SUMMARY
Recent findings have demonstrated that mitochondria can be transferred between cells to control metabolic homeostasis. Although the mitochondria of brown adipocytes comprise a large component of the cell volume and undergo reorganization to sustain thermogenesis, it remains unclear whether an intercellular mitochondrial transfer occurs in brown adipose tissue (BAT) and regulates adaptive thermogenesis. Herein, we demonstrated that thermogenically stressed brown adipocytes release extracellular vesicles (EVs) that contain oxidatively damaged mitochondrial parts to avoid failure of the thermogenic program. When re-uptaken by parental brown adipocytes, mitochondria-derived EVs reduced peroxisome proliferator-activated receptor-γ signaling and the levels of mitochondrial proteins, including UCP1. Their removal via the phagocytic activity of BAT-resident macrophages is instrumental in preserving BAT physiology. Depletion of macrophages in vivo causes the abnormal accumulation of extracellular mitochondrial vesicles in BAT, impairing the thermogenic response to cold exposure. These findings reveal a homeostatic role of tissue-resident macrophages in the mitochondrial quality control of BAT.
Graphical Abstract
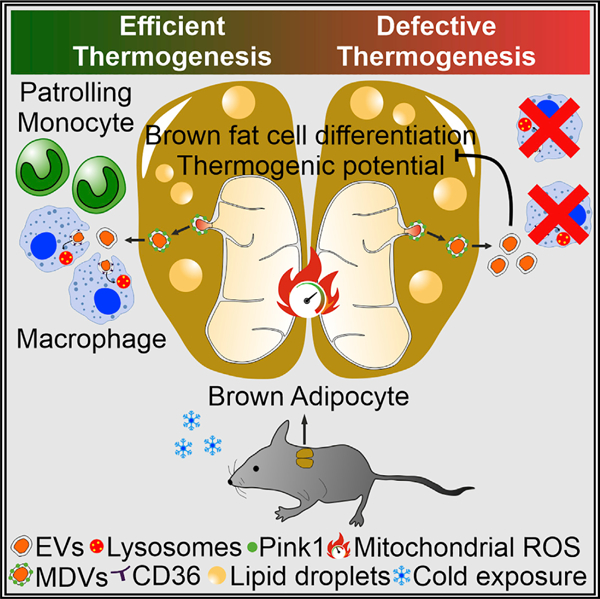
In brief
The mitochondria of brown adipose tissue undergo a deep rearrangement to produce heat. Here, Rosina et al. report that adrenergically stressed adipocytes eject damaged components of mitochondria via extracellular vesicles. Adipose tissue-resident macrophages remove extracellular mitochondria, ensuring thermogenesis. Defective clearance of extracellular mitochondria causes thermogenic failure in brown adipocytes.
INTRODUCTION
Brown adipose tissue (BAT) is involved in heat generation during cold exposure. Cold exposure induces a coordinated activation of mitochondrial metabolism and dynamics, as well as oxidative stress, which requires BAT to mediate mitochondrial quality control processes, such as the selective degradation of nonfunctional mitochondria to maintain mitostasis. In other organs with high metabolic activity, such as the heart, mitostasis is also achieved through the extrusion of mitochondrial components via the generation and release of mitochondrial-derived vesicles (MDVs) (Cadete et al., 2016). This pathway is induced by the generation of mild oxidative stress and supports the biogenesis of a population of MDVs, which bud off from mitochondria and contain a repertoire of specific cargo proteins (Abuaita et al., 2018; Cadete et al., 2016; McLelland et al., 2014). Selective packaging of mitochondrial parts into extracellular vesicles (EVs) has been observed in several physiological settings and is induced during mitochondrial damage (Clement et al., 2020; D’Acunzo et al., 2021; Peruzzotti-Jametti et al., 2021; Todkar et al., 2021). Recent studies suggest that tissue-resident macrophages are involved in actively taking up and removing extracellular mitochondria released by various cell types (Brestoff et al., 2021; D’Acunzo et al., 2021; Nicolás-Ávila et al., 2020; Phinney et al., 2015). However, it remains unknown whether this process occurs in BAT or regulates adaptive thermogenesis.
Recently, it was shown that adipocytes in white adipose tissue (WAT) can transfer mitochondria to macrophages and that this process may contribute to the maintenance of metabolic homeostasis, thereby limiting WAT expansion and obesity pathogenesis (Brestoff et al., 2021). Mice with impaired mitochondrial transfer to macrophages exhibited lower energy expenditure (Brestoff et al., 2021), and macrophage depletion impaired thermogenesis in subcutaneous adipose depots (Henriques et al., 2020; Qiu et al., 2014). In the heart, tissue-resident macrophage depletion or deficient phagocytic capacity led to a defective elimination of extracellular mitochondria from tissues, resulting in tissue inflammation, accumulation of anomalous mitochondria, and metabolic alterations (Nicolás-Ávila et al., 2020). Although these observations suggest that the transfer of mitochondria to macrophages regulates tissue homeostasis, evidence of this immunometabolic crosstalk in BAT is lacking.
This study shows that brown adipocytes release oxidatively damaged mitochondrial parts via EVs to control mitochondrial integrity and maintain thermogenic potential. Brown adipocyte-derived EVs are taken up by BAT-resident macrophages in vivo. Macrophage depletion induces the accumulation of mitochondrial EVs, reduces the expression of mitochondrial proteins, and suppresses BAT thermogenic response to cold exposure. Our data delineate an adipocyte-to-immune cell material transfer axis that is involved in mitochondrial quality control to preserve BAT function and homeostasis.
RESULTS
Brown adipocytes release damaged mitochondrial components via EVs
Brown adipocytes have high metabolic activity and endure extreme mitochondrial stress during cold exposure, which requires these cells to have efficient mitochondrial turnover. To develop a comprehensive overview of mitochondrial dynamics in BAT, we interrogated repository datasets reporting differentially expressed genes between interscapular BAT (highest thermogenic capacity) and visceral WAT (lowest thermogenic capacity). Functional enrichment analysis of overlapping genes revealed that they pertained to the mitochondrion, mitochondrial inner membrane, and extracellular exosome compartments (Figures 1A and 1B). We hypothesized that EVs containing mitochondrial proteins are distinct functional elements in BAT biology; thus, we assessed the potential incorporation of mitochondria in vesicles and their release from brown adipocytes. We found that the EVs collected from BAT and T37i brown adipocytes were positive for the mitochondrial probe MitoTracker Green (MTG) (Figure 1C). Based on the strong capacity of BAT to reorganize the mitochondrial compartment during thermogenesis, we evaluated whether cold stress could further enhance the release of mitochondrial material via EVs. The functional enrichment analysis of the up-regulated genes (Log2FC > 0.77; p < 0.05) in the BAT of cold-exposed mice (4°C, 20 h) revealed that they pertained to extracellular exosomes and mitochondria (Figure 1D). We next profiled the proteome of BAT EVs, and among the 1,779 identified proteins, almost 50% were ascribed as components of extracellular exosome and mitochondrion (Figures 1E and S1A). By comparing the proteome of BAT EVs with mitochondrial proteins (GO: 0005739) and MitoCarta 3.0, we found 148 overlapping proteins (Figure 1F), of which 134 were identified as EV-cargo proteins using the Vesiclepedia database (Figure 1G). The analysis of the top 100 most represented (Figure 1H) and up-regulated proteins (Figure 1I) revealed enriched terms for mitochondrial components and metabolism in BAT EVs 4°C. Remarkably, the proteins identified in BAT EVs 4°C were mostly restricted to brown adipocytes (Figure 1I).
Figure 1. BAT releases mitochondrial parts via EVs.
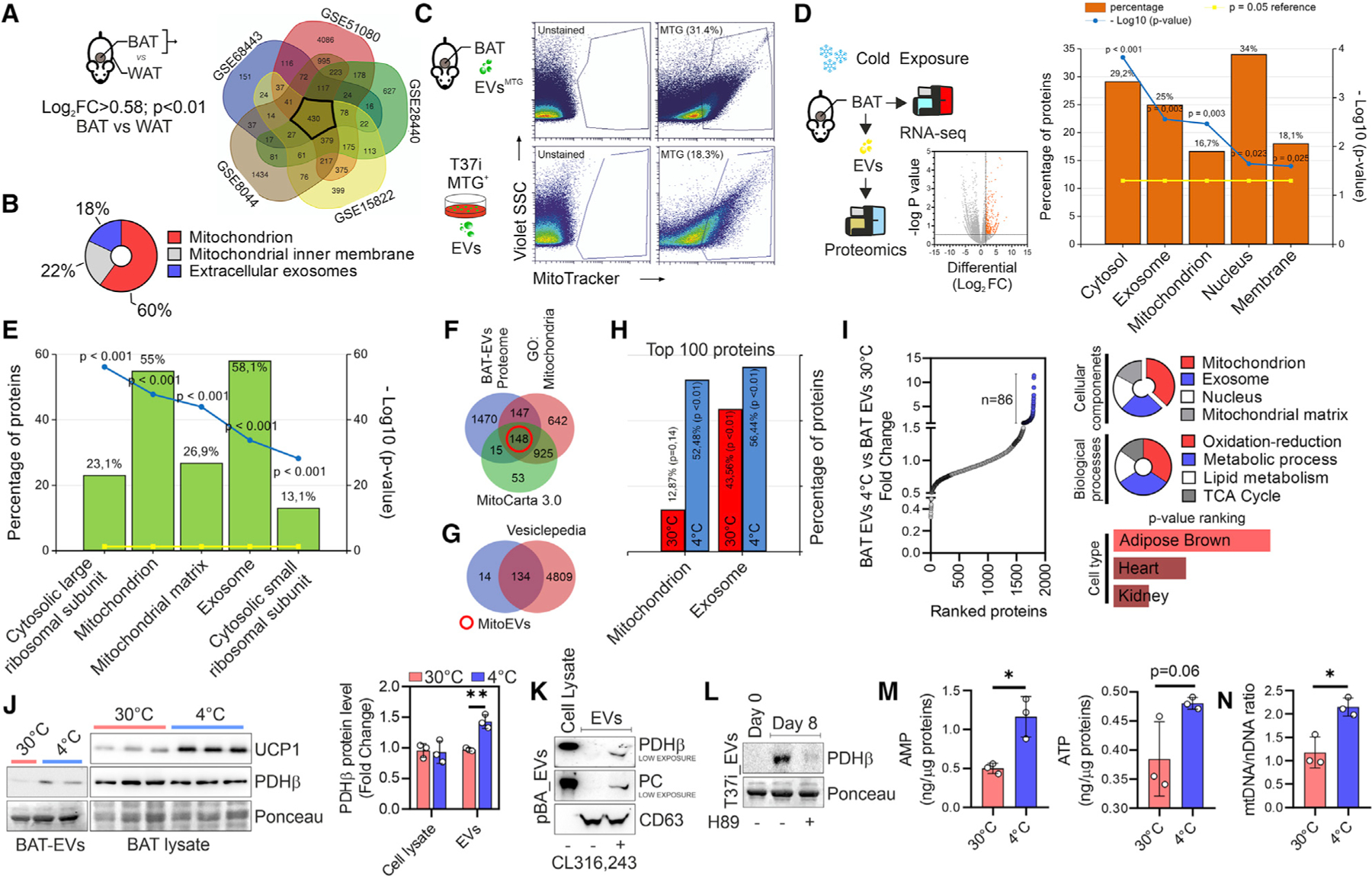
(A) Experimental design and Venn diagram of over-represented genes in BAT with respect to WAT, which were obtained by analyzing different GEO datasets.
(B) The three most representative cellular components of the 430 overlapping genes in (A) are shown (p < 0.05).
(C) Experimental design and flow-cytometry analysis of BAT EVs labeled with MitoTracker Green (MTG) (upper panel) and EVs isolated from MTG-labeled T37i adipocytes (lower panel).
(D) Experimental design and volcano plot (left panel) of differentially expressed genes in the BAT of mice at 4°C versus mice at 30°C (n = 4 mice/group). The five most representative cellular components (p < 0.05) are shown (right panel).
(E) Proteomic analysis of BAT EVs of mice at 4°C versus mice at 30°C. The five most representative cellular components (p < 0.001) are shown. Data are obtained from a pool of BAT EVs (n = 6 mice/group).
(F and G) Venn diagram of the BAT EV proteome with proteins reported in mouse MitoCarta 3.0 and GO terms for mitochondria (F). The 148 overlapping proteins (red circle) were integrated with the Vesiclepedia database (G) after filtering ‘‘mouse’’ species and ‘‘mass spectrometry’’ for analysis.
(H) Proteomic analysis of BAT EVs. The 100 most represented proteins were compared for mitochondrial and extracellular exosome compartments. Data are obtained from a pool of BAT EVs (n = 6 mice/group).
(I) Up-regulated proteins in BAT EVs at 4°C (n = 86; FC > 2.0, blue dots) (left panel). The most significant enriched terms (p < 0.05) for cellular components, biological processes, and cell type are shown (right panel).
(J) Immunoblots of BAT EVs and BAT homogenates (left panel). Densitometric analyses of immunoreactive bands (right panel) are reported as a ratio between PDHβ and Ponceau (loading control). Student’s t test, **p < 0.01, n = 6 mice/group or n = 6 mice/group pooled in pairs for EVs.
(K) Immunoblots of EVs released from primary murine brown adipocytes (pBA) treated with vehicle or CL316,243. CD63 was used as an EV loading control.
(L) Immunoblot of EVs released from undifferentiated (day 0), untreated, or H89-treated T37i adipocytes (day 8). Ponceau was used as loading control.
(M and N) Targeted metabolomics (M) and qPCR analysis of the mtDNA/nDNA ratio (N) in BAT EVs. Data are obtained from a pool of BAT EVs (n = 6 mice/group pooled in pairs). Student’s t test, *p < 0.05.
See also Figure S1.
To validate the proteomic data, we selected pyruvate dehydrogenase E1 subunit beta (PDHβ), which was among the most highly represented proteins in BAT EVs and brown adipocytes (Figure S1B). Increased levels of PDHβ were detected in BAT EVs at 4°C, although there was no difference between 4°C BAT homogenates and 30°C BAT homogenates with respect to their content (Figure 1J). Conversely, despite not being detected in EVs (Table S1), UCP1 was significantly increased in BAT homogenates following cold exposure (Figure 1J). The same enrichment was observed in the EVs isolated from primary (pBAs) and T37i brown adipocytes stimulated with the β3-adrenoreceptor agonist CL316,243 (CL) (Figures 1K and S1C). To explore whether the increased metabolic activity of brown adipocyte mitochondria could be the trigger of mitochondrial protein release, we isolated EVs before and at the end of the adipogenic program and after inhibiting adipocyte thermogenic potential. As shown in Figure 1L, T37i adipocytes released a higher level of PDHβ than undifferentiated T37i cells, and the inhibition of protein kinase A (PKA) signaling by H89 reduced the release of PDHβ via EVs. Targeted metabolomics showed that the metabolites of tricarboxylic acid (TCA) cycle were present in BAT EVs and unexpectedly reduced at 4°C (Figure S1D). By contrast, the levels of AMP, ATP, and mtDNA were higher in the BAT EVs at 4°C than in the BAT EVs at 30°C (Figures 1M and 1N).
The measurement of EV diameter by dynamic light scattering (DLS) revealed two main populations that were approximately 350 nm and 50 nm in size (Figure S1E), which likely represented microvesicles and exosomes, respectively. The filtration of EVs using a 0.22-µm pore-sized filter abrogated the detection of PDHβ and pyruvate carboxylase (PC) in the EVs released from both unstimulated and CL-treated brown adipocytes (Figure S1F), suggesting that extracellular mitochondria were present in the largest fraction of EVs and were not included within exosomes. To confirm this, we inhibited exosome biogenesis by GW4869 or by downregulating the neutral sphingomyelinase 2 (nSMase2, SMPD3), which is involved in exosome biogenesis (Trajkovic et al., 2008), and found increased protein levels of PDHβ and PC in the EVs (Figure S1C). Analysis of various EV fractions obtained by sequential centrifugation confirmed that PC and PDHβ were detectable only in the largest fraction (Figure S1G).
It has been demonstrated that metabolically stressed white adipocytes release damaged mitochondrial proteins through EVs (Crewe et al., 2021). Hence, we questioned whether the release of mitochondrial proteins into EVs could be a consequence of mitochondrial damage due to cold-induced metabolic stress. To demonstrate this, we treated pBAs with mitochondrial stressors, i.e., the cold-mimicking drug CL or low doses of trifluoromethoxy carbonylcyanide phenylhydrazone (FCCP), which provokes mitochondrial damage by inducing uncoupled respiration without adipocyte death. Proteomic analysis revealed distinctive protein clustering between the proteome of EVs and their originating brown adipocytes (Figures 2A and 2B). The mitochondrial proteins that were over-represented in EVs and brown adipocytes following CL or FCCP treatment (Figure 2C) were then integrated with MitoCarta 3.0. Of note, n = 63 mitochondrial proteins were over-represented selectively in EVs when compared with the proteins over-represented in the originating cells following both CL and FCCP treatment (Figure 2D). Several mitochondrial proteins detected in EVs of pBAs were also observed in the EVs released from T37i treated with FCCP (Figure 2E). Notably, pBAs treated with isoproterenol or another selective mitochondrial stressor, i.e., antimycin A, increased mitochondrial proteins mainly in the EV compartment (Figure 2F), confirming the proteomic data (Figure 2G).
Figure 2. Brown adipocytes release oxidized mitochondrial proteins through an MDVs/PINK1-dependent mechanism.
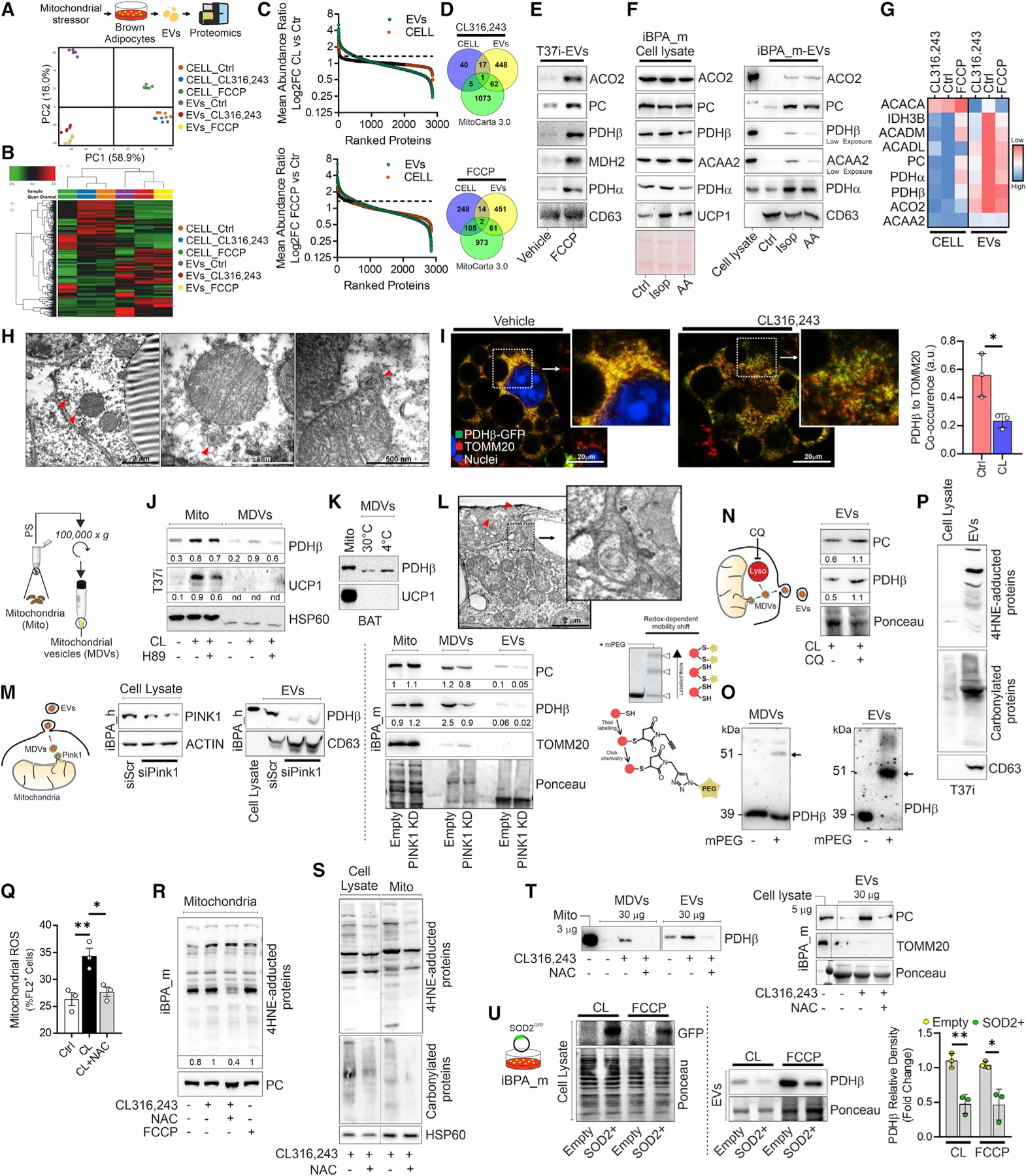
(A and B) Experimental design and PCA (A) and hierarchical clustering heatmap (B) obtained by proteomics of primary adipocytes (pBAs) treated with CL316,243 or FCCP and the related EVs.
(C) Mean abundance ratio of proteins detected in pBAs following CL (upper panel) or FCCP treatment (lower panel) and related EVs.
(D) Venn diagram integrating mitochondrial proteins (MitoCarta 3.0) with up-regulated proteins (FC > 1.3) in pBAs treated with CL316,243 (upper panel) or FCCP (lower panel) and the related EVs.
(E) Immunoblots of EVs released from T37i treated with FCCP. CD63 was used as an EV loading control.
(F) Immunoblots of iBPA_m treated with isoproterenol (Iso) or antimycin A (AA) and the related EVs. Ponceau and CD63 were used as lysate and EV loading controls, respectively.
(G) Heatmap of the mean abundance of mitochondrial proteins in pBAs and the related EVs under basal conditions (Ctr) and the following CL or FCCP treatment.
(H) Representative TEM images of BAT from mice at 4°C. Red arrows indicate mitochondrial buddings.
(I) Representative confocal microscopy images of iBPA_m transduced with lentiviral particles carrying PDHβ -GFP (green) and labeled with TOMM20 (red) antibodies. Histogram shows colocalization between PDHβ -GFP and TOMM20 under basal conditions or after CL treatment. Student’s t test, *p < 0.05, n = 3. (J and K) Immunoblots of crude mitochondria or MDVs isolated from T37i adipocytes (J) or BAT (K) following CL treatment or cold exposure, respectively. H89 was used to inhibit cAMP/PKA signaling. HSP60 was used as loading control. Densitometric analyses (J) were reported below as the ratio between PDHβ or UCP1 and HSP60.
(L) Representative TEM images of BAT at 4°C. Red arrows indicate a multivesicular body containing mitochondria. Image magnification reports the presence of intercellular EVs.
(M) Immunoblots of iBPA_h transiently transfected with siRNA against PINK1 (PINK1-KD) and the related EVs. Actin and CD63 were used as cell lysate and EV loading controls (left panels). Immunoblots of crude mitochondria, MDVs, and EVs isolated from iBPA_m transduced with PINK1 shRNA lentiviral particle (PINK1-KD). Ponceau was used as loading control. Densitometric analyses were reported below as ratio between protein and loading control (right panel).
(N) Experimental design and immunoblots of iBPA_m treated with CL and cotreated with vehicle or chloroquine (CQ). Ponceau was used as loading control. Densitometric analysis was reported below as ratio between PC or PDHβ and loading control.
(O) Immunoblots of T37i MDVs and EVs. Protein oxidation was determined after protein cysteine derivatization by MalPEG (mPEG). Black arrows indicate the mobility shift caused by mPEG conjugation.
(P) Immunoblots of T37i EVs. CD63 was used as loading control.
(Q) Flow-cytometry analysis of mitochondrial ROS production in iBPA_m treated with CL or cotreated with NAC. ANOVA test with multiple comparison correction, *p < 0.05; **p < 0.01, n = 3.
(R) Immunoblots of mitochondria isolated from iBPA_m treated with CL or FCCP and cotreated with vehicle or NAC. PC was used as loading control.
(S) Immunoblots of cell lysates and mitochondria isolated from T37i treated with CL and cotreated with NAC. HSP60 was used as loading control.
(T) Immunoblots of MDVs and EVs isolated from T37i (left panel) and EVs isolated from iBPA_m (right panel) after treatment with CL and co-treatment with vehicle or NAC. TOMM20 and Ponceau were used as EV purity and loading control, respectively.
(U) Experimental design and immunoblots of iBPA_m transduced with lentiviral particles carrying SOD2-GFP (left panel) and related EVs (right panel). Densitometric analyses were reported as ratio between PDHβ in EVs and loading control (Ponceau staining). Student’s t test, *p < 0.05; **p < 0.01.
See also Figure S1.
Transmission electron microscopy (TEM) revealed that the mitochondria of the BAT at 4°C developed buddings that contained both an inner and outer mitochondrial membrane (Figure 2H). These ultrastructural features were reminiscent of MDVs, which bud off from mitochondria and contain a specific repertoire of cargo proteins (e.g., PDH positive or TOM complex-positive, but not both) (Cadete et al., 2016; Sugiura et al., 2014; Todkar et al., 2021). To track MDVs, we performed confocal microscopy analyses in immortalized murine primary brown adipocytes (iBPA_m) transduced with lentiviral particles carrying PDHβ-GFP. Following treatment with CL, we observed increased levels of PDHβ-GFP+ dots with a significant decrease in the colocalization index between PDHβ and TOMM20 (PDHβ+/TOMM20−) (Figure 2I). To verify the packaging of PDHβ into TOM complex-negative MDVs, we isolated them, and an increased level of PDHβ and PC was observed in the activated BAT (Figure S1H). Remarkably, PDHβ was observed in MDVs already under basal conditions, and its content increased or decreased upon thermogenic induction or inhibition, respectively (Figures 2J and 2K). In accordance with the selective incorporation of mitochondrial proteins, UCP1 was undetected in MDVs (Figures 2J and 2K). MDVs are part of an alternative mitochondrial quality control system delivering mitochondrial components to EVs (Crewe et al., 2021; D’Acunzo et al., 2021; Rabas et al., 2021; Todkar et al., 2021; Vasam et al., 2021). Consistent with this, TEM analyses of the BAT at 4°C showed that MDVs were localized either close to the plasma membrane or in the intercellular space (Figures 2H and 2L).
Several findings reported that PTEN-induced kinase 1 (PINK1) regulates PDH+/TOMM20− MDV generation via a reactive oxygen species (ROS)-dependent and mitophagy-independent mechanism (Cairó et al., 2019; Matheoud et al., 2016; McLelland et al., 2016, 2014; Rabas et al., 2021). Thermogenic stimuli induced an increase in PINK1 levels (Figures S1I and S1J); thus, we downregulated PINK1 expression, and an impaired release of PDHβ and PC was observed both in MDVs and EVs (Figure 2M). This event was accompanied by augmented levels of protein oxidation in mitochondria, as assessed by evaluating 4-hydroxynonenal (4-HNE) protein adduct formation (Figure S1K). Conversely, the downregulation of the master regulator of mitochondrial fragmentation, dynamin-related protein-1 (DRP1) (Amari and Germain, 2021), did not affect the PC content in EVs (Figure S1L), excluding its involvement in EV release. MDVs can be targeted to lysosome for degradation or extruded via EVs (Amari and Germain, 2021). Pharmacological limitation of lysosomal degradation by chloroquine (CQ) increased PDHβ and PC protein levels in brown adipocyte EVs (Figure 2N), suggesting that PINK1-mediated MDVs and EVs formation is an additional mechanism to remove damaged mitochondrial proteins and assure mitochondrial homeostasis under mitochondrial stress conditions.
Brown adipocytes physiologically produce mitochondrial ROS (mtROS) to sustain thermogenesis (Chouchani et al., 2016; Lettieri-Barbato, 2019; Mills et al., 2018). Hence, we supposed that brown adipocytes eject oxidatively damaged mitochondrial proteins through MDVs into EV pathway. We evaluated cysteine oxidation of PDHβ in both MDVs and EVs, and a higher rate of oxidation was found in EVs than MDVs (Figure 2O). Other forms of protein oxidation (4-HNE and carbonyl protein adduct formation) were observed in EVs (Figure 2P) that further accumulated following CL treatment (Figure S1M). To implicate oxidative stress in the sorting of mitochondrial protein, we used N-acetylcysteine (NAC), which was effective in limiting oxidative damage in mitochondria, MDVs and EVs (Figures 2Q–2S and S1N). NAC was also able to restrain mitochondrial protein sorting into MDVs and EVs in CL-treated brown adipocytes (Figure 2T). A reduced release of PDHβ was also achieved by more specifically limiting mtROS production via SOD2 overexpression (Figure 2U).
BAT-derived EVs have a deleterious effect on brown adipocyte metabolism
Extracellular mitochondria, either as whole functional structures or as fragments, regulate many functions, including cell-to-cell communication, danger sensing, and immune responses (Boudreau et al., 2014; Brestoff et al., 2021; Peruzzotti-Jametti et al., 2021; Phinney et al., 2015; Todkar et al., 2021). If not adequately removed by macrophages, extracellular mitochondria may negatively affect tissue homeostasis (Nicolás-Ávila et al., 2020). Thus, we explored whether brown adipocyte EVs can alter cell metabolism in recipient brown adipocytes. We analyzed the differentially expressed genes of pBAs treated with the BAT EVs isolated from mice at 30°C or 4°C. The top 200 downregulated genes were included in a Venn diagram (Figure 3A; Table S2), and the overlapping genes (p < 0.01) were mapped by the KEGG pathway. Peroxisome proliferator-activated receptor (PPAR) signaling was significantly disrupted in the pBAs treated with BAT EVs (Figure 3B). qPCR analysis of T37i adipocytes treated with BAT EVs confirmed their ability to reduce the expression of typical PPARγ target genes (Figure 3C). PPARγ is involved in brown fat cell differentiation and mitochondrial thermogenic function (Shen et al., 2020); therefore, we predicted that BAT EVs would alter mitochondrial-dependent thermogenesis and metabolism in brown adipocytes. Accordingly, the brown adipocytes treated with the BAT EVs at 4°C showed a decrease in UCP1 (Figure S2A), an accumulation of metabolites pertaining to pyruvate metabolism and TCA cycle (Figure 3D), and a substantial reduction in mitochondrial respiration (Figure S2B). Similarly, EVs isolated from cultured brown adipocytes, once recaptured by the originating cells (Figure 3E), led to a downregulated expression of brown fat genes (Figures 3F, 3G, and S2C) and reduced mitochondrial activity (Figures 3H and S2D). EVs from brown adipocytes also inhibited adipogenesis in differentiating cells (Figures S2E and S2F), confirming their inhibitory role in PPAR-dependent brown fat cell differentiation.
Figure 3. BAT EVs affect PPARγ signaling in brown adipocytes.
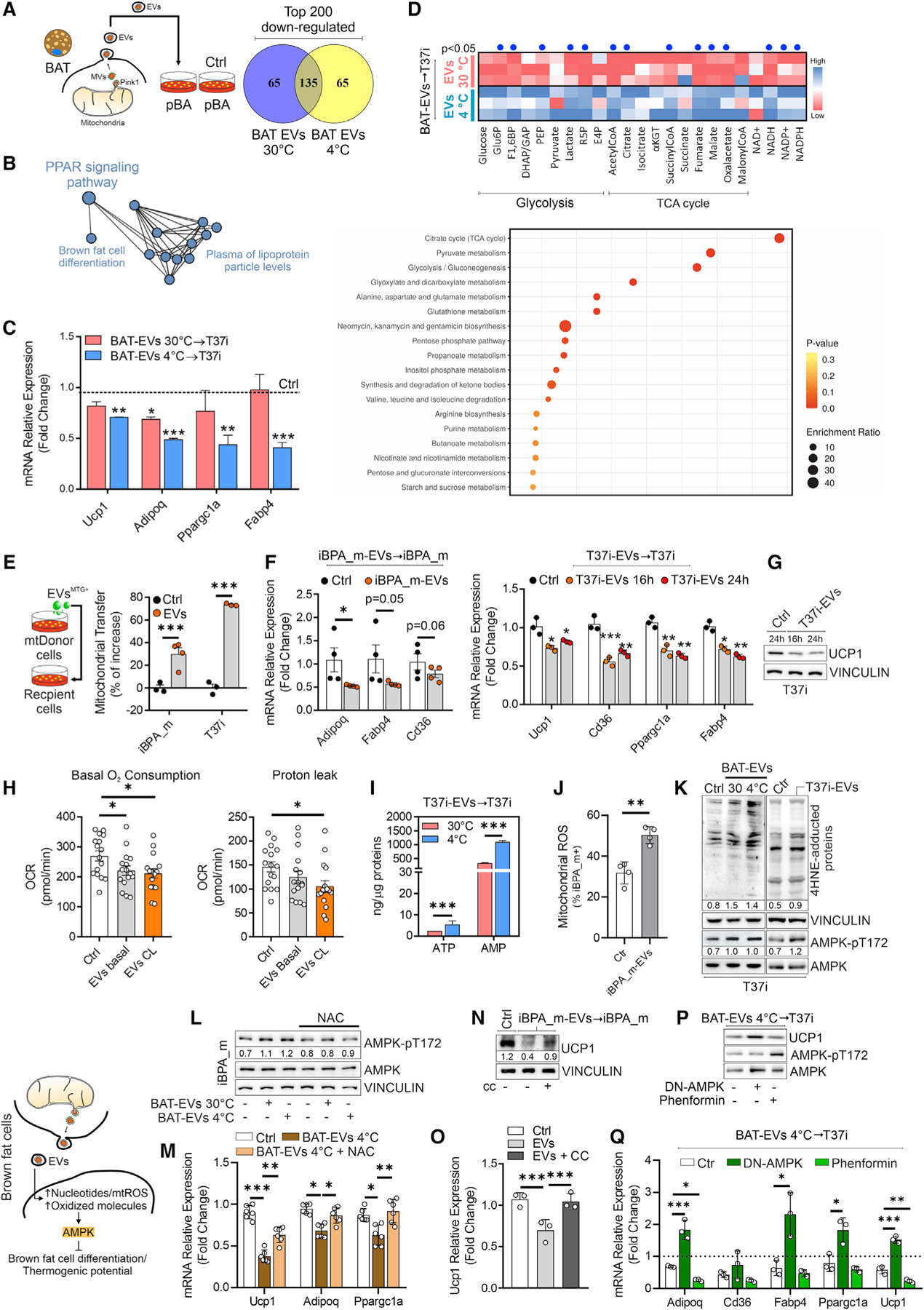
(A and B) Experimental design and Venn diagram (A) of the top 200 downregulated genes in pBAs treated (16 h) with BAT EVs. The 135 overlapping genes in (A) are represented by a functional protein association network (STRING) (B).
(C) s of PPARγ target genes in T37i treated (16 h) with BAT EVs. Student t test, *p < 0.05; **p < 0.01 vs. Ctrl, n = 3.
(D) Targeted metabolomics of T37i treated with BAT EVs (16 h). Results are reported as a heatmap (upper panel) and an enrichment pathway analysis (lower panel) of differentially modulated metabolites. Student’s t test, p < 0.05 (blue dots).
(E) Experimental design and flow-cytometry analysis of iBPA_m and T37i treated (4 h) with MTG-labeled EVs isolated from the same cell type (1:1 cell ratio). Student’s t test, ***p < 0.001, n = 3.
(F) qPCR analysis of PPAR-related genes in iBPA_m (left panel) and T37i (right panel) treated with EVs isolated from the same cell type (1:1 cell ratio). Student’s t test, *p < 0.05; **p < 0.01; ***p < 0.001, n = 3.
(G) Immunoblots of T37i treated with EVs isolated from the same cell type (1:1 cell ratio). VINCULIN was used as loading control.
(H) Basal oxygen consumption and proton leak analyzed by Sea-horse technology in iBPA_m treated (16 h) with EVs isolated from the same cell type (1:1 cell ratio). Two-way ANOVA test, *p < 0.05, n = 9.
(I) AMP and ATP quantitation in T37i treated (16 h) with BAT EVs. Student’s t test, ***p < 0.01, n = 3.
(J) Flow-cytometry analysis of mitochondrial ROS in iBPA_m treated with EVs isolated from the same cell type (1:1 cell ratio). Student’s t test, **p < 0.01, n = 3.
(K) Immunoblots of T37i treated (16 h) with BAT EVs (n = 3 mice/group) or EVs isolated from the same cell type (1:1 cell ratio). VINCULIN was used as loading control. Densitometric analyses were reported as ratio between AMPKpT172 and AMPK or 4-HNE protein adducts and VINCULIN.
(L) Experimental design and immunoblots of iBPA_m treated (16 h) with BAT EVs and cotreated with NAC (n = 3 mice/group). VINCULIN was used as loading control. Densitometric analyses were reported as ratio between AMPKpT172 and AMPK.
(M) qPCR analysis of iBPA_m treated with BAT EVs at 4°C and cotreated with NAC. Two-way ANOVA test, *p < 0.05; **p < 0.01; ***p < 0.001, n = 6.
(N and O) UCP1 protein (N) and mRNA (O) in iBPA_m treated with EVs (16 h) isolated from the same cell type (1:1 cell ratio) and cotreated with compound-c (CC). VINCULIN was used as loading control. Densitometric analysis was reported as ratio between UCP1 and VINCULIN. Two-way ANOVA test, ***p < 0.001, n = 3). (P and Q) Immunoblots (P) and qPCR analysis (Q) of T37i treated with BAT EVs (16 h). Phenformin or DN-AMPK was used as AMPK agonist or antagonist, respectively. Two-way ANOVA test, *p < 0.05; **p < 0.01; ***p<0.001, n = 3).
See also Figure S2.
As reported, brown adipocytes EVs deliver ATP, AMP, and oxidatively damaged components (Figures 1M, 2P, and S1M). Intriguingly, treatment with EVs increased AMP levels as well as mtROS production and protein oxidation in recipient brown adipocytes (Figures 3I–3K). These data led us to postulate that EV re-uptake perturbs the energy and redox sensor 5́ AMP-activated protein kinase (AMPK). AMPK limits adipogenesis and lipogenesis by downregulating several adipocyte-specific transcription factors, including PPARγ (Ahmad et al., 2020; Giri et al., 2006; Habinowski and Witters, 2001). As expected, EVs activated AMPK in recipient brown adipocytes (Figures 3K and 3L). NAC treatment blunted AMPK activation and limited the downregulation of brown fat gene expression (Figures 3L and 3M). To test whether AMPK signaling regulates the effects of EVs, we inhibited or activated it, and a recovery or downregulation of brown fat gene expression was observed, respectively (Figures 3N–3Q).
Macrophages are involved in the elimination of mitochondrial parts ejected from brown adipocytes and maintain BAT thermogenic program
Next, we investigated whether phagocytic immune cells resident in BAT could remove extracellular damaged mitochondria to preserve BAT phenotype. Through mass cytometry (CyTOF), we identified distinct clusters of immune cells in BAT following temperature changes (Figures 4A and 4B). We found that CD45+ CD11b+ F4/80+ BAT-macrophages (bMACs) were the most represented immune cells in BAT and further accumulated following cold exposure (Figures 4C and 4D). Bringing back cold-exposed mice to thermoneutrality, we observed a reduced proportion of bMACs in BAT (Figures 4B and S3A). BAT macrophage pool requires constant replenishment from monocytes, which have a key role in BAT physiology (Gallerand et al., 2021). Remarkably, during cold exposure, blood monocytes are increased in both mice and humans, mirroring the degree of BAT activation (Williams et al., 2017). Murine monocytes are generally classified into Ly6Chigh inflammatory monocytes, which migrate to the sites of inflammation and further differentiate into Ly6Clow patrolling monocytes, promoting the cleaning of debris and tissue regeneration (Kimball et al., 2018; Yang et al., 2014). Although monocyte subsets (CD45+/CD11b+/Ly6Chigh/CD62L− and CD45+/CD11b+/Ly6Clow/CD62L+ or CD45+/CD11b+/CCR2+/CD115+ and CCR2−/CD115−) and dynamics remained almost unchanged between the two states (Figure S3B), a significant increase in CD45+/CD11b+/Ly6Clow/CX3CR1+-patrolling monocytes was observed in the BAT at 4°C (Figure 4E). These results suggest that during cold exposure, inflammatory monocytes transit to patrolling monocytes to finally differentiate into bMACs. The analysis of bMAC phenotype revealed an increased expression of M2-like markers (i.e., CD206 and CD200R) and unaffected levels of M1-like markers (i.e., MHC-II and CD80/86) following cold stress (Figures 4F, S3A, and S3C). Next, to give more insights into the Mono-to-bMACs transition, we interrogated a scRNA-sequencing repository dataset (GSE160585; Figure S4A). By subsetting CD45+ cells, we observed an increased recruitment of Mono/bMACs following cold stress (Figures S4B–S4D), which was confirmed by fluorescence microscopy analysis (Figure S4E). Plotting genes sensitive to cold along the activation-induced macrophage differentiation index (AMDI) by MacSpectrum tool, we detected an increased proportion of ‘‘naïve’’-like and less inflammatory bMACs at 4°C (region D) (Figures S4F and S4G). To evaluate whether EVs take part in bMACs transition, bone marrow-derived macrophages (BMDM) were treated with T37i EVs and a less inflammatory phenotype (region D) was observed when compared with LPS-treated BMDM (region B) (Figure S4H). Consistently, EVs pretreatment limited the LPS-mediated inflammatory responses in macrophages (Figure S4I). Trajectory analyses of bMACs at room temperature (RT) and 4°C displayed distinct patterns along pseudotime distribution (Figure S4J), which was well-aligned with the mapping on MacSpectrum (A, B, and D) (Figure S4G). bMACs at 4°C showed a higher expression of genes controlling the oxidative stress response than bMACs at RT (Figure S4K) that was in line with the increased mtROS production and antioxidant gene expression in cultured macrophages treated with brown adipocyte EVs (Figures S4L and S4M).
Figure 4. Macrophage dynamics in BAT during temperature changes.
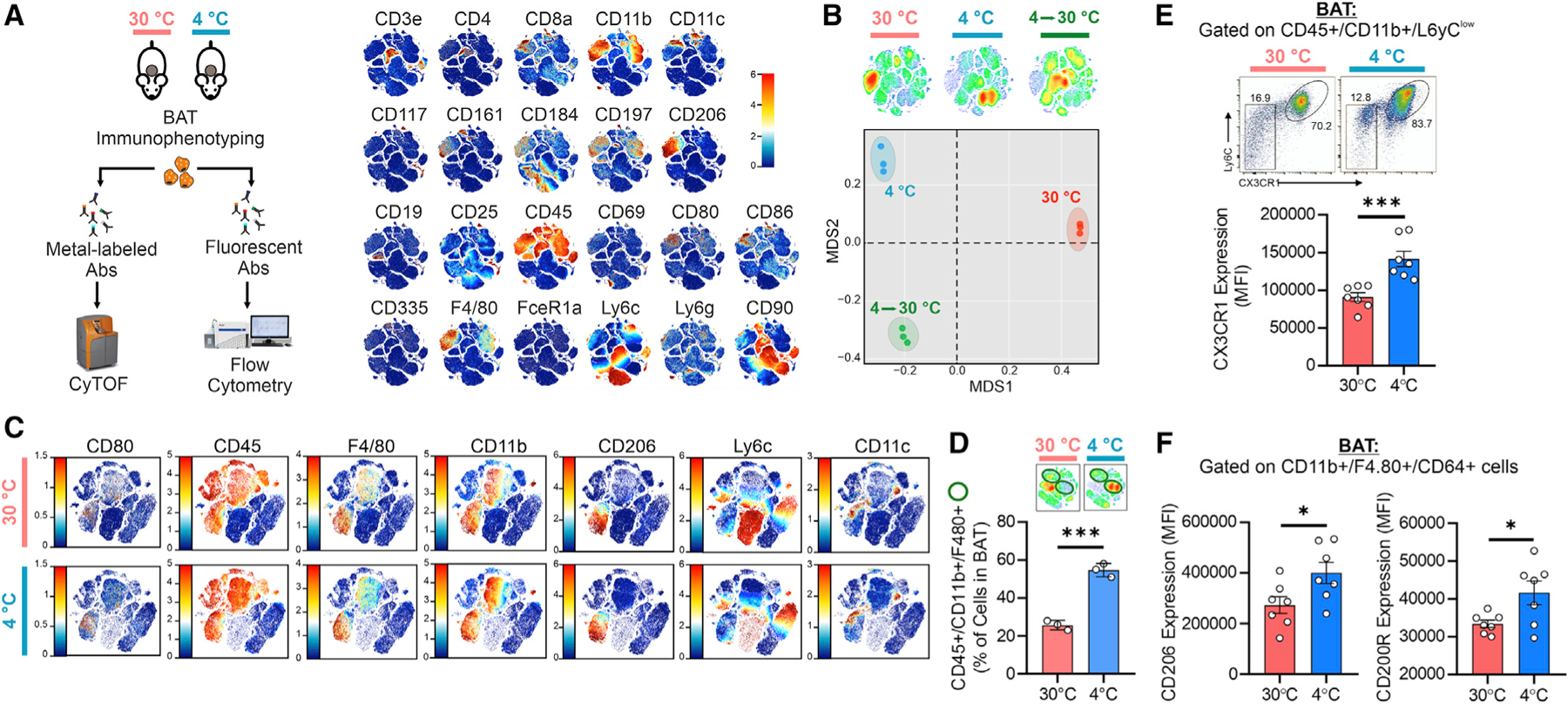
(A) Experimental design and merged single t-SNE maps obtained by CyTOF analysis (n = 6 mice/group pooled in pairs).
(B) CyTOF t-SNE maps of antigen redistribution (upper panel) and multidimensional plot of antigen expression (bottom panel) (n = 6 mice/group pooled in pairs).
(C) CyTOF t-SNE maps of the antigen subsets (n = 6 mice/group pooled in pairs).
(D) CyTOF density map (upper panel). Percentage of macrophages (lower panel). Student’s t test, ***p < 0.001, n = 3.
(E and F) Flow-cytometry density plot and quantitation of CX3CR1 expression in monocytes (E), and CD206 and CD200R expression in macrophages (F). Student’s t test, **p < 0.01, n = 7 mice/group.
See also Figures S3 and S4.
To test whether macrophages take up mitochondrial material originating in brown adipocytes in vivo, we performed bone marrow transplants of congenic CD45.1 wild-type (WT) bone marrow into host CD45.2 mitochondria reporter mice expressing the fluorescent protein Dendra2 coupled with a mitochondria-targeting sequence (mtD2) (Figure 5A). WT CD45.1 bone marrow was also transplanted into WT CD45.2 recipients as controls. The large majority of immune cells in BAT were WT donor-derived CD45.1+ CD45.2− cells, with chimerism of approximately 85% (Figure 5A). We then gated on WT CD45.1+ CD45.2− donor-derived BAT immune cells and found that approximately 30% were mtD2+ when isolated from mtD2 recipients (Figure 5B). Most of the WT donor-derived mtD2+ cells were macrophages (54.8%), with monocytes, CD11c+ cells, eosinophils, neutrophils, and undefined cells representing the remainder (Figures 5C and S5A). Next, we gated on WT donor-derived macrophages and found that approximately 36% were mtD2+ in WT → mtD2 mice (Figure 5D). To demonstrate the direct transfer from brown adipocytes to bMACs, we used adipocyte-specific mitochondria reporter mice (MitoFat) (Brestoff et al., 2021). Approximately 37% of the bMACs contained a mitochondrial signal (Figure 5E), which represented approximately 6% of the live cells (Figure 5F). Of note, cold exposure further enhanced mitochondria transfer from brown adipocytes to bMACs (Figure 5G). The mitochondrial transfer from brown adipocytes to macrophages was also effective in vitro (Figures S5B and S5C).
Figure 5. BAT-resident macrophages regulate the removal of brown adipocytes EVs via the CD36-lysosomal pathway.
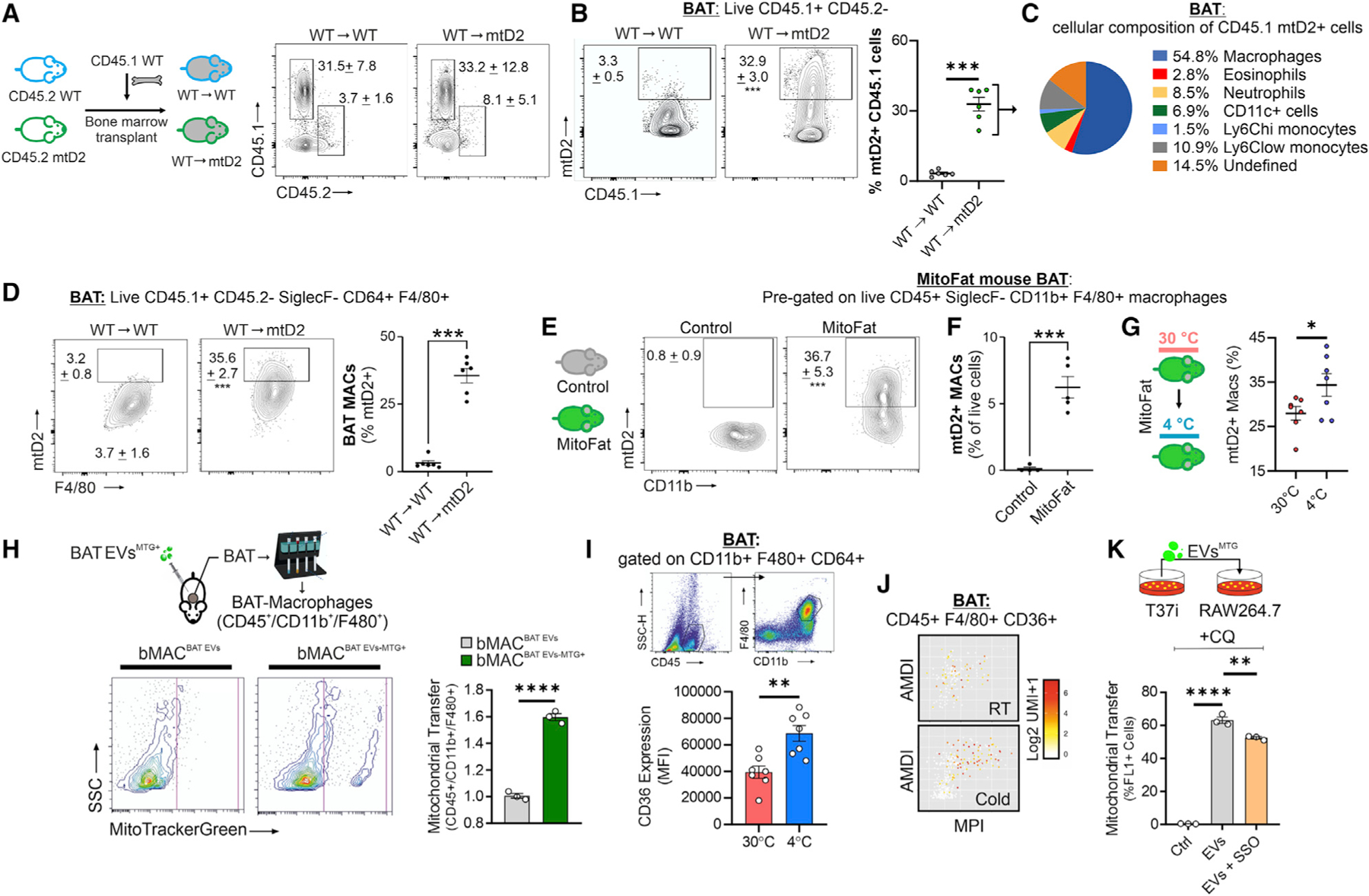
(A) Experimental design for panels (A–D), in which CD45.1 wild-type (WT) bone marrow was transplanted into either CD45.2 WT or CD45 mtD2 mitochondria reporter mice. Flow cytometry plots of CD45.1 WT donor cells and radioresistant CD45.2 host immune cells in BAT from chimeras after 12 weeks of engraftment.
(B) Flow cytometry plots and percentages of WT CD45.1 donor cells that are mtD2+.
(C) Cellular composition of CD45.1 WT donor cells that are mtD2+.
(D) Flow cytometry plots with the percentage of macrophages that are mtD2+.
(E) Flow cytometry plots of macrophages from BAT of MitoFat adipocyte-specific mitochondria reporter mice and negative controls. Percentages shown represent macrophages that are mtD2+.
(F) Percentage of live cells in BAT that are mtD2+ macrophages.
(G) Experimental design and percent of mtD2+ macrophages in BAT from MitoFat mice at 4°C or 30°C. Data are expressed as mean ± SEM. Student’s t tests, *p < 0.05, **p < 0.01, ***p < 0.001, n = 7 mice/group.
(H) Experimental design and flow-cytometry analysis of bMACs isolated from BAT of mice injected with BAT EVs labeled with MTG (bMACBAT EVs-MTG) or unlabeled BAT EVs (bMACBAT EVs). Student’s t test, ****p < 0.0001, n = 3 mice/group.
(I) Experimental design, flow cytometry density plot and quantitation of CD36 expression in bMACs. Student’s t test, **p < 0.01, n = 7 mice/group.
(J) MacSpectrum plots of Cd36 expression in bMACs extrapolated from scRNA-seq (GSE160585).
(K) Flow-cytometry analysis of RAW264.7 treated with T37i EVs labeled with MTG. Sulfo-N-succinimidyl oleate (SSO) was added 1 h prior to EV treatment to inhibit CD36. CQ treatment was used to inhibit lysosome activity. Two-way ANOVA test, **p < 0.01, ****p < 0.0001; n = 3.
See also Figure S5.
As our mitochondria reporter mouse models could not distinguish the type of phagocytosed mitochondrial material (e.g., nude whole or fragmented mitochondria or dead adipocytes versus BAT EVs), we took an alternative approach to determine whether BAT EVs can be phagocytized in vivo. Mice were injected with MTG-labeled BAT EVs, and bMACs were isolated. Flow-cytometry analysis revealed that bMACs became positive for MTG (Figure 5H). The inhibition of lysosomal activity by CQ showed that the MTG-labeled EVs colocalized with macrophage lysosomes (Figures S5D and S5E) and increased the accumulation of incorporated EVs in macrophages (Figure S5F). This was consistent with the increased protein levels of PC and PDHβ within macrophages without any modulation of their expression (Figure S5G).
Lipidomic analysis showed that EVs contained phosphatidylserine (PS), the main ‘‘eat-me’’ signal described in EVs that precedes microvesicle shedding (MacKenzie et al., 2001; Meldolesi, 2018; Nicolás-Ávila et al., 2020; Wei et al., 2018) (Figure S5H). We found that PS was present on EV surface (Figure S5I), suggesting that it may act as a signal for macrophage phagocytosis. To test this idea, we performed a transcriptomic analysis of RAW264.7 treated with EVs, and the top 200 up-regulated genes (FC > 4.0; p < 0.05) were integrated with the GO term of phagocytosis (GO: 0006909). Nine genes related to phagocytosis were identified including Cd36 (Figure S5J), which is related to phagocytosis via the PS receptor (Fadok et al., 1998; Grajchen et al., 2020; Woo et al., 2016). Remarkably, the upregulation of Cd36 was observed in EVs-treated macrophages (Figure S5K) and bMACs at 4°C (Figures 5I and 5J). To assess the role of CD36 in mediating EV internalization, we used a CD36 inhibitor, which partially limited the uptake of EVs by macrophages (Figure 5K).
To explore if phagocytic activity of bMACs supports BAT homeostasis and function by removing brown adipocyte EVs, we assessed the effects of bMAC depletion in the context of BAT physiology. We employed CD169DTR mice to deplete bMACs, as CD169+ macrophages were increased in BAT following cold exposure (Figures 6A, 6B, and S6A). In BAT of CD169DTR mice, we observed an increase in lipid droplet size and accumulation of PLIN1 protein, whereas mitochondrial OxPHOS subunits, PC, PPARγ, and UCP1 were strongly reduced (Figures 6B, 6C, and S6C). To deplete bMACs, we alternatively used clodronate liposomes (CLDNT), which efficiently reduced the number and expression level of macrophage markers in BAT (Figures 6D and 6E). Following cold exposure, BAT of CLDNT-treated mice exhibited a lower tissue density (Figure S6D), suppressed induction of key thermogenic genes (Figures 6E, 6F and S6E), and reduced cold tolerance (Figure 6G). In accordance with the role of bMACs in removing extracellular mitochondria, CLDNT treatment increased the amount of total BAT EVs as well as MTG+ BAT EVs and caused the accumulation of PDHβ and PC in EVs following cold exposure (Figures 6H–6J).
Figure 6. Macrophages control the thermogenic capacity of BAT.
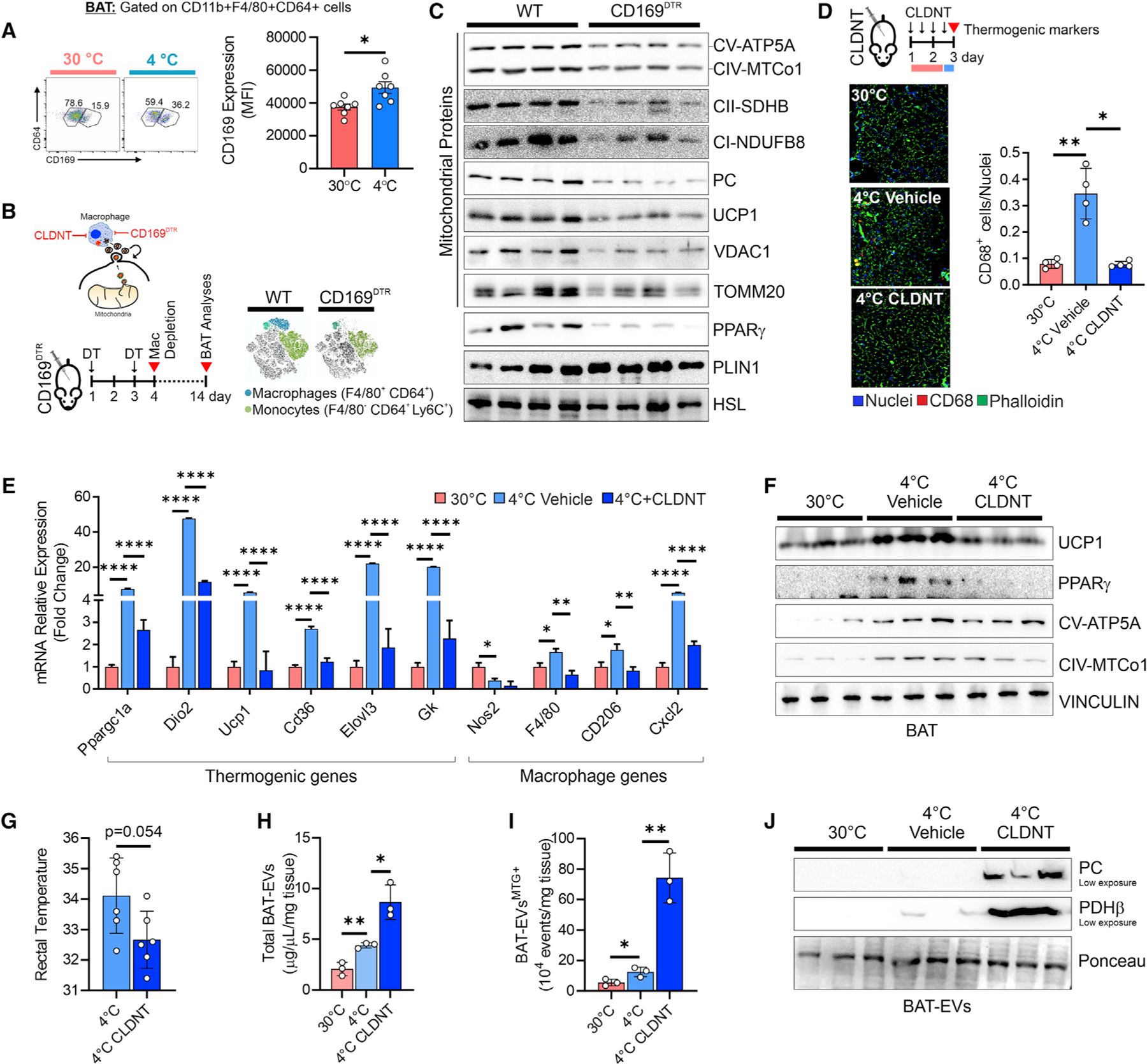
(A) Flow cytometry density plot and quantitation of CD169+ bMACs. Student’s t test, *p < 0.05, n = 7 mice/group.
(B) Experimental design and t-SNE map of antigen redistribution in BAT of WT and CD169DTR mice 4 days after DT injection.
(C) Immunoblots of BAT from WT and CD169DTR mice 14 days after DT injection. HSL was used as loading control (n = 4 mice/group).
(D) Experimental design and representative fluorescence micrographs of BAT stained with the CD68 antibody (red) to detect bMACs. Quantification of CD68+ bMACs is reported (right panel). ANOVA, *p < 0.05, **p < 0.01, n = 4 mice/group.
(E–G) qPCR (E), immunoblot (F) of BAT and rectal temperature (G) of mice at 30°C and 4°C. Mice were pre-treated with liposome clodronate (CLDNT) prior to cold exposure. ANOVA, *p < 0.05, **p < 0.01, ****p < 0.001; n = 3 or n = 6 mice/group.
(H and I) Flow-cytometry quantitation of total EVs (H) and MTG+ EVs (I) released from BAT. Student’s t test, *p < 0.05, **p < 0.01; n = 3 mice/group.
(J) Representative iunoblots of BAT EVs. Ponceau was used as loading control (n = 3 mice/group). See also Figure S6.
DISCUSSION
In this study, we highlighted the role of bMACs in regulating the thermogenic function of BAT by removing damaged mitochondria released from brown adipocytes during thermogenesis. Brown adipocytes respond to mild mitochondrial stress by producing MDVs to deliver oxidized mitochondrial components into EVs. In cardiac cells, proteomic findings recently revealed that mtROS-producing drugs promote the enrichment of oxidized mitochondrial proteins in MDVs, and part of these proteins was predicted to be ejected by EVs (Vasam et al., 2021). In line with these findings, by enhancing the antioxidant potential in brown adipocytes, we limited the release of mitochondrial and oxidized proteins in MDVs and EVs following mitochondrial stress. Although we observed an increase in the number of mitochondrial proteins in EVs, we did not find any modulation of the same proteins in the originating cells. These data are consistent with the evidence that MDVs are part of the selective mitochondrial protein sorting into EVs (Crewe et al., 2021; Todkar et al., 2021; Vasam et al., 2021). Recent work by Crewe et al. (2021) showed that oxidatively stressed white adipocytes are highly prone to releasing damaged mitochondrial proteins via the MDVs-EVs route. The mechanism of recognition and extrusion of such damaged mitochondrial components seems to involve Parkin as, when it is ablated in the originating adipocytes, a decrease in mitochondrial proteins is found in EVs (Crewe et al., 2021). We demonstrated that PINK1 is involved in the release of mitochondrial proteins specifically packaged into PDHβ+/TOMM20− MDVs for extracellular ejection, in line with previous evidence that PINK1 mediates the formation of MDVs in a mechanism that is distinct from canonical mitophagy and is induced by the onset of oxidative stress conditions within mitochondria (McLelland et al., 2014). By contrast, a work by Todkar et al. (2021) demonstrated that damaged mitochondrial proteins sorted into MDVs are preferentially directed to lysosomes rather than to EVs via the same Parkin-dependent system to avoid the propagation of extracellular inflammatory signals. Such a discrepancy could be due to the completely different cellular systems used, i.e., U2OS cells, which, being tumor cells, could have developed a mechanism forcing the intracellular elimination of oxidatively damaged mitochondrial protein to evade the immune response. Otherwise, we can postulate that at least in highly metabolically active cells with a very large amount of mitochondria, such as brown adipocytes or cardiac cells, the PINK1-dependent efflux of mitochondrial proteins into EVs could reflect enhanced mitochondrial damage and/or the overload of MDVs containing oxidized proteins and their incapacity to clear them via lysosomal degradation. In support of this hypothesis, upon the inhibition of lysosomal degradation by CQ, we were able to impinge on the extracellular release of mitochondrial proteins by EVs.
Intriguingly, one of the most abundant mitochondrial proteins, brown adipocyte-specific UCP1, was detected neither in MDVs nor in EVs, which could corroborate the specificity of the mitochondrial protein sorting and the insusceptibility of this protein to being removed by the MDVs-to-EVs pathway. This could be linked to the important protective role played by UCP1 against cold-induced oxidative stress (Stier et al., 2014) and to its tolerance to certain types of oxidations (i.e., on cysteine residues) (Chouchani et al., 2016; Mills et al., 2022) that are actually essential to elevate its uncoupling activity.
In this study, the mechanisms mediating the extrusion of MDVs carrying damaged mitochondrial parts were not explored. According to the literature (Teng and Fussenegger, 2020), it is likely that MDVs, after converging into multivesicular bodies, can fuse with the plasma membrane and be released into the extracellular space as EVs. Further investigations are needed to delineate whether the alteration of the pathways involved in EV biogenesis may be involved in the impairment of the mitochondrial quality system in BAT.
The release of damaged mitochondria into EVs during acute cold exposure was accompanied by patrolling monocytes and macrophages recruitment/expansion. Interestingly, patrolling monocytes differentiate into less inflammatory bMACs that are functional to maintain the thermogenic capacity of BAT via the elimination of extracellular mitochondrial material derived from metabolically stressed adipocytes. Different modalities of mitochondria uptake by macrophages have been reported, such as phagocytosis of exophers (Nicolás-Ávila et al., 2020) or tunneling nanotubes (Jackson et al., 2016); hence, it is conceivable that, concomitant to BAT EV phagocytosis, some degree of the observed mitochondrial transfer events to bMACs in our mitochondria reporter mouse models could be due to these processes. The injection in mice of BAT EVs labeled with MTG and the observed uptake by bMACs supports the idea that BAT EV release is a mechanism of mitochondria transfer from brown adipocytes to macrophages in vivo to preserve BAT function. Accordingly, in vivo depletion of bMACs led to the accumulation of BAT EVs containing mitochondrial protein, disruption of mitochondrial proteins, accumulation of lipids, and failure of the thermogenic response to cold exposure. Following warming, the number of bMACs was reduced, which may be causative of the decreased removal of EV-containing damaged mitochondria that, in turn, could contribute to the shutdown of the PPAR-mediated thermogenic program. Consistent with this assumption, treating brown adipocytes with BAT EVs containing damaged mitochondrial material led to the inhibition of PPAR signaling and reduced mitochondrial respiration.
We found that EVs contain oxidized material that, together with the delivered high levels of AMP, could be causative of the activation of the energy and redox sensor AMPK, a key modulator of lipid metabolism that may have opposite effects with respect to PPARγ by limiting adipogenesis/lipogenesis (Ahmad et al., 2020). Notwithstanding, many studies suggest a link between AMPK and thermogenic BAT activation; however, evidence identifying AMPK as a causal factor for BAT activation and the mechanisms that regulate BAT function via AMPK still remain controversial (van der Vaart et al., 2021). It has been reported that the pharmacological activation of AMPK downregulates several adipocyte-specific transcription factors, including PPARγ (Giri et al., 2006; Habinowski and Witters, 2001). Herein, we have shown that EV treatment activated AMPK, and the modulation of its activity limited the perturbing effects induced by EVs. In line with our results, AMPK activating drugs, such as metformin, induce the lowering of oxygen consumption in unstimulated brown adipocytes (Breining et al., 2018). These results are in contrast with the previous results from other research groups and our laboratory showing that AMPK activation is functional in promoting a brown-like phenotype in white/beige adipose cells and tissues (Aquilano et al., 2020; Lettieri Barbato et al., 2013; van der Vaart et al., 2021). Such a discrepancy could be explained by the limited role that this kinase has in BAT thermogenesis. Indeed, AMPK activation has not been observed during acute cold exposure in BAT (Aquilano et al., 2020; Wu et al., 2018). Based on this evidence, we can, therefore, postulate that the EVs-mediated activation of AMPK in recipient brown adipocytes is part of a stress response mechanism against the harmful consequence of their oxidized/damaged cargo internalization. Namely, AMPK activation and the downstream inhibitory effects on PPAR signaling seem to be functional in limiting mitochondrial activity and thus the metabolic gridlock that could derive from the uptake of damaged extracellular mitochondria when they are not efficiently cleared by macrophages (e.g., due to low or defective phagocytosis and/or macrophage recruitment). EVs contain a very large number of other molecular species, including lipids, nucleic acids, and metabolites. Hence, it could not be excluded that other factors could contribute to the dampening of PPARγ via AMPK-independent mechanisms.
Several studies have demonstrated that the communication between macrophages and brown adipocytes is crucial for thermogenic adaptation (Cereijo et al., 2018; Sanchez-Alavez et al., 2020; Wolf et al., 2017). However, the role of macrophages in BAT is still a debatable issue and is being actively researched (Boulet et al., 2021; Cereijo et al., 2018; Fischer et al., 2017; Polyák et al., 2016; Wolf et al., 2017). Herein, we revealed a homeostatic role for bMACs that is essential for maintaining the mitochondrial quality control and BAT thermogenic potential. Mechanistically, macrophages take up extracellular mitochondria via various routes (Brestoff et al., 2021; Nicolás-Ávila et al., 2020). In this study, we found that bMACs internalize oxidized adipocyte mitochondrial waste in a CD36-dependent manner and that this material is eliminated by lysosomes. However, other internalization routes, such as heparan sulfates and MERTK (Brestoff et al., 2021; Nicolás-Ávila et al., 2020), cannot be excluded, as CD36 inhibition could only partly inhibit EV internalization by macrophages. We also found that, due to the uptake of brown adipocyte EVs, macrophages induced an adaptive response comprising the induction of antioxidant genes, which may support the defense against the pro-oxidant load derived by phagocytosed material.
Noteworthy, adult humans hardly have ‘‘classical’’ BAT but rather small amounts of brown and beige adipocytes interspersed in discrete sites mainly in the cervical and supraclavicular region (Cypess et al., 2013). Considering the relatively high thermogenic activity of human BAT, it is possible that the removal of damaged mitochondrial components may occur via our proposed EV-macrophage system. Based on our results, it is reasonable to postulate that a failure of EV production and/or macrophage phagocytic activity could be one of the causes of the progressive loss of BAT function during aging or obesity (Lee et al., 2013; Saito et al., 2009).
BAT has an important metabolic significance in the context of obesity and its related metabolic complications. For example, BAT activity is associated with reduced visceral adiposity and improved cardiometabolic health (Becher et al., 2021). For this reason, our work could offer interesting clues on how to enhance thermogenic energy expenditure in disease models like obesity. Collectively, our results demonstrate an alternative mechanism for removing damaged mitochondria from brown adipocytes that is characterized by the extracellular elimination of oxidized mitochondrial fragments. bMACs remove mitochondrial waste and create a favorable environment that optimizes the thermogenic program of BAT.
Limitations of the study
A significant limitation of these studies is that brown adipocyte-derived EVs are heterogenous, with some that do and some that do not contain mitochondrial parts. Additional studies are needed to determine how EVs with or without mitochondria differentially regulate BAT function. Using bone marrow transplants and MitoFat mice, we found that BAT-resident macrophages appear to be the dominant recipients of adipocyte-derived mitochondria in BAT. However, these in vivo systems cannot distinguish mechanisms or routes of mitochondrial transfer. Furthermore, our studies involving the depletion of macrophages in vivo could have affected BAT metabolism and function through pathways that are unrelated to the clearance of the damaged brown adipocyte mitochondria in EVs. Genetic tools to specifically disrupt the release or capture of EVs containing mitochondria are lacking and need to be developed to understand how this process regulates not only BAT function but also metabolic alterations observed in obesity, type 2 diabetes, cancer, and aging.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact: Daniele Lettieri-Barbato (daniele.lettieri.barbato@uniroma2.it).
Materials availability
This study did not generate unique reagents.
Data and code availability
All raw data that support the findings of this study are available from the lead contact upon reasonable request. RNA-sequence datasets produced in this study are available from ArrayExpress: -E-MTAB-10655. The newly generated proteomic data have been deposited into the ProteomeXchange database with the following accession number ProteomeXchange: PXD026080.
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Animal experiments
Wild type mice
WT C57BL/6 J male mice were obtained from Charles River Laboratories. mtDendra2, CD45.1, and Adipoq-Cre+/− mice were obtained from Jackson Laboratories and were on a C57BL6/J background. Unless otherwise indicated, all experiments were performed with adult male mice kept on an inverted 12-h:12-h dark:light cycle, fed ad libitum with a chow diet. For cold stimulation, each thermoneutrality-acclimated mouse was housed in a single cage at 4°C for 20 h, whereas warming (4°C → 30°C) was performed by exposing cold-stimulated mice at 30°C for 3 h. For EV treatment, 2.5×3 104/g mice vesicles isolated from BAT were labeled with MTG (EVsMTG), and resuspended in 300 μL of sterile phosphate-buffered saline (PBS) containing 50 μM CQ prior to BAT injection (interscapular injection). One hour after injection, BAT was explanted, and macrophages (CD45+/CD11b+/F480+ cells) were isolated using magnetic cell sorting. Rectal temperature was measured using a high-precision (±0.1°C) rectal probe for mice (RET-3, ThermoWorks, Alpine, UT, USA). Mice were sacrificed by cervical dislocation, and the explanted BAT was directly used for ex vivo experiments or tissue processing. Experiments were approved by University of Rome Tor Vergata Animal Care Italian Ministry of Health Committee (protocol 378/2017-PR).
CD169DTR mice
Macrophages were depleted by i.p. injection of 150 μL/mice of clodronate-containing liposomes two times/day for two days prior to cold exposure. For the control group, the same protocol was applied using PBS control liposomes. Immunofluorescence and gene expression analysis for macrophage markers were performed to validate macrophage depletion.
CD169DTR mice were injected with i.p. 10 μg/kg diphtheria toxin (DT; Sigma-Aldrich, Saint Louis, MO, USA) on alternating days for 4 days (flow cytometry) or 14 (−80°C) days. At the end of the treatment, mice were sacrificed by cervical dislocation, and BAT was directly frozen at −80°C or used for flow cytometry analysis. For fluorescence-activated cell sorting (FACS), BAT was digested in Hanks’ balanced salt solution (HBSS) with liberase (1 U/mL, Roche Diagnostics, Basel, Switzerland) and DNase I (10 mU/mL, Sigma-Aldrich) for 45 min at 37°C and centrifuged at 600 rpm. After digestion, single-cell suspensions were obtained by gentle pipetting and mechanical dissociation of the remaining pieces through cell strainers (BD Falcon, Glendale, AZ, USA). Single-cell suspensions were incubated with the indicated antibodies for 10 min at RT under dark conditions (CD11c APC Cy7–Biolegend, San Diego, CA, USA, CD45 PerCP Cy5.5–BD, CD11b BV510–Biolegend, CCR2 PE–R&D, ST BV711–Biolegend, Ly6C FITC–Biolegend, CD64 Biotin–Biolegend, F4/80 PE/Cy7–Biolegend, and TIM-4 Alexa Fluor 647–Biolegend). Samples were acquired in an LSRII Fortessa (BD Biosciences, Franklin Lake, NJ, USA) equipped with DIVA software (BD). FlowJo software (FlowJo LLC, Ashland, OR, USA) was used to analyze the data. Experiments were approved by Animal Care and Ethics Committee of CNIC and the regional authorities of Comunidad de Madrid.
Bone marrow transplanted and MitoFat mice
Young adult male CD45.2 wildtype or CD45.2 mtDendra2 mice were lethally irradiated with 1000 rad by X-ray irradiation. The next day the mice received intravenous administration of bone marrow (5 × 106 cells) from CD45.1 wildtype mice. Donor bone marrow was collected by flushing tibias and femurs from 5 donors and pooling the cells. Recipients were allowed to engraft for 12 weeks prior to tissue harvest. Chimerism in BAT was determined by flow cytometric analysis of the stromal vascular fraction (SVF) and is equal to the proportion of total immune cells that are CD45.1+ CD45.2−. MitoFat mice were generated as described previously (Brestoff et al., 2021). Experiments were carried out under the guidelines of the Washington University School of Medicine Institutional Animal Care and Use Committee (protocol 19–0786).
Cell culture
Murine T37i and primary brown adipocytes
T37i murine preadipocytes were kindly provided by Professor Marc Lombes (INSERM U1185, Paris, France) and cultured in Dulbecco’s modified Eagle’s medium Nutrient Mixture F-12 (DMEM/12), 10% heat-inactivated fetal bovine serum (FBS), and 100 U/mL penicillin and 10 mg/mL streptomycin (1% P/S) (Life Technologies, Carlsbad CA, USA) until 100% confluence. For differentiation, cells were maintained at complete confluency. After 2 days, 2 nM triiodothyronine (T3, Sigma-Aldrich), 1 μM rosiglitazone, and 1 μg/ml insulin (Sigma-Aldrich) were added to fresh medium every 2 days up to 8 days. pBAs were isolated and differentiated as described by Oeckl et al. (2020). Interscapular BAT from 6-week-old male mice were excised and submerged in washing buffer (HBSS with 3.5% w/v BSA, 1% P/S, 40 μg/mL gentamicin, and 500 ng/mL amphotericin B). After mincing, BAT was digested in a collagenase solution (HBSS with 3.5% w/v BSA, 1% P/S, 0.1% w/v collagenase type 1, 0.1% w/v collagenase type 2, 40 μg/mL gentamicin, and 500 ng/mL amphotericin B) by incubating at 37°C with shaking at 150 rpm for 1 h. BAT homogenates were then centrifuged twice at 250 × g for 5 min at RT, and the resulting pellet was resuspended in 3 mL of red blood cell lysis buffer (ThermoFisher Scientific; Rockford, IL, USA) for 4 min. Successively, 10 mL of washing buffer was added, and the cell suspension was filtered through a 40-μm cell strainer. The cell suspension was centrifuged at 500 × g for 5 min at RT, and the pellet was resuspended in growth medium (DMEM/12 with 10% FBS, 1% P/S, 40 μg/mL gentamicin, and 500 ng/mL amphotericin B) until 60% confluency was attained and then plated. Twenty-four hours after attaining 100% confluence, brown adipocytes were differentiated by treatment with an induction medium (DMEM/12 with 10% FBS, 1% P/S, 40 ug/mL gentamicin, 500 ng/mL amphotericin B, 850 nM insulin, 1 nM T3, 1 μM rosiglitazone,1 μM dexamethasone, 500 μM 3-isobutyl-1-methylxanthine (IBMX), and 125 μM indomethacin) for 48 h. On day 2, the induction medium was removed, and a differentiation medium (DMEM/12 with 10% FBS, 1% P/S, 40 ug/mL gentamicin, 500 ng/mL amphotericin B, 850 nM insulin, 1 nM T3, 1 and μM rosiglitazone) was added and maintained for 6 days. Prior to treatments or collection of EVs, mature brown adipocytes (day 8) were treated with 10 μM CL316,243; 1 μM trifluoromethoxy carbonylcyanide phenylhydrazone (FCCP); 5 mM N-acetyl cysteine (NAC) or 25 μM CQ. To collect EVs from BAT, 125 mg of freshly isolated BAT was cultured for 6 h in serum-free DMEM/F12 containing 1% P/S, 40 μg/mL gentamicin, and 500 ng/mL amphotericin B. EVs were isolated from mature brown adipocytes at the end of the treatments. For gene silencing, mature T37i cells were transfected with 200 pmol of small interfering RNA (nSMase2 or scramble sequence) using Lipofectamine™ 2000 transfection reagent, according to manufacturer’s instructions (ThermoFisher). EVs were collected after 48 h from transfection.
Conditionally immortalized human and mouse brown adipocytes
Cultures of conditionally immortalized human and mouse brown adipocytes (iBPA_h and iBPA_m, respectively) were prepared and maintained as described by Liu et al. (Liu et al., 2019). For differentiation of iBPAs, cells were seeded on 0.2% gelatin-coated wells. After attaining confluency, a doxycycline-containing (100 ng/ml) proliferation medium (DMEM/F-12, GlutaMAX supplemented with 10% heat-inactivated FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin) was replaced with a doxycycline-free proliferation medium. After 2 days, the differentiation of human iBPAs was induced by exposure to a proliferation medium supplemented with 150 nM human insulin (Sigma-Aldrich), 1 μM dexamethasone (Sigma-Aldrich), 1 μM rosiglitazone, 1 nM triiodothyronine (T3; Sigma-Aldrich), 500 μM IBMX (Sigma-Aldrich), and 125 μM indomethacin (Sigma-Aldrich), which was refreshed after 48 h and replaced on day 4 with a proliferation medium supplemented with 150 nM insulin, 1 nM T3, and 1 μM rosiglitazone. The latter medium was replaced every alternate day up to day 16. Differentiation of mouse iBPAs was induced by supplementing the proliferation medium with 5.6 nM bovine insulin (Sigma-Aldrich), 126 μM sodium ascorbate (Sigma-Aldrich), 1 μM rosiglitazone (Sigma-Aldrich) and 10 mM HEPES, which was adjusted to a pH of 7.4 with NaOH. This medium was replaced every alternate day up to 14 days. Human iBPAs were transfected with 50 nM Pink1 siRNA or DN-Drp1 by Lipofectamine 2000 (ThermoFisher Scientific). For lentiviral transduction, iBPA_m cells were seeded at a density of 2 × 104 cells/well in a 6-well plate and cultures overnight in complete growth medium. PDHβ (MR205484L4V, Origene, Rockville, MD, USA), SOD2 (MR201642L2V, Origene, Rockville, MD, USA), PINK1 (sc-44599-V, Santa Cruz Biotechnolgies, Dallas, TX, USA) or empty vector (pLenti-C-mGFP-P2A-Puro) was inoculated at 10 multiplicities of infection (MOI) with 2 μg/mL polybrene reagent in complete growth medium and then incubated for 48 h. Cells were selected with 2 μg/mL puromycin for three passages and maintained in 1 μg/mL puromycin. Mature iBPA_m were treated with 10 μM isoproterenol (Iso), 5 μM compound C, 10 μM CL316,243, 1 μM antimycin A and 5 mM NAC for several time points.
Murine RAW 264.7 and bone marrow derived macrophages
Murine RAW 264.7 macrophages were cultured in DMEM supplemented with 10% FBS and 1% P/S (Life Technologies) at 37°C ina humidified incubator containing 5% CO2. Twenty-four hours after plating, RAW264.7 cells were treated with EVs collected from brown adipocytes, maintaining a cell ratio of 1:5. A similar ratio was maintained by co-culturing RAW264.7 cells with brown adipocytes. RAW264.7 cells were plated at a density of 3 × 105 cells/mL for treatment with EVs isolated from BAT (50 μg tissue/mL). Macrophages were classically activated using 100 ng/mL LPS. RAW264.7 cells were pretreated with 50 μM CQ or 100 μM SSO for 1 h and maintained throughout the experiment. The treatments and co-culture system were conducted in serum-free media. Bone marrow was extracted from the limbs of 8-week-old male mice by perfusion with PBS and 1% P/S. Bone marrow-derived cells were plated at a density of 3 × 105 cells/mL in alpha-MEM supplemented with 10% FBS, 1% P/S, and 1% GlutaMAX in a humidified incubator containing 5% CO2 at 37°C. Macrophage differentiation was induced by adding M-CSF (20 ng of cells/mL) in the culture medium for 5 days. Following adhesion, unattached cells were removed, and bone marrow-derived macrophages (BMDM) were treated in a serum-free media as described for RAW264.7 cells. At the end of the treatments, macrophages were harvested, and proteins or RNA were extracted as described below.
METHOD DETAILS
Seahorse analysis
T37i/iBPA_m mature adipocytes were seeded at 1.5 ×104 cells/well in a Seahorse XF 96-well plate. Mito-stress test was performed according to Agilent Technologies recommendations with minor adaptations for adipocyte cell types (Reggio et al., 2020a, 2020b). In particular, 1 μM oligomycin A, 1.5 μM FCCP and 1 μM /1 μM rotenone/antimycin were used to perturbe mitochondria respiration. Basal oxygen consumption rate and proton leak were calculated using the Desktop Wave Software (proprietary of Agilent Technolgies). Cells were treated for 16 h with EVs prior to Seahorse analysis.
Untargeted proteomics
For quantitative proteomics of EVs released from BAT derived from cold-exposed mice (4°C, 20 h) or mice acclimatized at thermoneutrality (30°C), protein concentration of samples was determined using the Pierce BCA Protein Assay Kit™ (ThermoFisher Scientific), according to the manufacturer’s instructions. An aliquot of each protein sample (100 µg) was adjusted to a final volume of 100 μL with 100 mM triethylammonium bicarbonate (TEAB). Proteins were reduced with 5 μL of 200 mM tris(2-carboxyethyl) phosphine for 60 min at 55°C and then alkylated by adding 5 μL of 375 μM iodoacetamide in the dark for 30 min at 25°C. Alkylated proteins were precipitated by adding six volumes of cold acetone, pelleted by centrifugation at 8,000 × g for 10 min at 4 °C, and then air dried. Each sample was digested overnight with freshly prepared trypsin (enzyme to protein ratio, 1:50) in 100 mM TEAB at 37°C. The resulting peptides were labeled with the Tandem Mass Tag (TMT) Label Reagent Set (ThermoFisher Scientific) at 25°C according to the manufacturer’s instructions and following the reported relationships: BAT EVs, 4°C: TMT6–126; BAT EVs, 30°C: TMT6–127. After 1 h, the reaction was quenched by adding 8 μL of 5% w/v hydroxylamine in each tube, and the reaction was vortexed for 15 min. For a set of comparative experiments, tagged peptide mixtures were mixed in equal molar ratios (1:1) and vacuum-dried under rotation. Then, pooled TMT-labeled peptide mixtures were suspended in 0.1% trifluoroacetic acid and fractionated using a Pierce™ High pH Reversed-Phase Peptide fractionation kit (ThermoFisher Scientific) to remove unbound TMT reagents according to the manufacturer’s instructions. After fractionation, eight fractions of TMT-labeled peptides were collected, vacuum-dried, and finally reconstituted in 0.1% FA. TMT-labeled peptide fractions were analyzed on a NanoLC-ESI-Q-Orbitrap-MS/MS platform consisting of high-performance liquid chromatography (HPLC) with an UltiMate™ 3000 RSLCnano System (Dionex, USA) coupled to a Q-ExactivePlus mass spectrometer through a Nanospray Flex Ion Source (ThermoFisher Scientific). Peptides were loaded onto an Acclaim™ PepMap™ RSLC C18 column (150 mM × 75 μM ID, 2 µm particles, 100 Å pore size) (ThermoFisher Scientific) and eluted with a gradient of solvent B (19.92/80/0.08 [v/v/v] water/acetonitrile/formic acid) in solvent A (99.9/0.1 [v/v] water/formic acid) at a flow rate of 300 nL/min. The gradient of solvent B started at 5%, increased to 60% over 125 min, increased to 95% over 1 min, remained at 95% for 8 min, and finally returned to 5% in 1 min, with a column equilibrating step of 20 min before the subsequent chromatographic run. The mass spectrometer was operated in a data-dependent mode, using a full scan (m/z range 375–1500, nominal resolution of 70,000), followed by tandem mass spectrometry (MS/MS) scans of the 10 most abundant ions. MS/MS spectra were acquired in a scan m/z range of 110–2,000 using a normalized collision energy of 32%, an automatic gain control target of 100,000, a maximum ion target of 120 ms, and a resolution of 17,500. A dynamic exclusion of 30 s was used. All MS and MS/MS raw data files of each sample were merged for protein identification and relative protein quantification into Proteome Discoverer version 2.1 software (Thermo Scientific), enabling a database search in Mascot Server version 2.4.2 (Matrix Science, UK) using the following criteria: UniProtKB protein database (Mus musculus, 17,030 protein sequences, 10/2019), including the most common protein contaminants; carbamidomethylation of Cys and TMT6plex modification of lysine and peptide N-terminal as fixed modifications; oxidation of Met; deamidation of Asn/Gln; pyroglutamate formation of Gln; and phosphorylation of Ser/Thr/Tyr as variable modifications. The peptide mass tolerance was set to ±10 ppm, and the fragment mass tolerance was set to ±0.02 Da. The proteolytic enzyme was trypsin, and the maximum number of missed cleavages was set to 2. Protein candidates assigned on the basis of at least two sequenced peptides and an individual Mascot Score ≥25 were considered to have been confidently identified. For quantification, the ratios of TMT reporter ion intensities in the MS/MS spectra from raw datasets were used to calculate FCs between samples. Results were filtered to a false discovery rate of 1%.
For quantitative proteomics of cell lysates or EVs released from pBAs untreated (control) or treated with CL316,243 or FCCP, we adopted the whole procedure reported above. In this case, resulting peptides from cells or EVs obtained with different experimental conditions (as reported above) were labeled with TMT Label Reagent Set (ThermoFisher Scientific) according to the following relationships: EVs_Ctrl: TMT6–126; EVs_CL316,243: TMT6–127; EVs_FCCP: TMT6–128; CELL_Ctrl: TMT6–129; CELL_CL316,243: TMT6–130; CELL_FCCP: TMT6–131.
Cellular components and cell types were analyzed using FunRich version 3.1.3 (http://www.funrich.org), EnrichR (https://amp.pharm.mssm.edu/Enrichr) or David 6.8 (https://david.ncifcrf.gov).
Transmission electron microscopy
BAT was fixed with 2.5% glutaraldehyde for 24 h, and sample preparation was performed at the Core Facility for Integrated Microscopy, University of Copenhagen, Denmark. Images were acquired with a CM 100 BioTWIN electron microscope operating with an 80 kV accelerating tension and equipped with an Olympus Veleta digital camera. Image processing was performed using Olympus ITEM software. The brightness and contrast of images were adjusted using Fiji ImageJ (Schindelin et al., 2012).
Magnetic cell and mitochondrial sorting
Stromal vascular cells were resuspended in 500 mL of magnetic bead buffer (MBB) (PBS 1× without calcium and magnesium, 0.5% w/v BSA, and 2 μM ethylenediaminetetraacetic acid [EDTA]) and filtered through a 30-μM pre-separation filter (Miltenyi, Bergisch Gladbach, Germany) following three filter washes to remove large particles and debris. The cell suspension was separated at 300 × g for 5 min at 4°C and resuspended in 90 mL of MBB plus 10 mL of anti-CD45 magnetic beads-conjugated antibody (Miltenyi) for each 107 cell sample. After 15 min of incubation at 4°C, the cell suspension was diluted with 2 mL of MBB and centrifuged. The cell pellet was resuspended in 500 mL of MBB, applied on hydrated MS-columns (Miltenyi), washed three times with 500 mL of MBB, and collected with 1 mL of MBB through piston elution. Incubation with anti-CD11b and anti-F4/80 magnetic beads-conjugated antibodies (Miltenyi) was performed in a unique incubation, as was performed for CD45. BAT was homogenized using a Potter–Elvehjem polytetrafluoroethylene pestle and a glass tube and TO μM 22+ mitochondria were magnetically isolated by a commercially available kit (Miltenyi).
scRNA-seq and MacSpectrum analyses
Single-cell suspensions were prepared for scRNA-seq immediately after cell sorting using a Chromium™ (V3V2) Single-Cell Reagent Kit according to the manufacturer’s protocol (10× Genomics). Following capture and lysis, cDNA was synthesized for each droplet of captured cells and amplified (12 cycles). The amplified cDNA from each channel of the Chromium™ system was used to construct an Illumina sequencing library and was sequenced on a HiSeq 4000 system with 150-cycle sequencing (asymmetric reads per 10× Genomics). Illumina base call files (*.bcl) were converted to FASTQs using CellRanger version 1.3, which uses bcl2fastq version 2.17.1.14. Then, FASTQ files were aligned to the mm10 mouse reference genome and transcriptome using the CellRanger version 1.3 software pipeline with default parameters (10× Genomics); this demultiplexed the samples and generated a gene versus cell expression matrix based on the barcodes and assigned unique molecular identifiers that enable the determination of an individual cell from which the RNA molecule originated. For clustering, genes were selected based on normalized dispersion analysis. Dimensionality reduction was performed using the Cell Ranger pipeline with PCA and was passed to t-SNE or Uniform Manifold Approximation and Projection with default parameters.
Macrophages were selected from the scRNA-seq dataset by filtering cells with CD11b and Adgre1/(F4/80) detection. The profiles of select macrophages were further analyzed using MacSpectrum program for activation and inflammatory responses. The macrophage polarization index (MPI) and activation-induced macrophage differentiation index (AMDI) were calculated for each cell using previously described methods (Li et al., 2019). In brief, each cell was first evaluated using 500 polarization signature genes for rm1 and rm2, followed by linear regression to develop a polarization axis. The projection of each cell onto the polarization axis was then scaled and assigned to a proportional MPI value. A higher MPI value represented a stronger inflammatory response in the cell. Accordingly, each individual cell was evaluated using a set of 435 activation-induced macrophage differentiation signature genes for rm0 and rescaled to assign an AMDI value. A higher AMDI value suggested a greater activated status relative to the naïve (pre-activated) status. Monocle 3 program (https://cole-trapnell-lab.github.io/monocle3/docs/trajectories/) along the pseudotime distribution, was used to perform the trajectory analyses of bMACs
Bulk RNA-sequencing and functional enrichment analysis
Total adipose RNA was isolated using TRIzol reagent (Invitrogen, Waltham, MA, USA) and purified using the RNeasy mini kit protocol (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Isolated RNA was sequenced using an Illumina NextSeq500, and the indexed libraries were prepared from 1 µg of purified RNA with TruSeq-stranded mRNA (Illumina) Library Prep Kit according to the manufacturer’s instructions. The quality of the single-end reads was evaluated using FastQC version 0.11.5 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc). All FastQC files were filtered to remove low-quality reads and adapters using Trimmomatic version 0.36 (Bolger et al., 2014). The resulting reads were mapped to the Mus musculus genome (GRCm38) using HISAT2 version 2.1.0 (Kim et al., 2015) using default parameters, and StringTie version 1.3.4d (Pertea et al., 2015) was applied to the BAM files obtained using HISAT2 to generate expression estimates and to quantify the transcript abundance as transcripts per kilobase per million of mapped reads. The count matrices generated by StringTie were imported in R, in which differential expression analysis was performed using Deseq2 to compare the two different conditions. Functional annotation was performed using the AnnotationDbi R library (http://bioconductor.org). Differentially expressed genes were selected with a threshold of Log2FC > 0.58 (p < 0.05). Functional enrichment analyses, including GO and the KEGG pathway, were performed using FunRich version 3.1.3 (http://www.funrich.org), EnrichR (https://amp.pharm.mssm.edu/Enrichr), or David 6.8 (https://david.ncifcrf.gov).
qPCR gene expression analysis
Total adipose RNA was isolated using TRIzol reagent and purified using the RNeasy mini kit protocol according to the manufacturer’s instructions. Then, 1 mg of RNA was treated with genomic DNase and retro-transcripted using PrimeScript RT Reagent Kit (Takara Bio Inc., Japan). qPCR was performed in triplicate on 50 ng of cDNA using validated qPCR primers (BLAST), Applied Biosystems™ Power™ SYBR™ Green Master Mix, and the QuantStudio3 Real-Time PCR System (Thermo Fisher, Waltam, MA, USA), as described by Turchi et al. (2020). The primers used for qPCR are listed below:
Nos2; FWD 5’-GCCTTCAACACCAAGGTTGTC-3’; REV 5’-ACCACCAGCAGTAGTTGCTC-3’
Adgre1 (F4/80); FWD 5’-CCAGGAGTGGAATGTCAAGATGT-3’; REV 5’-GCAGACTGAGTTAGGACCACA-3’
Mrc1 (Cd206); FWD 5’-GGCTGATTACGAGCAGTGGA-3’; REV 5’-CATCACTCCAGGTGAACCCC-3’
Cxcl2; FWD 5’-ATCCAAAAGATACTGAACAAAGGCA-3’; REV 5’-CTCTTTGGTTCTTCCGTTGAGG-3’
PDHβ; FWD 5’-AGTTGCCCAGTATGACGGTG-3’; REV 5’-TCTGAGATGGGGGTGTCGAT-3’
Pc; FWD 5’-GCTTATCCCGAACATCCCGT-3’; REV 5’-GGCCACCTCACAGAACTTGAA-3’
Il1b; FWD 5’-GCACTGGGTGGAATGAGACT-3’; REV 5’-GGACATCTCCCACGTCAATCT-3’
Ucp1; FWD 5’-ACCACCCTGGCAAAACAGA-3’; REV 5’-GAGGCAGGTGTTTCTCTCCC-3’
Adipoq; FWD 5’-GACCTGGCCACTTTCTCCTC-3’; REV 5’-TCCTGAGCCCTTTTGGTGTC-3’
PPARγ c1a; FWD 5’-CTGGAACTGCAGGCCTAACT-3’; REV 5’-GCAAGAGGGCTTCAGCTTTG-3’
Fabp4; FWD 5’-AAATCACCGCAGACGACAGG-3’; REV 5’-TCCATCCCACTTCTGCACCT-3’
Cd36; FWD 5’-AAGGCCATCTCTACCATGCC-3’; REV 5’-TGTGGCTAAATGAGACTGGGAC-3’
Dio2; FWD 5’-CAAACAGGTTAAACTGGGTGAAGA-3’; REV 5’-GGTCAGGTGGCTGAACCAAA-3’
Elovl3; FWD 5’-TCCCTACCCCAAGCTCTGTAA-3’; REV 5’-AATGAGCTTACCCAGTACTCCTC-3’
Gk; FWD 5’-TGTCACAATGGAGCGGTTTG-3’; REV 5’-TGGGATACCACTTTCTGGAGAC-3’
Rpl8; FWD 5’-GGAGCGACACGGCTACATTA-3’; REV 5’-CCGATATTCAGCTGGGCCTT-3’
Actb; FWD 5’-CACACCCGCCACCAGTTCGC-3’; REV 5’-TTGCACATGCCGGAGCCGTT-3’
The reaction was performed according to the manufacturer’s protocol using QuantStudio 3 Real-Time PCR System. Data were analyzed following the 2—∆∆Ct method.
Targeted metabolomics
For metabolomic analyses, EVs and T37i cells were resuspended in 250 mL of methanol:acetonitrile 1:1 (v/v) containing 1 ng/mL D-Glucose-13C6 (internal standard, Sigma-Aldrich, 389374) and centrifuged at 4°C. The supernatant was passed through cellulose filter and used for subsequent analysis. For aminoacids and biogenic amines analyses, samples were incubated with phenyl isothiocyanate solution for 20 min at 25°C, dried, and resuspended in 5 mM ammonium acetate in methanol:water 1:1 (v/v). Metabolomics data were obtained on an API-4000 triple quadrupole mass spectrometer (AB SCIEX) coupled with an HPLC system (Agilent) and CTC Analytics HTS PAL LC Autosampler (PAL System). The identity of all metabolites was confirmed using pure standards. Quantification of different metabolites was performed with liquid chromatography/MS/MS using a cyano-phase LUNA column (50 mm × 4.6 mm, 5 mm; Phenomenex, Torrance, CA, USA). Metabolites were analyzed through a 10-min run in negative ion mode in a 5-min run. The mobile phase for the negative ion mode analysis was 5 mM ammonium acetate at a pH of 7 in methanol (solvent A). An isocratic elution at 100% solvent A was performed at a flow rate of 500 µL/min. MultiQuant™ software version 3.0.2 was used for data analysis and peak review of chromatograms. Quantitative evaluation of all metabolites was performed based on calibration curves with pure standards; then, data were normalized on micrograms of proteins.
Phospholipidomics
Quantification of the different phospholipid families was performed by flow injection analysis-tandem mass spectrometry (FIA-MS/MS) method using an ESI source connected with an API 4000 triple quadrupole instrument (AB Sciex, Framingham, MA, USA) as previously described (Tatulli et al., 2018). MultiQuant™ software version 3.0.2 was used for data analysis and peak review of chromatograms.
BAT immune cell isolation and high dimensional flow cytometry
The SVF was isolated from BAT by mincing tissues finely with scissors and incubating in high-glucose DMEM containing 0.1% collagenase II (Sigma-Aldrich) at 37°C in an orbital shaker at ~150 rpm for 1 h. The cell suspension was filtered through a 100-µm nylon mesh. Cells were collected in a 50-mL conical tube and centrifuged at 500 × g for 5 min at 4°C. The supernatants and floating adipocytes were aspirated, and the SVF pellet was resuspended in 1 mL of ACK RBC lysis buffer (Gibco) and incubated at RT for 2 min. The RBC lysis reaction was quenched by adding 10 mL of cold wash media (high-glucose DMEM containing 5% heat-inactivated FBS, L-glutamine, and penicillin/streptomycin). The cells were centrifuged at 500 × g for 5 min at 4°C, supernatants were aspirated, and cells were resuspended in 200 µL of wash media for subsequent staining on a polystyrene 96-well round bottom tissue-culture treated plate. The cells were centrifuged at 500 × g for 5 min at 4°C, and the cell pellet was washed in PBS once and centrifuged again as described above. The cells were stained with 50 µL of ZombieUV (1:600 in PBS for analysis on a BD X20 flow cytometer) or ZombieNIR (1:1,000 in PBS for analysis on a Cytek Aurora spectral flow cytometer) for 5 min on ice. FACS buffer (200 µL; PBS containing 2.5% heat-inactivated FBS and 2.5 μM EDTA) was added, and the cells were centrifuged as described above, followed by washing once with 200 µL of FACS buffer. The cells were incubated with 25–50 µL of FcBlock (1:100, BD Pharmigen in FACS buffer) on ice for 10 min, and then an equal volume of 2× stain cocktail was added and mixed. After 30 min of staining on ice (protected from light), the cells were washed 2–3 times as described above. For monocyte and macrophage dynamics, cells were finally resuspended in 200 µL of FACS buffer for high dimensional flow cytometry analysis and stained with different panels of 20 different cell surface markers, with CD45 as the common maker to identify leukocytes. Macrophages were identified as CD11b+F4/80+ and further as CD64+CCR5+ cells and inside this gate, the expression of several markers defining cell subsets, inflammatory status and phagocytosis ability was evaluated by staining with anti-CD169, anti-CD68, anti-CD80, anti-CD86, anti-MHC-II, anti-CD36, anti-CD206 and anti-CD200R. Monocytes were identified with several pan and subset markers, such as anti-Ly6C, anti-CD11b, anti-F4/80, anti-CX3CR1, anti-CCR2, anti-CD115, anti-CD62L. Samples were acquired on a 21-color Cytoflex (Beckman Coulter) and for each analysis, at least 0.5×106 live cells were acquired by gating on aqua Live/Dead negative cells (Leuti et al., 2021; Talamonti et al., 2017).
Analysis of mitochondrial mass, membrane potential and ROS
For cytofluorimetric analyses, EVs were incubated with MTG (M7514, ThermoFisher Scientific) or Annexin V-FITC (A35111, ThermoFisher Scientific) for 30 min at 37°C. To eliminate unincorporated dye from EVs, exosome spin columns were used to remove any low MW (≤3,000 Da) admixtures (4484449, ThermoFisher Scientific). To stain mitochondria and mitochondrial ROS, mature brown adipocytes were loaded with MitoTracker Green (M7514, ThermoFisher Scientific), MitoTracker Red CMXRos (M7512, ThermoFisher Scientific) or MitoSOX (M36008, ThermoFisher Scientific) for 1 h. Flow cytometry analyses were performed using FACSCalibur (BD, USA) and analyzed using FlowJo software version 4.14.
Transcriptomic data mining strategy
Gene expression data from the repository datasets that showed transcriptomic profiles in iBAT and eWAT were downloaded from the public Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov). Raw transcriptomic data were log-transformed without background signal extraction, and a bulk Loess normalization was applied to all datasets using Babelomics 5 online software (http://babelomics.bioinfo.cipf.es). Statistical significance was set at p < 0.05 to identify differentially expressed genes (0.5 < FC > 1.5).
Isolation of mitochondria, MDVs and EVs
To isolate mitochondria, BAT and brown adipocytes were homogenized using a Potter–Elvehjem polytetrafluoroethylene pestle and a glass tube with Mitochondria Isolation Buffer (1 M sucrose, 0.1 M Tris/MOPS, 0.1 M EGTA/Tris, pH 7.4) containing protease inhibitor cocktail (Roche Diagnostics). The homogenate was centrifuged at 600 × g, for 10 min at 4°C to remove nuclei and unbroken cells. The supernatant was then centrifuged twice at 9,000 × g for 20 min at 4°C (crude mitochondrial fraction). Trypsin (0.01 μg/µL) was added to the resulting post-mitochondrial supernatant, followed by incubation for 10 min on ice. Successively, the supernatant was centrifuged at 100,000 × g for 1 h at 4°C to collect MDVs.
To isolate EVs, finely minced BAT or brown adipocytes were cultured in serum-free DMEM and after 24 h, culture media were collected and centrifuged at 600 × g for 10 min at 4°C to remove dead cells. Successively, the supernatant was centrifuged at 17,000 × g at 4°C to isolate large EV fractions or ultracentrifuged at 100,000 × g for 16 h at 4°C (Rotor SW28 or SW40, according to sample volume; Beckman Coulter, CA, USA) to recover the total EVs fraction. EVs pellets were washed once with PBS or radioimmunoprecipitation assay (RIPA) buffer for treatments or immunoblotting, respectively. Protein determination was performed using the Lowry method.
Immunofluorescence and confocal microscopy
Cells were incubated with a permeabilization solution (PBS/Triton X100 0.2% [v/v]), blocked for 1 h with a blocking solution (PBS/BSA 5% [v/v]), and then incubated for 18 h with anti-PDHβ (gtx-119625, GeneTex, Irvine, CA, USA), anti-Cyt c (sc-13156, Santa Cruz Biotechnology, Dallas, TX, USA), and anti-TOMM20 (FL-145, Santa Cruz Biotechnology). Cells were then washed with cold PBS and incubated for 1 h with AlexaFluor-488 or −568-conjugated secondary antibodies (ThermoFisher Scientific). Nuclei were stained with 10 μg/mL Hoechst 33342 (Thermo Fisher Scientific).
Live cells were incubated for 30 min with LysoTracker Deep Red (L12492, ThermoFisher Scientific) and MitoTracker™ Red CMXRos (M7512, ThermoFisher Scientific) to stain lysosomes and mitochondria, respectively.
Sections of frozen BAT were incubated with anti-CD68 (ab125212, Abcam, Cambridge, UK) and the appropriate AlexaFluor 568-conjugated secondary antibody (ThermoFisher Scientific). AlexaFluor 488 Phalloidin (A12379, ThermoFisher Scientific) was used to stain actin.
Confocal micrographs were acquired using an Olympus IX-81 confocal microscope at 60× magnitude. Representative regions of interest were acquired using a digital 3× zoom. Fluorescence intensities were set for the control samples and were maintained for all samples. Micrographs were acquired at 8 µs/pixel speed and 800 × 800-pixel resolution. Colocalization analysis was performed on z-gallery with the JACoP plugin (Bolte and Cordelières, 2006) implemented in Fiji software (Schindelin et al., 2012). Channel thresholds were set for the control samples and were maintained for all analyzed samples (tetramethylrhodamine isothiocyanate = 37; FITC = 17). Pearson’s coefficient and Manders’s M1 and M2 coefficients (Manders et al., 1992) were used to calculate colocalization between PDHβ and TOMM20.
Immunoblotting
Tissues or cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 12 mM deoxycholic acid, 0.5% Nonidet P-40, and protease and phosphatase inhibitors). Then, 10 µg of proteins were loaded on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to immunoblotting. Nitrocellulose membranes were incubated with anti-PDHβ (gtx-119625, GeneTex); anti-UCP1 (ab23841, Abcam); anti-MDH2 (GTX105870, GeneTex); anti-HSP60 (sc-13966, Santa Cruz Biotechnology); anti-PC (sc-271493, Santa Cruz Biotechnology); anti-TOMM20 (sc-11415, Santa Cruz Biotechnology); anti-VDAC1 (sc-390996, Santa Cruz Biotechnology); anti-CD63 (cd63 A-1 System Bioscience); anti-phospho-DRP1 (#48675, Cell Signaling); anti-PKA-serine substrates (#9621S, Cell Signaling); anti-TUBULIN (10094–1-AP, Proteintech, Rosemont, IL, USA); anti-4HNE (ab46545, Abcam); anti-CD36 (ab124515, Abcam); anti-ACO2 (sc-130677, Santa Cruz Biotechnology); anti-ACAA2 (GTX115417, GeneTex); anti-PDHα (GTX104015, GeneTex); anti-NOS2 (ab205529, Abcam); anti-PINK1 (sc-517353, Santa Cruz Biotechnology); anti-ACTIN (20536-i-ap, Proteintech) and anti-VINCULIN (MA5–11690, Invitrogen) primary antibodies at a dilution of 1:1000. The membranes were then incubated with the appropriate horseradish peroxidase-conjugated secondary antibodies. Immunoreactive bands were detected using a FluorChem FC3 System (Protein-Simple, San Jose, CA, USA) after incubating the membranes with ECL Select Western Blotting Detection Reagent (GE Healthcare, Pittsburgh, PA, USA). Densitometric analyses of the immunoreactive bands were performed using FluorChem FC3 Analysis software. Representative immunoblots of at least n=3 independent experiments or mice/group are shown.
PEG-switch assay
Cysteine oxidation in MDVs and EVs was evaluated using the method described by Burgoyne et al. (Burgoyne et al., 2013). Briefly, vesicle pellets were lysed into alkylating buffer (1% SDS, 100 mM maleimide, 100 mM Tris-HCl, pH 7.4) and heated at 50°C for 25 min. After incubation, samples were desalted to remove free maleimide using Zeba Spin Desalting Columns (Pierce, Waltham, MA, USA). Immediately after desalting, 50 mM dithiothreitol (DTT) was added to each sample, which was incubated for 20 min at RT to reduce reversibly oxidized cysteine thiols. After incubation, samples were desalted to remove excess DTT using Zeba Spin Desalting Columns. Reduced desalted samples were labeled with 2 mM PEG-maleimide (MW = 5,000 Da, Sigma-Aldrich) in the presence of 0.5% SDS for 2 h at RT. After further incubation for 2 h at RT, samples were prepared for SDS-PAGE by the addition of a sample buffer containing 5% β-mercaptoethanol and immunoblotted as described above.
Single-cell mass cytometry
For single-cell analysis performed with a CyTOF2 instrument, 2 × 106 SVCs isolated from BAT as described above were used. To label dead cells, incubation with a DNA intercalator (Rhodium-103, Rh103; Fluidigm, CA, USA) was conducted for 15 min at 37°C. Cells were then centrifuged at 800 × g for 5 min and washed in Dulbecco’s PBS without calcium and magnesium (BioWest, Riverside, MO, USA). To reduce inter-sample staining variation, a mass-tag barcoding protocol on fixed cells was used. Cells were fixed with 1 mL of Fix I buffer and incubated for 10 min at RT, followed by quenching with Barcode Perm Buffer (Fluidigm, South San Francisco, CA, USA). The cells under the different conditions were barcoded with the appropriate combination of palladium isotopes from the Cell-ID™ 20-Plex Pd Barcoding Kit in Barcode Perm Buffer for 30 min at RT. The staining was then quenched with MaxPar Cell Staining Buffer (Fluidigm). After mass-tag cellular barcoding, the pooled samples were subsequently stained with metal-tagged antibodies directed against surface antigens. The samples were then collected into a unique tube, and the surface antibody-staining protocol was performed according to the manufacturer’s instructions for 30 min at RT. After staining, cells were washed twice with MaxPar Cell Staining Buffer and stained for 1 h at RT with an intercalation solution composed of Cell-ID Intercalator-Ir (191Ir and 193 Ir) (Fluidigm) present in MaxPar Fix and Perm Buffer at a final concentration of 125 nM. Cells were washed twice with MaxPar Cell Staining Buffer and MaxPar Water, respectively. For mass cytometry acquisition, cells were resuspended at a final concentration of 2.5 × 105 cells/mL in ddH2O containing 10% EQ™ Four Element Calibration Beads and filtered through a 30-µm filter-cap FACS tube. Samples were kept on ice prior to acquisition using the mass cytometry platform CyTOF2 (Fluidigm). Data were collected as.fcs files. Following data acquisition, the channel intensity was normalized using calibration beads, as described by Finck et al. (Finck et al., 2013), and the normalized.fcs file was de-barcoded using Debarcoder software (Fluidigm). Data were preprocessed through the Cytobank software platform (Kotecha et al., 2010). Cells were then manually gated from debris based on DNA content, followed by the incorporation of the Iridium (Ir) intercalator. Doublets were then eliminated according to the event length parameter, and single live cells were manually gated using the Rh103 intercalator signal. Manually gated singlet (191Ir+ 193Ir+), viable (195Pt—) events were analyzed using Cytofkit and dedicated R scripts (Chen et al., 2016). Cytofkit parameters were set as follows: 22 biomarkers were included for clustering all detected live cells per sample to merge all events. Data were transformed using the cytofAsinh transformation method; FlowSOM was used as the clustering algorithm, with k = 10, t-SNE perplexity set to 30 with 1,000 iterations, and seed set to 42.
Cell populations were assigned based on the expression level of distinct biomarkers across the clusters identified by FlowSOM. The expression levels of each cell population were quantified by setting the mean ± standard deviation (SD) as the threshold, depending on the expression value distribution of the specific marker.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses
Data were expressed as the mean ± SD unless otherwise stated. The exact numbers of replicates are given in each figure legend. A two-tailed unpaired Student’s t-test was performed to assess the statistical significance between two groups. Analysis of variance (ANOVA) followed by Dunnett’s (comparisons relative to controls) or Tukey’s (multiple comparisons among groups) post hoc tests was used to compare three or more groups. Venn diagrams were constructed using Draw Venn Diagram or Venny 2.1.0 software. Statistical analyses were performed using GraphPad Prism 9 (GraphPad Software Inc., San Diego, CA, USA). In all cases, a p value of 0.05 was set as the significance threshold.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies—western blot and immunofluorescence | ||
| CD68 antibody, Abcam | Abcam | Cat# ab125212; RRID:AB_10975465 |
| UCP1 antibody, Abcam | Abcam | Cat# ab23841; RRID:AB_2213764 |
| 4 Hydroxynonenal antibody, Abcam | Abcam | Cat# ab46545; RRID:AB_722490 |
| Anti-CD36 antibody | Abcam | Cat# ab124515 |
| Recombinant Anti-iNOS antibody [EPR16630] | Abcam | Cat# ab205529 |
| Phospho-DRP1 (Ser637) Antibody, Cell Signaling TechnologySA | Cell Signaling Technology | Cat# 4867; RRID:AB_10622027 |
| Phospho-(Ser/Thr) PKA Substrate Antibody, Cell Signaling Technology | Cell Signaling Technology | Cat# 9621; RRID:AB_330304 |
| Pyruvate Dehydrogenase E1 beta subunit antibody, GeneTex | GeneTex | Cat# GTX119625; RRID:AB_11163683 |
| MDH2 antibody, GeneTex | GeneTex | Cat# GTX105870; RRID:AB_1950902 |
| ACAA2 antibody, GeneTex | GeneTex | Cat# GTX115417; RRID:AB_2885361 |
| Pyruvate Dehydrogenase E1 alpha antibody, GeneTex | GeneTex | Cat# GTX104015; RRID:AB_1951155 |
| Beta Tubulin antibody, Proteintech | Proteintech | Cat# 10094–1-AP; RRID:AB_2210695 |
| Beta Actin Polyclonal antibody | Proteintech | Cat# 20536-i-ap |
| PINK1 (38CT20.8.5) | Santa Cruz Biotechnology | Cat# sc-517353 |
| cytochrome c (A-8) antibody, Santa Cruz Biotechnology | Santa Cruz Biotechnology | Cat# sc-13156; RRID:AB_627385 |
| Tom20 (FL-145) Antibody, Santa Cruz Biotechnology | Santa Cruz Biotechnology | Cat# sc-11415; RRID:AB_2207533 |
| HSP 60 (H-300) Antibody, Santa Cruz Biotechnology | Santa Cruz Biotechnology | Cat# sc-13966; RRID:AB_2121457 |
| PCB (H-2) antibody, Santa Cruz Biotechnology | Santa Cruz Biotechnology | Cat# sc-271493; RRID:AB_10649369 |
| VDAC1 Antibody (B-6), Santa Cruz Biotechnology | Santa Cruz Biotechnology | Cat# sc-390996; RRID:AB_2750920 |
| Rabbit Anti-ACO2 Polyclonal Antibody, Unconjugated, Clone A-22, Santa Cruz Biotechnology | Santa Cruz Biotechnology | Cat# sc-130677; RRID:AB_2221514 |
| Mass-Cytometry antibodies | Table S4 | |
| Anti-CD63 Antibody, System Biosciences | System Biosciences | Cat# EXOAB-CD63A-1; RRID:AB_2561274 |
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 555, Thermo Fisher Scientific | Thermo Fisher Scientific | Cat# A-21422; RRID:AB_2535844 |
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488, Thermo Fisher Scientific | Thermo Fisher Scientific | Cat# A-11001; RRID:AB_2534069 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488, Thermo Fisher Scientific | Thermo Fisher Scientific | Cat# A-11008; RRID:AB_143165 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 555, Thermo Fisher Scientific | Thermo Fisher Scientific | Cat# A-21428; RRID:AB_2535849 |
| Vinculin Monoclonal Antibody (VLN01), Thermo Fisher Scientific | Thermo Fisher Scientific | Cat# MA5–11690; RRID:AB_10976821 |
|
Antibodies—flow cytometry | ||
| Rat Anti-CD45 Monoclonal Antibody, PerCP-Cy5.5 Conjugated, Clone 30-F11, BD Biosciences | BD Biosciences | Cat# 550994; RRID:AB_394003 |
| CD195 (CCR5) antibody, BD Biosciences | BD Biosciences | Cat# 743697; RRID:AB_2741679 |
| APC anti-mouse CD11c antibody, BioLegend | Biolegend | Cat# 117309; RRID:AB_313778 |
| PE anti-mouse/human CD11b antibody, BioLegend | BioLegend | Cat# 101207; RRID:AB_312790 |
| FITC anti-mouse Ly-6C antibody, BioLegend | BioLegend | Cat# 128005; RRID:AB_1186134 |
| Purified anti-mouse CD64 (FcgammaRI) antibody, BioLegend | BioLegend | Cat# 139301; RRID:AB_10612757 |
| Purified anti-mouse F4/80 antibody, BioLegend | BioLegend | Cat# 123101; RRID:AB_893504 |
| Alexa Fluor(R) 647 anti-mouse Tim-4 antibody, BioLegend | BioLegend | Cat# 130007; RRID:AB_2201842 |
| FITC anti-mouse/human CD11b antibody, BioLegend | BioLegend | Cat# 101206; RRID:AB_312789 |
| PE/Cyanine7 anti-mouse/human CD11b antibody, BioLegend | BioLegend | Cat# 101216; RRID:AB_312799 |
| PerCP/Cyanine5.5 anti-mouse CD36 antibody, BioLegend | BioLegend | Cat# 102619; RRID:AB_2750187 |
| Brilliant Violet 510(TM) anti-mouse CD45 antibody, BioLegend | BioLegend | Cat# 103138; RRID:AB_2563061 |
| CD62L BV650 | BioLegend | Cat# 1122265 |
| Brilliant Violet 711(TM) anti-mouse CD64 (FcgammaRI) antibody, BioLegend | BioLegend | Cat# 139311; RRID:AB_2563846 |
| FITC anti-mouse CD80 antibody, BioLegend | BioLegend | Cat# 104705; RRID:AB_313126 |
| PE anti-mouse CD115 (CSF-1R) antibody, BioLegend | BioLegend | Cat# 135505; RRID:AB_1937254 |
| PerCP/Cyanine5.5 anti-mouse CD169 (Siglec-1) antibody, BioLegend | BioLegend | Cat# 142409; RRID:AB_2563908 |
| PE/Cyanine7 anti-mouse CD206 (MMR) antibody, BioLegend | BioLegend | Cat# 141720; RRID:AB_2562248 |
| FITC anti-mouse CD192 (CCR2) antibody, BioLegend | BioLegend | Cat# 150608; RRID:AB_2616980 |
| Brilliant Violet 421(TM) anti-mouse CX3CR1 antibody, BioLegend | BioLegend | Cat# 149023; RRID:AB_2565706 |
| APC anti-mouse F4/80 antibody, BioLegend | BioLegend | Cat# 123116; RRID:AB_893481 |
| APC/Cyanine7 anti-mouse F4/80 antibody, BioLegend | BioLegend | Cat# 123117; RRID:AB_893489 |
| PE/Cyanine7 anti-mouse Ly-6C antibody, BioLegend | BioLegend | Cat# 128018; RRID:AB_1732082 |
| PE/Dazzle(TM) 594 anti-mouse Ly-6G antibody, BioLegend | BioLegend | Cat# 127648; RRID:AB_2566319 |
| Brilliant Violet 650(TM) anti-mouse CD62L antibody, BioLegend | BioLegend | Cat# 104453; RRID:AB_2800559 |
| CD11b MicroBeads, human and mouse | Miltenyi Biotec | Cat#130–049-601 |
| Anti-F4/80 MicroBeads UltraPure, mouse antibody, Miltenyi Biotec | Miltenyi Biotec | Cat# 130–110-443; RRID:AB_2858241 |
| CD11b Antibody, anti-human/mouse, APC-Vio® 770, Miltenyi Biotec | Miltenyi Biotec | Cat# 130–113-232; RRID:AB_2726043 |
| CD11c-APC, mouse antibody, Miltenyi Biotec | Miltenyi Biotec | Cat# 130–102-800; RRID:AB_2660155 |
| CD68 Antibody, anti-mouse, PE, Miltenyi Biotec | Miltenyi Biotec | Cat# 130–102-923; RRID:AB_2659048 |
| CD86 Antibody, anti-mouse, FITC, Miltenyi Biotec | Miltenyi Biotec | Cat# 130–123-672; RRID:AB_2889633 |
| CD200R Antibody, anti-mouse, PE, REAfinity™, Miltenyi Biotec | Miltenyi Biotec | Cat# 130–112-458; RRID:AB_2725830 |
| MHC Class II (I-Ak) Antibody, anti-mouse, APC, REAfinity™, Miltenyi Biotec | Miltenyi Biotec | Cat# 130–109-273; RRID:AB_2652875 |
| Mouse CCR2 PE-conjugated Antibody | R&D Systems | Cat# FAB5538P |
|
Antibodies—magnetic cells sorting | ||
| Miltenyi Biotec, Inc. CD45 MICROBEADS | Miltenyi Biotec | Cat#130–052-301;RRID:AB_2877061 |
|
Antibodies—mass cytometry | ||
| CD3e | Fluidigm | Cat# 3152004b |
| CD4 | Fluidigm | Cat# 3145002b |
| CD8a | Fluidigm | Cat# 3153012b |
| CD11b | Fluidigm | Cat# 3148003b |
| CD11c | Fluidigm | Cat# 3142003b |
| CD19 | Fluidigm | Cat# 3149002b |
| CD25 | Fluidigm | Cat# 3150002b |
| CD45 | Fluidigm | Cat# 3147003b |
| CD69 | Fluidigm | Cat# 3143004b |
| CD80 | Fluidigm | Cat# 3171008b |
| CD86 | Fluidigm | Cat# 3172016b |
| CD90 | Fluidigm | Cat# 3156006b |
| CD117 | Fluidigm | Cat# 3166004b |
| CD161 | Fluidigm | Cat# 3170002b |
| CD184 | Fluidigm | Cat# 3159030b |
| CD197 | Fluidigm | Cat# 3164013a |
| CD206 | Fluidigm | Cat# 3169021b |
| CD335 | Fluidigm | Cat# 3167008b |
| F4/80 | Fluidigm | Cat# 3146008b |
| FceR1a | Fluidigm | Cat# 3176006b |
| Ly6c | Fluidigm | Cat# 3162014b |
| Ly6g | Fluidigm | Cat# 3141008b |
|
Bacterial and virus strains | ||
| Pdhb (NM_024221) Mouse Tagged ORF Clone Lentiviral Particle | OriGene Technologies | Cat# MR205484L3V |
| Sod2 (BC066063) Mouse Tagged ORF Clone Lentiviral Particle | OriGene Technologies | Cat# MR201642L2V |
| Lenti ORF control particles of pLenti-C-mGFP-P2A-Puro | OriGene Technologies | Cat# PS100093V |
| PINK1 shRNA (m) Lentiviral particles | Santa Cruz Biotechnologies | Cat# sc-44599-V |
|
Chemicals, peptides, and recombinant proteins | ||
| Recombinant Mouse MCSF (Animal-Free) | Cell Guidance Systems | Cat# GMF8AF |
| Clophosome anionic liposomal clodronate for macrophage depletion | FormuMax Scientific | Cat# F70101C-A |
| Placebo control liposomes for Clophosome-A (anionic) | FormuMax Scientific | Cat# F70101-A |
| Puromycin | Invivogen | Cat# ant-pr-1 |
| Chloroquine diphosphate salt | Sigma Aldrich | # C6628 |
| DNase I | Sigma Aldrich | Cat# 10104159001 |
| 3,3′,5-Triiodo-L-thyronine sodium salt powder, BioReagent, suitable for cell culture 3,3′,5-Triiodo-L-thyronine sodium salt | Sigma Aldrich | Cat# T6397 |
| Rosiglitazone,≥98% (HPLC) | Sigma Aldrich | Cat# R2408 |
| Insulin solution human | Sigma Aldrich | Cat# I9278 |
| 3-Isobutyl-1-methylxanthine,BioUltra, ≥99% | Sigma Aldrich | Cat# I7018 |
| Indomethacin | Sigma Aldrich | Cat# I7378 |
| Cl 316,243 hydrate, ≥98% (HPLC), powder | Sigma Aldrich | Cat# C5976 |
| Carbonyl cyanide 4-(trifluoromethoxy)pheny lhydrazone ≥98% (TLC), powder Carbonyl cyanide 4-(trifluoromethoxy)pheny lhydrazone | Sigma Aldrich | Cat# C2920 |
| N-Acetyl-L-cysteine, cell culture tested, BioReagent, CAS 616–91-1, Sigma-Aldrich | Sigma Aldrich | Cat# A9165 |
| Doxycycline hyclate, ≥98% (HPLC) | Sigma Aldrich | Cat# D9891 |
| (R)-(—)-Isoproterenol crystalline (R)-(—)-Isoproterenol | Sigma Aldrich | Cat# I6379 |
| AMPK Inhibitor, Compound C – CAS 866405–64-3 – Calbiochem | Sigma Aldrich | Cat# 171260 |
| Antimycin a from Streptomyces sp. | Sigma Aldrich | Cat# A8674 |
| Lipopolysaccharides from Escherichia coli O111:B4,gamma-irradiated, BioXtra | Sigma Aldrich | Cat# L4391 |
| Sulfo-N-succinimidyl Oleate sodium | Sigma Aldrich | Cat# SML2148 |
| Rotenone | Sigma Aldrich | Cat# R8875 |
| Oligomycin A | Sigma Aldrich | Cat# 75351 |
| Collagenase, Type I, powder | Thermo Fisher Scientific | Cat# 17100017 |
| Collagenase, Type II, powder | Thermo Fisher Scientific | Cat# 17101015 |
| eBioscience™ 1X RBC Lysis Buffer | Thermo Fisher Scientific | Cat# 00–4333-57 |
|
Critical commercial assays | ||
| Seahorse XFe96 FluxPak | Agilent Technologies | Cat# 102416–100 |
| Seahorse XF DMEM assay medium pack, pH 7.4 | Agilent Technologies | Cat# 103680–100 |
| Zombie UV™ Fixable Viability Kit | Biolegend | Cat# 423107 |
| Zombie NIR™ Fixable Viability Kit | Biolegend | Cat# 423105 |
| Maxpar 10X Barcode Perm Buffer | Fluidigm | Cat# 201057 |
| Cell-ID™ 20-Plex Pd Barcoding Kit | Fluidigm | Cat# 201060 |
| Maxpar® Cell Staining Buffer | Fluidigm | Cat# 201068 |
| Cell-ID™ Intercalator-Ir | Fluidigm | Cat# 201192A |
| EQ Four Element Calibration Beads | Fluidigm | Cat# 201078 |
| OctoMACS™ Starting Kit | Miltenyi Biotec | Cat# 130–042-108 |
| QuadroMACS™ Starting Kit (LS) | Miltenyi Biotec | Cat# 130–091-051 |
| MS Columns | Miltenyi Biotec | Cat# 130–042-201 |
| LS Columns | Miltenyi Biotec | Cat# 130–042-401 |
| Pre-Separation Filters (30 μm) | Miltenyi Biotec | Cat# 130–041-407 |
| Mitochondria Isolation Kit, mouse tissue | Miltenyi Biotec | Cat# 130–096-946 |
| RNeasy Lipid Tissue Mini Kit | Qiagen | Cat# 74804 |
| Mito Tracker Green | Thermo Fisher Scientific | Cat# M7514 |
| Lipofectamine™ 2000 Transfection Reagent | Thermo Fisher Scientific | Cat# 11668019 |
| Annexin V Conjugates for Apoptosis Detection | Thermo Fisher Scientific | Cat# A35111 |
| Exosome Spin Columns (MW 3000) (DISCONTINUED PRODUCT) | Thermo Fisher Scientific | Cat# 4484449 |
| MitoTracker™ Red CMXRos | Thermo Fisher Scientific | Cat# M7512 |
| MitoSOX™ Red Mitochondrial Superoxide Indicator, for live-cell imaging | Thermo Fisher Scientific | Cat# M36008 |
| LysoTracker™ Deep Red | Thermo Fisher Scientific | Cat# L12492 |
| Alexa Fluor™ 488 Phalloidin | Thermo Fisher Scientific | Cat# A12379 |
|
Deposited data | ||
| RNA-seq data | This paper | ArrayExpress: E-MTAB-10655 |
| Proteomics data | This paper | ProteomeXchange: PXD026080 |
|
Experimental models: Cell lines | ||
| iBPA_m | (Liu et al., 2019) | N/A |
| iBPA_h | (Liu et al., 2019) | N/A |
| RAW 264.7 cell line, ATCC | ATCC | Cat# TIB-71, RRID:CVCL_0493 |
| T37i murine preadipocytes | Professor Marc Lombes (INSERM U1185, Paris, France) | |
|
Oligonucleotides | ||
| mtDendra2 | (Brestoff et al., 2021) | N/A |
| CD45.1 | (Brestoff et al., 2021) | N/A |
| Adipoq Cre +/− | (Brestoff et al., 2021) | N/A |
| MitoFat | (Brestoff et al., 2021) | N/A |
| CD45.2 WT | (Brestoff et al., 2021) | N/A |
| CD45.2 DENDRA2 | (Brestoff et al., 2021) | N/A |
| CD169 MICE | (Chow et al., 2011) | N/A |
| See Real-time qPCR section for qPCR primers | N/A | N/A |
| Smpd3 (Mouse) - 3 unique 27mer siRNA duplexes - 2 nmol each | OriGene Technologies | Cat# SR427338 |
| PINK1 siRNA (human) | OriGene Technologies | CAT#: SR324912 |
| C57BL/6J | The Jackson Laboratory | #000664; RRID: IMSR_JAX:000664 |
|
Software and algorithms | ||
| Trimmomatic version 0.36 | (Bolger et al., 2014) | N/A |
| JACoP plugin | (Bolte and Cordeliè res, 2006) | N/A |
| HISAT2 version 2.1.0 | (Kim et al., 2015) | N/A |
| Cytobank software platform | (Kotecha et al., 2010) | N/A |
| StringTie version 1.3.4d | (Pertea et al., 2015) | N/A |
| Desktop Wave Software | Agilent Technologies | N/A |
| Debarcoder software | Fluidigm | N/A |
| Babelomics 5 online software | http://babelomics.bioinfo.cipf.es | N/A |
| AnnotationDbi R library | http://bioconductor.org | N/A |
| FunRich version 3.1.3 | http://www.funrich.org | N/A |
| EnrichR | https://amp.pharm.mssm.edu/Enrichr | N/A |
| MultiQuant™ software version 3.0.2 | https://download.sciex.com/MultiQuant_302_Software_Release_Notes.pdf | N/A |
| Fiji Image J | https://imagej.net/software/fiji/downloads | N/A |
| FastQC version 0.11.5 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc | N/A |
| Gene Expression Omnibus database | https://www.ncbi.nlm.nih.gov | N/A |
| Olympus ITEM software | OLYMPUS | N/A |
|
Other | ||
| Mini Rectal Probe Type T, 0.1°C (RET-3) | ThermoWorks | Cat# RET-3 |
Highlights.
Brown adipocytes eliminate damaged mitochondrial parts through EVs
Thermogenic stimuli increase the release of mitochondrial EVs
EVs exert a negative autocrine action on brown adipocyte thermogenesis
bMACs actively take up mitochondrial EVs ensuring optimal BAT thermogenesis
ACKNOWLEDGMENTS
This work was partially supported by the European Foundation for the Study of Diabetes (EFSD/Lilly, 2017 and EFSD/Boehringer Ingelheim European Research Programme on ‘‘Multi-System Challenges in Diabetes’’) and the Italian Ministry of Health (GR-2018–12367588) to D.L.-B.; Associazione Italiana per la Ricerca sul Cancro (AIRC) under IG 2019 - ID. 23562 project to K.A.; MIUR ‘‘Progetto Eccellenza’’ to Dipartimento di Scienze Farmacologiche e Biomolecolari, Università degli Studi di Milano, and NUTRAGE (CNR FOE 2019, DSB.AD004.271) to A.S; Italian Foundation of Multiple Sclerosis (grant 2017/R/8), the Italian Ministry of Health (grant GR-2016–02362380) and the MAI Award grant to V. Chiurchiù; National Institutes of Health (NIH) common fund (DP5 OD028125) and the Burroughs Wellcome Fund (1019648) to J.R.B.; NIH K01DK125608 to F.S.; R01DK102898 and R01DK122808 to Y.-H.T.; and NIH RO1 DK121805 and AHA 19TPA34910079 to B.Z. A.H. was supported by RTI2018–095497-B-I00 from MICINN, HR17_00527 from La Caixa Foundation, and TNE-18CVD04 from the Leducq Foundation. M.S-D was supported by a fellowship PRE2019–08746 from the Ministerio de Ciencia e Innovación. M.R. was partially supported by a fellowship from AIRC (IG 2019 - ID. 23562) and by the Italian Ministry of Health (SG-2019–12368589).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.cmet.2022.02.016.
DECLARATION OF INTERESTS
J.R.B. has a pending patent application related to mitochondria transfer. The other authors declare no competing interests.
REFERENCES
- Abuaita BH, Schultz TL, and O’Riordan MX (2018). Mitochondria-derived vesicles deliver antimicrobial reactive oxygen species to control phagosome-localized Staphylococcus aureus. Cell Host Microbe 24, 625–636.e5. 10.1016/j.chom.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad B, Serpell CJ, Fong IL, and Wong EH (2020). Molecular mechanisms of adipogenesis: the anti-adipogenic role of AMP-activated protein kinase. Front. Mol. Biosci 7, 76. 10.3389/fmolb.2020.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amari L, and Germain M (2021). Mitochondrial extracellular vesicles—origins and roles. Front. Mol. Neurosci 14, 767219. 10.3389/fnmol.2021.767219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquilano K, Sciarretta F, Turchi R, Li BH, Rosina M, Ceci V, Guidobaldi G, Arena S, D’Ambrosio C, Audano M, et al. (2020). Low-protein/high-carbohydrate diet induces AMPK-dependent canonical and non-canonical thermogenesis in subcutaneous adipose tissue. Redox Biol 36, 101633. 10.1016/j.redox.2020.101633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher T, Palanisamy S, Kramer DJ, Eljalby M, Marx SJ, Wibmer AG, Butler SD, Jiang CS, Vaughan R, Schöder H, et al. (2021). Brown adipose tissue is associated with cardiometabolic health. Nat. Med 27, 58–65. 10.1038/s41591-020-1126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, and Cordelières FP (2006). A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc 224, 213–232. 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Boudreau LH, Duchez AC, Cloutier N, Soulet D, Martin N, Bollinger J, Paré A, Rousseau M, Naika GS, Lévesque T, et al. (2014). Platelets release mitochondria serving as substrate for bactericidal group IIA-secreted phospholipase A2 to promote inflammation. Blood 124, 2173–2183. 10.1182/blood-2014-05-573543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet N, Luijten IHN, Cannon B, and Nedergaard J (2021). Thermogenic recruitment of brown and Brite/beige adipose tissues is not obligatorily associated with macrophage accretion or attrition. Am. J. Physiol. Endocrinol. Metab 320, E359–E378. 10.1152/ajpendo.00352.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breining P, Jensen JB, Sundelin EI, Gormsen LC, Jakobsen S, Busk M, Rolighed L, Bross P, Fernandez-Guerra P, Markussen LK, et al. (2018). Metformin targets brown adipose tissue in vivo and reduces oxygen consumption in vitro. Diabetes Obes. Metab 20, 2264–2273. 10.1111/dom.13362. [DOI] [PubMed] [Google Scholar]
- Brestoff JR, Wilen CB, Moley JR, Li Y, Zou W, Malvin NP, Rowen MN, Saunders BT, Ma H, Mack MR, et al. (2021). Intercellular mitochondria transfer to macrophages regulates white adipose tissue homeostasis and is impaired in obesity. Cell Metab 33, 270–282.e8. 10.1016/j.cmet.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne JR, Oviosu O, and Eaton P (2013). The PEG-switch assay: a fast semi-quantitative method to determine protein reversible cysteine oxidation. J. Pharmacol. Toxicol. Methods 68, 297–301. 10.1016/j.vascn.2013.07.001. [DOI] [PubMed] [Google Scholar]
- Cadete VJ, Deschênes S, Cuillerier A, Brisebois F, Sugiura A, Vincent A, Turnbull D, Picard M, McBride HM, and Burelle Y (2016). Formation of mitochondrial-derived vesicles is an active and physiologically relevant mitochondrial quality control process in the cardiac system. J. Physiol 594, 5343–5362. 10.1113/JP272703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairó M, Campderrós L, Gavaldà-Navarro A, Cereijo R, Delgado-Anglé s A, Quesada-López T, Giralt M, Villarroya J, and Villarroya F (2019). Parkin controls brown adipose tissue plasticity in response to adaptive thermogenesis. EMBO Rep 20, e46832. 10.15252/embr.201846832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereijo R, Gavaldà-Navarro A, Cairó M, Quesada-López T, Villarroya J, Morón-Ros S, Sánchez-Infantes D, Peyrou M, Iglesias R, Mampel T, et al. (2018). CXCL14, a brown adipokine that mediates brown-fat-to-macrophage communication in thermogenic adaptation. Cell Metab 28, 750–763.e6. 10.1016/j.cmet.2018.07.015. [DOI] [PubMed] [Google Scholar]
- Chen H, Lau MC, Wong MT, Newell EW, Poidinger M, and Chen J (2016). Cytofkit: a bioconductor package for an integrated mass cytometry data analysis pipeline. PLoS Comput. Biol 12, e1005112. 10.1371/journal.pcbi.1005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchani ET, Kazak L, Jedrychowski MP, Lu GZ, Erickson BK, Szpyt J, Pierce KA, Laznik-Bogoslavski D, Vetrivelan R, Clish CB, et al. (2016). Mitochondrial ROS regulate thermogenic energy expenditure and sulfenylation of UCP1. Nature 532, 112–116. 10.1038/nature17399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow A, Lucas D, Hidalgo A, Méndez-Ferrer S, Hashimoto D, Scheiermann C, Battista M, Leboeuf M, Prophete C, van Rooijen N, et al. (2011). Bone marrow CD169+ macrophages promote the retention of hematopoietic stem and progenitor cells in the mesenchymal stem cell niche. J. Exp. Med 208, 261–271. 10.1084/jem.20101688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement E, Lazar I, Attané C, Carrié L, Dauvillier S, Ducoux-Petit M, Esteve D, Menneteau T, Moutahir M, Le Gonidec S, et al. (2020). Adipocyte extracellular vesicles carry enzymes and fatty acids that stimulate mitochondrial metabolism and remodeling in tumor cells. EMBO J 39, e102525. 10.15252/embj.2019102525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crewe C, Funcke JB, Li S, Joffin N, Gliniak CM, Ghaben AL, An YA, Sadek HA, Gordillo R, Akgul Y, et al. (2021). Extracellular vesicle-based interorgan transport of mitochondria from energetically stressed adipocytes. Cell Metab 33, 1853–1868.e11. 10.1016/j.cmet.2021.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess AM, White AP, Vernochet C, Schulz TJ, Xue R, Sass CA, Huang TL, Roberts-Toler C, Weiner LS, Sze C, et al. (2013). Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat. Med 19, 635–639. 10.1038/nm.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Acunzo P, Pérez-González R, Kim Y, Hargash T, Miller C, Alldred MJ, Erdjument-Bromage H, Penikalapati SC, Pawlik M, Saito M, et al. (2021). Mitovesicles are a novel population of extracellular vesicles of mitochondrial origin altered in Down syndrome. Sci. Adv 7, eabe5085. 10.1126/sciadv.abe5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok VA, Warner ML, Bratton DL, and Henson PM (1998). CD36 is required for phagocytosis of apoptotic cells by human macrophages that use either a phosphatidylserine receptor or the vitronectin receptor (alpha v beta 3). J. Immunol 161, 6250–6257. [PubMed] [Google Scholar]
- Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, Pe’er D, Nolan GP, and Bendall SC (2013). Normalization of mass cytometry data with bead standards. Cytometry A 83, 483–494. 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K, Ruiz HH, Jhun K, Finan B, Oberlin DJ, van der Heide V, Kalinovich AV, Petrovic N, Wolf Y, Clemmensen C, et al. (2017). Alternatively activated macrophages do not synthesize catecholamines or contribute to adipose tissue adaptive thermogenesis. Nat. Med 23, 623–630. 10.1038/nm.4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallerand A, Stunault MI, Merlin J, Luehmann HP, Sultan DH, Firulyova MM, Magnone V, Khedher N, Jalil A, Dolfi B, et al. (2021). Brown adipose tissue monocytes support tissue expansion. Nat. Commun 12, 5255. 10.1038/s41467-021-25616-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri S, Rattan R, Haq E, Khan M, Yasmin R, Won JS, Key L, Singh AK, and Singh I (2006). AICAR inhibits adipocyte differentiation in 3T3L1 and restores metabolic alterations in diet-induced obesity mice model. Nutr. Metab. (Lond.) 3, 31. 10.1186/1743-7075-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajchen E, Wouters E, van de Haterd B, Haidar M, Hardonnière K, Dierckx T, Van Broeckhoven J, Erens C, Hendrix S, Kerdine-Römer S, et al. (2020). CD36-mediated uptake of myelin debris by macrophages and microglia reduces neuroinflammation. J. Neuroinflammation 17, 224. 10.1186/s12974-020-01899-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habinowski SA, and Witters LA (2001). The effects of AICAR on adipocyte differentiation of 3T3-L1 cells. Biochem. Biophys. Res. Commun 286, 852–856. 10.1006/bbrc.2001.5484. [DOI] [PubMed] [Google Scholar]
- Henriques F, Bedard AH, Guilherme A, Kelly M, Chi J, Zhang P, Lifshitz LM, Bellvé K, Rowland LA, Yenilmez B, et al. (2020). Single-cell RNA profiling reveals adipocyte to macrophage signaling sufficient to enhance thermogenesis. Cell Rep 32, 107998. 10.1016/j.cel-rep.2020.107998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MV, Morrison TJ, Doherty DF, McAuley DF, Matthay MA, Kissenpfennig A, O’Kane CM, and Krasnodembskaya AD (2016). Mitochondrial transfer via tunneling nanotubes is an important mechanism by which mesenchymal stem cells enhance macrophage phagocytosis in the in vitro and in vivo models of ARDS. Stem Cells 34, 2210–2223. 10.1002/stem.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball A, Schaller M, Joshi A, Davis FM, denDekker A, Boniakowski A, Bermick J, Obi A, Moore B, Henke PK, et al. (2018). Ly6C(Hi) blood monocyte/macrophage drive chronic inflammation and impair wound healing in diabetes mellitus. Arterioscler. Thromb. Vasc. Biol 38, 1102–1114. 10.1161/ATVBAHA.118.310703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotecha N, Krutzik PO, and Irish JM (2010). Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom Chapter 10, Unit 10. 17. 10.1002/0471142956.cy1017s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P, Swarbrick MM, and Ho KK (2013). Brown adipose tissue in adult humans: a metabolic renaissance. Endocr. Rev 34, 413–438. 10.1210/er.2012-1081. [DOI] [PubMed] [Google Scholar]
- Lettieri Barbato D, Tatulli G, Aquilano K, and Ciriolo MR (2013). FoxO1 controls lysosomal acid lipase in adipocytes: implication of lipophagy during nutrient restriction and metformin treatment. Cell Death Dis 4, e861. 10.1038/cddis.2013.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettieri-Barbato D (2019). Redox control of non-shivering thermogenesis. Mol. Metab 25, 11–19. 10.1016/j.molmet.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuti A, Talamonti E, Gentile A, Tiberi M, Matteocci A, Fresegna D, Centonze D, and Chiurchiù V (2021). Macrophage plasticity and polarization are altered in the experimental model of multiple sclerosis. Biomolecules 11, 837. 10.3390/biom11060837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Menoret A, Farragher C, Ouyang Z, Bonin C, Holvoet P, Vella AT, and Zhou B (2019). Single cell transcriptomics based-MacSpectrum reveals novel macrophage activation signatures in diseases. JCI Insight 5, e126453. 10.1172/jci.insight.126453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Kuipers EN, Sips HCM, Dorleijn JC, van Dam AD, Christodoulides C, Karpe F, Zhou G, Boon MR, Rensen PCN, et al. (2019). Conditionally immortalized brown preadipocytes can switch between proliferative and differentiated states. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1864, 158511. 10.1016/j.bbalip.2019.08.007. [DOI] [PubMed] [Google Scholar]
- MacKenzie A, Wilson HL, Kiss-Toth E, Dower SK, North RA, and Surprenant A (2001). Rapid secretion of interleukin-1beta by microvesicle shedding. Immunity 15, 825–835. 10.1016/s1074-7613(01)00229-1. [DOI] [PubMed] [Google Scholar]
- Manders EM, Stap J, Brakenhoff GJ, van Driel R, and Aten JA (1992). Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J. Cell Sci 103, 857–862. [DOI] [PubMed] [Google Scholar]
- Matheoud D, Sugiura A, Bellemare-Pelletier A, Laplante A, Rondeau C, Chemali M, Fazel A, Bergeron JJ, Trudeau LE, Burelle Y, et al. (2016). Parkinson’s disease-related proteins PINK1 and Parkin repress mitochondrial antigen presentation. Cell 166, 314–327. 10.1016/j.cell.2016.05.039. [DOI] [PubMed] [Google Scholar]
- McLelland GL, Lee SA, McBride HM, and Fon EA (2016). Syntaxin-17 delivers PINK1/parkin-dependent mitochondrial vesicles to the endolysosomal system. J. Cell Biol 214, 275–291. 10.1083/jcb.201603105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLelland GL, Soubannier V, Chen CX, McBride HM, and Fon EA (2014). Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J 33, 282–295. 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldolesi J (2018). Exosomes and Ectosomes in intercellular communication. Curr. Biol 28, R435–R444. 10.1016/j.cub.2018.01.059. [DOI] [PubMed] [Google Scholar]
- Mills EL, Harmon C, Jedrychowski MP, Xiao H, Gruszczyk AV, Bradshaw GA, Tran N, Garrity R, Laznik-Bogoslavski D, Szpyt J, et al. (2022). Cysteine 253 of UCP1 regulates energy expenditure and sex-dependent adipose tissue inflammation. Cell Metab 34, 140–157.e8. 10.1016/j.cmet.2021.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EL, Pierce KA, Jedrychowski MP, Garrity R, Winther S, Vidoni S, Yoneshiro T, Spinelli JB, Lu GZ, Kazak L, et al. (2018). Accumulation of succinate controls activation of adipose tissue thermogenesis. Nature 560, 102–106. 10.1038/s41586-018-0353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolás-Ávila JA, Lechuga-Vieco AV, Esteban-Martínez L, Sánchez-Díaz M, Díaz-García E, Santiago DJ, Rubio-Ponce A, Li JL, Balachander A, Quintana JA, et al. (2020). A network of macrophages supports mitochondrial homeostasis in the heart. Cell 183, 94–109.e23. 10.1016/j.cell.2020.08.031. [DOI] [PubMed] [Google Scholar]
- Oeckl J, Bast-Habersbrunner A, Fromme T, Klingenspor M, and Li Y (2020). Isolation, Culture, and Functional Analysis of Murine Thermogenic Adipocytes. STAR Protoc 1, 100118. 10.1016/j.xpro.2020.100118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, and Salzberg SL (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol 33, 290–295. 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peruzzotti-Jametti L, Bernstock JD, Willis CM, Manferrari G, Rogall R, Fernandez-Vizarra E, Williamson JC, Braga A, van den Bosch A, Leonardi T, et al. (2021). Neural stem cells traffic functional mitochondria via extracellular vesicles. PLoS Biol 19, e3001166. 10.1371/journal.pbio.3001166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phinney DG, Di Giuseppe M, Njah J, Sala E, Shiva S, St Croix CM, Stolz DB, Watkins SC, Di YP, Leikauf GD, et al. (2015). Mesenchymal stem cells use extracellular vesicles to outsource mitophagy and shuttle microRNAs. Nat. Commun 6, 8472. 10.1038/ncomms9472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyák Á, Winkler Z, Kuti D, Ferenczi S, and Kovács KJ (2016). Brown adipose tissue in obesity: fractalkine-receptor dependent immune cell recruitment affects metabolic-related gene expression. Biochim. Biophys. Acta 1861, 1614–1622. 10.1016/j.bbalip.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Nguyen KD, Odegaard JI, Cui X, Tian X, Locksley RM, Palmiter RD, and Chawla A (2014). Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell 157, 1292–1308. 10.1016/j.cell.2014.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabas N, Palmer S, Mitchell L, Ismail S, Gohlke A, Riley JS, Tait SWG, Gammage P, Soares LL, Macpherson IR, et al. (2021). PINK1 drives production of mtDNA-containing extracellular vesicles to promote invasiveness. J. Cell Biol 220, e202006049. 10.1083/jcb.202006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggio A, Rosina M, Krahmer N, Palma A, Petrilli LL, Maiolatesi G, Massacci G, Salvatori I, Valle C, Testa S, et al. (2020a). Metabolic reprogramming of fibro/adipogenic progenitors facilitates muscle regeneration. Life Sci. Alliance 3, e202000646. 10.26508/lsa.202000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggio A, Rosina M, Palma A, Cerquone Perpetuini A, Petrilli LL, Gargioli C, Fuoco C, Micarelli E, Giuliani G, Cerretani M, et al. (2020b). Adipogenesis of skeletal muscle fibro/adipogenic progenitors is affected by the WNT5a/GSK3/beta-catenin axis. Cell Death Differ 27, 2921–2941. 10.1038/s41418-020-0551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Okamatsu-Ogura Y, Matsushita M, Watanabe K, Yoneshiro T, Nio-Kobayashi J, Iwanaga T, Miyagawa M, Kameya T, Nakada K, et al. (2009). High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes 58, 1526–1531. 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Alavez M, Bortell N, Basova L, Samad F, and Marcondes MCG (2020). Macrophages and brown adipocytes cross-communicate to modulate a thermogenic program following methamphetamine exposure. Int. J. Hyperthermia 37, 1368–1382. 10.1080/02656736.2020.1849822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Su Y, Silva FJ, Weller AH, Sostre-Colón J, Titchenell PM, Steger DJ, Seale P, and Soccio RE (2020). Shared PPARalpha/gamma target genes regulate brown adipocyte thermogenic function. Cell Rep 30, 3079–3091.e5. 10.1016/j.celrep.2020.02.032. [DOI] [PubMed] [Google Scholar]
- Stier A, Bize P, Habold C, Bouillaud F, Massemin S, and Criscuolo F (2014). Mitochondrial uncoupling prevents cold-induced oxidative stress: a case study using UCP1 knockout mice. J. Exp. Biol 217, 624–630. 10.1242/jeb.092700. [DOI] [PubMed] [Google Scholar]
- Sugiura A, McLelland GL, Fon EA, and McBride HM (2014). A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J 33, 2142–2156. 10.15252/embj.201488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talamonti E, Pauter AM, Asadi A, Fischer AW, Chiurchiù V, and Jacobsson A (2017). Impairment of systemic DHA synthesis affects macrophage plasticity and polarization: implications for DHA supplementation during inflammation. Cell. Mol. Life Sci 74, 2815–2826. 10.1007/s00018-017-2498-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatulli G, Mitro N, Cannata SM, Audano M, Caruso D, D’Arcangelo G, Lettieri-Barbato D, and Aquilano K (2018). Intermittent fasting applied in combination with rotenone treatment exacerbates dopamine neurons degeneration in mice. Front. Cell. Neurosci 12, 4. 10.3389/fncel.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, and Fussenegger M (2020). Shedding light on extracellular vesicle biogenesis and bioengineering. Adv. Sci. (Weinh) 8, 2003505. 10.1002/advs.202003505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todkar K, Chikhi L, Desjardins V, El-Mortada F, Pépin G, and Germain M (2021). Selective packaging of mitochondrial proteins into extracellular vesicles prevents the release of mitochondrial DAMPs. Nat. Commun 12, 1971. 10.1038/s41467-021-21984-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brügger B, and Simons M (2008). Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247. 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- Turchi R, Tortolici F, Guidobaldi G, Iacovelli F, Falconi M, Rufini S, Faraonio R, Casagrande V, Federici M, De Angelis L, et al. (2020). Frataxin deficiency induces lipid accumulation and affects thermogenesis in brown adipose tissue. Cell Death Dis 11, 51. 10.1038/s41419-020-2253-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart JI, Boon MR, and Houtkooper RH (2021). The role of AMPK signaling in brown adipose tissue activation. Cells 10, 1122. 10.3390/cells10051122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasam G, Nadeau R, Cadete VJJ, Lavallée-Adam M, Menzies KJ, and Burelle Y (2021). Proteomics characterization of mitochondrial-derived vesicles under oxidative stress. FASEB J 35, e21278. 10.1096/fj.202002151R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Malcor JM, and Harper MT (2018). Lipid rafts are essential for release of phosphatidylserine-exposing extracellular vesicles from platelets. Sci. Rep 8, 9987. 10.1038/s41598-018-28363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JW, Elvington A, Ivanov S, Kessler S, Luehmann H, Baba O, Saunders BT, Kim KW, Johnson MW, Craft CS, et al. (2017). Thermoneutrality but not UCP1 deficiency suppresses monocyte mobilization Into blood. Circ. Res 121, 662–676. 10.1161/CIRCRESAHA.117.311519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf Y, Boura-Halfon S, Cortese N, Haimon Z, Sar Shalom H, Kuperman Y, Kalchenko V, Brandis A, David E, Segal-Hayoun Y, et al. (2017). Brown-adipose-tissue macrophages control tissue innervation and homeostatic energy expenditure. Nat. Immunol 18, 665–674. 10.1038/ni.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo MS, Yang J, Beltran C, and Cho S (2016). Cell surface CD36 protein in monocyte/macrophage contributes to phagocytosis during the resolution phase of ischemic stroke in mice. J. Biol. Chem 291, 23654–23661. 10.1074/jbc.M116.750018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhang L, Li B, Jiang H, Duan Y, Xie Z, Shuai L, Li J, and Li J (2018). AMP-activated protein kinase (AMPK) regulates energy metabolism through modulating thermogenesis in adipose tissue. Front. Physiol 9, 122. 10.3389/fphys.2018.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang L, Yu C, Yang XF, and Wang H (2014). Monocyte and macrophage differentiation: circulation inflammatory monocyte as biomarker for inflammatory diseases. Biomark. Res 2, 1. 10.1186/2050-7771-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data that support the findings of this study are available from the lead contact upon reasonable request. RNA-sequence datasets produced in this study are available from ArrayExpress: -E-MTAB-10655. The newly generated proteomic data have been deposited into the ProteomeXchange database with the following accession number ProteomeXchange: PXD026080.


