Abstract
MARCH1 and MARCH8 are ubiquitin ligases that control the expression and trafficking of critical immunoreceptors. Understanding of their function is hampered by three major knowledge gaps: (i) it is unclear which cell types utilize these ligases; (ii) their level of redundancy is unknown; and (iii) most of their putative substrates have been described in cell lines, often overexpressing MARCH1 or MARCH8, and it is unclear which substrates are regulated by either ligase in vivo. Here we address these questions by systematically analyzing the immune cell repertoire of MARCH1- or MARCH8-deficient mice, and applying unbiased proteomic profiling of the plasma membrane of primary cells to identify MARCH1 and MARCH8 substrates. Only CD86 and MHC II were unequivocally identified as immunoreceptors regulated by MARCH1 and MARCH8, but each ligase carried out its function in different tissues. MARCH1 regulated MHC II and CD86 in professional and “atypical” antigen presenting cells of hematopoietic origin, including neutrophils, eosinophils and monocytes. MARCH8 only operated in non-hematopoietic cells, such as thymic and alveolar epithelial cells. Our results establish the tissue-specific functions of MARCH1 and MARCH8 in regulation of immune receptor expression and reveal that the range of cells constitutively endowed with antigen-presentation capacity is wider than generally appreciated.
Keywords: Antigen presentation and processing, Regulation of surface stimulatory molecules, MARCH E3 ubiquitin ligases, Primary cell analysis
1. Introduction
Ubiquitination is a major mechanism for the regulation of membrane proteostasis. In brief, covalent attachment of ubiquitin (Ub) chains to the cytosolic tail of transmembrane proteins promotes endosomal trafficking to multivesicular bodies for subsequent degradation in lysosomes (Komander and Rape, 2012). This post-translational modification enables the fine-tuning of surface protein expression levels. Ub is attached to substrates by E3 Ub ligases. Membrane Associated RING-CH Finger (MARCH, gene symbol Marchf) is a family of eleven E3 ligases, all of which possess two or more transmembrane domains, with the exception of MARCH7 and MARCH10 (Liu et al., 2019). They were initially identified as the mammalian homologues of herpesvirus immunoevasins that ubiquitinate host molecules involved in anti-viral immunity to subvert immune responses (Coscoy et al., 2001; Ishido et al., 2000). MARCH E3 Ub ligases are thought to be specialized at ubiquitinating immunoregulatory receptors, but their physiological substrates remain largely unknown (Liu et al., 2019; Samji et al., 2014). It is also unclear if their expression and function is restricted to cells of the immune system and, if so, which.
MARCH1 and MARCH8 are the most studied members of the MARCH family. As they share approximately 60% overall sequence homology (Liu et al., 2019), they are thought to also share substrate specificity. Indeed, both ubiquitinate major histocompatibility complex class II (MHC II) molecules, the receptor employed by antigen presenting cells (APCs) to display peptide antigens to CD4+ T cells. MARCH1 and MARCH8 regulate the surface expression of MHC II (Young et al., 2008; Cho et al., 2015; Bannard et al., 2016) and thereby play key roles in the activation of naïve CD4+ T cells in the periphery (Ishikawa et al., 2014), as well as their development in the thymus (Liu et al., 2016; von Rohrscheidt et al., 2016; Oh et al., 2013). Furthermore, they have been involved in complex immune reactions such as inflammation (Galbas et al., 2017), infection (Wu et al., 2020; Zhang et al., 2019), cancer (Xie et al., 2019), allergy and autoimmunity (Kishta et al., 2018; Toyomoto et al., 2011). This raises the question whether both ligases regulate the expression of other immune receptors, some of which reportedly include CD44 (Eyster et al., 2011), CD71 (Fujita et al., 2013), CD86 (Corcoran et al., 2011), CD95 (Bartee et al., 2004) and CD98 (Eyster et al., 2011; Ablack et al., 2015) among others (Samji et al., 2014). However, to date CD86 is the only membrane protein apart from MHC II that has been shown to be regulated by MARCH1 in vivo (Baravalle et al., 2011), and it is not known if it can also be regulated by MARCH8. All other putative MARCH1 or MARCH8 substrates have been described in cell lines and/or overexpression studies. MARCH proteins are expressed at very low levels in primary cells (Liu et al., 2019; Jabbour et al., 2009; Kaul et al., 2018; Thibodeau et al., 2008), and since E3 ligase overexpression can cause off-target effects, it remains unclear which, if any of the MARCH1 and MARCH8 substrates described in transfected cell lines are ubiquitinated by these ligases in physiological settings. To summarize, the repertoire of MARCH1 and MARCH8 substrates in vivo remains largely unknown. This is an important shortcoming because ubiquitination is amenable to pharmacological manipulation (Zhao et al., 2020; Edelmann et al., 2011), and development of drugs targeting MARCH1 or MARCH8 might have therapeutic potential provided their substrates are identified.
Another important knowledge gap in MARCH1 and MARCH8 biology pertains to their expression pattern. Quantitating MARCH1 or MARCH8 protein expression is unfeasible due to their low abundance (Liu et al., 2019) and fast turn-over (Bourgeois-Daigneault and Thibodeau, 2012; Lei et al., 2018), and even their transcription levels are poor predictors of function (Jabbour et al., 2009; Kaul et al., 2018; Thibodeau et al., 2008). Identification of MARCH1- or MARCH8-expressing cells thus relies on analysis of surface expression of membrane protein substrates as a surrogate of activity. MARCH1 ubiquitinates MHC II and CD86 in B cells and conventional and plasmacytoid dendritic cells (cDCs and pDCs, respectively) (Young et al., 2008; Cho et al., 2015; Bannard et al., 2016), but it is not functional in thymic epithelial cells (TECs) (Liu et al., 2016; von Rohrscheidt et al., 2016). Whether it is active in other hematopoietic or non-hematopoietic cells remains unknown. In contrast, MARCH8 ubiquitinates MHC II in TECs, not in B cells or DCs (Liu et al., 2016; von Rohrscheidt et al., 2016), but it is not known if it ubiquitinates other receptors in these cells, and whether it is also expressed in other cells. Incomplete understanding of the pattern of MARCH expression again limits the development and potential application of ubiquitination-modulating agents as immunomodulatory drugs.
Here, we present a systematic analysis of the pattern of activity of MARCH1 and MARCH8 in multiple hematopoietic and non-hematopoietic cells isolated from Marchf1-/- and Marchf8-/- mice. We have also carried out quantitative proteomic comparisons of wild type (WT) vs Marchf1-/- or Marchf8-/- plasma membrane purified from cDCs and B cells. Our results define physiological substrates regulated by these two ligases and demonstrate functional specializations of MARCH1 and MARCH8 in two ontogenically distinct compartments.
2. Materials and methods
2.1. Mice
Wild type (WT, C57BL/6), Marchf1-/- (Matsuki et al., 2007), Marchf8-/- (Liu et al., 2016) and I-Aα-/- (Köntgen et al., 1993) mice were bred and maintained in specific pathogen-free conditions within the Melbourne Bioresources Platform at the Bio21 Molecular Science and Biotechnology Institute. Analyses were undertaken with male or female mice aged between 6 and 14 weeks and performed in accordance with the Institutional Animal Care and Use Committee guidelines of the University of Melbourne. All procedures were approved by the Animal Ethics Committee at the University of Melbourne (#1714238 and #1513472).
2.2. Isolation of mouse primary cells and analytical flow cytometry
Single cell suspensions from blood, spleen, subcutaneous lymph nodes (LN), thymus, peritoneal cavity and lung were generated for analysis of B cells, T cells, DCs, macrophages, monocytes, neutrophils, eosinophils and thymic or alveolar epithelial cells. Blood was collected from submandibular veins and red blood cells were lysed. Whole single cell suspensions from spleen and subcutaneous LN (axillary and inguinal) were generated by digestion with 0.1% DNase I (Roche) and 1 mg/ml collagenase type III (Worthington) and red blood cell lysis. DCs from spleen and LN were further enriched by selection of low-density cells by density gradient centrifugation in 1.077 g/cm3 Nycodenz® (Axis shield). Thymi were digested in 0.1% DNase I (Roche) and 0.5 U/ml liberase (Roche) and thymic cDCs were further enriched by 1.077 g/cm3 Nycodenz® density gradient centrifugation. Cells from the peritoneal cavity were harvested by injection and aspiration of PBS. Lungs were perfused with PBS and digested with 50 μg/ml DNase I (Roche) and 0.25 mg/ml liberase (Roche) and red blood cells were lysed.
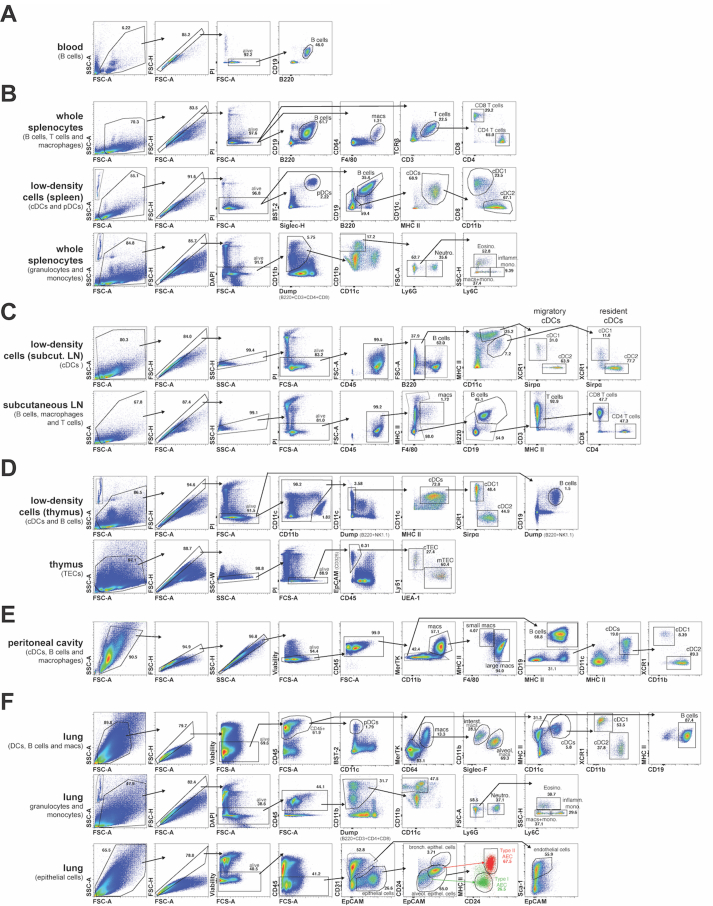
For flow cytometry, cells were incubated with FcR blocking reagent (Miltenyi Biotec), prior to staining with mAb detecting B220/CD45R (RA3-6B2), CD19 (6D5), CD64 (X54-5/7.1), F4/80 (F4/80, Walter Eliza Hall Institute (WEHI) Antibody Facility), CD3 (KT3-1.1, WEHI Antibody Facility), TCRβ (Η57-597, WEHI Antibody Facility), CD4 (GK1.5), CD8 (YTS169.4 WEHI Antibody Facility), CD8 (53–6.7), BST-2 (927), Siglec-H (551), MHC II (M5/114), CD11c (N418), CD11b (M1/70), Ly6G (1A8), Ly6C (HK1.4), NK1.1 (PK136, BD Biosciences), Sirpα (P84), XCR1 (ZET), CD45 (30-F11), EpCAM (G8.8), Ly51 (6C3), UEA-1 (Vector Laboratories), MerTK (2B10C42), Siglec-F (E50-2440 BD Biosciences), CD31 (390), CD24 (M1/69, WEHI Antibody Facility), Sca-1 (D7), CD86 (GL-1), CD40 (FGK45.5, Miltenyi Biotec), CD80 (16-10A1, BD Biosciences), CD44 (IM7.81), CD71 (R17217, eBiosciences), CD95 (15A7, eBiosciences), CD98 (RL388), PD-L1 (10F.9G2), PD-L2 (TY25), ICOS-L (HK5.3), B7–H3 (MIH35) or B7–H4 (HMH4-5G1), conjugated to fluorochromes BUV395, BUV805, FITC, PE, PE-Cy7, PerCP/Cy5.5, APC, APC-Cy7, AF700, BV785, BV650, BV510 or BV421 (all from BioLegend, if not stated differently). Cell viability was determined with Fixable Viability Dye eFluor™ 780 (eBiosciences), propidium iodide (PI) or diamidino phenylindole (DAPI) staining. Analysis was performed using a LSRFortessa (BD Biosciences) or CytoFLEX LX (Beckman Coulter) in the Melbourne Cytometry Platform (University of Melbourne). Data was analyzed with FlowJo (Tree Star) and GraphPad Prism. Supplementary Figs. 1 and 2 summarize gating strategies for cells from blood, spleen, subcutaneous lymph nodes (LN), thymus, peritoneal cavity and lung.
2.3. Isolation of primary immune cells for proteomic analysis
B cells were purified from spleens using Ficoll® Paque Plus (GE Healthcare) gradient centrifugation and negative depletion with FITC-conjugated mAb specific for CD4 (GK1.5), Ly-76 (TER119) and CD43 (S7) and magnetic anti-FITC MicroBeads (Miltenyi Biotec). Preparations were approximately 95–98% pure for CD19+ B220+ B cells. Splenic cDCs were purified from mice subcutaneously injected with Flt3L-secreting melanoma cells (Mach et al., 2000), nine days before purification. cDCs were purified from spleens of Flt3L-expanded mice following spleen digestion with DNase I (Roche) and collagenase type III and Nycodenz® density gradient centrifugation with subsequent negative depletion using rat mAb specific for CD3 (KT3-1.1), Thy1 (T24/31.7), Ly-76 (Ter119), B220 (RA3-6B2) and Ly-6C/G (RB6-8C5) and anti-rat IgG-coupled magnetic beads (Qiagen) as previously described (Vremec, 2010). Preparations were approximately 90–95% pure for CD11c+ MHC II+ cDCs.
2.4. Preparation of subcellular fractions enriched in plasma membrane and intracellular compartments for proteomics
Subcellular fractionation was performed as previously described (Segura et al., 2009). In brief, purified B cells (4–5 x 107 cells, 95–98% purity) and cDCs (4–5 x 107 cells, 90–95% purity) from spleens of WT, Marchf1-/- and Marchf8-/- mice were incubated with FITC-conjugated anti-CD19 and anti-B220 mAb (B cells) or anti-CD11c, anti-CD45.2, anti-CD49d and anti-MHC I mAb (cDCs). mAb-labelled cells were homogenized in the presence of cOmplete™ protease inhibitors (Roche) by mechanical disruption using a cell-cracker (HGM Laboratory equipment). Homogenized preparations were centrifuged at low speed to obtain post-nuclear supernatant (PNS). Surface-labelled plasma membrane (PM) microsomes were isolated by magnetic immunoaffinity using anti-FITC mAb-coated magnetic beads (Miltenyi Biotec) and concentrated by ultracentrifugation in thickwall polycarbonate tubes (Beckman Coulter). PNS, with the PM fraction removed, was likewise ultracentrifuged to sediment the “intracellular compartments” (IC) fraction.
2.5. Proteomic profiling of differentially expressed PM proteins
Subcellular fractions (PM and IC) from equal cell numbers of Marchf1-/- and Marchf8-/- cDCs and B cells were prepared for mass spectrometry analysis from three independent cell preparations using FASP protein digestion (Protein Discovery) as previously described (Wiśniewski et al., 2009), with the following modifications. Proteins were reduced and digested with sequence-grade modified Trypsin Gold (Promega). Peptides were eluted with ammonium bicarbonate and acidified peptide mixtures from each biological replicate were analyzed in technical triplicates by nanoflow reverse-phase liquid chromatography tandem mass spectrometry (LC-MS/MS) on a nanoAcquity system (Waters) coupled to a Q-Exactive mass spectrometer equipped with a nanoelectrospray ion source for automated MS/MS (Thermo Fisher Scientific). High-resolution MS/MS spectra were processed with MaxQuant (version 1.6.7.0) for feature detection and protein identification using the Andromeda search engine (Cox et al., 2011). Extracted peak lists were searched against the UniProtKB/Swiss-Prot Mus musculus database (Oct-2019) and a separate reverse decoy database to empirically assess the false discovery rate (FDR) using a strict trypsin specificity allowing up to two missed cleavages. The minimum required peptide length was seven amino acids. The “match between runs” option in MaxQuant was used (Cox and Mann, 2008). PSM and protein identifications were filtered using a target-decoy approach at a FDR of 1%. LFQ quantification was performed, with a minimum ratio of two. Protein relative quantitative analysis was performed in R using MaxQuant's proteinGroups.txt and LFQ intensities. Missing values were imputed using a random normal distribution of values derived from the measured distribution of intensities (Cox et al., 2014) using a mean with a negative shift of 1.8 standard deviations and a standard deviation equal to 0.3 of the standard deviation of the measured intensities. The probability of differential expression was calculated using the function lmFit from the Bioconductor package limma (Ritchie et al., 2015) followed by eBayes using the default settings (Phipson et al., 2016) and false-discovery rate correction using the Benjamini–Hochberg method. The output included P value, confidence interval and ratio estimate. GO-term enrichment analysis was performed using the enrichr function in the Bioconductor clusterProfiler package (Yu et al., 2012). Enrichment was calculated separately for the proteins overrepresented in each fraction, relative to all proteins identified in collected fractions across all the LC-MS/MS runs, and GO term association was filtered to include only experimental and high throughput evidence. Enrichment P values were corrected for multiple testing using the function's ‘fdr’ method. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD023115.
3. Results
3.1. MARCH1, but not MARCH8, is functional in professional APCs
The first objective of this study was to establish which mouse cells express MARCH1 or MARCH8. Their low level of transcription combined with fast turn-over contribute to maintain the two proteins at non-detectable levels in primary cells, hampering definition of their expression pattern. We reasoned that MHC II and/or CD86 could be used as reporters of MARCH1 and MARCH8 activity because in all primary or transformed cells analyzed so far, the surface level of these two receptors decreases by expression of either ligase (Liu et al., 2019). Cells that express MHC II or CD86 and either MARCH1 or MARCH8 should therefore display higher levels of the receptor(s) in Marchf1-/- or Marchf8-/- mice.
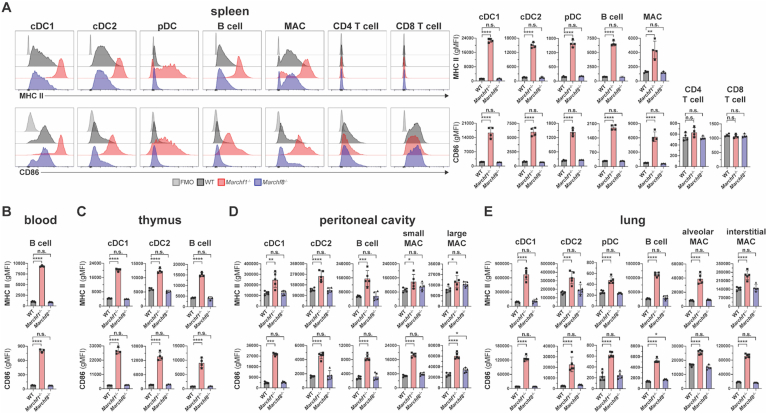
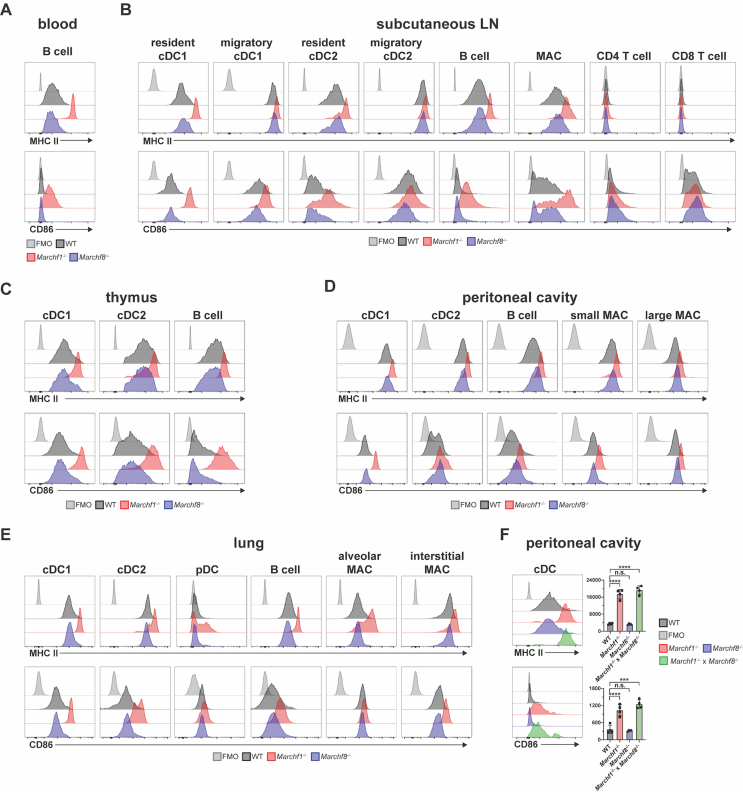
First, we examined professional APCs (defined as cells that express detectable levels of MHC II in the steady-state (Lassila et al., 1988; Kambayashi and Laufer, 2014) and T cells across various tissues. B cells, cDC1s, cDC2s, pDCs and macrophages from spleen, blood, thymus, peritoneal cavity, lung, and subcutaneous lymph nodes (LN) of Marchf1-/- mice displayed elevated surface MHC II and CD86 relative to WT cells, while no changes were observed in their Marchf8-/- counterparts (Fig. 1A–E and Supplementary Fig. 3A–E). CD4+ and CD8+ T cells in spleen and LN showed no detectable surface MHC II and their CD86 expression was not altered by MARCH1- nor MARCH8-deficiency (Fig. 1A and Supplementary Fig. 3B). MHC II and CD86 expression in peritoneal cDCs deficient in both MARCH1 and MARCH8 (Marchf1-/- x Marchf8-/-) was not elevated above that of Marchf1-/- cells (Supplementary Fig. 3F). These results indicate that MARCH1 is expressed and active in all professional APCs across various organs/tissues whereas MARCH8 is not or, if it is, does not display enough activity to compensate for the loss of MARCH1.
Fig. 1.
Ubiquitination of MHC II and CD86 by MARCH1 and MARCH8 in haemopoietic professional APCs. Surface expression levels of MHC II and CD86 in (A) splenic cDC1s, cDC2s, pDCs, B cells, macrophages (MAC), and CD4+/CD8+ T cells, (B) blood B cells, (C) thymic cDC1s, cDC2s, and B cells, (D) peritoneal cDC1s, cDC2s, B cells, and small/large MAC, and (E) lung cDC1s, cDC2s, pDC, B cells, and alveolar/interstitial MAC, all purified from WT mice or mice deficient in either MARCH1 or MARCH8. Bars represent mean ± SD with each symbol representing an individual mouse (n = 4–5). Statistical analysis was performed using one-way ANOVA followed by Sidak's multiple comparisons test. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.0002, ∗∗p < 0.002, ∗p < 0.03, n.s. not significant.
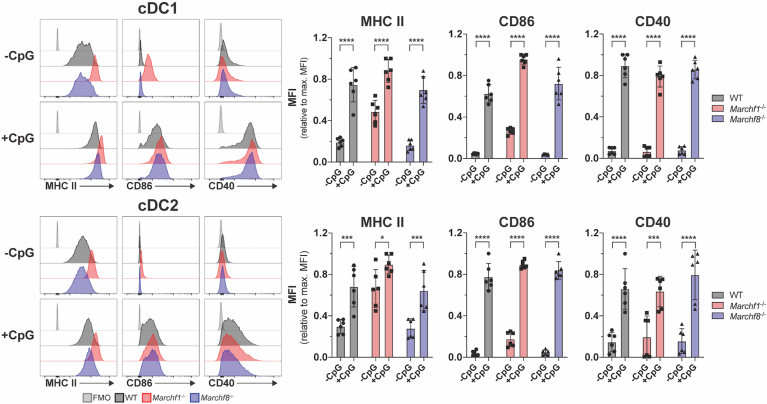
Next, we assessed the contribution of MARCH1 to activation-dependent regulation of MHC II and CD86 expression in cDCs, the archetypical professional APCs. Toll-like receptor (TLR) ligands trigger an activation program in DCs, known as DC maturation, that includes up-regulation of MHC II and CD86 expression on the plasma membrane, among other receptors (Wilson et al., 2004). Activation also leads to down-regulation of Marchf1 transcription which, combined with fast turn-over of MARCH1, results in negligible expression of the protein in activated DCs (Young et al., 2008; Vega-Ramos et al., 2014; De Gassart et al., 2008; Galbas and Thibodeau, 2012). It has been assumed that this change is responsible for the accumulation of MHC II and CD86 on the plasma membrane during cDC activation, but this has not been directly examined. If ubiquitination were the dominant mechanism controlling how much MHC II and CD86 is displayed on cDCs, it would be expected that the expression of these two molecules would not vary during activation of Marchf1-/- cDCs. However, activation of Marchf1-/- cDCs further increased surface expression of MHC II by ∼1.5 times, and increased CD86 by ∼4 times, when compared with their resting counterparts (Fig. 2). CD40, which also increases in expression during activation, though it is not a MARCH1 substrate, was expressed at equivalent levels in WT and Marchf1-/- cDCs at both resting and activated states, so up-regulation of MHC II and CD86 in Marchf1-/- cDCs could not be attributed to overall dysregulation of surface receptor expression (Fig. 2). These results indicate that deposition of newly synthesized molecules on the cell surface (Villadangos et al., 2001; Wilson et al., 2004) is the main contributor to MHC II and, especially, CD86 up-regulation during DC activation. cDCs lacking MARCH8 were indistinguishable from WT cDCs in these experiments, again indicating it has no role in resting or activated cDCs (Fig. 2).
Fig. 2.
The role of ubiquitination of MHC II and CD86 by MARCH1 and MARCH8 in DC maturation. Surface expression of MHC II and CD86 in CpG-activated cDCs purified from the spleen of WT mice or mice deficient in either MARCH1 or MARCH8. Purified splenic cDCs (2x105 cells) were kept on ice (-CpG) or incubated for 16 hours ex vivo with 50 nm CpG in 96-well plates, then washed and analyzed by flow cytometry for MHC II and CD86 surface expression. A fluorescence-minus-one (FMO) control was included, for which cells were incubated with the corresponding multi-colour staining panel, excluding the fluorescently labelled antibody species of interest (i.e. anti-CD86 or anti-MHC II mAb). Bars represent mean ± SD with each symbol representing an individual mouse (n = 6). Statistical analysis was performed using one-way ANOVA followed by Sidak's multiple comparisons test. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.0002, ∗p < 0.03.
3.2. Neutrophils, eosinophils and monocytes express MHC II and CD86, but MARCH1 ubiquitination maintains their surface expression at negligible levels
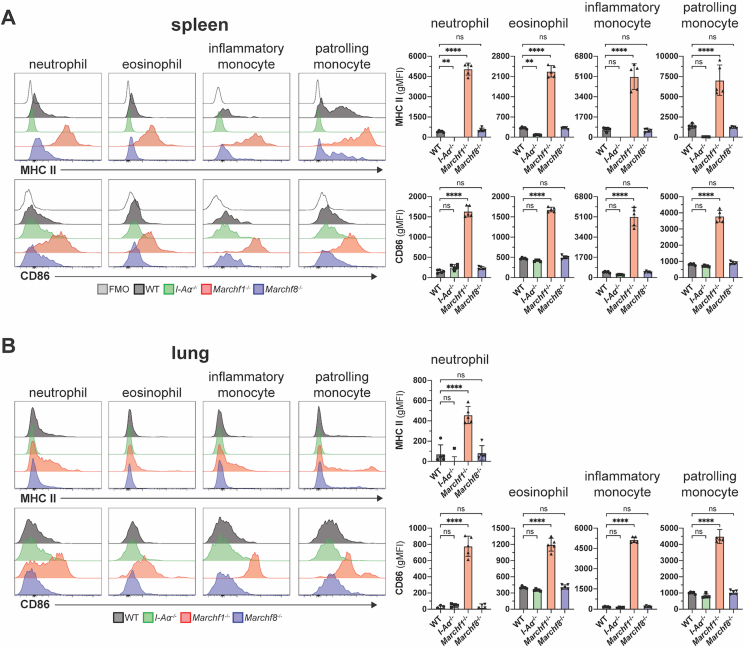
Next we assessed MARCH1 and MARCH8 activity in “atypical” APCs, that is, immune cells that are not considered professional APCs but have been suggested to play antigen-presenting roles under certain conditions (Kambayashi and Laufer, 2014). These include neutrophils, eosinophils and “inflammatory” (Ly6C+) and “patrolling” (Ly6C-) monocytes. While monocytes have the potential to develop into macrophages or DCs in inflamed sites (Auffray et al., 2009), they are not thought to perform antigen presenting functions in their undifferentiated state (Jakubzick et al., 2017). We examined these “atypical” APCs in spleen and lung. MHC II expression in WT neutrophils, eosinophils and monocytes was barely detectable by flow cytometry, staining at just above the background level observed in cells of mice that do not express any surface MHC II at all (Fig. 3). Strikingly, all four cell types deficient in MARCH1 expressed MHC II at levels comparable to WT B cells or cDCs (compare Fig. 1, Fig. 2, Fig. 3), though expression was higher in spleen than it was in their lung counterparts (Fig. 3A and B). CD86 was also highly expressed at the surface of all four MARCH1-deficient cell types, in this case both in spleen and lungs (Fig. 3). MARCH8-deficient cells did not display altered MHC II or CD86 expression, confirming this member of the MARCH family is not expressed and/or active in hematopoietic cells (Fig. 3). Of note, MARCH1-deficient T cells lacked surface MHC II and did not exhibit enriched CD86 expression when deficient in MARCH1 (Fig. 1A), so neither mutation caused ectopic or increased expression of either molecule. We conclude that neutrophils, eosinophils, monocytes and possibly other atypical APC types (Kambayashi and Laufer, 2014) produce receptors for antigen presentation and T cell stimulation constitutively. While MARCH1 ubiquitination maintains the surface expression of these proteins at barely detectable levels, these “atypical” APCs might be capable of CD4+ T cell priming under certain conditions.
Fig. 3.
Ubiquitination of MHC II and CD86 by MARCH1 and MARCH8 in granulocytes and monocytes. Surface expression of MHC II and CD86 in neutrophils, eosinophils and inflammatory and patrolling monocytes purified from (A) spleen and (B) lung of WT mice or mice deficient in I-Aα, MARCH1, or MARCH8. Bars represent mean ± SD with each symbol representing an individual mouse (n = 5). Statistical analysis was performed using one-way ANOVA followed by Sidak's multiple comparisons test. ∗∗∗∗p < 0.0001, ∗∗p < 0.002, n.s. not significant.
3.3. Previously predicted MARCH1 substrates display normal expression in Marchf1-/- mice
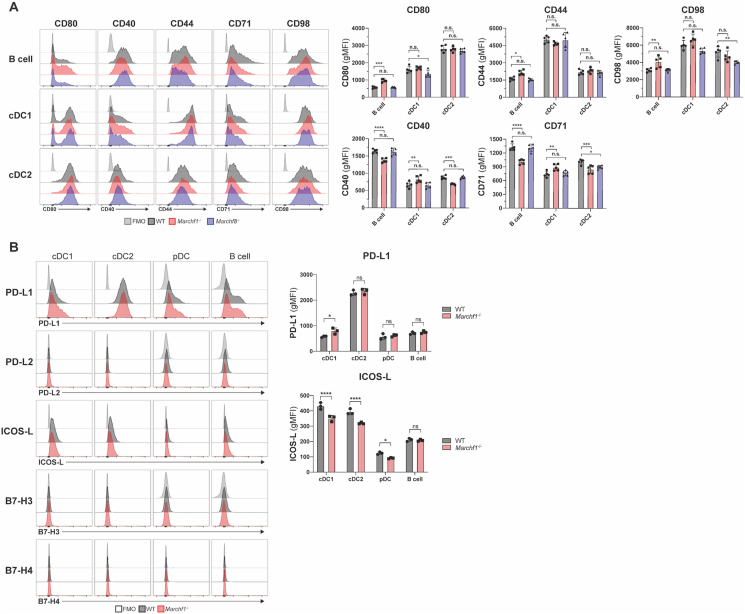
The second objective of this study was to identify which of the receptors found to be ubiquitinated by MARCH1 or MARCH8 in (transfected) cell lines are also substrates in vivo under physiological conditions. Such receptors include CD44, CD71, CD95 and CD98 (reviewed in (Samji et al., 2014)). Carrying out this analysis also allowed us to address the possibility that, contrary to our conclusions above, MARCH8 might be expressed and active in these cells but dedicated to ubiquitinate these receptors rather than MHC II and CD86. This was not the case; expression of CD44, CD71, and CD98 was unaltered in Marchf8-/- cDCs and B cells compared to WT cells (Fig. 4A). Furthermore, Marchf1-/- cDCs and B cells also expressed normal levels of the three receptors (Fig. 4A). We extended our analysis to other regulatory receptors of T cell activation, including CD40 and members of the B7 family to which CD86 (B7.2) belongs: CD80 (B7.1), CD274 (PD-L1), CD273 (PD-L2), CD275 (ICOS-L), CD276 (B7–H3) and B7–H4. No surface expression for PD-L2, B7–H3, and B7–H4 was detected on cDC1s, cDC2s, pDCs, and B cells, and PD-L1 and ICOS-L expression was unaltered in the absence of MARCH1 (Fig. 4B).
Fig. 4.
Analysis of putative MARCH1 and MARCH8 substrates in haemopoietic APCs. (A) Surface expression of CD80, CD40, CD44, CD71, and CD98 in splenic B cells, cDC1s, and cDC2s from WT, Marchf1-/-, and Marchf8-/- mice. (B) Surface expression of B7 costimulatory molecules, PD-L1, PD-L2, ICOS-L, B7–H3 and B7–H4, in splenic cDC1s, cDC2s, pDCs, and B cells purified from WT or Marchf1-/- mice. In all cases a fluorescence-minus-one (FMO) control was included, for which cells were incubated with the corresponding multi-colour staining panel, excluding the fluorescently labelled antibody species of interest. Bars represent mean ± SD with each symbol representing an individual mouse (n = 3–6). Statistical analysis was performed using one-way ANOVA followed by Sidak's multiple comparisons test. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.0002, ∗∗p < 0.002, ∗p < 0.03, n.s. not significant.
3.4. Proteomic profiling of the plasma membrane of MARCH1- and MARCH8-deficient cDCs and B cells
To more comprehensively address the role of MARCH1 and MARCH8 in APC membrane proteostasis, we performed an unbiased proteomic screen where we compared the proteomes of subcellular microsomal fractions enriched in plasma membrane (PM) of WT versus Marchf1-/- or Marchf8-/- cDCs and B cells. We have previously shown this is a robust approach to identify differentially expressed PM proteins between closely related cell populations such as the two major cDC subtypes, cDC1s and cDC2s (Segura et al., 2010). To obtain sufficient numbers of primary cDCs for this purpose, they were expanded in WT, Marchf1-/- and Marchf8-/- mice bearing a melanoma cell line that secretes the DC growth factor, Flt3L (Mach et al., 2000). The cDCs expanded using this approach are phenotypically and functionally equivalent to their counterparts in untreated mice (Segura et al., 2010). Splenic B cells were purified from untreated mice. The protein profiles of each fraction were identified by semi-quantitative mass spectrometry from three biological replicates, each measured in technical triplicates.
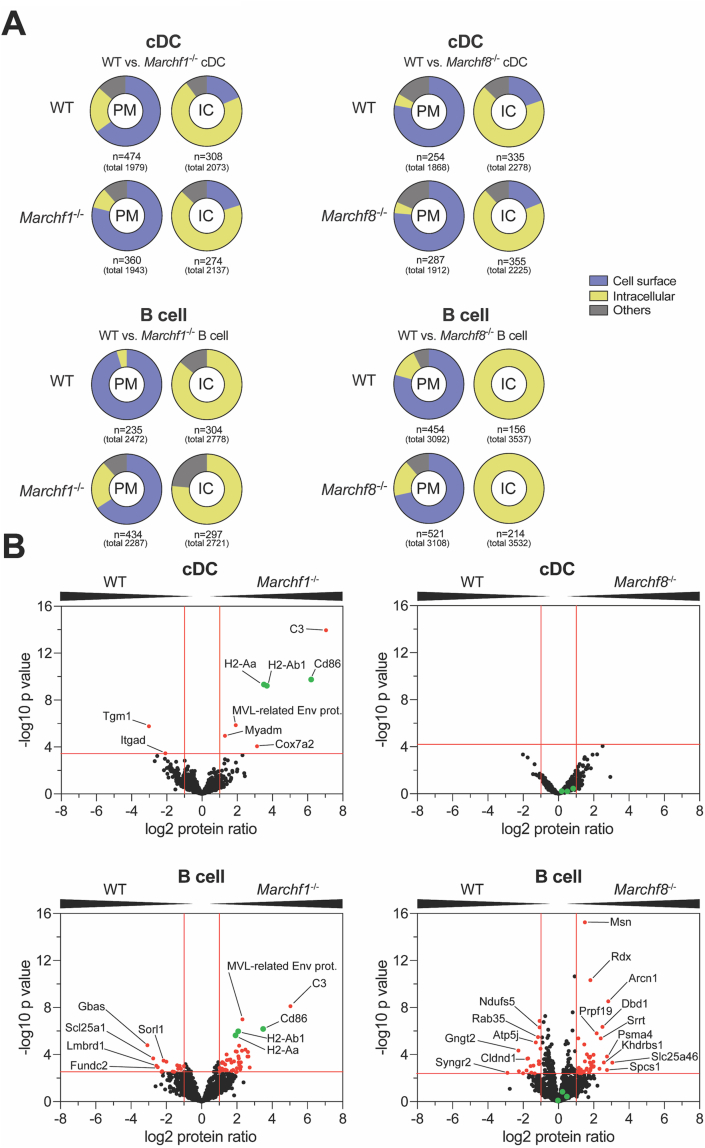
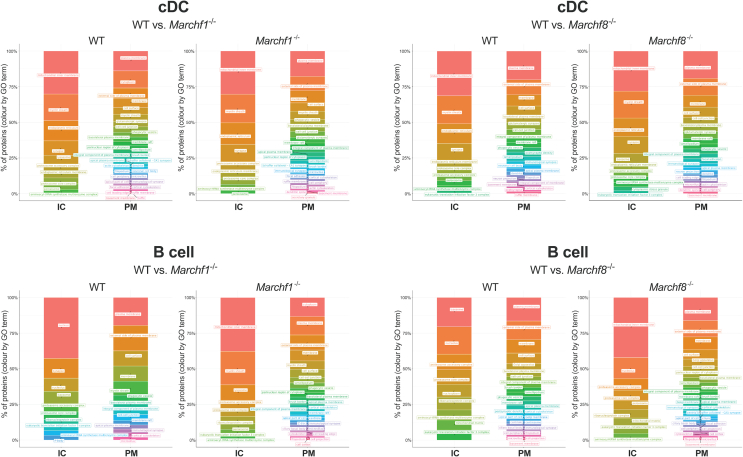
We identified 1868–3108 proteins in the PM fraction of each cell type (Supplementary Table 1, total number of identified proteins regardless of any restrictions). Of note, the subcellular PM fractions are comprised of microsomes generated during mechanical homogenization of cells, so their composition includes PM but also cytosolic and extracellular content ‘trapped’ inside microsomes or tethered to the cell surface. This method enables analysis of proteins loosely associated with the inner or outer leaflet of the PM. To test the efficiency of the PM-enrichment method, we also sedimented and analyzed in parallel the compartments that remained in the post-nuclear supernatant (PNS) of homogenized cells after retrieval of the PM fraction (mitochondria, endosomes, etc., henceforth termed intracellular compartments, IC). We identified 2073–3537 proteins in the IC fraction of each cell type (Supplementary Table 2, total number of identified proteins regardless of any restrictions). In order to assess enrichment of the PM by this methodology, we compared Gene Ontology (GO) terms/annotations of the proteins identified in the PM and IC fractions of each cell type. This comparison clearly demonstrated enrichment of proteins known to be expressed at the cell surface in the PM fractions, and enrichment of proteins known to occur in intracellular compartments in the IC fractions, validating the subcellular fractionation protocol (Fig. 5A and Supplementary Fig. 4).
Fig. 5.
Proteomic analysis of differentially expressed proteins in the plasma membrane between WT and Marchf1-/-or Marchf8-/-cDCs and B cells. PM fractions were purified from PNS of mAb surface stained cDCs and B cells via magnetic immunoaffinity and analyzed by semi-quantitative mass spectrometry from three biological replicates (in total 3x 8 samples; WT vs. Marchf1-/- and WT vs. Marchf8-/- cDCs as well as WT vs. Marchf1-/- and WT vs. Marchf8-/- B cells). The remaining compartments (mitochondria, endosomes, etc.) sedimented from the PNS of homogenized cells following PM fraction retrieval was termed intracellular compartment (IC). (A) Enrichment analysis (performed using the function enricher included in the Bioconductor clusterProfiler package (Yu et al., 2012) of detected proteins via MS from PM or IC fractions from cDCs and B cells of WT versus Marchf1-/- and WT versus Marchf8-/- mice. Only experimentally verified Gene Ontology (GO) annotations were used (n = XY) from all detected proteins (total number in parenthesis), and grouped into categories of ‘Cell surface’, ‘Intracellular Compartment (IC)’ and ‘others’. ‘Cell surface’ category included the GO terms ‘plasma membrane’, ‘external side of plasma membrane’ and ‘cell surface’ among others, while the categories ‘Intracellular Compartment (IC)’ and ‘others’ included GO terms such as ‘mitochondrial membrane’ and ‘endoplasmic reticulum’ as well as ‘myelin sheath’, respectively. A detailed list of all annotated GO terms of all fractions can be found in Supplementary Fig. 4. (B) Detection of differentially expressed proteins in the PM fraction of cDCs and B cells of WT versus Marchf1-/- and WT versus Marchf8-/- mice. Equivalent amounts of PM fractions (based on cell count) of three biological replicates were analyzed by mass spectrometry and semi-quantitative proteomics in three technical replicates, for which the foldchange of the label-free quantification (LFQ) peptide peak intensities of each protein was used. Proteins detected in both WT and Marchf1-/- or Marchf8-/- cDCs or B cells were displayed in volcano plots (1020 proteins for WT vs. Marchf1-/- cDCs, 922 proteins for WT vs. Marchf8-/- cDCs, 1275 proteins for WT vs. Marchf1-/- B cells and 1819 proteins for WT vs. Marchf8-/- B cells) with differentially expressed proteins [red dots] identified based on two-fold ratio (log2 protein ratio >1 or <1) and significance (5% FDR) across three biological replicates, each measured in technical triplicates. The known MARCH1 substrates, MHC II (H2-Aa and H2-Ab1) and CD86 in B cells and cDCs, are highlighted in green in each volcano plot.
Comparison of the PM proteomes of WT and Marchf1-/- cDCs showed that, as expected, most proteins were present at similar levels in the two preparations (1020 proteins in total, Supplementary Table 3). Nine proteins were differentially expressed between WT and Marchf1-/- cDC PM [log2 protein ratio >1 or <1 and -log10 adjusted p value > 3.47 (5% FDR)] (Fig. 5B and Supplementary Table 5). These included MHC IIα and β chains (H2-Aa and H2-Ab1), as well as CD86, confirming the validity of our approach to detect MARCH1 substrates. Surprisingly, the protein that appeared most significantly overexpressed in the PM of Marchf1-/- cDCs was complement component 3 (C3) (Fig. 5B, Supplementary Table 5). This was due to covalent binding of constitutively activated C3 to MHC II molecules (Schriek et al., 2021). The remaining three proteins appearing over-expressed in the Marchf1-/- cDC PM fraction are not known to be immunoreceptors expressed at the PM: Cox7a2 is a mitochondrial protein, Myadm a component of the cytoskeleton, and MLV-related proviral Env polyprotein, a protein endogenously encoded by a retrovirus integrated in the genome of commonly used mouse strains (Stocking and Kozak, 2008). As our main goal was to identify immunoregulatory MARCH1 substrates, we did not investigate further whether these were true or false “hits” of the proteomic analysis. Comparison of the PM fractions of WT and Marchf8-/- cDCs did not reveal any differentially expressed proteins (Fig. 5B, 922 proteins in total, Supplementary Table 3), supporting the previous results indicating that MARCH8 is not expressed or active in cDCs.
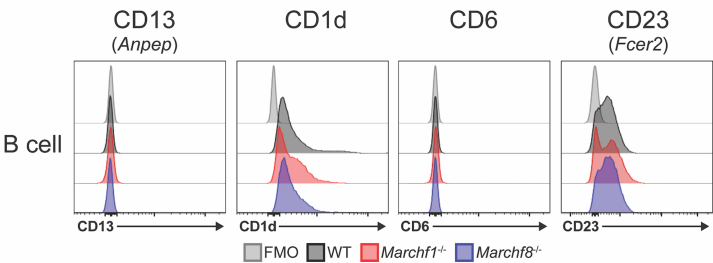
Marchf1-/- and Marchf8-/- B cells exhibited 45 and 40 enriched and 15 and 17 reduced proteins, respectively, in their PM fractions [log2 protein ratio >1 or <1. and -log10 adjusted p value > 2.5 and > 2.36 for Marchf1-/- and Marchf8-/-, respectively (both 5% FDR)] (Fig. 5B, Supplementary Tables 6 and 7, 1275 and 1819 proteins in total, Supplementary Table 3). MHC IIα and β chains (H2-Aa and H2-Ab1), as well as CD86 and C3 were the most significantly enriched proteins in the PM fraction of Marchf1-/- B cells (Fig. 5B and Supplementary Table 6), but neither of the four were enriched in Marchf8-/- B cells (Fig. 5B and Supplementary Table 7). Only 14 of the 60 proteins differentially expressed in the PM fraction of Marchf1-/- B cells, and 10 of the 57 proteins differentially expressed in the PM fraction of Marchf8-/- B cells, were immunoreceptors and/or proteins known to be expressed at the plasma membrane (Supplementary Tables 6 and 7). They included aminopeptidase N (CD13, gene Anpep), antigen-presenting glycoprotein CD1d, T cell differentiation antigen CD6 and the immunoglobulin epsilon Fc receptor CD23 (gene Fcer2). However, analysis by flow cytometry did not confirm differential expression in either Marchf1-/- or Marchf8-/- B cells (Supplementary Fig. 5). The most likely explanation for detection of these “false positives” is that they were caused by subtle differences in the purity of the B cell preparations or their subcellular fractions. In conclusion, MHC class II and CD86 were the only membrane proteins that we could unequivocally confirm as MARCH1 substrates in B cells, and while we cannot discard the possibility that some of the “hits” found in the proteomic screen of Marchf8-/- B cells are indeed MARCH8 substrates, it is more likely that MARCH8 is not active in B cells, just as it is not in DCs.
3.5. MARCH8, not MARCH1, is active in non-hematopoietic cells
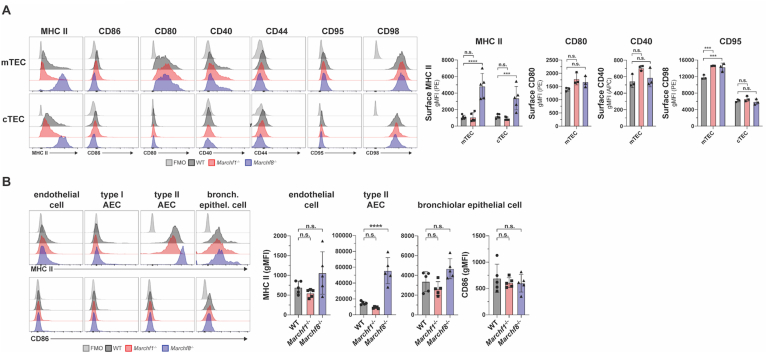
The only cell type in which MARCH8 activity has been demonstrated is thymic epithelial cells (TECs), where it regulates MHC II surface expression but not CD86 (Liu et al., 2016; von Rohrscheidt et al., 2016). Analysis of CD40, CD44, CD95 and CD98 expression in WT and Marchf8-/- medullary and cortical TECs showed that neither of these receptors, which have been shown to be ubiquitinated in cell lines overexpressing MARCH8, are physiological substrates (Fig. 6A).
Fig. 6.
Ubiquitination of MHC II, CD86 and putative substrates by MARCH1 and MARCH8 in non-haemopoietic APCs. (A) Surface expression of MHC II, CD86, CD80, CD40, CD44, CD95, and CD98 in medullary and cortical thymic epithelial cells (mTECs and cTECs) purified from WT, Marchf1-/- and Marchf8-/- mice. (B) Surface expression of MHC II and CD86 in endothelial cells, type I and type II alveolar epithelial cells (AECs) as well as bronchiolar epithelial cells, purified from the lung of WT, Marchf1-/- and Marchf8-/- mice. In all cases a fluorescence-minus-one (FMO) control was included, for which cells were incubated with the corresponding multi-colour staining panel, excluding the fluorescently labelled antibody species of interest. Bars represent mean ± SD with each symbol representing an individual mouse (n = 5). Statistical analysis was performed using one-way ANOVA followed by Sidak's multiple comparisons test. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.0002, n.s. not significant.
Although TECs constitutively present antigens via MHC II, they are not hematopoietic cells, but of endodermal origin (Gordon et al., 2004). Therefore, we asked the question whether other cells ontogenically related to TECs also use MARCH8 to regulate surface MHC II expression. Epithelial cells in the respiratory tract are known to express MHC II, with the highest level found on type II alveolar epithelial cells (AECs) (Wosen et al., 2018; Nakano et al., 2018; Hasegawa et al., 2017). We found that MARCH8-deficient endothelial cells, bronchial epithelial cells and type I and II AECs showed enriched MHC II surface expression (Fig. 6B), though only in type II AECs this difference was clear and statistically significant (Fig. 6B). Neither cell type displayed increased CD86 expression in the absence of MARCH8, and lack of MARCH1 did not affect MHC II nor CD86 expression in any of the cell types analyzed (Fig. 6B). In conclusion, not all epithelial cells regulate MHC II expression via ubiquitination to the same extent, but those that do employ MARCH8.
4. Discussion
Determining which cells utilize MARCH1 and MARCH8 has been hampered by their low level of expression, but analysis of MHC II and CD86 as surrogate markers of activity has allowed us to establish the role of MARCH1 as a master regulator of MHC II and CD86 expression in all hematopoietic cells. MARCH8 plays an equivalent role in the two major types of TECs, in type II AECs and, probably, other epithelial cells, where it ubiquitinates MHC II. We did not observe high CD86 expression in any Marchf8-/- cell, but this could be because these cells do not ubiquitinate CD86 or because they do not express it.
There are at least two precedents for ontogeny-specific differences in the use of components of MHC II antigen presentation machinery. Expression of CIITA, which directs transcription of the genes for MHC II and for several accessory molecules involved in antigen presentation, is driven by distinct promoters in hematopoietic and non-hematopoietic cells (Reith and Mach, 2001). Proteolysis of the chaperone invariant chain, a critical step in the MHC II antigen presentation pathway, is carried out by cathepsin S in hematopoietic cells and by cathepsin L in non-hematopoietic cells (Villadangos and Ploegh, 2000). It is unclear why this dichotomy exists, which is probably caused by the establishment of cell lineage-specific gene programs during embryonic development.
While our finding that MARCH1 is operative in professional APCs confirmed previous observations, we were surprised to observe high MHC II and CD86 expression in non-professional APCs lacking MARCH1. This was not caused by ectopic induction or overexpression of either molecule because MARCH1-deficient T cells maintained WT levels of MHC II (negative) and CD86 (low) expression. As MARCH1 ubiquitinates substrates that have already trafficked through the cell surface, this finding implies that “atypical” APCs express and deposit on their PM larger amounts of MHC II and CD86 than is usually appreciated, but their steady-state levels are kept low by virtue of MARCH1 ubiquitination and accelerated turn-over. Eosinophils are associated with inflammatory responses during allergy or parasitic infections, while neutrophils are recruited in abundant numbers to sites of tissue damage or infection. The role of MHC II antigen presentation by either cell type is controversial. While there is evidence for both purified eosinophils and neutrophils that demonstrates their capacity to present antigen via MHC II (Kambayashi and Laufer, 2014), it is difficult to exclude the possibility of DC contamination in these assays. In vivo evidence of their antigen presentation capacity is scarce but there are reported examples where both eosinophils (Padigel et al., 2006) and neutrophils (Abi Abdallah et al., 2011; Vono et al., 2017; Lin and Loré, 2017) contribute to enhancing antigen-specific CD4+ T cell responses. The realization that these cells regulate MHC II and CD86 via ubiquitination utilizing the same mechanism as professional APCs lends weight to the notion that they perform antigen presentation in vivo.
One of the functions attributed to MARCH8 in humans is to ubiquitinate viral proteins deposited on the PM of infected cells and that will be incorporated in the envelop of the virion upon budding (Tada et al., 2015; Zhang et al., 2020; Kumar et al., 2019). The reduction of viral protein expression that ensues inhibits spread of the infection, protecting the host. This activity has not been described in mice, but our results suggest that if it occurs in this species, it is unlikely to be operative in hematopoietic cells, where perhaps other members of the MARCH family replace the function of MARCH8.
While several membrane proteins have been identified for MARCH1 and MARCH8 based on studies using overexpression and/or cell lines, our flow cytometry analysis rules out CD44, CD71, CD95 and CD98 as bona fide MARCH1 or MARCH8 substrates in all primary cells examined. This highlights that caution needs to be taken when interpreting studies that rely on E3 ligase overexpression. Our unbiased proteomic profiling of B cells and DCs unequivocally confirmed the role of MARCH1 in MHC II and CD86 ubiquitination in both cell types, but did not reveal any other MARCH1 substrate that we could validate by flow cytometry with the exception of complement C3. We have recently shown that C3 is not a MARCH1 substrate but constitutively forms covalent complexes with MHC II on the surface of APCs (Schriek et al., 2021). Its enhanced level on MARCH1-deficient DCs and B cells is therefore an indirect consequence of increased MHC II expression. We have also shown that lack of MHC II ubiquitination induces higher or lower expression of other surface receptors that are not direct MARCH1 substrates (Wilson et al., 2018). However, the magnitude of these changes (∼less than 2-fold) is below the level of resolution afforded by high-throughput, unbiased proteomic analysis of subcellular fractions. Analyses of whole spleen cell lysates by western blot revealed increased expression of the cytosolic proteins MAVS, STING, TRAF3 and TRAF6 in MARCH1-deficient mice (Wu et al., 2020). We could not replicate these findings by mass spectrometry because our analysis focused on proteins associated with microsomal subcellular fractions, excluding the cytosol. MARCH ligases recognize their substrates via transmembrane region interactions (Liu et al., 2019; Trenker et al., 2021) so cytosolic proteins appear unlikely targets of MARCH1 ubiquitination. It is possible their altered abundance in whole spleens of mutant mice is due to changes in cellular composition or to indirect effects of MARCH1 deficiency on transcription programs in APCs (Kim et al., 2021). Confirmation that MAVS, STING, TRAF3 and TRAF6 are MARCH1 substrates will require direct assessment of their ubiquitination status in cells that do or do not express MARCH1.
We did not observe changes in expression of any protein on the PM of Marchf8-/- DCs. The “hits” detected in Marchf8-/- B cell PM by proteomics could be attributed to contamination with other subcellular compartments because they were not classified as PM proteins and/or could not be validated by flow cytometry as differentially expressed. The proteomic analysis thus confirmed that neither B cells nor DCs express functional MARCH8.
In summary, MHC II is the only membrane protein unequivocally regulated by MARCH1 and MARCH8 in primary mouse cells, with each ligase playing its role in haemopoietic and non-haemopoietic cells, respectively. CD86 is also a MARCH1 substrate in hematopoietic cells. These results help to predict the potential effects of genetic or pharmacological manipulation of MARCH1 or MARCH8 activities as a treatment for immunological disorders.
CRediT authorship contribution statement
Patrick Schriek: Methodology, Investigation, Visualization, Writing – original draft, Writing – review & editing. Haiyin Liu: Methodology, Investigation. Alan C. Ching: Methodology, Investigation. Pauline Huang: Methodology, Investigation. Nishma Gupta: Methodology, Investigation. Kayla R. Wilson: Methodology, Investigation. MinHsuang Tsai: Methodology, Investigation. Yuting Yan: Methodology, Investigation. Christophe F. Macri: Methodology, Investigation. Laura F. Dagley: Methodology, Investigation. Giuseppe Infusini: Methodology, Investigation. Andrew I. Webb: Methodology, Investigation. Hamish E.G. McWilliam: Methodology, Investigation, Funding acquisition. Justine D. Mintern: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. Jose A. Villadangos: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Jose A. Villadangos Justine D. Mintern and Hamish E.G. McWilliam report financial support was provided by National Health and Medical Research Council. Jose A. Villadangos and Justine D. Mintern report financial support was provided by Australian Research Council. Jose A. Villadangos and Satoshi Ishido reports financial support was provided by Human Frontier Science Program. Patrick Schriek reports financial support was provided by Australian Government Department of Education Skills and Employment.
Acknowledgements
We thank the Antibody Services Facility and Genomics Hub (Walter and Eliza Hall Institute) and the Melbourne Cytometry Platform (The University of Melbourne) for expert assistance. This research was funded with National Health and Medical Council of Australia (NHMRC) Fellowships 1058193 (JAV) and 1154502l (JAV), NHMRC Program Grant 1016629 (JAV), NHMRC Project Grant 1161101 (JDM) and 1129672 (JDM), 2003192 (HEGMcW), Australian Research Council Discovery Projects 190101242 (JDM), 180100844 (JDM), 160101373 (JDM), 180100521 (JDM), 160103134 (JAV) and 110101383 (JAV), Human Frontiers Science Program Grant 0064/2011 (SI and JAV), and Australian Research Training Programme Scholarship provided by the Australian Commonwealth Government and the University of Melbourne (PS).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crimmu.2021.10.004.
Contributor Information
Justine D. Mintern, Email: jmintern@unimelb.edu.au.
Jose A. Villadangos, Email: j.villadangos@unimelb.edu.au.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. s1.
Representative flow cytometry gating strategies for the identification of cell populations of interest in blood, spleen, subcutaneous lymph nodes (LN), thymus, peritoneal cavity and lung. In all cases cell doublets and dead cells were identified and excluded based on forward and side scatter (FSC and SSC) as well as staining with propidium iodide (PI), diamidino phenylindole (DAPI) or Fixable Viability Dye eFluor™780 (Viability). (A) Blood B cells were identified as CD19+ B220+. (B) Splenic B cells, macrophages and T cells were identified from whole splenocyte suspensions as CD19+ B220+, F4/80+ CD64+ and TCRβ+ CD3+ respectively with further discrimination of CD4+ and CD8+ T cells. Splenic DCs were identified from low-density splenocyte suspensions, with pDCs identified as Siglec-H+ BST-2+ and cDC as B220- CD19- CD11c+ MHC II+, with further discrimination of cDC1s as CD11b- CD8+ and cDC2s as CD11b+ CD8-. Splenic granulocytes and monocytes were identified from whole splenocyte suspensions as B220- CD3- CD4- CD8- CD11clow-mid CD11bhigh with neutrophils identified as Ly6G+, eosinophils as Ly6G- SSC-Hhigh Ly6Clow-mid, patrolling monocytes as Ly6G- Ly6Clow-mid SSC-Hlow and inflammatory monocytes as Ly6G- Ly6Chigh SSC-Hlow (as described in Liyanage et al. (2016)). (C) cDCs from subcutaneous LN were identified from low-density cell suspensions, with resident cDCs identified as CD11chigh MHC IImid and migratory cDCs identified as CD11cmid MHC IIhigh and further discrimination of cDC1s as Sirpα- XCR1+ and cDC2s as Sirpα+ XCR1-. B cells, macrophages and T cells from subcutaneous LN were identified from whole cell suspensions as CD19+ B220+, F4/80+ MHC II+ and CD3+ respectively with further discrimination of CD4+ and CD8+ T cells. (D) Thymic cDCs were identified from low-density cell suspensions as B220– NK1.1- CD11c+ MHC II+, with further discrimination of cDC1s as Sirpα- XCR1+ and cDC2s as Sirpα+ XCR1- (as described in Ardouin et al. (2016)). Thymic epithelial cells (TECs) were identified from whole thymocyte suspensions as CD45- EpCAM+, with further discrimination of cortical TECs (cTECs) as UEA-1- Ly51+ and medullary TECs (mTECs) as UEA-1+ Ly51- (as described in Liu et al. (2016)). (D) Peritoneal macrophages were identified as CD11b+ MerTK+, with further discrimination of small peritoneal macrophages as F4/80low MHC IIhigh and large peritoneal macrophages as F4/80high MHC IImid-high (as described in Bain et al. (2016)). Peritoneal B cells were identified as MerTK- MHC II+ CD19+ and cDCs as MerTK- CD19- CD11c+ MHC II+ with further discrimination of cDC1s as CD11b- XCR1+ and cDC2s as CD11b+ XCR1-. (E) Haemopoietic cells in the lung were identified as CD45+ with pDC as CD11clow-mid BST-2+ and macrophages as CD64+ MerTK+, with further discrimination of interstitial macrophages as Siglec-Flow CD11bhigh and alveolar macrophages as Siglec-Fhigh CD11bmid (as described in Svedberg et al. (2019)). Lung B cells were identified as CD64- MerTK- CD11c- MHC II+ CD19+ and cDCs as CD64- MerTK- CD11c+ MHC II+ with further discrimination of cDC1s as CD11b- XCR1+ and cDC2s as CD11b+ XCR1-. Lung granulocytes and monocytes were identified as B220- CD3- CD4- CD8- CD11clow-mid CD11bhigh with neutrophils identified as Ly6G+, eosinophils as Ly6G- SSC-Hhigh Ly6Clow-mid, patrolling monocytes as Ly6G- Ly6Clow-mid SSC-Hlow and inflammatory monocytes as Ly6G- Ly6Chigh SSC-Hlow (as described in Liyanage et al., 2016). Non-haemopoietic cells in the lung were identified as CD45- with endothelial cells identified as EpCAMlow-mid CD31+ Sca-1+ and epithelial cells as EpCAMmid-high CD31-. Further discrimination of epithelial cells was carried out based of CD24, EpCAM and MHC II expression (as described in Nakano et al. (2018) and Hasegawa et al. (2017)) with bronchiolar epithelial cells identified as EpCAMhigh CD24high, type II alveolar epithelial cells (AECs) identified as EpCAMhigh CD24mid MHC IIhigh (red) and type I AECs identified as EpCAMmid CD24low MHC IIlow (green).
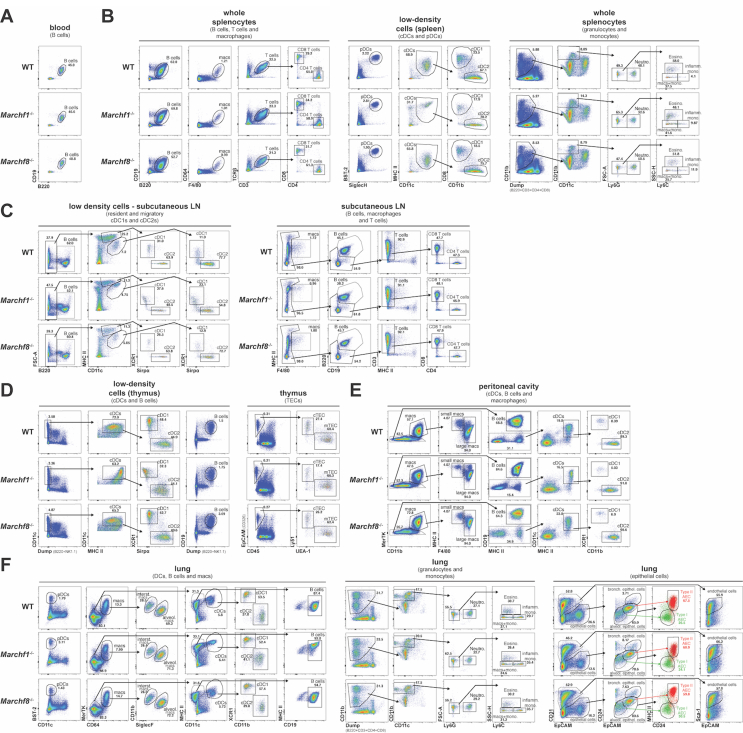
A comparison of the representative flow cytometry gating strategies for the identification of all cell populations of interest between WT, Marchf1-/- and Marchf8-/- mice is shown in Supplementary Fig. 2.
Fig. s2.
Comparison of representative flow cytometry gating strategies for the identification of various cell populations in (A) blood, (B) spleen, (C) subcutaneous lymph node, (D) thymus, (E) peritoneal cavity and (F) lung from WT, Marchf1-/- and Marchf8-/- mice. A detailed description of the gating strategies for each individual cell population of interest is presented in Supplementary Fig. 1.
Fig. s3.
Representative flow cytometry histograms for the surface expression of MHC II and CD86 in (A) blood B cells, (B) resident and migratory cDC1s and cDC2s, as well as B cells, macrophages and CD4+/CD8+ T cells in subcutaneous (axillary + inguinal) lymph nodes, (C) thymic cDC1s, cDC2s, and B cells, (D) peritoneal cDC1s, cDC2s, B cells, and small/large macrophages and (E) lung cDC1s, cDC2s, pDC, B cells, and alveolar/interstitial macrophages, all purified from WT mice or mice deficient in either MARCH1 or MARCH8. (F) Surface expression of MHC II and CD86 in peritoneal cDCs from WT, Marchf1-/- and Marchf8-/- mice or from mice deficient in both MARCH1 and MARCH8 (Marchf1-/- x Marchf8-/-). In all cases (A-F) a fluorescence-minus-one (FMO) control was included, for which cells were incubated with the corresponding multi-colour staining panel, excluding the fluorescently labelled antibody species of interest (i.e. anti-CD86 or anti-MHC II mAb) Bar plots in (F) represent mean ± SD with each symbol representing an individual mouse (n = 4). Statistical analysis was performed using one-way ANOVA followed by Sidak's multiple comparisons test. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.0002, n.s. not significant.
Fig. s4.
Gene ontology (GO) enrichment analysis of proteins detected in plasma membrane (PM)-enriched and intracellular compartment (IC)-enriched microsome fractions of splenic cDCs and B cells purified from WT, Marchf1-/-, or Marchf8-/- mice. PM fractions were purified from post-nuclear supernatants of mAb surface stained cDCs and B cells via magnetic immunoaffinity. IC (intracellular compartments) was retrieved from the post-nuclear supernatant of homogenized cells following PM fraction extraction. IC and PM fraction were analyzed using semi-quantitative mass spectrometry and GO term enrichment analysis was performed using the Bioconductor clusterProfiler package (Yu et al., 2012) with GO-IDs grouped based on experimentally verified Gene Ontology (GO) annotations.
Fig. s5.
Representative flow cytometry histograms for the surface expression of CD13, CD1d, CD6 and CD23 in B cells from WT, Marchf1-/- and Marchf8-/- mice. A fluorescence-minus-one (FMO) control was included, for which cells were incubated with the corresponding multi-colour staining panel, excluding the fluorescently labelled antibody species of interest.
References
- Abi Abdallah D.S., Egan C.E., Butcher B.A., Denkers E.Y. Mouse neutrophils are professional antigen-presenting cells programmed to instruct Th1 and Th17 T-cell differentiation. Int. Immunol. 2011;23:317–326. doi: 10.1093/intimm/dxr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablack J.N.G., Cantor J.M., Metz P.J., Chang J.T., Ginsberg M.H. Ubiquitination of CD98 limits cell proliferation and clonal expansion. J. Cell Sci. 2015;128:4273–4278. doi: 10.1242/jcs.178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardouin L., Luche H., Chelbi R., Carpentier S., Shawket A., Montanana Sanchis F., Santa Maria C., Grenot P., Alexandre Y., Grégoire C., Fries A., Vu Manh T.-P., Tamoutounour S., Crozat K., Tomasello E., Jorquera A., Fossum E., Bogen B., Azukizawa H., Bajenoff M., Henri S., Dalod M., Malissen B. Broad and largely concordant molecular changes characterize tolerogenic and immunogenic dendritic cell maturation in thymus and periphery. Immunity. 2016;45:305–318. doi: 10.1016/j.immuni.2016.07.019. [DOI] [PubMed] [Google Scholar]
- Auffray C., Sieweke M.H., Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu. Rev. Immunol. 2009;27:669–692. doi: 10.1146/annurev.immunol.021908.132557. [DOI] [PubMed] [Google Scholar]
- Bain C.C., Hawley C.A., Garner H., Scott C.L., Schridde A., Steers N.J., Mack M., Joshi A., Guilliams M., Mowat A.M.I., Geissmann F., Jenkins S.J. Long-lived self-renewing bone marrow-derived macrophages displace embryo-derived cells to inhabit adult serous cavities. Nat. Commun. 2016;7 doi: 10.1038/ncomms11852. ncomms11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannard O., McGowan S.J., Ersching J., Ishido S., Victora G.D., Shin J.-S., Cyster J.G. Ubiquitin-mediated fluctuations in MHC class II facilitate efficient germinal center B cell responses. J. Exp. Med. 2016;213:993–1009. doi: 10.1084/jem.20151682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baravalle G., Park H., McSweeney M., Ohmura-Hoshino M., Matsuki Y., Ishido S., Shin J.-S. Ubiquitination of CD86 is a key mechanism in regulating antigen presentation by dendritic cells. J. Immunol. 2011;187:2966–2973. doi: 10.4049/jimmunol.1101643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee E., Mansouri M., Hovey B.T., Gouveia K., Früh K., Nerenberg B.T.H., Fru K. Downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. J. Virol. 2004;78:1109–1120. doi: 10.1128/JVI.78.3.1109-1120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois-Daigneault M.-C., Thibodeau J. Autoregulation of MARCH1 expression by dimerization and autoubiquitination. J. Immunol. 2012;188:4959–4970. doi: 10.4049/jimmunol.1102708. [DOI] [PubMed] [Google Scholar]
- Cho K.-J., Walseng E., Ishido S., Roche P.A. Ubiquitination by March-I prevents MHC class II recycling and promotes MHC class II turnover in antigen-presenting cells. Proc. Natl. Acad. Sci. Unit. States Am. 2015;112:10449–10454. doi: 10.1073/pnas.1507981112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran K., Jabbour M., Bhagwandin C., Deymier M.J., Theisen D.L., Lybarger L. Ubiquitin-mediated regulation of CD86 protein expression by the ubiquitin ligase membrane-associated RING-CH-1 (MARCH1) J. Biol. Chem. 2011;286:37168–37180. doi: 10.1074/jbc.M110.204040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscoy L., Sanchez D.J., Ganem D. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J. Cell Biol. 2001;155:1265–1274. doi: 10.1083/jcb.200111010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- De Gassart A., Camosseto V., Thibodeau J., Ceppi M., Catalan N., Pierre P., Gatti E. MHC class II stabilization at the surface of human dendritic cells is the result of maturation-dependent MARCH I down-regulation. Proc. Natl. Acad. Sci. Unit. States Am. 2008;105:3491–3496. doi: 10.1073/pnas.0708874105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann M.J., Nicholson B., Kessler B.M. Pharmacological targets in the ubiquitin system offer new ways of treating cancer, neurodegenerative disorders and infectious diseases. Expet Rev. Mol. Med. 2011;13:1–17. doi: 10.1017/S1462399411002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyster C.A., Cole N.B., Petersen S., Viswanathan K., Früh K., Donaldson J.G. MARCH ubiquitin ligases alter the itinerary of clathrin-independent cargo from recycling to degradation. Mol. Biol. Cell. 2011;22:3218–3230. doi: 10.1091/mbc.E10-11-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita H., Iwabu Y., Tokunaga K., Tanaka Y. Membrane-associated RING-CH (MARCH) 8 mediates the ubiquitination and lysosomal degradation of the transferrin receptor. J. Cell Sci. 2013;126:2798–2809. doi: 10.1242/jcs.119909. [DOI] [PubMed] [Google Scholar]
- Galbas T., Raymond M., Sabourin A., Bourgeois-Daigneault M.-C., Guimont-Desrochers F., Yun T.J., Cailhier J.-F., Ishido S., Lesage S., Cheong C., Thibodeau J. MARCH1 E3 ubiquitin ligase dampens the innate inflammatory response by modulating monocyte functions in mice. J. Immunol. 2017;198:852–861. doi: 10.4049/jimmunol.1601168. [DOI] [PubMed] [Google Scholar]
- Galbas T., Thibodeau J. Cell-type specific regulation of MARCH1 E3 ubiquitin ligase by the anti-inflammatory cytokine IL-10. Open J. Immunol. 2012;2:161–167. [Google Scholar]
- Gordon J., Wilson V.A., Blair N.F., Sheridan J., Farley A., Wilson L., Manley N.R., Blackburn C.C. Functional evidence for a single endodermal origin for the thymic epithelium. Nat. Immunol. 2004;5:546–553. doi: 10.1038/ni1064. [DOI] [PubMed] [Google Scholar]
- Hasegawa K., Sato A., Tanimura K., Uemasu K., Hamakawa Y., Fuseya Y., Sato S., Muro S., Hirai T. Fraction of MHCII and EpCAM expression characterizes distal lung epithelial cells for alveolar type 2 cell isolation. Respir. Res. 2017;18:150. doi: 10.1186/s12931-017-0635-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishido S., Wang C., Lee B.-S., Cohen G.B., Jung J.U. Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J. Virol. 2000;74:5300–5309. doi: 10.1128/jvi.74.11.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R., Kajikawa M., Ishido S. Loss of MHC II ubiquitination inhibits the activation and differentiation of CD4 T cells. Int. Immunol. 2014;26:283–289. doi: 10.1093/intimm/dxt066. [DOI] [PubMed] [Google Scholar]
- Jabbour M., Campbell E.M., Fares H., Lybarger L. Discrete domains of MARCH1 mediate its localization, functional interactions, and posttranscriptional control of expression. J. Immunol. 2009;183:6500–6512. doi: 10.4049/jimmunol.0901521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubzick C.V., Randolph G.J., Henson P.M. Monocyte differentiation and antigen-presenting functions. Nat. Rev. Immunol. 2017;17:349–362. doi: 10.1038/nri.2017.28. [DOI] [PubMed] [Google Scholar]
- Kambayashi T., Laufer T.M. Atypical MHC class II-expressing antigen-presenting cells: can anything replace a dendritic cell? Nat. Rev. Immunol. 2014;14:719–730. doi: 10.1038/nri3754. [DOI] [PubMed] [Google Scholar]
- Kaul S., Mittal S.K., Roche P.A. A major isoform of the E3 ubiquitin ligase March-I in antigen-presenting cells has regulatory sequences within its gene. J. Biol. Chem. 2018;293:4478–4485. doi: 10.1074/jbc.RA118.001775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Bandola-Simon J., Ishido S., Wong N.W., Koparde V.N., Cam M., Roche P.A. Ubiquitination of MHC class II by march-I regulates dendritic cell fitness. J. Immunol. 2021;206:494–504. doi: 10.4049/jimmunol.2000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishta O.A., Sabourin A., Simon L., McGovern T., Raymond M., Galbas T., Majdoubi A., Ishido S., Martin J.G., Thibodeau J. March1 E3 ubiquitin ligase modulates features of allergic asthma in an ovalbumin-induced mouse model of lung inflammation. J. Immunol. Res. 2018;2018:1–17. doi: 10.1155/2018/3823910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Köntgen F., Süss G., Stewart C., Steinmetz M., Bluethmann H. Targeted disruption of the MHC class II Aa gene in C57BL/6 mice. Int. Immunol. 1993;5:957–964. doi: 10.1093/intimm/5.8.957. [DOI] [PubMed] [Google Scholar]
- Kumar S., Barouch-Bentov R., Xiao F., Schor S., Pu S., Biquand E., Lu A., Lindenbach B.D., Jacob Y., Demeret C., Einav S. MARCH8 ubiquitinates the hepatitis C virus nonstructural 2 protein and mediates viral envelopment. Cell Rep. 2019;26:1800–1814. doi: 10.1016/j.celrep.2019.01.075. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassila O., Vainio O., Matzinger P. Can B cells turn on virgin T cells? Nature. 1988;334:253–255. doi: 10.1038/334253a0. [DOI] [PubMed] [Google Scholar]
- Lei L., Bandola-Simon J., Roche P.A. Ubiquitin-conjugating enzyme E2 D1 (Ube2D1) mediates lysine-independent ubiquitination of the E3 ubiquitin ligase March-I. J. Biol. Chem. 2018;293:3904–3912. doi: 10.1074/jbc.RA117.001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A., Loré K. Granulocytes: new members of the antigen-presenting cell family. Front. Immunol. 2017;8:1–8. doi: 10.3389/fimmu.2017.01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Jain R., Guan J., Vuong V., Ishido S., La Gruta N.L., Gray D.H., Villadangos J.A., Mintern J.D. Ubiquitin ligase MARCH 8 cooperates with CD83 to control surface MHC II expression in thymic epithelium and CD4 T cell selection. J. Exp. Med. 2016;213:1695–1703. doi: 10.1084/jem.20160312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Mintern J.D., Villadangos J.A. MARCH ligases in immunity. Curr. Opin. Immunol. 2019;58:38–43. doi: 10.1016/j.coi.2019.03.001. [DOI] [PubMed] [Google Scholar]
- Liyanage S.E., Gardner P.J., Ribeiro J., Cristante E., Sampson R.D., Luhmann U.F.O., Ali R.R., Bainbridge J.W. Flow cytometric analysis of inflammatory and resident myeloid populations in mouse ocular inflammatory models. Exp. Eye Res. 2016;151:160–170. doi: 10.1016/j.exer.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach N., Gillessen S., Wilson S.B., Sheehan C., Mihm M., Dranoff G. Differences in dendritic cells stimulated in vivo by tumors engineered to secrete granulocyte-macrophage colony-stimulating factor or Flt3-ligand. Cancer Res. 2000;60:3239–3246. [PubMed] [Google Scholar]
- Matsuki Y., Ohmura-Hoshino M., Goto E., Aoki M., Mito-Yoshida M., Uematsu M., Hasegawa T., Koseki H., Ohara O., Nakayama M., Toyooka K., Matsuoka K., Hotta H., Yamamoto A., Ishido S. Novel regulation of MHC class II function in B cells. EMBO J. 2007;26:846–854. doi: 10.1038/sj.emboj.7601556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano H., Nakano K., Cook D.N. In: Methods in Molecular Biology. Springer New York, New York, NY. Reinhardt R.L., editor. 2018. Isolation and purification of epithelial and endothelial cells from mouse lung; pp. 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J., Wu N., Baravalle G., Cohn B., Ma J., Lo B., Mellman I., Ishido S., Anderson M., Shin J.-S. MARCH1-mediated MHCII ubiquitination promotes dendritic cell selection of natural regulatory T cells. J. Exp. Med. 2013;210:1069–1077. doi: 10.1084/jem.20122695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padigel U.M., Lee J.J., Nolan T.J., Schad G.A., Abraham D. Eosinophils can function as antigen-presenting cells to induce primary and secondary immune responses to strongyloides stercoralis. Infect. Immun. 2006;74:3232–3238. doi: 10.1128/IAI.02067-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz Ş., Tiwary S., Cox J., Audain E., Walzer M., Jarnuczak A.F., Ternent T., Brazma A., Vizcaíno J.A. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B., Lee S., Majewski I.J., Alexander W.S., Smyth G.K. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann. Appl. Stat. 2016;10:946–963. doi: 10.1214/16-AOAS920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reith W., Mach B. The bare lymphocyte syndrome and the regulation of MHC expression. Annu. Rev. Immunol. 2001;19:331–373. doi: 10.1146/annurev.immunol.19.1.331. [DOI] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samji T., Hong S., Means R.E. The membrane associated RING-CH proteins: a family of E3 ligases with diverse roles through the cell. Int. Sch. Res. Not. 2014;2014:1–23. doi: 10.1155/2014/637295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura E., Albiston A.L., Wicks I.P., Chai S.Y., Villadangos J.A. Different cross-presentation pathways in steady-state and inflammatory dendritic cells. Proc. Natl. Acad. Sci. U. S. A. 2009;106:20377–20381. doi: 10.1073/pnas.0910295106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura E., Kapp E., Gupta N., Wong J., Lim J., Ji H., Heath W.R., Simpson R., Villadangos J.A. Differential expression of pathogen-recognition molecules between dendritic cell subsets revealed by plasma membrane proteomic analysis. Mol. Immunol. 2010;47:1765–1773. doi: 10.1016/j.molimm.2010.02.028. [DOI] [PubMed] [Google Scholar]
- Schriek P., Ching A.C., Moily N.S., Moffat J., Beattie L., Steiner T.M., Hosking L.M., Thurman J.M., Holers V.M., Ishido S., Lahoud M.H., Caminschi I., Heath W.R., Mintern J.D., Villadangos J.A. Marginal zone B Cells trogocytose from dendritic cells and present to T cells MHC II-C3 complexes. Science. 2021 doi: 10.1126/science.abf7470. (in press) [DOI] [PubMed] [Google Scholar]
- Stocking C., Kozak C.A. Endogenous retroviruses. Cell. Mol. Life Sci. 2008;65:3383–3398. doi: 10.1007/s00018-008-8497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svedberg F.R., Brown S.L., Krauss M.Z., Campbell L., Sharpe C., Clausen M., Howell G.J., Clark H., Madsen J., Evans C.M., Sutherland T.E., Ivens A.C., Thornton D.J., Grencis R.K., Hussell T., Cunoosamy D.M., Cook P.C., MacDonald A.S. The lung environment controls alveolar macrophage metabolism and responsiveness in type 2 inflammation. Nat. Immunol. 2019;20:571–580. doi: 10.1038/s41590-019-0352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Zhang Y., Koyama T., Tobiume M., Tsunetsugu-Yokota Y., Yamaoka S., Fujita H., Tokunaga K. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins. Nat. Med. 2015;21:1502–1507. doi: 10.1038/nm.3956. [DOI] [PubMed] [Google Scholar]
- Thibodeau J., Bourgeois-Daigneault M.-C., Huppé G., Tremblay J., Aumont A., Houde M., Bartee E., Brunet A., Gauvreau M., de Gassart A., Gatti E., Baril M., Cloutier M., Bontron S., Früh K., Lamarre D., Steimle V. Interleukin-10-induced MARCH1 mediates intracellular sequestration of MHC class II in monocytes. Eur. J. Immunol. 2008;38:1225–1230. doi: 10.1002/eji.200737902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomoto M., Ishido S., Miyasaka N., Sugimoto H., Kohsaka H. Anti-arthritic effect of E3 ubiquitin ligase, c-MIR, expression in the joints. Int. Immunol. 2011;23:177–183. doi: 10.1093/intimm/dxq470. [DOI] [PubMed] [Google Scholar]
- Trenker R., Wu X., Nguyen J.V., Wilcox S., Rubin A.F., Call M.E., Call M.J. Human and viral membrane–associated E3 ubiquitin ligases MARCH1 and MIR2 recognize different features of CD86 to downregulate surface expression. J. Biol. Chem. 2021;297:100900. doi: 10.1016/j.jbc.2021.100900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Ramos J., Roquilly A., Zhan Y., Young L.J., Mintern J.D., Villadangos J.A. Inflammation conditions mature dendritic cells to retain the capacity to present new antigens but with altered cytokine secretion function. J. Immunol. 2014;193:3851–3859. doi: 10.4049/jimmunol.1303215. [DOI] [PubMed] [Google Scholar]
- Villadangos J.A., Cardoso M., Steptoe R.J., van Berkel D., Pooley J., Carbone F.R., Shortman K. MHC class II expression is regulated in dendritic cells independently of invariant chain degradation. Immunity. 2001;14:739–749. doi: 10.1016/s1074-7613(01)00148-0. [DOI] [PubMed] [Google Scholar]
- Villadangos J.A., Ploegh H.L. Proteolysis in MHC class II antigen presentation: who's in charge? Immunity. 2000;12:233–239. doi: 10.1016/s1074-7613(00)80176-4. [DOI] [PubMed] [Google Scholar]
- von Rohrscheidt J., Petrozziello E., Nedjic J., Federle C., Krzyzak L., Ploegh H.L., Ishido S., Steinkasserer A., Klein L. Thymic CD4 T cell selection requires attenuation of March8-mediated MHCII turnover in cortical epithelial cells through CD83. J. Exp. Med. 2016;213:1685–1694. doi: 10.1084/jem.20160316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vono M., Lin A., Norrby-Teglund A., Koup R.A., Liang F., Loré K. Neutrophils acquire the capacity for antigen presentation to memory CD4+ T cells in vitro and ex vivo. Blood. 2017;129:1991–2001. doi: 10.1182/blood-2016-10-744441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vremec D. In: Methods in Molecular Biology. Clifton N.J., editor. 2010. The isolation of mouse dendritic cells from lymphoid tissues and the identification of dendritic cell subtypes by multiparameter flow cytometry; pp. 205–229. [DOI] [PubMed] [Google Scholar]
- Wilson K.R., Liu H., Healey G., Vuong V., Ishido S., Herold M.J., Villadangos J.A., Mintern J.D. MARCH1-mediated ubiquitination of MHC II impacts the MHC I antigen presentation pathway. PLoS One. 2018;13 doi: 10.1371/journal.pone.0200540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson N.S., El-Sukkari D., Villadangos J.A. Dendritic cells constitutively present self antigens in their immature state in vivo and regulate antigen presentation by controlling the rates of MHC class II synthesis and endocytosis. Blood. 2004;103:2187–2195. doi: 10.1182/blood-2003-08-2729. [DOI] [PubMed] [Google Scholar]
- Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Wosen J.E., Mukhopadhyay D., Macaubas C., Mellins E.D. Epithelial MHC class II expression and its role in antigen presentation in the gastrointestinal and respiratory tracts. Front. Immunol. 2018;9:1–14. doi: 10.3389/fimmu.2018.02144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Xia L., Yao X., Yu X., Tumas K.C., Sun W., Cheng Y., He X., Peng Y.C., Singh B.K., Zhang C., Qi C.F., Bolland S., Best S.M., Gowda C., Huang R., Myers T.G., Long C.A., Wang R.F., Su X.Z. The E3 ubiquitin ligase MARCH1 regulates antimalaria immunity through interferon signaling and T cell activation. Proc. Natl. Acad. Sci. U. S. A. 2020;117:16567–16578. doi: 10.1073/pnas.2004332117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Dai H., Li M., Yang W., Yu G., Wang X., Wang P., Liu W., Hu X., Zhao M. MARCH1 encourages tumour progression of hepatocellular carcinoma via regulation of PI3K-AKT-β-catenin pathways. J. Cell Mol. Med. 2019;23:3386–3401. doi: 10.1111/jcmm.14235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L.J., Wilson N.S., Schnorrer P., Proietto A., ten Broeke T., Matsuki Y., Mount A.M., Belz G.T., O'Keeffe M., Ohmura-Hoshino M., Ishido S., Stoorvogel W., Heath W.R., Shortman K., Villadangos J.A. Differential MHC class II synthesis and ubiquitination confers distinct antigen-presenting properties on conventional and plasmacytoid dendritic cells. Nat. Immunol. 2008;9:1244–1252. doi: 10.1038/ni.1665. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Tada T., Ozono S., Kishigami S., Fujita H., Tokunaga K. MARCH8 inhibits viral infection by two different mechanisms. Elife. 2020;9:1–14. doi: 10.7554/eLife.57763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Tada T., Ozono S., Yao W., Tanaka M., Yamaoka S., Kishigami S., Fujita H., Tokunaga K. Membrane-associated RING-CH (MARCH) 1 and 2 are MARCH family members that inhibit HIV-1 infection. J. Biol. Chem. 2019;294:3397–3405. doi: 10.1074/jbc.AC118.005907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Tsai Y.C., Jin B., Wang B., Wang Y., Zhou H., Carpenter T., Weissman A.M., Yin J. Protein engineering in the ubiquitin system: tools for discovery and beyond. Pharmacol. Rev. 2020;72:380–413. doi: 10.1124/pr.118.015651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.