Abstract
In recent years, advances in tissue engineering and microfabrication technologies have enabled rapid growth in the areas of in vitro organoid development as well as organoid-on-a-chip platforms. These 3D model systems often are able to mimic human physiology more accurately than traditional 2D cultures and animal models. In this review, we describe the progress that has been made to generate organ-on-a-chip platforms and, more recently, more complex multi-organoid body-on-a-chip platforms and their applications. Importantly, these systems have the potential to dramatically impact biomedical applications in the areas of drug development, drug and toxicology screening, disease modeling, and the emerging area of personalized precision medicine.
Introduction
Drug development models in their current state are inadequate for the development of new pharmaceuticals to treat the many diseases that afflict humans. There is a considerable need for more accurate human-representative systems to model the effects of drug candidate compounds on the body [1,2]. Currently, animal models serve as gold standards for testing, but the drawbacks associated with such models are high costs and uncertainties in interpretation of the results in many pathologies. Animal models are not always representative of results in humans. In vitro systems that use human tissues and are accurate with respect to the human body would be preferable; however, for these systems to serve as legitimate drug discovery tools, key physiological features and toxicology endpoints need to be validated. Traditional in vitro 2D cultures (the norm for early-stage drug compound screening) fail to recapitulate the 3D microenvironment of in vivo tissues [3,4]. As a result, 2D culture can place a selective pressure on cells, significantly altering their phenotypic properties. Drug diffusion kinetics are not accurately modeled in 2D tissue cultures, drug doses effective in 2D are often ineffective when scaled to patients, and the lack of cell–cell and/or cell–matrix interactions in 2D often lead to the loss of cell function [3,5,6]. Instead, ‘organ-on-a-chip’ devices that can recapitulate 3D tissue architectures and the physiological fluid flow conditions that support normal tissues are better options [7]. These engineering platforms facilitate robust hardware systems, capability for scale up, high throughput, and control over physical factors, such as fluid shear stress and mechanical deformations. Many organ-on-chip systems have been developed [8–10]. Likewise, a variety of on-chip disease models have been investigated [9]. To make significant strides in organ-on-a-chip technologies, the next challenge is to combine multiple organs in the same platform to model a reductionist organism-on-a-chip for more advanced and accurate drug and therapeutic studies. This is a crucial feature; as in the human body, tissues and organs are interdependent on one another in a more complicated fashion than achievable in traditional cell cultures or current organ-on-a-chip systems. In this review, we highlight a variety of organoid-on-a-chip systems for applications such as drug screening and disease models, and look to the future of multi-organoid body-on-a-chip systems and applications in personalized precision medicine.
Advance of in vitro organoid development: progression from 2D to 3D models
Development of novel drugs that are effective therapies in humans has been significantly limited because of the inability to accurately model human physiology, including tissue phenotype, function, and signaling mechanisms, in controlled environments that facilitate experimental manipulation. Animal models, although useful over the years and traditionally regarded as the gold standard for drug testing, allow only limited manipulation and study of cellular mechanisms and responses, and experimental results are not always predictive of results in humans. The second traditional type of model system, in vitro 2D cultures, fails to recapitulate the 3D microenvironment, and often function, of in vivo tissues [3,11]. Drug diffusion kinetics varies dramatically, drug doses effective in 2D are often ineffective when scaled to patients, and cell–cell and/ or cell–matrix interactions are inaccurate [5,6]. Tissue culture dishes differ in major ways from the tissue where cells were originally isolated: their surface topography, surface stiffness, oxygen tension, mechanical loading, biochemical composition, differences in local tissue density, and most importantly, a 2D rather than 3D architecture. These dramatically unnatural characteristics can significantly alter the molecular and phenotypic properties of many cell types. Over the past decade, the functional differences between 2D cultures and 3D constructs have been demonstrated repeatedly in many tissue types and diseases. In general, 3D systems outperform 2D cultures in many aspects, including recapitulating in vivo function and response to drugs and toxins [12]. In current drug development pipelines (Fig. 1a), the lack of in vivo accuracy in traditional 2D cultures has contributed to countless discrepancies between in vitro drug screening outcomes and later performance in patients during or even after clinical trials [13]. As an example, it was recently demonstrated that, on 2D tissue culture dishes, metastatic colon carcinoma cells appeared epithelial, but, when transitioned into a tumor foci form factor inside a 3D liver organoid host environment, they ‘switched’ to a phenotype that appeared more mesenchymal and metastatic, which is more representative of a malignant tumor in the human body [14]. These kinds of documented difference force us ask to why 2D cell cultures are still being used in drug and toxicology screening when they cannot recapitulate the basic morphology, phenotype, or function of cells and tissues inside the body.
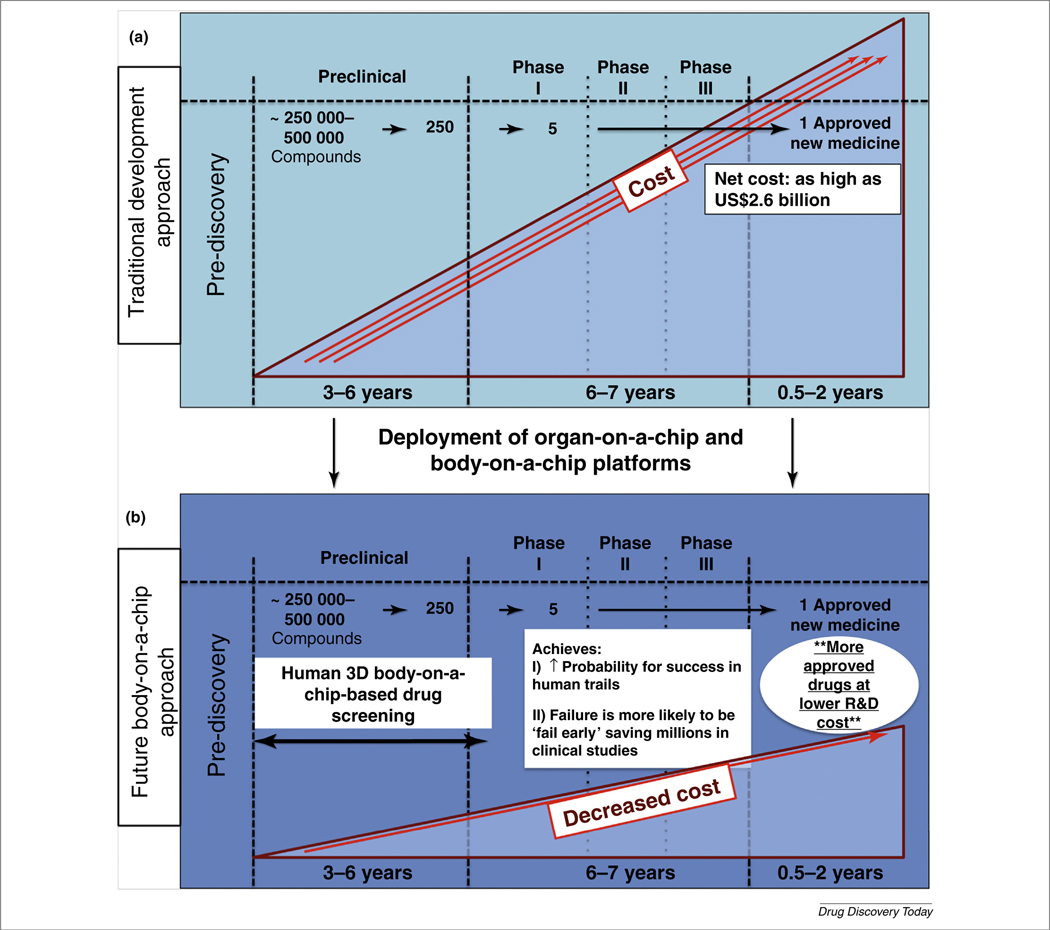
FIGURE 1.
Potential improvements in the drug development pipeline as a result of the deployment of organ-on-a-chip and body-on-a-chip technologies into pharmaceutical research and development. (a) The current drug development pipeline requires many years and multiple billions of dollars to bring a drug to market. (b) Plugging in human-based biofabricated on-a-chip platforms into preclinical stages could drastically improve the efficiency of the drug development pipeline.
Fortunately, bioengineered construct technologies have evolved to the point that they can better mimic the structure, cellular heterogeneity, and function of in vivo tissue. These model organs can often be maintained in viable states for longer periods of time and are cultured to develop functional properties similar to native tissues. They can also often recapitulate the dynamic role of cell–cell, cell–extracellular matrix (ECM), and mechanical interactions that cells experience inside tissues. Oxygenation can be a concern in 3D tissue models. If they are too large in bulk, an oxygen gradient and, therefore, viability gradients can develop, leading to necrotic regions. However, oxygen gradients exist in vivo. As such, as long these gradients are taken into consideration and controlled, either by limiting the size of a 3D organoid, or by creating perfusion channels or other features, 3D systems can in this respect also better represent in vivo tissues. In general, these relatively new model systems are superior to their 2D predecessors for drug and toxicology testing.
The general concept of performing research using 3D versus 2D cultures has gained significant traction over the past decade. However, hurdles and challenges remain. 2D cell culture is an established practice that will remain a widely used tool for many years, because it is simply too easy and too inexpensive in comparison to many 3D culture systems. Additionally, in most cases, using 3D systems in the laboratory is more complicated, requiring learning new technologies, such as biomaterial development and biofabrication techniques. Furthermore, once 3D culture environments have been established, processes regarded as simple in 2D culture, such as cell harvesting and cell passaging, can be difficult and, in some cases, not possible without harming the cells. For example, if cells are cultured within a 3D hydrogel construct, one must effectively dissolve the matrix to isolate or harvest the cells. Some biomaterials support cell isolation by building specific features into the material [15], but most do not, instead requiring enzymatic dissolution that can, in some cases, influence cell viability or phenotype. Also, most cell imaging techniques were developed for 2D cell cultures, environments in which cells exist in a narrow focal plane. In 3D, cells reside in many focal planes. Consequently, high-quality imaging in 3D might require confocal or macroconfocal microscopes and cameras, expensive equipment that many laboratories do not have. Additionally, there are a variety of assays that can be significantly more difficult to run on 3D models, or that require significant effort in adapting for compatibility with 3D models. Some on-a-chip device hardware (polydimethylsiloxane, for example) is susceptible to fouling and drug and protein adsorption, but advances in materials for device hardware are working to solve this problem.
However, when one comprehensively evaluates the data, it is generally clear that 3D systems or dynamic on-a-chip platforms often surpass static 2D environments in achieving the accurate modeling of human physiology [12]. As a result, these more capable 3D platforms have immense potential to influence the drug development pipeline, by decreasing development costs and increasing the success of drug candidates compounds in clinical trials (Fig. 1b). Perhaps just as important, these models can be used to identify nonoptimal drug candidates early, before human trials.
Organs-on-a-chip and their applications
In recent years, advances in biotechnology areas, such as tissue engineering [16], biomaterials [17], and micro- and biofabrication [18], have allowed the derivation of new biological systems with massive potential as test platforms. Researchers have developed a variety of human-derived in vitro models that can be used as specific normal tissues for testing drugs, toxins, and drug candidates [10,19–21]. Furthermore, through advancements in genetics paired with tissue-engineering technologies, these platforms can be used as specific disease models [9,22–24]. Further combinations are possible with other technologies, such as microfabrication, and microfluidic technology organoid-on-a-chip devices that support cell and organoid culture, fluid flow, high-throughput testing, and environmental sampling and biosensing. These organs-on-a-chip vary widely, represent a range or tissue types, and are currently being explored and, in some cases, implemented in drug discovery [8], and purport to significantly impact the future of medicine. Here, we specifically highlight examples of liver-on-a-chip, vessel-on-a-chip, lung-on-a-chip, and cancer-on-a-chip systems. However, many varieties of other promising tissue type platforms exist, including brain, gut, heart, and others.
Liver-on-a-chip
Early tissue- or organoid-on-a-chip devices were devices or hardware with patterns or wells geometrically designed to drive cell aggregation, thereby creating multicellular organoids. For example, devices were designed with microwells of various shapes and sizes containing cell-adherent collagen or nonadherent polyethylene glycol. Based on the well parameters, HepG2 cells or rat hepatocytes could be formed into either spheroids or cylindrical constructs in a highly controlled manner. These 3D constructs maintained better liver function than the 2D controls [25,26]. In another example, spheroids were created from HepG2 cells using an array of channels and pyramid-shaped microwells, which could then be used in the same piece of hardware for increased throughput drug screening [27].
In recent years, liver-on-a-chip devices have become more complex. Now they are often used to control fluid flow for circulating nutrients, drugs, or toxins, aliquot sampling, or even connecting liver constructs to other tissue types to form multi-organoid devices, as we discuss below. In one such liver-on-a-chip, hydrogels were used to encapsulate HepG2 and NIH-3T3 cells. These arrays of 3D organoids had increased liver function compared with 2D controls, and responded to acetaminophen in a drug-screening experiment [28]. A versatile photopolymerizable hyaluronic acid biopolymer system was recently used to in situ photopattern HepG2 cells in 3D liver constructs inside parallel channel fluidic devices fabricated by soft lithography and molding of PDMS [29]. The authors showed how this system could be used for toxicity screening by administering multiple alcohol concentrations per chip. As expected, alcohol administration resulted in dose-dependent decreases in viability and function with increasing doses [29]. There is now a focus on miniaturizing this and other systems further to increase throughput in experiments on single devices that are the size of traditional microscope slides. Miniaturization and microfabrication approaches can be used to generate more intricate liver structures, such as liver sinusoids. Precise seeding and layering of hepatocytes and endothelial cell co-cultures with fluid flow can generate sinusoid-like models [30]. In another example, a device with two distinct chambers separated by a porous membrane with human hepatocytes and endothelial cells was shown to generate higher albumin and urea secretion under flow conditions compared with traditional static cultures [31].
Vessel-on-a-chip
The term ‘microfluidics’ indicates that this technology in general should be naturally capable of fluid routing. Thus, microfluidic devices are effective for serving as vascular models and systems. Moreover, since many drugs are introduced directly to the blood stream rather than orally, and most other drugs arrive in the drug stream shortly after oral or airway introduction, fluidic systems that mimic vasculature are an important component of drug-screening technologies that can be used when assessing drug delivery and transport. Many vascular-like fluidic devices have been developed, including both straight channels devices [32,33] and fluidic devices with more complex branched features [34,35]. One of the major roles of the vascular system in the body, beyond transport of blood and nutrients from one tissue to another, is the transport of drugs and other molecules through the endothelium and vascular wall into adjacent tissues. Many microfluidic devices have been developed to model endothelial transport. In one example, a device comprising two individual channels passing perpendicular to one another was developed [36]. At the Passover point, a semipermeable membrane on which endothelial cells were seeded formed a cellular barrier between the two channels. Using fluorescently labeled albumin, mass transport through the endothelial monolayer could be quantified by a laser in the outflow channel [36]. Another endothelial device, capable of regulating shear stress conditions, was used to assess the influence of fluid shear stress levels on nanoparticle uptake by the endothelial cells, demonstrating the capability to control flow and shear stress to mimic various zones of vasculature in the body that might respond differently to administered drug agents [37]. Likewise, other microfluidic endothelial devices have been used to assess how changing the geometry and size of drugs and nanoparticles can change the rate that they adhere to and pass through endothelium [38]. As stated above, one function of vasculature is to allow transport between tissues and organs throughout the body. This function is being implemented in multi-organoid body-on-a-chip devices currently in development in several laboratories.
Lung-on-a-chip
The lungs, which are exposed to air, serve as one of the most common ports of entry in the human body for drugs, toxins, pathogens, and other agents. Therefore, modeling the lung as an organ-on-a-chip system is important if one is interested in assessing the administration and mass transport of aerosol-based drugs. Lung-on-a-chip devices have been under development for about a decade [39]. Most devices comprise lung epithelial–endothelial monolayers that are generally on either side of a semipermeable membrane. These interfaces often separate an airway channel and a fluid channel, forming a barrier that can facilitate transport across the interface under the appropriate conditions. Additionally, lung epithelial cells only differentiate correctly when exposed to air. In several more complex devices, additional pneumatic channels were built into the devices, and cyclic wall stretching paired with fluid dynamics in the fluid channel can provide cues to the cells that resemble the mechanics of breathing [40,41]. These semifunctional models have proven to be versatile, being able to serve as mimics of several lung pathologies, including inflammation, pulmonary edema, mucus plug rupture, and epithelial cell damage, and have begun to be used in drug-screening applications [42–45]. These on-a-chip lung models typically rely on this bilayer construction, rather than on a 3D bulk construct morphology.
Cancer-on-a-chip
In addition to typical organoid types, such as liver and heart, tumor organoids have also been combined with microfluidic platforms to form tumor-on-a-chip devices for more substantial in vitro applications with more capabilities. The microenvironments of tumors are complex, with varying levels of vascularization, pressures, and mass transport. These physical characteristics are parameters that can be monitored and controlled using microfluidic and microfabrication strategies. Cancer-on-a-chip systems can be used for general drug development screening and dose testing, and to test treatment regimens on a patient-by-patient basis [46,47].
Recent advancements in tumor-on-a-chip include the development of devices designed for integration with additional highly complex, advanced technologies, such as imaging or microarray analyses, allowing more novel investigations and detailed observations and quantifications of tumor cell behavior to be performed. The small size of on-a-chip systems has been shown to have a significant role in cell metabolism, based on the bioavailability of oxygen. This metabolism analysis platform demonstrated that the microfluidic environment provided more access to oxygen compared with Petri dish cultures, resulting in increases in Krebs cycle activity and decreased expression of hypoxia-regulated factor 1 [48]. A device comprising multiple drug gradient mixers and parallel cell culture chambers was developed to support multi-dose drug screens paired with cell labeling and high content imaging data collection on-chip [49]. In another example, microscale bioreactors were prepared that housed hepatocytes, nonparenchymal cells (NPCs), and breast cancer cells with the goal to model the hepatic liver niche. The device contained oxygen sensors, micropumps for controlling nutrient distribution, and real-time sampling capabilities [50]. This study resulted in the documentation of the spontaneous dormancy of the breast cancer cells when inside the hepatic niche, which was believed to be a result of the microenvironment cytokine profiles that were created by the other cells. Additionally, breast cancer has been studied using a recently developed mammary duct-on-a-chip and breast cancer-on-a-chip systems [51,52]. In another device, HCT-116 colon carcinoma cells and HepG2 cells (used as a liver model), were encapsulated in Matrigel in separate chambers, while myeloblasts were formed with alginate gels an additional chamber to simulate bone marrow, such that the cytotoxic effects of the 5-fluorouracil (5-FU) prodrug tegafur could be tested on each cell type. Interestingly, in 3D, the liver constructs were able to metabolize tegafur to 5-FU, resulting in cell death in the other 3D constructs, whereas cells in 2D could not metabolize the prodrug to its activated form [53].
Lung tumor models have also been evaluated in microfluidic device platforms. Human non-small cell lung cancer was assessed for sensitivity to several common chemotherapy agents in a 3D organoid form versus 2D controls in parallel channel devices for increasing throughput and simplifying drug administration [54]. In another example, lung cancer spheroids were formed from either cell lines or patient-derived lung cancer cells, with and without pericyte co-cultures, and screened for susceptibility to the drug cisplatin. Co-culture systems were observed to demonstrate higher levels of chemoresistance, demonstrating the important results that can be attained by using multi-cell type systems for drug screening [55]. It is unlikely that these same results would be attainable in single cell type cultures.
These examples of tumor-on-a-chip platforms demonstrate the kinds of experimentation, screening protocol, and finding that are possible as on-a-chip systems gain acceptance and become the norm in cancer research.
Body-on-a-chip: multi-organ systems and future applications
On-a-chip technologies have gained significant momentum in recent years. Although relatively new technologies, many are already showing promise for applications in research and development. However, over the past few years, systems of increased biological complexity have begun to emerge that feature more than one organoid [56–59]. These two-organoid [60] or multi-organoid devices, sometimes referred to as ‘body-on-a-chip’ systems, have immense potential beyond that of single organoid platforms, but, until recently, primarily comprised cell lines and even animal cells [53,61], rather than fully functional human primary cells or fully differentiated cells derived from stem or progenitor populations; thus, they required additional advancement to accurately mimic human physiology and responses to factors such as drugs and toxins. However, within the past year, there have been several notable published studies demonstrating more complex multi-tissue representative systems. In one such example, a four-tissue system was developed in a perfusion platform that did not require a pump, in which 2D tissue cultures of liver, cardiac, skeletal muscle, and neuronal components were integrated within a single device. This platform was then deployed as a screening tool to assess cell toxicity in experiments using doxorubicin, atorvastatin, valproic acid, acetaminophen and N-acetyl-m-aminophenol [62]. This pumpless concept has also been used in a two-tissue gut and liver system that incorporated trans-epithelial electrical resistance sensors for gut epithelial barrier function monitoring and supported basic liver function [63]. In another study, a microfluidic platform was developed containing microphysiological intestine and skin constructs, liver spheroids, and an epithelial kidney barrier tissue model, in which basic function, gene expression, and viability were maintained for 28 days [57]. These examples represent important steps towards systems that can mimic complex responses and interactions between tissues during drug and toxicology screens.
The importance of multi-organoid integration
In vitro models that accurately recapitulate human tissues and model disease are limited, and there are fewer still in which multiple tissues are integrated in a single platform. This is an important biological limitation, because tissues do not exist in isolation in the body. Moreover, it is essential that tissues receive signals and support, such as vascular, neural, metabolic, and hormonal cues from other locations, for normal tissue viability and function. With respect to drugs, for example, effects in secondary tissues can be as important as effects at the target site, particularly if they induce toxicity. If undetected, these secondary effects can lead to failure or withdrawal from commercial or clinical use because these adverse effects. Likewise, in cancer metastasis, in which some malignant tumor cells are able to migrate from one location to another, multiple tissue or organ sites and often a circulatory system (vascular or lymphatic) are involved. As such, although useful for many applications, single organoid models have limited efficacy for recapitulating the many types of interaction between multiple tissues that occur in the human body. Here, we describe several examples of phenomena that demonstrate the importance of multi-organoid platforms and their ability to potentially model these phenomena.
Cancer
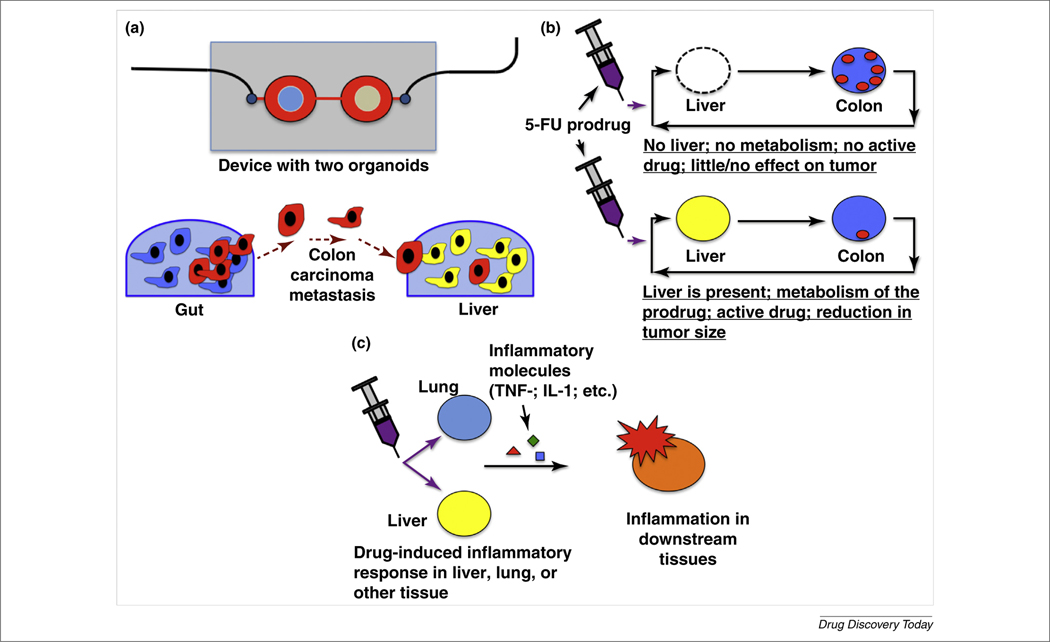
As described above, one medical phenomenon that calls for the use of more than one organoid in a model is cancer metastasis. In metastasis, after often undergoing epithelial-to-mesenchymal transition, cells in a tumor proliferate rapidly in the primary tumor site and intravasate through endothelium into the blood stream or lymphatic system, after which they extravasate and invade a secondary tissue site, often downstream from the primary site. Few in vitro systems have been developed that use a multi-organoid approach to model the kinetics of metastasis. However, they are in development and it has been demonstrated that it is possible to recapitulate metastasis in vitro, although in a reductionist manner. A metastasis-on-a-chip platform was created in such a way to facilitate tracking of the migration of metastatic tumor cells from a bioengineered colon organoid to a bioengineered liver organoid within a simple microfluidic device under recirculating fluid flow (Fig. 2a) [64]. It was shown that metastatic colorectal cancer cells were able to migrate and disseminate out of the colon organoid into the circulating perfusion system and engraft in the downstream liver organoid. Conversely, a non-metastatic colorectal cancer cell type proliferated at the primary site, but never migrated to the liver within the study timeframe [64]. The authors are currently advancing this system in several ways to try to model metastasis more accurately.
FIGURE 2.
Examples of multi-organ interactions that cannot be modeled with single organoid systems. (a) Migration and metastasis of tumor cells from one organ or organoid site to another, demonstrated in vitro in a metastasis-on-a-chip device in which colorectal carcinoma metastasizes from the colon to the liver [64]. (b) Reliance of a prodrug therapy, such as the anticancer 5-fluorouracil (5-FU) prodrug tegafur, on liver metabolism to activate the drug to generate a positive effect by successfully targeting tumor cells. (c) Inflammatory molecules secreted from organs such as the liver and lung upon drug injury can cause detrimental inflammatory responses and cell injury in downstream tissues. Abbreviations: IL, interleukin; TNF, tumor necrosis factor.
Drug testing/toxicology
As we have discussed throughout this article, a broad area of importance in medicine is understanding how multiple organs and tissues respond to administration of particular drugs. A variety of examples demonstrate this concept. For example, 5-FU is a common chemotherapy agent used to treat colorectal cancer. Unfortunately, 5-FU can induce a variety of adverse effects in patients, including cell damage in the gastrointestinal tract. In an attempt to reduce toxicity, several prodrugs have been developed, such as tegafur. Tegafur and other prodrugs are inactive in the administered form, only becoming activated after metabolism, generally by hepatocytes in the liver. Consequently, without including a metabolically active liver organoid in one’s in vitro prodrug drug studies, results are likely to be completely meaningless. Building a platform with a liver and other organoids or tumors would allow metabolism of the prodrug followed by assessment of the activated drug on the downstream tissues or tumors (Fig. 2b).
Disease modeling
Another example that shows the importance of multi-organoid systems incorporates a variety of organs and the vasculature. There are many drugs that are known to cause inflammatory responses. For example, large doses of the very common analgesic acetaminophen (known to many as Tylenol™) cause significant inflammation and toxicity in the liver. Other drugs, such as many chemotherapeutics, including bleomycin, cause inflammation, toxicity, and irreversible fibrosis in the lungs or other organs. In most of these cases, toxicity and apoptosis result in the release of inflammatory molecules, such as tumor necrosis factor (TNF)-α and interleukin (IL)-1 into the blood. Subsequently, these molecules can cause disruption and loss of integrity or inflammation in other tissues, such as the blood vessel endothelium (Fig. 3c). Integrated multi-organoid model systems, if designed correctly, can detect these complex and multi-organ drug effects in a predictive and physiologically relevant manner, whereas single organoid systems cannot.
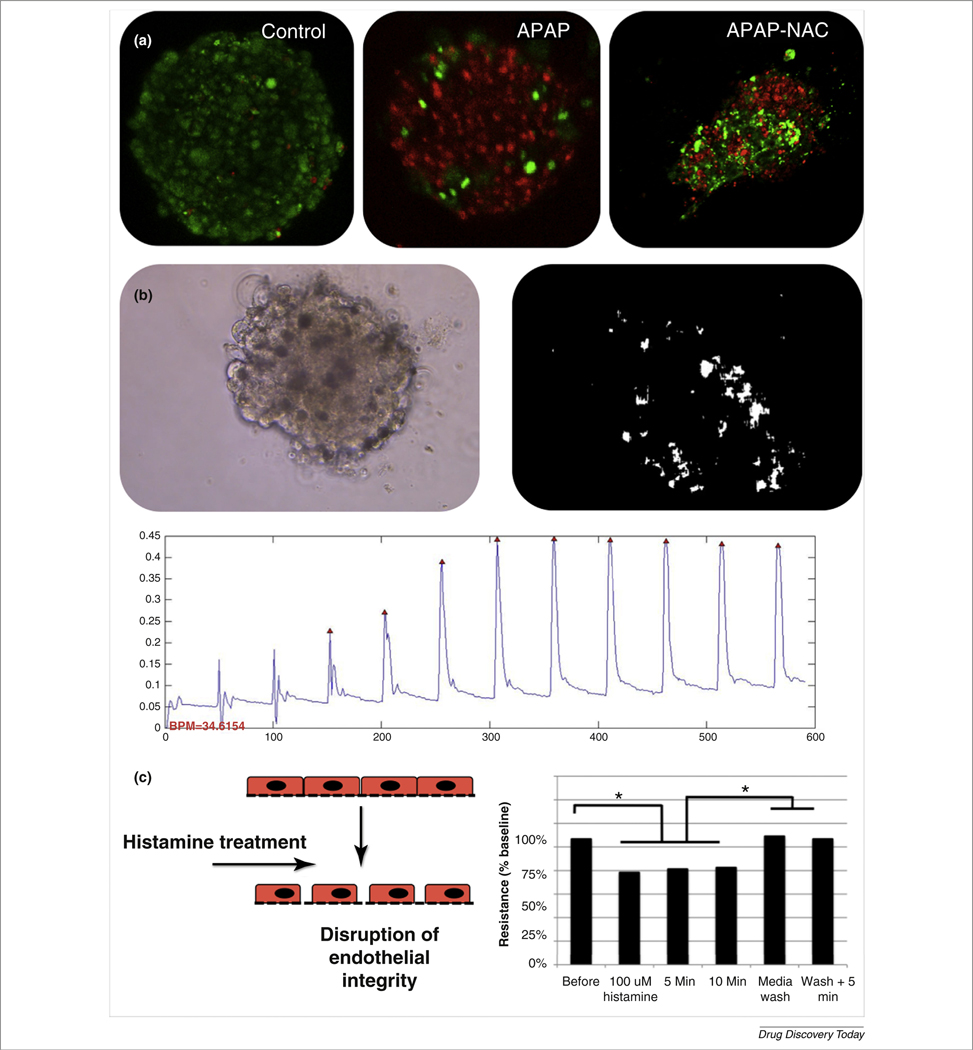
FIGURE 3.
Highly functional organoids for a multi-organoid body-on-a-chip platform. (a) Acetaminophen (APAP) toxicity in liver organoids and reduction in toxicity by N-acetyl-L-cysteine (NAC). (b) Cardiac organoids remained viable long-term and supported the transport of fluorescent dyes [Lucifer yellow (yellow stain) and fluorescein (green stain)] through interconnected ion channels, suggesting high levels of cell–cell communication. (c) Beating analysis of cardiac organoids: an onboard camera captured a video of beating organoids, after which beating rates can be calculated by quantifying pixel movement, generating beat plots. (d) Vascular endothelium devices responded to changes in endothelium integrity as measured by a trans-endothelium electrical resistance sensor. Reproduced, with permission, from (A. Skardal, et al. Unpublished data).
Where are we? The first highly functional multi-organoid systems
As we have described, there is a current lack of a significant number of multi-organ systems, despite the clear need for such systems to sufficiently model and test the complex responses of the human body to drugs, toxins, and disease. However, there is progress being made, primarily within the past several years.
Cancer metastasis
Multi-site metastasis-on-a-chip platforms, as described above, comprising multiple organoids that allow for tumor cells to metastasize from one location to another, have not yet been explored widely. However, there have been on-a-chip devices that assess certain discrete aspects of metastasis. For example, one recent system is a microfluidic device that can help model the process by which multicellular tumor aggregates migrate through a collagen gel and an endothelial layer [65]. Another device that has been developed includes an endothelial cell layer that acts as a barrier to a chamber that mimics 3D bone, allowing researchers to model extravasation of circulating breast cancer cells into bone [66,67]. Other recent devices include a system for assessing the effects of interstitial pressure on cell migration [68] and a system for screening antiangiogenic drugs [69]. These systems illustrate the potential that on-a-chip cancer technologies can have.
However, there is still a major lack of platforms integrating both primary and metastatic sites, and the zones in between (i.e., circulation and endothelium) in a single platform. By providing circulating flow through a system with multiple organoids, one that contains tumor cells, we can recapitulate the dissemination of tumor cells from the primary site organoid into the circulating cell culture media, after which metastatic cells can colonize one or more organoids downstream. As we have described, these are only some of the kinetics of metastasis, but it has been possible to show them in colorectal cancer cells traveling between a colon organoid and liver organoid onboard a metastasis-on-a-chip device. It has also been demonstrated that this platform is highly amenable to experimentation and drug screening [64].
The ECHO platform
Support from the Department of Defense via the Defense Threat Reduction Agency (DTRA) has enabled the development of a multi-organoid integrated body-on-a-chip system for use in bioweapon and chemical weapon assessment and countermeasure development, as well as drug and toxicology screening. During this program, the research team developed and tested the multi-organoid body-on-a-chip platform, termed ECHO (Ex vivo Console of Human Organoids). This platform initially comprised four engineered tissue organoid types (liver, cardiac, vascular, and lung), which were developed independently and integrated into a single system that could provide more complex physiological responses to toxic agents and pharmaceuticals. To date, the team has development a robust three-organoid system, with several other organoids, including lung, in initial stages of integration into the overall system.
With the ECHO system, a comprehensive data set has been compiled showcasing the characteristics of these organoids. In general, the 3D organoids in the system are integrated into the microfluidic platform by using bioprinting of tissue-supportive hydrogels to create 3D ECM-derived environments for organoid maintenance [70,71]. Liver organoids fabricated using liver ECM-derived hydrogels maintained viability and function in vitro for 4 weeks [71], and the presence of key liver markers (e.g., albumin, multiple cytochrome P450 proteins, epithelial cell–cell adhesion markers, dipeptidyl peptidase IV, and organic solute transporter-α) has been demonstrated. These organoids produce albumin and urea, respond to toxins, such as acetaminophen (APAP) in a dose-dependent manner, and can be rescued from such insults with N-acetyl-L-cysteine, a common clinical treatment for APAP overdose (Fig. 3a). Cardiac organoids also remained viable beyond 4 weeks; support transport of fluorescent dye molecules throughout the organoids, suggesting high levels of cell–cell communication; beat spontaneously; and change their beating rates in response to a variety of drugs. These kinetics were captured using an onboard camera system [72,73] and custom software for analysis (Fig. 3c). Additionally a blood vessel endothelium device has been incorporated that responds to agents such as histamine by disruption of its cell monolayer (Fig. 3d), which can allow increased cross-endothelium transfer of larger molecular weight molecules that are in solution.
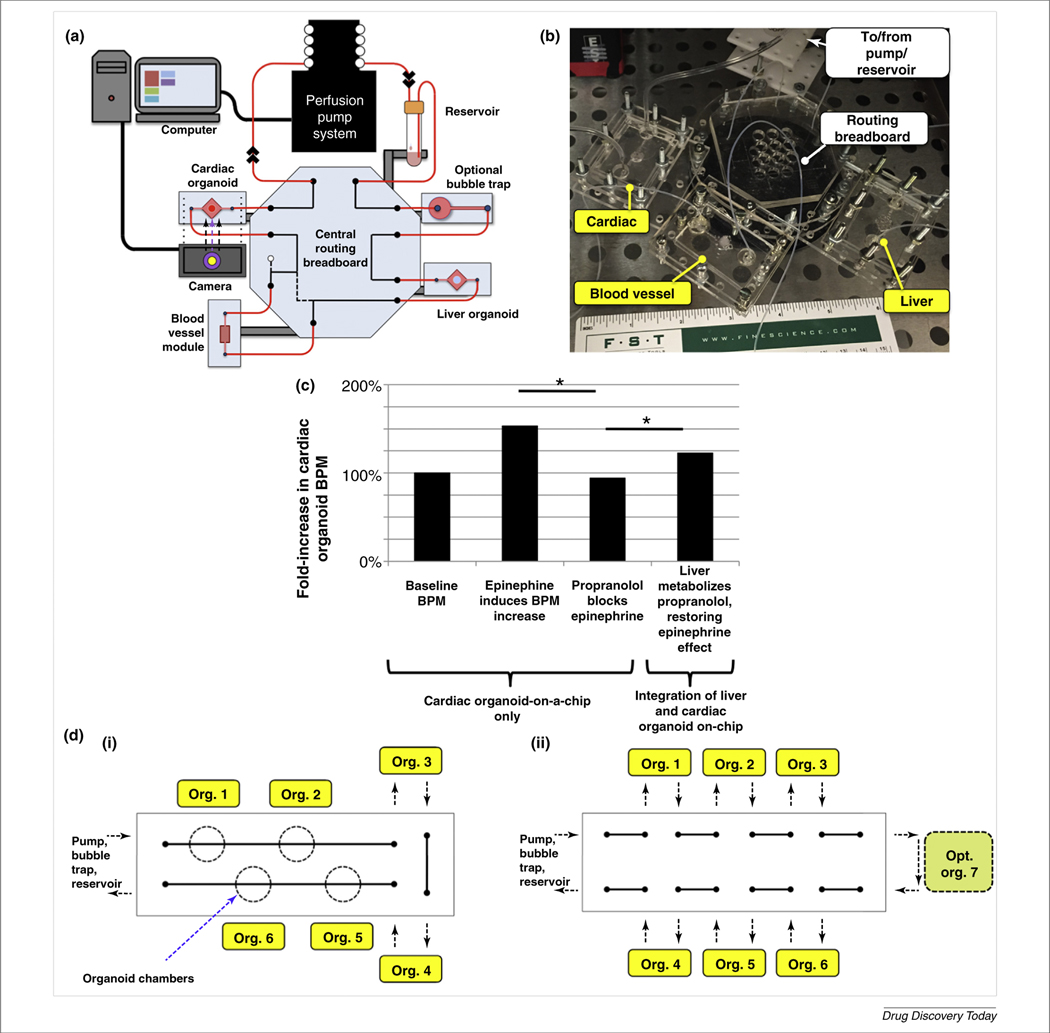
As described above, to date, few research programs in the organ-on-a-chip filed use multiple organ models within one platform. Instead, most focus on a single tissue of interest. This can be shortsighted in many applications, because organs actively interact inside the body constantly. Therefore, multi-organoid interactions should be recapitulated in those platforms that are being developed. Liver organoids, cardiac organoids, and endothelial modules have been integrated in microfluidic devices (Fig. 4a,b) under common media, have shown sufficient viability under these integrated conditions, and have demonstrated multi-organoid responses to drugs, very similar to those encountered in humans. For example, Fig. 4c describes the effects of propranolol and epinephrine on cardiac organoids, with or without liver organoids present in the microfluidic platform. Normally, epinephrine induces an increase in beating rate in cardiomyocytes. Without liver, propranolol, a beta-blocker, blocks the β1- and β2-adrenergic receptors, preventing cardiac beating increases. However, with both organoids linked together in the system, propranolol can be metabolized by the liver component, resulting in significant epinephrine-induced increases in beating rates (A. Skardal, et al. Unpublished data). To our knowledge, these experiments are some of the first interdependent multi-organoid studies to have been performed successfully. Fig. 4d describes additional form factors for expanding the system to additional organoid types and miniaturizing the platform footprint.
FIGURE 4.
A multi-organoid body-on-a-chip. (a) A depiction of a liver, cardiac, and vascular organoid-containing body-on-a-chip platform. (b) Photograph of the three-organoid system. (c) Description of the effects of propranolol and epinephrine on cardiac organoids, with or without liver organoids, illustrating the importance of multi-organoid systems. Without liver, propranolol, a beta-blocker, blocks cardiac-beating increases by epinephrine. However, with both organoids present, propranolol is metabolized by the liver organoid, resulting in a measurable epinephrine-induced increase in beating rates. (di,ii) Future body-on-a-chip platforms for increased capabilities for linking multiple organoids within a single circulatory system. Reproduced, with permission, from (A. Skardal, et al. Unpublished data). Abbreviation: BPM, beats per minute.
Given the lack of optimal models for drug screening, a surprisingly large number of drugs have passed through preclinical studies and clinical trials, after which they remained on the commercial market and were used clinically for years in some cases, before being recalled by the US Food and Drug Administration (FDA) for causing toxic effects in humans. Approximately 90% of these drugs that have been removed from market are done so because of toxic effects in the liver and the heart. Therefore, to demonstrate the capabilities of the ECHO platform, screening studies of a panel of these withdrawn drugs have been performed. These drugs included troglitazone (Rezulin™), an antidiabetic and anti-inflammatory that was recalled for causing liver failure, and mibefradil, an ion channel blocker that was recalled for having fatal interactions with other drugs, including antibiotics. In the ECHO platform, troglitazone and mibefradil both resulted in liver toxicity. Also screened were rofecoxib (Vioxx™), an nonsteroidal anti-inflammatory drug (NSAID) that was recalled because of cardiovascular adverse effects, such as heart attack, stroke, as well as skin reactions and gastrointestinal bleeding; astemizole, an antihistamine that caused slowing of potassium channels, torsade de pointes, and QT prolongation; terodiline, a drug for bladder incontinence that caused QT prolongation and toxicity; and the anticancer drug 5-FU and isoproterenol, a beta-adrenergic agonist, both of which are known to induce cardiac toxicity. In the cardiac organoids, all of these drugs resulted in increased levels of dead cells as doses increased [73]. More importantly, however, using the onboard camera, beating effects were also observed to decrease with dose increases. This is an important distinction, because drugs withdrawn from the market for cardiac toxicity are generally not withdrawn for killing cells in the heart, but rather for causing changes in heartbeat kinetics [87].
ATHENA, DARPA, and NIH initiatives
In addition to the ECHO platform initiative, a variety of other high-profile programs have been underway over the past several years. In particular, the DTRA funded the ATHENA (Advanced Tissue-engineered Human Ectypal Network Analyser) program and the Defense Advanced Research Projects Agency (DARPA) funded a ten-organoid project [74].
These projects are notable and differ from many of the established organ-on-a-chip technologies that we described above in that, similar to ECHO, the explicit goal of the programs was to create integrated multi-organ platforms. Specifically, the ATHENA program, based out of the Los Alamos National Labs, developed a system comprising four organs: liver, heart, lung, and kidney [75]. The DARPA program, based out of Harvard’s Wyss Institute, is working a collection of ten organoids, including representations of endocrine, gastrointestinal, immune, musculoskeletal, and reproductive tissues [76].
Additionally, the National Institutes of Health (NIH) are supporting an equally major organ-on-a-chip program through the National Center for Advancing Translational Science, the National Institute for Biomedical Imaging and Bioengineering, the National Cancer Institute, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institute of Environmental Health Sciences, NIH Common Fund, and NIH Office of Research on Women’s Health. However, the NIH initiative differs in that the funding is divided among a variety of individual research laboratories, which, between them, are working on organoid representations of a range of tissue types [77]. As such, how integration of multi-organoid systems will be performed efficiently remains to be seen.
Organ-on-a-chip systems for personalized precision medicine
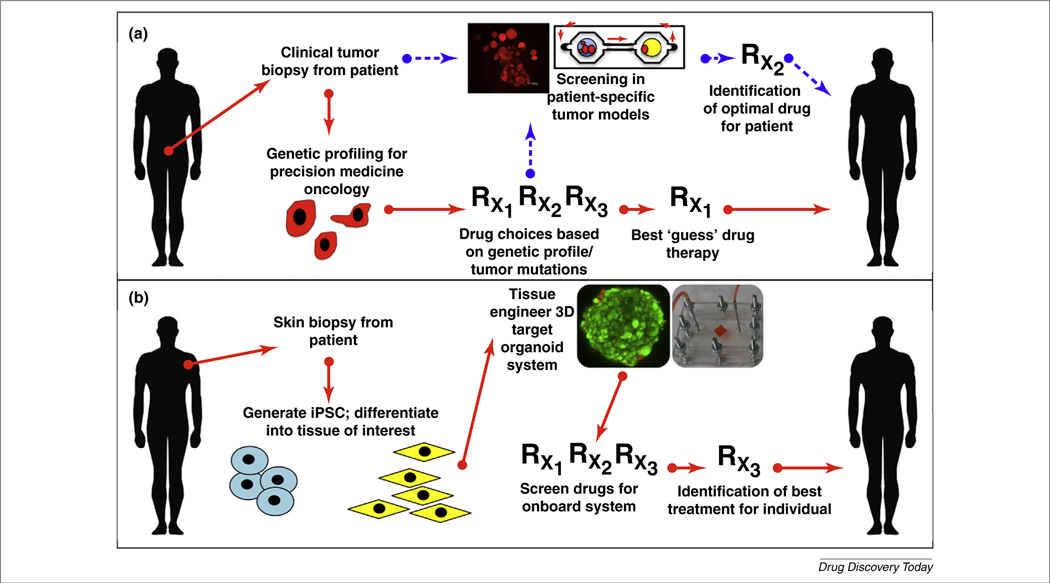
Although numerous in vitro systems are being developed for general drug development screening, few exist for clinical deployment to benefit specific patients. This is an unmet clinical need, because the prescription of therapies to some patients with certain diseases is often a trial and error process. With personalized organoid models (Fig. 5a,b), therapies can be screened using a patient’s own cells in sufficiently physiologically accurate 3D organoid systems. For example, accurate prediction of a patient’s tumor progression and response to therapy is one of the most challenging areas in oncology. Prescribed treatments are often made based on a general success rate of a drug, not on how a specific individual might respond to a drug. Recently, the concept of precision, or personalized, medicine has evolved to address these problems by using the patient’s genetic profile to identify ‘druggable’ targets for treatment [78–80]. However, in real-world practice, the effectiveness of personalized medicine is somewhat less straightforward [81]. After identification of key mutations through genetic profiling, physicians can still be left with a variety of drug options, with no concrete data about potential adverse effects or actual drug effectiveness in the patient. As such, there is a clear need to develop tools that can help predict the response of individual patients to drugs [82,83]. Work is currently underway to develop a multi-organoid platform that contains patient-specific tumor organoids in which drug therapies designed based on genetic profiling can be tested for efficacy. By using the microfluidic devices, it is possible to provide a circulatory system and multiple tissue organoid sites as described above, and thereby visualize and track the kinetics of tumor progression and metastasis to a distant site in vitro [64]. Pairing this with more complex body-on-a-chip systems as described above will further advance these personalized on-a-chip platforms. We believe that it will be possible to perform personalized screens of drugs before treatment, while monitoring all organ systems in the platform for adverse effects, dramatically transforming patient care and improving treatment outcomes. Such powerful and comprehensive technology does not currently exist and would represent an incredible leap forward in how cancer and other diseases are treated.
FIGURE 5.
Use of biofabricated tissues in personalized medicine. (a) In precision medicine for patients with cancer, a potential list of drugs is currently determined based on mutations found in the tumor genetic profile, from which best-guess therapies are prescribed. In the future, cells from tumor biopsies could be used to create in vitro tumor models specific to a given patient. Potentially, effective drug therapies can then be screened in the models, thereby identifying the optimal drug therapy for that patient, both in terms safety and effectiveness. (b) In genetic diseases, cells can be harvested from alternative tissues, such as skin, translated into induced pluripotent stem cells (iPSC), differentiated into cells of the tissue of interest (e.g., lung or heart), and bioengineered into 3D organoids and organoid-on-a-chip systems, after which generic and genome-specific drug therapies can be screened for the original patient.
Conclusions and perspectives
Whereas the rationale for multi-organoid systems is clear, several hurdles remain to be overcome before their acceptance and deployment in actual drug development pipelines and applications, such personalized medicine. Currently, most multi-organoid systems, as well as single organoid systems, do a passable job at mimicking certain aspects in vivo physiology, and can be used for simple drug-testing protocols [84]. However, they are generally created and maintained in low-throughput settings. This currently places a significant limit on the number of experimental compounds or conditions that can be screened. Fortunately, many research groups are working on miniaturizing these devices and platforms to be able to place many organoids and many multi-organoid systems within single devices with small footprints [10]. Significant reduction of size, together with improvements in biosensing and diagnostic technology [72,73,85], (S.A.M. Shaegh et al., unpublished data), has the potential to significantly increase the number of replicates during each screening study, thereby fully realizing the potential of multi-organoid screening platforms in biomedical research and clinical and commercial applications. Another common concern for multi-organoid, and even simple multi-cell type, cultures is the requirement of a common medium. Typically, most high-functioning cell types, such as human primary cells and induced pluripotent stem cell (iPSC)-derived cells, require complex, highly specialized media formulations. However, there is growing evidence that, by transitioning to 3D systems, cells might be able to support one another rather than rely on expensive media supplements and serum. It has been possible to maintain a variety of cell types in viable states in cancer models [64,14] and the use of biomaterial microenvironment customization to support cells in place of the media formulation, including in serum-free conditions, has been investigated [70,71,86]. In ongoing body-on-a-chip work, up to six organoid types in culture have been maintained simultaneously in serum-free media, most of which were biofabricated from primary cells or iPSC. Development of versatile media formulations has the potential to commercially impact the catalog of cell culture media and supplement products. Reducing the variety of cell culture media formulations would help to standardize experimental conditions and reduce the overall cost of many laboratories, thereby allowing researchers to focus on platform development and organoid functional testing, rather than on basic media-screening studies. Multi-organoid body-on-a-chip systems are rapidly advancing and are positioned to be deployed into drug screening in the very near future [7]. These platforms have significant utility in many areas, and will likely dramatically change the way that precision medicine, cancer modeling, common cell culture media development, and drug development are performed.
Biographies

Aleksander Skardal received his BSc in biomedical engineering from Johns Hopkins University and his PhD in bioengineering from the University of Utah. He joined the Wake Forest Institute for Regenerative Medicine in 2010 initially focusing on the application of hydrogel biomaterials for wound-healing cell therapies and fabrication of environments for modulating stem and primary cell viability and function in vitro. He is currently an assistant professor with appointments in regenerative medicine, biomedical engineering, and cancer biology, and is a member of the Comprehensive Cancer Center and Wake Forest Baptist Medical Center. His current research continues to apply hydrogel biomaterials within the framework of several areas including: biofabrication of microtissue organoids for diagnostics and personalized medicine (aka ‘body-on-a-chip’), host tissue organoids for in vitro modeling of metastasis phenomena and mechanisms, integration with microfluidic systems, bio-ink design and bioprinting, and hydrogels for wound-healing therapies.

Dr Thomas Shupe has accumulated 20 years of experience working with in vivo and in vitro models for liver disease and liver regeneration. His postgraduate training was administered by Stewart Sell and Bryon Petersen, both of whom are considered world leaders in the fields of liver physiology, liver progenitor cells, and liver cancer. Dr Shupe was instrumental in the characterization of several molecular factors that govern the phenotypes of liver cells in both normal and disease states. More recently, he was the first to publish a method for the decellularization of intact liver. His current research interests focus on liver tissue engineering and liver- and organ-on-a-chip platforms for drug and toxicology testing. Dr Shupe is currently an assistant professor at the Wake Forest Institute for Regenerative Medicine.

Anthony Atala, MD, is the Director of the Wake Forest Institute for Regenerative Medicine, and the W.H. Boyce Professor and Chair of the Department of Urology at Wake Forest University. Dr Atala is a practicing surgeon and a researcher in the area of regenerative medicine. His current work focuses on growing new human cells, tissues, and organs, and he is considered a pioneer of tissue engineering and regenerative medicine. He is the editor of 12 books, has published more than 400 journal articles, and has applied for, or received, over 200 national and international patents.
References
- 1.Jamieson LE et al. (2015) Chemical analysis of multicellular tumour spheroids. Analyst 140, 3910–3920 [DOI] [PubMed] [Google Scholar]
- 2.Sung JH et al. (2014) Using physiologically-based pharmacokinetic-guided ‘body-on-a-chip’ systems to predict mammalian response to drug and chemical exposure. Exp. Biol. Med. (Maywood) 239, 1225–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kunz-Schughart LA et al. (2004) The use of 3-D cultures for high-throughput screening: the multicellular spheroid model. J. Biomol. Screen. 9, 273–285 [DOI] [PubMed] [Google Scholar]
- 4.Pasirayi G et al. (2014) Low cost microfluidic cell culture array using normally closed valves for cytotoxicity assay. Talanta 129, 491–498 [DOI] [PubMed] [Google Scholar]
- 5.Ho WJ et al. (2010) Incorporation of multicellular spheroids into 3-D polymeric scaffolds provides an improved tumor model for screening anticancer drugs. Cancer Sci. 101, 2637–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drewitz M et al. (2011) Towards automated production and drug sensitivity testing using scaffold-free spherical tumor microtissues. Biotechnol. J. 6, 1488–1496 [DOI] [PubMed] [Google Scholar]
- 7.Marx U et al. (2012) ‘Human-on-a-chip’ developments: a translational cutting-edge alternative to systemic safety assessment and efficiency evaluation of substances in laboratory animals and man? Altern. Lab. Anim. 40, 235–257 [DOI] [PubMed] [Google Scholar]
- 8.Polini A et al. (2014) Organs-on-a-chip: a new tool for drug discovery. Expert Opin. Drug Discov. 9, 335–352 [DOI] [PubMed] [Google Scholar]
- 9.Benam KH et al. (2015) Engineered in vitro disease models. Annu. Rev. Pathol. 10, 195–262 [DOI] [PubMed] [Google Scholar]
- 10.Skardal A et al. (2015) In situ patterned micro 3D liver constructs for parallel toxicology testing in a fluidic device. Biofabrication 7, 031001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Messner S et al. (2013) Multi-cell type human liver microtissues for hepatotoxicity testing. Arch. Toxicol 87, 209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nam KH et al. (2015) Biomimetic 3D tissue models for advanced high-throughput drug screening. J. Lab. Autom. 20, 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKim JM Jr (2010) Building a tiered approach to in vitro predictive toxicity screening: a focus on assays with in vivo relevance. Comb. Chem. High Throughput Screen. 13, 188–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skardal A et al. (2015) Liver-tumor hybrid organoids for modeling tumor growth and drug response in vitro. Ann. Biomed. Eng. 43, 2361–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J et al. (2008) Engineered extracellular matrices with cleavable crosslinkers for cell expansion and easy cell recovery. Biomaterials 29, 4521–4531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy SV and Atala A (2013) Organ engineering – combining stem cells, biomaterials, and bioreactors to produce bioengineered organs for transplantation. Bioessays 35, 163–172 [DOI] [PubMed] [Google Scholar]
- 17.Williams D (2011) The continuing evolution of biomaterials. Biomaterials 32, 1–2 [DOI] [PubMed] [Google Scholar]
- 18.Kang HW et al. (2016) A 3D bioprinting system to produce human-scale tissue constructs with structural integrity. Nat. Biotechnol. 34, 312–319 [DOI] [PubMed] [Google Scholar]
- 19.Skardal A et al. (2010) The generation of 3-D tissue models based on hyaluronan hydrogel-coated microcarriers within a rotating wall vessel bioreactor. Biomaterials 31, 8426–8435 [DOI] [PubMed] [Google Scholar]
- 20.Prestwich GD (2008) Evaluating drug efficacy and toxicology in three dimensions: using synthetic extracellular matrices in drug discovery. ACC Chem. Res. 41, 139–148 [DOI] [PubMed] [Google Scholar]
- 21.Prestwich GD et al. (2007) 3-D culture in synthetic extracellular matrices: new tissue models for drug toxicology and cancer drug discovery. Adv. Enzyme Regul. 47, 196–207 [DOI] [PubMed] [Google Scholar]
- 22.Barrila J et al. (2010) Organotypic 3D cell culture models: using the rotating wall vessel to study host-pathogen interactions. Nat Rev Microbiol 8 (11), 791–801 [DOI] [PubMed] [Google Scholar]
- 23.Nickerson CA and Ott CM (2004) A new dimension in modeling infectious disease. ASM News 70, 169–175 [Google Scholar]
- 24.Nickerson CA et al. (2007) Studying host–pathogen interactions in 3-D: organotypic models for infectious disease and drug development. J. Neuroimmune Pharmacol. 2, 26–31 [DOI] [PubMed] [Google Scholar]
- 25.Mori R et al. (2008) Micropatterned organoid culture of rat hepatocytes and HepG2 cells. J. Biosci. Bioeng. 106, 237–242 [DOI] [PubMed] [Google Scholar]
- 26.Fukuda J et al. (2006) Novel hepatocyte culture system developed using microfabrication and collagen/polyethylene glycol microcontact printing. Biomaterials 27, 1061–1070 [DOI] [PubMed] [Google Scholar]
- 27.Torisawa YS et al. (2007) A multicellular spheroid array to realize spheroid formation, culture, and viability assay on a chip. Biomaterials 28, 559–566 [DOI] [PubMed] [Google Scholar]
- 28.Au SH et al. (2014) Hepatic organoids for microfluidic drug screening. Lab Chip 14, 3290–3299 [DOI] [PubMed] [Google Scholar]
- 29.Skardal A et al. (2015) In situ patterned micro 3-D liver constructs for parallel toxicology testing in a fluidic device. Biofabrication 7, 031001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang YB et al. (2015) Liver sinusoid on a chip: long-term layered co-culture of primary rat hepatocytes and endothelial cells in microfluidic platforms. Biotechnol. Bioeng. 112 (12), 2571–2582 [DOI] [PubMed] [Google Scholar]
- 31.Prodanov L et al. (2015) Long term maintenance of a microfluidic 3-D human liver sinusoid. Biotechnol. Bioeng. 113 (1), 241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D et al. (2014) On-chip evaluation of platelet adhesion and aggregation upon exposure to mesoporous silica nanoparticles. Analyst 139, 906–913 [DOI] [PubMed] [Google Scholar]
- 33.Korin N et al. (2012) Shear-activated nanotherapeutics for drug targeting to obstructed blood vessels. Science 337, 738–742 [DOI] [PubMed] [Google Scholar]
- 34.Lamberti G et al. (2013) Adhesive interaction of functionalized particles and endothelium in idealized microvascular networks. Microvasc. Res. 89, 107–114 [DOI] [PubMed] [Google Scholar]
- 35.Doshi N et al. (2010) Flow and adhesion of drug carriers in blood vessels depend on their shape: a study using model synthetic microvascular networks. J. Control. Release 146, 196–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young EW et al. (2010) Technique for real-time measurements of endothelial permeability in a microfluidic membrane chip using laser-induced fluorescence detection. Anal. Chem. 82, 808–816 [DOI] [PubMed] [Google Scholar]
- 37.Samuel SP et al. (2012) Multifactorial determinants that govern nanoparticle uptake by human endothelial cells under flow. Int. J. Nanomedicine 7, 2943–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolhar P et al. (2013) Using shape effects to target antibody-coated nanoparticles to lung and brain endothelium. Proc. Natl. Acad. Sci. U. S. A. 110, 10753–10758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huh D et al. (2007) Acoustically detectable cellular-level lung injury induced by fluid mechanical stresses in microfluidic airway systems. Proc. Natl. Acad. Sci. U. S. A. 104, 18886–18891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Douville NJ et al. (2011) Combination of fluid and solid mechanical stresses contribute to cell death and detachment in a microfluidic alveolar model. Lab Chip 11, 609–619 [DOI] [PubMed] [Google Scholar]
- 41.Huh D et al. (2010) Reconstituting organ-level lung functions on a chip. Science 328, 1662–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benam KH et al. (2015) Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat. Methods 13, 151–157 [DOI] [PubMed] [Google Scholar]
- 43.Huh D et al. (2012) A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci. Transl. Med. 4, 159ra147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tavana H et al. (2011) Epithelium damage and protection during reopening of occluded airways in a physiologic microfluidic pulmonary airway model. Biomed. Microdevices 13, 731–742 [DOI] [PubMed] [Google Scholar]
- 45.Hu Y et al. (2015) A microfluidic model to study fluid dynamics of mucus plug rupture in small lung airways. Biomicrofluidics 9, 044119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wlodkowic D and Cooper JM (2010) Tumors on chips: oncology meets microfluidics. Curr. Opin. Chem. Biol. 14, 556–567 [DOI] [PubMed] [Google Scholar]
- 47.Young EW (2013) Cells, tissues, and organs on chips: challenges and opportunities for the cancer tumor microenvironment. Integr. Biol. 5, 1096–1109 [DOI] [PubMed] [Google Scholar]
- 48.Ouattara DA et al. (2012) Metabolomics-on-a-chip and metabolic flux analysis for label-free modeling of the internal metabolism of HepG2/C3A cells. Mol. Biosyst. 8, 1908–1920 [DOI] [PubMed] [Google Scholar]
- 49.Ye N et al. (2007) Cell-based high content screening using an integrated microfluidic device. Lab Chip 7, 1696–1704 [DOI] [PubMed] [Google Scholar]
- 50.Wheeler SE et al. (2013) All-human microphysical model of metastasis therapy. Stem Cell Res. Ther. 4 (Suppl. 1), S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vidi PA et al. (2014) Disease-on-a-chip: mimicry of tumor growth in mammary ducts. Lab Chip 14, 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang Y et al. (2015) Evaluation of photodynamic therapy efficiency using an in vitro three-dimensional microfluidic breast cancer tissue model. Lab Chip 15, 735–744 [DOI] [PubMed] [Google Scholar]
- 53.Sung JH and Shuler ML (2009) A micro cell culture analog (microCCA) with 3-D hydrogel culture of multiple cell lines to assess metabolism-dependent cytotoxicity of anti-cancer drugs. Lab Chip 9, 1385–1394 [DOI] [PubMed] [Google Scholar]
- 54.Xu Z et al. (2013) Application of a microfluidic chip-based 3D co-culture to test drug sensitivity for individualized treatment of lung cancer. Biomaterials 34, 4109–4117 [DOI] [PubMed] [Google Scholar]
- 55.Ruppen J et al. (2015) Towards personalized medicine: chemosensitivity assays of patient lung cancer cell spheroids in a perfused microfluidic platform. Lab Chip 15, 3076–3085 [DOI] [PubMed] [Google Scholar]
- 56.Atac B et al. (2013) Skin and hair on-a-chip: in vitro skin models versus ex vivo tissue maintenance with dynamic perfusion. Lab Chip 13, 3555–3561 [DOI] [PubMed] [Google Scholar]
- 57.Maschmeyer I et al. (2015) A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 15, 2688–2699 [DOI] [PubMed] [Google Scholar]
- 58.Materne EM et al. (2015) The multi-organ chip–a microfluidic platform for long-term multi–tissue coculture. J. Vis. Exp. 98, e52526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner I et al. (2013) A dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3D human liver and skin tissue co-culture. Lab Chip 13, 3538–3547 [DOI] [PubMed] [Google Scholar]
- 60.Kim JY et al. (2015) 3D spherical microtissues and microfluidic technology for multi-tissue experiments and analysis. J. Biotechnol. 205, 24–35 [DOI] [PubMed] [Google Scholar]
- 61.Miller PG and Shuler ML (2016) Design and demonstration of a pumpless 14 compartment microphysiological system. Biotechnol. Bioeng 10.1002/bit.25989 [DOI] [PubMed] [Google Scholar]
- 62.Oleaga C et al. (2016) Multi-organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci. Rep. 6, 20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Esch MB et al. (2016) Modular, pumpless body-on-a-chip platform for the co-culture of GI tract epithelium and 3D primary liver tissue. Lab Chip 16 (14), 2719–2729 [DOI] [PubMed] [Google Scholar]
- 64.Skardal A et al. (2016) A reductionist metastasis-on-a-chip platform for in vitro tumor progression modeling and drug screening. Biotechnol. Bioeng. Published online February 16, 2016. 10.1002/bit.25950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Niu Y et al. (2014) Validating antimetastatic effects of natural products in an engineered microfluidic platform mimicking tumor microenvironment. Mol. Pharm. 11, 2022–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bersini S et al. (2014) A microfluidic 3D in vitro model for specificity of breast cancer metastasis to bone. Biomaterials 35, 2454–2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bersini S et al. (2014) In vitro models of the metastatic cascade: from local invasion to extravasation. Drug Discov. Today 19, 735–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Polacheck WJ et al. (2014) Mechanotransduction of fluid stresses governs 3D cell migration. Proc. Natl. Acad. Sci. U. S. A. 111, 2447–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim C et al. (2015) A quantitative microfluidic angiogenesis screen for studying anti-angiogenic therapeutic drugs. Lab Chip 15, 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Skardal A et al. (2015) A hydrogel bioink toolkit for mimicking native tissue biochemical and mechanical properties in bioprinted tissue constructs. Acta Biomater. 25, 24–34 [DOI] [PubMed] [Google Scholar]
- 71.Skardal A et al. (2012) Tissue specific synthetic ECM hydrogels for 3-D in vitro maintenance of hepatocyte function. Biomaterials 33, 4565–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim SB et al. (2012) A mini-microscope for in situ monitoring of cells. Lab Chip 12, 3976–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang YS et al. (2015) A cost-effective fluorescence mini-microscope for biomedical applications. Lab on a Chip 15, 3661–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reardon S (2015) Scientists seek ‘Homo chippiens’. Nature 518, 285–286 [DOI] [PubMed] [Google Scholar]
- 75.Roark K (2015) Project ATHENA Creates Surrogate Human Organ Systems. Los Alamos National Laboratory [Google Scholar]
- 76.Anon (2015) Towards a body-on-a-chip. The Economist June 13 [Google Scholar]
- 77.Anon (2014) NIH Funds Next Phase of Tissue Chip for Drug Screening Program. National Institutes of Health [Google Scholar]
- 78.Tran NH et al. (2015) Precision medicine in colorectal cancer: the molecular profile alters treatment strategies. Ther. Adv. Med. Oncol. 7, 252–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miles G et al. (2015) Genetic testing and tissue banking for personalized oncology: analytical and institutional factors. Semin. Oncol. 42, 713–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bando H and Takebe N (2015) Recent innovations in the USA National Cancer Institute-sponsored investigator initiated Phase I and II anticancer drug development. Jpn. J. Clin. Oncol. 45 (11), 1001–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hayes DF and Schott AF (2015) Personalized medicine: genomics trials in oncology. Trans. Am. Clin. Climatol. Assoc. 126, 133–143 [PMC free article] [PubMed] [Google Scholar]
- 82.Cantrell MA and Kuo CJ (2015) Organoid modeling for cancer precision medicine. Genome Med. 7, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao D et al. (2014) Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Esch EW et al. (2015) Organs-on-chips at the frontiers of drug discovery. Nat. Rev. Drug Discov. 14, 248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim SB et al. (2011) A cell-based biosensor for real-time detection of cardiotoxicity using lensfree imaging. Lab Chip 11, 1801–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Skardal A et al. (2016) Bioprinting cellularized constructs using a tissue-specific hydrogel bioink. J. Vis. Exp. 21, 2016 Published online April; 10.3791/53606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peters MF et al. (2015) Human stem cell-derived cardiomyocytes in cellular impedance assays: bringing cardiotoxicity screening to the front line. Cardiovasc. Toxi. 15 (2), 127–139 [DOI] [PubMed] [Google Scholar]