prospermatogonial specification in vitro from human induced pluripotent stem cells. Nat. Commun. 2020;11:5656. doi: 10.1038/s41467-020-19350-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie N., Weinberger L., Tang W.W., Kobayashi T., Viukov S., Manor Y.S., Dietmann S., Hanna J.H., Surani M.A. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253–268. doi: 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikura Y., Ohta H., Sato T., Murase Y., Yabuta Y., Kojima Y., Yamashiro C., Nakamura T., Yamamoto T., Ogawa T., Saitou M. In vitro reconstitution of the whole male germ-cell development from mouse pluripotent stem cells. Cell Stem Cell. 2021;28:2167–2179 e2169. doi: 10.1016/j.stem.2021.08.005. [DOI] [PubMed] [Google Scholar]
- Kojima Y., Yamashiro C., Murase Y., Yabuta Y., Okamoto I., Iwatani C., Tsuchiya H., Nakaya M., Tsukiyama T., Nakamura T., et al. GATA transcription factors, SOX17 and TFAP2C, drive the human germ-cell specification program. Life Sci. Alliance. 2021;4 doi: 10.26508/lsa.202000974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Dong J., Yan L., Yong J., Liu X., Hu Y., Fan X., Wu X., Guo H., Wang X., et al. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell. 2017;20:891–892. doi: 10.1016/j.stem.2017.05.009. [DOI] [PubMed] [Google Scholar]
- Mitsunaga S., Odajima J., Yawata S., Shioda K., Owa C., Isselbacher K.J., Hanna J.H., Shioda T. Relevance of iPSC-derived human PGC-like cells at the surface of embryoid bodies to prechemotaxis migrating PGCs. Proc. Natl. Acad. Sci. U S A. 2017;114:E9913–E9922. doi: 10.1073/pnas.1707779114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsunaga S., Shioda K., Hanna J.H., Isselbacher K.J., Shioda T. Production and analysis of human primordial germ cell-like cells. Methods Mol. Biol. 2021;2195:125–145. doi: 10.1007/978-1-0716-0860-9_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsunaga S., Shioda K., Isselbacher K.J., Hanna J.H., Shioda T. Generation of human primordial germ cell-like cells at the surface of embryoid bodies from primed-pluripotency induced pluripotent stem cells. J. Vis. Exp. 2019 doi: 10.3791/58297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi N., Stel J.M., Shioda K., Qu N., Odajima J., Mitsunaga S., Zhang X., Nagano M., Hochedlinger K., Isselbacher K.J., Shioda T. Erasure of DNA methylation, genomic imprints, and epimutations in a primordial germ-cell model derived from mouse pluripotent stem cells. Proc. Natl. Acad. Sci. U S A. 2016;113:9545–9550. doi: 10.1073/pnas.1610259113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase Y., Yabuta Y., Ohta H., Yamashiro C., Nakamura T., Yamamoto T., Saitou M. Long-term expansion with germline potential of human primordial germ cell-like cells in vitro. EMBO J. 2020;39 doi: 10.15252/embj.2020104929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls P.K., Schorle H., Naqvi S., Hu Y.C., Fan Y., Carmell M.A., Dobrinski I., Watson A.L., Carlson D.F., Fahrenkrug S.C., Page D.C. Mammalian germ cells are determined after PGC colonization of the nascent gonad. Proc. Natl. Acad. Sci. U S A. 2019;116:25677–25687. doi: 10.1073/pnas.1910733116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta H., Kurimoto K., Okamoto I., Nakamura T., Yabuta Y., Miyauchi H., Yamamoto T., Okuno Y., Hagiwara M., Shirane K., et al. In vitro expansion of mouse primordial germ cell-like cells recapitulates an epigenetic blank slate. EMBO J. 2017;36:1888–1907. doi: 10.15252/embj.201695862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterhuis J.W., Looijenga L.H.J. Human germ cell tumours from a developmental perspective. Nat. Rev. Cancer. 2019;19:522–537. doi: 10.1038/s41568-019-0178-9. [DOI] [PubMed] [Google Scholar]
- Saitou M. Mammalian germ cell development: from mechanism to in vitro reconstitution. Stem Cell Rep. 2021;16:669–680. doi: 10.1016/j.stemcr.2021.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K., Nakamura T., Okamoto I., Yabuta Y., Iwatani C., Tsuchiya H., Seita Y., Nakamura S., Shiraki N., Takakuwa T., et al. The germ cell fate of cynomolgus monkeys is specified in the nascent amnion. Dev. Cell. 2016;39:169–185. doi: 10.1016/j.devcel.2016.09.007. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Yokobayashi S., Nakamura T., Okamoto I., Yabuta Y., Kurimoto K., Ohta H., Moritoki Y., Iwatani C., Tsuchiya H., et al. Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell. 2015;17:178–194. doi: 10.1016/j.stem.2015.06.014. [DOI] [PubMed] [Google Scholar]
- Seisenberger S., Andrews S., Krueger F., Arand J., Walter J., Santos F., Popp C., Thienpont B., Dean W., Reik W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y., Yamaji M., Yabuta Y., Sano M., Shigeta M., Matsui Y., Saga Y., Tachibana M., Shinkai Y., Saitou M. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development. 2007;134:2627–2638. doi: 10.1242/dev.005611. [DOI] [PubMed] [Google Scholar]
- Talbot N.C., Sparks W.O., Powell A.M., Kahl S., Caperna T.J. Quantitative and semiquantitative immunoassay of growth factors and cytokines in the conditioned medium of STO and CF-1 mouse feeder cells. In Vitro Cell Dev Biol Anim. 2012;48:1–11. doi: 10.1007/s11626-011-9467-7. [DOI] [PubMed] [Google Scholar]
- Tang W.W., Dietmann S., Irie N., Leitch H.G., Floros V.I., Bradshaw C.R., Hackett J.A., Chinnery P.F., Surani M.A. A unique gene regulatory network resets the human germline epigenome for development. Cell. 2015;161:1453–1467. doi: 10.1016/j.cell.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyser R.C.V., Mahammadov E., Nakanoh S., Vallier L., Scialdone A., Srinivas S. Single-cell transcriptomic characterization of a gastrulating human embryo. Nature. 2021;600:285–289. doi: 10.1038/s41586-021-04158-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro C., Sasaki K., Yabuta Y., Kojima Y., Nakamura T., Okamoto I., Yokobayashi S., Murase Y., Ishikura Y., Shirane K., et al. Generation of human oogonia from induced pluripotent stem cells in vitro. Science. 2018;362:356–360. doi: 10.1126/science.aat1674. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bulk RNA-seq, single-cell RNA-seq, WGBS, and digital karyotyping whole genome sequencing data generated in this study are available at NCBI Gene Expression Omnibus and Sequence Read Archive: GSE174485.