Summary
Scientists in disciplines such as neuroscience and bioinformatics are increasingly relying on science gateways for experimentation on voluminous data, as well as analysis and visualization in multiple perspectives. Though current science gateways provide easy access to computing resources, datasets and tools specific to the disciplines, scientists often use slow and tedious manual efforts to perform knowledge discovery to accomplish their research/education tasks. Recommender systems can provide expert guidance and can help them to navigate and discover relevant publications, tools, data sets, or even automate cloud resource configurations suitable for a given scientific task. To realize the potential of integration of recommenders in science gateways in order to spur research productivity, we present a novel “OnTimeRecommend” recommender system. The OnTimeRecommend comprises of several integrated recommender modules implemented as microservices that can be augmented to a science gateway in the form of a recommender-as-a-service. The guidance for use of the recommender modules in a science gateway is aided by a chatbot plug-in viz., Vidura Advisor. To validate our OnTimeRecommend, we integrate and show benefits for both novice and expert users in domain-specific knowledge discovery within two exemplar science gateways, one in neuroscience (CyNeuro) and the other in bioinformatics (KBCommons).
Keywords: science gateway, recommender system, chatbot guided user interface, knowledge discovery, microservices
1 |. INTRODUCTION
Recent science and engineering research tasks are increasingly becoming data-intensive and thus relying on workflows to automate integration and analysis of voluminous data to test hypotheses. For example, research and training in neural science and engineering increasingly deal with diverse and voluminous multi-parameter data1, posing unique challenges outlined in an NSF iNeuro report2 as limited access to: multi-omics data archives3, heterogeneous software4 and computing resources (Neuroscience Gateway5, Amazon Web Services (AWS)), and multi-site interdisciplinary expertise (e.g., engineering, biology and psychology). Existing distributed high-performance computing resources (HPC) and other cyberinfrastructure (CI) tools for data management support the related data analysis and visualization capabilities. However, to fully utilize such capabilities, neuroscientists (often with limited CI skills) are required to take valuable time away from the focus of knowledge discovery in neuroscience, in order to learn about how to use the various technologies.
In addition to the challenges in identifying the relevant CI resources to expedite research and discovery, scientists are often hindered by limited exchange of ideas, sharing of data, and collaboration within the community. Factors such as reproducibility, usability, automation, and distributed cognition6 have been significant barriers for use of CI resources in scientific tasks. Labs pursue their individual research independently, distribute their research via journal articles, and reinvent the computing pipelines used in their analysis7 time and again. Such practices often results in redundant analysis scripts from these independent labs, which are often difficult to reproduce, due to: poor coding skills, lack of version control and manual implementation of certain tasks8. Moreover, additional data at all levels of science continues to accumulate at rapid rates and volumes9,10. However, the community lacks effective CI tools to harness such data sets. CI tools are also necessary to foster effective interdisciplinary interactions to advance the ‘team science’ research in a scalable, reproducible, sustainable, and efficient manner. Hence, scientists require guided knowledge discovery at various levels to help them navigate through relevant publications, tools, data sets, or even automate cloud resource configurations suitable to accomplish important research and education activities.
In this paper, we address the above challenges by presenting OnTimeRecommend, a novel recommender system, to spur research productivity and interdisciplinary collaborations in scientific communities. OnTimeRecommend system requirements are motivated by science application drivers within a CyNeuro science gateway11 for neuroscientists, and a KBCommons science gateway12 for bioinformaticians. In the example case of CyNeuro, users use CI resources (e.g., Neuroscience Gateway, JetStream, XSEDE, AWS) to integrate data, analytics tools, computing, and visualization with cyber and software automation. In the case of KBCommons, users use CI resources (e.g., CyVerse, XSEDE, AWS) for similar purposes. Through OnTimeRecommend integration, such science gateways can increase their effectiveness of serving novice/expert scientist needs that involve knowledge discovery from openly accessible publications or data sets, or leveraging CI resources via web services in workflow management within next-generation science gateways.
As shown in Figure 1, OnTimeRecommend comprises of multiple integrated recommender modules implemented as microservices that can be integrated into any science gateway in the form of a recommender-as-a-service. The recommender modules help scientists perform knowledge discovery through data mining of relevant publications, datasets. It can help guide training tasks through recommendations on Jupyter notebooks. In addition, it can help automate the deployment of distributed HPC resources for a given set of user CI needs. Further, it can help scientists find relevant scholars who can collaborate on scientific tasks where there are knowledge gaps that require multi-disciplinary expertise. To this end, OnTimeRecommend features specific recommender modules: Domain-specific Topic Recommender, Publication Recommender, Cloud Solution Template Recommender, Jupyter Notebook Recommender and Scholar Recommender. OnTimeRecommend can be customized within a science gateway through a set of support services to ensure the personalization and sustainability of the recommender modules. We detail the support service capabilities through descriptions of application programming interfaces (APIs) for creation of end-to-end application pipelines with distributed CI resources, and user interface functions that help in integration, execution and management of customization of the recommender modules. The guidance for use of the recommender modules in a science gateway is aided by a chatbot plug-in viz., Vidura Advisor. The Vidura is context-aware and features a user interface that can be embedded in a science gateway to helps users with step-by-step navigational support and interactive text responses. Based on users’ inputs in a questionnaire, Vidura uses a quadrant-based analysis that profiles user proficiency in a scientific domain as well as CI domain. Based on user’s intent, Vidura generates distinct responses from the various recommender modules to help users to accomplish research and educational tasks. The Vidura chatbot design is motivated by recent works on distributed cyber-robots for scientific workflows such as AVA13 and IRIS14, and leverages Google’s Dialogflow natural language processing framework15 to enhance conversational understanding and interaction with users.
FIGURE 1.
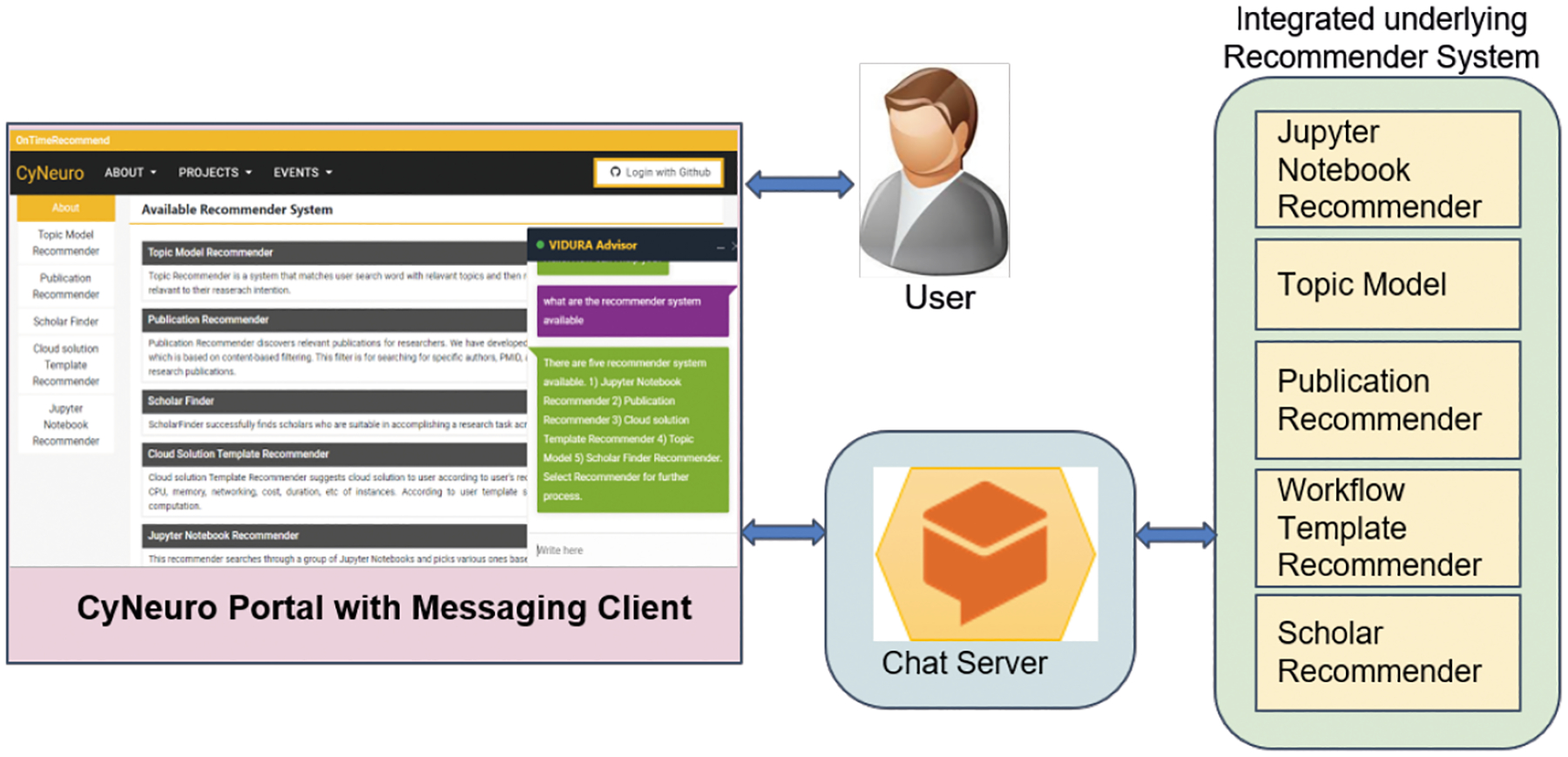
Recommender system interacting with CyNeuro portal through Vidura Advisor and chat server.
To validate our OnTimeRecommend, we integrate and show benefits for two exemplar science gateways, one in neuroscience (CyNeuro)11 and the other in bioinformatics (KBCommons)12. The neuroscience research use case involves simulation of a single cell neuron modeling in order to quantify its electrophysiological properties given specific model parameters. The bioinformatics research use case involves an RNAseq data analytics workflow management that uses Pegasus16 and HTCondor17 to automate various data processing steps. Through a usability study for both CyNeuro and KBCommons users, we demonstrate how the OnTimeRecommend with Vidura can interactively assist both novice and expert users in domain-specific knowledge discovery based on an profiling of users’ expertise level, their science gateway needs and available computing resources.
The remainder of the paper is organized as follows: Section 2 discusses related work. Section 3 presents our OnTimeRecommend system architecture, middleware design and the Vidura Advisor implementation. Section 4 details our integrated recommender modules in OnTimeRecommend that can be used in science gateways. Section 5 presents implementation and results from OnTimeRecommend integration in exemplar neuroscience (CyNeuro) and bioinformatics (KBCommons) science gateways. Section 6 concludes the paper.
2 |. RELATED WORK
2.1 |. User Guidance in Science Gateways
The idea of using a science gateway for easy access to HPC or High Throughput Computing (HTC) resources is not novel. However, creating a science gateway for local HPC/HTC resources at campus-level that is federated with national resources is an emerging concept in academia. In the past few years, many science gateways have been developed to provide users easier access to CI resources such as storage, computing, data analysis, and visualization. Some of the well known gateways that include SciServer18, UltraScan19, CyVerse20, TreeGenes21, and NeuroScience Gateway (NSG)22 address needs in research disciplines such as physics, chemistry, material science, and biology. However, most of these gateway platforms do not provide interactive guidance to assist users with their tasks needs in an intelligent and automatic manner. Science gateways need to support capabilities that allow users to easily and quickly execute workflows and algorithms or to use existing tools in a guided manner. Lack of necessary tools to provide such capabilities poses challenges for researchers, especially who are novice in a scientific domain (e.g., neuroscience, bioinformatics) or in the HPC domain.
In this work, we develop a novel collaborative recommender system with a chatbot guided knowledge discovery to over come these challenges in science gateways by providing user guidance on their queries through a step-by-step navigational support that fulfills the needs of novice/expert users. Our efforts are motivated by science gateways: CyNeuro11 and KBCommons12, which have been established to serve neuroscience and bioinformatics educators and researchers. These science gateway efforts are in collaboration with large-scale CI resource providers such as NeuroScience Gateway (NSG) and CyVerse.
2.2 |. User Profiling and Recommender System
The quality of recommendations could be improved with the usage of artificial intelligence (AI) as well as storing profile information of each and every user who interacts with the system. Some studies have demonstrated this point and provided insights into user profiling methods. Authors in23 created a keyword map based method to acquire user profiling based on their access to the learning materials and the processes of learning on a web based learning platform. Authors in24 developed a technique to predict user’s interests and preferences on a web platform by matching a user to other users with similar profiles. Also, authors in25 studied the changes in the patterns of user’s profile and interests over time, and developed an evolutionary view of user’s profile using historical data to predict the user’s profile in the future. In e-learning platforms, the user proficiency is one of the most significant factors that influences the absorption of learning content26.
In this work, a questionnaire related to domain knowledge is provided to the student to assess his/her knowledge level and a Mamdani approach27 is used to decide how much content should be presented to a student according to his/her intellectual level. We also demonstrate a conversational agent acting as a recommender. Further, we leverage an users’ profile to provide sustainable and scalable support in science gateways for neuroscience and bioinformatics use cases.
2.3 |. Automating Data Analysis with Chatbots
Recently, natural language processing has advanced to an extent that industry is widely enabling users to interact with virtual assistants such as Apple Siri, Amazon Alexa, and Google Personal Assistant to accomplish day-to-day life chores (e.g., playing music, appointment scheduling). Recent bibliometric study on chatbots28 reveals the scope of contributions in the state-of-the-art for chatbot applications in many domains with a high number of alternatives. This also confirms the novelty of our contribution to science gateways, since there is no prior work in this domain to the best of our knowledge. In recent development, a new category of chatbot enables natural language processing to map suitable commands for their execution to provide convenience for user’s requirements. An example is implemented in29,30,31 for a conversational visualization service to extract keywords from the conversation and to apply filters to the visualizations. Chatbots for data analysis workflow management such as AVA13 and more recently IRIS14 are promising solutions that motivate our work. The chatbot interface AVA is designed to work on Jupyter Notebooks, where users can interact with a bot to execute Python code and build their data model. In contrast, IRIS uses linguistic theory to handle complex requests interactively by combining commands through a nested conversation.
In this work, instead of just enabling users with a few pre-configured data workflows, our Vidura Advisor (chatbot) uses integrated recommender systems to generate custom responses and provides recommendations to users on resources, such as text, simulation tools, related publications, datasets, experts, and cloud resources. Vidura also guides users through the execution of their workflows by answering user queries in a step-by-step manner during the entire process. A major contribution of our approach is the capability to analyze users’ proficiency based on initial conversation, and then generate customized responses based on evaluation of their expertise levels.
3 |. THE ONTIMERECOMMEND SYSTEM
Recommender systems with strong algorithms are at the core of today’s most successful online companies such as Amazon, Google, Netflix and Spotify. With the growing amount of information on the Internet and with a significant rise in the number of users, it is a difficult task for science gateways to search, map and provide the users with the relevant information according to the users’ preferences and needs. In our OnTimeRecommend system, we have developed and integrated several recommender modules with functionality to serve target users with access to data and computing resources through a knowledge-driven approach that is based on their needs and preferences. By providing recommendations that suit users’ requirement, our integrated recommender modules provide a personalized and expert guidance across diverse computing platforms and data archives. The OnTimeRecommend design features a multi-layer recommender architecture as shown in Figure 2 to enable novice/expert user interfaces for knowledge discovery within science gateways, as well as template-driven control of federated CI resources.
FIGURE 2.
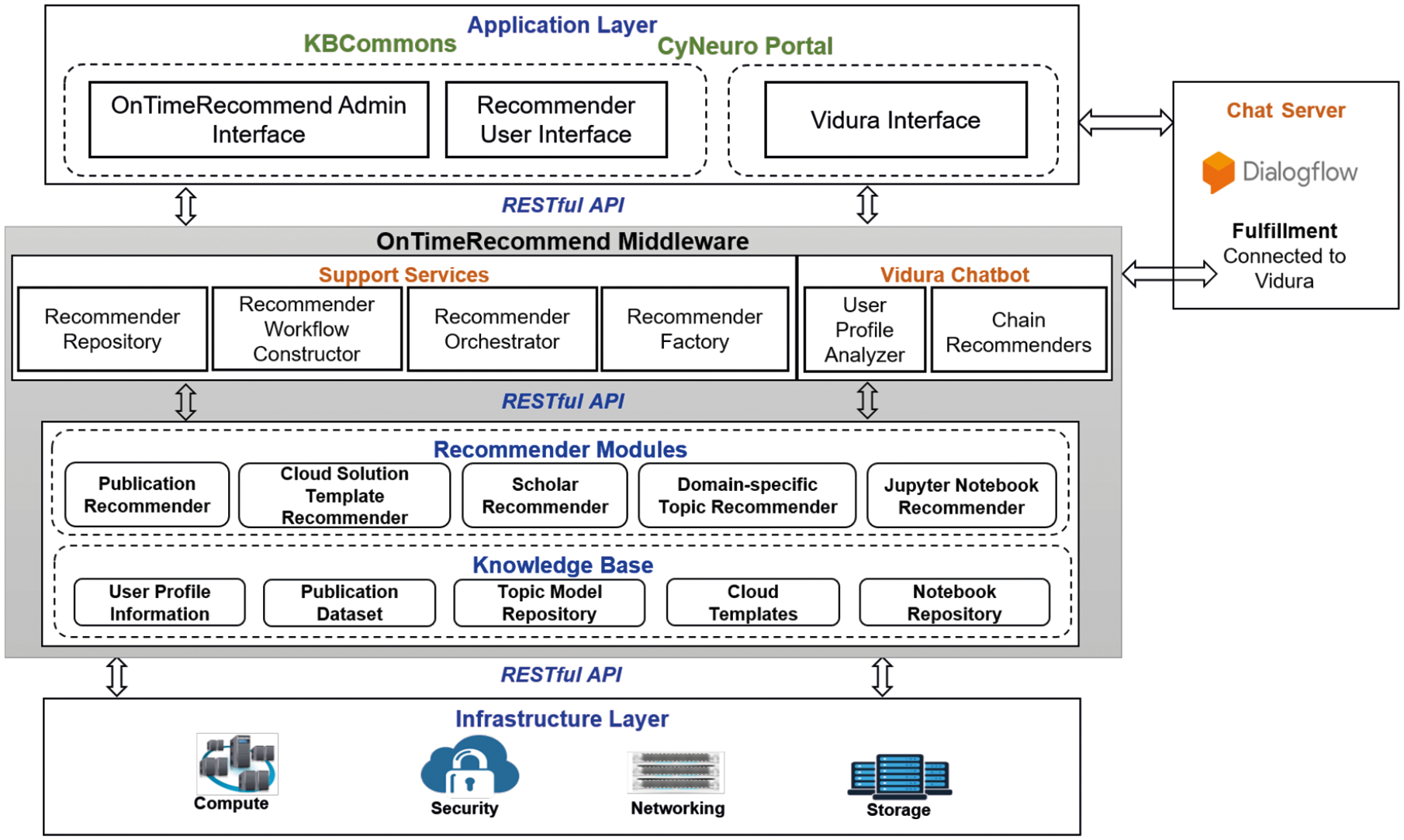
Multi-layered OnTimeRecommend architecture with application-facing and infrastructure-facing RESTful APIs.
3.1 |. System Architecture
The OnTimeRecommend system comprises of a four-layer architecture that includes: Application layer, Middleware layer, Recommender layer, and the Infrastructure layer. These layers are interconnected and communicate to other layers via RESTful APIs during users’ specific tasks/processes in the system.
Application Layer:
Using science gateway portals, scientists build custom interfaces and controls to submit jobs and query results in their workflows at the Application Layer. OnTimeRecommend user interface provides a central resource hub via a web application to enable guided user experience to domain novice/expert researchers. OnTimeRecommend Admin Interface capability provides the ability to integrate and manage recommender modules with relevant support services. The Recommender User Interface provide recommendations through RESTful web services on various resources (publications, tools, datasets, experts database, cloud resources) using Support Services of underlying integrated recommender modules. Context-aware Vidura Advisor interface provides intelligent guidance using the Google Dialogflow and related Vidura services.
OnTimeRecommend Middleware Layer:
OnTimeRecommend prototype was designed to provide microservices to the Application and Infrastructure Layers via RESTful APIs. The OnTimeRecommend middleware features several customizable web services that seamlessly interface with the Application Layer, Recommender Modules and the Infrastructure Layer to accomplish user’s tasks. Details of the OnTimeRecommend Middleware Layer are presented in Section 3.2.
Recommender Module Layer:
The novelty of our proposed OnTimeRecommend is that it integrates existing recommender modules with relevant knowledge bases to serve the Application Layer by leveraging resources at the Infrastructure Layer in a seamless manner, as shown in Figure 2. The Recommender Modules features customizable modules (based on application needs) that include: (i) Domain-specific Topic Recommender, (ii) Publication Recommender (iii) Cloud Solution Template Recommender, (iv) Jupyter Notebook Recommender, and (v) Scholar Recommender. Table 1 provides details for integrated recommender modules such as use cases, algorithm details, expected (sample) input parameters, and sample recommendation outputs. Each individual recommender module has its own knowledge base with datasets used for training machine learning/deep learning algorithms and providing recommendations based on predicted classification results. The knowledge base and algorithms of all the recommender modules will be explained in more detail in Section 4.
TABLE 1.
Summary of Recommender Modules integrated within OnTimeRecommend Middleware.
| Recommender Module | Motivating Factors | Output of Recommender | Nature of User Input | Algorithm |
|---|---|---|---|---|
| Cloud Solution Template Recommender32 33 | Due to problems using distributed resources, limited CI skills, distributed infrastructure deployment is manual and guesswork prone | CI Templates (node type, CPU, memory, GPU, networking specifications) | User specification regarding application type, HPC site, performance, cost expectation | K-Nearest Neighbor & Integer Linear Programming |
| Domain-specific Topic Recommender34 | It is hard to keep up with growing literature, and rapid development of analysis tools | Datasets, software tools; correlation between topics, trend analysis | Scientific topics, text corpus / publication archives | Hierarchical Bayesian model |
| Publication Recommender35 36 | Users have difficulty in rapid search of domain-specific publications | Articles, potential collaborators, social network connections | Search keywords | Content-based filtering, deep learning |
| Jupyter Notebook Recommender37 | Users want just-in-time training to familiarize themselves with skills using relevant Jupyter Notebooks | Jupyter Notebook for training | Keywords of learning topic | Document indexing & query |
| Scholar Recommender38 | Users want to find scholars who could potentially collaborate in a multi-domain research task | List of scholars with Top 3 scholars and their expertise | Scientific topics and keywords | Deep generative model-Variational Autoencoder |
Infrastructure Layer:
The Infrastructure layer mainly provides computing, storage, and networking resources to support the workflow needs of data-intensive applications. Our OnTimeRecommend system can be hosted in any cloud platform e.g., it can be hosted on the AWS cloud, and a science gateway application (such as e.g., CyNeuro) instances can connect to the recommender modules via REST web service calls.
3.2 |. OnTimeRecommend Middleware
The recommender modules can guide researchers and educators to discover relevant resources, enhance interdisciplinary knowledge sharing and help collaboration match-making. The implementation of OnTimeRecommend provides capability of ‘recommender as a service’ to the science gateways. For the implementation, we followed the software design of a leading science gateway, “Apache Airavata”39. Airavata provides a software suite to compose, manage, execute, and monitor large-scale applications and workflows on computational resources. By following their design principles, we implemented Support Services of OnTimeRecommend to provide a user interface and recommender microservice40 suite to integrate, manage and execute different recommender modules, as well as configuring machine learning pipelines on cloud resources through the microservices.
The recommender modules can be developed in different languages or scripts (such as Java, Python), and library packages (such as MATLAB, R). The process of integrating and running a recommender module on a science gateway through OnTimeRecommend middleware consists of a number of steps: registering the recommender module; preparing the environment; downloading & processing of datasets; training recommender module with a suitable model parameters; handling errors and exceptions; and generating & and transferring a knowledge base to a desired location or repository; and finally running the recommender module to provide recommendations to user queries through a science gateway user interface.
These steps are mainly components of a machine learning pipeline for individual recommender modules. Execution of these processes requires different algorithms implementation, knowledge bases, dataset files, command-line arguments, and environment variables. The outputs include recommendations in JSON format as well as on the UNIX standard output and error. The OnTimeRecommend uses the microservice architectural style to decouple services in the form of a set of small services that can run as separate processes. Major benefits of using the microservice architecture is the modularization of the application to ensure agility, flexibility, portability and scalability. Our approach leverages this feature by dividing the entire machine learning workflow into different processes for individual recommender modules and implementing microservices for each process. Moreover, we have implemented microservices that follow the standard practices of RESTful API design for each of the steps for researchers to customize the recommender modules. Using these APIs, we generate software package in Java and python as part of the Support Services to integrate OnTimeRecommend in a science gateway. Science gateways accesses recommender modules through these OnTimeRecommend middleware APIs.
In the following, we detail how the Support Service APIs connect different the components and processes of recommender modules in OnTimeRecommend with a science gateway:
Recommender Registry: It is the repository for all recommender modules related metadata. This includes descriptions of recommenders, science gateway client details, OnTimeRecommend middleware customization details. Our current implementation of this registry uses the Hibernate framework for object-relational mapping over a relational database i.e., a MySQL backend. This registry is designed and developed to efficiently demarcate individual recommender modules for any science gateway. This registry metadata includes information such as recommender module related: knowledge base details, algorithm details, and request/response parameters; configuration to automate machine learning workflows for recommender modules, and recommender modules executions; and, user profile information for capturing user quadrants for the Vidura Chatbot.
Recommender Workflow Constructor: It is used to create recommender module workflows for OnTimeRecommend. The entire machine learning workflow/pipeline building and execution can be broken down into multiple processes such as gathering data, processing data, modeling recommender, generating knowledge base, and run recommender model. For all the processes, we implemented microservices that interact with the OnTimeRecommend middleware to configure and execute workflows for specific recommender modules. Example details of microservices for one of the recommender modules i.e., Domain-specific Topic Recommender is described in Table 2. A client component interacts with the Recommender Registry for persistently storing workflow details, metadata related to workflow execution parameters of the recommender module processes.
Recommender Orchestrator: It configures the queuing layer for OnTimeRecommend. More specifically, it configures execution parameters for different processes of a given ML pipeline. Subsequently, it uses RabbitMQ to queue an entire ML pipeline by queuing all processes. This component is also responsible as a client to the Registry for persistently storing all parameter configuration and scheduling requests for processes in a queue format.
Recommender Factory: It is responsible for execution of an ML pipeline and user recommendation requests. It interacts with the recommender modules through microservices generated by Recommender Workflow Constructor for workflow processes execution of individual recommender modules. This component is responsible as a client to the Recommender Orchestrator and invokes queue service execution for workflows by using specified configuration parameters. This components also provide metadata information to dynamically generate user interface related information on a science gateway. After a knowledge base is generated, the Recommender Factory provides recommendations to users on science gateway user interface using RESTful API of specific recommender modules. Our approach is to provide a central user interface for the recommender systems that can be leveraged using REST APIs that follow the standard practices of RESTful design. Examples of RESTful APIs for individually integrated recommender modules in OnTimeRecommend are described in Table 3.
TABLE 2.
Microservices for Domain-specific Topic Recommender module in OnTimeRecommend.
| Workflow Process Name | URL | Request Parameters | Response | Detail |
|---|---|---|---|---|
| Data Collection | /services/topicmodel/datacollection | {“domain”:String} | Publication dataset generated | Dataset generated by collecting papers from different websites for specific domain |
| Data Processing | /services/topicmodel/dataprocess | {“domain”:String, “operation”:vocabulary|bag-of-words|tool|dataset} | Generated bag-of-words, vocabulary, tool-to-doc, dataset-to-doc | Raw text dataset transformed into bag-of-words format, generated vocabulary, tool-to-doc, & dataset-to-doc tables |
| Modeling Recommender | /services/topicmodel/modelinference | {“data_source”:String, “mode”:demo|est|inf “run_mode”:String, “num_iterations”:Int, “num_topics”:Int, “save”:String} | Trained knowledge base generated | Parameters estimation used to estimate latent variables and pattern between variables. Gibbs sampling algorithm used to infer latent patterns |
| Recommendations to user queries | /service/topic-model/recommendation | {“text”: string} | [{“id”:int, “summary”:string, “tools”:string, “datasets”:string},{..}] | Receives topic as query input from end-user & returns JSON array of recommended topics that contain topic id, summary, tools &datasets that are relevant to the topic |
TABLE 3.
RESTful web services for the individual recommender modules in OnTimeRecommend.
| Recommender Modules | URL | Methods | Parameters | Response | Description |
|---|---|---|---|---|---|
| Topic Recommender | /service/topic-model/recommendation/ | POST | “text”: string | [{“id”:int, “summary”:string, “tools”:string, “datasets”:string},{..}] | Receives topic as query input from user and returns JSON array of recommended topics that contain topic id, summary, tools and datasets that are relevant to the topic |
| Publication Recommender | /service/publication-model/recommendation | POST | “type”:“title|author|pmid”, “keyword” : string | [{“id”:int, “abstract”:string, “pmid”:string, “title”:string},{..}] | Receives title as query input from user and returns JSON array with pubMed ID, abstract, title, authors |
| Jupyter Notebook Recommender | /service/jupyter-recommender/recommendation/ | POST | “keyword”: string | [{“filename”:string, “cell_no”:string, “filepath”:string},{..}] | Receives text as query input from user, searches & returns JSON array with Jupyter Notebooks related to a given keyword |
| Cloud Solution Template Recommender | /TestingWeb/rest/getTemplateCatalog | POST | “app_type”: string, “site”: string, “memory”: string, “cpu”: int, “networking”:string | {“red”:[{“cloudprovider”:string, “duration”:int,“cost”:int}],“green”:[{..}], “gold”: [{..}]} | Receives cpu, memory and other functional parameters as query input from user and returns JSON array of CI resources divided in red, green, gold templates with expected cost |
| Scholar Recommender | /service/scholar-finder/recommendation/ | POST | “domain”: string | [{“name”:string, “coordinates”:Array, “expertise”:Array},{..}] | Receives research task as query input from user and returns JSON array of scholar recommendations that are relevant to perform the task |
Any recommender module can be integrated on science gateway interface using ‘recommender-as-a-service’ capability on the OnTimeRecommend platform. Our middleware supports two major roles: end user scientists and middleware/gateway administrators. The steps for a middleware/gateway administrator are the following:
Step-1: register different recommender modules in OnTimeRecommend using the web application interface. This interface uses ‘Recommender Registry’ services to register information related to recommender modules for a science gateway.
Step-2: register scientific plug-in processes such as data collection/processing parameters, and knowledge base, such that all the necessary information is provided to execute relevant recommenders in OnTimeRecommend using the ‘Recommender Workflow Constructor’.
Step-3: add a science gateway (i.e., CyNeuro in this work) client and link recommender modules with the client to allow users to use recommenders on their science gateway interface.
Step-4: configure parameters of recommender processes and queue processes of recommender modules for a specific science gateway client.
Step-5: execute all recommender processes using provided configurations and publish recommender outputs to end users on their science gateway interface.
3.3 |. Vidura Advisor
A context-aware chatbot viz., Vidura Advisor is integrated with the OnTimeRecommend through Vidura Services to provide guided user interfaces i.e., step-by-step navigational support to use the recommender modules. The Vidura Advisor implementation is divided into three different components: Vidura widget on User Interface, Google Dialogflow - Chat server, and Vidura Services as shown in Figure 2. Vidura generates distinct responses for users based on their proficiency and intent. Two components in Vidura provide the functionality: One is the User Profile Analyzer that is used to categorize user profiles into quadrants (Quadrant 1 ‘Novice users’, Quadrant 2 ‘Domain Experts’, Quadrant 3 ‘HPC Experts’, Quadrant 4 ‘Domain and HPC Experts’). We also use the updates of user profile information as feedback to improve the recommendations. Another component is the Chain Recommenders that executes chaining of microservices of different recommender modules (output of one influences the input of the other) to use a series of recommenders based on user intent and profile.
Throughout a user’s workflow on our system, the user interacts with a context-aware chatbot, which is embedded within OnTimeRecommend and provides guided user interface, step-by-step navigational support, and generates distinct responses for users based on user’s proficiency and intent. By utilizing concept of our previous work27 to determine user’s proficiency, we developed a questionnaire related to HPC/CI in the neuroscience and bioinformatics science domains to evaluate the user’s overall proficiency. For this purpose, we used many-valued logic in the fuzzy approach41. Fuzzy logic can capture an accurate and detailed representation of the questionnaire information, thereby providing a realistic analysis of the real-time user data. Intuitionistic fuzzy logic42 extends fuzzy logic and deals with uncertainty. It assigns each element a membership degree and a non-membership degree and allows assessment of the elements by membership and non-membership functions that belong to the real unit interval [0, 1] with the sum also belonging to the same interval. It helps to move closer to human thinking and work with linguistic variables and terms. We developed our Vidura chatbot based on SG challenges and user requirements for multi-disciplinary neuroscience and bioinformatics applications, and categorize users into the following four user quadrants: Quadrant 1: ‘Novice users’ with limited knowledge of neuroscience or bioinformatics domain and the computational resources. For example, typically IT/CS freshmen; Quadrant 2: ‘Domain Experts’ have a high domain or subject knowledge, but have limited knowledge about SGs and HPC resources. For example, typical neuroscience or bioinformatics Master students; Quadrant 3: ‘HPC Experts’ have low domain or subject knowledge but have high computational resource usage proficiency. For example, IT/CS Master Students; Quadrant 4: ‘Domain and HPC Experts’ have expertise in domain/subject as well as the usage of computational resources. For example, neuroscience or bioinformatics faculty and PhD students.
Our Vidura Advisor uses an attached knowledge base and fuzzy logic implementation as part of a questionnaire process to evaluate the proficiency of users in a specific science domain, and utilizes set of pre-defined set of rules created to drive the questionnaire. The questionnaire features questions to evaluate the proficiency of users in the HPC domain and the neuroscience or bioinformatics workflow domains. Example knowledge base dataset contains multiple choice questions, with 7 in the HPC/CI domain and 7 each in the bioinformatics and neuroscience workflow domains. The knowledge base also stores the user responses to these questions as part of the user profile data. Example questions involve “Rate your familiarity/understanding about” each of the items that are listed in Table 4. Each question is measured on a four-point Likert scale, ranging from: (i) little to none, (ii) below average, (iii) above average, and (iv) mastery. While answering the questionnaire, the user can assign a weight between 0 and 1 for every option in each question to convey their respective degree of vagueness in the option selection. The sum of weights given by a user of all options for each question should not exceed 1. After the user has completed the questionnaire, we calculate the sum of weights of all options from all questions belonging to a specific category. For example, we add weights of option 1 and option 2 to find the ‘Novice’ membership value. Similarly, we add weights of option 3 and option 4 to find ‘Expert’ membership value. We have implemented details about this procedure of categorizing users into the four quadrants. We have implemented a “User Proficiency Analyzer” to categorize users into the four quadrants and store user profile data.
TABLE 4.
Example Questionnaire for HPC, Neuroscience, and Bioinformatics Domains to determine the User Quadrant.
| Example Questionnaire for HPC/CI Domain | Example Questionnaire for Neuroscience Domain | Example Questionnaire for Bioinformatics Domain |
|---|---|---|
| National high-performance computing(HPC) resources such as XSEDE(Extreme Science & Engineering Discovery Environment) | Concept of time constant as it relates to Neuron cell membranes | Concept of conducting transcriptomics RNAseq or single cell RNAseq (scRNAseq) analysis |
| Using Linux Operating System like (Ubuntu, Fedora, Redhat) | Ionic currents give rise to an Action Potential | Concept of gene expression analysis for various experimental conditions |
| Proficiency in installing required software for your re-search simulations on local machine | Neuron modeling using the Hodgkin-Huxley formulation | Proficiency in using NGS experimental datasets |
Our Vidura implementation uses Google Dialogflow, which is a conversational artificial intelligence service that performs natural language processing to find the intentions in the user queries. Google Dialogflow has the ability to create intelligent agents that hold definitions for various functions involving ‘intents’, ‘entities’, ‘knowledge bases’, and ‘fulfillments’. Intents are used to categorize the user intentions for one conversational turn with the agent. Multiple intents can be constructed and combined to develop fluency in conversations. The Dialogflow agent trains each intent to match the end-user’s expression by using a rule-based grammar that is capable of correcting, then categorizing syntactic structure of a sentence or query–and ML matching algorithms for giving indicative responses. We utilized a pre-trained agent of Dialogflow, and re-trained the agent by following the prescribed setup process to create intents for the user dialogue around our recommender module entities such as templates, notebooks, publications, tools, and datasets. As shown in Figure 3, Dialogflow allows the use of external web services to generate custom responses by using the ‘fulfillment’ feature, which then allows Dialogflow chat server to interact with our fuzzy-based conversational recommender to recommend resources to the users. Further, Vidura Advisor calls REST APIs based on the prediction of the user’s intent to connect to a specific recommender module that provides recommendations and displays responses to the users through the fulfillment feature of Dialogflow. After receiving Vidura Advisor response, the users will follow the guidance to acquire the information they need or continue with their workflow tasks.
FIGURE 3.
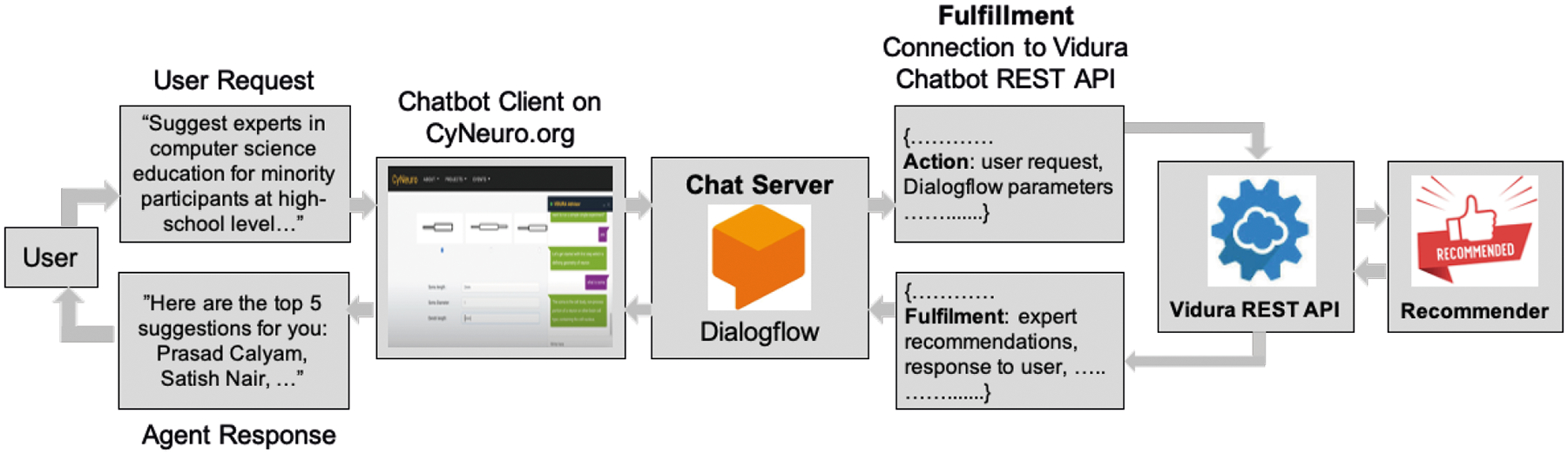
Workflow logic of the Vidura Advisor, which seamlessly integrates the components of an example use case, the CyNeuro science gateway. The workflow begins with an accept of a user’s request from the web portal, which is then analyzed to understand user’s intent using the Dialogflow algorithm. Finally, a customized response is generated for the interactions of the Vidura REST API and the corresponding underlying recommender module(s).
Vidura Advisor has learning update capabilities based on user profiling and intent prediction that is possible through the Dialogflow integration, and through the additional training data collected in the knowledge base for either the neuroscience or bioinformatics domain. During our workflow handling, the OnTimeRecommend system keeps records of all users’ session information as well as the input requests along with any prior user-specific recommendations. This meta data is updated in an on-going basis within the knowledge base and is used for re-training of the Vidura Advisor for the quadrant analysis and users’ intent analysis. In the context of the recommender modules that support the Vidura Advisor, individual knowledge bases (i.e., trained models) of recommenders are updated separately on a periodic basis in order to keep up with the most current scientific knowledge for more relevant recommendations.
Once a user quadrant is identified based on the questionnaire, Vidura executes microservices for “Chain Recommenders” functionality in the chatbot interface. The chaining allows a series of recommendations from the recommender modules to be output for the user to answer specific queries based on user’s profile. In this context, we use Google Dialogflow Fulfillment feature to integrate custom dynamic responses from the OnTimeRecommend system. For a novice user’s (Quadrant 1) query on “Help me with the publication and tools for neuron simulation”, Vidura first identifies user’s intent (e.g., here user wants to get information on entities ‘tools’ and ‘publications’ for the intent of ‘neuron simulation’) using Google Dialogflow APIs. Based on this identificaiton, it provides a custom text response. In addition, based on intent and entities details from Dialogflow APIs, Vidura executes “Chain Recommenders” functionality in the chatbot interface to internally execute publication and domain-specific topic recommenders. Such a chaining provides recommendations for knowledge discovery on e.g., trends of tools within relevant publications for neuron simulation over a period of several years in the past.
4 |. RECOMMENDER MODULES
In this section, we detail the recommender modules that are integrated into our OnTimeRecommend system to support user queries in science gateways.
4.1 |. Domain-specific Topic Recommender
The Domain-specific Topic Recommender guides the novice/expert users to identify topics, tools, and datasets that are generally used in specific domains, such as bioinformatics and neuroscience domain sciences, from existing databases. It can also analyze the latent correlations between topics spanning the different application domains. This helps researchers to get started on research topics without having to manually sort through a large amount of information.
The core logic for Topic Recommender is to provide suggestions relevant to research topics and then recommend tools and datasets for each topic based on the user’s search term, as shown in Table 3. The Vidura chatbot integrated within the OnTimeRecommend, shown in Figure 4b, leverages a domain-specific topic model in answering researcher questions such as e.g., What are the best tools to handle particular modeling problems with high accuracy? Which types of datasets have been used previously to evaluate a certain hypothesis? Data used in the domain-specific topic model has to be trained by parsing through published scientific papers (e.g., in neuroscience and bioinformatics) and this allows for discovery of the relationship among the topics, tools, and datasets. The Topic model recommender implements a novel “domain-specific topic model” (DSTM), which is a hierarchical Bayesian model that extends the Latent Dirichlet Allocation (LDA) model for text modeling and classification. It also uses a Markov Chain Monte Carlo (MCMC) algorithm to infer latent patterns within a specific domain in an unsupervised manner34. For training and testing of our ML model, the knowledge base of this recommender contains over >20,000 full papers that have been published in well-known journals in computational neuroscience and bioinformatics domains for the years 2009 to 2019. Each paper was represented as a “bag of words” in our model. The full papers provided a vocabulary size of over 2.5 million word tokens. For tools, we collected the most commonly used tools in neuroscience and bioinformatics domains that cover a wide range of research efforts in computation, simulation, database and visualization. For example, we used common tools such Matlab, Python, TensorFlow, R, Python, Neuron, BLAST, and Pegasus. Our knowledge base includes ~50 tools for neuroscience domain and ~70 for bioinformatics domain. In addition, we have collected >170 datasets used in neuroscience experiments and >45 datasets used in bioinformatics sequencing analytics and benchmarking. Given that computational and data intensive science is expensive and time consuming, availability of domain-specific topic models to provide useful guidance through data mining of massive open information within a domain or across domains can significantly benefit domain scientists.
FIGURE 4.
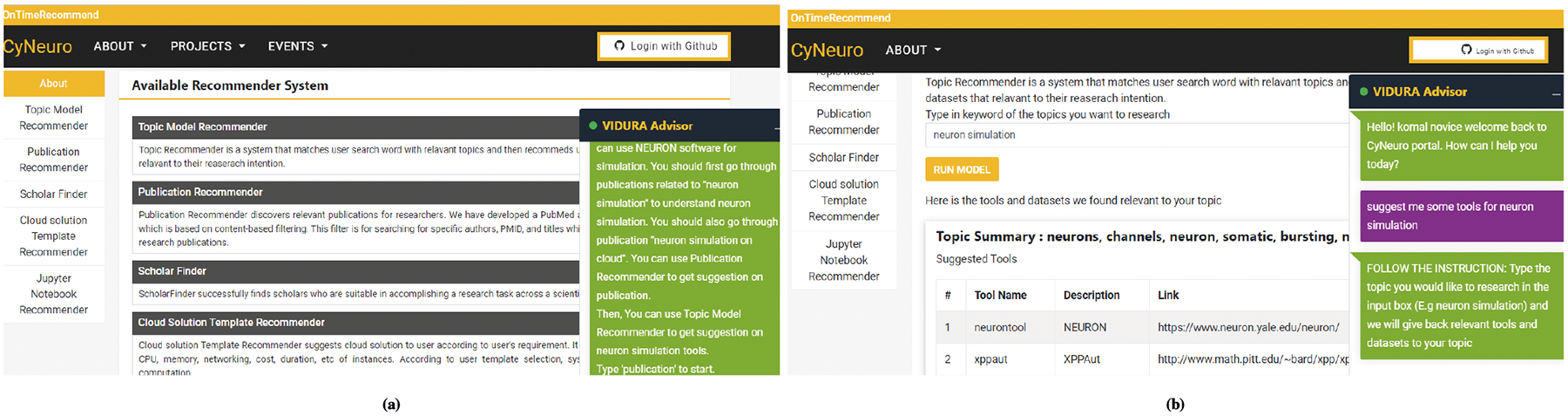
Example CyNeuro UI of: (a) Vidura Advisor interface to utilize the different recommender modules, (b) Domain-specific Topic Recommender inputs/outputs guided through the Vidura Advisor interface.
4.2 |. Publication Recommender
Finding relevant publications is essential for scientists who have to cope with exponentially increasing numbers of scholarly material. However, discovering relevant publications for researchers is a non-trivial task. The publication recommender in our OnTimeRecommend system can reduce the effort required to find relevant publications.
Our Publication Recommender35 36 suggests most relevant publications that are related to the user’s query in terms of PMID (PubMed ID) / author/ title. This recommender is based on an efficient hybrid model using term frequency and inverse document frequency43, chi-square feature selection44 and softmax regression. In this recommender, pretrained word2vec is used to construct the start-up feature space, followed by a deep convolutional neural network (CNN) that was constructed to achieve a high-level representation of abstracts. Further, we adopted a fully connected softmax model to recommend the best journals to the user. The knowledge base of this recommender contains ~880,000 collectively downloaded papers from over 1,130 journals from PubMed Central for the years 2007 to 2017. The abstracts and full texts of all these papers were used to induce vectors of words using word embedding methods and the word2vec tool. Thus obtained word vectors representing abstracts were input to a deep CNN model to extract semantic information. Finally a fully connected softmax layer was used for classification and providing publication recommendations. Our publication recommender functionality steps consists of: (i) user interface to obtain the input data and present recommendation results to the user; (ii) data preprocessing to perform data filtering and word segmentation; (iii) abstract representation to embed abstracts with pre-trained word vectors that are fed into a CNN model to achieve more abstract representation; and (iv) classification and ranking phase to finally classify output and provide recommendations.
By using the publication recommender, a user can find out about researchers and faculty who are doing similar work with the possibility for future collaborations. The recommender system takes a user’s input with an author, title, or PubMed reference number (PMID) using the REST service via CyNeuro recommender module UI, and then finds the best results that match the keyword. The RESTful service returns the result as JSON and the UI displays the list of publications to the user, as shown in Table 3. Our publications repository is stored on a MongoDB Atlas Cloud database. After the user receives the recommendations, the user could click on a title link to read full version of a publication.
4.3 |. Jupyter Notebook Recommender
Jupyter Notebooks45 are shared documents that contain computer code, equations and rich text elements, which can be run to perform data analysis and visualization. Our Jupyter Notebook Recommender27 takes as input of a list of keywords that a user provides, and searches through all the notebooks in a repository to extract the visible text from it. The Jupyter Notebook Recommender uses an embedded Whoosh Python module to index the notebook documents in the database, and then queries the index by using the user-provided input parameters. It uses a standard Term Frequency and Inverse Document Frequency (TF-IDF) statistics to score and sort the search results by ranking the importance of a keyword in a notebook document. The recommender module finally returns the candidate notebooks with the highest TF-IDF scores for a given user input. The recommender can disseminate code and improve scientists’ productivity by appropriate notebook suggestions. The user inputs a keyword that is sent to the REST service as shown in Table 3. The REST service searches through a group of Jupyter Notebooks and finds cells where the keyword is located. The recommender returns a JSON array of notebooks and the cell number where the keyword is found as output. The resulting Jupyter Notebooks are made downloadable by the API.
The REST service in this recommender essentially comprises of two parts. One is the web service that parses an incoming request and uses python search engine. A search engine using Python Whoosh that indexes the Jupyter Notebooks and performs extensive search queries based on the request keyword from the user. The second part is the file server hosting the Jupyter Notebooks. In order to allow the user to see the notebooks recommended to them, the notebook files are hosted as static files along with the API. By hosting them in the same location, any updates or additions made to the list of notebooks will be reflected back to the API.
4.4 |. Cloud Solution Template Recommender
Lack of ‘repeatable’ and ‘reusable’ cyberinfrastructure (CI) resource templates along with unique capabilities of multitude of heterogeneous components makes traditional infrastructure deployment practices manual and guesswork prone. This in turn leads to sub-optimal infrastructure configurations, which impacts user experience when performing complex analytic workflows. Our Cloud Solution Template Recommender suggests CI resources such as federated cloud infrastructure templates, which users can customize as per their simulations and workflow requirements. The main advantages of using such a template is that it provides the ability to ‘re-use’ or customize these artifacts and ‘re-purpose’ them for building a whole new cloud solution.
This Cloud Solution Template Recommender uses k-Nearest Neighbors (KNN) Machine learning algorithm and Integer Linear Programming (ILP) to recommend a suitable cloud template based on user requirements32 33 (both functional and nonfunctional requirements).It features a Component Abstraction Model to implement intelligent resource ‘abstractions’ coupled with ‘reusable’ hardware and software configurations in the form of “custom templates” to simplify heterogeneous resource management efforts. In this recommender, an offline initial recommender module improves the user productivity by narrowing down cloud resource compositions to the most appropriate options. Subsequently, an online iterative recommender module regularly monitors the application behavior, and dynamically provides automated resource adaptions based on user application demands. Recommendations seek to match the user requirements to available resources, and thus assist novice/expert users to choose between a narrow list of configuration selections with a suitable CSP.
We evaluate the recommender in a bioinformatics science domain with a catalog of application workflows through consideration of four CSP resources (i.e., Amazon AWS, Google GCP, Microsoft AZURE, and NSF GENI cloud) featuring a knowledge base with more than 300 different instance configurations, including computing resources such as CPU, memory, storage, and network bandwidth. The instances used in knowledge base are discrete instances and are available on pay as you go basis. A user should provide functional information about CPU, memory, networking, cost, duration etc. as per user’s application requirements and assign resources for computation, which are passed to the recommender API. The recommender takes user’s inputs and offers a list of cloud solution templates a JSON array that meet desired non-functional requirements of users (e.g., performance, cost), as shown in Table 3. The recommended templates are divided into three groups: Red Solutions provide templates with strict user defined constraints; Green solutions are templates with amplified user defined resources up to threshold limit; and Gold solutions provide templates which has priority user defined resources amplified up to threshold limit, as illustrated in Figure 5a. The user can then choose from these templates based on their preferences of performance and cost. The selected cloud template will be deployed in real time and the custom user workflow will be launched on the deployed resources. The user has the facility to monitor status of the deployed cloud resources as well as the workflow on the science gateway e.g., on the CyNeuro portal.
FIGURE 5.
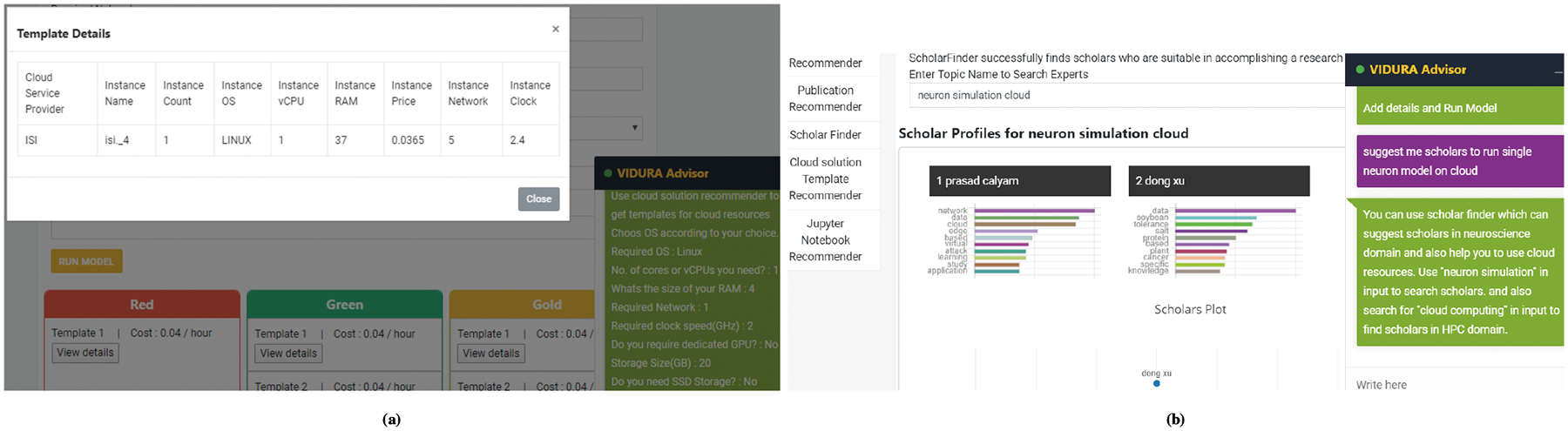
Example CyNeuro UI of: (a) Cloud Solution Template Recommender, and (b) Scholar Recommender.
4.5 |. Scholar Recommender
Research today involves working on bold problems that require experts developing solutions that are multi-disciplinary in nature. Existing methods to identify relevant research experts/collaborators are based on specific domains or high-level interests by querying existing databases. However, these methods do not always provide suitable recommendations of experts for specific multi-domain research tasks. Our novel Scholar Recommender38 addresses this problem by recommending scholars in the similar research field for potential collaboration opportunities.
The Scholar Recommender uses a deep generative model such as a Variational Autoencoder (VAE) to embed knowledge that is learned from authored publications. Latent knowledge representations of scholars are used to discover an individual scholar who is qualified to execute tasks that require proficiency in certain domains. The recommender uses a new ‘Knowledge Embedding’ technique to learn/quantify scholars’ expertise that is then used to generate recommendations for a given research task. Our model also uniquely uses a ‘follow-the-money’ approach that involves expert recommendations based on analysis of a corpus of competitive funding awards evidence that strongly reflects the latent expertise. Our knowledge base of this recommender contains a large collection of NSF (National Science Foundation) grant awards dataset collected over the last twenty years that contains more than 20,000 award records with project abstracts and names of 15,074 scholars who received grants. Our Scholar Recommender has five major components in the system architecture: (i) web crawler that will extract publication abstracts from Google Scholar using RESTful APIs for building knowledge embedding for scholars. Users can custom define the policy that decides when to update datasets for capturing the latest knowledge about candidate scholars; (ii) data pre-processing and (iii) data generation that use basic natural language processing algorithms to pre-process text datasets (such as, lowercasing, the stop-words removing, lemmatization and tokenization) in order to generate target dataset structures for training with e.g., bag-of-words, sequence; (iv) deep generative learning model that serves as a core component in our model for checking whether a candidate scholar is suitable for a certain multi-disciplinary research task or not; and (v) visualization is used to drill-down on the performance results using plots of e.g., embedding in a 2D space, and monitoring the model training status (such as plotting the loss or checking the reconstruction errors).
The user provides input query, i.e., research task to our model via chatbot that is passed through the REST web services. The research task, i.e. the user query, is translated to numerical format and fed to our model. Our model then returns a list of top N scholars who are suitable to execute the task. Such knowledge generation uses a generative model in order to recommend scholars with top three scholars in a particular domain. As shown in Figure 5b, a scholar map using scholars coordinates can be visualized based on scholar’s knowledge through their publications and funding records that are publicly accessible. The new generated knowledge thus helps a science gateway user to compare scholars that have the potential to generate new knowledge with a collaboration task based on multi-disciplinary expertise.
5 |. EVALUATION STUDIES
In this section, we discuss in detail - how OnTimeRecommend can be used into two exemplar science gateways. First science gateway relates to CyNeuro11 in the neuroscience domain, and the second science gateway relates to KBCommons12 in the bioinformatics domain. For both these science gateways, we also detail the usability studies we conducted with science gateway users on the Vidura integration as well as the customization of recommender modules of the OnTimeRecommend system.
5.1 |. OnTimeRecommend Use Cases in Exemplar Science Gateways
5.1.1 |. Neuroscience Use Case
In this section, we illustrate an exemplar computational neuroscience use case that shows utility and benefits owing to our OnTimeRecommend system. For a neuroscience researcher who wants to build biophysical realistic single cell model, they typically have to perform extensive parameter searches to develop a model cell using biologically measured properties. We have two methods that can help researchers accomplish such a parameter search.
The first part is to simulate a single neuron model and quantify its electrophysiology properties given specific model parameters. The user can use the Publication Recommender to discover that a Hodgkin-Huxley type of neuron model can be used as an example. A corresponding literature search reveals the parameters such as membrane capacitance, maximum conductance and reversal potentials of channels, etc., that determine the electrophysiological properties such as resting membrane potential, membrane time constant and rheobase current, etc. Next, the user inputs model parameters such as gbar_na, gbar_k, gbar_leak to the Juypter Notebook Recommender, and relevant notebooks will returned with quantified properties for running simulations.
The second part is to predict the model parameters using a machine learning tool and related data sets given desired electrophysiology properties. To find the relevant tool and data set, the user relies on the Domain-specific Topic Recommender. Using inputs such as membrane time constant, membrane resistance, resting membrane potential, and sample points on the FIR curve, the tool leverages a trained neural network to predict the model parameters. The tool also helps to compare the FIR curve given by predicted model parameters and the target FIR curve in one plot. All other passive properties and spiking properties will be generated using the predicted parameters for validation.
For both the above methods, the researcher has access to the relevant notebooks, tools, data sets required to run the simulations to meet the research/education objectives. To scale the simulations, the researcher may or may not have required CI resources to run the simulations on special hardware such as GPUs. In this scenario, the user can use the Cloud Solution Template Recommender to obtain a relevant solution as per the user’s preferred functional and nonfunctional requirements such as performance or cost. The user can then deploy suggested cloud templates, install relevant tools and run notebooks over the resources to get relevant outputs. Based on the results, users can have their own evaluations and can further deep dive into their research with the feedback obtained by again using the Publication Recommender.
5.1.2 |. Bioinformatics Use Case
In RNAseq analysis, there are many different tools and NGS data sets available for the users to select based on their research field and data of interests. Instead of spending a lot of efforts to search from online resources or many existing publications, users can utilize the Domain-specific Topic Recommender to acquire relevant details about existing tools and which are the most suitable ones for RNAseq analysis, e.g. Tophat2 and HISAT for sequence alignment, Cufflinks or HTseq for gene read counts, Cuffcompare and Cuffdiff/EdgeR for finding differentially expressed genes. The users can also use the Publication Recommender to find the most relevant publications that discuss similar research problems by querying with the key words of their research interest, which will help the user in terms of research ideas and methods, etc. In addition, users can leverage the Scholar Recommender to help them identify researchers who are experts in the same research area and potential collaborators to work on similar scientific research problems.
During execution of users’ scientific workflows, users can use the Cloud Solution Template Recommender to assist them with the configuration and running of the workflows. For example, in the scRNAseq analysis workflow, our Cloud Solution Template Recommender provides a workflow that uses the Cell Ranger program to automate sequence analysis processes. The recommended workflow uses a series of Cellranger commands to guide the users starting from annotating raw sequence reads to alignment of sequences, and finally, counting and generating feature-barcode count matrix of experimental samples. For the downstream analysis, we use Seurat tools guided by the Jupyter Notebook Recommender to explore parameter configurations to process the count matrices generated by Cell Ranger,including QC and data filtration, calculation of high-variance genes, dimensional reduction, graph-based clustering, and the identification of cluster markers to help address biological research problems.
During running the RNAseq workflow, the user will gain access to the relevant distributed computing resources (through the Cloud Solution Template Recommender), notebooks (through the Jupyter Notebook Recommender), and tools, data sets (through the Domain-specific Topic Recommender and Publication Recommender) and multi-disciplinary expert guidance (through the Scholar Recommender) required to run the analysis to meet the research/education objectives.
5.2 |. Usability Study
In this section, we present the evaluation of our OnTimeRecommend system with integrated Vidura Advisor and the underlying recommender modules customized in exemplar science gateways. We detail the evaluation results of our user profiling method in the neuroscience and bioinformatics science domains, and demonstrate the recommendations for single neuron cell simulation in the neuroscience use case, and the RNAseq analysis workflow simulation in the bioinformatics use case.
5.2.1 |. User Quadrant Questionnaire
While performing user profiling, the reliability of a questionnaire is determined by how cohesive each set of assessment questions cluster together, with a high internal consistency, to measure the user’s knowledge level on neuroscience/bioinformatics and HPC. For this purpose, we calculate the Cronbach’s alpha for our user quadrant questionnaire using the SPSS (Statistical Package for Social Sciences) software46. According to47, this index value should be above 0.70 to be deemed reliable (i.e., have high internal consistency). We compiled responses to these questions from a human subjects group of 25 users, with diverse domain backgrounds and levels of expertise. We found that the HPC oriented questions achieved a Cronbach’s alpha of 0.88, the neuroscience domain knowledge questions achieved 0.89, and the bioinformatics domain knowledge questions achieved 0.92. All these three scores are greater than the minimum required index value of 0.7, which supports our claim that our user quadrant questionnaires are reliable and consistent for the HPC, neuroscience and bioinformatics domains.
5.2.2 |. Custom User Query Responses
To evaluate user-specific response generator results, we supply Vidura Advisor with example queries for neuroscience and bioinformatics use cases, and then compare the responses with annotations from domain experts. In our experiments, domain experts attested the quality of auto-generated response and its variation that suits with user proficiency. Table 5 shows the responses generated for a sample question “Help me with Neuron Simulation” for different users based on their proficiency. Similarly, Table 6 shows the responses generated for a sample question “Help me withRNAseq analysis sub-workflow”, for different users based on their proficiency. The responses for users in different quadrants are summarized as follows:
Quadrant 1 users receive mostly information about the both domain, subject and HPC to support their low level knowledge about HPC/CI resources and scientific domains (Neuroscience or Bioinformatics) along with instructions to user different recommender modules. These users also get extensive publication and scholars recommendations related to their research problems to help them gain more background knowledge on both the domains. User also get recommendation on tools, datasets, Jupyter notebooks and cloud resources. In addition, the chatbot prompts them to use the guided user interface with significant chatbot support and also chains different recommender systems based on user’s request and proficiency. This in turn leads to chaining of microservices of recommenders, which chains different recommender modules so that they can be used in conjunction to provide a single domain-specific user interface based on user intent and profile, as shown in the Tables 5 and 6.
For the same question, Quadrant 2 users (subject domain experts with low HPC knowledge), are given less subject domain related information and more HPC related information to compensate for the knowledge gap. These users will also get recommendation on publications, experts and cloud resources to run simulation on cloud platforms as they have low HPC knowledge. In addition, they will get more guidance on HPC but minimum guidance on subject science domains.
Quadrant 3 users (HPC experts with low subject domain knowledge) receive publication recommendations, scholar recommendations, Jupyter Notebook recommendations and recommendation on tools and datasets from Domain-specific Topic Recommender. Rather than using the basic graphical user interface, which will slow down their productivity, using Jupyter Notebooks is more a suitable response, and is much faster because of the users’ prior HPC and programming skill sets. In addition, cloud templates are given to the user to explore alternative options to run their workflows or simulations on cloud resources.
Quadrant 4 users who are experts in both HPC and subject domains require minimum guidance for running their workflows and simulations. Their main focus is to successfully accomplish complex experiments in a productive manner and hence, continuous chatbot interruptions may slow down their processes and reduce productivity.
TABLE 5.
Responses generated by Vidura for the sample user query “help me with neuron simulation”.
| User Proficiency | Vidura Response Text | Chained Recommender Systems |
|---|---|---|
| Novice User (Quadrant I) | Neuron is a single cell of our brain. Follow below instruction to getmstarted with neuron simulation. Type ‘publication’ to get publication related to neuron simulation. Type ‘tools’ or ‘datasets’ to get suggestion on tools and datasets. Type ‘Jupyter Notebooks’ to get relevant Jupyter Notebooks. Type ‘scholars’ to get suggestion on experts. Type ‘cloud’ to get suggestion on cloud resources. |
Guided User Interface with below recommender chained with chatbot Publication Recommender (Suggestion for ‘neuron simulation’, ‘Hodgkin-Huxley type of neuron model’, ‘neuron simulation on cloud resources’) Jupyter Notebook Recommender (Suggestion for ‘neuron simulation’) Domain-specific Topic Recommender (Suggestion for ‘simulation tools’, ‘membrane time constant, membrane resistance, resting membrane potential’) Scholar Recommender (Suggestion for ‘neuron simulation’, ‘cloud resources’) Cloud Solution Template Recommender (Suggestion for ‘neuron simulation on cloud’) |
| Neuroscience Expert (Quadrant II) | Follow below instruction to get started with neuron simulation. Type ‘publication’ to get publication related to cloud resources. Type ‘scholars’ to get suggestion on experts on cloud resources. Type ‘cloud’ to get suggestion on cloud resources. |
Guided User Interface with below recommender chained with chatbot Publication Recommender (Suggestion for ‘neuron simulation on cloud resources’) Scholar Recommender (Suggestion for ‘cloud resources’) Cloud Solution Template Recommender Suggestion for ‘neuron simulation on cloud’) |
| HPC Experts (Quadrant III) | Neuron is a single cell of our brain. Follow below instruction to get started with neuron simulation. Type ‘publication’ to get publication related to neuron simulation. Type ‘tools’ or ‘datasets’ to get suggestion on tools and datasets. Type ‘Jupyter Notebooks’ to get relevant Jupyter Notebooks. Type ‘scholars’ to get suggestion on experts on neuron simulation. |
Guided User Interface with below recommender chained with chatbot Publication Recommender (Suggestion for ‘neuron simulation’, ‘Hodgkin-Huxley type of neuron model’) Jupyter Notebook Recommender (Suggestion for ‘neuron simulation’) Domain-specific Topic Recommender (Suggestion for ‘simulation tools’, ‘membrane time constant, membrane resistance, resting membrane potential’) Scholar Recommender (Suggestion for ‘neuron simulation’) |
| Neuroscience & HPC Experts (Quadrant IV) | You can directly start neuron simulation using cloud resources. Type ‘cloud’ to get suggestion on cloud resources. |
Directly start neuron simulation using cloud resources |
TABLE 6.
Responses generated by Vidura for the sample user query “help me with RNAseq analysis sub-workflow”.
| User Proficiency | Vidura Response Text | Chained Recommender Systems |
|---|---|---|
| Novice User (Quadrant I) | RNAseq analysis sub-workflow is used to perform quantification of gene expression from RNAseq transcriptomics NGS datasets & conducting statistical analysis to discover differential expressed genes. Follow below instruction to get started with it Type ‘publication’ to get publication related to RNAseq analysis. Type ‘tools’ or ‘datasets’ to get suggestion on tools & datasets. Type ‘Jupyter Notebooks’ to get relevant Jupyter Notebooks. Type ‘scholars’ to get suggestion on experts. Type ‘cloud’ to get suggestion on cloud resources. |
Guided User Interface with below recommender chained with chatbot Publication Recommender (Suggestion for ‘RNAseq analysis sub-workflow’,’single cell RNAseq analysis’, ‘second single cell RNAse’, ‘gene expression analysis’, ‘RNAseq analysis sub-workflow on cloud resources’) Jupyter Notebook Recommender (Suggestion for ‘Cell Ranger’) Domain-specific Topic Recommender (Suggestion for ‘scRNAseq tools’, ‘RNAseq analysis tools’) Scholar Recommender (Suggestion for ‘scRNAseq expert’, ‘cloud resources’) Cloud Solution Template Recommender (Suggestion for ‘scRNAseq workflow on cloud’) |
| Bioinformatics Expert (Quadrant II) | Follow below instruction to get started with RNAseq analysis sub-workflow. Follow below instruction to get started with it. Type ‘publication’ to get publication related to cloud resources. Type ‘scholars’ to get suggestion on experts on cloud resources. Type ‘cloud’ to get suggestion on cloud resources. |
Guided User Interface with below recommender chained with chatbot Publication Recommender (Suggestion for ‘RNAseq analysis sub-workflow on cloud resources’) Scholar Recommender (Suggestion for ‘cloud resources’) Cloud Solution Template Recommender (Suggestion for ‘scRNAseq workflow on cloud’) |
| HPC Experts (Quadrant III) | RNAseq analysis sub-workflow is used to perform quantification of gene expression from RNAseq transcriptomics NGS datasets & conducting statistical analysis to discover differential expressed genes. Follow below instruction to get started. Type ‘publication’ to get publication related to RNAseq analysis. Type ‘tools’ or ‘datasets’ to get suggestion on tools & datasets. Type ‘Jupyter Notebooks’ to get relevant Jupyter Notebooks. Type ‘scholars’ to get suggestion on experts. |
Guided User Interface with below recommender chained with chatbot Publication Recommender (Suggestion for ‘RNAseq analysis sub-workflow’, ‘single cell RNAseq analysis’, ‘second single cell RNAse’, ‘gene expression analysis’) Jupyter Notebook Recommender (Suggestion for ‘QC and data filtration’) Domain-specific Topic Recommender (Suggestion for “scRNAseq tools”, “RNAseq analysis tools”) Scholar Recommender (Suggestion for “scRNAseq expert”) |
| Bioinformatics & HPC Experts (Quadrant IV) | You can directly start RNAseq analysis sub-workflow using cloud resources. Type ‘cloud’ to get suggestion on cloud resources. |
Directly start RNAseq analysis sub-workflow cloud resources |
Overall, the integrated recommender system acting as a single source of knowledge discovery, as illustrated in Figure 2, can be navigated by a chatbot interface using a dialog design methodology for identifying user’s intent and proficiency profile. Thus, our integrated recommender system with a chatbot interface can cut down the manual/tedious work of knowledge discovery, and overall enhance the user’s research/education productivity with convenience.
5.2.3 |. Evaluation of Recommenders in the CyNeuro Science Gateway
In this section, we present two evaluation experiments conducted using the Domain-specific Topic Recommender and Scholar Recommender in the CyNeuro science gateway. Through these experiments, we demonstrate how our OnTimeRecommend system with specific recommender modules can be used to maximize users’ quality of experience by providing highly accurate recommendations with relevant data or resources based on user request inputs.
In the neuroscience community, we use our Domain-specific Topic Recommender34 integrated in the OnTimeRecommend system to recommend possible tools and datasets based on the research topics for the neuroscience researchers. Figure 6(a) shows results involving 4 samples of topics from 70 topics learned by DSTM for the neuroscience dataset. These samples were extracted from a single chain at the 80th iteration of the Gibbs sampler. Each sub-table in Figure 6(a) shows the top 10 words that are most likely to be generated conditioned on that topic; the top 4 most likely tools to be used for the topic; the top 4 most likely types of datasets that arise from the topic. Note that we have provided the topic heading annotation to each topic for demonstration in the sub-tables. These knowledge patterns were confirmed to be valid and helpful by neuroscience experts. The evaluation thus demonstrates that our DSTM effectively captures the salient domain knowledge patterns, and can provide helpful knowledge patterns from publications, tools, and datasets for computational and data-intensive scientific domains.
FIGURE 6.
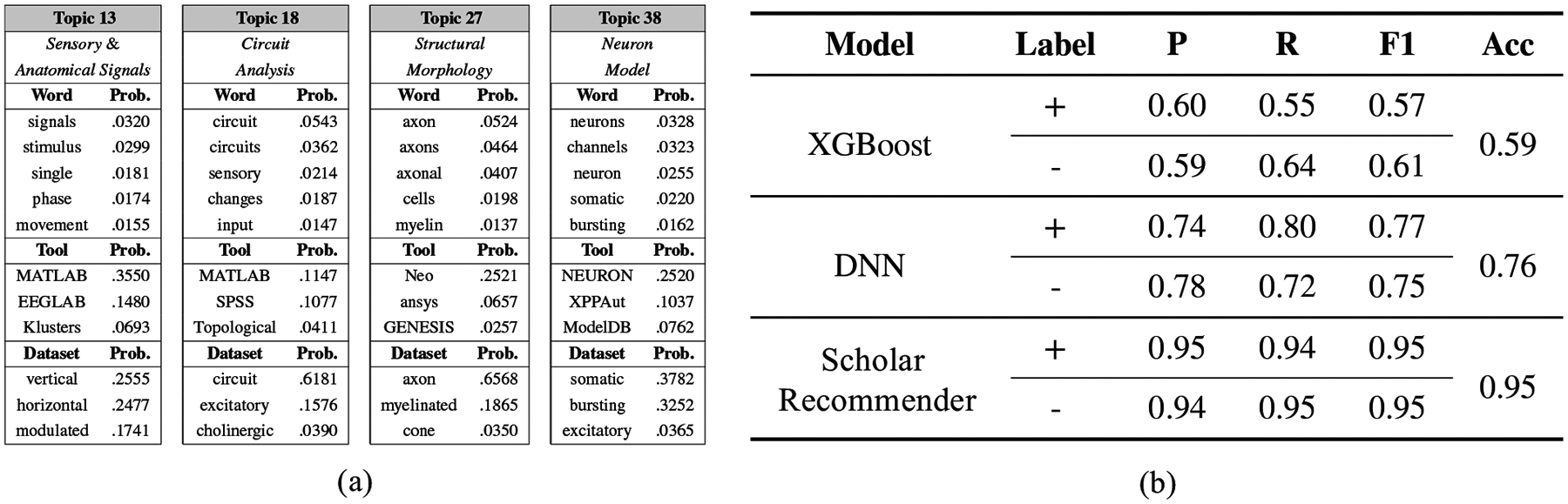
Evaluation results to show OnTimeRecommend benefits to users: (a) Results of DSTM showing 4 sample topics (out of 70 topics in total) extracted from the neuroscience publications from 2009 to 2019. Each topic is associated with 10 most likely words, 4 most likely tools and datasets that have the highest probability conditioned on that topic; (b) Results of Scholar Recommender model compared with state-of-the-art XGBoost and DNN models in terms of precision, recall, F1 score and accuracy metrics.
We also evaluated the effectiveness of our Scholar Recommender integrated in the OnTimeRecommend system in the CyNeuro science gateway. In related evaluation experiments, we use the NSF award dataset as detailed in Section 4.5 to evaluate whether a scholar is suitable for an NSF award funding or not. Given the lack of negative samples in our dataset, we apply a negative sampling scheme to generate the same amount of negative samples for training and testing phrases. We compare the performance of our Scholar Recommender model with state-of-the-art XGBoost and Deep Neural Network (DNN) models. As evident from the precision, recall and F1-scores in Figure 6(b), we demonstrate how our Scholar Recommender significantly outperforms state-of-the-art models in finding suitable scholars to help solve multi-disciplinary research tasks in the neuroscience domain.”
5.2.4 |. Evaluation of Recommenders in the KBCommons Science Gateway
In this section, we present evaluation experiments using the Cloud Solution Template Recommender33 in the KBCommons bioinformatics science gateway. Our experiments show the Cloud Solution Template Recommender’s consistency in composing cost-effective templates solutions based on user requirements, in comparison to the state-of-the-art k-NN approach32. For experimental simulation, three bioninformatics application workflows were selected, namely Fastqc (small-scale resources), RNASeq (medium-scale resources), and PGen (high-scale resources). Every workflow was then executed with diverse user-defined resource requirements, including size of data, number of virtual CPUs (vCPUs), size of memory (RAM), and network bandwidth, and CPU clock frequency, as shown in Figure 7(a). Based on the requirements collected from user, our Vidura Advisor interacts with the Cloud Solution Template Recommender module and provides the recommended resource composition using our expert system based template recommender algorithm by choosing from the machine configuration datasets stored in the knowledge base as detailed in Section 4.4.
FIGURE 7.
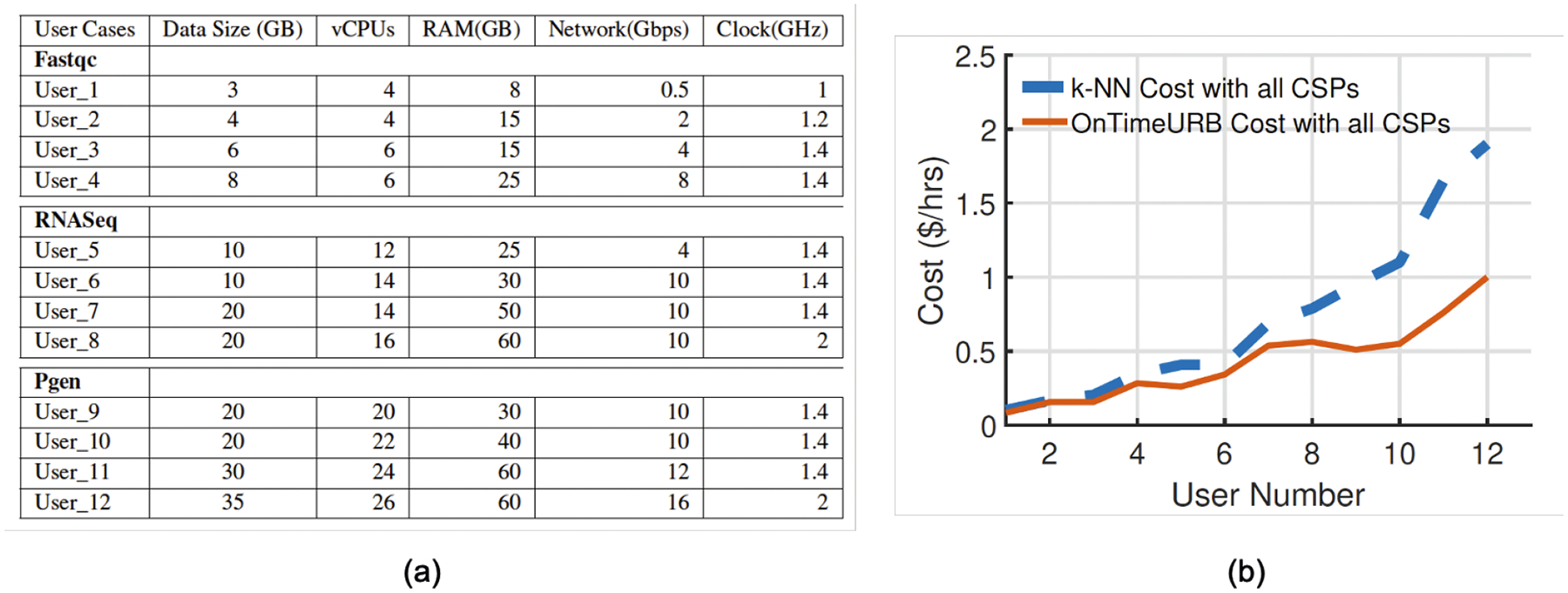
(a) Increasing user specifications for the workflows; (b) Comparison of cost-to-template with our template recommender and k-NN approach with all four cloud service provider (Amazon AWS, Google GCP, Microsoft AZURE, and NSF GENI cloud platforms).
Figure 7(b) shows comparison results of our Cloud Solution Template Recommender’s cost-to-template solution for the user specifications shown in Figure 7(a) against the k-NN approach32. From these cost comparisons, it is evident that the our Cloud Solution Template Recommender outperforms k-NN for all user requirements specified in Figure 7(a) for composing template solutions in a multiple cloud CSP case. Evaluation results demonstrate that our OnTimeRecommend system with integrated Cloud Solution Template Recommender provides recommendations of cloud computing template with lower cost while fulfilling user’s requirements in computing resources. This will in turn save users’ expense on using cloud computing resources, especially when users are running analysis of large datasets such as in bioinformatics applications that requires relatively longer running times.
6 |. CONCLUSION AND FUTURE WORK
In this paper, we have developed an integrated recommender system, the OnTimeRecommend, that comprises of five recommender modules along with a guided user interface and a chatbot functionality for neuroscience and bioinformatics researchers and educators/students.
The OnTimeRecommend integration within next-generation science gateways can provide promising advantages and convenience. The recommender modules can intelligently guide researchers and educators to discover relevant resources, enhance interdisciplinary knowledge sharing and train on Jupyter Notebook enabled learning exercises. A leading-edge multi-layer recommender architecture in OnTimeRecommend is designed to increase the effectiveness of novice/expert neuroscientists using CI resources in workflow management. Using text mining methods and topic modeling, the proposed recommender modules aim at fostering interdisciplinary collaborations around distributed databases, parallel and distributed computing resources for analysis and visualization pertaining to different scientific user tasks in a science gateway.
In addition, the OnTimeRecommend design and integration within the CyNeuro science gateway and KBCommons bioinformatics gateway were described supporting exemplar neuroscience research and bioinformatics research use cases. Our evaluation through usability studies of intelligent user profiling and Vidura chatbot guided user workflow/simulation indicate that our OnTimeRecommend can significantly enhance user’s productivity and provide a customized experience to user queries based on their proficiency.
In the future, one can focus on CI providers’ (e.g., CyVerse, AWS) perspectives, and integrate their knowledge bases into our OnTimeRecommend system to further improve benefits to science gateway users in their research and education tasks. To generalize the benefit of OnTimeRecommend for real interactions of diverse user needs, one can conduct a focused usability study to qualitatively and quantitatively measure subjective impact (e.g., user satisfaction scores), as well as objective impact (e.g., problem-solving time, degree of problem-solving effectiveness). Such a study can characterize how much time is consumed by real users to successfully complete their tasks with and without our Vidura Advisor. The study can also document the frequency of users coming back to the Vidura Advisor for performing additional new tasks. Lastly, the study can collect feedback on the social aspects of user engagement (e.g., does Vidura Advisor feel human-like) in order to build trust for on-going benefical use and sustained use of the OnTimeRecommend system in science gateways that have large user community adoption.
Funding Information
This research was supported by the National Science Foundation under award numbers OAC-1730655 and OAC-2006816. Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
References
- 1.NSF (2016) Understanding the Brain. [Online] Available at https://nsf.gov/about/budget/fy2016/pdf/42_fy2016.pdf.
- 2.NSF (2015) Ineuro conference report. [Online] Available at https://mdcune.psych.ucla.edu/modules/ineuro/reports/ineuro_conferencereport_20150804.pdf.
- 3.Neuro-Databases ((2019)) List of neurosciene databases. [Online] Available at https://en.wikipedia.Org/wiki/list_of_neuroscience_databases.
- 4.OCNS (2016) Organization for computational neurosciences software. [Online] Available at https://www.cnsorg.org/software.
- 5.Majumdar A, Sivagnanam S, Carnevale N, et al. Neuroscience Gateway–Cyberinfrastructure Providing Supercomputing Resources for Large Scale Computational Neuroscience Research. Neuroinformatics 2016. [Google Scholar]
- 6.Rule A Design and Use of Computational Notebooks. PhD thesis. UC San Diego, 2018. [Google Scholar]
- 7.Freeman J Open source tools for large-scale neuroscience. Current opinion in neurobiology 2015; 32: 156–163. [DOI] [PubMed] [Google Scholar]
- 8.Kubilius J A framework for streamlining research workflow in neuroscience and psychology. Frontiers in Neuroinformatics 2014; 7: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Usoskin D, Furlan A, Islam S, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nature neuroscience 2015; 18(1): 145. [DOI] [PubMed] [Google Scholar]
- 10.Poulin JF, Tasic B, Hjerling-Leffler J, Trimarchi JM, Awatramani R. Disentangling neural cell diversity using single-cell transcriptomics. Nature neuroscience 2016; 19(9): 1131. [DOI] [PubMed] [Google Scholar]
- 11.Nair S, Calyam P, Xu D, Joshi T, Zeng S, Zhang Y. Cyber and software automation in neuroscience. [Online] Available at http://cyneuro.org.
- 12.Zeng S, Lyu Z, Narisetti SRK, Xu D, Joshi T. Knowledge Base Commons (KBCommons) v1.0: A multi OMICS web-based data integration framework for biological discoveries. IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 2018: 589–594. [Google Scholar]
- 13.John RJL, Potti N, Patel JM. Ava: From Data to Insights Through Conversations. CIDR 2017. [Google Scholar]
- 14.Fast E, Chen B, Mendelsohn J, Bassen J, Bernstein MS. Iris: A conversational agent for complex tasks. Proceedings of the 2018 CHI Conference on Human Factors in Computing Systems 2018: 473. [Google Scholar]
- 15.Google Dialogflow. [Online] Available at https://dialogflow.com.
- 16.Deelman E, Vahi K, Juve G, et al. Pegasus, a workflow management system for science automation. Future Generation Computer Systems 2015; 46: 17–35. [Google Scholar]
- 17.HTCondor. [Online] Available at https://research.cs.wisc.edu/htcondor/index.html.
- 18.Medvedev D, Lemson G, Rippin M. Sciserver compute: Bringing analysis close to the data. Proceedings of the 28th international conference on scientific and statistical database management 2016: 1–4. [Google Scholar]
- 19.Demeler B UltraScan: a comprehensive data analysis software package for analytical ultracentrifugation experiments. Modern analytical ultracentrifugation: techniques and methods 2005: 210–229. [Google Scholar]
- 20.Merchant N, Lyons E, Goff S, et al. The iPlant collaborative: cyberinfrastructure for enabling data to discovery for the life sciences. PLoS biology 2016; 14(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wegrzyn JL, Staton M, Street N, et al. Cyberinfrastructure to improve forest health and productivity: The role of tree databases in connecting genomes, phenomes, and the environment. Frontiers in plant science 2019; 10: 813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sivagnanam S, Majumdar A, Yoshimoto K, et al. Introducing the Neuroscience Gateway. IWSG 2013. [Google Scholar]
- 23.Wan X, Jamaliding Q, Anma F, Okamoto T. Applying keyword map based learner profile to a recommender system for group learning support. Second International Workshop on Education Technology and Computer Science 2010; 1: 3–6. [Google Scholar]
- 24.Ouaftouh S, Sassi I, Zellou A, Anter S. Flat and hierarchical user profile clustering in an e-commerce recommender system. 1st International Conference on Smart Systems and Data Science (ICSSD) 2019: 1–5. [Google Scholar]
- 25.Lu Z, Pan SJ, Li Y, Jiang J, Yang Q. Collaborative Evolution for User Profiling in Recommender Systems. IJCAI 2016: 3804–3810. [Google Scholar]
- 26.Goyal M, Yadav D, Tripathi A. Intuitionistic fuzzy approach for adaptive presentation in an E-learning environment. IEEE International Conference on Research in Computational Intelligence and Communication Networks (ICRCICN) 2015: 108–113. [Google Scholar]
- 27.Chandrashekara AA, Talluri RKM, Sivarathri SS, et al. Fuzzy-Based Conversational Recommender for Data-intensive Science Gateway Applications. IEEE International Conference on Big Data (Big Data) 2018: 4870–4875. [Google Scholar]
- 28.Io H, Lee C. Chatbots and conversational agents: A bibliometric analysis. IEEE International Conference on Industrial Engineering and Engineering Management (IEEM) 2017: 215–219. [Google Scholar]
- 29.Bieliauskas S, Schreiber A. A conversational user interface for software visualization. IEEE Working Conference on Software Visualization (VISSOFT) 2017: 139–143. [Google Scholar]
- 30.Siangchin N, Samanchuen T. Chatbot Implementation for ICD-10 Recommendation System. International Conference on Engineering, Science, and Industrial Applications (ICESI) 2019: 1–6. [Google Scholar]
- 31.Papaioannou I, Dondrup C, Novikova J, Lemon O. Hybrid chat and task dialogue for more engaging hri using reinforcement learning. 26th IEEE International Symposium on Robot and Human Interactive Communication (RO-MAN) 2017: 593–598. [Google Scholar]
- 32.Antequera RB, Calyam P, Chandrashekara AA, Mitra R. Recommending heterogeneous resources for science gateway applications based on custom templates composition. Future Generation Computer Systems 2019; 100: 281–297. [Google Scholar]
- 33.Pandey A, Lyu Z, Joshi T, Calyam P. OnTimeURB: Multi-Cloud Resource Brokering for Bioinformatics Workflows. 2019: 466–473. doi: 10.1109/BIBM47256.2019.8983386 [DOI] [Google Scholar]
- 34.Zhang Y, Calyam P, Joshi T, Nair S, Xu D. Domain-specific Topic Model for Knowledge Discovery through Conversational Agents in Data Intensive Scientific Communities. IEEE International Conference on Big Data (Big Data) 2018: 4886–4895. [Google Scholar]
- 35.Wang D, Liang Y, Xu D, Feng X, Guan R. A content-based recommender system for computer science publications. Knowledge-Based Systems 2018; 157: 1–9. [Google Scholar]
- 36.Feng X, Zhang H, Ren Y, et al. The Deep Learning–Based Recommender System “Pubmender” for Choosing a Biomedical Publication Venue: Development and Validation Study. Journal of Medical Internet Research 2019; 21(5): e12957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sivarathri SS, Calyam P, Zhang Y, others. Chatbot Guided Domain-science Knowledge Discovery in a Science Gateway Application. Proceedings of Gateways 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Sivarathri SS, Calyam P. ScholarFinder: Knowledge Embedding based Recommendations using a Deep Generative Model. IEEE BigDataService 2020. [Google Scholar]
- 39.Pierce ME, Marru S, Gunathilake L, et al. Apache Airavata: design and directions of a science gateway framework. Concurrency and Computation: Practice and Experience 2015; 27(16): 4282–4291. [Google Scholar]
- 40.Newman S Building microservices: designing fine-grained systems. O’Reilly Media, Inc. 2015. [Google Scholar]
- 41.Lin YH, Wu B. Fuzzy mode and its applications in survey research. Proceedings of the 10th WSEAS International Conference on Applied Mathematics 2006: 286–291. [Google Scholar]
- 42.Goyal M, Yadav D, Tripathi A. Intuitionistic fuzzy approach for adaptive presentation in an E-learning environment. IEEE International Conference on Research in Computational Intelligence and Communication Networks (ICRCICN) 2015: 108–113. [Google Scholar]
- 43.Chen B, He H, Guo J. Constructing maximum entropy language models for movie review subjectivity analysis. Journal of Computer Science and Technology 2008; 23(2): 231–239. [Google Scholar]
- 44.Jin C, Ma T, Hou R, et al. Chi-square statistics feature selection based on term frequency and distribution for text categorization. IETE journal of research 2015; 61(4): 351–362. [Google Scholar]
- 45.Jupyter Notebooks. [Online] Available at https://jupyter.org/documentation.
- 46.Agresti A, Finlay B. Statistical methods for the social sciences: With SPSS from A to Z: A brief step-by-step manual. 2009. [Google Scholar]
- 47.DeVellis RF. Scale development: Theory and applications. 26. Sage publications. 2016. [Google Scholar]


