Abstract
Recently, it has been shown that a new mutational pattern (the E44D/A and/or V118I mutation) confers moderate phenotypic lamivudine resistance in the absence of the M184V mutation. The E44D/A and/or the V118I mutation does not exist in drug-naive patients, and the prevalence increases with the number of treatment regimens and lamivudine experience. The mutations can preexist in nucleoside-experienced but lamivudine-naive patients. They are always associated with zidovudine resistance-associated mutations, even in the absence of M184V. These mutations are more stable than the M184V substitution during antiretroviral treatment interruptions.
The emergence of drug-resistant human immunodeficiency virus type 1 (HIV-1) is frequently observed during the course of treatment of patients with the use of antiretroviral drugs (7). Mutation M184V in the HIV-1 reverse trancriptase (RT) has been shown to be specifically associated with high-level (>50-fold) phenotypic resistance to lamivudine (3TC) in vitro and in vivo (1, 9, 12). Previously, some mutation patterns have been described to be associated with moderate levels of phenotypic resistance (4- to <50-fold) to 3TC. This observation has been associated with the selection of mutations implicated in cross-resistance to other nucleoside analogs, such as in the case of the nucleoside multidrug resistance complex of mutations (Q151M, F77L, F116Y, A62V, and V75I) (6, 8, 10, 11). A moderate level of 3TC resistance has also been reported in strains harboring either the insertion in position 69 of RT (2, 13) or the K65R mutation (3, 4).
Recently, a novel mutational pattern in HIV-1 RT, associated with a moderate level of phenotypic resistance to 3TC in the absence of the characteristic replacement of methionine by valine at position 184, was described. Thus, genotypic and phenotypic analyses of clinical isolates revealed the presence of moderate levels of phenotypic resistance to 3TC in a subset of isolates that did not harbor the M184V substitution. Mutational cluster analysis and comparison with the phenotypic data revealed a significant correlation between a moderate level of phenotypic 3TC resistance and an increased incidence of amino acid changes at RT codons 44 (glutamic acid to aspartic acid or alanine) and 118 (valine to isoleucine). This occurred predominantly in isolates harboring zidovudine (ZDV) resistance-associated mutations (M41L, T215Y). Moreover, the results from the site-directed mutagenesis experiment confirmed that mutations at codons 44 and 118 are indeed associated with moderate levels of phenotypic resistance to 3TC when they are present with ZDV resistance-associated mutations (5).
In this study, we evaluated the prevalence and the conditions of selection of the E44D or E44A (E44D/A) and/or V118I RT mutation in clinical practice. Plasma samples obtained from 344 HIV-1-infected individuals from a routine clinical practice were analyzed by genotypic resistance testing. Genotyping analysis were performed by automated population-based full sequence analysis (ABI 377), and the results were reported as the amino acid sequence compared to that for the RT gene sequence of the HXB2 wild-type reference strain. Patients analyzed were naive for treatment with antiretroviral drugs (n = 83) or had been previously treated with antiretroviral drugs (n = 261). Among the previously treated patients, we defined as moderately experienced patients who had received one to five antiretroviral regimens (n = 151) and as heavily experienced patients who had received more than five antiretroviral regimens (n = 110). An antiretroviral regimen was defined in this study as a combination of antiretroviral drugs received during a minimum of 1 month and should be different from the last antiretroviral combination used.
(This work was presented in part at the 7th Conference on Retroviruses and Opportunistic Infections, San Francisco, Calif., 2000 and at the 4th International Workshop on Drug Resistance and Treatment Strategies, Sitgès 2000.)
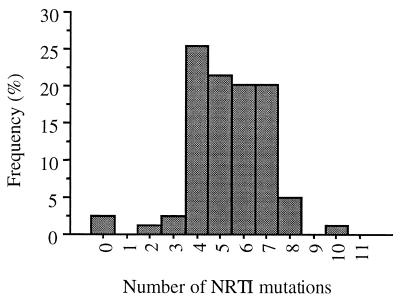
Prevalences of E44D/A and V118I in the population studied.
The E44A, E44D, and V118I mutations occurred in the clinical samples studied at overall frequencies of 1.5, 10, and 21%, respectively. Forty-three percent of the clinical isolates with the V118I mutation simultaneously harbored the E44D or E44A mutation. In contrast, 82% of the clinical isolates carrying E44D/A mutation simultaneously harbored the V118I mutation. Statistical analysis showed a significant association between the frequencies of the E44D/A and/or the V118I mutation and the number of nucleoside analog resistance mutations, especially ZDV resistance mutations (P < 0.0001 by the Fisher exact test) (Fig. 1). The E44D/A and V118I mutations were also be observed in association with the Q151M multidrug resistance pattern (n = 3) and with the insertion at RT codon 69 (n = 2).
FIG. 1.
Frequency of the E44D/A and/or V118I mutation according to the number of nucleoside analog resistance-associated mutations.
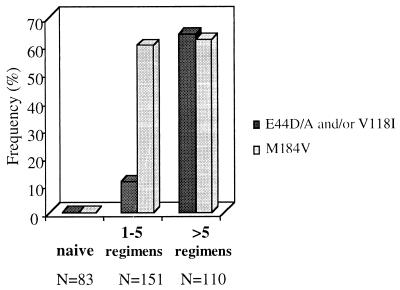
Among the subgroups studied, the E44D/A and/or V118I mutation was not observed in antiretroviral-naive patients and was found in 78 of 261 (30%) previously treated patients. This frequency was statistically higher when the patients were heavily previously treated (more than five regimens) than when the patients were moderately experienced (one to five regimens) (P < 0.001 by the Fisher exact test). This difference was observed whether the M184V mutation was present or not (Fig. 2).
FIG. 2.
Frequencies of E44D/A and/or V118I mutation pattern and the M184V mutation among drug-naive patients and previously treated patients who were moderately (one to five regimens) and heavily (more than five regimens) experienced.
Details of association between E44D/A and/or V118I pattern and M184V mutation among previously treated patients.
As can be deduced from the data in Table 1, changes at RT positions 44 and 118 can exist in clinical samples with or without the presence of the M184V substitution, but their prevalence was higher when the M184V mutation was present (P = 0.03 by the Fisher exact test). In the absence of the M184V mutation, the E44D/A and/or V118I pattern was observed among 23 previously treated patients (9%), including 17 heavily experienced patients (more than five regimens) and 6 moderately experienced patients (one to five regimens). In these patients, the E44D/A and/or V118I mutation was always associated with a median of four ZDV resistance-associated mutations (range, two to six mutations), including in all patients the ZDV major resistance-associated mutation T215Y.
TABLE 1.
Prevalence of E44D/A and/or V118I pattern in association with 3TC resistance-associated mutation M184V in RT in clinical samples of previously treated patients
| E44D/A and/or V118I pattern | M184V | No. of samples with the pattern/total no. of samples tested | % |
|---|---|---|---|
| Present | Present | 55/261 | 21 |
| Present | Absent | 23/261 | 9 |
| Absent | Present | 102/261 | 39 |
| Absent | Absent | 81/261 | 31 |
Conditions of selection and reversion of E44D/A and V118I mutations.
We carefully investigated a subset of 3TC-naive patients who had received ZDV, didanosine, and zalcitabine (n = 22). The E44D/A and/or V118I mutation existed prior to 3TC therapy in 14% of these patients, but always in association with ZDV resistance-associated mutations. It is interesting that in this group of patients, the selection of the M184V substitution was observed in all patients when 3TC was added to the regimen.
Among 19 previously treated patients who had discontinued an antiretroviral regimen for 2 to 6 months, the reversion of E44D/A to wild-type codon E (acid glutamic) and V118I to wild-type codon V (valine) was observed for only two patients. The reversion of these mutations was seen exclusively in a context of a complete shift of the mutations to genotypic wild-type virus. Thus, this reversion of the E44D/A and V118I mutations seems to happen only when codons with nucleoside analog RT inhibitor (NRTI)-associated resistance mutations shift to wild-type codons. The E44D/A and V118I mutations seem to be more stable than the 3TC-associated M184V substitution that reverts to the wild-type codon rapidly and individually from all RT gene mutations.
In conclusion, our results confirm that the E44D/A and V118I substitutions in HIV-1 RT can occur with or without the M184V mutation but always occur in a ZDV resistance-associated mutation background. The E44D/A and/or V118I mutation rarely if ever exists in drug-naive patients, and the prevalence increases with the number of treatment regimens and 3TC experience. Their prevalence also increases with the number of mutations in the RT gene, like nucleoside analog associated resistance mutations and especially the ZDV-resistance-associated mutations, but their selection seems to be independent of the presence or the absence of the M184V mutation. Thus, when the antiretroviral treatment is totally interrupted, the codons with the E44D/A and/or V118I mutations shift to wild-type codons only in the context of a complete shift to wild-type virus codons and particularly the shift of nucleoside analog resistance-associated mutations. These mutations seem to be more stable in the viral genome than the classic M184V substitution. This suggests that the fitness impairment induced by the M184V mutation is higher than the one induced by E44D/A and/or V118I. Finally, whereas the 3TC resistance-associated mutation M184V appears independently of a ZDV treatment background and clearly appears to be due to 3TC selection pressure, the accumulation of E44D/A and/or V118I appears to be selected by ZDV or other nucleoside analogs and, fortuitously, confers only a moderate level of resistance to 3TC. The fact that the prevalence of the E44D/A and/or V118I mutation increases with the number of antiretroviral treatments suggests that the mutation might be involved in a more broad-spectrum nucleoside resistance.
REFERENCES
- 1.Boucher C A, Cammack N, Schipper P, Schuurman R, Rouse P, Wainberg M A, Cameron J M. High-level resistance to (−) enantiomeric 2′-deoxy-3′-thiacytidine in vitro is due to one amino acid substitution in the catalytic site of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1993;37:2231–2234. doi: 10.1128/aac.37.10.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Jong J J, Goudsmit J, Lukashov V V, Hillebrand M E, Baan E, Huismans R, Danner S A, ten Veen J H, de Wolf F, Jurriaans S. Insertion of two amino acids combined with changes in reverse transcriptase containing tyrosine-215 of HIV-1 resistant to multiple nucleoside analogs. AIDS. 1999;13:75–80. doi: 10.1097/00002030-199901140-00010. [DOI] [PubMed] [Google Scholar]
- 3.Gu Z, Fletcher R S, Arts E J, Wainberg M A, Parniak M A. The K65R mutant reverse transcriptase of HIV-1 cross-resistant to 2′,3′-dideoxycytidine, 2′,3′-dideoxy-3′-thiacytidine, and 2′,3′-dideoxyinosine shows reduced sensitivity to specific dideoxynucleoside triphosphate inhibitors in vitro. J Biol Chem. 1994;269:28118–28122. [PubMed] [Google Scholar]
- 4.Gu Z, Gao Q, Fang H, Salomon H, Parniak M A, Goldberg E, Cameron J, Wainberg M A. Identification of a mutation at codon 65 in the IKKK motif of reverse transcriptase that encodes human immunodeficiency virus resistance to 2′,3′-dideoxycytidine and 2′,3′-dideoxy-3′-thiacytidine. Antimicrob Agents Chemother. 1994;38:275–281. doi: 10.1128/aac.38.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hertogs K, Bloor S, De Vroey V, van Den Eynde C, Dehertogh P, van Cauwenberge A, Sturmer M, Alcorn T, Wegner S, van Houtte M, Miller V, Larder B A. A novel human immunodeficiency virus type 1 reverse transcriptase mutational pattern confers phenotypic lamivudine resistance in the absence of mutation 184V. Antimicrob Agents Chemother. 2000;44:568–573. doi: 10.1128/aac.44.3.568-573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iversen A K, Shafer R W, Wehrly K, Winters M A, Mullins J I, Chesebro B, Merigan T C. Multidrug-resistant human immunodeficiency virus type 1 strains resulting from combination antiretroviral therapy. J Virol. 1996;70:1086–1090. doi: 10.1128/jvi.70.2.1086-1090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schinazi R, Larder B, Mellors J. Mutations in retroviral genes associated with drug resistance. Int Antivir News. 1997;5:1–14. [Google Scholar]
- 8.Schmit J C, Van Laethem K, Ruiz L, Hermans P, Sprecher S, Sonnerborg A, Leal M, Harrer T, Clotet B, Arendt V, Lissen E, Witvrouw M, Desmyter J, De Clercq E, Vandamme A M. Multiple dideoxynucleoside analogue-resistant (MddNR) HIV-1 strains isolated from patients from different European countries. AIDS. 1998;12:2007–2015. doi: 10.1097/00002030-199815000-00012. [DOI] [PubMed] [Google Scholar]
- 9.Schuurman R, Nijhuis M, van Leeuwen R, Schipper P, de Jong D, Collis P, Danner S A, Mulder J, Loveday C, Christopherson C, et al. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC) J Infect Dis. 1995;171:1411–1419. doi: 10.1093/infdis/171.6.1411. [DOI] [PubMed] [Google Scholar]
- 10.Shirasaka T, Kavlick M F, Ueno T, Gao W Y, Kojima E, Alcaide M L, Chokekijchai S, Roy B M, Arnold E, Yarchoan R, et al. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc Natl Acad Sci USA. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shirasaka T, Yarchoan R, O'Brien M C, Husson R N, Anderson B D, Kojima E, Shimada T, Broder S, Mitsuya H. Changes in drug sensitivity of human immunodeficiency virus type 1 during therapy with azidothymidine, dideoxycytidine, and dideoxyinosine: an in vitro comparative study. Proc Natl Acad Sci USA. 1993;90:562–566. doi: 10.1073/pnas.90.2.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tisdale M, Kemp S D, Parry N R, Larder B A. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci USA. 1993;90:5653–5656. doi: 10.1073/pnas.90.12.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winters M A, Coolley K L, Girard Y A, Levee D J, Hamdan H, Shafer R W, Katzenstein D A, Merigan T C. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J Clin Investig. 1998;102:1769–1775. doi: 10.1172/JCI4948. [DOI] [PMC free article] [PubMed] [Google Scholar]