Abstract
Autoimmune skin diseases are complex and are thought to arise from a combination of genetics and environmental exposures, which trigger an ongoing immune response against self-antigens. Companion animals including cats and dogs are known to develop inflammatory skin conditions similar to humans and share the same environment, providing opportunities to study spontaneous disease that encompasses genetic and environmental factors with a One Health approach. A strength of comparative immunology approaches is that immune profiles may be assessed across different species to better identify shared or conserved pathways that might drive inflammation. Here, we performed a comparative study of skin from canine discoid lupus erythematosus (DLE) using NanoString nCounter technology. We compared these gene expression patterns to those of human DLE and a mouse model of cutaneous lupus. We found strong interferon signatures, with CXCL10, ISG15, and an S100 gene family member among the highest, most significant DEGs upregulated across species. Cell type analysis revealed marked T-cell and B-cell infiltration. Interestingly, canine DLE samples also recapitulated downregulated skin homeostatic genes observed in human DLE. We conclude that spontaneous DLE in dogs captures many features that are present in human disease and may serve as a more complete model for conducting further genomic and/or transcriptomic studies.
Keywords: Cutaneous lupus erythematosus, Discoid lupus, Canine (dog), Chemokine, Cytokine, Skin, Comparative immunology, Immunopathogenesis
Graphical abstract
Highlights
-
•
Canine DLE lesions express known drivers of pathogenesis including CXCL10, IFNG, FAS.
-
•
Enrichment of key cell types, including T, B, NK cells, is observed in canine DLE.
-
•
Canine, mouse and human DLE share similar proinflammatory profiles.
-
•
Canine DLE exhibits downregulated skin homeostatic and immune regulatory genes.
1. Introduction
Discoid lupus erythematosus (DLE), the most common subtype of chronic cutaneous lupus erythematosus (CCLE), is an autoimmune disease characterized by erythema, telangiectasias, follicular plugging, and coin shaped or ‘discoid’ lesions which begin as atrophy and resolve to scars (Yu et al., 2013). DLE lesions mostly affect the scalp, face and neck, but can present on other areas of the body. DLE exhibits a female sex bias, and has an estimated incidence of 4.2–4.3 per 100,000 (Jarukitsopa et al., 2015; Durosaro et al., 2009; Hejazi and Werth, 2016; George and Tunnessen, 1993). Approximately 58% of patients who present with DLE exhibit lupus band reaction in their skin (Shahidullah et al., 1995), which consists of IgG and IgM antibodies deposited at the dermal-epidermal junction (DEJ) (Weigand, 1986; Verdelli et al., 2019). A minority of patients exhibit high anti-nuclear antibody (ANA) titers, and there is a lower risk of DLE progressing to systemic lupus as compared to other CLE clinical subtypes (Merola et al., 2013).
Dogs also develop DLE and share similar clinical and immunopathological characteristics with the human form. Facial DLE (FDLE), the head-limited form of canine DLE, disproportionately affects German Shepherd Dogs and their crosses, but can affect all breeds. Generalized DLE (GDLE) in dogs tends to affect the face, front limbs, neck, and abdomen, and breed preference is currently unknown (Banovic et al., 2016). DLE lesions in dogs appear annular (discoid) to polycyclic plaques with dyspigmentation (depigmentation or hyperpigmentation), adherent scaling, follicular plugging and central alopecia. Lesions progress into dyspigmented scars, much like lesion progression in human DLE patients. Lymphocyte rich interface dermatitis is a common histopathological feature in both species (Olivry et al., 2018; Rothfield et al., 2006).
An advantage of studying spontaneous DLE in dogs is that it eliminates the layer of potential bias inherent in constructing an inducible animal model. Dogs have served as a comparative model for human cancers and immune dysregulation, particularly for bone and blood cancers and allergic skin disease (Freudenberg et al., 2019; Lee et al., 2018; Todhunter et al., 2019; Khanna et al., 2006). While gene expression patterns in canine lymphomas, sarcomas and atopic dermatitis have been compared to their human orthologs, most of the established relationships between human and canine autoimmune diseases have relied on clinical and histological comparisons. Elucidating molecular associations between human, induced and spontaneous models of autoimmunity are necessary for incorporating spontaneous models of disease with existing inducible models. To begin to understand these relationships, we performed a retrospective gene expression analysis study of canine DLE, human DLE, and a mouse model of cutaneous lupus which recapitulates elements of DLE. Here we show similar gene expression patterns across species and confirm protein-level expression of key genes in canine skin tissue by immunohistochemistry. Importantly, canine DLE and human DLE shared downregulated gene signatures for skin homeostatic genes. We hope that our studies will provide the basis for development of novel and more targeted treatments for both human and veterinary DLE patients.
2. Materials and methods
Clinical samples: The animal study was reviewed and approved by Cummings School of Veterinary Medicine at Tufts University IACUC. At the time the veterinary patients were seen at the Cummings School of Veterinary Medicine, samples were retained in the biorepository with written owner consent.
Skin biopsies from the biorepository at Cummings School of Veterinary Medicine at Tufts (NAR, CPM) between the years 2011 and 2019 were queried for “interface dermatitides”, and potential cases were reviewed based on prior histological reports and clinical history before being categorized into their appropriate diagnoses. Inclusion criteria for DLE samples included interface dermatitis on H&E and clinical findings consistent with DLE. Control tissue from healthy leg margin skin were also selected from the biorepository. These samples were derived from marginal skin from leg amputations (e.g. if a dog was hit by a motor vehicle and needed a limb amputation). The surgeons always remove an extra margin of skin to check for malignancy, necrosis, etc., much like what is done for human surgeries (panniculectomies, etc.). Slides and pathology reports from all cases were reexamined by a board-certified veterinary pathologist and dermatologist (NAR, RMA) to confirm diagnoses and absence of obvious infectious disease. Clinical characteristics are summarized in Table 1.
Table 1.
Clinical characteristics of canine DLE cases and healthy leg margin controls.
| Case & Diagnosis | Signalmenta | Breed | Duration & Past Medical History | Prior treatment |
|---|---|---|---|---|
| DLE 1 | 7 yo, MN | German Shepherd cross | Nasal discharge for 4 months | Antibiotics (Cephalexin) with partial and transient improvement |
| DLE 2 | 3 yo, MN | Saint Bernard | ND | None |
| DLE 3 | 11 yo, FS | Pitbull | Recurrent facial ulceration with partial and temporarily responded to antibiotics and steroids | Antibiotics and steroids |
| DLE 4 | 5 yo, Male | Mixed | 1 year history of progressive nasal lesions | Topical antibiotics |
| DLE 5 | 6 yo, MN | Coonhound | 4 month history of nasal depigmentation and crusting | None |
| DLE 6 | 13 yo, MN | German Shorthair Pointer | 10 month history of nasal crusting and ulceration, allergic rhinitis | Carprofen, Apoquel, Tramadol, Cetirizine, Amoxicilin-clavulanate |
| DLE 7 | 1 yo, FS | Boxer | ND | ND |
| Healthy 1 | 8 yo, FS | Labrador Retriever | NA | None |
| Healthy 2 | 11 yo, FS | Siberian Husky cross | NA | None |
| Healthy 3 | 11 yo, MN | Golden Retriever | NA | None |
| Healthy 4 | 12 yo, MN | German Shepherd cross | NA | None |
| Healthy 5 | 6 yo, FS | Alaskan Malamute | NA | None |
at time of biopsy; FS = female spayed, MN = male neutered, yo = year old, ND = not determined/information not available, NA = not applicable.
Isolation of RNA from FFPE blocks: 30 μm curls were cut from the blocks and stored in Eppendorf tubes at ambient temperature. RNA was isolated using the Qiagen FFPE RNeasy kit per the manufacturer directions. Briefly, razor blades were treated with RNase, excess paraffin was removed, and tissues were sliced into thin strips (5 μm) to create more surface area prior to incubation with deparaffinization solution (Qiagen). The protocol was followed and RNA was quantified using a nanodrop.
NanoString cartridge and processing: A custom NanoString canine gene panel of 160 genes including cytokine, chemokine, and immune genes, as well as skin and immune cell specific transcripts was created. RNA was hybridized using a BioRad C1000 touch machine, and samples were loaded into NanoString cartridges and analyzed with a Sprint nCounter. Raw data are deposited on Gene Expression Omnibus (GEO) Database under accession #GSE160260.
nSolver analysis: NanoString’s software, nSolver was used for all normalization and fold change calculations of canine samples. We used B2m, Rpl13a, cg14980, and hprt as housekeeping genes for this study. Advanced analysis was used for DEG calculation and the “Cell Type Score”, which is a summary statistic of the expression of the marker genes for each cell type (Danaher et al., 2017). Any counts under log2 of 5 (y axis) on the cell type scores is considered undetected. P-values for cell type scores were generated using the ‘Ttest_ind’ function from the ‘stats’ module in SciPy (v1.4.1) with the equal variance parameter set to False. Heatmap of "Cell Type Score" was created with Morpheus software, https://software.broadinstitute.org/morypheus.
Reanalysis of Murine CLE samples: The mouse model of CLE is on a lupus-prone background (TLR9−/−) and uses a model autoantigen, ovalbumin (OVA), which is expressed under the control of a tet response element in MHC Class II + cells. Administration of doxycycline chow turns on OVA expression in antigen presenting cells, and i.v. injection of OVA-specific DO11+ T cells induces an inflammatory response that mimics many features of CLE, including alopecia, erythema, crusting, interface dermatitis, mucin deposition, lupus band reaction, ANAs and mild proteinuria (Mande et al., 2018). Mouse CLE skin NanoString expression data (Mande et al., 2018) were normalized to the housekeeping genes provided in the murine cancer immune codeset, and were then truncated to match as many of the 160 genes used in the canine custom probeset as possible, which encompassed 92 overlapping genes. Data comparisons for dog versus mouse were analyzed with nSolver.
H&E and IHC: Biopsy samples were formalin fixed and routinely processed to be paraffin embedded. Blocks were cut to 5 μm sections and stained with H&E on a Sakura Tissue-Tek DRS autostainer before being coverslipped.
IHC was performed on unstained 5 μm sections using rabbit-anti-canine CXCL10, IFNγ (US Biological) or isotype control (Biolegend) at 1:50 or 1:100 dilution using a Dako automated slide staining machine with Dako EnVision + Dual Link System-HRP for anti-rabbit secondary antibody to visualize staining with brown chromogen. All sections were counterstained with hematoxylin. For canine Ig stain, slides were shipped to the histopathology lab at the University of Minnesota (fee-for-service) and were visualized with red chromogen.
Images were taken using an Olympus BX51 microscope with Nikon NIS Elements software version 3.10.
Reanalysis of GSE95474 dataset: DLE patient samples from GSE95474 series matrix data (Scholtissek et al., 2017) were downloaded using the getGEO function in the R package GEOquery. Data was then log2 transformed, and differential expression was calculated with the topTable function in the R package limma.
Python Visualization: Data for heatmaps and volcano plots were created using Pandas (v1.0.3). Volcano plots were produced using the scatterplot function from matplotlib (v3.2.1) and heatmaps were created using the heatmap or the clustermap function in Seaborn (v0.10.0). All numerical transformations (e.g. log transformation, z-scoring) performed for heatmaps and volcano plots were done using NumPy (v1.18.1).
Canine, Mouse and Human Dataset alignment and analysis: The names from the 160 gene custom canine dataset and the 755 gene murine cancer immune dataset (Mande et al., 2018) were queried in the GSE95474 dataset (Scholtissek et al., 2017) in Excel using the formula “ = IF(ISERROR(VLOOKUP(A2,$B$2:$B$1001,1,FALSE)),FALSE, TRUE)”. Values from the original canine and mouse datasets that were retrieved as FALSE were then queried for alternate gene names using GeneCards.org. Missing values were renamed to match the naming conventions used across all species, which yielded 160 overlapping genes for canine versus human (entire canine codeset), 730 out of 755 overlapping genes for mouse versus human, and 92 genes overlapping all 3 species (Table S1). Genes that did not have a human homologue were excluded from the mouse dataset, namely several chemokine family members and Klr family members. Data comparisons for all three species (dog, mouse, human) were analyzed with ClustVis (Metsalu and Vilo, 2015) and BioVenn (Hulsen et al., 2008).
Statistics: Statistical analyses for box plots and generation of volcano plots was performed in GraphPad Prism version 9. Shapiro-Wilk normality tests were performed, and normally distributed data were analyzed with two-tailed student’s t-test, and non-normally distributed data were analyzed with two-tailed Mann Whitney U test, to compare healthy to DLE lesional skin. P values < 0.05 were considered significant, with P < 0.01 as highly significant.
3. Results
3.1. Canine gene expression analysis reveals key immune and skin genes upregulated in DLE versus healthy controls
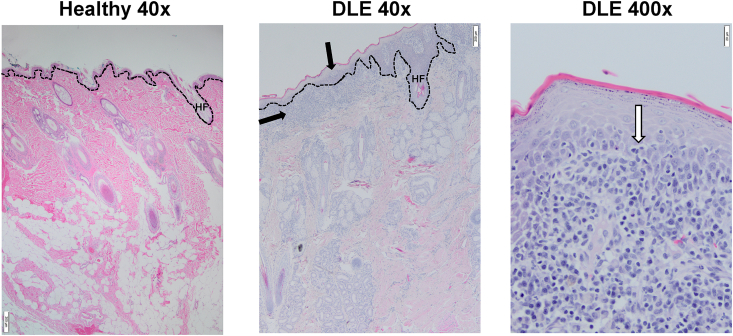
We isolated RNA from FFPE skin curls from six canine DLE and five healthy canine patients (clinical characteristics in Table 1). All cases exhibited marked interface dermatitis, and some exhibited basement membrane thickening (Fig. 1a). Vacuolization of basal keratinocytes, melanocyte dropout, and basal epidermal layer atrophy were also observed.
Fig. 1.
H&E staining of canine DLE and healthy canine skin reveals interface dermatitis consistent with human DLE. Healthy (left, 40x mag) and DLE (center 40x mag, right 400x mag) skin stained with H&E. DLE exhibits a number of cutaneous lupus hallmarks, including basement membrane thickening, basal cell degeneration (vertical black arrow), obfuscation of DEJ (white arrow), and lymphocyte infiltration (horizontal black arrow). Dashed lines represent the DEJ; HF = hair follicle. Scale bars 40x images = 200 μm, scale bars 400x image = 20 μm.
We chose NanoString technology to analyze RNA from these FFPE samples for several reasons. First, NanoString works on difficult samples such as fragmented RNA which is isolated from FFPE tissue due to the use of hybridization technology. Second, there are no enzymatic steps (RT, amplification) which could introduce a bias in data acquisition. Additionally, NanoString analysis has a fast turnaround time (24 h) while providing robust data which are concordant with RNA seq (Veldman-Jones et al., 2015).
Utilizing the high-fidelity NanoString nCounter platform, we probed gene expression for 160 targets including a range of chemokines and cytokines, immune related genes previously reported as significant to lupus pathogenesis, immune cell marker genes, skin associated genes, and neuroendocrine genes. Hierarchical clustering of gene expression for the whole panel discriminates between healthy and DLE canine skin (Supplemental Fig. 1). Principal component analysis (PCA) revealed that the DLE cases and healthy cases all fell within the 95% confidence interval for predicting disease status (Supplemental Fig. 2). Of the genes in our NanoString panel, 47 genes were differentially expressed using a significance cutoff of p < 0.01. Thirty-three genes were upregulated with a log fold change of greater than 2, mainly consisting of inflammation related genes. Fourteen genes were downregulated with a log fold change less than −1.5, some related to lipid metabolism and hormone response and others were homeostatic skin genes.
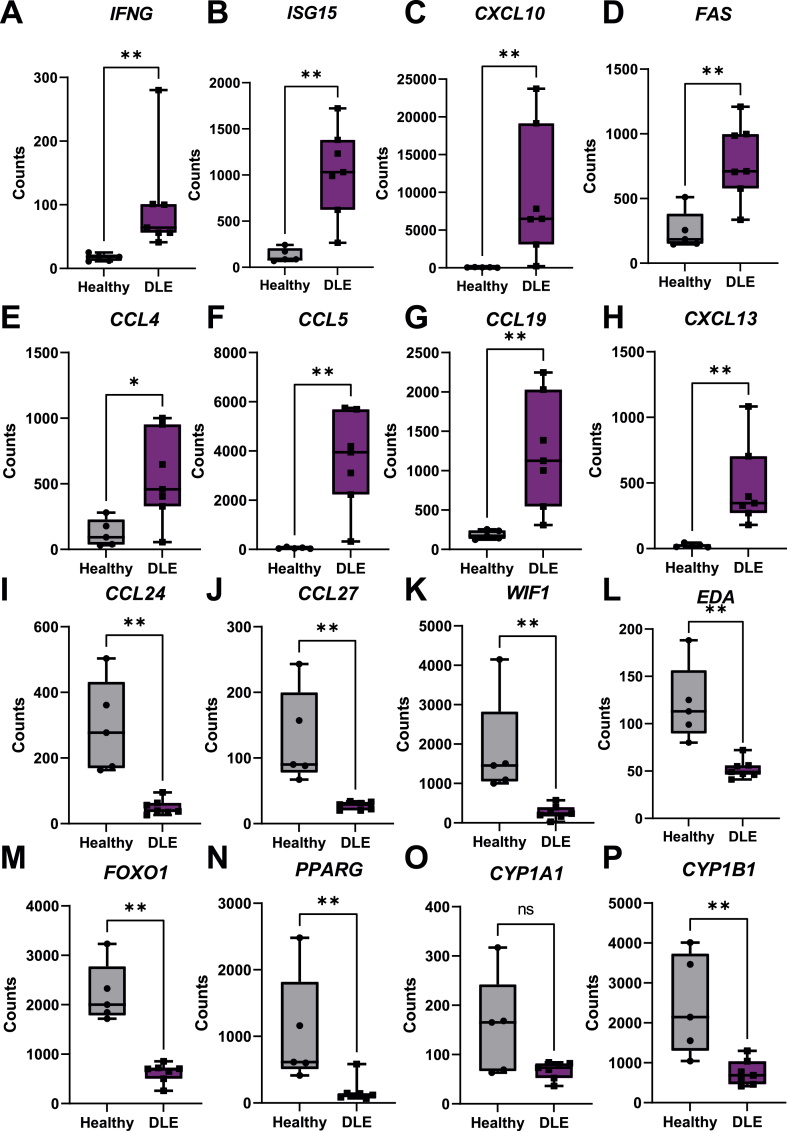
Next, we examined known immune mediators of DLE. Interferons are well characterized drivers of lupus, with IFNG uniquely driving DLE (Yu et al., 2013; Berthier et al., 2019; Ferrigno et al., 2019). IFNG was significantly upregulated in canine DLE (Fig. 2A), however IFNA was not. The IFN inducible gene ISG15 (Fig. 2B) as well as the IFN inducible chemokine CXCL10 (Fig. 2C) were upregulated, supporting IFN-mediated responses. TNF was upregulated in canine DLE, while IFNB1 was significantly downregulated 1.3 fold compared to healthy controls (P = 0.0011, Table S1). Cytokines IL-16 and TSLP were downregulated compared to healthy controls (Table S1). FASL and its receptor FAS (Fig. 2D), which have been implicated in lupus pathogenesis (Wu et al., 1996; Nakajima et al., 1997), were significantly upregulated in lupus cases compared to healthy controls. The chemokines CCL4 (Fig. 2E), CCL5 (Fig. 2F), CCL19 (Fig. 2G), and CXCL13 (Fig. 2H) were significantly upregulated, while the homeostatic chemokines CCL24 (Fig. 2I) and CCL27 (Fig. 2J), and the CXCR4 ligand CXCL12 were downregulated compared to healthy controls (Table S1). VGLL3, a factor that has been implicated in driving the female dominant sex disparity in CLE and autoimmune diseases as a whole (Billi et al., 2019), was downregulated.
Fig. 2.
Upregulated genes associated with inflammation and lupus, and downregulated genes associated with skin homeostasis in canine DLE skin. Boxplots of significantly upregulated genes in DLE versus healthy canine tissue previously associated with inflammation and lupus are presented as follows: (A) IFNG, (B) ISG15, (C) CXCL10, (D) FAS, (E) CCL4, (F) CCL5, (G) CCL19 and (H) CXCL13. Significantly downregulated genes associated with keratinocyte and skin homeostasis are presented as follows: (I) CCL24, (J) CCL27, (K) WIF1, (L) EDA, (M) FOXO1, (N) PPARG. (O) CYP1A1 trended lower and (P) CYP1B1 exhibited significantly lower expression in DLE lesional skin. (n=7 DLE and n=5 healthy controls, student’s t test and/or Mann Whitney U tests significant as indicated).
Several skin homeostatic and immune regulatory genes were significantly downregulated in DLE lesions, including WIF1 (Fig. 2K), a WNT signaling inhibitory protein. EDA (Fig 2L), an inducer of non-canonical NF-kB signaling, was downregulated. FOXO1 (Fig 2M), FOXO3A, RXRG, and PPARG (Fig 2N) were all downregulated to varying degrees, and are implicated in cellular steroid hormone response (GO:0071383). CYP1B1, but not CYP1A1 (Fig. 2O–P), and TAC1 were all downregulated to varying degrees, and are implicated in cellular steroid hormone response (GO:0071383). Lastly, analysis of neuroendocrine genes revealed that ADRB1, MC5R, and CPA3 were downregulated in DLE versus healthy controls (Table S1).
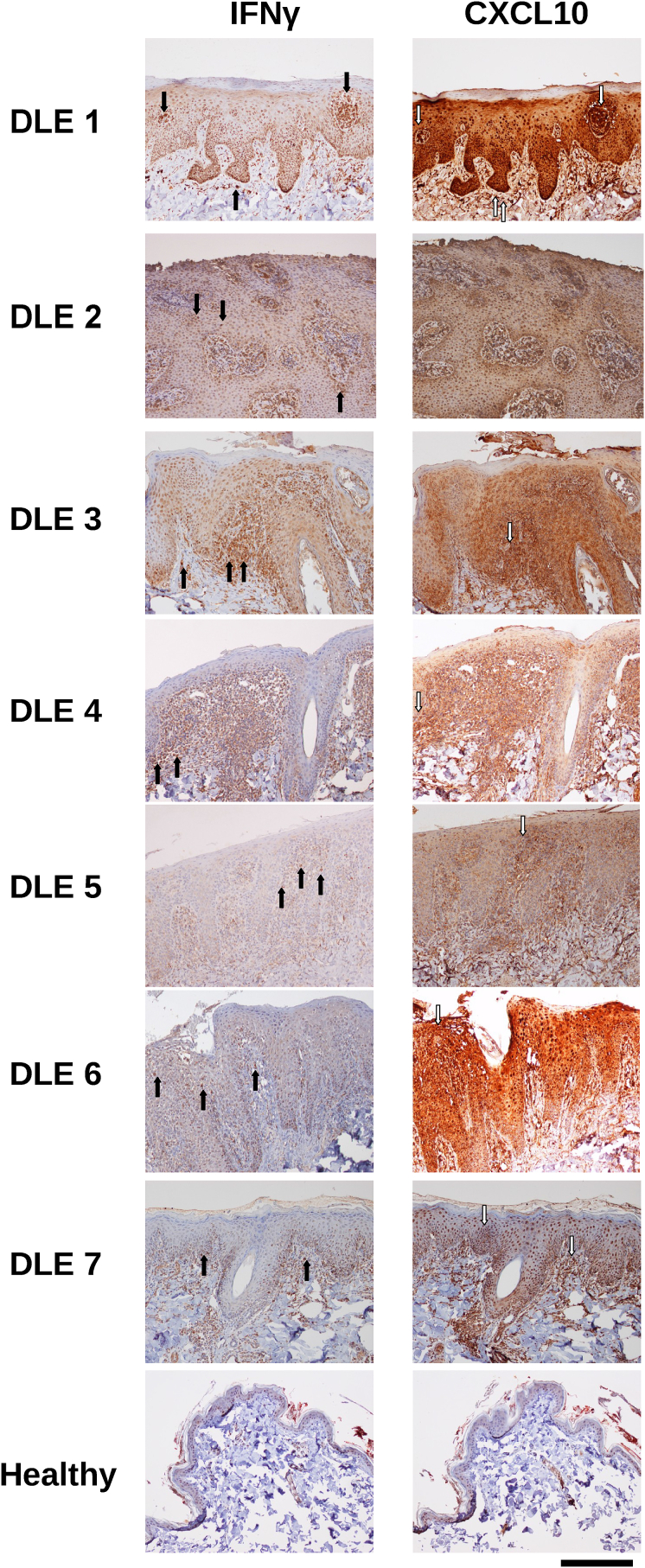
3.2. CXCL10 and IFNγ are highly expressed at the protein level in canine DLE lesions
To confirm protein level expression of key genes identified in our NanoString codeset, we performed immunohistochemistry for IFNγ and CXCL10. IFNγ staining was strongest in infiltrating lymphocytes near the basal epidermis (Fig. 3). CXCL10 was expressed throughout the epidermis, and staining was strongest in keratinocytes.
Fig. 3.
Immunohistochemical confirmation of protein-level expression of key immunopathogenesis genes in canine DLE. Immunohistochemistry staining of interferon gamma (IFNγ) and CXCL10 in canine DLE and healthy skin (10x). IFNγ exhibits punctate/cellular expression (black arrows), and is most prevalent around the dermoepidermal junction. CXCL10 is expressed throughout the tissue, and is most prominently expressed by keratinocytes in the epidermis (white arrows; scale bar = 300 μm).
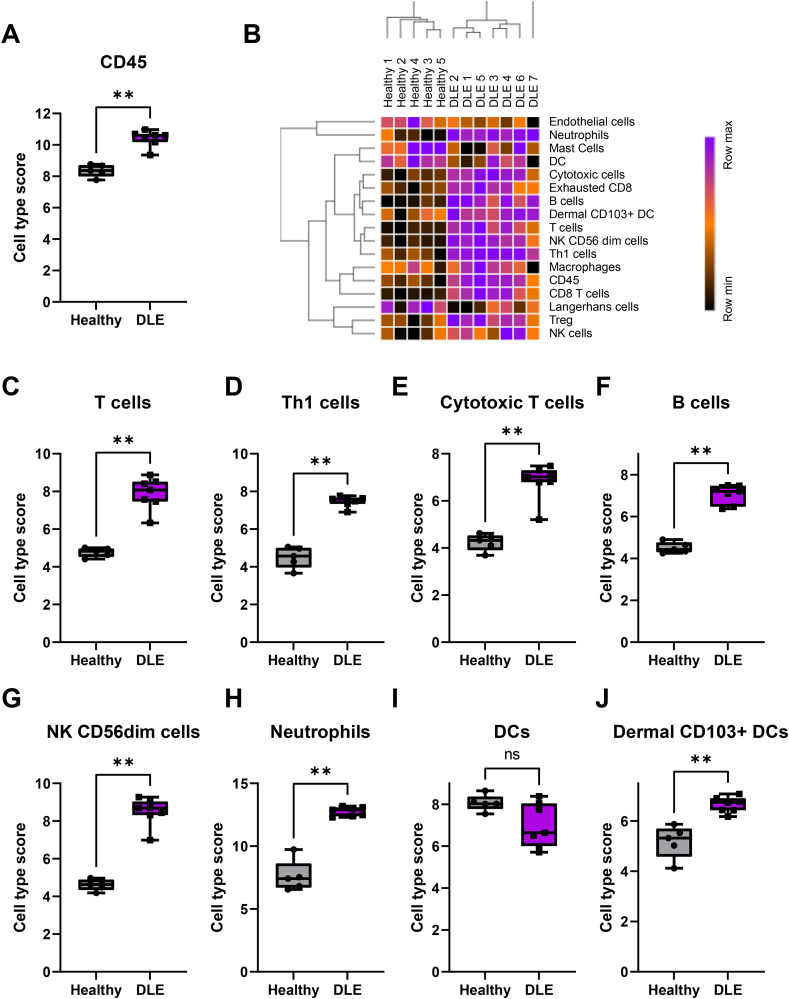
3.3. Cell type analysis reveals infiltration of activated T, B, and NK cells, neutrophils, and positive staining for IgG + plasma cells and lupus band reaction in canine DLE lesions
There was a significant influx of CD45+ immune cells in the skin of DLE dogs compared to healthy controls (Fig. 4A). We performed advanced cell type analysis to determine which cellular gene signatures were present, which is indicative of which cells have infiltrated the tissue (heatmap of all cell types in Fig. 4B). T-cell (CD3E, SH2D1A, TRAT1) (Fig. 4C), including cytotoxic T cells (CD8A, EOMES, LAG3) and Th1 cells (Tbet/TBX21) (Fig. 4D–E), and B-cell gene signatures (CD19, MS4A1, FCRL2) (Fig. 4F) were higher in DLE compared to healthy controls. Markers for activated T-cells (CD6, CD27) were present, as well as the canonical T-regulatory cell marker FOXP3. A marker for mature B-cells, TNFRSF17, was also significantly upregulated (Table S1). Upregulation of GZMA, GZMB, KLRD1, CTSW, and PRF1 suggests the presence of activated cytotoxic T-cells and natural killer (NK) cells (Fig. 4G). Two neutrophil markers, S100A12 and CEACAM1 (canine ortholog of human CEACAM3), were significantly upregulated (Fig. 4H). A potential macrophage marker, CD84, was also enriched but was just below statistical significance. CPA3, a mast cell maker, was downregulated (Table S1). Interestingly, we found a trend towards a reduction in dendritic cell score and a significant increase in dermal CD103+ dendritic cell score (Fig. 4I–J), which matches observations in the literature of a loss of Langerhans cells and increases of inflammatory dendritic cells in lupus skin (Shipman et al., 2018).
Fig. 4.
Cell type analysis reveals influx of a number of both adaptive and innate immune cell subtypes in canine DLE lesions. (A) Total CD45 infiltrate as approximated by NanoString nSolver. (B) Z-scored cell type scores from NanoString Advanced Analysis. Heatmap generated with Morpheus software. Cell type scores by disease state for (C) T-cells (p-value = 6.72 × 10-7), (D) Th1 cells (p-value = 1.09 × 10-9), (E) cytotoxic cells (p-value = 6.21 × 10-7), (F) B-cells (p-value = 3.12 × 10-6), (G) NK CD56dim cells, (H) neutrophils (p-value = 6.51 × 10-4), (I) dendritic cells and (J) dermal CD103+ dendritic cells. (n = 6 DLE and n = 5 healthy controls, student’s t tests and/or Mann Whitney U tests significant as indicated).
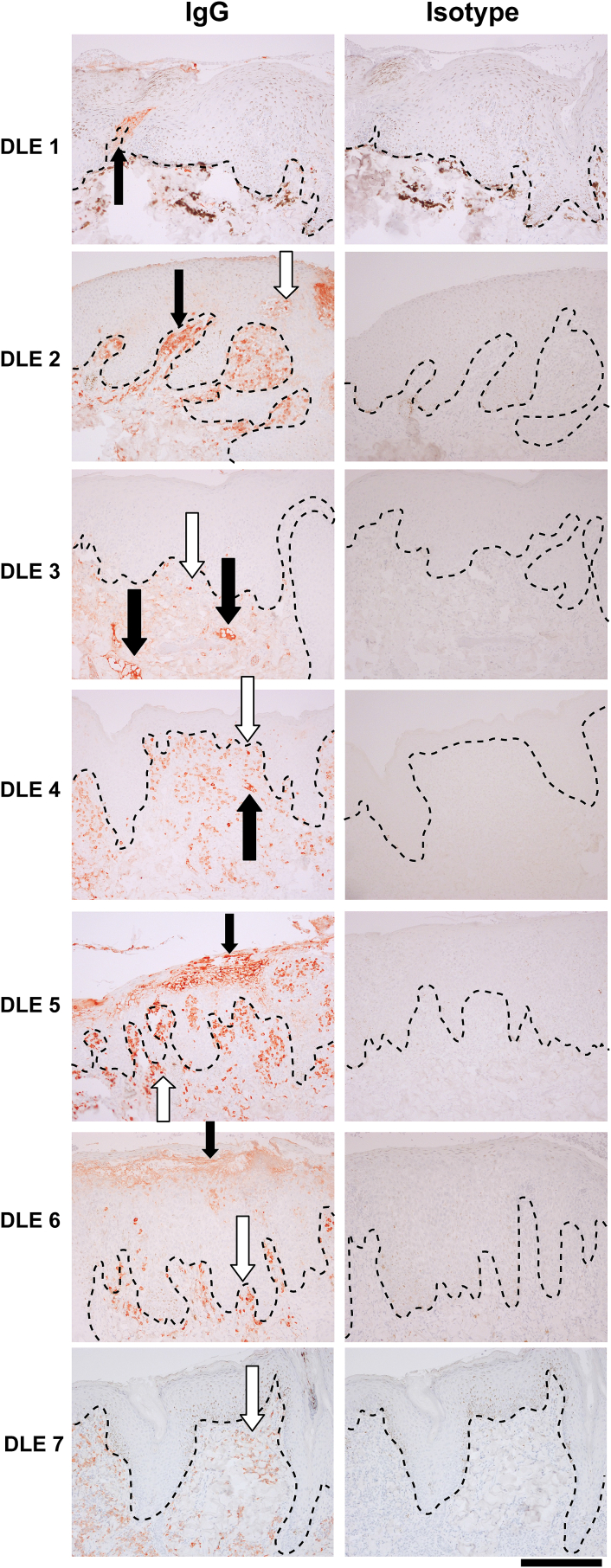
To confirm the presence of IgG in canine skin, which is a hallmark of human cutaneous lupus and a product secreted by plasma cells, we performed IgG staining. Several cases revealed lupus band reaction (IgG deposition at the DEJ), with some brightly staining cells in the superficial dermis which are consistent with plasma cells (Fig. 5).
Fig. 5.
Canine IgG staining reveals lupus band reaction and plasma cells in DLE lesions. IgG staining of canine DLE skin reveals lupus-band reaction, autoantibody deposition at the dermoepidermal junction, in several samples (black arrows). Infiltrating plasma cells are also present in several samples (white arrows; 10x, scale bar = 300 μm; dashed lines = DEJ).
3.4. Comparative gene expression analysis of canine, human and mouse DLE reveals shared inflammatory gene expression signatures
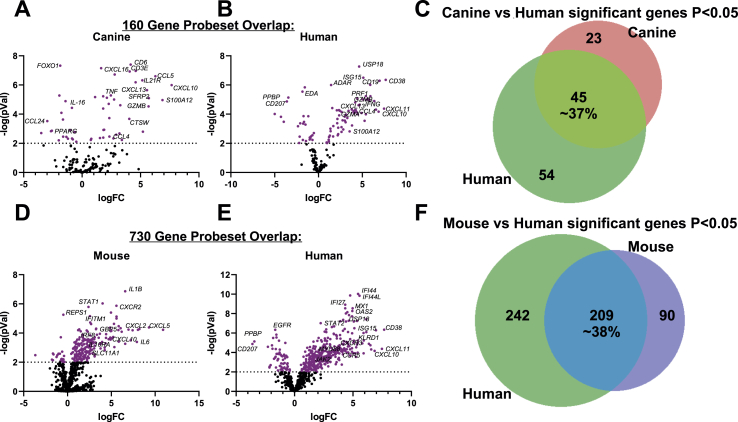
To begin to examine conserved elements of DLE immunopathogenesis, we compared our dog NanoString dataset to mouse NanoString (Mande et al., 2018) and human DLE microarray (GSE95474) (Scholtissek et al., 2017) datasets. We first truncated the human dataset to match each of the corresponding datasets for mouse and canine DLE so a common denominator was used for cross-species comparisons. We created volcano plots of each species to examine overall up and downregulated genes in each species’ dataset. Next, we compared pair-wise species expression of significant genes (examining both P < 0.05 and P < 0.01 for DLE versus healthy control). Comparison of canine and human DLE revealed approximately 37% overlap, or 45 significant genes in the 160 gene set excluding 4 housekeeping genes (B2M, RPL13A, CG14980, HPRT; Fig. 6A–C). Truncation of the human dataset to match mouse resulted in 730 out of 755 genes in the mouse NanoString cancer immune codeset (genes that were excluded were chemokines and Klr family members for which no human homologues have been identified). This resulted in approximately 38% gene overlap excluding housekeeping genes (Table S2), with human DLE uniquely expressing 242 genes and mouse uniquely expressing 90 genes (Fig. 6D–F, Table S2).
Fig. 6.
Gene expression in canine DLE, human DLE, and the TLR9KO mouse model of DLE. Volcano plots of (A) canine and (B) human DLE differential gene expression in the 160 gene set. (C) Venn diagrams comparing shared probes between canine and human that are significant DEGs for healthy versus DLE. Volcano plots of (D) mouse and (E) human DLE differential gene expression in the 730 gene set. (F) Venn diagrams comparing shared probes between human and mouse that are significant DEGs for healthy versus DLE.
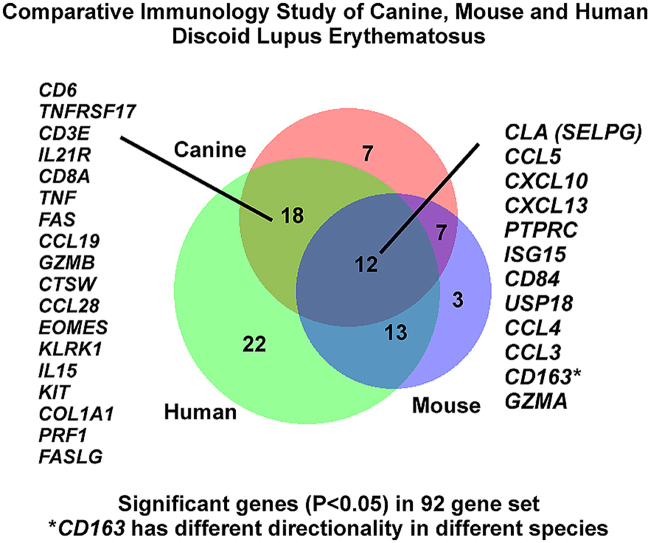
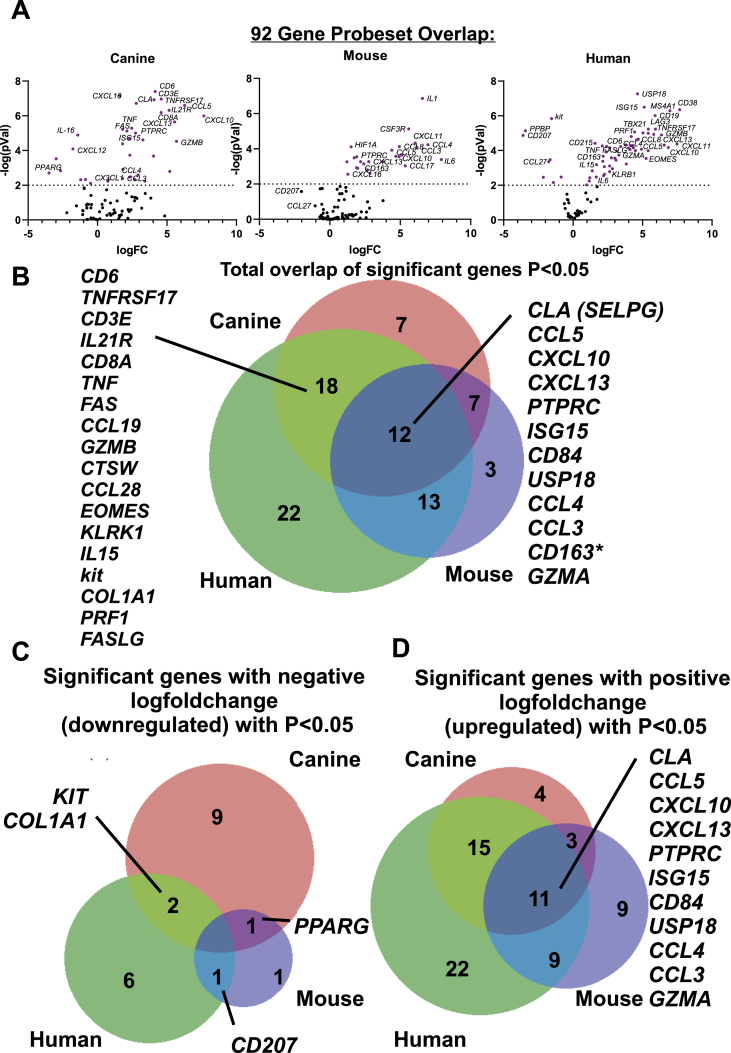
To determine total overlap of significant differentially expressed genes across all 3 species (regardless of directionality, up or down), we truncated the datasets to match the gene list across species, which resulted in a 92 gene set (Table S2). Using a P < 0.05 cutoff, we annotated which genes were significantly differentially expressed (up or down versus healthy controls) in each species and compared the gene lists using Interactive Heat Map Builder (Ryan et al., 2019) and BioVenn (Hulsen et al., 2008). We created volcano plots for all genes in a 92 gene set across canine, human, and mouse datasets (Fig. 7A). Next, we examined shared gene expression patterns across the three datasets. This analysis revealed approximately a 12 gene-overlap between all 3 species, with an 18 gene overlap between dog and human, 13 gene overlap between mouse and human, and 7 gene overlap between dog and mouse (Fig. 7B). Notably, CD163 was upregulated in human and mouse, but downregulated in canine skin.
Fig. 7.
Comparison of gene expression and directionality in DLE across canine, mouse and human datasets. (A) Volcano plots of canine, mouse and human in the 92 gene set. (B) Total overlap of significant (P < 0.05 for DLE vs healthy) genes in the 92 gene matched set across canine (red), mouse (blue) and human (green) datasets. The 12 significant overlapping genes are indicated in the figure. Analysis of significant genes with (C) negative or (D) positive logfoldchange with cutoff P < 0.05.
3.5. Examination of directionality of genes reveals that canine DLE, but not mouse, recapitulates downregulated genes observed in human DLE
We next compared the directionality of the significant genes from the 92 gene set in dog, mouse and human DLE using P < 0.05 as a cutoff. While there were no significantly downregulated genes conserved across the 3 datasets, human and dog shared significantly downregulated genes, namely KIT and COL1A1, human and mouse shared CD207 and mouse and dog shared PPARG (Fig. 7C). All 3 species shared 11 significantly upregulated genes: CLA (SELPG), CCL5, CXCL10, CXCL13, PTPRC, ISG15, CD84, USP18, CCL4, CCL3 and GZMA (Fig. 7D). Taken together, these results suggest that spontaneous DLE in canines recapitulates certain aspects of human DLE that may warrant further study.
4. Discussion
Here we characterized canine DLE gene expression with a curated set of genes related to immunopathogenesis, skin homeostasis and known drivers of cutaneous lupus. Infiltration of adaptive and innate immune cells mirrored similar patterns in canine and human DLE, specifically T, B and NK cell infiltrates (Wenzel, 2019; Garelli et al., 2020). Consistent with B cells and plasma cells in CLE, we noted IgG punctate staining in the majority of cases. It is possible that cases without an obvious band of IgG had different antibody isotypes deposited at the DEJ, such as IgM or IgA, as has been noted in other CLE histopathology studies (Crowson and Magro, 2009). Additionally, for canine FDLE, a IgG positive lupus band test has been reported in 40% of cases (Shahidullah et al., 1995). We also noted increased LAG3 and EOMES, which are indicative of exhausted CD8 T-cells, a T-cell phenotype typically associated with chronic viral infection (Yi et al., 2010; Andrews et al., 2017). These phenotypes may be related to DLE immunopathogenesis, as autoantigens are also chronically available to the immune system. CD163 was upregulated in human and mouse, but downregulated in canine skin. Therefore, we hypothesize that there may be an alternate antigen presenting cell population responsible for immunopathogenesis in canines compared to mice and humans.
A number of dysregulated cytokines and chemokines strongly suggest similar inflammatory states in canine, mouse and human DLE. The overlap of proinflammatory genes between mouse and other species indicates that preclinical mouse models may be of use for developing anti-inflammatory agents for lupus with both veterinary and human medicine applications.
Lesional skin from dogs with DLE exhibited downregulation of genes involved in skin homeostasis and immune tolerance. FOXO1, a known immunoregulatory gene which can repress TBX21-mediated effector functions to promote memory CD8+ T cell formation and Treg function (Rao et al., 2012; Ouyang et al., 2009, 2012). PPARG, which promotes T cell differentiation and survival (Wohlfert et al., 2007; Yang et al., 2002), and FOXO3, a transcription factor that is an important regulator of the magnitude of CD8 T cell memory (Sullivan et al., 2012), were decreased in our cases compared to healthy controls. These decreases may indicate reduced ability of Tregs to function to suppress autoreactive T cells in DLE lesions. WIF1, another significantly downregulated gene, is involved in skin homeostasis, and alterations in Wnt signaling have been reported in psoriasis and UV-exposed skin, and in hair cycle arrest in dogs (Brunner et al., 2017), where it may synergize with IFN production (Gudjonsson et al., 2010; Michalczyk et al., 2018; Romanowska et al., 2009). Ectodysplasin, also known as EDA, is most well known as a mediator of ectodermal appendage (e.g. nails, hair, teeth) development (Mustonen et al., 2003). In adults, EDA is involved in several processes in the skin, including sebaceous gland growth and development, hair follicle maintenance, wound healing, and barrier function (Li et al., 2018). Downregulation of EDA indicates loss of hair follicles and sebaceous glands and may cause dysregulated wound healing response. Loss of PRLR is indicative of loss of hair follicles (Foitzik et al., 2009; Langan et al., 2010), and downregulation of Wnt pathway impaires anagen onset (Brunner et al., 2017). While these differences may be attributed to hair follicle density of disease samples (nasal planum) and leg margins (densely haired skin), loss of hair follicles is a characteristic of DLE lesions and may be relevant to canine DLE pathogenesis.
Interestingly, overexpression of VGLL3 in K5+ cells in mouse skin resulted in a cutaneous lupus-like reaction (Billi et al., 2019). A previous study characterized VGLL3 as a driver of female bias in CLE. We observed that VGLL3 was in fact decreased in lesional skin, though many of the animals tested had been spayed or neutered, potentially impacting sex hormones.
Limitations in our study include the small sample of genes analyzed (160 genes), differences in sampling site (facial DLE versus healthy leg margins) and small sample size. Other limitations include scratching behavior in dogs, which may increase neutrophil recruitment to the skin (Walsh et al., 2019), though neutrophil infiltration is also observed in mouse and human CLE. Dogs have hair or fur covering their skin, which may reduce UV light exposure in some dogs, a known trigger of CLE and a potent inducer of type I IFN (Skopelja-Gardner et al., 2020). However, nasal planum involvement in dogs, which has less fur and may experience UV light, may mirror photosensitive CLE in humans. Further studies dissecting molecular and cellular events on hair-bearing and hair-spare skin are warranted across species. Despite these caveats, we observed many DEGs that match previously published literature regarding potential drivers of DLE immunopathogenesis. Larger scale comparative analyses and whole transcriptome sequencing and/or single cell RNA sequencing will ultimately allow for a better understanding of the pathogenesis of DLE across species.
5. Conclusion
There are more than 77 million dogs and up to 48% of American households have pet dogs, yet canines are rarely considered for clinical studies (How many Americans have p, 2019). Furthermore, canine companions may be exposed to similar environmental triggers for DLE as humans: for example, UV exposure and cigarette smoking are highly associated with DLE and reduces the efficacy of some treatments (Böckle and Sepp, 2015; Rahman et al., 1998). Pet dogs whose owners have SLE are significantly more likely to have circulating ANAs and to develop lupus (Chiou et al., 2004; Jones et al., 1992). Our study demonstrates that canines share similar proinflammatory features and molecular pathway signatures as mice and humans with DLE, and that dogs uniquely capture downregulated genes observed in human disease. Future studies examining interactions of genes and environment on DLE immunopathogenesis are warranted and may pave the way for drug repurposing in human and veterinary medicine. For example, JAK inhibitors have been reported to be efficacious in case studies of cutaneous lupus in humans (Klaeschen et al., 2017; Wenzel et al., 2016), in some dogs [unpublished personal communication RMA], and in preclinical studies in mice (Furumoto et al., 2017; Chan et al., 2015). Oclacitinib is a veterinary JAK inhibitor currently marketed for allergy and itch relief (Gonzales et al., 2014; Cosgrove et al., 2013a, 2013b; Little et al., 2015) and has been reported efficacious in the treatment of other interface dermatitis diseases (High et al., 2020; Levy et al., 2019). Thus, it would be interesting to perform studies to explore oclacitinib as a potential treatment for CLE in dogs.
Data availability statement
The canine dataset presented in this study can be found on Gene Expression Omnibus (GEO) Database under Accession # GSE160260.
Author contributions
Conceptualization: JMR. Methodology: NAR, JMR. Software: CND, CJG. Validation: CP-M, RMA, JMR. Formal analysis: CJG, NBW, JMR. Investigation: CJG, CP-M, LMW, GS, NAR, JMR, CND. Resources: NAR, JMR. Data curation: NAR, CP-M, RMA, CJG, JMR. Writing—original draft: CJG, JMR. Writing—review and editing: all authors. Visualization: CJG, CP-M, NBW, JMR. Supervision: JMR, NAR. Project administration: JMR. Funding acquisition: JMR. All authors contributed to the article and approved the submitted version.
Funding
JMR is supported by a Women’s Health Career Development Award from the Dermatology Foundation; and a Target Identification in Lupus Award from the Lupus Research Alliance. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: JMR is an inventor on patent application #15/851,651, “Anti-human CXCR3 antibodies for the Treatment of Vitiligo” which covers targeting CXCR3 for the treatment of vitiligo; and on patent #62489191, “Diagnosis and Treatment of Vitiligo” which covers targeting IL-15 and Trm for the treatment of vitiligo. NAR and GS are employees of bluebird bio. CND is an employee of NanoString Technologies.
Acknowledgments
We thank Yu Liu from UMass for technical assistance. We thank Ann Rothstein for sharing the raw counts from the published mouse NanoString dataset, and Shruti Sharma and Lindsey Bierfeldt for insightful discussions during curation of the canine lupus gene probe set. We thank the Finberg lab at UMass for use of their microscope. Immunohistochemistry studies were performed in collaboration with the UMass DERC morphology core. The NanoString Sprint machine is maintained by the Silverman lab in the UMass Department of Medicine.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crimmu.2021.03.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Andrews L.P., Marciscano A.E., Drake C.G., Vignali D.A.A. LAG3 (CD223) as a cancer immunotherapy target. Immunol. Rev. 2017;276:80–96. doi: 10.1111/imr.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banovic F., Linder K.E., Uri M., Rossi M.A., Olivry T. Clinical and microscopic features of generalized discoid lupus erythematosus in dogs (10 cases) Vet. Dermatol. 2016;27 doi: 10.1111/vde.12389. 488–e131. [DOI] [PubMed] [Google Scholar]
- Berthier C.C., Tsoi L.C., Reed T.J., Stannard J.N., Myers E.M., Namas R., Xing X., Lazar S., Lowe L., Kretzler M., et al. Molecular profiling of cutaneous lupus lesions identifies subgroups distinct from clinical phenotypes. J. Clin. Med. Res. 2019;8 doi: 10.3390/jcm8081244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi A.C., Gharaee-Kermani M., Fullmer J., Tsoi L.C., Hill B.D., Gruszka D., Ludwig J., Xing X., Estadt S., Wolf S.J., et al. The female-biased factor VGLL3 drives cutaneous and systemic autoimmunity. JCI Insight. 2019;4 doi: 10.1172/jci.insight.127291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böckle B.C., Sepp N.T. Smoking is highly associated with discoid lupus erythematosus and lupus erythematosus tumidus: analysis of 405 patients. Lupus. 2015;24:669–674. doi: 10.1177/0961203314559630. [DOI] [PubMed] [Google Scholar]
- Brunner M.A.T., Jagannathan V., Waluk D.P., Roosje P., Linek M., Panakova L., Leeb T., Wiener D.J., Welle M.M. Novel insights into the pathways regulating the canine hair cycle and their deregulation in alopecia X. PloS One. 2017;12 doi: 10.1371/journal.pone.0186469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E.S., Herlitz L.C., Jabbari A. Ruxolitinib attenuates cutaneous lupus development in a mouse lupus model. J. Invest. Dermatol. 2015;135:1912–1915. doi: 10.1038/jid.2015.107. [DOI] [PubMed] [Google Scholar]
- Chiou S.-H., Lan J.-L., Lin S.-L., Chen D.-Y., Tsai N.-Y., Kuan C.-Y., Lin T.-Y., Lin F.-J., Lee W.-M., Chang T.-J. Pet dogs owned by lupus patients are at a higher risk of developing lupus. Lupus. 2004;13:442–449. doi: 10.1191/0961203303lu1039oa. [DOI] [PubMed] [Google Scholar]
- Cosgrove S.B., Wren J.A., Cleaver D.M., Martin D.D., Walsh K.F., Harfst J.A., Follis S.L., King V.L., Boucher J.F., Stegemann M.R. Efficacy and safety of oclacitinib for the control of pruritus and associated skin lesions in dogs with canine allergic dermatitis. Vet. Dermatol. 2013;24 doi: 10.1111/vde.12047. 479–e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove S.B., Wren J.A., Cleaver D.M., Walsh K.F., Follis S.I., King V.I., Tena J.-K.S., Stegemann M.R. A blinded, randomized, placebo-controlled trial of the efficacy and safety of the J anus kinase inhibitor oclacitinib (A poquel®) in client-owned dogs with atopic dermatitis. Vet. Dermatol. 2013;24 doi: 10.1111/vde.12088. 587–e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowson A.N., Magro C.M. Cutaneous histopathology of lupus erythematosus. Diagn. Histopathol. 2009;15:157–185. [Google Scholar]
- Danaher P., Warren S., Dennis L., D’Amico L., White A., Disis M.L., Geller M.A., Odunsi K., Beechem J., Fling S.P. Gene expression markers of tumor infiltrating leukocytes. J Immunother Cancer. 2017;5:18. doi: 10.1186/s40425-017-0215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durosaro O., Davis M.D.P., Reed K.B., Rohlinger A.L. Incidence of cutaneous lupus erythematosus, 1965-2005: a population-based study. Arch. Dermatol. 2009;145:249–253. doi: 10.1001/archdermatol.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrigno A., Hoover K., Blubaugh A., Rissi D., Banovic F. Treatment of exfoliative cutaneous lupus erythematosus in a German shorthaired pointer dog with mycophenolate mofetil. Vet. Dermatol. 2019 doi: 10.1111/vde.12747. 0. [DOI] [PubMed] [Google Scholar]
- Foitzik K., Langan E.A., Paus R. Prolactin and the skin: a dermatological perspective on an ancient pleiotropic peptide hormone. J. Invest. Dermatol. 2009;129:1071–1087. doi: 10.1038/jid.2008.348. [DOI] [PubMed] [Google Scholar]
- Freudenberg J.M., Olivry T., Mayhew D.N., Rubenstein D.S., Rajpal D.K. The comparison of skin transcriptomes confirms canine atopic dermatitis is a natural homologue to the human disease. J Invest Dermatol 04/ 2019;139:968–971. doi: 10.1016/j.jid.2018.10.018. [DOI] [PubMed] [Google Scholar]
- Furumoto Y., Smith C.K., Blanco L., Zhao W., Brooks S.R., Thacker S.G., Zarzour A., Sciumè G., Tsai W.L., Trier A.M., et al. Tofacitinib ameliorates murine lupus and its associated vascular dysfunction. Arthritis & Rheumatology. 2017;69:148–160. doi: 10.1002/art.39818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garelli C.J., Refat M.A., Nanaware P.P., Ramirez-Ortiz Z.G., Rashighi M., Richmond J.M. Current insights in cutaneous lupus erythematosus immunopathogenesis. Front. Immunol. 2020;11:1353. doi: 10.3389/fimmu.2020.01353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George P.M., Tunnessen W.W., Jr. Childhood discoid lupus erythematosus. Arch. Dermatol. 1993;129:613–617. [PubMed] [Google Scholar]
- Gonzales A.J., Bowman J.W., Fici G.J., Zhang M., Mann D.W., Mitton-Fry M. Oclacitinib (APOQUEL®) is a novel Janus kinase inhibitor with activity against cytokines involved in allergy. J. Vet. Pharmacol. Therapeut. 2014;37:317–324. doi: 10.1111/jvp.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudjonsson J.E., Johnston A., Stoll S.W., Riblett M.B., Xing X., Kochkodan J.J., Ding J., Nair R.P., Aphale A., Voorhees J.J., et al. Evidence for altered Wnt signaling in psoriatic skin. J. Invest. Dermatol. 2010;130:1849–1859. doi: 10.1038/jid.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejazi E.Z., Werth V.P. Cutaneous lupus erythematosus: an update on pathogenesis, diagnosis and treatment. Am. J. Clin. Dermatol. 2016;17:135–146. doi: 10.1007/s40257-016-0173-9. [DOI] [PubMed] [Google Scholar]
- High E.J., Linder K.E., Mamo L.B., Levy B.J., Herrmann I., Bizikova P. Rapid response of hyperkeratotic erythema multiforme to oclacitinib in two dogs. Vet. Dermatol. 2020;31 doi: 10.1111/vde.12852. 330–e86. [DOI] [PubMed] [Google Scholar]
- How Many Americans Have Pets? an Investigation of Fuzzy Statistics. The Washington Post; 2019. [Google Scholar]
- Hulsen T., de Vlieg J., Alkema W. BioVenn--a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genom. 2008;9:1–6. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarukitsopa S., Hoganson D.D., Crowson C.S., Sokumbi O., Davis M.D., Michet C.J., Jr., Matteson E.L., Maradit Kremers H., Chowdhary V.R. Epidemiology of systemic lupus erythematosus and cutaneous lupus erythematosus in a predominantly white population in the United States. Arthritis Care Res. 2015;67:817–828. doi: 10.1002/acr.22502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.R.E., Hopkinson N.D., Powell R.J. Autoantibodies in pet dogs owned by patients with systemic lupus erythematosus. Lancet. 1992;339:1378–1380. doi: 10.1016/0140-6736(92)91197-g. [DOI] [PubMed] [Google Scholar]
- Khanna C., Lindblad-Toh K., Vail D., London C., Bergman P., Barber L., Breen M., Kitchell B., McNeil E., Modiano J.F., et al. The dog as a cancer model. Nat. Biotechnol. 2006;24:1065–1066. doi: 10.1038/nbt0906-1065b. [DOI] [PubMed] [Google Scholar]
- Klaeschen A.S., Wolf D., Brossart P., Bieber T., Wenzel J. JAK inhibitor ruxolitinib inhibits the expression of cytokines characteristic of cutaneous lupus erythematosus. Exp. Dermatol. 2017;26:728–730. doi: 10.1111/exd.13253. [DOI] [PubMed] [Google Scholar]
- Langan E.A., Foitzik-Lau K., Goffin V., Ramot Y., Paus R. Prolactin: an emerging force along the cutaneous-endocrine axis. Trends Endocrinol. Metabol. 2010;21:569–577. doi: 10.1016/j.tem.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Lee K.-H., Park H.-M., Son K.-H., Shin T.-J., Cho J.-Y. Transcriptome signatures of canine mammary gland tumors and its comparison to human breast cancers. Cancers. 2018;10:317. doi: 10.3390/cancers10090317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy B.J., Linder K.E., Olivry T. The role of oclacitinib in the management of ischaemic dermatopathy in four dogs. Vet. Dermatol. 2019;30 doi: 10.1111/vde.12743. 201–e63. [DOI] [PubMed] [Google Scholar]
- Li S., Zhou J., Zhang L., Li J., Yu J., Ning K., Qu Y., He H., Chen Y., Reinach P.S., et al. Ectodysplasin A regulates epithelial barrier function through sonic hedgehog signalling pathway. J. Cell Mol. Med. 2018;22:230–240. doi: 10.1111/jcmm.13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little P.R., King V.L., Davis K.R., Cosgrove S.B., Stegemann M.R. A blinded, randomized clinical trial comparing the efficacy and safety of oclacitinib and ciclosporin for the control of atopic dermatitis in client-owned dogs. Vet. Dermatol. 2015;26 doi: 10.1111/vde.12186. 23–e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mande P., Zirak B., Ko W.-C., Taravati K., Bride K.L., Brodeur T.Y., Deng A., Dresser K., Jiang Z., Ettinger R., et al. Fas ligand promotes an inducible TLR-dependent model of cutaneous lupus-like inflammation. J. Clin. Invest. 2018;128:2966–2978. doi: 10.1172/JCI98219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merola J.F., Prystowsky S.D., Iversen C., Gomez-Puerta J.A., Norton T., Tsao P., Massarotti E., Schur P., Bermas B., Costenbader K.H. Association of discoid lupus erythematosus with other clinical manifestations among patients with systemic lupus erythematosus. J. Am. Acad. Dermatol. 2013;69:19–24. doi: 10.1016/j.jaad.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsalu T., Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalczyk T., Biedermann T., Böttcher-Haberzeth S., Klar A.S., Meuli M., Reichmann E. UVB exposure of a humanized skin model reveals unexpected dynamic of keratinocyte proliferation and Wnt inhibitor balancing. J Tissue Eng Regen Med. 2018;12:505–515. doi: 10.1002/term.2519. [DOI] [PubMed] [Google Scholar]
- Mustonen T., Pispa J., Mikkola M.L., Pummila M., Kangas A.T., Pakkasjärvi L., Jaatinen R., Thesleff I. Stimulation of ectodermal organ development by Ectodysplasin-A1. Dev. Biol. 2003;259:123–136. doi: 10.1016/s0012-1606(03)00157-x. [DOI] [PubMed] [Google Scholar]
- Nakajima M., Nakajima A., Kayagaki N., Honda M., Yagita H., Okumura K. Expression of Fas ligand and its receptor in cutaneous lupus: implication in tissue injury. Clin. Immunol. Immunopathol. 1997;83:223–229. doi: 10.1006/clin.1997.4352. [DOI] [PubMed] [Google Scholar]
- Olivry T., Linder K.E., Banovic F. Cutaneous lupus erythematosus in dogs: a comprehensive review. BMC Vet. Res. 2018;14 doi: 10.1186/s12917-018-1446-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W., Beckett O., Flavell R.A., Li M.O. An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity. 2009;30:358–371. doi: 10.1016/j.immuni.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W., Liao W., Luo C.T., Yin N., Huse M., Kim M.V., Peng M., Chan P., Ma Q., Mo Y., et al. Novel Foxo1-dependent transcriptional programs control T reg cell function. Nature. 2012;491:554–559. doi: 10.1038/nature11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman P., Gladman D.D., Urowitz M.B. Smoking interferes with efficacy of antimalarial therapy in cutaneous lupus. J. Rheumatol. 1998;25:1716–1719. [PubMed] [Google Scholar]
- Rao R.R., Li Q., Bupp M.R.G., Shrikant P.A. Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8+ T cell differentiation. Immunity. 2012;36:374–387. doi: 10.1016/j.immuni.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowska M., Evans A., Kellock D., Bray S.E., McLean K., Donandt S., Foerster J. Wnt5a exhibits layer-specific expression in adult skin, is upregulated in psoriasis, and synergizes with type 1 interferon. PloS One. 2009;4:e5354. doi: 10.1371/journal.pone.0005354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield N., Sontheimer R.D., Bernstein M. Lupus erythematosus: systemic and cutaneous manifestations. Clin. Dermatol. 2006;24:348–362. doi: 10.1016/j.clindermatol.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Ryan M.C., Stucky M., Wakefield C., Melott J.M., Akbani R., Weinstein J.N., Broom B.M. vol. 8. 2019. p. F1000Res. (Interactive Clustered Heat Map Builder: an Easy Web-Based Tool for Creating Sophisticated Clustered Heat Maps). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtissek B., Zahn S., Maier J., Klaeschen S., Braegelmann C., Hoelzel M., Bieber T., Barchet W., Wenzel J. Immunostimulatory endogenous nucleic acids drive the lesional inflammation in cutaneous lupus erythematosus. J. Invest. Dermatol. 2017;137:1484–1492. doi: 10.1016/j.jid.2017.03.018. [DOI] [PubMed] [Google Scholar]
- Shahidullah M., Lee Y.S., Khor C.J., Ratnam K.V. Chronic discoid lupus erythematosus: an immunopathological and electron microscopic study. Ann. Acad. Med. Singapore. 1995;24:789–792. [PubMed] [Google Scholar]
- Shipman W.D., Chyou S., Ramanathan A., Izmirly P.M., Sharma S., Pannellini T., Dasoveanu D.C., Qing X., Magro C.M., Granstein R.D., et al. A protective Langerhans cell-keratinocyte axis that is dysfunctional in photosensitivity. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aap9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopelja-Gardner S., An J., Tai J., Tanaka L., Sun X., Hermanson P., Baum R., Kawasumi M., Green R., Gale M., Jr., et al. The early local and systemic Type I interferon responses to ultraviolet B light exposure are cGAS dependent. Sci. Rep. 2020;10:7908. doi: 10.1038/s41598-020-64865-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J.A., Kim E.H., Plisch E.H., Peng S.L., Suresh M. FOXO3 regulates CD8 T cell memory by T cell-intrinsic mechanisms. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todhunter R.J., Garrison S.J., Jordan J., Hunter L., Castelhano M.G., Ash K., Meyers-Wallen V., Krotscheck U., Hayward J.J., Grenier J. Gene expression in hip soft tissues in incipient canine hip dysplasia and osteoarthritis. J. Orthop. Res. 2019;37:313–324. doi: 10.1002/jor.24178. [DOI] [PubMed] [Google Scholar]
- Veldman-Jones M.H., Brant R., Rooney C., Geh C., Emery H., Harbron C.G., Wappett M., Sharpe A., Dymond M., Barrett J.C., et al. Evaluating robustness and sensitivity of the NanoString Technologies nCounter platform to enable multiplexed gene expression analysis of clinical samples. Canc. Res. 2015;75:2587–2593. doi: 10.1158/0008-5472.CAN-15-0262. [DOI] [PubMed] [Google Scholar]
- Verdelli A., Coi A., Marzano A.V., Antiga E., Cozzani E., Quaglino P., La Placa M., Benucci M., De Simone C., Papini M., et al. Autoantibody profile and clinical patterns in 619 Italian patients with cutaneous lupus erythematosus. J. Eur. Acad. Dermatol. Venereol. 2019;33:742–752. doi: 10.1111/jdv.15147. [DOI] [PubMed] [Google Scholar]
- Walsh C.M., Hill R.Z., Schwendinger-Schreck J., Deguine J., Brock E.C., Kucirek N., Rifi Z., Wei J., Gronert K., Brem R.B., et al. Neutrophils promote CXCR3-dependent itch in the development of atopic dermatitis. Elife. 2019;8 doi: 10.7554/eLife.48448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigand D.A. Lupus band test: anatomic regional variations in discoid lupus erythematosus. J. Am. Acad. Dermatol. 1986;14:426–428. doi: 10.1016/s0190-9622(86)70052-2. [DOI] [PubMed] [Google Scholar]
- Wenzel J. Cutaneous lupus erythematosus: new insights into pathogenesis and therapeutic strategies. Nat. Rev. Rheumatol. 2019;15:519–532. doi: 10.1038/s41584-019-0272-0. [DOI] [PubMed] [Google Scholar]
- Wenzel J., van Holt N., Maier J., Vonnahme M., Bieber T., Wolf D. JAK1/2 inhibitor ruxolitinib controls a case of chilblain lupus erythematosus. J. Invest. Dermatol. 2016;136:1281–1283. doi: 10.1016/j.jid.2016.02.015. [DOI] [PubMed] [Google Scholar]
- Wohlfert E.A., Nichols F.C., Nevius E., Clark R.B. Peroxisome proliferator-activated receptor γ (PPARγ) and immunoregulation: enhancement of regulatory T cells through PPARγ-dependent and-independent mechanisms. J. Immunol. 2007;178:4129–4135. doi: 10.4049/jimmunol.178.7.4129. [DOI] [PubMed] [Google Scholar]
- Wu J., Wilson J., He J., Xiang L., Schur P.H., Mountz J.D. Fas ligand mutation in a patient with systemic lupus erythematosus and lymphoproliferative disease. J. Clin. Invest. 1996;98:1107–1113. doi: 10.1172/JCI118892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.Y., Wang L.H., Mihalic K., Xiao W., Chen T., Li P., Wahl L.M., Farrar W.L. Interleukin (IL)-4 indirectly suppresses IL-2 production by human T lymphocytes via peroxisome proliferator-activated receptor γ activated by macrophage-derived 12/15-lipoxygenase ligands. J. Biol. Chem. 2002;277:3973–3978. doi: 10.1074/jbc.M105619200. [DOI] [PubMed] [Google Scholar]
- Yi J.S., Cox M.A., Zajac A.J. T-cell exhaustion: characteristics, causes and conversion. Immunology. 2010;129:474–481. doi: 10.1111/j.1365-2567.2010.03255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Chang C., Zhang J. Immunologic and genetic considerations of cutaneous lupus erythematosus: a comprehensive review. J. Autoimmun. 2013;41:34–45. doi: 10.1016/j.jaut.2013.01.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The canine dataset presented in this study can be found on Gene Expression Omnibus (GEO) Database under Accession # GSE160260.