Highlights
-
•
Bovine insulin growth factor 1 (IGF-1) was estimated in different cow milk samples.
-
•
In house validation of a LC-MS/MS IGF-1 investigation method in milks obtained by different technological treatments.
-
•
Development of a sample treatment for the extraction of IGF-1 from different types of cow milk.
-
•
IGF-1 level in cow’s milk was not dependent form milk technological processing.
Keywords: Insulin like growth factor-1 distribution, Cow milk samples, Method validation, Urea-based extraction
Abstract
A simple and reliable targeted liquid chromatography-electrospray-tandem mass spectrometry (LC-MS/MS) method was developed and validated through the selection of two biomarker peptides for the identification and determination of bovine insulin like growth factor-1 (IGF-1) in milk samples. Two urea-based sample extraction procedures were tested. The validation results provided detection limits at the 1–5 ng IGF-1/mL level as a function of the milk matrix, precision ranged from 3 to 8% and the method accuracy in the different milk matrices was assured. Finally, IGF-1 was measured in milk samples obtained by treatment with eleven different technological processes: IGF-1 concentrations were spread over a wide range from 11.2 ± 0.3 ng/mL to 346 ± 8 ng/mL with a median of 57.0 ± 0.2 ng/mL. The highest amount of IGF-1 was found in fresh whole milk samples and no significant correlation was found between the total milk protein content and the IGF-1 concentration level.
1. Introduction
Insulin like growth factor-1 (IGF-1) is the growth hormone mediator, a strong mitogenic factor capable of stimulating various cellular responses, including cell growth, proliferation and differentiation by activation of IGF-1 receptor (IGF1R) and the subsequent upregulation of phosphoinositol-3-kinase-protein kinase B signaling cascade (Fig. 1), (Guidi et al., 2001, Melnik, 2009). IGF-1 receptors are present in almost every cell in the human body (Melnik, 2009) and IGF-1 acting as hormone, circulates in serum with more than 90% of molecules that forms a ternary complex bound to its principal carrier/binding proteins, IGF-binding protein-3 and acid labile subunit (Ketha & Singh, 2015). IGF-1 synthesis is subject to other hormones, nutrition, age, sex and genetic variability (Ketha et al., 2015). Researches have shown that a diet rich in milk and dairy products increases serum IGF-1 levels; milk exerts its signaling mechanisms by inducing long-lasting increase in serum IGF-1 levels and postprandial fast upregulation of insulin secretion (Clatici, Voicu, Voaides, Roseanu, Icriverzi, & Jurcoane, 2018; Melnik, John, & Schmitz, 2011). It is also interesting to note that in milk, regardless the processed treatment (pasteurization, homogenization, and digestion), there are high concentrations of growth-stimulating hormones such as IGF-1 (Clatici et al., 2018). Mammals produce structurally highly similar IGF-1 and the bovine and human molecules have the same amino acid sequence (Juskevich & Guyer, 1990), therefore bovine IGF-1 can bind to the human IGF receptor (Francis, Upton, Ballard, McNeil, & Wallace, 1988). In addition, IGF-1 remains active in serum after milk consumption because IGF-1 digestion in the gut is being protected by milk’s proteins (Melnik, 2009). Interestingly, it’s the hepatic IGF-1 production stimulation via amino acid transfer induced by milk that increase the consumer’s serum IGF-1 level (Melnik, John, & Schmitz, 2013). Although benefits related to milk consumption are known such as increased bone mineral content, reduced risk of protein-deficiency malnutrition and rickets and protects against dental caries and fractures, milk consumption has also been associated with numerous diseases due to the presence of growth factors such as IGF-1 (Clatici et al., 2018, Furstenberger and Senn, 2002). Circulating IGF-1 can penetrate the blood–brain barrier suggesting the possibility that reduced IGF-1 signaling in the brain can led to an extended mammalian life span (Taguchi & White, 2008). IGF-1 is also a key element required for differentiation of pre-adipocytes into adipocytes (Blüher, Kratzsch, & Kiess, 2005) so milk intake may also be a risk factor for obesity (Olsen et al., 2007). In addiction, IGF-1 is involved in the pathogenesis of acne, a chronic inflammatory disease that affect almost 9.4% of the global population (Rodighiero et al. 2020). In terms of food-safety and nutrition, little is known about the so-called threshold doses, i.e. the minimum amount of an active ingredient which is able to cause a reaction. Concerns about the potential health risk of IGF-1 have drawn increasing attention to the development of rapid, sensitive and accurate IGF-1 detection assay (Guidi et al., 2001) in order to improve food labeling directives and to increase consumer protection. Numerous assays have been developed over the years for the identification and quantification of IGF-1, especially in plasma: ELISA assay (Ginjala & Pakkanen, 1998), electrochemiluminescent assay (McGrath, Bogosian, Fabellar, Staub, Vicini, & Widger, 2008), automated surface plasmon resonance-based biosensor system (BIA-technology) (Guidi et al., 2001), immunometric assays (Anderson, Tamayose, & Garcia, 2018), radioimmunoassay (RIA) (Mann et al., 2020, Mann et al., 2020). RIA methodology is still used today thanks to its high specificity and sensitivity but there are several difficulties in the automation of this very expensive assay such as specialized structures for the storage and handling of radioactive material, the high cost of waste disposal, long analyzes and IGFBPs should be dissociated from IGF-1 as immunoassay interferents (Ketha and Singh, 2015, Salameh et al., 2014). A study published by Chanson et al., (2016) compared the analytical performances of six different immunoassays for the detection of IGF-1 in the serum of the same large population of healthy subjects. The data obtained evidenced that the reference intervals differed somewhat from one assay to another, and agreement between assays was moderate to good. Moreover, the concordances between the manufacturers' reference intervals and those obtained in the study were generally poor (Chanson et al., 2016). An alternative to immunoassays is liquid chromatography-tandem mass spectrometry (LC-MS/MS), already used for IGF-1 quantification mainly in serum (Ketha and Singh, 2015, Cox et al., 2014, Such-Sanmartín et al., 2015, Lopes et al., 2014, Bronsema et al., 2018, Thevis et al., 2011, Cox et al., 2014, Mongongu et al., 2021, Seo et al., 2021, Coppieters et al., 2020). In particular, Bronsema et al. (2018) developed and validated an LC-MS/MS targeted method for the detection of IGF-1 in serum in the 10–1000 ng/mL concentration range by selecting two signature peptides; an urea-based sample preparation procedure was proposed and no analyte enrichment step was required. Tanna, Lame and Wrona (2020) developed and validated a sensitive (LOQ 5 ng/mL) and reliable (mean accuracy of 101.76%) ultra-pressure (UP)LC-MS/MS targeted method for the detection of intact IGF-1 in clinical samples. Pratt, van Faassen, Remmelts, Bischoff and Kema (2021) recently published an antibody free LC-MS/MS method for the detection of intact IGF-1 and IGF-2 in human plasma with LOQ values ranging between 5.9 and 8.4 ng/mL. The performances of the LC-MS/MS method were compared to the results obtained by immunoassay for the detection of these two molecules in human plasma samples. Even if a good data correlation was observed (R2 = 0.97) between the two techniques, a statistically significant overestimation of the concentration levels detected by immunoassay was observed (Pratt et al., 2021). In the last years, increasing emphasis has been put on the development of confirmatory and screening LC–ESI-MS/MS based-methods for the identification and determination of active compounds in foods (Miranda, Bianchi, Krupova, Trossat, & Martin, 2020). Welsh et al. (2019) used LC-MS/MS for detection of contaminants (pesticides, antibiotics and hormones) in organic and conventionally produced milk samples sold in the USA and they detected IGF-1 in milk via LC–high-resolution linear ion trap (LTQ Orbitrap). By considering that the direct application of immunoassay kits to the detection of IGF-1 in different types of milks can fail to provide reliable quantitative data due to antibody-antigen binding variability in complex matrices and most of the LC-MS/MS methods were developed for plasma analysis, here we propose a developed and validated LC-MS/MS assay for IGF-1 identification and quantification in cow’s milk of different brands and subjected to different technological treatments. By using a shotgun proteomic approach, IGF-1 was enzymatically degraded to peptides and three selected biomarker peptides were then analyzed under SRM mode in a triple quadrupole (QqQ) mass analyzer. The quantitation of IGF-1 was carried out by selecting one quantifier biomarker peptide and evaluated in terms of sensitivity, selectivity, accuracy, recovery, limits of detection and quantitation. Moreover, the IGF-1 extraction procedure from milk must be reproducible and throughput in order to identify and quantify this very low abundant peptide hormone in such different matrices. Here, we tested two urea-based extraction protocols and after validation, the LC-MS/MS method was applied to the detection of IGF-1 in commercially available bovine milk samples and the results were discussed.
Fig. 1.
Bovine IGF-1 protein. (A) 3D structure of bovine IGF-1 protein. (B) IGF-1 experimentally determined interaction networks mainly involving IGF-binding proteins (BP), IGF-receptor (R) (www.uniprot database; accession number: P07455. STRING interaction network analysis). (C) IGF-1 primary sequence (in bold the cysteine involved in the disulfide bonds); the three biomarker peptides selected for the LC-MS/MS analysis are underlined.
2. Materials and methods
2.1. Chemicals
Ammonium bicarbonate (ABC), acetonitrile (HPLC purity), formic acid (analytical reagent grade), urea were purchased from Carlo Erba (Milan, Italy). Bovine insulin growth factor-1 (white powder, purity >95%), protein assay dye reagent concentrate were from Bio-Rad (Hercules, California, USA). Isotopically labeled IGF-1 peptide (LEMYCAPLKPA{K*} {K*} = Lys [13C6, 15 N2] isotopic enrichment: >99%) was from Pepscan (Presto AB Lelystad, NL). Bovine serum albumin (BSA), Tris-HCL, trypsin from bovine pancreas, iodoacetamide (IAA), dithiotreitol (DTT) were from Sigma-Aldrich® (Darmstadt, Germany).
2.2. IGF-1 extraction and sample preparation
Eleven cow milk types coming from different providers (fresh whole milk, UHT whole milk, fresh semi-skimmed milk, UHT semi-skimmed milk, fresh whole microfiltered milk, fresh semi-skimmed microfiltered milk, UHT skimmed milk, UHT semi-skimmed microfiltered milk, UHT lactose free milk, UHT lactose free low-fat milk, UHT low-fat milk, detailed in the Results section) for a total of 25 different samples were obtained from local food stores, aliquoted in 10 mL Falcon tubes and stored at −20 °C until processing. All milk pH were measured and ranged from 6.61 to 6.74 pH unit (average pH 6.68 ± 0.04). Initially, two different extraction methods were tested on a fresh whole milk sample. In the first case, IGF-1 extraction was performed by adding 1 mL of a 8 M urea-Tris-HCl buffer (pH = 8) solution to 1 mL of milk. In the second case, IGF-1 extraction was performed by dissolving directly urea in 700 µL of each milk sample in order to obtain a final concentration of 8 M urea in each sample. Subsequently, for both methods, the samples were immersed in an ultrasonic bath (Bransonic ultrasonic cleaner 2510E –DTH) at 4 °C for 10 min at 42 kHz and centrifuged (15000 rpm × 10 min); after centrifugation, it is necessary to remove the fat layer formed on each sample surface, and transfer the supernatants to new eppendorf (Eppendorf Srl, Milan, Italy). The proteins contained in each supernatant were assayed according to Bradford’s method. The Bradford assay was performed by adding 10 µL of each diluted sample / standard to 500 µL of previously diluted (1: 5) Bradford reagent (Absorbance = 595 nm). Calibration was achieved using the BSA protein standard with a final concentration of 1 mg/mL from which serial dilutions were performed to cover the 25–400 μg/mL range. By comparing the total milk protein content obtained from the two protocols, the second method (approx. 24 mg/mL) was more effective with respect to the first one (approx. 8 mg/mL) and it has been selected to perform all milk sample extraction. Subsequently, the supernatant was diluted with 50 mM ammonium bicarbonate buffer (pH 8.0) to decrease urea concentration under 2 M; the proteins were reduced and alkylated using 100 mM DTT (30 min at 37° C) followed by 200 mM IAA (30 min at room temperature in the dark), the reaction was stopped with 100 mM DTT (15 min at room temperature). Finally, all samples were digested by incubation with trypsin (1μg/μL) at 37 °C (protein:enzyme ratio 50:1) overnight. To stop the digestion, 1 μL of formic acid was added to each sample. Finally, samples were dried under nitrogen flow/lyophilized and reconstituted with 50 μL of a water / acetonitrile / formic acid aqueous solution (49.95 / 49.95 / 0.1, v/v/v).
2.3. Liquid chromatography–triple quadrupole mass spectrometry
LC separation was carried out on a C18 Luna (150 mm × 2.1 mm, 5 μm particles) (Phenomenex, CA, USA) column thermostated at 25 °C using a gradient solvent elution system [(A) aqueous formic acid 0.1% solution (v/v)/(B) 0.08% (v/v) formic acid in acetonitrile]. Gradient elution was as follows: solvent B was set at 5% for 2 min, then delivered by a linear gradient from 5% to 80% in 60 min. Solvent B was maintained at 80% for 1 min before column re-equilibration (5 min). The flow-rate was 200 μL/min. The mobile phase was delivered by the Agilent HP 1200 chromatographic system (Agilent Technologies, Germany) equipped with a 100-vial capacity sample tray. Injection volume was 10 μL. A QTRAP 4000 instrument (ABSCIEX, CA, USA) equipped with a pneumatically assisted ESI interface was used. The system was controlled by the Analyst software. The sheath gas (nitrogen, 99.999% purity) and the auxiliary gas (nitrogen, 99.998% purity) were delivered at flow-rates of 45 and 5 arbitrary unit, respectively. Optimized conditions of the interface were as follows: ESI voltage 5.5 kV, capillary voltage 20 V, capillary temperature 350 °C. MS/MS experiments were performed under both product-ion and SRM conditions with a collision gas (nitrogen) pressure of 2.1 × 10−3 mbar in the collision cell. The SRM transitions monitored were reported in Table 1. For quantitative purposes, the most intense SRM transition was monitored for IGF-1, whereas the other transitions reported in Table 1 were monitored for confirmatory purposes.
Table 1.
List of IGF-1 peptides and SRM transitions monitored with their m/z values and fragment ions. Sequences chosen respective as qualifier transition and quantifier biomarker are in bold.
| Peptide sequence | Precursor ion [M + 2H]2+m/z | Product ion m/z | Fragment ion |
|---|---|---|---|
| GFYFNKPTGYGSSSR | 834.4 | 1153.6 | y11 |
| GFYFNKPTGYGSSSR (selected qualifier transition) | 834.4 | 1039.5 | y10 |
| GFYFNKPTGYGSSSR | 834.4 | 911.4 | y9 |
| GFYFNKPTGYGSSSR | 834.4 | 757.4 | b6 |
| GFYFNKPTGYGSSSR | 834.4 | 854.4 | b7 |
| GFYFNKPTGYGSSSR | 834.4 | 955.5 | b8 |
| GFYFNKPTGYGSSSR | 834.4 | 1012.5 | b9 |
| APQTGIVDEC[+57]C[+57]FR | 776.8 | 1481.6 | y12 |
| APQTGIVDEC[+57]C[+57]FR | 776.8 | 1098.5 | y8 |
| APQTGIVDEC[+57]C[+57]FR | 776.8 | 985.8 | y7 |
| APQTGIVDEC[+57]C[+57]FR | 776.8 | 886.3 | y6 |
| APQTGIVDEC[+57]C[+57]FR | 776.8 | 782.4 | b8 |
| APQTGIVDEC[+57]C[+57]FR | 776.8 | 911.4 | b9 |
| APQTGIVDEC[+57]C[+57]FR | 776.8 | 1071.5 | b10 |
| LEMYC[+57]APLKPAK | 710.9 | 1047.6 | y9 |
| LEMYC[+57]APLKPAK | 710.9 | 884.5 | y8 |
| LEMYC[+57]APLKPAK | 710.9 | 724.5 | y7 |
| LEMYC[+57]APLKPAK | 710.9 | 653.4 | y6 |
| LEMYC[+57]APLKPAK (selected quantifier biomarker) | 710.9 | 315.2 | y3 |
| LEMYC[+57]APLKPAK | 710.9 | 768.3 | b6 |
| LEMYC[+57]APLKPAK | 710.9 | 865.4 | b7 |
| LEMYC[+57]APLKPAK | 710.9 | 978.4 | b8 |
| LEMYC[+57]APLKPAK | 710.9 | 1106.5 | b9 |
2.4. Method validation
Validation of the whole analytical procedure was performed under MS/MS mode on fortified milk samples according to Eurachem guidelines (Eurachem, 1998). For this purpose, milk samples were fortified with different IGF-1 amounts and measurements were carried out by monitoring the most abundant MS/MS transition (Table 1). The detection limits (LOD) and the quantitation limits (LOQ) were calculated from the calibration curve as:
where s is the standard deviation of the blank signal obtained by performing 10 independent blank measurements. Linearity was assessed over one order of magnitude in a suitable ng IGF-1/mL matrix range [50–500 ng/mL], starting from LOQ values. Six equispaced concentration levels were tested and three replicated analysis were performed for each level. The Levene test was used to check homoschedasticity. The Mandel test was performed to check linearity. The first order calibration curve (y = b0 + b1x) was compared with the second order calibration curve (y2 = b0 + b1x + b2x2) by using the following equations:
| ΔS2= (N-2)s2y1 – (N-3)s2y2 |
| F = ΔS2/s2y2 |
where N is the total number of observations and sy is the residual standard deviation for each equation.
The significant of the intercept was tested by using the two-tailed Student’s t-test (95% significant level).
Precision was evaluated as relative standard deviation (RSD) in terms of intra-day repeatability and intermediate precision (inter-day repeatability). For this purpose, the within day repeatability was evaluated by performing three independent extractions of the matrix fortified with 10 and 50 ng IGF-1/mL matrix and three LC–MS/MS injections for each extract in the same day and the inter-day repeatability was calculated on five days by performing five independent extractions of the fortified matrix and three LC–MS/MS injections for each extract. The matrix effect was assessed by using the recovery function. The matrix-matched calibration curve were obtained by analyzing the matrix extracts fortified with IGF-1 at six concentration levels [50–500 ng/mL] and treated applying the whole analytical procedure. Each level was analyzed three times. The recovery of the protein extraction procedure was evaluated by performing the extraction on the matrix fortified at two different concentration levels (10 and 50 ng IGF-1/mL). The total protein content was determined by using a Bradford assay. Relative quantitation of IGF-1 peptides in the milk samples was achieved through normalization of data with respect total protein content. Absolute quantitation was obtained by using the isotopically-labeled internal standard (IS) method, by adding IS (final concentration 30 ng/mL) to each extract before LC-MS/MS analysis.
Differences in ng/mL between milk groups were calculated using a one-way ANOVA (95% significant level). All results were considered significant if p < 0.05.
2.5. Bioinformatic analysis and mass spectrometry (MS) data processing
IGF-1 protein from cow milk was considered for the assay. FASTA protein sequence was obtained from Uniprot Bovine proteome database and SRM transitions were simulated by Skyline (version 20.1, SCIEX) (Redwood City, CA, USA), setting trypsin as digestion mode with no missed cleavage and carbamidomethylation of cysteins as structural modification (Table 1). Uniqueness of candidate peptide sequences was assessed by BLASTp tool (basic local alignment search tool; www.ncbi.nml.nih.gov link NCBI BLAST) search (algorithm: blastp; ATRIX PA 30; GAP COASTS: existence 10, extension:1; DATABASE: non redundant protein sequences) from NCBI (National Center for Biotechnology Information) (Bethesda, MA, USA). LC-MS/MS SRM data were analyzed using the Analyst v 1.4 software and the peaks areas were obtained by the MultiQuant program (version 2.1, ABSCIEX).
3. Results
3.1. In silico selection of biomarker peptides
Bovine IGF-1 is a small protein composed by 154 amino acid residues (MW 17066 Da) with three disulfide bonds (55 ↔ 97; 67 ↔ 110; 66 ↔ 101) as post translational modifications (PTMs) (Fig. 1) and a 95.56% homology with the human IGF-1. In this work the peptides targeting the bovine IGF-1 protein were selected on the basis of the published sequences present in the protein database (http://www.uniprot.org/). As for biomarker peptide selection, different criteria, such as absence of missed cleavages, good ESI sensitivity, no PTMs and sequence specificity, were considered. Thereby, by analyzing a tryptic digest extract from raw milk a subset of peptides was directly identified by the mass spectrometer in a data-dependent acquisition mode to characterize IGF-1 protein with a sequence coverage average of ∼65%. Those peptides providing good ESI-MS responses and unequivocally identifying the target protein were selected (Table 1). To develop a reliable qualitative and quantitative method, three peptides present in all the eight (1–8) bovin IGF-1 isoforms were selected and monitored (Table 1). It should be noted that these three peptides are all located in the second half of the aminoacid sequence close to C-term, have 100% homology with the human IGF-1 sequences and are present in all its four (1–4) human isoforms. These findings indicate that they could be successful candidates for the detection of bovin and human IGF-1 in different matrices. In a further step, product-ion MS/MS measurements were carried out on the [M + 2H]2+ peptide signals, as the most abundant, by varying collision energy (from 20 to 45 eV) to select specific SRM transitions for each biomarker. The MS/MS spectra exhibited several fragments of the y- and b-series to cover and confirm their sequences (Table 1). All SRM transition tested for the two peptides GFYFNKPTGYGSSSR and LEMYC[+57]APLKPAK were detected in all kind of milk samples with excellent signal intensity and stability. On the other hand, only one transition of the APQTGIVDEC[+57]C[+57]FR peptide (m/z 776.8/911.4, b9 fragment) was detected in all milk samples (Table 1). Since this peptide contain a disulfide bridge, considered as PTM, even if reduction and alkylation step are carried out during the sample treatment protocol, it was no more considered a suitable IGF-1 biomarker because of its poor signal stability. Attention was thus paid to the selection of those fragment ions that provide optimal signal intensity and that could discriminate the targeted peptides from other species present in the sample. The assay was constituted of a series of transitions (precursor/fragment ion pairs) in combination with the retention time for each targeted peptide as reported in Table 1.
3.2. Chromatographic separation and LC–MS/MS method validation
Milk is a complex matrix rich in proteins and taking into account that the chromatographic separation plays a fundamental role in the MS analysis of complex peptide mixtures, suitable separation methods have to be developed to improve resolution, sensitivity, analysis time and reduce matrix effects. By analyzing a IGF-1 protein aqueous solution a LC-MS/MS elution profile (within 8 min) of the peptides was obtained (Fig. 2). The chromatographic profile of the two peptides on the C18 column showed excellent peak shape (in-run-peak width (FWHM, average on the peptides) = 15.80 ± 0.07 s) and retention time stability (RSD < 1.0%). The primary goal of this work was to validate a sensitive and robust LC–MS/MS method for the analysis of IGF-1 at trace levels in foods. As for MS acquisition mode, the non-scanning nature of selected reaction monitoring (SRM) analysis, usually performed by a triple quadrupole mass spectrometer allows to obtain excellent sensitivity and enables the detection of low-abundance proteins in highly complex mixtures. For this purpose, studies on linearity, trueness, precision, selectivity and recovery were performed and the LEMYCAPLKPAK [M + 2H]2+/y3 biomarker m/z transition was selected for quantitative purposes as providing the most intense signal. Even if little is known about the minimum concentration level able to provide reaction, detection limits between 1 and 5 ng of IGF-1 per mL of sample were determined from the matrix-matched calibration curves. LOQ values were found in the 3–12 ng protein/mL of milk sample. Generally, the LC–ESI-MS/MS linear range was explored over one order of magnitude of concentration [y = 36.9(±0.3)x, r2 = 0.999; concentration range: 50–500 ng/mL]. Homogeneity of variance of replicate measurements at different concentration levels was proved at the 95% confidence level (p > 0.18). After testing significance of the intercept (p value lower than 0.05 at 95% confidence level), linearity was mathematically verified by applying the Mandel fitting test. A p value of 0.781 demonstrated that the best data fit could be obtained using a first order regression model. The method accuracy was then tested both in terms of precision and trueness. Excellent precision in terms of intra-day repeatability was calculated providing RSD% in the 3–8% (n = 9) range. The intermediate precision results were found not exceed 15% (n = 15), confirming good method precision. As for trueness, a calculation of the recovery function was performed to ascertain the influence of the matrix for the determination of the IGF-1 biomarker peptide. For this purpose, an aqueous tryptic digest and different milk sample (fresh whole milk, fresh semi-skimmed microfiltrated milk, UHT semi-skimmed milk and UHT skimmed milk; n = 3, n = number of independent milk type) fortified with the tryptic digest (at six concentration levels n = 6) were analyzed. The slope and the intercept of the recovery functions calculated for the analytes were compared respectively with 1 and 0 by means of a t-test. The t-test performed on the intercept provided a p value at the 95% confidence level higher than 0.05 (p > 0.05) demonstrating that all the calibration equations were in the y = b1 × form and thus the absence of constant systematic errors. In the case of the slopes, since the t-calculated resulted to be higher than the t-tabulated at the 95% confidence level (1.86), it can be inferred that the calibration curves obtained by spiking samples are significantly different from that obtained using standard solution as a function of the different milk matrices. In all the cases a signal suppression for the selected biomarker between 85% and 89%, was observed. These findings suggest that a matrix effect able to cause significant systematic error in the IGF-1 quantitation is present, but even that the percentage of matrix effect is very similar among different type of milks, independently from the technological treatment applied. To overcome matrix effect isotopically labeled internal standard was used to build up the calibration curve as quantification method. As for recovery, fresh whole milk sample, selected as the most complex matrix, was enriched with a isotopically labeled IGF-1 and values ranging from 89 % (SD = 4%) to 98 % (SD = 3%) were obtained.
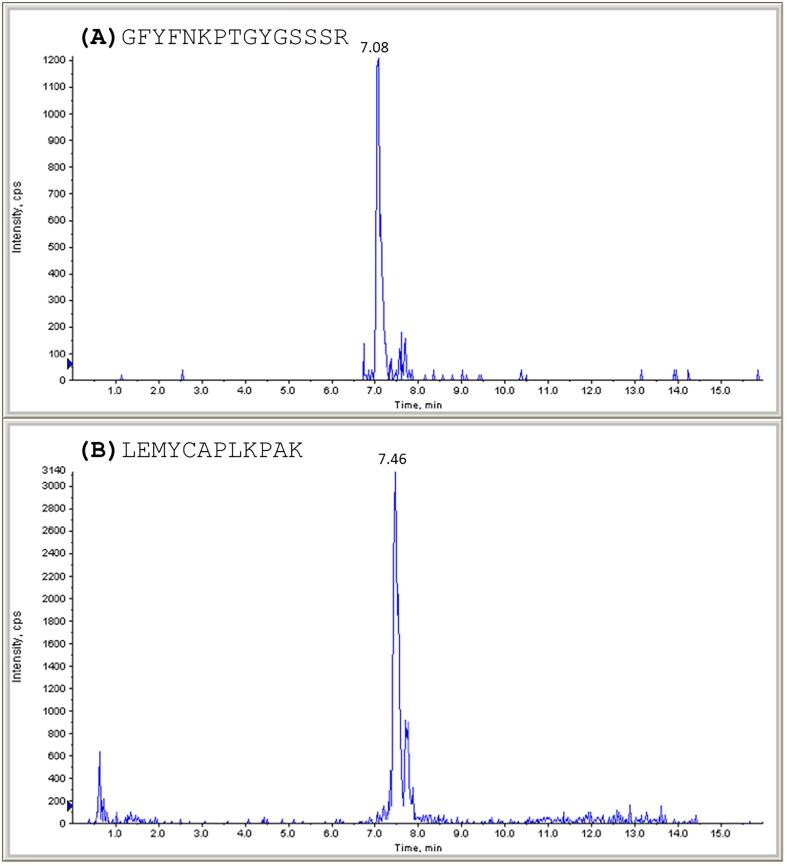
Fig. 2.
Liquid chromatography separation. LC-ESI-MS/MS SRM profile of the two IGF-I biomarkers (A) GFYFNKPTGYGSSSR and (B) LEMYCAPLKPAK identified in a IGF-1 protein aqueous solution (IGF-1 conc. 50 ng/mL) respectively with retention time of 7.08 and 7.46 min.
3.3. Sample analysis
In the last step of the work the LC–MS/MS method was applied to a variety of commercial milk products to evaluate the presence of the IGF-1 under investigation. It must be considered that milk is a complex matrix whose proteome is made of very abundant proteins such as casein and medium to low abundance whey proteins such as ß-lactoglobulin, lactoferrin, immunoglobulins, glycoproteins, enzymes and peptide hormones (Vincent et al., 2016). The urea-based protocol 2 was applied and no sample purification or enrichment was required. All samples analyzed, under MS/MS mode, exhibited naturally occurring IGF-1 levels (Table 2). As for confirmatory purposes, these samples were run under MS/MS SRM mode in order to acquire all biomarker peptides that univocally verified the presence of IGF-1. No isobaric interferents and carry-over effects were detected. The quantitative results are shown in Fig. 3 and Table 2.
Table 2.
LC-ESI-MS/MS SRM determination of IGF-1 in different milk samples. Total protein content (mg/mL) and IGF-1 concentration ± standard deviation (±SD) (ng/mL) are reported for the investigated milk samples.
| Sample | Milk Types | Milk Brand | Total protein content (mg/mL) | IGF-1 concentration (±SD) (ng/mL) |
|---|---|---|---|---|
| 1 | Fresh whole | Conad | 10.6 | 346 ± 5* |
| 2 | Esselunga | 32.1 | 29.2 ± 0.9 | |
| 3 | Granarolo | 25.8 | 34.6 ± 0.5 | |
| 4 | Granarolo | 21.1 | 26.7 ± 0.3 | |
| 5 | UHT whole | Coop | 17.4 | 91.4 ± 0.4 |
| 6 | Esselunga | 23.4 | 11.1 ± 0.4** | |
| 7 | Parmalat | 21.7 | 23.9 ± 0.8 | |
| 8 | Fresh semi-skimmed | Conad | 24.4 | 89 ± 6 |
| 9 | Granarolo | 23.6 | 68.8 ± 0.8 | |
| 10 | UHT semi-skimmed | Coop | 19.3 | 153 ± 5 |
| 11 | Granarolo | 23.5 | 57.2 ± 0.4 | |
| 12 | Parmalat | 37.9 | 17.7 ± 0.6 | |
| 13 | Esselunga | 29.7 | 20.4 ± 0.9 | |
| 14 | Fresh whole microfiltered | Conad | 30.3 | 121 ± 7 |
| 15 | Esselunga | 27.3 | 54.8 ± 0.4 | |
| 16 | Fresh semi-skimmed microfiltered | Conad | 29.2 | 87 ± 2 |
| 17 | Esselunga | 27.4 | 124 ± 3 | |
| 18 | Parmalat | 24.0 | 27.5 ± 0.9 | |
| 19 | UHT skimmed | Coop | 18.5 | 166 ± 7 |
| 20 | Esselunga | 23.6 | 74 ± 2 | |
| 21 | UHT semi-skimmed microfiltered | Coop | 20.5 | 147 ± 4 |
| 22 | UHT lactose free | Parmalat | 28.8 | 29.7 ± 0.8 |
| 23 | UHT lactose free low-fat | Parmalat | 27.8 | 17.8 ± 0.2 |
| 24 | Granarolo | 31.2 | 11.7 ± 0.4** | |
| 25 | UHT low-fat | Granarolo | 22.9 | 61.8 ± 0.5 |
*Highest IGF-1 concentration. ** Lowest IGF-1 concentration.
Fig. 3.
IGF-1 concentration (ng/mL) in different milk types. Selective distribution of IGF-1 in 25 milk samples of 11 different types: fresh whole, UHT whole, fresh semi-skimmed, UHT semi-skimmed, UHT skimmed, fresh whole microfiltered, fresh semi-skimmed microfiltered, UHT semi-skimmed microfiltered, UHT lactose free, UHT lactose free low-fat, UHT low-fat. Data are expressed as mean ± standard deviation (±SD).
4. Discussion
Daily cow milk consumption is a diffused aspect of human diet and generally recommended by different nutritional guidelines as important supplier of calcium, magnesium, iodine, vitamin B12, proteins and other important nutrients (Givens, 2020). However, the study of the health benefits and risks associate to milk intake is still a hot topic under investigation, involving different scientific disciplines such as microbiology, anatomy, endocrinology, cardiology, dermatology and biochemistry. Among these studies, several papers deal with the regulation level of the hormone like protein IGF-1 in milk and its correlation between milk consumption and health effects (Guidi et al., 2001, Clatici et al., 2018, Melnik et al., 2011). IGF-1 is usually secreted from bovine mammary gland at high levels during the last 2 weeks ante partum followed by a rapid decrease during the first milkings post-partum. IGF-1 appeared mainly in the bound form (91%) at days 40–2 ante partum, whereas free IGF-1 preponderated in the first milkings post-partum (73%) and changed again to about 85% in the bound form after day 4 post- partum (Einspanier & Schams, 1991). Before to reach the consumers, raw milk usually undergoes different heat treatments commonly used as processing technologies in the dairy industry. In order to better understand the effects of milk intake on the human health the development and validation of reliable analytical methods for sample characterization plays a pivotal role.
Therefore, IGF-1 determination is usually based on a common and standard radioimmunoassay approach for quick and reliable assay. RIA analysis presents various advantages (i.e. simplicity, sensitivity) but, remarkably, even several disadvantages (use of antibodies, use of radioactive compounds, false positive results). These reasons stimulated the development of alternative analytical approaches such as the use of LC-MS/MS as highly consolidated, sensitive and selective technique for the qualitative and quantitative analysis of analytes in complex matrices even at the trace levels.
Here, we developed and validated in house an SRM targeted LC-ESI-MS/MS method for the unambiguous identification and determination of total IGF-1 content in milk, as reliable alternative to immunoassay. Welsh et al. (2019) reported the use of LC-MS/MS technique for the identification of IGF-1 in milk samples, but no complete validation was reported and an external standard method was used for quantitative analysis. Other methods (Kirsch et al., 2007) proposed the GFYFNKPTGYGSSSR peptide as biomarker for human IGF-1, because it was the only one that did not contain a cysteine residue. In our case both the GFYFNKPTGYGSSSR and LEMYC[+57]APLKPAK peptides were monitored and the last one exhibited the most intense (>70% relative intensity over all monitored SRM transitions) and stable signal (chromatographic peak area n = 6; RSD% <1%).
The analytical procedure developed and validated in our laboratory is able to identify and quantify total IGF-1 presents in variable milk matrices, the complexity strongly depending on the sample origin and the technological treatments performed. The accuracy was assured by the use of the isotopically labelled internal standard. All the milk samples investigated exhibited a comparable total protein content (25 ± 5 mg/mL by Bradford assay) but, by using the internal standard method, IGF-1 concentrations in the 25 analyzed samples were spread over a wide range from 11.2 ± 0.3 ng/mL to over 346 ± 8 ng/mL with a median of 57 ± 2 ng/mL. The highest amount of IGF-1 was found in the fresh milk samples (average value 96 ng/mL) with respect to UHT treated milks (average value 66 ng/mL), however the high data variability (in the 84–110% range), probably due to the different origin of the milks, did not allow to demonstrate a statistically significant difference (p > 0.05) among technological treatments. Moreover, different average amounts of IGF-1 were found in the milk samples having different milk fat content (average value in the whole milks: 82 ng/mL; average value in the semi-skimmed milks: 74 ng/mL; average value in the skimmed and low-fat milks: 66 ng/mL). However even in this case the high data variability (between 66 and 128%) did not allow to statistically (p > 0.05) confirm a significant trend.
5. Conclusions
As the demand to accurately measure active compounds in food is increasing, here a new effective LC-ESI-MS/MS method for the detection of IGF-1 in milk with different technological treatments was developed and successfully validated. By operating under SRM conditions, the validation procedure demonstrate the fit-to-purpose of the method to identify and detect IGF-1 through biomarker peptides with LODs from 1 and 5 ng IGF-1/mL matrix, precision from 3 to 8%, high accuracy an recovery in different types of milk. Our study has enlightened the very selective detection of IGF-1 in all kinds of milk samples at the levels of interest without any sample purification, preconcentration treatment or use of antibodies, in order to provide an accurate detection method useful for food control analysis. This method can be easy transferred to other laboratories equipped with a LC-triple quadrupole mass analyzer for routine quantitative analysis and does not required expensive high resolution MS instruments. Moreover, after methodical evaluation of the sample treatment procedure, this approach can be extended to the simultaneous and specific detection of IGF-1 at the trace level in other milk-based products and, as a consequence, it might represent a very flexible tool able to gain insights into the study of milk composition.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Giulia Remaggi: Writing – original draft, Investigation. Roberta Saleri: Supervision. Melania Andrani: Supervision, Investigation. Francesca Satolli: Supervision. Eleonora Rodighiero: Supervision. Lisa Elviri: Conceptualization, Project administration, Methodology, Data curation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Giulia Remaggi, Email: giulia.remaggi@unipr.it.
Roberta Saleri, Email: roberta.saleri@unipr.it.
Melania Andrani, Email: melania.andrani@unipr.it.
Francesca Satolli, Email: fra.satolli@libero.it.
Lisa Elviri, Email: lisa.elviri@unipr.it.
References
- Anderson L.J., Tamayose J.M., Garcia J.M. Use of growth hormone, IGF-I, and insulin for anabolic purpose: Pharmacological basis, methods of detection, and adverse effects. Molecular and Cellular Endocrinology. 2018;464:65–74. doi: 10.1016/j.mce.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blüher S., Kratzsch J., Kiess W. Insulin-like growth factor I, growth hormone and insulin in white adipose tissue. Best Practice & Research. Clinical Endocrinology & Metabolism. 2005;19(4):577–587. doi: 10.1016/j.beem.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Bronsema K.J., Klont F., Schalk F.B., Bischoff R., Kema I.P., van de Merbel N.C. A quantitative LC-MS/MS method for insulin-like growth factor 1 in human plasma. Clinical Chemistry and Laboratory Medicine. 2018;56(11):1905–1912. doi: 10.1515/cclm-2017-1042. [DOI] [PubMed] [Google Scholar]
- Chanson, P., Arnoux, A., Mavromati, M., Brailly-Tabard, S., Massart, C., Young, J., Piketty, M. L., Souberbielle, J. C., & VARIETE Investigators (2016). Reference Values for IGF-I Serum Concentrations: Comparison of Six Immunoassays. The Journal of Clinical Endocrinology and Metabolism, 101(9), 3450–3458. 10.1210/jc.2016-1257. [DOI] [PMC free article] [PubMed]
- Clatici, V. G., Voicu, C., Voaides, C., Roseanu, A., Icriverzi, M., & Jurcoane, S. (2018). Diseases of Civilization - Cancer, Diabetes, Obesity and Acne - the Implication of Milk, IGF-1 and mTORC1. Maedica, 13(4), 273–281. 10.26574/maedica.2018.13.4.273. [DOI] [PMC free article] [PubMed]
- Coppieters G., Judák P., Van Haecke N., Van Renterghem P., Van Eenoo P., Deventer K. A high-throughput assay for the quantification of intact Insulin-like Growth Factor I in human serum using online SPE-LC-HRMS. Clinica Chimica Acta. 2020;510:391–399. doi: 10.1016/j.cca.2020.07.054. [DOI] [PubMed] [Google Scholar]
- Cox H.D., Hughes C.M., Eichner D. Sensitive quantification of IGF-1 and its synthetic analogs in dried blood spots. Bioanalysis. 2014;6(19):2651–2662. doi: 10.4155/bio.14.109. [DOI] [PubMed] [Google Scholar]
- Cox, H. D., Lopes, F., Woldemariam, G. A., Becker, J. O., Parkin, M. C., Thomas, A., Butch, A. W., Cowan, D. A., Thevis, M., Bowers, L. D., & Hoofnagle, A. N. (2014). Interlaboratory agreement of insulin-like growth factor 1 concentrations measured by mass spectrometry. Clinical chemistry, 60(3), 541–548. doi: 10.1373/clinchem.2013.208538. [DOI] [PubMed]
- Einspanier R., Schams D. Changes in concentrations of insulin-like growth factor 1, insulin and growth hormone in bovine mammary gland secretion ante and post partum. The Journal of Dairy Research. 1991;58(2):171–178. doi: 10.1017/s002202990002971x. [DOI] [PubMed] [Google Scholar]
- Eurachem. (1998). The Fitness for Purpose of Analytical Methods. In Eurachem Guide, ISBN: 0-94948926-12-0. http://www.eurachem.org/images/stories/Guides/pdf/valid.pdf.
- Francis G.L., Upton F.M., Ballard F.J., McNeil K.A., Wallace J.C. Insulin-like growth factors 1 and 2 in bovine colostrum. Sequences and biological activities compared with those of a potent truncated form. The Biochemical Journal. 1988;251(1):95–103. doi: 10.1042/bj2510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furstenberger G., Senn H.J. Insulin-like growth factors and cancer. Lancet Oncology. 2002;3(5):298–302. doi: 10.1016/S1470-2045(02)00731-3. [DOI] [PubMed] [Google Scholar]
- Ginjala V., Pakkanen R. Determination of transforming growth factor-β1 (TGF-β1) and insulin-like growth factor 1 (IGF-1) in bovine colostrum samples. Journal of Immunoassay. 1998;19(2–3):195–207. doi: 10.1080/01971529808005480. [DOI] [PubMed] [Google Scholar]
- Givens D.I. MILK Symposium review: The importance of milk and dairy foods in the diets of infants, adolescents, pregnant women, adults, and the elderly. Journal of Dairy Science. 2020;103(11):9681–9699. doi: 10.3168/jds.2020-18296. [DOI] [PubMed] [Google Scholar]
- Guidi A., Laricchia-Robbio L., Gianfaldoni D., Revoltella R., Del Bono G. Comparison of a conventional immunoassay (ELISA) with a surface plasmon resonance-based biosensor for IGF-1 detection in cows’ milk. Biosensors & Bioelectronics. 2001;16(9–12):971–977. doi: 10.1016/S0956-5663(01)00245-7. [DOI] [PubMed] [Google Scholar]
- Juskevich J.C., Guyer C.G. Bovine growth hormone: Human food safety evaluation. Science (New York, N.Y.) 1990;249(4971):875–884. doi: 10.1126/science.2203142. [DOI] [PubMed] [Google Scholar]
- Ketha H., Singh R.J. Clinical assays for quantitation of insulin-like-growth-factor-1 (IGF1) Methods. 2015;81:93–98. doi: 10.1016/j.ymeth.2015.04.029. [DOI] [PubMed] [Google Scholar]
- Lopes F., Cowan D.A., Thevis M., Thomas A., Parkin M.C. Quantification of intact human insulin-like growth factor-I in serum by nano-ultrahigh-performance liquid chromatography/tandem mass spectrometry. Rapid Communications in Mass Spectrometry: RCM. 2014;28(13):1426–1432. doi: 10.1002/rcm.6908. [DOI] [PubMed] [Google Scholar]
- Mann S., Curone G., Chandler T.L., Moroni P., Cha J., Bhawal R., Zhang S. Heat treatment of bovine colostrum: I. Effects on bacterial and somatic cell counts, immunoglobulin, insulin, and IGF-I concentrations, as well as the colostrum proteome. Journal of Dairy Science. 2020;103(10):9368–9383. doi: 10.3168/jds.2020-18618. [DOI] [PubMed] [Google Scholar]
- Mann S., Curone G., Chandler T.L., Sipka A., Cha J., Bhawal R., Zhang S. Heat treatment of bovine colostrum: II. Effects on calf serum immunoglobulin, insulin, and IGF-I concentrations, and the serum proteome. Journal of Dairy Science. 2020;103(10):9384–9406. doi: 10.3168/jds.2020-18619. [DOI] [PubMed] [Google Scholar]
- McGrath M.F., Bogosian G., Fabellar A.C., Staub R.L., Vicini J.L., Widger L.A. Measurement of bovine somatotropin (bST) and Insulin-like Growth Factor-1 (IGF-1) in bovine milk using an electrochemiluminescent assay. Journal of Agriculture and Food Chemistry. 2008;56(16):7044–7048. doi: 10.1021/jf800696d. [DOI] [PubMed] [Google Scholar]
- Melnik B.C. Milk – The promoter of chronic Western diseases. Medical Hypotheses. 2009;72(6):631–639. doi: 10.1016/j.mehy.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Melnik B.C., John S.M., Schmitz G. Over-stimulation of insulin/IGF-1 signaling by western diet may promote diseases of civilization: Lessons learnt from laron syndrome. Nutrition and Metabolism. 2011;8:2–5. doi: 10.1186/1743-7075-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnik B.C., John S.M., Schmitz G. Milk is not just food but most likely a genetic transfection system activating mTORC1 signaling for postnatal growth. Nutrition Journal. 2013;12:103. doi: 10.1186/1475-2891-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda G., Bianchi L., Krupova Z., Trossat P., Martin P. An improved LC–MS method to profile molecular diversity and quantify the six main bovine milk proteins, including genetic and splicing variants as well as post-translationally modified isoforms. Food Chemistry: X. 2020;5 doi: 10.1016/j.fochx.2020.100080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongongu C., Coudoré F., Domergue V., Ericsson M., Buisson C., Marchand A. Detection of LongR 3 -IGF-I, Des(1–3)-IGF-I, and R 3-IGF-I using immunopurification and high resolution mass spectrometry for antidoping purposes. Drug Testing and Analysis. 2021;13(7):1256–1269. doi: 10.1002/dta.3016. [DOI] [PubMed] [Google Scholar]
- Olsen, S. F., Halldorsson, T. I., Willett, W. C., Knudsen, V. K., Gillman, M. W., Mikkelsen, T. B., Olsen, J., & NUTRIX Consortium (2007). Milk consumption during pregnancy is associated with increased infant size at birth: prospective cohort study. The American journal of clinical nutrition, 86(4), 1104–1110. doi: 10.1093/ajcn/86.4.1104. [DOI] [PubMed]
- Pratt M.S., van Faassen M., Remmelts N., Bischoff R., Kema I.P. An antibody-free LC-MS/MS method for the quantification of intact insulin-like growth factors 1 and 2 in human plasma. Analytical and Bioanalytical Chemistry. 2021;413(8):2035–2044. doi: 10.1007/s00216-021-03185-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodighiero E., Bertolani M., Sareri R., Pedrazzi G., Torello L., Feliciani C., Satolli F. Do acne treatment affect insulin-like growth factor serum levels? A clinical and laboratory study on patients with acne vulgaris. Dermatologic Therapy. 2020;33 doi: 10.1111/dth.13439. [DOI] [PubMed] [Google Scholar]
- Salameh W.A., Redor-Goldman M.M., Clarke N.J., Mathur R., Azziz R., Reitz R.E. Specificity and Predictive Value of Circulating Testosterone Assessed By Tandem Mass Spectrometry for the Diagnosis of Polycystic Ovary Syndrome by the NIH 1990 Criteria. Fertility and Sterility. 2014;101(4):1135–1141.e2. doi: 10.1016/j.fertnstert.2013.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo Y., Park J., Kim M., Sung C., Kwon O.S., Lee H.J., Min H. Optimization, validation, and comparison of a rapid method for the quantification of insulin-like growth factor 1 in serum using liquid chromatography-high-resolution mass spectrometry. Drug Testing and Analysis. 2021;13(2):451–459. doi: 10.1002/dta.v13.210.1002/dta.2944. [DOI] [PubMed] [Google Scholar]
- Such-Sanmartín G., Bache N., Callesen A.K., Rogowska-Wrzesinska A., Jensen O.N. Targeted mass spectrometry analysis of the proteins IGF1, IGF2, IBP2, IBP3 and A2GL by blood protein precipitation. Journal of Proteomics. 2015;113:29–37. doi: 10.1016/j.jprot.2014.09.013. [DOI] [PubMed] [Google Scholar]
- Taguchi A., White M.F. Insulin-like signaling, nutrient homeostasis, and life span. Annual Review of Physiology. 2008;70(1):191–212. doi: 10.1146/physiol.2008.70.issue-110.1146/annurev.physiol.70.113006.100533. [DOI] [PubMed] [Google Scholar]
- Tanna N.N., Lame M.E., Wrona M. Development of an UPLC/MS-MS method for quantification of intact IGF-I from human serum. Bioanalysis. 2020;12(1):53–65. doi: 10.4155/bio-2019-0234. [DOI] [PubMed] [Google Scholar]
- Thevis M., Thomas A., Schänzer W. Doping control analysis of selected peptide hormones using LC-MS(/MS) Forensic Science International. 2011;213(1–3):35–41. doi: 10.1016/j.forsciint.2011.06.015. [DOI] [PubMed] [Google Scholar]
- Vincent D., Ezernieks V., Elkins A., Nguyen N., Moate P.J., Cocks B.G., Rochfort S. Milk bottom-up proteomics: Method optimization. Frontiers in Genetics. 2016;6(JAN) doi: 10.3389/fgene.2015.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J.A., Braun H., Brown N., Um C., Ehret K., Figueroa J., Barr D.B. Production-related contaminants (pesticides, antibiotics and hormones) in organic and conventionally produced milk samples sold in the USA. Public Health Nutrition. 2019;22(16):2972–2980. doi: 10.1017/S136898001900106X. [DOI] [PMC free article] [PubMed] [Google Scholar]