Abstract
CD8 T cells have multiple functional properties that mediate acute phase and long-term immune protection. Several effector and memory CD8 T cell subsets have been described with diverse functionalities and marker profiles. In contrast to the many comprehensive mouse studies, most human studies lack samples from the acute infection phase, a major reason why current knowledge of human T cell subsets and differentiation remains incomplete, particularly with regard to the T cell heterogeneity early during the immune response. Here we analysed the human CD8 T cell response to yellow fever vaccination as the best-known model to study the human immune response to acute viral infection. We performed flow cytometry on 21 markers conventionally used in mice and in humans to describe differentiation, activation, cycling, and so-called effector functions. We found clearly distinct ‘acute traits’ at the peak of the response that are shared amongst all non-naïve antigen-specific subsets, including memory-differentiated cells. These acute traits were low BCL-2 and high KI67, CD38, HLA-DR, as well as increased Granzyme B and Perforin, previously attributed only to effector cells at the peak of the response. Furthermore, analysis of chromatin accessibility at the single cell level revealed that memory- and effector-differentiated cells clustered together specifically in the acute phase. Altogether, we demonstrate ‘acute traits’ across differentiation subsets, and point out the need to discriminate the differentiation states when studying human CD8 T cells that undergo an acute response.
Keywords: Human CD8 T cells, Acute traits, Effector, Memory, YF-17D vaccination
Graphical abstract

Highlights
-
•
YF-17D vaccination to study human CD8 T cell differentiation.
-
•
Comparing acute versus long-term phases of the immune response.
-
•
Multidimensional single cell analyses by flow cytometry and ATAC sequencing.
-
•
Discovery that acute phase traits are shared in all differentiated human CD8 T cells.
-
•
Several traits previously attributed only to effector are shared with memory cells.
1. Introduction
The differentiation of CD8 T cells reflects the cellular specialisation that guarantees the diverse and complementary features of the adaptive immune response. These features include tissue-specific homing, effector cell functions (cytotoxicity and cytokine production) and memory cell functions (Farber et al., 2014; Mahnke et al., 2013). Subsets of CD8 T cells with distinct differentiation states have been thoroughly characterized, particularly more in-depth in the last decade with the venue of techniques for broad transcriptomic and epigenomic profiling (Crompton et al., 2016). CD8 T cell differentiation subsets such as “Naïve”, “effector” and “memory” are distinguished based on well accepted markers. The activation, differentiation and memory formation of CD8 T cells is typically studied during viral infection (Kaech and Wherry, 2007; Wiesel et al., 2009). In mice, acute immune responses have been widely studied with prototypic models such as LCMV-Armstrong infection. Murine Naïve cells express CD62L and no CD44, while differentiated cells express CD44: memory cells preserve CD62L (CD44+ CD62L+) whereas effector cells lose CD62L (CD44+ CD62L-). At the initiation of the acute murine CD8 T cell response, two subsets have been described and termed Short-Lived Effector Cells (SLECs) with a KLRG1+ IL7Rα- profile and, conversely, Memory-Precursor Effector Cells (MPECs) that are KLRG1- IL7Rα+ (Lefrançois and Obar, 2010; Yuzefpolskiy et al., 2015).
The evidence on acute immune responses has been much more limiting in humans, and studies are mostly done using peripheral blood. Acutely infected humans are not readily traceable and are therefore lacking in the majority of studies. Classic human CD8 T cell subsets have been coined based on resting phenotypes as found in healthy individuals. Naïve cells are double positive for CCR7+ and CD45RA+, Central Memory (CM) cells are CCR7+ CD45RA-, Effector Memory (EM) cells are CCR7- and CD45RA- and Effector Memory RA (EMRA) cells are CCR7- and CD45RA+ (Sallusto et al., 2004). This terminology particularly distinguished tissue localisation between CM and EM, in that CM cells (CCR7+) are called to lymph nodes for ‘reactive memory’ (proliferation and progeny potential) while EM cells migrate to the sites of inflammation for ‘tissue memory’ (with immediate so-called effector functions such as antigen-specific cytolysis and cytokine production).
In this terminology, the EM subset combines “effector” and “memory” for the least in the resting states: it is “effector” because it is capable of immediate so-called effector functions, and it is “memory” because it is found outside an ongoing acute response. Further characterisation of subsets more recently described the Stem Cell-like Memory (SCM) subset, a population of cells that are CD95+ CD58+ within the classically Naïve phenotype cells (CCR7+ CD45RA+) (Gattinoni et al., 2009, 2011; Lugli et al., 2013). To date, all differentiated human CD8 T cell subsets are considered CD95+ CD58+ and Naïve cells are thus CCR7+ CD45RA+ CD95- CD58-. The fact that markers conventionally used to distinguish T cell subsets in mice differ from markers used in humans (e.g. SCA-1 has no ortholog in humans) challenges knowledge transfer from murine models to the human system.
Vaccination with the replicating live-attenuated Yellow Fever (YV) virus YF-17D vaccine stands out as the best-known model to study a human immune response longitudinally, in an experimental setting with controlled timing of antigen exposure, enabling to monitor the entire immune response including the acute phase (Ahmed and Akondy, 2011; Pulendran, 2009). We have previously elaborated longitudinal clinical studies in healthy volunteers receiving the YF-17D vaccine and found that a YF-specific CD8 T SCM population remarkably persists stably for at least two decades (Fuertes Marraco et al., 2015). More recently, we reported that this SCM phenotype is clearly detectable early, and expanding at the peak of the response i.e. at 14 days post-vaccine (Fuertes Marraco et al., 2019). Here, we analysed the various human CD8 T cell subsets at the peak of the acute response to YF-17D vaccination, as well as in long-term phases, assessing the protein expression of 21 markers as well as genome-wide chromatin accessibility, at the single cell level. At the peak of the response, we found that distinct ‘acute traits’ previously attributed only to effector cells are shared across all differentiated subsets, including the SCM subset. We discuss the semantic and marker distinction in the acute and long-term phases.
2. Methods
2.1. Study design, population and ethics statement
Peripheral blood samples used in this study originated from healthy volunteers aged 18–65 years (metadata in Table S1) that participated in one of two observational study protocols on YF-17D vaccination (Stamaril, Sanofi Pasteur), previously described (“YF1” cross-sectional study protocol 329/12 and “YF2” longitudinal study protocol 324/13 (Fuertes Marraco et al., 2015; Fuertes Marraco et al., 2019). The study protocols were approved by the Cantonal Ethics Committee on research involving humans of the Canton of Vaud (CER-VD). Control resting peripheral blood samples from healthy donors were blood donations (apheresis) obtained from the Blood Transfusion Center, Epalinges. All study participants and blood donors provided written informed consent.
2.2. Peripheral blood processing
Peripheral Blood Mononuclear Cells (PBMC) were obtained by density gradient fractionation using Ficoll gradients and immediately cryopreserved in complete medium with 10% dimethylsulfoxide awaiting experimental use as previously described (Fuertes Marraco et al., 2015; Fuertes Marraco et al., 2019).
2.3. Flow cytometry staining, acquisition and analysis
Markers were studied across different flow cytometry panels and staining conditions. Human CD8 T cells were enriched from PBMC using the negative selection kit from Stem Cell. Samples were either stained directly after thawing or stimulated with PMA and ionomycin in presence of Brefeldin A to reveal cytokines by intracellular cytokine staining. The staining conditions were optimised based on setup experiments testing various culture conditions (data not shown) concluding that i) overnight resting results in 5-times less A2/LLW multimer+ cells and loss of positivity for KI67, CXCR3 and CD38, while BCL-2+ cells are increased; ii) IFNγ, TNFα and CD107a are not visible in acutely activated cells (day 14 samples) whether stained directly after thawing or after resting (nor 5 h nor overnight) in medium with Brefeldin A, these rather require restimulation in culture with Brefeldin A, and iii) although resting overnight before stimulation enhances the potential for cytokine production, cytokines are also detectable with direct stimulation (without prior resting). Preliminary experiments using LLW peptide-loaded T2 cells also showed drastic loss of detection of multimer+ CD8 T cells (presumably due to TCR internalisation or TCR occupation with the LLW peptide that precludes multimer binding). Thus, we chose the condition of direct polyclonal stimulation: a brief (5 h) culture of freshly thawed cells with PMA and Ionomycin and Brefeldin A (without overnight resting). This allowed to assess cytokine production while preserving expression of KI67, BCL-2, CD38 and CXCR3. To this end, PBMC were thawed and cultured in complete medium (RPMI supplemented with 10% FCS, 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mm l-glutamine, all Invitrogen) with 10 μg/ml Brefeldin A (Sigma-Aldrich), 20 ng/ml of phorbol-12-myristat-13-acetat and 250 ng/ml of ionomycin. CD107a staining was done by adding the antibody during stimulation in the culture medium. For all experiments, frozen vials of PBMC were thawed in RMPI containing 10 μg/ml of DNAse I (Sigma). Fluorescence-activated cell sorting buffer (FACS) buffer was made with PBS supplemented with 5 mM EDTA, 0.2% Bovine Serum Albumin and 0.2% sodium azide.
Flow cytometry reagents are listed in Table S2. Stainings were stepped as follows: 1) staining with multimers for 30 min at 4 °C in FACS buffer and wash in FACS buffer, 2) surface antibodies in FACS buffer with 10% human Fc block reagent (Miltenyi) for 30 min at 4 °C and wash with PBS, 3) staining with fixable viability dye in PBS 30 min at 4 °C and wash with PBS, 4) fixation 1 h at 4 °C and wash in permeabilization buffer, 5) intracellular staining in permeabilization buffer at 4 °C (for TCF1 staining, the primary rabbit anti-TCF1 and the secondary fluorochrome-conjugated anti-rabbit IgG were stained in two subsequent steps). The fixation and permeabilization buffers were from the Foxp3 staining kit from eBioscience. Washes were made by centrifugation at 450g for 5 min. Samples were resuspended in FACS buffer for acquisition. Cytometers were the LSR II Special Order Research Product and Fortessa (Beckton Dickinson, 5 laser including UV, 13- or 14-color). Flow cytometry FCS data files were analysed in FlowJo 9.7.7, except for t-distributed Stochastic Neighbour Embedding (tSNE) for which the plugin in FlowJo 10.4.2 was used. Downsampling and concatenation of specific populations across samples for tSNE were performed as indicated in the figure legends, in Fig. S1 and in Table S3. The detection threshold for multimer positive populations was 0.01% of total CD8 T cells and at least 10 events, based on control stainings using HLA-A*02 negative samples stained with multimer and unstained controls. The positivity threshold for each marker was set according to distinct negative and positive populations in bulk CD8 T in resting and/or activated samples. Flow cytometry data analysed with FlowJo was quantified based on tabulated exports of the events in the gates of interest. Calculations and data display thereafter was performed using the software: Microsoft Excel 15.21.1, GraphPad prism 7.0c and R studio. Statistical values were obtained as detailed in each figure legend, with p-values scaled as * = p < 0.05, ** = p < 0.01, *** = p < 0.001 and ns = not significant.
2.4. Single cell sequencing assay for transposase-accessible chromatin (scATACseq)
2.4.1. Sample sorting and preparation of nuclei libraries
Several CD8 T cell populations were isolated by flow cytometry, from six different YF vaccinees. This included YF-specific CD8 T cell subsets and reference CD8 T cell subsets (from total CD8 T cells), as indicated in Fig. S2. The various CD8 T cell populations were sorted and pooled into 5 libraries according to the scheme in Table S4. This pooling was done at sorting, operating two cytometers: donor samples were run through the sorters (two samples in parallel, one per cytometer, to reduce experimental timespan) and the populations were gated and isolated directly into their corresponding library tubes according to the aforementioned library scheme. Cells were sorted into DNA low-binding Eppendorf tubes containing PBS supplemented with 0.2% BSA (nuclease-/protease-free, Calbiochem). Immediately after sorting, libraries were spun down at 300 g in a pre-chilled benchtop centrifuge at 4 °C for 5 min. The supernatants were discarded and the cell pellets were resuspended in 50 μL of PBS supplemented with 0.2% BSA and transferred to 0.2 mL PCR tubes. The tubes were spun down at 300 g at 4 °C for 5 min using the tube adapters. After carefully removing 45 μL of supernatant the pellets were resuspended in 45 μL of cold Lysis buffer (10 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 5 mM NaCl, 1% BSA, 0.1% Tween-20, 0.1% NP-40, 0.01% Digitonin), mixed by pipetting up and down 5 times and incubated 3 min on ice. Then, 50 μL of cold Wash buffer (10 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 5 mM NaCl, 1% BSA, 0.1% Tween-20) was added without disturbing the cell pellets. The tubes were spun down at 500 g in the cold centrifuge for 5 min and 95 μL of supernatant were removed. After that, 45 μL of cold Nuclei buffer (10x Genomics) was carefully layered into each tube without disturbing the pellets and spun down at 500 g for 5 min. After removing the supernatant, the resulting nuclei pellets were resuspended in 7 μL of Nuclei buffer. From this resuspension, 2 μL were used for trypan blue staining and nuclei counting. The target count for the libraries was 5’000 nuclei, with a correction factor of 1.53x, that is, samples were adjusted to 6’500 nuclei in 5 μL of Nuclei buffer for each library. Table S4 A shows the sample composition of each of the five 5’000 cell libraries.
2.4.2ATAC library preparation and sequencing
The Chromium chip loading and the ATAC library preparation were performed with the Chromium Single Cell ATAC kit (v1.0 Chemistry) strictly following the 10X guidelines (CG000168, revB). Briefly, nuclei were diluted to around 1’300 nuclei per microliter to target a recovery of 5000 nuclei with 5 μL loading. The initial linear amplification was performed with 12 cycles (GEM incubation step), and the subsequent index PCR step for 12 cycles. Libraries were quantified with the QubIT dsDNA HS kit (ThermoFisher) and their size pattern verified on Fragment analyzer with the HS NGS Fragment Kit (Agilent). An equimolar pool of each library was prepared and loaded at 1.7 nM (with 10% PhiX spike) for a paired end run on a HiSeq4000 (Illumina) following the manufacturer's recommendations. Sequencing data were demultiplexed and further processed with CellRanger ATAC (version 1.2.0) using default parameters.
A unique peak/cell matrix of chromatin accessibility, and a fragment file, were obtained after libraries were aggregated using the cellranger-atac aggr command with default parameters. Normalization was automatically performed by equalizing sensitivity before the merging. In silico analysis of the scATACseq data was based on the peak/cell matrix of chromatin accessibility and fragment file output by CellRanger and was performed in R using various functions from the Signac package version 1.1.0 (Stuart et al., 2020). Table S4 B shows the barcode yields for each of the processed 5’000 cell libraries. Only barcodes unequivocally assigned to single donors (singlets) were considered for downstream analyses: 11’990 cells (82.1% of barcodes). The dataset is accessible in NCBI GEO repository, accession GSE148223.
2.4.3. Genotype demultiplexing
In order to subsequently attribute each nucleus of the libraries to its corresponding donor, genotyping arrays were performed. Genomic DNA samples were isolated using the Qiagen DNeasy Tissue and Blood kit on aliquots of 106 PBMC from each donor. Genotyping was performed using the Illumina Infinium Global Screening Array v3.0 + MD (provided by Life and Brain GmbH, Bonn, Germany), at the iGE3 Genomics Platform of the University of Geneva, Switzerland. Arrays were processed on an iScan according to manufacturer's protocol. Genotype calls were generated using Illumina GenomeStudio software.
Single nucleotide polymorphisms (SNPs) with a GenCall <0.5, minor allele frequency (MAF) < 0.01 and called in less than five of the six samples, were removed. To impute additional SNPs, genotypes were uploaded to the Michigan Imputation Server (Das et al., 2016), and the 1000 Genomes Phase 3 (version 5) reference panel for the European population was used, with a threshold of 0.3 set for the Rsq filter. Imputed and non-imputed genotypes were then used to deconvolute donor identities from the pooled samples using Demuxlet (Kang et al., 2018), with the error rate parameter set to 0.01. A list of SNPs previously confirmed to work well with single cell ATACseq data was used to pre-filter the SNPs (https://github.com/statgen/demuxlet/blob/master/tutorial/YeLab_scRNAseq_exome_variants.bed).
2.4.4. Quality control and cell filtering
Data quality was assessed based on the following metrics, computed using specific functions from the Signac R package: nucleosome signal (approximate ratio of mononucleosomal to nucleosome-free fragments, NucleosomeSignal function); transcriptional start site (TSS) enrichment score (TSSEnrichment function); total number of fragments in peaks; fraction of fragments in peaks; and ratio of reads in genomic blacklisted regions, downloaded from the ENCODE project (Amemiya et al., 2019).
Downstream analyses included only cells with the following criteria (quality control, QC in Fig. S4): a total number of fragments between 3’000 and 20’000 (2’661 cells excluded), more than 15% of fragments in peaks (31 cells excluded), less than 5% of reads in blacklisted regions (no cells excluded), a TSS enrichment score superior to two (13 cells excluded) and a nucleosome signal less than 4 (no cells excluded). Out of the 11’990 singlets, 2’676 cells were excluded (22.3%) and 9’314 cells were included (77.7%).
Selected cells were associated to a specifically sorted CD8 T cell population based on the output from Demuxlet (donor identity) and library identity. Out of the 9’314 cells that passed the aforementioned QC filters, 1’262 cells (13.5%) were not associated to a cell population and were not considered for downstream analysis. The remaining 8’050 cells (86.4%) were associated with a CD8 T cell population assigned to a specific donor. Table S4 C shows the final numbers of cells used for downstream analyses in each of the 30 different samples, after QC filtering and single cell identification based on association to library and donor (genetic demultiplexing).
Coverage plots for specific protein coding genes (pseudo-bulk accessibility tracks by cell population) were produced using the CoveragePlot function and gene coordinates obtained from Ensembl (Yates et al., 2020).
2.4.5. Hierarchical clustering of cell populations based on peak accessibility
Cell populations were clustered based on peak accessibility using the FindAllMarkers function (to identify peaks with differential accessibility between cell populations) and the BuildClusterTree function (to produce a phylogenetic tree relating the ‘average’ cell accessibility for these peaks in each population). Cluster (clade) support was computed by performing 1’000 bootstraps using the boot.phylo function from the R package ape v5.4-1 (Paradis and Schliep, 2019). Support was computed as the proportion of times a given cluster is present in the bootstraps.
2.4.6. Dimension reduction and visualization
For dimension reduction of the peak/cell accessibility matrix, a latent semantic indexing (LSI) analysis was performed, consisting of two steps: first, the matrix was normalized both across cells and across peaks using the term frequency-inverse document frequency (TF-IDF) algorithm (RunTFIDF function); afterwards, singular value decomposition (SVD) was run on the TF-IDF matrix with all the available peaks as features (RunSVD function). The first component of the resulting SVD was removed from downstream analyses after verifying its correlation with sequencing depth (technical variation rather than biological) using the DepthCor function. Boxplots of LSI dimensions two and three by cell population were obtained using custom R functions.
To visualise the cells embedded in the LSI space, a Uniform Manifold Approximation and Projection (UMAP) non-linear dimension reduction was performed based on dimensions 2–10 using the RunUMAP function. The number of neighbouring points used in local approximations of manifold structure was set to 20 and the minimum distance to compress points was set to 0.1. Graphs of the output of the UMAP were obtained using the DimPlot function.
2.4.7. Graph-based clustering of cells
To perform a graph-based clustering of cells, a shared nearest neighbour (SNN) modularity optimisation algorithm was used. First, the SNN graph was produced using the FindNeighbors function (with the same parameter values used in RunUMAP), followed by the identification of clusters of cells using the FindClusters function (with algorithm for modularity optimisation set to SLM and resolution set to 0.7).
The overlap between graph-based clusters of cells and cell populations was evaluated using pairwise Jaccard similarity scores computed using the PairWiseJaccardSetsHeatmap function from the scclusteval R package (Tang et al., 2020). An adjusted Rand index comparing the two classifications was computed using the adjustedRandIndex function from the mclust v5.4.7 R package (Scrucca et al., 2016).
2.4.8. Chromatin accessibility computation
Chromatin accessibility was computed using the GeneActivity() function from the Signac version 1.1.0 R package, summing the fragments intersecting the gene body and promoter region (coordinates were extended to include the 2 kb upstream region, as promoter accessibility is often correlated with gene expression). Chromatin accessibility was then log normalized using the NormalizeData() function from Seurat(), using the median of the number of fragments as a scaling factor. To perform a t-test, the function used was the compare_means() function from the ggpubr version 0.04 R package. Symbols indicating statistical significance refer to p-values, as follows: * ≤0.05; ** ≤0.01; *** ≤0.001; and **** ≤0.0001.
3. Results
3.1. Expression of conventional murine CD8 T cell subset markers in human counterparts
We analysed a comprehensive set of differentiation markers during the acute CD8 T cell response in humans, including markers that are typically used both in humans as well as in mouse studies. First, we profiled the expression of conventional markers used to distinguish differentiation subsets in murine CD8 T cells (i.e. CD44, CD62L, KLRG1 and IL7Rα) by ortholog stainings in resting human PBMC, gating human CD8 T cell subsets conventionally based on CCR7, CD45RA, CD95 and CD58 (Fig. 1 A). As expected, IL7Rα and CD62L were high on Naïve and diminished progressively with differentiation (being lowest in EMRA) (Fig. 1 B and C). KLRG1 was negative in Naïve, intermediate in SCM and CM, and high in EM and EMRA. Lastly, the CD44 staining showed poor discrimination yet as expected being lowest in Naïve and shifted to more positive in SCM, CM and EM. EMRA however showed intermediate levels that could not be clearly separated by gating versus the remainder of subsets. Altogether, conventional markers of murine CD8 T cell subsets show overall agreement in expression profiles in human CD8 T cell subsets, however some mouse markers are not optimal for discriminating human CD8 T cell subsets in full correspondence with the human conventional markers CCR7, CD45RA, CD95 and CD58 (e.g. EMRA are intermediate for CD44, and there are CCR7+ populations that are negative for CD62L and positive for KLRG1). We retained KLRG1, CD62L and IL7Rα for subsequent stainings.
Fig. 1.
Analysis of murine CD8 T cell subset markers in resting human CD8 T cells. A. Human CD8 T cell subsets were analysed from resting peripheral blood of healthy donors gating on CD8b+ live singlets, then gating for the differentiation subsets as shown using CCR7, CD45RA, CD95 and CD58. Total cells were studied, independently of antigen specificity. B. Representative off-set overlay histograms showing expression of the indicated markers across subsets from resting human total CD8 T cells. C. Quantification of the frequencies of positive cells for each marker across subsets, with p-values from Tukey's multiple comparisons tests (N = 3 donors).
3.2. Acute phase traits are shared by all effector and memory CD8 T cell subsets at the peak of the immune response
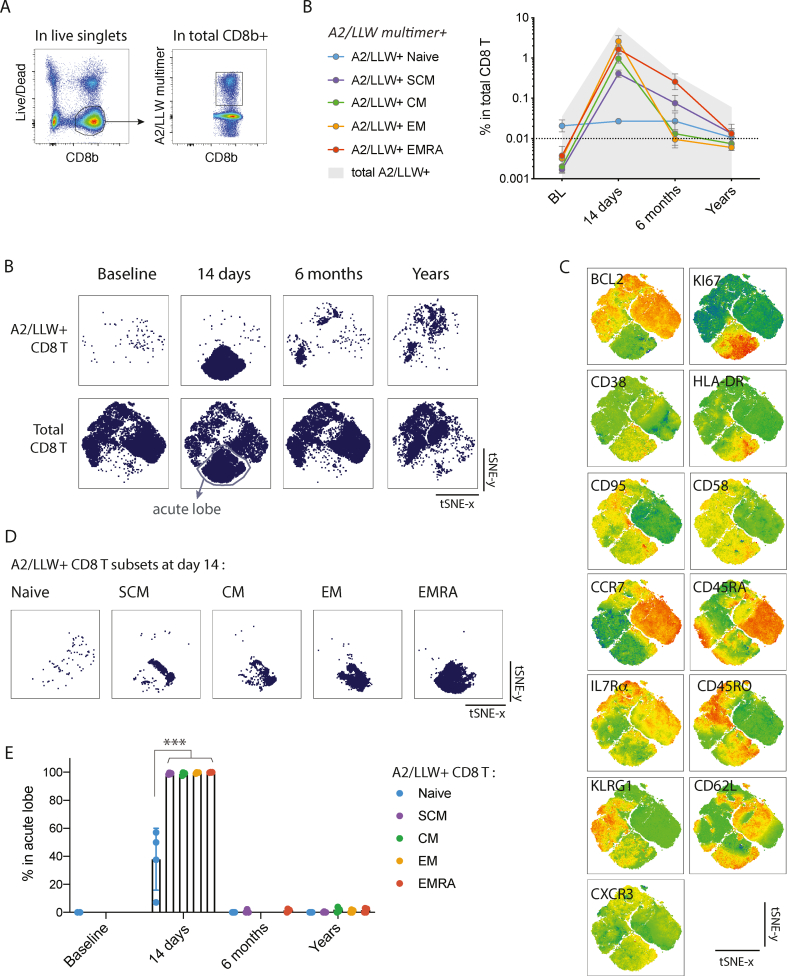
We next analysed markers in human YF-specific CD8 T cells at various time-points in the course of YF-17D vaccination. Recently, we reported that CCR7+ memory phenotype subsets including CM and SCM appear as early as effector-phenotype subsets in response to YF-17D. For this analysis, the time-course included baseline (‘BL’, before vaccination), ‘14 days’ (at the peak of the CD8 T cell response) and ‘6 months’ longitudinally from n = 4 individuals, and samples from n = 4 individuals having received the vaccine between 1.24 and 8.66 years ago (‘Years’) (metadata in Table S1). As previously described, all YF-specific CD8 T cell subsets as detected by the A2/LLW multimer had sharply expanded by day 14 post-vaccine, and the SCM and EMRA subsets particularly persisted in the longer term (Fig. 2A) (Fuertes Marraco et al., 2015; Fuertes Marraco et al., 2019; Akondy et al., 2009).
Fig. 2.
Longitudinal 13-parameter tSNE analysis of human CD8 T cells responding to YF-17D vaccination. A. Enriched CD8+ fractions from PBMC of YF-17D vaccinees were stained and gated for CD8β+ live singlets and A2/LLW multimer+ populations. Differentiation subsets were subsequently gated as in Fig. 1A. B. Quantification of the frequencies of A2/LLW multimer+ subsets in total CD8 T cells. The dotted line shows the 0.01% detection threshold as detailed in methods. C to F. 13-parameter tSNE was ran on a concatenate of all 16 samples (4 time-points, n = 4 donors per time-point), pooling all A2/LLW+ cells together with a downsample of 5’000 CD8β+ live singlet counterparts per sample as internal references. The tSNE analysis strategy is depicted in Fig. S1. C. The concatenate was gated for A2/LLW-specific CD8 T cells (top panels) or total CD8 T cells (bottom panels) across the 4 time-points, plotting the two tSNE dimensions. The “acute lobe” gating is indicated. D. The tSNE plots of the full concatenate showing each of the 13 parameters, heatmapping one marker per plot. E. Display of each of the indicated A2/LLW-specific CD8 T cell subsets at 14 days of vaccination in the tSNE analysis. F. Quantification of the % of cells that fall in the acute lobe gate, in each A2/LLW-specific CD8 T cell subset, at each time-point, with p-values from Tukey's multiple comparisons tests.
Using flow cytometry, we analysed YF-specific CD8 T cells responding to YF-17D vaccination to determine the expression of human differentiation markers (CD45RA, CD45RO, CCR7, CD95, CD58, CXCR3, TCF1, PD1) as well as markers for activation (HLA-DR, CD38), cell cycling (KI67), pro-survival (Bcl-2), cytokines (TNFα, IFNγ, Granzyme B, Perforin), degranulation (CD107a) and mouse-related conventional differentiation markers (KLRG1, CD62L, IL7Rα). These markers were studied by different flow cytometry panels and staining conditions: human CD8 T cells were enriched from cryopreserved PBMC and either stained directly (immediately after thawing) or stimulated in presence of Brefeldin A to reveal cytokines.
We first analysed YF-specific CD8 T cells stained directly with a panel that included 13 markers of activation, proliferation, pro-survival and differentiation: CD45RA, CD45RO, CCR7, CD95, CD58, IL7Rα, KLRG1, CD62L, CXCR3, HLA-DR, CD38, KI67 and BCL-2, combined with the A2/LLW multimer to gate on YF-specific CD8 T cells. In order to perform dimensionality reduction analysis of these markers, we first concatenated the 16 samples, with n = 4 samples from each the time-point: baseline, 14 days, 6 months, and years after vaccination. In the concatenate, we included all events from the A2/LLW+ multimer gate and 5’000 cells from total (multimer negative) CD8 T cells as internal references (Figure S1 A). We then ran tSNE on this concatenate, selecting for the 13 aforementioned parameters (Figure S1 B).
We found that human CD8 T cells undergoing acute activation, i.e. the A2/LLW-specific CD8 T cells at 14 days of vaccination, distinctly clustered from cells at any other time-point, into a hereafter called ‘acute lobe' (Fig. 2 B). This acute lobe had distinctly low BCL-2 expression and was highly enriched for KI67 and the activation markers CD38 and HLA-DR (Fig. 2 C). At baseline, YF-specific CD8 T cells were sparse in a naïve-like lobe (CCR7+ CD45RA+ CD45RO- CD95- CD58-, on the right-hand area of the tSNE), and relocated predominantly in the more resting and differentiated areas (top and left-hand of the tSNE, non-activated CD95+ CD58+) after the peak of the response, in the ‘6 months’ or ‘Years’ samples (Fig. 2 B and C). In order to control for a potential bias due to the unequal relative proportions of multimer+ events across samples in the latter tSNE analysis, we ran a second tSNE on a concatenation of only multimer+ events, downsampling to 100 events and keeping all events for those populations below 100 events (Table S3, Fig. S1C). This second tSNE analysis showed again that YF-specific CD8 T cells locate in distinct areas at baseline, at the peak of the response (14 days) and at later time-points (6 months and years after vaccination).
Importantly, we found that the acute lobe contained a heterogeneity of subset markers as defined by CCR7, CD45RA, CD95 and CD58 (Fig. 2 C). By gating on the various differentiation subsets in A2/LLW+ CD8 T cells, we quantified the fraction of cells that located in the acute lobe in each differentiation subset, at each time-point (Fig. 2 D and E). At day 14, all non-naïve subsets were practically entirely located in the acute lobe (all around 99%), and the naïve-like subset was substantially (on average at 37%) located in the acute lobe (Fig. 2 D and E). At all other time-points, the detected A2/LLW+ populations were at 0% or close to 0% in the acute lobe. This analysis shows that the acute lobe contained CCR7+ memory phenotype CD8 T cells including both CM and SCM, displaying similar traits as CCR7- effector CD8 T cells in terms of the profile HLA-DR+ CD38+ KI67+ BCL-2.
We next quantified the expression frequencies of a total of 17 markers of differentiation, activation, cycling, cytokine production and degranulation, by gating on the 5 conventional differentiation subsets in A2/LLW multimer+ CD8 T cells. These 17 markers included those shown in Fig. 2, and were complemented with TCF1 and PD1 from previous analyses (preprinted in (Fuertes Marraco et al., 2019)) (Fig. 3 A) as well as intracellular cytokine staining and degranulation from stimulation experiments (Fig. 3 B). The latter assay consisted in polyclonal stimulation to stain intracellularly for IFNγ, TNFα, IL-2 as well as CD107a degranulation. Since this assay required high cell numbers, it could only be performed with samples in the acute phase, which had sufficiently high frequencies of YF-specific CD8 T cells.
Fig. 3.
Flow cytometry analysis of 17 markers of activation, differentiation, cycling, survival and cytokines in human CD8 T cells responding to YF-17D vaccination. A. Overlay histograms of the indicated markers, showing representative flow data from A2/LLW-specific CD8 T cells at 14 days (open) or total resting CD8 T cells (at baseline, grey fill). B. Overlay histograms of the indicated markers. Day 14 samples were either stimulated with PMA & ionomycin (open) or left in medium (grey fill) for 5 h in presence of Brefeldin A (BrefA). C and D. The frequencies of cells positive for the 17 markers across the five differentiation subsets, at various time-points following YF-17D vaccination. Populations above the detection limit (>10 events) in at least 2 donors were retained. N = 4 donors were analysed per time-point, except for PD1 and TCF1 with n = 3 at BL, 14 days and 6months. To assess CD107a, IFNγ, TNFα and IL-2, cells from N = 2 donors at Day 14 were stimulated with PMA & ionomycin for 5 h. Positive expression frequencies were compiled, averaged and heatmapped for A2/LLW-specific CD8 T cells alone (in C), or in combination with data from total CD8 T cell counterparts with unsupervised hierarchical clustering by R studio (in D). In D, sample names are formulated as “subset_population_time-point”, where “population” can be A2/LLW multimer+ or total CD8 T cells. For the time-points, BL = Baseline, D14 = 14 days, M6 = 6 months. Day 14 samples are in bold and subsets are color-coded. The full dataset of expression frequencies from C and D is listed in Table S5. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In this 17-marker analysis, the profiles of YF-specific naïve CD8 T cells remained largely stable over time, while all differentiated subsets displayed shared ‘acute traits’ at the peak of the response that faded in the long-term (Fig. 3 C; the full dataset of positive expression frequencies for each marker in each detectable population is listed in Table S5). These acute traits specific to day 14 samples included low BCL-2 and high HLA-DR, CD38 and KI67 (as previously visualised in the ‘acute’ lobe of the tSNE analysis, Fig. 2), as well as increased CD45RO, Granzyme B and to a lesser extent Perforin (Fig. 3 C). Importantly, these traits were seen in all non-naïve subsets including CM and SCM phenotype cells. Stimulation in vitro further revealed that all non-naïve YF-specific cells could produce IFNγ and low levels of TNFα, IL-2 and degranulation (in contrast to no signals in YF-specific naïve cells). The remaining markers, including IL7Rα, TCF1, CD62L, CXCR3, KLRG1, PD1 (conventionally considered differentiation markers) showed profiles that were relatively stable over-time and in line with described differentiation phenotypes (Mahnke et al., 2013) and our previous studies (Fuertes Marraco et al., 2015)(Fuertes Marraco et al., 2019).
In order to interrogate the relatedness of expression profiles, we performed unsupervised hierarchical clustering across samples (subsets and time-points) and across markers, in combination with expression frequencies in total CD8 T cell counterpart populations as internal references (Fig. 3 D). This clustering highlighted that non-naïve YF-specific populations from Day 14 samples cluster together and apart from all other populations. Outside this ‘acute’ non-naïve cluster, populations clustered together into 3 major groups: 1) naïve cells, 2) SCM and CM cells, and 3) EM and EMRA cells; this was independent of YF-specificity and time-point. Altogether, clustering of this 17-marker data compilation reveals that all non-naïve subsets (including memory-phenotype cells) show distinct acute traits that are independent of conventional distinction of differentiation subsets.
3.3. Chromatin accessibility is distinct in the acute phase in both memory and effector human CD8 T cells
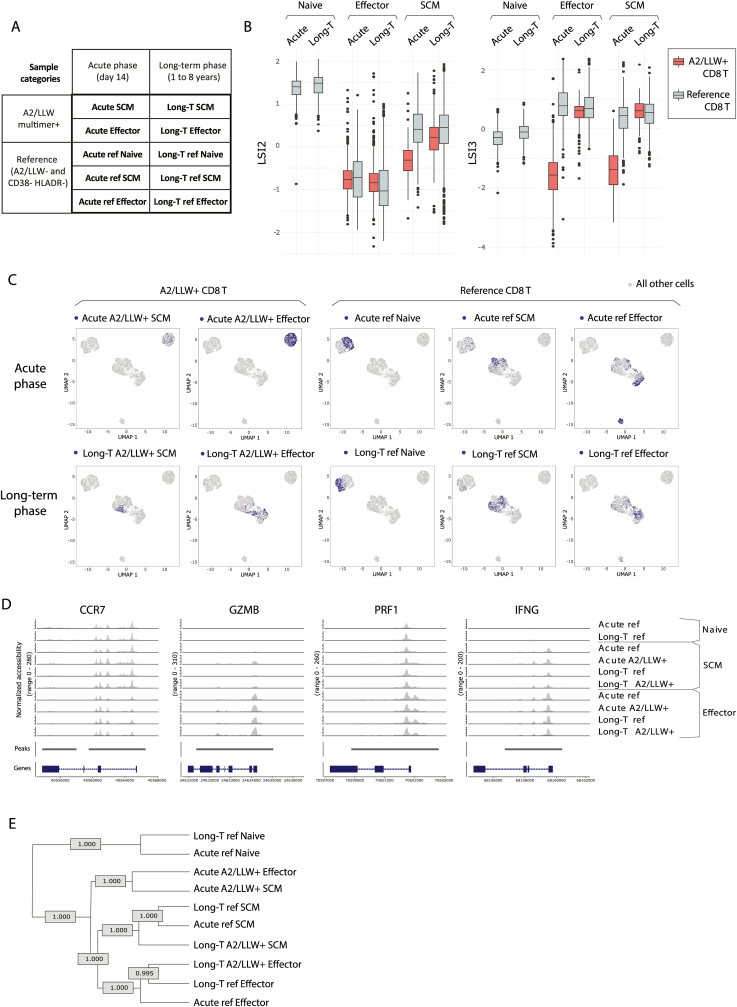
To further assess the molecular properties of CD8 T cells responding to YF-17D vaccination, we profiled their epigenetic landscape, by assessing chromatin accessibility, at the single cell level. We aimed to compare YF-specific CD8 T cells with a memory or effector phenotype, either in the acute phase or in the long-term after YF-17D vaccination (Fig. 4 A).
Fig. 4.
Single cell chromatin accessibility in human CD8 T cell effector and memory populations, in acute and long-term phases following YF-17D vaccination. A. Table recapitulating the characteristics of the 10 different populations analysed by single cell ATACseq (described in Fig. S2). B. Boxplots of the two main dimensions (LSI2 and LSI3) following linear dimensional reduction by Singular Value Decomposition considering all peak features, displaying the 10 experimental populations (A2/LLW multimer+ populations in red, as indicated). C. Clustering analyses based on UMAP 2D-plots following non-linear reduction of dimensions 2–10, showing all cells (in grey) and each of the 10 experimental populations individually mapped in blue, one per plot as indicated in the plot titles. D. Gene coverage plots for the indicated markers showing profiles of the 10 populations, arranged in rows. E. Clustering tree of the 10 populations based all the differentially-accessible peaks (logFC >0.25, 11200 peaks). Numbers indicate the confidence level of the branching (>0.95 = over 95% confidence). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To this end, we sorted A2/LLW+ CD8 T cell populations either with an effector-like phenotype (A2/LLW+ CCR7-) or with a stem cell-like memory phenotype (A2/LLW+ CCR7+ CD45RA+ CD58+ CD95+, Fig. S2). These YF-specific Effector or SCM populations were sorted from donors at day 14 after vaccination (‘acute’ samples) or from donors 1–8 years after vaccination (‘long-term’ samples) (Fig. 4 A, Fig. S2 B to D). As reference populations for the analyses, we also sorted A2/LLW multimer negative populations from total CD8 T cells that showed no activation based on double negative staining for CD38 and HLA-DR. These reference populations were Naïve (CCR7+ CD45RA+ CD95- CD58-), SCM (CCR7+ CD45RA+ CD95+ CD58+) and Effector (CCR7-), all sorted from total CD38- HLA-DR- CD8 T cells, from each donor (Fig. 4 A, Fig. S2).
Given the scarce biomaterial and the technical challenge to isolate sufficient YF-specific CD8 T cells for scATACseq, we designed libraries that cross-pool different populations from each donor once. The experimental design included 5 different sorted CD8 T cell populations from 6 different donors (n = 3 per time-point either ‘acute’ or ‘long-term’) (Table S4 A). Populations were mixed into five libraries during sorting, and each library contained 6 samples (one per donor) with a determined variety of the 5 sorted CD8 T cell populations. Including each donor once per library allowed us to associate each single cell to both, a specific donor based on genotyping information, and a specific CD8 T cell population based on library identity.
The sequencing data was processed as described in the methods, including QC filtering and donor identification (Fig. S3 and Table S4). It yielded 8’050 single cells associated with a donor (time-point) and a specifically sorted CD8 T cell population. Final yields of single cells per sample ranged between 42 cells for the rarest (donor LAU 5094, SCM) to 494 cells (LAU 5048, Effector) for the largest sample. One of the 30 populations (LAU 5048, SCM) was lost due to an incident at sorting (Table S4 C). We found no data quality bias based on individual nor on library identity (data not shown).
Linear dimensional reduction revealed distinct and complementary observations. First, concerning the reference populations, the three subsets were distinct to each other, with differences especially marked in the first dimension (LSI2) (Fig. 4 B). Per subset, reference populations did not differ comparing ‘acute’ versus ‘long-term’ samples. A second series of observations concerned the A2/LLW-specific populations. In the LSI2 dimension, all four effector populations were comparable. In contrast, SCM populations showed that long-term A2/LLW+ SCM resemble reference SCM populations, but acute A2/LLW+ SCM were intermediate between Effector populations and reference SCM populations. In the second dimension (LSI3), the sharpest contrast was found specifically in the acute A2/LLW+ populations both effector and SCM, which distant apart from long-term and reference populations.
In agreement with these observations, additional 2-D UMAP clustering analyses confirmed that A2/LLW+ SCM and Effector clustered together specifically in the acute phase, visible by colour-mapping A2/LLW multimer populations in UMAP plots (Fig. 4C, Fig. S4 A and B). We further confirmed that A2/LLW+ SCM cluster with A2/LLW+ Effector specifically in the acute phase and not in the long-term, by two means: i) unsupervised cluster identification followed by cluster distribution analyses across populations (Fig. S4 D and E); and ii) clustering tree analyses of the 10 populations (Fig. 4 E). In the long-term, A2/LLW+ SCM and A2/LLW+ Effector populations distanced from each other, and each rather clustered close to their reference counterparts (Fig. 4 C and E, Fig. S4). As in the LSI2 dimension, Naïve populations clustered far apart from all other populations, while SCM and Effector reference populations were closer together (Fig. 4 C and E, Figure S4 C).
The data also revealed that a fraction of acute reference Effector cells formed a specific and distant cluster (cluster 7 in Fig. S4 D and E), not present in A2/LLW+ cells, and absent in the long-term phase. We believe this may be acutely activated cells carried over in spite of the HLA-DR and CD38 double positive exclusion during sorting of reference populations (Fig. S2).
In the analysis of discrete markers, the acute A2/LLW+ SCM population showed open chromatin regions at the proximal promoters of subset-defining markers such as CCR7, but also for major effector-associated molecules, such as Granzyme B (GZMB), Perforin (PFN1), and IFNγ (IFNG) (Fig. 4 D). For instance, chromatin accessibility was significantly different between acute and long-term A2/LLW+ SCM for GZMB and IFNG (not for PFN1) (Fig. S5). Finally, clustering tree analysis across the 10 populations again reinforced the relatedness of A2/LLW+ SCM and Effector populations particularly in the acute phase, while all other experimental populations rather clustered based on subset identity (Naïve references together, long-term and/or reference SCM together, long-term and/or reference Effector together) (Fig. 4 E).
Altogether, the scATACseq landscapes of YF-specific SCM and effector CD8 T cells, comparing the acute versus long-term phases of YF-17D vaccination, also supported the evidence that there exist distinct acute traits in both memory and effector subsets, independently of conventionally defined differentiation states.
4. Discussion
In this study, we provide indepth evidence on the profiles of human CD8 T cell subsets in the acute phase of the response versus long-term after vaccination, combining multi-parameter flow cytometry and ATACseq at the single cell level. These experimental assays were performed on samples obtained from YF-17D vaccination studies in humans, including both a longitudinal study covering the first six months and a cross-sectional study covering years after vaccination. As described in the results section, we successfully distinguished and sorted cells according to the established differentiation subset phenotypes. This was technically challenging yet fundamental to further characterise profiles specifically comparing across CD8 T cell subsets and temporal phases of the response. To our knowledge, this is the first time that human CD8 T cell subsets are distinctly characterized with such detail in the course of an acute immune response in humans.
Based on multi-parameter flow cytometry, we found that all differentiated subsets including CM and SCM cells expressed a number of markers specifically in the acute phase, which we refer to as ‘acute traits’. These were BCL2 low (pro-apoptosis), KI67+ (cycling), HLA-DR+ and CD38+ (activation), and several cytokines and cytotoxic molecules (IFNγ, TNFα, Perforin, Granzyme B). To date, these markers have not been associated with memory phenotypes but rather been previously attributed to the effector phenotype and the so-called effector stage in both humans and in mice (Crauste et al., 2017; Miller et al., 2008). The attribution of markers is inherently linked with the study of T cell differentiation pathways, which has been intensely studied and largely debated over the last decades. Mouse studies supporting linear differentiation pathways, where memory cells arise from an obligate effector stage, suggested that biphasic expression of markers such as Bcl2, KI67 and Granzyme B are key to distinguish between effector and memory cells (Crauste et al., 2017; Farber et al., 2014). The concept of an obligate effector phase is for instance supported based on expression of cytotoxic molecules including Granzyme B that have been uniquely attributed to effector cells (Lefrançois and Obar, 2010; Yuzefpolskiy et al., 2015). Direct evidence on differentiation pathways and ontology often requires genetic manipulation and alternatives remain scarce and challenging to apply in human research. We previously discussed how the detection of SCM early in the acute response to YF-17D vaccination in humans may contribute to the debate on differentiation pathways (Fuertes Marraco et al., 2019).
In addition to the detection of memory and effector CD8 T cell subsets early in the YF-17D response, we focused on the profiles that distinguish cells in the acute versus long-term phases (Fig. 5). We performed dimensionality reduction and clustering analyses on single cell datasets, from multi-parameter flow cytometry as well as ATACseq data, specifically adding the separation of subsets in the analyses. We revealed that memory-like subsets (SCM and CM) and effector-like subsets (CCR7- subsets) clustered together during the acute phase of the response. This is direct evidence that there is a considerable number of ‘acute traits’ that are shared across all differentiated subsets in the early, acute phase of the human CD8 T cell response.
Fig. 5.
Human CD8 T cell response to YF-17D vaccination. Modelling of the human CD8 T cell response to YF-17D vaccination considering temporal phases, subset phenotypes and acute traits. The magnitudes shown (y-axis) correspond to values on a logarithmic scale. This modelling is based on our published (Fuertes Marraco et al., 2015; Fuertes Marraco et al., 2019) and present study data.
We and others commonly use the terms “effector” and “memory” for T cell subsets based on the widely accepted molecular features. However, these two terms have historically been used with regard to the temporality of the immune response, whereby “effector” referred to the acute response phase, and “memory” to the following phase in which CD8 T cells persist long-term. Consequently, some studies assigned the term “effector cells” to all cells in the acute phase, and “memory cells” to all cells that persist long-term. Such discrimination of CD8 T cell populations based on the temporality of the response has been used to investigate ontology and differentiation of CD8 T cell subsets, including with the YF-17D vaccination model in humans (Akondy et al., 2017). In the latter study, an ATACseq experiment was performed where “memory” YF-specific CD8 T cells were sorted as total YF tetramer+ cells years after vaccination, while “effector” YF-specific CD8 T cells were sorted as total YF tetramer+ cells 14 days after vaccination. However, we have demonstrated that there is a large heterogeneity in subset composition in YF-specific CD8 T cells at any time-point, with both effector and memory phenotypes that are detectable in both the acute phase and long-term following YF-17D vaccination (Fuertes Marraco et al., 2015; Fuertes Marraco et al., 2019). A more recent study has focused on the analysis of clonal repertoires and trajectories, also sorting total YF-specific dextramer+ CD8 T cells in the acute and long-term phases of YF-17D vaccination (Mold et al., 2021). While this study showed cells with SCM/CM phenotype in the acute phase, and index sorting was included, the analysis is focused on clonal diversity over time and expansion experiments were done on cells in the “memory” (long-term) phase. Heterogeneity of memory phenotypes is indeed observed but authors stated this observation “after resolution of the acute phase” (Mold et al., 2021). To our knowledge, experimental layouts in previous studies on CD8 T cells during YF-17D vaccination (such as the aforementioned) rely on sorting total YF-specific cells and do not include specific sorting of subsets or phenotypic populations. Although heterogeneity of memory CD8 T cells is well studied, only a few papers explicitly consider activated states on top of the consideration of CD8 T cell subset phenotypes particularly in the acute phase (Mahnke et al., 2013). One recent article refers to “hybrid states” with evidence including single cell RNA sequencing on stem and effector properties in human memory CD8 T cells in healthy volunteers (i.e. resting PBMC) (Lugli et al., 2020). Subset sorting (or for the least indexing during sorting) may be critical to draw conclusions since we know that the SCM-like population is around 10-times less frequent than effector-phenotype (EM/EMRA) populations in the acute phase, while SCM and EMRA subsets are on average relatively equal in the long-term (months to years after vaccination; Fig. 2 B and Fuertes Marraco et al., 2015).
Altogether, we believe that our data highlight that there is the need to experimentally and semantically distinguish between ‘temporality’ and ‘phenotype’ in studying the CD8 T cell response. For the least, we reveal the need to clarify and convene on how to distinguish the reference to cells based on the temporal phase of the response (early versus long-term phases) and based on the subset phenotype (effector versus memory phenotypes). There is a perhaps historical confusion that has been nourished by a lack of distinction between temporality and functionality in the description of different CD8 T cell subsets. At the centre of this clarification is to split the assumption that cells in the so-called “effector stage” (which would be the early, acute, activated phase) are the equivalent of cells with an effector phenotype. Indeed, based on the commonly agreed markers to distinguish human CD8 T cell subsets (Mahnke et al., 2013; Sallusto et al., 2004), we showed that there are cells with a memory phenotype (both CM and SCM) that exist in the early acute phases, and that these memory-like cells show the aforementioned ‘acute traits’ previously attributed only to effector cells (Fig. 5). Conversely, there are human CD8 T cells with an effector-like phenotype (in particular EMRA) that can persist long-term together with SCM cells, and these long-term EMRA and long-term SCM no longer show ‘acute traits’. We believe that these facts will increasingly be considered, enabled by the use of modern multiparameter single cell analysis techniques.
CRediT authorship contribution statement
SAFM, DA, SL, AB and DES conceived and designed experiments.
SAFM, HMEH, and DES elaborated the clinical study protocols.
HMEH supervised and coordinated the clinical studies.
SAFM, DA, SL and AB performed experiments.
SAFM, DA, SL, JL and AB analysed data.
JL performed and wrote the in silico methods.
SAFM designed and wrote the manuscript.
DES edited the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are grateful to Federica Sallusto for expert advice. We thank the Flow Cytometry Facility of the University of Lausanne (CH) for cytometer instrument configuration and maintenance, as well as the Genomic Technologies Facility of the University of Lausanne for sustaining the single cell ATACseq procedure and the iGE3 Genomics Platform of the University of Geneva for genotyping (CH). We thank Laurène Cagnon, Samia Abed Maillard, Paula Marcos Mondéjar and Nicole Montandon for participating in the study coordination and sample collection. We thank Blaise Genton, Francine Widmer, Pierrette Meige and the personnel of the ‘Centre de vaccination et médecine des voyages’ at the PMU for collaboration and clinical care of the volunteers of the longitudinal study. Finally, we warmly thank all donors that volunteered and thus preciously contributed to our findings. This study was funded by the University of Lausanne, Ludwig Cancer Research and the Cancer Research Institute (both N.Y., U.S.A), and the Swiss National Science Foundation (grants: 320030-152856 and 310030-179459).
Footnotes
One sentence summary: Acute traits in memory and effector human CD8 T cells.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crimmu.2021.12.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Ahmed R., Akondy R.S. Insights into human CD8+ T-cell memory using the yellow fever and smallpox vaccines. Immunol. Cell Biol. 2011 doi: 10.1038/icb.2010.155. [DOI] [PubMed] [Google Scholar]
- Akondy R.S., Fitch M., Edupuganti S., Yang S., Kissick H.T., Li K.W., Youngblood B.A., Abdelsamed H.A., McGuire D.J., Cohen K.W., Alexe G., Nagar S., McCausland M.M., Gupta S., Tata P., Haining W.N., McElrath M.J., Zhang D., Hu B., Ahmed R. Origin and differentiation of human memory CD8 T cells after vaccination. 2017. Naturefile:///Library/Application Support/Papers2/Articles/2017/Akondy/Nature 2017 Akondy-1.Pdf. [DOI] [PMC free article] [PubMed]
- Akondy R.S., Monson N.D., Miller J.D., Edupuganti S., Teuwen D., Wu H., Quyyumi F., Garg S., Altman J.D., Del Rio C., Keyserling H.L., Ploss A., Rice C.M., Orenstein W.A., Mulligan M.J., Ahmed R. The yellow fever virus vaccine induces a broad and polyfunctional human memory CD8 + T cell response. J. Immunol. 2009 doi: 10.4049/jimmunol.0803903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya H.M., Kundaje A., Boyle A.P. The ENCODE blacklist: identification of problematic regions of the genome. Sci. Rep. 2019 doi: 10.1038/s41598-019-45839-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crauste F., Mafille J., Boucinha L., Djebali S., Gandrillon O., Marvel J., Arpin C. Identification of nascent memory CD8 T cells and modeling of their ontogeny. Cell Systems. 2017 doi: 10.1016/j.cels.2017.01.014. [DOI] [PubMed] [Google Scholar]
- Crompton J.G., Narayanan M., Cuddapah S., Roychoudhuri R., Ji Y., Yang W., Patel S.J., Sukumar M., Palmer D.C., Peng W., Wang E., Marincola F.M., Klebanoff C.A., Zhao K., Tsang J.S., Gattinoni L., Restifo N.P. Lineage relationship of CD8+ T cell subsets is revealed by progressive changes in the epigenetic landscape. Cell. Mol. Immunol. 2016 doi: 10.1038/cmi.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M., Schlessinger D., Stambolian D., Loh P.R., Iacono W.G., Swaroop A., Scott L.J., Cucca F., Kronenberg F., Boehnke M., Fuchsberger C. Next-generation genotype imputation service and methods. Nat. Genet. 2016 doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber D.L., Yudanin N.A., Restifo N.P. Nature Reviews Immunology. 2014. Human memory T cells: generation, compartmentalization and homeostasis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes Marraco S.A., Soneson C., Cagnon L., Gannon P.O., Allard M., Maillard S.A., Montandon N., Rufer N., Waldvogel S., Delorenzi M., Speiser D.E. Long-lasting stem cell-like memory CD8+ T cells with a naïve-like profile upon yellow fever vaccination. Sci. Transl. Med. 2015 doi: 10.1126/scitranslmed.aaa3700. [DOI] [PubMed] [Google Scholar]
- Fuertes Marraco S., Bovay A., Nassiri S., Maby-El Hajjami H., Ouertatani-Sakouhi H., Held W., Speiser D. The human CD8 T stem cell-like memory phenotype appears in the acute phase in Yellow Fever virus vaccination. 2019. [DOI]
- Gattinoni L., Lugli E., Ji Y., Pos Z., Paulos C.M., Quigley M.F., Almeida J.R., Gostick E., Yu Z., Carpenito C., Wang E., Douek D.C., Price D.A., June C.H., Marincola F.M., Roederer M., Restifo N.P. A human memory T cell subset with stem cell-like properties. Nat. Med. 2011 doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L., Zhong X.S., Palmer D.C., Ji Y., Hinrichs C.S., Yu Z., Wrzesinski C., Boni A., Cassard L., Garvin L.M., Paulos C.M., Muranski P., Restifo N.P. Wnt signaling arrests effector T cell differentiation and generates CD8 + memory stem cells. Nat. Med. 2009 doi: 10.1038/nm.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S.M., Wherry E.J. Immunity. 2007. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.M., Subramaniam M., Targ S., Nguyen M., Maliskova L., McCarthy E., Wan E., Wong S., Byrnes L., Lanata C.M., Gate R.E., Mostafavi S., Marson A., Zaitlen N., Criswell L.A., Ye C.J. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol. 2018 doi: 10.1038/nbt.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrançois L., Obar J.J. Once a killer, always a killer: from cytotoxic T cell to memory cell. Immunol. Rev. 2010 doi: 10.1111/j.0105-2896.2010.00895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli E., Galletti G., Boi S.K., Youngblood B.A. Stem, effector, and hybrid states of memory CD8+ T cells. Trends Immunol. 2020;41(1):17–28. doi: 10.1016/j.it.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli E., Gattinoni L., Roberto A., Mavilio D., Price D.A., Restifo N.P., Roederer M. Identification, isolation and in vitro expansion of human and nonhuman primate T stem cell memory cells. Nat. Protoc. 2013 doi: 10.1038/nprot.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahnke Y.D., Brodie T.M., Sallusto F., Roederer M., Lugli E. European Journal of Immunology. 2013. The who's who of T-cell differentiation: human memory T-cell subsets. [DOI] [PubMed] [Google Scholar]
- Miller J.D., van der Most R.G., Akondy R.S., Glidewell J.T., Albott S., Masopust D., Murali-Krishna K., Mahar P.L., Edupuganti S., Lalor S., Germon S., Del Rio C., Mulligan M.J.J., Staprans S.I., Altman J.D., Feinberg M.B., Ahmed R. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008 doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Mold J.E., Modolo L., Hård J., Zamboni M., Larsson A.J.M., Stenudd M., Eriksson C.J., Durif G., Ståhl P.L., Borgström E., Picelli S., Reinius B., Sandberg R., Réu P., Talavera-Lopez C., Andersson B., Blom K., Sandberg J.K., Picard F., Frisén J. Divergent clonal differentiation trajectories establish CD8+ memory T cell heterogeneity during acute viral infections in humans. Cell Rep. 2021 doi: 10.1016/j.celrep.2021.109174. [DOI] [PubMed] [Google Scholar]
- Paradis E., Schliep K. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019 doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- Pulendran B. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nat. Rev. Immunol. 2009 doi: 10.1038/nri2629. [DOI] [PubMed] [Google Scholar]
- Sallusto F., Geginat J., Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Ann. Rev. Immunol. 2004 doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- Scrucca L., Fop M., Murphy T.B., Raftery A.E. Mclust 5: clustering, classification and density estimation using Gaussian finite mixture models. R J. 2016 doi: 10.32614/rj-2016-021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Srivastava A., Lareau C., Satija R. Multimodal single-cell chromatin analysis with Signac. 2020. bioRxiv. [DOI]
- Tang M., Kaymaz Y., Logeman B.L., Eichhorn S., Liang Z.S., Dulac C., Sackton T.B. Evaluating single-cell cluster stability using the Jaccard similarity index. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesel M., Walton S., Richter K., Oxenius A. APMIS. 2009. Virus-specific CD8 T cells: activation, differentiation and memory formation. [DOI] [PubMed] [Google Scholar]
- Yates A.D., Achuthan P., Akanni W., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R., Bhai J., Billis K., Boddu S., Marugán J.C., Cummins C., Davidson C., Dodiya K., Fatima R., Gall A., Flicek P. Ensembl 2020. Nucl. Acids Res. 2020 doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzefpolskiy Y., Baumann F.M., Kalia V., Sarkar S. Cellular and Molecular Immunology. 2015. Early CD8 T-cell memory precursors and terminal effectors exhibit equipotent in vivo degranulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.