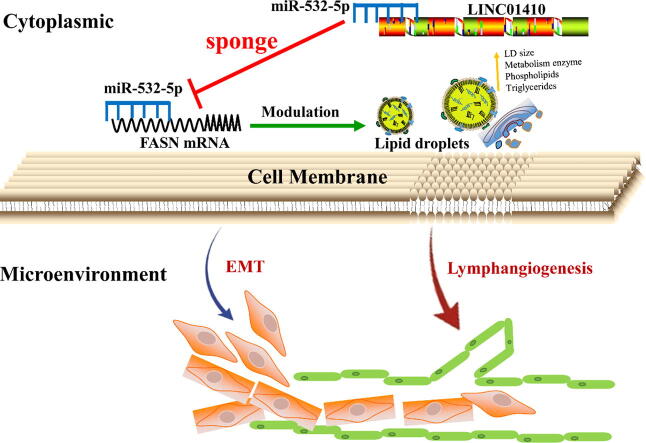
Graphical abstract
The prognostic miR-532-5p inhibited epithelial-mesenchymal transition and lymphangiogenesis by regulating lipid droplets accumulation. Mechanistically, a miR-532-5p-correlated ceRNA network in which LINC01410 directly bound to miR-532-5p effectively functioned as a sponge for miR-532-5p to disinhibit its target gene-FASN. Combined therapy with miR-532-5p and FASN inhibitor-orlistat blocked lymph node metastasis and tumor growth.
Keywords: miRNA prognostic model, Lipid droplet, FASN, Lymph node metastasis, Cervical cancer
Abbreviations: CC, Cervical cancer; LDs, Lipid droplets; miRNAs, MicroRNAs; LNM, lymph node metastasis; LINC01410, Long non-coding RNA 01410; TCGA, The Cancer Genome Atlas; LASSO, The least absolute shrinkage and selection operator; FA, Fatty acid; TGs, Triglycerides; PLs, Phospholipids; ncRNAs, Noncoding RNAs; LncRNAs, Long noncoding RNAs; CircRNAs, circular RNAs; ceRNA, Competing endogenous RNA; MREs, MiRNA response elements; EMT, Epithelial-mesenchymal transition; ATCC, The American Type Culture Collection; HLECs, Human lymphatic endothelial cells; STR, short tandem repeat; ISH, In situ hybridization; IHC, immunohistochemistry; RIP, RNA immunoprecipitation; HE, Hematoxylin-eosin; GSEA, Gene set enrichment analysis; ES, enrichment score; MES, Maximum enrichment score; SNR, signal-to-noise ratio; GEPIA, The Gene Expression Profiling Interactive Analysis; ROC, The receiver operating characteristic; OS, Overall survival; DFS, Disease free survival; FASN, Fatty acid synthase; ACC1, Acetyl-CoA carboxylase 1; ACOX1, Acyl-CoA oxidase 1; CPT1A, Carnitine palmitoyltransferase-1A; PLIN2, Perilipin 2; Mfe, The minimum free energy; CPAT, The coding potential assessment tool; BMI, Body mass index; NCF2, Neutrophil cytosolic factor 2
Highlights
-
•
The prognostic miR-532-5p inhibited epithelial-mesenchymal transition and lymphangiogenesis by regulating lipid droplets accumulation.
-
•
miR-532-5p-correlated ceRNA network in which LINC01410 directly bound to miR-532-5p effectively functioned as a sponge for miR-532-5p to disinhibit its target gene-FASN.
-
•
Combined therapy with miR-532-5p and FASN inhibitor-orlistat blocked lymph node metastasis and tumor growth.
Abstract
Introduction
The prognosis for cervical cancer (CC) patients with lymph node metastasis (LNM) is extremely poor. Lipid droplets (LDs) have a pivotal role in promoting tumor metastasis. The crosstalk mechanism between LDs and LNM modulated in CC remains largely unknown.
Objectives
This study aimed to construct a miRNA-dependent progonostic model for CC patients and investigate whether miR-532-5p has a biological impact on LNM by regualting LDs accumulation.
Methods
LASSO-Cox regression was applied to establish a prognostic prediction model. miR-532-5p had the lowest P-value in RNA expression (P < 0.001) and prognostic prediction (P < 0.0001) and was selected for further study. The functional role of the prognostic miR-532-5p-correlated competing endogenous RNA (ceRNA) network was investigated to clarify the crosstalk between LDs and LNM. The underlying mechanism was determined using site-directed mutagenesis, dual luciferase reporter assays, RNA immunoprecipitation assays, and rescue experiments. A xenograft LNM model was established to evaluate the effect of miR-532-5p and orlistat combination therapy on tumor growth and LNM.
Results
A novel 5-miRNAs prognostic signature was constructed to better predict the prognosis of CC patient. Further study demonstrated that miR-532-5p inhibited epithelial-mesenchymal transition and lymphangiogenesis by regulating LDs accumulation. Interestingly, we also found that LDs accumulation promoted cell metastasis in vitro. Mechanistically, we demonstrated a miR-532-5p-correlated ceRNA network in which LINC01410 was bound directly to miR-532-5p and effectively functioned as miR-532-5p sponge to disinhibit its target gene-fatty acid synthase (FASN). Combined therapy with miR-532-5p and FASN inhibitor-orlistat further inhibited tumor growth and LNM in vivo.
Conclusion
Our findings highlight a LD accumulation-dependent mechanism of miR-532-5p-modulated LNM and support treatment with miR-532-5p/orlistat as novel strategy for treating patients with LNM in CC.
Introduction
Cervical cancer (CC) is among the top four cancers affecting women worldwide.[1]. Metastasis, especially lymph node metastasis (LNM), is frequent in CC patients, even in disease after early stage and is the leading causes of CC-associated mortality [2]. The 5-year survival rate of CC patients without LNM is 80–90%, while in those with LNM, it is reduced to 50–65% [3], [4]. To improve the prognosis of CC patients with LNM, exploration of molecular mechanisms and effective targets to prevent LNM are urgently needed.
Metabolic reprogramming has been recognized as a hallmark of cancer metastasis [5]. Although the Warburg effect is the best characterized metabolic phenotype of tumor, growing evidence indicates that the metabolic phenotype of carcinogenesis is not consistent with those in cancer metastasis [6], [7]. Lipid metabolic abnormalities may confers a selective metastatic advantage for cancer cells and fatty acid (FA) metabolism reprogramming is involved in modulating tumor metastasis [8], [9]. Lipid droplets (LDs), the main storage site of intracellular lipids, are multifunctional dynamic organelles. Studies have shown that LDs are composed of a neutral lipid core surrounded by a monolayer of phospholipids with a diverse array of associated proteins involved in lipid metabolism [10], [11]. LDs are not only the main building blocks of membrane biosynthesis but also critical central regulators of inflammatory and metastatic pathways [12], [13]. However, the relative contribution of LD accumulation to nodal metastasis in CC remains incompletely understood.
Noncoding RNAs (ncRNAs) have been defined as the new central dogma of cancer biology [14], [15]. With rapid advancement of research on the mechanisms of ncRNA interactions in tumor progression, it has become increasingly difficult to view ncRNA functions in isolation. The “competing endogenous RNA” (ceRNA) hypothesis provides an exciting novel aspect of RNA biology [16], [17]. According to this hypothesis, ceRNAs are gene-regulatory ncRNAs that harbor miRNA response elements (MREs) [17]. Indeed, numerous studies have demonstrated the value of ceRNA network in cancer progression [18], [19], [20]. We have characterized LNMICC as an lncRNA that promotes metastasis in CC through the reprogramming of fatty acid metabolism [9]. Unfortunately, it is still unclear whether and how the ceRNA network is involved in modulating the crosstalk between LD accumulation and nodal metastasis in CC.
Here, we report that miR-532-5p is downregulated in CC patients with LNM based on the TCGA (The Cancer Genome Atlas) database for the first time. miR-532-5p suppressed epithelial-mesenchymal transition (EMT) signaling pathway by targeting fatty acid synthase (FASN). We also demonstrate that miR-532-5p specifically inhibits LNM by modulating LD accumulation and metabolism. Importantly, we verified that LINC01410 acted in a ceRNA-dependent manner to positively regulate FASN expression through sponging miR-532-5p. Moreover, our results suggest that LINC01410/miR-532-5p/FASN ceRNA network is a prognostic marker for poor survival and correlates positively to LNM. Collectively, these findings may provide a novel avenue for exploring the mechanism of LNM and highlight the potential clinical utility of the ceRNA network as a promising therapeutic target in CC.
Materials and methods
Ethics statement
The study was in accordance with the Ethics Committee of Peking University Third Hospital and Helsinki declaration. Informed consent was obtained from each patient. All specimens were handled based on the legal and ethical standards. All animal experiments were approved by the Animal Ethical and Welfare Committee of Peking University. (Approval no. S2019142)
Clinical specimens
For total RNA isolation, all freshly frozen CC samples (n = 20) were obtained from Peking University Third Hospital. Normal uterine cervical tissues were obtained from hysterectomy specimens of 5 nonmalignant pathology patients and used used as control. All specimens were frozen immediately at −80 °C.
Cell lines
The HeLa, SiHa and MS751 cells were purchased from American Type Culture Collection. Human lymphatic endothelial cells (HLECs) were obtained from ScienCell Research Laboratories (Carlsbad, USA). The cells were cultured at 37 °C containing 5% CO2 according to the guidelines.
RNA extraction, qRT-PCR, in situ hybridization (ISH) and Western blot analysis
Total RNA was extracted using TRIzol reagent (TaKaRa, Japan). QRT-PCR was conducted on ABI 7500 Real-Time PCR system (Thermo Scientific) using SYBR Premix Ex Taq (TaKaRa). For the detection of miRNA and U6, RNA samples were reverse-transcribed (RT) into complementary DNA using PrimeScrip RT Reagent Kit (TaKaRa, Japan) by specific stem loop RT primers. The results were calculated using 2–ΔΔCT method. The ISH probe of LINC01410 was synthesized by TaKaRa Biotech Co. Primer sequences are listed in Supplementary Table 1. ISH and Western blot analysis were carried out using routine procedures as described in the Supplementary Methods and Supplementary Table 2.
Cytosolic and nuclear RNA fractionation
SiHa and HeLa cells were used to perform cytosolic and nuclear RNA extraction with the PARIS Kit (Invitrogen) following the manufacturer's instructions. HOTAIR and U6 and GAPDH and β-actin acted as cytoplasmic and nuclear RNAs controls, respectively.
Cell transfection
For transient transfection assays, miRNA mimics, miRNA inhibitors and LINC01410 siRNA synthesized by GeneCreate (Wuhan, China) were transfected into CC cells using Lipofectamine RNAiMAX Reagent (Invitrogen, USA). The transfection of FASN and LINC01410 plasmid synthesized by GeneCreate (Wuhan, China) was performed with X-tremeGENE HP DNA Transfection Reagent (Roche, Mannheim, Germany). The detailed description is provided in the Supplementary Methods and Supplementary Table 1.
Cell proliferation assays, transwell invasion assays and endothelial cell tube formation assay
For the cell proliferation assays, an MTT (Sigma-Aldrich) solution was added to each well containing an appropriate number of cells after transfection. The absorbance value was measured at 490 nm to evaluate cell viability. For transwell assays, the cells were seeded onto the upper chamber, while the lower chamber was filled with medium containing 10% bovine serum albumin. The cells that migrated on the lower surface were fixed, stained, and counted. For tube formation assay, HLECs were treated with the conditioned mediums from the cervical cancer cells for incubation 6 h. Tube structures was quantified according to the total tube length or the number of branch nodes using ImageJ software (NIH, Boston, MA, USA).
Quantification of LD accumulation
For cell LD quantification, the lipophilic fluorescence dye BODIPY 493/503 (Invitrogen) and Oil Red O (Sigma-Aldrich, USA) staining were employed to monitor LD accumulation in CC cells/tissues. Cells grown on coverslips were washed in Phosphate Buffered Saline (PBS), fixed with 4% paraformaldehyde, and stained with BODIPY 493/503 (1 µg/ml). After nuclei were counterstained with Hoechst (10 μg/ml), confocal analysis was performed with an Olympus FV-1000 confocal microscope (Tokyo, Japan), and LD numbers per cell were evaluated with ImageJ software. The Oil Red O staining procedure for cryosections was performed. In brief, frozen sections were fixed in formalin, rinsed with 60% isopropanol and stained with Oil Red O working solution (5:1 Oil Red O in 60% triethyl-phosphate (w/v)).
Quantification of triglycerides and phospholipids
The quantification of triglycerides (TGs) and phospholipids (PLs) were performed using EnzyChromTM Triglyceride and Phospholipid Assay Kits (BioAssay Systems), respectively, based on the manufacturer's instructions.
Dual luciferase assay
The luciferase assay was performed as previously described [9]. The targeting mutant LINC01410 and FASN were synthesized into the vector by GeneCreate (Wuhan, China), and the resulting constructs were named pmirGLO-LINC01410-Mut and pmirGLO-FASN-mut, respectively. A mixture of Renilla luciferase reporter vectors, luciferase reporter vectors (pmirGLO-LINC01410-wt, pmirGLO-LINC01410-mut, pmirGLO-FASN-wt, or pmirGLO-FASN-mut) and other transfection components (miRNA-Ctrl or miR-532-5p mimics) were transfected into cells for 48 h. Firefly and Renilla luciferase activities were detected using Dual Luciferase Reporter Assay kit (Promega).
RNA immunoprecipitation (RIP) assay
RIP experiments were performed with Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA). Briefly, cell lysates in RIP lysis buffer were conjugated with 6 µg of anti-AGO2 antibody or normal anti-IgG. Then, RNA-protein complexes were precleared with protein G Dynabeads. Subsequently, the retrieved RNA was assayed by qPCR.
Hematoxylin-eosin (HE) staining
In brief, the formalin-fixed paraffin-embedded sections were dipped in hematoxylin for 5 min, covered with 0.1% HCl and 0.1% NH4OH, and washed with running tap water. Then, the sections were stained with eosin. The stained sections were imaged and analyzed under microscope (Olympus, Tokyo).
Xenograft model
Xenograft models have been previously described using female BALB/c nude mice (age 4–5 weeks, n = 40) [9]. For subcutaneous xenograft tumor model, 1 × 107 cells/150 µl transfected with lentivirus containing miR-532-5p or control vector were injected subcutaneously into nude mice. After injection, tumor growth was measured in mice three times per week for 30 days. Tumor volume was calculated by a following formula: length × width2 × 0.52. For xenograft LNM model, 1 × 107 cells/50 µl were directly injected into the foot pads of mice. The lymph nodes from the popliteal fossa or other sites were removed and fixed for HE staining. The metastatic foci was counted and diagnosed by pathologists. For experiments with orlistat (an FASN inhibitor), when tumor sizes reached approximately 150–200 mm3, the mice were intraperitoneally injected with orlistat (240 mg/kg/d) or equal volume of vehicle (33% ethanol in PBS) for 3 weeks.
Bioinformatics analysis
OncomiR or OncoLnc was used to retrieve miRNAs associated with CC prognosis [21], [22]. miRWalk2.0 containing DIANAmT, miRanda, miRDB, miRWalk, RNAhybrid and PICTAR4 was used to predict potential miR-532-5p targets [23]. The predicted genes were used to perform functional enrichment studies using gene set enrichment analysis (GSEA) to reveal the biological function of miR-532-5p [24]. Reference gene collection is c2.cp.v7.2.symbols.gmt (curated). We randomly assign the original phenotype labels to samples, reorder all genes according to the difference in expression (using an appropriate metric, signal-to-noise ratio, SNR). We then measure the extent of association by a non-parametric, running sum statistic termed the enrichment score (ES) and record the maximum ES (MES) over all gene sets in the actual data from affected individuals. The entire above procedure is repeated 1000 times, using permuted diagnostic assignments and creating a histogram of the maximum ES.We used the RNAhybrid bioinformatics prediction tool to find the minimum free energy (mfe) structure of the hybridization of miR-532-5p and FASN or LINC01410 [25]. Differential expression and correlation analysis of the LINC01410/FASN/PLIN2 were analyzed by Gene Expression Profiling Interactive Analysis (GEPIA) database which including TCGA and GTEx samples [26]. The immunohistochemistry (IHC) analysis of FASN and PLIN2 was obtained from The Human Protein Atlas database [27], [28], [29].
Statistical analysis
All statistical analyses were performed with SPSS software (version 13.0) and Prism 5.0 (GraphPad software). Student’s t-test or one-way ANOVA was used for comparisons. The data are presented as the mean ± SD of at least three independent experiments. Least absolute shrinkage and selection operator (LASSO)-Cox regression were applied to build miRNA prognostic model. The optimal cutoff values for overall survival (OS) and disease free survival (DFS) was determined by the “sur_cutpoint” function of the “survminer” R package. It automatically calculated the segmentation point with the minimum P value, which is the threshold value of high-risk group and low-risk group. Multiple statistical packages were used in R software (version 4.0.4, R Core Team, 2021) to download TCGA data (including FIGO stage, lymph node status, OS and DFS, Supplementary Table 3) and generate forest plots, survival curves and the time-dependent receiver operating characteristic (ROC) curve. P < 0.05 were considered significant.
Results
Establishment of the prognostic miRNAs score model and expression study of candidate miRNAs
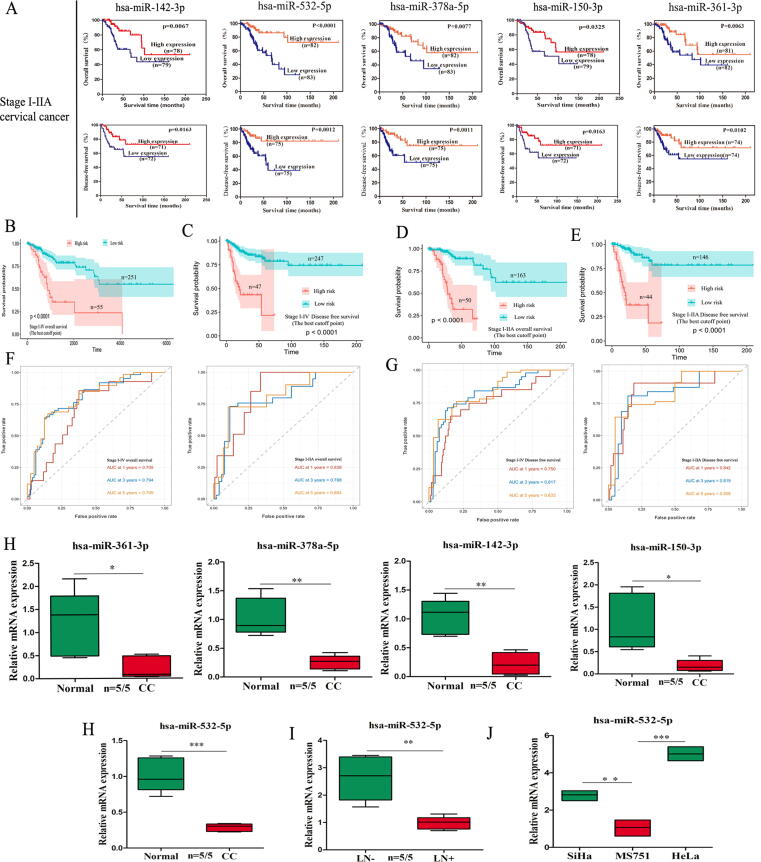
To identify miRNAs associated with prognosis, we obtained the expression profiles of miRNAs based on correlation between miRNAs and pathologic nodal metastasis or distant metastasis status, clinical stage, expression differences in different survival states and the significance threshold P < 0.01 using OncomiR and OncoLnc. The entire workflow is presented in Supplementary Fig. S1. Subsequently, these miRNAs were analyzed in a Lasso regression model (Supplementary Fig. S2A-B) and finally, 5 candidate prognostic miRNAs signature were identified, namely, miR-142-3p, miR-532-5p, miR-378a-5p, miR-150-3p, and miR-361-3p (Fig. 1A and Supplementary Fig. S2C). Next, the Risk Score of each sample was calculated based on the LASSO regression co-efficiency through the following formula:
Fig. 1.
Identification of a prognostic miRNA signature in CC. (A) KM analyses of OS and DFS for 5 candidate prognostic miRNAs in CC patients (stage I–IIA). (B-C) KM analyses in CC patients based on the risk score of 5 candidate prognostic miRNAs. (D-E) Kaplan-Meier analyses in CC patients (stage I–IIA) based on the risk score of 5 candidate prognostic miRNAs. (F-G) ROC curve for 1, 3, and 5 year survival prediction by the prognostic miRNA score model (H) Relative mRNA expression of 5 candidate prognostic miRNAs. (I) miR-532-5p expression was significantly downregulated in the primary site of CC with LNM compared with that without LNM. (J) miR-532-5p expression in three CC cell lines with different metastatic potentials.
Coefi is the risk coefficient of prognostic miRNAs calculated by the LASSO model and Xi is the expression value. we calculated a prognostic risk score with the following formula: Risk Score = [miR-142-3p*(−0.00002696198)] + [miR-532-5p*(−0.0001861752)] + [miR-378a-5p*(−0.0003280755)] + [miR-150-3p*(−0.03612045)] + [miR-361-3p*(−0.0000827455)] (Supplementary Table 4). The patients were divided into two groups according to median value. The Kaplan-Meier (KM) analysis showed that patients (stage I–IV) in the high-risk group had worse OS than patients in the low-risk group (Fig. 1B-C and Supplementary Fig. S2D-F). The difference was more significant than that with the single miRNA index. Then, all patients with early-stage (I–IIA) disease were further evaluated by the risk score. Similar significant predictive power of the 5-miRNAs signature was found in both OS and DFS (Fig. 1D-E and Supplementary Fig. S2G-I). Subsequently, the accuracy of the five-miRNAs signature was validated using the testing set from our patients corhorts. We found that the high-risk group resulted in poor OS and DFS (Supplementary Fig. S2J). The time-dependent ROC curve analysis of risk model demonstrated its promising predictive value for CC survival (Fig. 1F-G). Significantly, the 5-year OS was 31.944% in high risk group versus 88.783% in low risk group in stage I-IIA (Supplementary Table 4).
To validate the above analysis in terms of miRNA expression, we measured the expression levels of the 5 selected miRNAs. As shown in Fig. 1H, the 5 miRNAs expression in the candidate prognostic signature was markedly decreased in CC tissues compared to normal controls. miR-532-5p had the lowest P-value in RNA expression (P < 0.001) and prognostic prediction (P < 0.0001) and was selected for further study. The qPCR data showed that miR-532-5p expression was significantly lower in primary CC tumors with LNM than without LNM (Fig. 1I). Meanwhile, miR-532-5p expression in HeLa and SiHa cells which derived from primary sites were significantly higher than those in MS751 cells which derived from lymph node metastatic sites (Fig. 1J).
miR-532-5p suppresses CC cell metastasis, lymphangiogenesis and EMT in vitro
According to the correlation of expression level between miR-532-5p and mRNAs, The result of GSEA indicates that miR-532-5p maybe participate in the metastasis and arachidonic acid metabolism (Supplementary Fig. S3A). Therefore, to further explore the potential inhibitory effects of miR-532-5p on LNM, we chose SiHa and HeLa cell lines to perform gain-of-function and loss-of-function experiments. The efficiencies of interference and overexpression reagents were detected by qRT-PCR (Supplementary Fig. S3B).
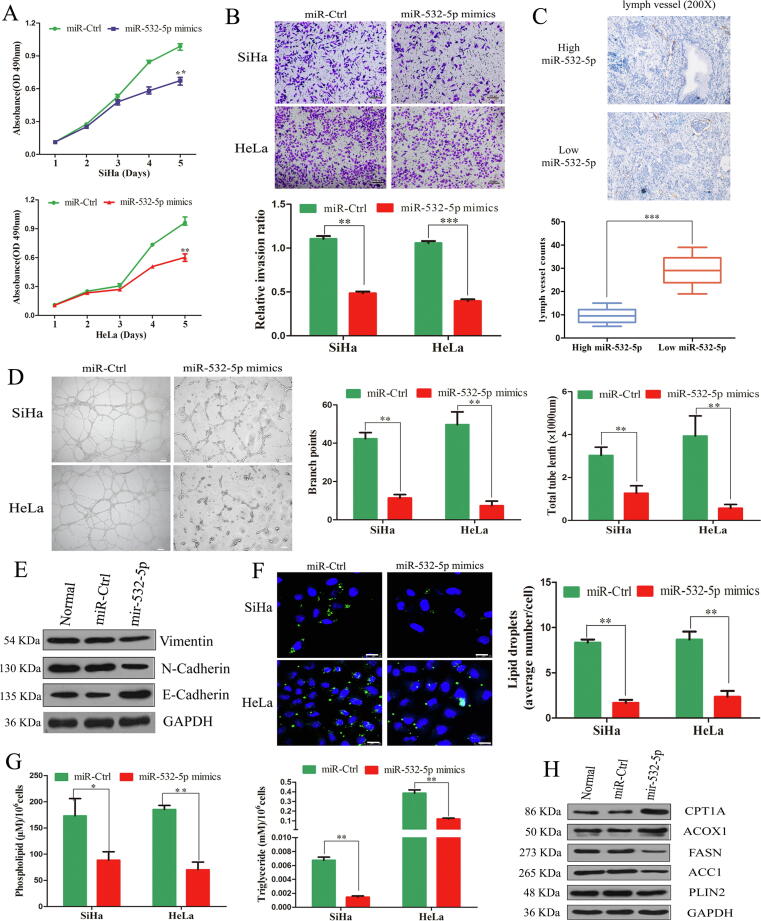
First, we investigated whether miR-532-5p inhibits CC cell proliferation since primary tumor size has been positively correlated with LNM in various solid tumors [30], [31]. Cell proliferation assays (with MTT) revealed that miR-532-5p overexpression significantly led to decreased proliferation (P < 0.01, Fig. 2A). Next, transwell assays indicated that miR-532-5p overexpression significantly hindered CC cells invasion (P < 0.01, Fig. 2B). Conversely, inhibition of miR-532-5p led to enhanced cell invasive and migratory behaviors (Supplementary Fig. S3C). Furthermore, lymphangiogenesis is a crucial factor for tumor LNM [32]. The D2-40-marked lymph vessels density in CC tissues with low miR-532-5p expression was significantly higher compared with that of tumors with high miR-532-5p expression (P < 0.001, Fig. 2C). Meanwhile, the culture supernatants of CC cells with ectopic miR-532-5p expression significantly inhibited HLEC tube formation compared with those of the corresponding control cells (Fig. 2D).
Fig. 2.
miR-532-5p inhibits CC cell metastasis and lymphangiogenesis via EMT and reduces intracellular LD accumulation and metabolic reprogramming. (A) miR-532-5p mimics significantly decreased cell proliferation compared with that of miR-Ctrl cells, as indicated by the MTT assay. (B) The effects of miR-532-5p on the cell invasion abilities (scale bars, 500 µm). (C) D2-40 staining for lymph vessel in CC tissues (representative image, n = 10, 200×). (D) The effects of miR-532-5p on the tube formation of HLECs (scale bars, 100 µm). (E) The protein levels of EMT markers. (F) The LDs staining with BODIPY 493/503 (Scale bars, 25 µm). (G) The cellular content of PLs and TGs. (H) The protein levels of key lipid metabolic enzymes. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In addition, cancer cells that undergo EMT have been verified to acquire an enhanced capacity to disseminate through the lymphatic system [33]. miR-532-5p overexpression resulted in significant downregulation of vimentin and N-cadherin, and upregulation of E-cadherin (Fig. 2E).
miR-532-5p inhibits LD accumulation and modulates fatty acid metabolism in CC cells
As described in Supplementary Fig. S3A, miR-532-5p was inversely associated with the arachidonic acid metabolism. LDs are particularly active sites for the metabolism of arachidonic acid [34]. Therefore, we further explored whether miR-532-5p regulates LD accumulation. Our data indicated that miR-532-5p overexpression strongly decreased, while silencing miR-532-5p increased, the number of LDs in CC cells (P < 0.01, Fig. 2F and Supplementary Fig. S3D).
To evaluate the effects of miR-532-5p on FA metabolism, we first measured the intracellular TGs and PLs levels in CC cell lines. Our data indicated that miR-532-5p mimics significantly reduced intracellular TAG and PLs levels, whereas miR-532-5p knockdown had opposite effects (P < 0.01, Fig. 2G and Supplementary Fig. S3E-F). Consistent with the above results, Oil Red O staining in CC tissues indicated that miR-532-5p significantly negatively correlated with LD accumulation (Supplementary Fig. S3G). Next, the key enzymes involved in FA oxidation and de novo FA synthesis, including carnitine palmitoyltransferase-1A (CPT1A), acyl-CoA oxidase 1 (ACOX1), acetyl-CoA carboxylase 1 (ACC1), FASN and perilipin 2 (PLIN2), were examined. Consequently, miR-532-5p mimics significant downregulated PLIN2, ACC1 and FASN, and upregulated CPT1A and ACOX1 in CC cells compared with the controls (Fig. 2H).
Identification of FASN as direct target of miR-532-5p
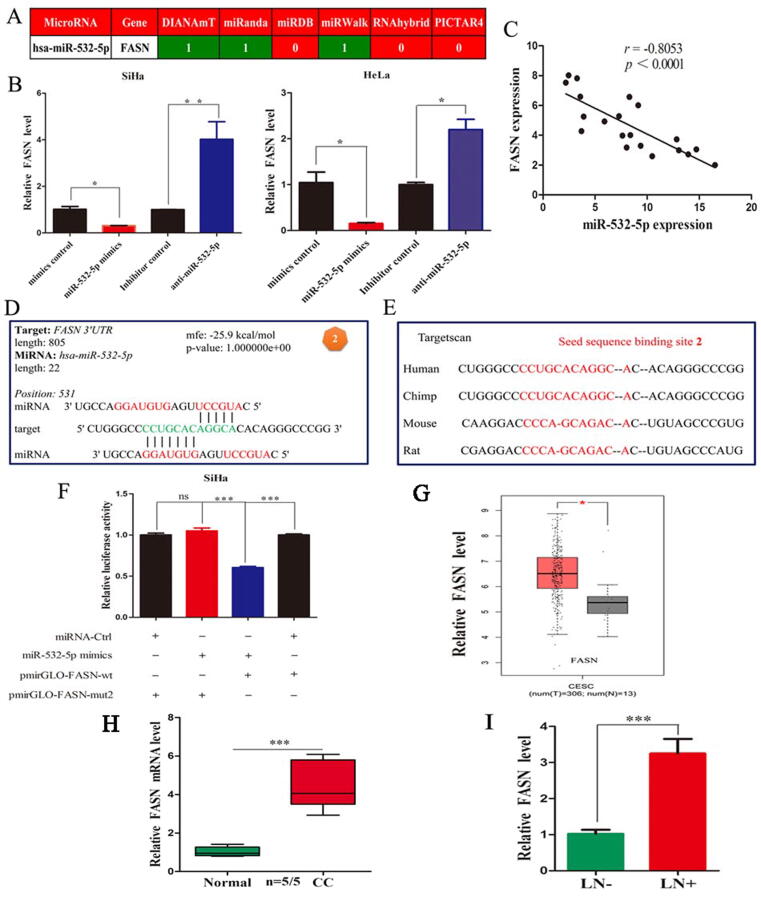
To investigate the potential targets of miR-532-5p, we searched the comprehensive database of predicted and validated miRNA-target interactions, named miRWalk2.0. Interestingly, FASN as the only metabolic gene among all candidate genes tested [35], was predicted to be potential direct target of miR-532-5p (Fig. 3A). As mentioned above, it was already confirmed that FASN protein levels dramatically decreased upon miR-532-5p overexpression (Fig. 2H). Then, we investigated whether miR-532-5p regulates FASN expression at the posttranscriptional level. As anticipated, miR-532-5p knockdown significantly increased FASN expression, while FASN expression was significantly inhibited after miR-532-5p overexpression (P < 0.05, Fig. 3B). Furthermore, miR-532-5p expression had a high inverse correlation coefficient with FASN expression (R = −0.8053, P < 0.0001, Fig. 3C).
Fig. 3.
FASN is a direct target of miR-532-5p. (A) Prediction of candidates using a comprehensive database of miRNA-target interactions, named miRWalk2.0. (B) FASN expression in cells transfected with miR-532-5p mimics and inhibitor. (C) Correlation analysis between FASN and miR-532-5p. (D) The predicted target sequence of miR-532-5p in the 3′-UTR of FASN. (E) All nucleotides of the seed sequence of the binding site are conserved in several species, including humans, chimps, rats and mice. (F) Luciferase activity in CC cells cotransfected with the pmirGLO-FASN-wt or pmirGLO-FASN-mut reporter plasmid and miR-532-5p mimics. (G) FASN was higher in CC tissues than in normal tissues from TCGA database. (H) qRT-PCR analysis of FASN. (I) FASN was significantly upregulated in the primary site of CC with LNM compared to that without LNM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To confirm whether miR-532-5p suppresses FASN expression through binding to seed sequences in the FASN 3′-untranslated region (UTR), RNAhybrid was used to predict their interaction binding sites. The mfe of two predicted hybridizations were −26.1 and −25.9 kcal/mol (Fig. 3D, Supplementary Fig. S4A). Furthermore, the binding sequences were within the range of authentic miRNA-target pairs and conserved across species (Fig. 3E, Supplementary Fig. S4B). Next, the predicted seed sequence binding sites were verified by luciferase assay. Unfortunately, for seed sequence binding site-1, miR-532-5p overexpression did not result in any reduction in luciferase activity compared to that in control cells (Supplementary Fig. S4C). Subsequently, for seed sequence binding site-2, miR-532-5p mimics significantly reduced the luciferase activity of the wild-type FASN reporter but not the activity of the mutated reporter (P < 0.001, Fig. 3F), indicating that binding site-2 strongly contributes to the miR-532-5p/FASN mRNA interaction.
We next investigated the FASN expression and its correlation with LNM in CC tissues to further determine whether miR-532-5p was truly inversely correlated with FASN. We discovered that FASN mRNA levels were markedly higher in CC tissues compared to normal tissues in TCGA dataset (P < 0.05, Fig. 3G) and our center (P < 0.001, Fig. 3H). By IHC analyses, we found that FASN was also higher in CC tissues and mainly located in the cell membrane and cytoplasm based on HPA database. Furthermore, FASN was significantly upregulated in patients with LNM compared to those without LNM (P < 0.001, Fig. 3I). The above results strongly indicate that miR-532-5p downregulates FASN at both transcript and protein levels by directly binding to its 3′-UTR.
miR-532-5p inhibits LNM through FASN-associated LD accumulation in vitro
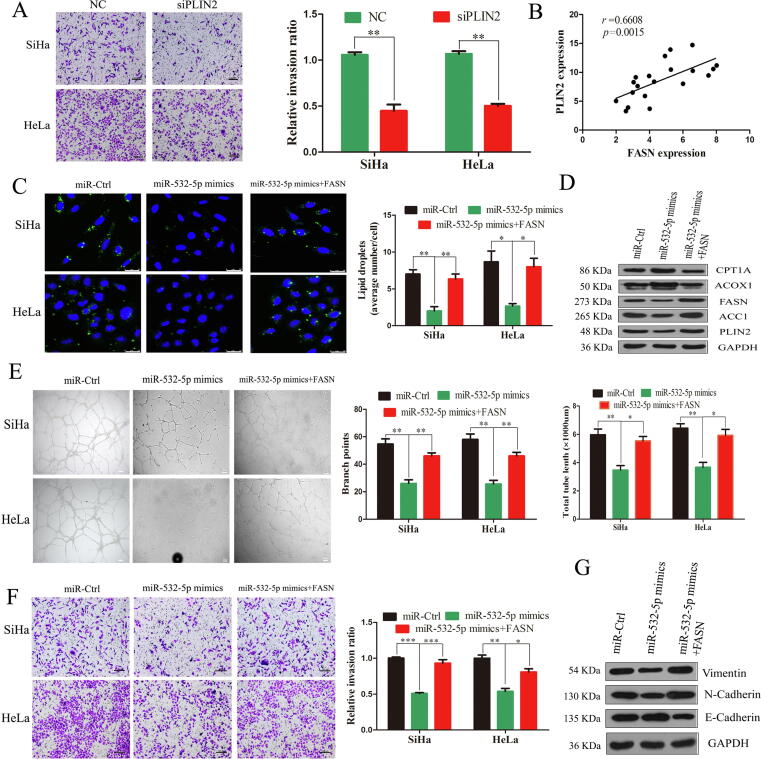
To confirm that FASN-associated LD accumulation was involved in the antimetastatic effects of miR-532-5p, we first explored the biological function of LDs in CC cells. The perilipin family are the major LD-associated proteins. Among them, PLIN2 as a LD marker, is universally expressed in nearly all tissues [36]. PLIN2 were relatively higher in CC tissues than normal tissues by IHC analysis based on HPA database. We next explored the potential roles of LDs in CC cell metastasis. We efficiently decreased PLIN2 levels in SiHa and HeLa cells with a PLIN2 siRNA (Supplementary Fig. S5A) and found that upon LD deprivation by PLIN2 siRNA treatment, SiHa and HeLa cells exhibited reduced invasiveness capability (P < 0.01, Fig. 4A). Furthermore, a positive correlation was observed between PLIN2 and FASN expression (P = 0.0015, Fig. 4B and Supplementary Fig. S5B).
Fig. 4.
miR-532-5p inhibits LNM through FASN-associated LD accumulation in vitro. (A) Transwell assays showed the effects of PLIN2 on the cell invasion abilities (scale bars, 500 µm). (B) Pearson correlation between FASN and PLIN2 expression in CC tissues (n = 20 tissues). (C) LD accumulation staining with BODIPY 493/503. Scale bars, 25 µm. (D) The protein levels of key lipid metabolic enzymes in CC cells. (E) The effects of miR-532-5p and FASN on the tube formation of HLECs (scale bars, 100 µm). (F) Transwell assays showed the effects of miR-532-5p and FASN on the cell invasion abilities (scale bars, 500 µm). (G) The expression of EMT markers in CC cells *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To further assess the crosstalk between LNM and LD accumulation, we explored whether FASN played an important role in miR-532-5p-induced LNM in vitro. First, we explored the role of FASN on LD accumulation using rescue assays. As expected, FASN overexpression dramatically reversed the effects of LD deprivation induced by ectopic miR-532-5p (Fig. 4C and Supplementary Fig. S5C). Moreover, increased ACOX1 and CPT1A protein expression and downregulation of FASN, ACC1, and PLIN2 induced by miR-532-5p overexpression were rescued by FASN treatment (Fig. 4D).
Next, the tube formation assay showed that the total lengths of tubules was significantly longer in the HLEC co-cultured with conditional medium of SiHa or Hela cells by cotransfecting with FASN overexpression plasmid and miR-532-5p mimics than those with the medium from the only miR-532-5p mimics group (Fig. 4E). Similarly, the effects of overexpressing miR-532-5p on SiHa and HeLa cell migration and invasion were partially reversed by overexpressing FASN (Fig. 4F). Moreover, miR-532-5p mimics and FASN plasmid cotransfection partly increased vimentin and N-cadherin compared the controls, while the E-cadherin showed the opposite response (Fig. 4G).
LINC01410 acts as ceRNA to sponge miR-532-5p
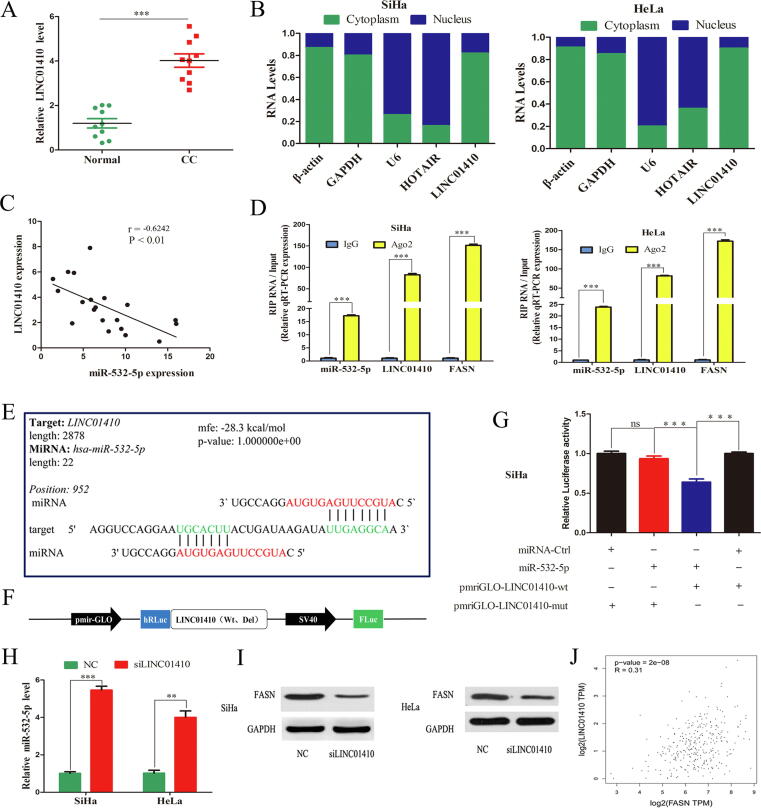
miRNA activity can be affected by the miRNA sponge transcripts, the so-called ceRNA in humans, and accumulating studys show that lncRNAs act as sponge for miRNAs [17]. Thus, lncRNAs having competitive binding site with miR-532-5p were predicted by LncBase Experimental v.2 online tool. The top 5 lncRNAs were selected to examine their expression level in CC (Supplementary Table 5). Interestingly, only LINC01410 was found to be upregulated in CC and validated by qRT-PCR (P < 0.001, Fig. 5A). In addition, the coding potential assessment tool (CPAT) was used to access the coding potential of LINC01410. Indeed, LINC01410 had a low coding probability, which lacks typical protein-coding ORF (Supplementary Table 6). Given that the molecular function of lncRNAs largely depends on their subcellular location [37], we performed cell nuclear/cytoplasm fractionation and found that LINC01410 predominantly localized in the cytoplasm (Fig. 5B). Furthermore, LINC01410 was also negatively associated with miR-532-5p (R = −0.6242, P < 0.01, Fig. 5C).
Fig. 5.
LINC01410 acts as a miRNA sponge for miR-532-5p. (A) The LINC01410 mRNA expression level (B) Nuclear/cytoplasmic fractionation assay showing the nuclear localization of LINC01410 in CC cells. (C) Correlation analysis between LINC01410 and miR-532-5p (D) miR-532-5p, LINC01410 and FASN were retrieved by the AGO2 antibody in SiHa and HeLa cells. (E) The potential binding sites between miR-532-5p and LINC01410. (F) Schematic of pmirGLO-LINC01410 reporter constructs. (G) Luciferase activity in CC cells cotransfected with the pmirGLO-LINC01410-wt or -mut reporter plasmid and miR-532-5p mimics. (H) miR-532-5p detection after LINC01410 knockdown. (I) The effects of si-LINC01410 on the protein levels of FASN in SiHa and HeLa cells. (J) Pearson correlation between LINC01410 and FASN *, P < 0.05; **, P < 0.01; ***, P < 0.001.
miRNAs binding to their target genes results in posttranscriptional repression in an AGO2-dependent manner [38]. To determine whether LINC01410 is a binding platform for miR-532-5p to regulate FASN, we conducted RIP analysis. LINC01410, miR-532-5p and FASN were all detected in Ago2 immunoprecipitates (P < 0.001, Fig. 5D). In addition, the direct targeting sequence between LINC01410 and miR-532-5p was confirmed by dual luciferase assay (mfe = −28.3 kcal/mol, Fig. 5E). We constructed two luciferase reporters to monitor miR-532-5p activity by inserting either a perfect LINC01410 sequence or a mutant sequence into the 3′-UTR of Renilla luciferase (Fig. 5F). Cotransfection of the luciferase reporter with the pmirGLO-LINC01410-wt vector substantially reduced the luciferase activity in miR-532-5p mimics group. Furthermore, these inhibitory effects were abrogated by site-directed mutagenesis of LINC01410 (Fig. 5G). Moreover, miR-532-5p upregulation was caused by siRNA-mediated LINC01410 knockdown (P < 0.01, Fig. 5H), while FASN expression was inhibited (Fig. 5I). In addition, LINC01410 was positively associated with FASN in CC tissues based on TCGA database (Fig. 5J).
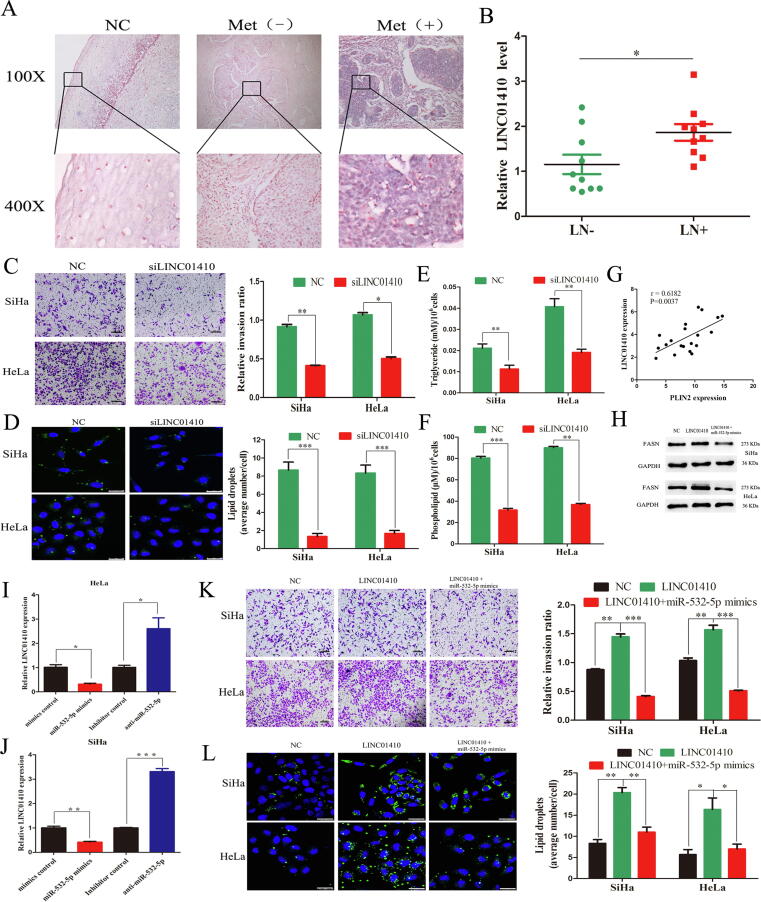
Knockdown of LINC01410 inhibits tumor cell migration and LD accumulation
To further explore the role of LINC01410, we first evaluated the expression of LINC01410. We found that LINC01410 expression at the primary site of CCs with LNM was higher than those without LNM or normal controls via ISH analyses (Fig. 6A). Additionally, a similar result was verified in other tissues based on qRT-PCR analysis (Fig. 5B and 6B). Next, we knocked down LINC01410 in CC cells (Supplementary Fig. S5D). Transwell assays indicated that LINC01410 siRNA significantly decreased the cell invasion and migration abilities (Fig. 6C). Moreover, we found that LINC01410 knockdown inhibited LD accumulation, as indicated by BODIPY staining (P < 0.001, Fig. 6D). Furthermore, the intracellular TAG and PLs levels were markedly reduced by LINC01410 knockdown (Fig. 6E-F). In addition, LINC01410 was positively associated with PLIN2 in CC tissues (P = 0.0037, Fig. 6G and Supplementary Fig. S5E).
Fig. 6.
LINC01410 inhibited CC metastasis and LD accumulation by directly targeting miR-532-5p. (A) ISH analysis of LINC01410 in tissues. (B) The qRT-PCR analysis of LINC01410 (n = 10 tissues/group). (C) Migrating ability of CC cells (scale bars, 500 µm). (D) LD accumulation staining with BODIPY 493/503 (Scale bars, 25 µm). The cellular contents of TGs (E) and PLs (F). (G) Pearson correlation between LINC01410 and PLIN2 expression in CC tissues (n = 20 tissues). (H) Western blot analysis of FASN. (I-J) LINC01410 expression in CC cells transfected with miR-532-5p mimics and inhibitor. (K) Transwell assays showed the effects of LINC01410 and miR-532-5p on the invasion abilities of the indicated cells (scale bars, 500 µm). (L) LD accumulation staining with BODIPY 493/503. Scale bars, 25 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
miR-532-5p mimics reverse the metastasis- and LD-promoting effects of LINC01410 in vitro
To assess whether LINC01410 induced CC cell migration and LD accumulation through the miR-532-5p/FASN axis, we ectopically expressed miR-532-5p in LINC01410-overexpressing SiHa and HeLa cells. The abnormal FASN expression induced by LINC01410 overexpression was reversed by miR-532-5p mimics (Fig. 6H). Moreover, LINC01410 expression was downregulated by miR-532-5p overexpression but increased after miR-532-5p inhibition (Fig. 6I-J). Next, the Transwell assay results revealed that miR-532-5p mimics counteracted the promoting effect of LINC01410 on cell invasion and migration (Fig. 6K). In parallel, LD numbers were significantly increased in cells overexpressing LINC01410, and this effect was counteracted by miR-532 mimics (Fig. 6L).
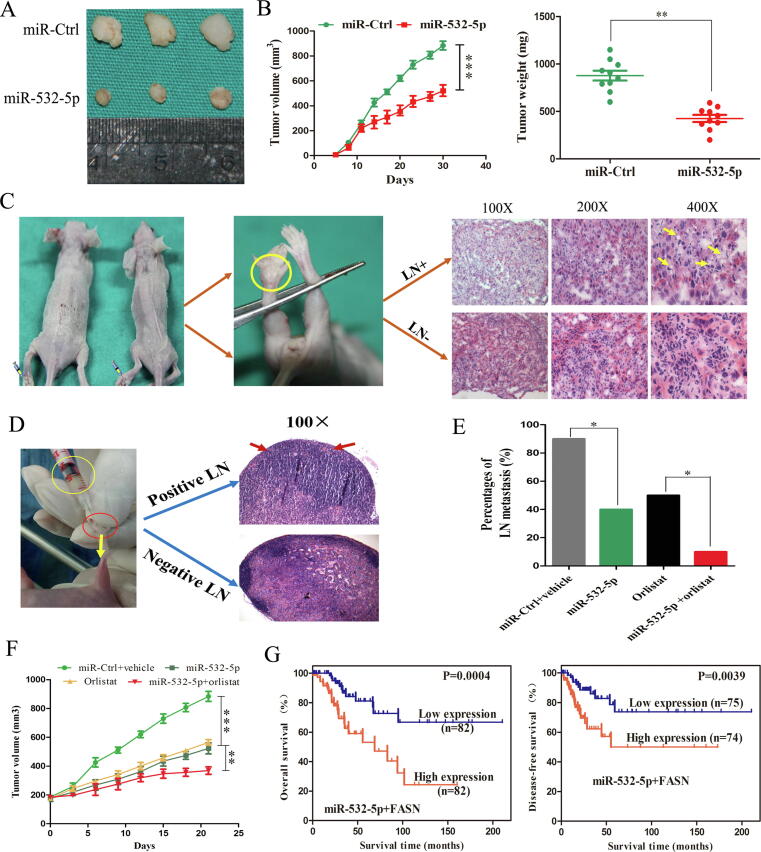
Targeting miR-532-5p versus miR-532-5p/orlistat combination therapy in vivo
Finally, we explored the role of miR-532-5p and FASN on LNM in CC xenografts using a humanized mouse tumor model. First, miR-532-5p overexpression decreased tumor growth in subcutaneous xenograft model (Fig. 7A). Furthermore, tumors in the miR-532-5p mimics group were of greater weight and volumes than those in the controls (P < 0.01, Fig. 7B). In addition, more fat vacuoles appeared in the foot pads of mice with LN-positive group compared with those in the LN-negative group (Fig. 7C), which indicated that LD accumulation may promote LNM in CC in vivo.
Fig. 7.
Combination therapy with miR-532-5p and orlistat has synergistic effects in vivo. (A) Representative images of dissected tumors. (B) Subcutaneous tumor growth curves of mice and the average weight of tumors in different treatment groups are depicted. (C) The enrichment of fat vacuoles. (D) Representative images of HE staining of lymph nodes from nude mice. (E) The LNM rate. (F) Mean tumor volume over time (n = 10). (G) KM analyses of OS and DFS in CC patients based on the risk score of miR-532-5p and FASN. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Orlistat, a FASN inhibitor, suppress the tumor growth and metastasis of various human cancers [39], [40], [41]. Thus, we evaluated the therapeutic efficacy of the orlistat/miR-532-5p combination using the lymph node metastatic xenograft mouse model (Fig. 7D). Mice that received miR-532-5p overexpression treatment had lower incidence of LNM (Fig. 7E). Notably, the miR-532-5p/ orlistat combination therapy had more prominent LNM-suppressing effects than either orlistat or miR-532-5p alone, corroborating the additive effect of the combination therapy in vivo (P < 0.05, Fig. 7E). Additionally, tumor weight and volumes were smaller in the group that received miR-532-5p and orlistat treatment than in the miR-532-5p only or orlistat only group (P < 0.01, Fig. 7F).
Next, we calculated a prognostic risk score with the following formula: Risk Score = [miR-532-5p*(−0.0022)] + [FASN*(1.1879)] based on the coefficient. The patients were divided into two groups according to the median value. The KM analysis revealed that patients with high-risk had poor OS and DFS (P = 0.0004, Fig. 7G).
Collectively, these results suggest that upregulation of miR-532-5p combined with orlistat treatment had synergistic effects on the inhibition of tumor LNM and increased survival compared with the effects of upregulation of miR-532-5p in CC cells or orlistat treatment alone.
The prognostic value of hub genes associated with the miR-532-5p signaling pathway
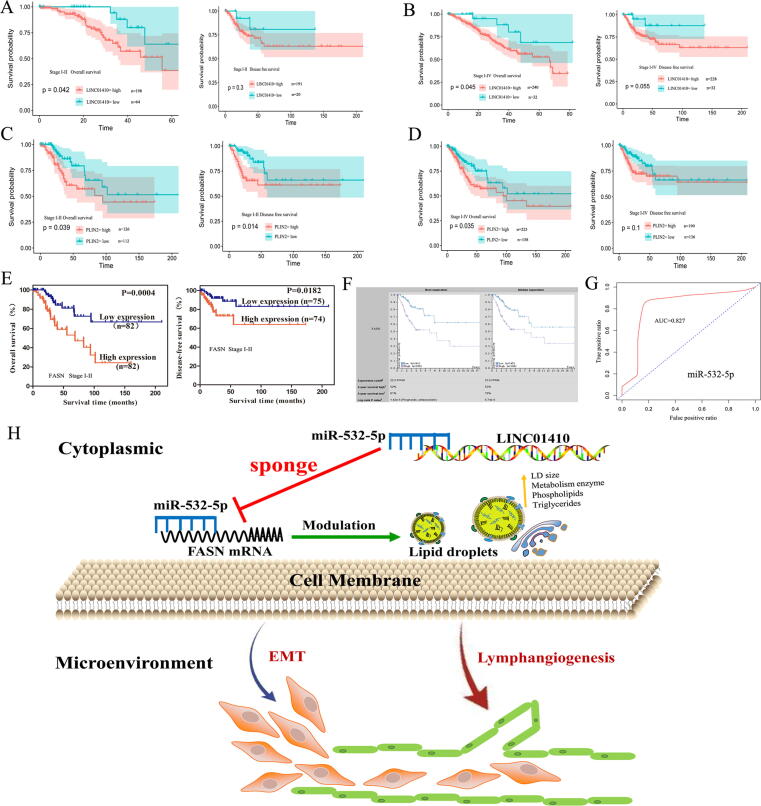
As mentioned above, we evaluated the critical role and prognostic value of miR-532-5p mRNA based on TCGA database. Next, we aimed to determine whether the hub genes associated with the miR-532-5p signaling pathway, including LINC01410, FASN and PLIN2, are also potential prognostic biomarkers in CC. Both high expression of LINC01410 (Fig. 8A-B, P = 0.042), PLIN2 (Fig. 8C-D, P = 0.039) and FASN (Fig. 8E-F, P = 0.0004) mRNA were associated with poor survival. Moreover, miR-532-5p was an independent protective predictor of OS and had good survival prediction potential (AUC = 0.827) (Fig. 8G and Supplementary Fig. S6). In addition, BMI (Body mass index), lymphovascular involvement and age were also the independent risk predictors of OS (Supplementary Table 7).
Fig. 8.
The prognostic value of the miR-532-5p-correlated ceRNA network. KM analyses of OS and DFS for PLIN2 (A-B), LINC01410 (C-D), and FASN (E-F) in CC patients at different stages. (G) ROC curve analyses based on miR-532-5p. (H) Schematic illustration of the prognostic miR-532-5p-correlated ceRNA network.
Discussion
LNM is a common occurrence in the natural progression of CC and is the critical cause of disease deterioration and even death. However, effective treatment modalities for these patients are still lacking. In-depth analysis of the detailed mechanism of LNM from multiple perspectives will provide preventive and therapeutic strategies for patients. Herein, we preliminarily identified a novel miRNA signature that affords an opportunity to improve the prognosis of CC. We propose a miR-532-5p/FASN regulatory ceRNA network to illustrate the crosstalk mechanism of LD accumulation in regulating LNM in CC and explored the potential prognostic value of the ceRNA network. Surprisingly, targeting miR-532-5p in combination with orlistat therapy achieved better tumor and LNM control than either treatment alone. Our findings provide a potential strategy for the treatment of LNM.
miRNAs usually act as tumor suppressor by targeting mRNAs that encode oncoproteins and take part in molecular dysfunctions in tumor [42]. In ovarian cancer, high miR-532-5p expression results in better prognosis [43]. In contrast, miR-532-5p acts as a tumor inducer to downregulate RUNX3 expression in malignant melanoma [44]. However, whether miR-532-5p promoting or inhibiting LNM in CC remains unknown. We first identified and constructed a miRNA signature of CC using a prognostic risk score model. miR-532-5p was downregulated in the primary tumors with LNM and significantly negatively associated with LNM. Consistent with the results obtained in ovarian cancer, miR-532-5p was an independent prognostic factor of CC and achieved 82.7% predictive accuracy. Moreover, miR-532-5p inhibited CC metastasis and lymphangiogenesis, and reduced the rate of LNM in vivo. These results provide strong evidence that miR-532-5p could serve as a novel promising prognostic biomarker for CC and may contribute to the clinical decision-making.
Perceived as mere cytoplasmic inclusions of fat for a long time, LDs have emerged as true organelles with key functions in energy homeostasis and cancer aggressiveness [45]. However, no direct links or full comprehensive mechanisms of intracellular LD accumulation in LNM have been provided thus far. Our results showed that miR-532-5p overexpression inhibited LD accumulation in CC cells and modulated lipid metabolism reprogramming. Moreover, we provide evidence that preventing the formation of LDs in cancer cells with a PLIN2 siRNA decreased the ability of the cells to metastasize. As evidenced by others [46], the formation of LDs in response to TGF-β2 activation has emerged as a major player in the shift toward a transitory invasive mesenchymal-like phenotype. Similarly, our study also addresses this issue by integrating ncRNAs. We previously reported that lncRNAs are involved in fatty acid metabolism reprogramming to regulate the metastatic process [9]. Now, we show that miR-532-5p-induced FASN downregulation inhibited both partial EMT and intracellular LD accumulation. These findings open up a new field for the biological role of LDs in cancer.
An enormous number of studies have highlighted that cytoplasmic lncRNAs act as decoys for miRNAs to influence miRNA activity and thus regulate mRNA stability or translation [47]. In our previous work, we also provided evidence to illustrate the potential role of lnRNA in LNM [9]. In our study, we demonstrated that LINC01410 performed their functions by directly binding to miR-532-5p from the perspective of ceRNA. Importantly, no research studies on the specific mechanism of miR-532-5p-correlated ceRNA network in regulating LD metabolism have been conducted to date. We observed that the abnormal LD accumulation and cell metastasis induced by LINC01410 overexpression were reversed after introduction of miR-532-5p mimics. Our study provides the first evidence that miR-532-5p-correlated ceRNA network can serve as a bridge to explore the crosstalk mechanism between LD metabolism and LNM, which has not been elucidated in CC previously. However, it is important to note that miRNAs compete for binding to several protein-coding genes or other ncRNAs by sharing miRNA response elements. Given this phenomenon, to some extent, it explains why the ceRNA can play only a partial role in the functional recovery of miR-532-5p. Therefore, such a discovery could open a new window to better understand the mechanisms of LNM.
FASN, the key enzyme for neoplastic lipogenesis, plays key role in tumor progression and survival. Accumulating evidence suggests that early small-molecule FASN inhibitors can cause tumor growth delay in vivo, making FASN an attractive target for cancer therapy [40]. Our findings also suggested that patients with higher FASN expression had poor prognosis. More importantly, we observed that overexpression of FASN could partially restore the metastasis and LD accumulation inhibited by miR-532-5p in CC cells. Similarly, in prostate cancer, the highest FASN expression was detected in hormone-independent bone metastases [49]. Thus, there is sufficient evidence to show that FASN upregulationn and FASN-catalyzed de novo fatty acid biogenesis accompanies the metastatic process.
Another important finding was that the combined treatment of tumor-bearing mice with miR-532-5p and orlistat dramatically suppressed LNM and tumor growth in vivo. The low prognostic risk score calculated with miR-532-5p and FASN showed a clear survival advantage for CC patients. These findings indicate that combination therapy with miR-532-5p and orlistat is a potential therapeutic option for CC, and highly promising results of miRNA-based therapeutics in phase I/II clinical trials has been produced [50], [51]. However, some obstacles remain to be overcome, including difficulties in tumor site-specific delivery, off-target effects, efficacious dose determination, and safety. Interestingly, endogenous exosomes-mediated circulating miRNAs originating from cancer cells may be a novel tool developed to deliver miRNA therapeutics [52]. It is important to explore miR-532-5p in exosomes derived from CC cells and may provide a novel theoretical basis for the future development of clinical applications.
In our study, we propose new ideas to explore prognostic risk models from the perspective of a miR-532-5p-correlated ceRNA network. As this article shows, LINC01410, FASN and PLIN2 were all unfavorable prognostic factors in CC patients. The results on LINC01410 are consistent with those of another study in CC tissues [53]. However, in this ceRNA network, only miR-532-5p could serve as an independent protective prognostic factor for both OS and DFS. The complexity of the miR-532-5p-correlated ceRNA network may explain this phenomenon. For example, RUNX3 and NCF2 have also been identified to be targeted by miR-532-5p, and LINC01410 could also sponge miR-124-3p [44], [48], [54]. In addition, the effects of sample size and technical factors should not be excluded. It is worth noting that BMI ≥ 25 (kg/m2) was an independent unfavorable prognostic indicator for OS. These findings are consistent with our previous study in CC patients at our center [9]. These findings lead us to believe that miR-532-5p-modulated LD metabolism contributes to CC progression.
Conclusion
In summary, we established a 5-miRNAs prognostic signature in CC to identify high- and low-risk patients. Our study demonstrated that miR-532-5p may be a promising independent protective prognostic biomarker for CC for the first time, and we identified miR-532-5p-correlated ceRNA network in which miR-532-5p is directly sponged by LINC01410, in turn inhibiting LNM by regulating FASN-modulated LD accumulation and metabolic reprogramming (Fig. 8H). As a result, combination therapy with miR-532-5p and the FASN inhibitor orlistat may be a novel potential antimetastatic therapeutic target in CC patients with LNM.
Compliance with Ethics requirements
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008 (5). Informed consent was obtained from all patients for being included in the study.
CRediT authorship contribution statement
Chunliang Shang: Data curation, Investigation, Methodology, Writing - original draft, Software. Yuan Li: Data curation, Investigation, Methodology, Writing - original draft. Tianhui He: Methodology, Software. Yuandong Liao: Methodology, Software. Qiqiao Du: Project administration, Supervision, Validation. Pan Wang: Project administration, Supervision, Validation. Jie Qiao: Conceptualization, Writing - review & editing. Hongyan Guo: Conceptualization, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the TCGA database for providing their platforms and contributing their valuable datasets. The authors thank Dr. Yuxiang Wang and Yan Liu (Department of Pathology, Peking university Third hospital) for help with the pathologic diagnoses and guidance. We thank prof. Huanran Tan (Department of Pharmacology, School of Basic Medical Sciences, Peking University Health Science Center) for providing their research platforms.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2021.09.009.
Contributor Information
Jie Qiao, Email: jie.qiao@263.net.
Hongyan Guo, Email: bysyghy@163.com.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Wright Jason D., Matsuo Koji, Huang Yongmei, Tergas Ana I., Hou June Y., Khoury-Collado Fady. Prognostic performance of the 2018 international federation of gynecology and obstetrics cervical cancer staging guidelines. Obstet Gynecol. 2019;134(1):49–57. doi: 10.1097/AOG.0000000000003311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noordhuis M.G., Fehrmann R.S., Wisman G.B., Nijhuis E.R., van Zanden J.J., Moerland P.D., et al. Involvement of the TGF-beta and beta-catenin pathways in pelvic lymph node metastasis in early-stage cervical cancer. Clin Cancer Res. 2011;17:1317–1330. doi: 10.1158/1078-0432.CCR-10-2320. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y., Zhang L., Tian J., Fu X., Ren X., Hao Q. Significance of the absolute number and ratio of metastatic lymph nodes in predicting postoperative survival for the International Federation of Gynecology and Obstetrics stage IA2 to IIA cervical cancer. Int J Gynecol Cancer. 2013;23(1):157–163. doi: 10.1097/IGC.0b013e3182778bcf. [DOI] [PubMed] [Google Scholar]
- 5.Pavlova N., Thompson C. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pascual G., Avgustinova A., Mejetta S., Martín M., Castellanos A., Attolini C.-O., et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017;541(7635):41–45. doi: 10.1038/nature20791. [DOI] [PubMed] [Google Scholar]
- 7.Nath A., Chan C. Genetic alterations in fatty acid transport and metabolism genes are associated with metastatic progression and poor prognosis of human cancers. Sci Rep-Uk. 2016;6(1) doi: 10.1038/srep18669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang C., Liao Y., Liu P., Du Q., Liang Y., Ooi S., et al. FABP5 promotes lymph node metastasis in cervical cancer by reprogramming fatty acid metabolism. Theranostics. 2020;10(15):6561–6580. doi: 10.7150/thno.44868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shang C., Wang W., Liao Y., Chen Y., Liu T., Du Q., et al. LNMICC promotes nodal metastasis of cervical cancer by reprogramming fatty acid metabolism. Cancer Res. 2018;78(4):877–890. doi: 10.1158/0008-5472.CAN-17-2356. [DOI] [PubMed] [Google Scholar]
- 10.Thiam A.R., Farese Jr R.V., Walther T.C. The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Bio. 2013;14(12):775–786. doi: 10.1038/nrm3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin S., Parton R.G. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7(5):373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- 12.Cruz A.L.S., Barreto E.d.A., Fazolini N.P.B., Viola J.P.B., Bozza P.T. Lipid droplets: platforms with multiple functions in cancer hallmarks. Cell Death Dis. 2020;11(2) doi: 10.1038/s41419-020-2297-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright H.J., Hou J., Xu B., Cortez M., Potma E.O., Tromberg B.J., et al. CDCP1 drives triple-negative breast cancer metastasis through reduction of lipid-droplet abundance and stimulation of fatty acid oxidation. Proc Natl Acad Sci. 2017;114(32):E6556–E6565. doi: 10.1073/pnas.1703791114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saw P.E., Xu X., Chen J., Song E.-W. Non-coding RNAs: the new central dogma of cancer biology. Sci China Life Sci. 2021;64(1):22–50. doi: 10.1007/s11427-020-1700-9. [DOI] [PubMed] [Google Scholar]
- 15.Anastasiadou E., Jacob L.S., Slack F.J. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karreth F.A., Pandolfi P.P. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3(10):1113–1121. doi: 10.1158/2159-8290.CD-13-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X.-Z., Cheng T.-T., He Q.-J., Lei Z.-Y., Chi J., Tang Z., et al. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway. Mol Cancer. 2018;17(1) doi: 10.1186/s12943-018-0874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J., Yu Y., Li H., Hu Q., Chen X., He Y., et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol Cancer. 2019;18(1) doi: 10.1186/s12943-019-0947-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Wong N.W., Chen Y., Chen S., Wang X. OncomiR: an online resource for exploring pan-cancer microRNA dysregulation. Bioinformatics. 2018;34:713–715. doi: 10.1093/bioinformatics/btx627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput Sci. 2016;2:67. [Google Scholar]
- 23.Dweep H., Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(697) doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 24.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhlen M., Zhang C., Lee S., Sjostedt E., Fagerberg L., Bidkhori G., et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357 doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 28.Thul P.J., Åkesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., et al. A subcellular map of the human proteome. Science. 2017;356(6340):eaal3321. doi: 10.1126/science:aal3321. [DOI] [PubMed] [Google Scholar]
- 29.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(12):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 30.Cairns R.A., Hill R.P. Acute hypoxia enhances spontaneous lymph node metastasis in an orthotopic murine model of human cervical carcinoma. Cancer Res. 2004;64(6):2054–2061. doi: 10.1158/0008-5472.can-03-3196. [DOI] [PubMed] [Google Scholar]
- 31.Hyslop T., Weinberg D.S., Schulz S., Barkun A., Waldman S.A. Occult tumor burden predicts disease recurrence in lymph node-negative colorectal cancer. Clin Cancer Res. 2011;17(10):3293–3303. doi: 10.1158/1078-0432.CCR-10-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bovay E., Sabine A., Prat-Luri B., Kim S., Son K., Willrodt A., et al. Multiple roles of lymphatic vessels in peripheral lymph node development. J Exp Med. 2018;215:2760–2777. doi: 10.1084/jem.20180217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlsson M.C., Gonzalez S.F., Welin J., Fuxe J. Epithelial-mesenchymal transition in cancer metastasis through the lymphatic system. Mol Oncol. 2017;11(7):781–791. doi: 10.1002/1878-0261.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dvorak A.M., Dvorak H.F., Peters S.P., Shulman E.S., MacGlashan D.J., Pyne K., et al. Lipid bodies: cytoplasmic organelles important to arachidonate metabolism in macrophages and mast cells. J Immunol. 1983;131:2965–2976. [PubMed] [Google Scholar]
- 35.Menendez J.A., Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7(10):763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 36.Itabe H., Yamaguchi T., Nimura S., Sasabe N. Perilipins: a diversity of intracellular lipid droplet proteins. Lipids Health Dis. 2017;16(1):83. doi: 10.1186/s12944-017-0473-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kopp F. Molecular functions and biological roles of long non-coding RNAs in human physiology and disease. J Gene Med. 2019;21(8) doi: 10.1002/jgm.v21.810.1002/jgm.3104. [DOI] [PubMed] [Google Scholar]
- 38.Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 39.Kuhajda F.P., Pizer E.S., Li J.N., Mani N.S., Frehywot G.L., Townsend C.A. Synthesis and antitumor activity of an inhibitor of fatty acid synthase. Proc Natl Acad Sci U S A. 2000;97(7):3450–3454. doi: 10.1073/pnas.050582897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kridel S.J., Axelrod F., Rozenkrantz N., Smith J.W. Orlistat is a novel inhibitor of fatty acid synthase with antitumor activity. Cancer Res. 2004;64(6):2070–2075. doi: 10.1158/0008-5472.can-03-3645. [DOI] [PubMed] [Google Scholar]
- 41.Li C.-F., Fang F.-M., Chen Y.-Y., Liu T.-T., Chan T.-C., Yu S.-C., et al. Overexpressed fatty acid synthase in gastrointestinal stromal tumors: targeting a progression-associated metabolic driver enhances the antitumor effect of imatinib. Clin Cancer Res. 2017;23(16):4908–4918. doi: 10.1158/1078-0432.CCR-16-2770. [DOI] [PubMed] [Google Scholar]
- 42.Tili E., Michaille J.-J., Croce C.M. MicroRNAs play a central role in molecular dysfunctions linking inflammation with cancer. Immunol Rev. 2013;253(1):167–184. doi: 10.1111/imr.12050. [DOI] [PubMed] [Google Scholar]
- 43.Wang F., Chang J.-H., Kao C.J., Huang R.S. High expression of miR-532-5p, a tumor suppressor, leads to better prognosis in ovarian cancer both in vivo and in vitro. Mol Cancer Ther. 2016;15(5):1123–1131. doi: 10.1158/1535-7163.MCT-15-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kitago M., Martinez S.R., Nakamura T., Sim M.-S., Hoon D.S.B. Regulation of RUNX3 tumor suppressor gene expression in cutaneous melanoma. Clin Cancer Res. 2009;15(9):2988–2994. doi: 10.1158/1078-0432.CCR-08-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olzmann J.A., Carvalho P. Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol. 2019;20(3):137–155. doi: 10.1038/s41580-018-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corbet C., Bastien E., Santiago de Jesus J.P., Dierge E., Martherus R., Vander Linden C., et al. TGFβ2-induced formation of lipid droplets supports acidosis-driven EMT and the metastatic spreading of cancer cells. Nat Commun. 2020;11(1) doi: 10.1038/s41467-019-14262-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmitt A.M., Chang H.Y. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J.-X., Chen Z.-H., Chen D.-L., Tian X.-P., Wang C.-Y., Zhou Z.-W., et al. LINC01410-miR-532-NCF2-NF-kB feedback loop promotes gastric cancer angiogenesis and metastasis. Oncogene. 2018;37(20):2660–2675. doi: 10.1038/s41388-018-0162-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Rossi S., Graner E., Febbo P., Weinstein L., Bhattacharya N., Onody T., et al. Fatty acid synthase expression defines distinct molecular signatures in prostate cancer. Mol Cancer Res. 2003;1:707–715. [PubMed] [Google Scholar]
- 50.van Zandwijk N., Pavlakis N., Kao S.C., Linton A., Boyer M.J., Clarke S., et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017;18(10):1386–1396. doi: 10.1016/S1470-2045(17)30621-6. [DOI] [PubMed] [Google Scholar]
- 51.Beg M.S., Brenner A.J., Sachdev J., Borad M., Kang Y.-K., Stoudemire J., et al. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs. 2017;35(2):180–188. doi: 10.1007/s10637-016-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torrano V., Royo F., Peinado H., Loizaga-Iriarte A., Unda M., Falcon-Perez J.M., et al. extracellular vesicles in liquid biopsy and cancer. Curr Opin Pharmacol. 2016;29:47–53. doi: 10.1016/j.coph.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu F., Wen C. LINC01410 knockdown suppresses cervical cancer growth and invasion via targeting miR-2467-3p/VOPP1 axis. Cancer Manag Res. 2020;12:855–861. doi: 10.2147/CMAR.S236832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang T., Wang C., Zhu Y., Han H. LINC01410 promotes cell proliferation and migration of cholangiocarcinoma through modulating miR-124-3p/SMAD5 axis. J Gene Med. 2020;22(6) doi: 10.1002/jgm.v22.610.1002/jgm.3162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.