Abstract
Recently, we showed how an early restriction of gut flora proliferation by Plasmodium vivax favors immune-suppression and Plasmodium survival in the gut lumen (Sharma et al., 2020). Here, we asked post gut invasion how P. vivax interacts with individual tissues such as the midgut, hemocyte, and salivary glands, and manages its survival in the mosquito host. Our data from tissue-specific comparative RNA-Seq analysis and extensive temporal/spatial expression profiling of selected mosquito transcripts in the uninfected and P. vivax infected mosquito’s tissues indicated that (i) a transient suppression of gut metabolic machinery by early oocysts; (ii) enriched expression of nutritional responsive proteins and immune proteins against late oocysts, together may ensure optimal parasite development and gut homeostasis restoration; (iii) pre-immune activation of hemocyte by early gut-oocysts infection via REL induction (p < 0.003); and altered expression of hemocyte-encoded immune proteins may cause rapid removal of free circulating sporozoites from hemolymph; (iv) while a strong suppression of salivary metabolic activities, and elevated expression of salivary specific secretory, as well as immune proteins together, may favor the long-term storage and survival of invaded sporozoites. Finally, our RNA-Seq-based discovery of 4449 transcripts of Plasmodium vivax origin, and their developmental stage-specific expression modulation in the corresponding infected mosquito tissues, predicts a possible mechanism of mosquito responses evasion by P. vivax. Conclusively, our system-wide RNA-Seq analysis provides the first genetic evidence of direct mosquito-Plasmodium interaction and establishes a functional correlation.
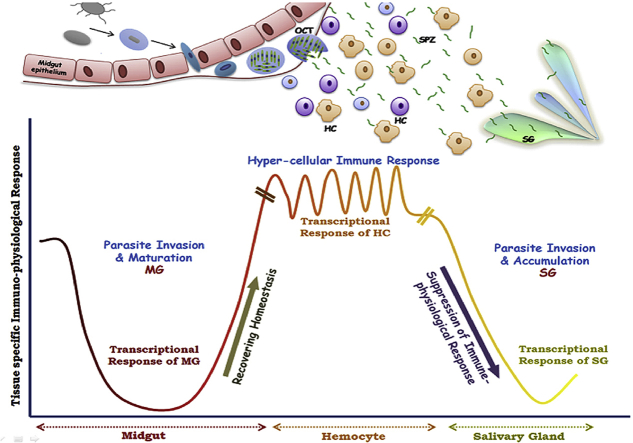
Graphical abstract
Tissue specific molecular interactions during complete development of P. vivax in the mosquito host: As the Plasmodium parasites enter the midgut (MG) through infected blood meal, it undergo fertilization and form zygotes, which further transform into ookinetes. These ookinetes then invade the midgut epithelium and reside beneath the epithelium, where they form oocysts (OCT) and get matured. During this period midgut metabolic machinery remains suppressed to fulfill parasites’ nutritional demand and as the parasite leaves the basal lamina in the form of free circulatory sporozoites (SPZ), MG starts regaining its homeostasis. After sporozoites get perfused in mosquito hemolymph they have to interact directly with hemocytes (HC) which shows hyper cellular immune response to delimit parasite population. Survived sporozoites proceed to invade the salivary glands and accumulate there for further transmission to next host, and during this period salivary gland transcriptional response remain suppressed. Ability of Plasmodium vivax to alter its gene expression profile during developmental transitions may help to evade tissue specific mosquito response, favoring successful survival and transmission.
Highlights
-
•
System-wide RNASeq analysis discloses direct mosquito-Plasmodium vivax interaction and establishes a functional correlation.
-
•
Discovery of 4449 transcripts specific to P. vivax developmental stages sheds light on possible immune evasion mechanisms.
-
•
Upregulation of nutritional related transcripts ensures optimal late oocyst development and midgut homeostasis.
-
•
Hemocyte activation by early oocysts ensure removal of free circulatory sporozoites by proliferating immune transcripts.
-
•
Heightened levels of salivary immune transcripts show that an active local immune response restricts salivary invaded sporozoite
1. Introduction
The evolution and adaptation of adult female mosquitoes to blood-feeding have a significant influence on their reproductive outcome and disease transmission dynamics. The sexual cycle of malaria-causing Plasmodium immediately begins after the ingestion of gametocytes containing blood meal by anopheline mosquitoes (Bennink et al., 2016; Kuehn and Pradel, 2010; Talman et al., 2004). Within the lumen of the midgut, male and female gametes fuse to form a zygote, which then transforms into motile ookinete (Aly et al., 2009; Bennink et al., 2016). Eventually, within 24h ookinetes traverse through the gut epithelium to reach the basal lamina either through intracellular and/or intercellular routes and transform into tiny oocysts (Baton and Ranford-Cartwright, 2004; Han et al., 2000). During this phase, mosquitoes impart early defense responses through nitration of the midgut and activation of signaling pathways (Ramphul et al., 2015). Midgut nitration results in modification of ookinete surfaces, which makes them perceptible to mosquito complement-like system (Garver et al., 2013; Shiao et al., 2006). Whereas signaling pathways show different responses towards different Plasmodium species, such as the IMD pathway, work more effectively against P. falciparum than P. berghei, and the Toll pathway is more responsive to P. berghei, and P. gallinaceum (Kumar et al., 2004; Cirimotich et al., 2010). Active pathways boost the rapid production of immune peptides i.e. AMPs (Cecropin, gambicin, etc.), which are responsible for parasite killing.
The majority of ookinetes killing is targeted at the basal side of midgut epithelium, through an indirect hemocyte-mediated cellular response such as lysis or melanization (Smith et al., 2016; Belachew, 2018). A recent study showed that during the immune active complement system, the hemocyte responds through microvesicles. (Castillo et al., 2017). Once they reached the basal lamina beneath the epithelium and enveloped by basal lamina, the surviving ookinetes rapidly transform into oocysts. These hidden young oocysts gradually mature to round shaped large oocysts and remain protected from any external response until they burst up into millions of sigmoid-shaped sporozoites. The underlying mechanism of how maturing oocysts (i) are least affected by any external responses such as immune responses caused by hemocytes; (ii) regulates their time-dependent developmental transformation to mature oocysts; (iii) trigger sporozoites release outside the basal lamina by rupturing the oocysts, is not well known.
Once released, millions of free circulatory sporozoites (fcSpz) are rapidly killed by the immune blood cells i.e. hemocytes. Despite a substantial reduction in the population of sporozoites (~85 fold) (Gouagna et al., 1998), a significant number of parasites succeed to specifically invade the salivary glands and survive within them. Several hemocyte-mediated mechanisms such as melanization, lysis, engulfment, and toxic anti-Plamsodium immune effector molecules have been proposed to kill the free circulating sporozoites, but the molecular nature of these interactions is unknown (De et al., 2018; Juli Hillyer et al., 2003; Smith et al., 2016). Even for salivary invasion, Plasmodium needs salivary gland proteins such as saglin, MAEBL, and PCRMP (Kariu et al., 2002). A recent study also showed that Plasmodium invasion induces several salivary immune proteins such as SP14D1, Ficolin, SP24D, PGRP, and LRIM (Roth et al., 2018). However, the genetic basis of how sporozoites (i) manage to avoid hemocyte-mediated killing responses, (ii) guide its movements for salivary invasion, and (iii) enhances their virulence in the salivary environment is not well understood (Mueller et al., 2010; Roth et al., 2018; Vanderberg et al., 1975; Mikolajczak et al., 2008; Sato et al., 2014).
In our preceding study (see Sharma et al., 2020) we demonstrated that in the gut lumen, Plasmodium vivax follows a unique strategy of immunosuppression by disabling gut flora proliferation. Here, we further decode and establish a functional correlation of how the post-gut invasion of P. vivax tempers the tissue-specific molecular responses. Using a comprehensive RNA-Seq analysis of P. vivax-infected midgut, hemocyte, and salivary glands, we not only identified mosquito transcripts but also captured a large pool of P. vivax transcripts expressed in respective tissues. Detailed annotation and transcriptional profiling revealed that P. vivax can avoid immuno-physiological responses by altering its genetic architecture. Manipulating tissue-specific immuno-physiology of mosquitoes may limit the Plasmodium development and hence the transmission.
2. Materials and methods
Technical overview and workflow of the project are presented in Supplementary Material Fig. S1 document (Data Sheet-1).
2.1. Mosquito rearing
The cyclic colony of the mosquito An. stephensi was maintained at 28 °C ± 2, RH = 80% in the insectary fitted with a simulated dawn and dusk machine with a 12 h Light/Dark cyclic period at NIMR. For egg maturation and regular gonotrophic cycle maintenance, mosquitoes were fed on rabbit blood. All protocols for rearing, maintenance of the mosquito culture were approved by the ethical committee of the institute (NIMR/IAEC/2017-1/07).
P. vivax infection and tissue collection: As described in our preceding study, essentially we followed the same artificial membrane feeding assay (AMFA) protocol to infect mosquitoes with P. vivax (Sharma et al., 2020). Briefly, venous blood was drawn either from P. vivax infected patient or healthy volunteers, into heparin containing tubes and kept at 37 °C till feeding. For infection studies, all protocols were duly approved by the Ethics committee of NIMR, Delhi (ECR/NIMR/EC/2012/41). An equal number of age-matched overnight starved 4–5 days old female A. stephensi mosquitoes were fed on uninfected or P. vivax infected blood samples, using-optimized artificial membrane feeding assay (AMFA). Fully engorged mosquitoes were maintained at optimal insectarium conditions and positive infection was confirmed by standard mercurochrome staining of gut oocysts readily observed under a compound microscope (Musiime et al., 2019). Post confirmation of P. vivax infection, ~20–25 mosquitoes were dissected at 3–6days post-infection (DPI) and 8–10 DPI for midgut, 9–12 DPI for hemocytes, and 12–14 DPI for salivary glands in PBS and collected in Trizol reagent.
Tissue-specific RNA-Seq library preparation & sequencing: Total RNA was isolated from each dissected tissue sample using the standard trizol method. Due to technical limitation, we followed a well-established and in-house optimized sample pool strategy, with the add-in of a SMART- PCR based mRNA pre-amplification protocol available with Clontech SMART™ kit (Dixit et al., 2009, 2011, 2015a; Palomares et al., 2019). Approximately, ~ 1 μg of purified total RNA, was subjected to construct a full-length double-stranded cDNA library. Chances of over-amplification and optimal normalization were achieved by PCR cycle numbers optimization for individual samples, followed by validation through Real-time PCR strategy (see Data sheet1). Quality of the ds-cDNA was checked on Bioanalyzer 2100 using Agilent DNA HS chip Sequencing of whole transcriptomes was performed on Illumina NextSeq, where a constant amount of 200 ng of each ds cDNA samples was subjected to Covaris shearing followed by end-repair of overhangs resulting from shearing. The paired-end cDNA sequencing libraries were prepared using Illumina TruSeq Nano DNA HT Library Preparation Kit as per the described protocol. The end-repair fragments were A-tailed, adapter-ligated, and then enriched by a limited number of PCR cycles. Library quantification and qualification was performed using DNA High Sensitivity Assay Kit and sequenced using 2 × 150 PE chemistry on NextSeq for generating ~1 GB of data per sample.
RNASeq database generation and assembly: Following sequencing raw data were filtered using Trimmomatic v0.30. All the reads with adaptor contamination and reads with low-quality value i.e. an average QV less than 20 (QV < 20) were filtered. Post filtration high-quality clean reads of all tissue-specific samples were assembled with Trinity software (release r2013-02-25) on optimized parameters. Due to poor annotation of mosquito An. stephensi reference genome, we followed the De-Novo strategy to assemble individual tissue-specific transcriptome databases. CD-HIT-EST (Version 4.6) was used to remove the shorter redundant transcripts when they were entirely covered by other transcripts with 90% identity. From assembled contigs/transcripts, CDS were predicted from the longest reading frame using the Trans-decoder. Subsequently, the predicted CDS were annotated as per the technical plan presented in Fig. S1 (see Data sheet1). To validate the quality of assembly, we randomly selected at least five transcripts of sizes ranging from >1000bp (large), 500-1000bp (medium), and 300-500bp (short) from each RNAseq database, and manually performed homology using BLASTX search analysis against both NCBI/NR database and BLASTn as well as mosquito specific Genome/transcripts databases, as described earlier (Sharma et al., 2015a). A maximum BLASTX homology (>80% identity BLASTn homology (>99% identity with full coverage), and global BLASTX based species distribution analysis showing the dominant match with anopheline mosquito species, together provide enough confidence to accept and process the denovo assembly database for functional annotation (see Data sheet1).
GO annotation and molecular cataloging: Post database assembly, the predicted CDS FASTA file of individual tissues of P. vivax infected or uninfected control samples, were processed for functional prediction analyses and molecular cataloging through using Blast2GO PRO 3.0 software (Conesa et al., 2005). A comprehensive functional annotation included BLASTx analysis against the NCBI/NR database as well as Gene Ontology databases. Basic BLASTx based homology search analysis was valuable to assign accession IDS and predict the closest species homology to the transcript encoded proteins. Next, GO mapping was carried out to retrieve GO terms for all the BLASTX functionally annotated CDS, where assignments were used to classify the functions of the predicted CDS and defined ontology terms representing gene product properties, which could be grouped into three main domains: Biological process, Molecular Function, and Cellular Component. To retrieve GO terms for annotated CDS, the BLASTX resulted in accession IDs were used to retrieve gene names or symbols, followed homology search in the species-specific entries of the gene-product tables or via direct accession IDs search against dbxref table of the database.
For immune cataloging, all predicted CDS were subjected to blast analysis against An. gambiae, D. melanogaster, Ae. aegypti and Culex pipiens Immunome database (http://cegg.unige.ch/Insecta/immunodb), as described earlier (Thomas et al., 2016). Then, transcripts with E-value < e−10 were shortlisted, cataloged, and compared to select transcripts for expression profiling. To catalog tissue-specific P. vivax genes from a pre-analyzed whole transcripts database (NR/BLASTX), the best match Plasmodium transcripts were retrieved as the FASTA file. A standard GO annotation was performed, and P. vivax transcripts were analyzed for functional prediction.
Digital Gene Expression analysis: The high-quality reads for each sample was mapped on their respective set of CDS using CLC Genomics Workbench for read count calculation. The common hit accessions based on BLAST against the Nr database were identified for differential gene expression analysis. The differential gene expression was carried out by using the DESeq R package. CDS were further classified as up and down-regulated based on their log fold change (FC) value calculated by formula, FC = Log 2 (Treated/Control). FC values greater than zero were considered up-regulated whereas less than zero were down-regulated. The level of expression is represented as a log2 ratio of transcript abundance between control (uninfected) and experimental samples (P. vivax infected). Differentially expressed genes identified in control and experimental conditions were analyzed by hierarchical clustering. A heat map was constructed using the log-transformed and normalized value in FPKM (fragments per kilobase of exon per million fragments mapped) of genes based on Pearson uncentered correlation distance as well as based on complete linkage method, and the significance for differentially expressed genes was calculated at a P-value of <0.05. We compared and estimated the tag dispersion, assuming each tissue-specific two samples from different conditions with comparable gene expression for common transcripts. As both the samples contained RNA extracted from tissues of approximately ~25 mosquitoes each and pooled to form one single sample, a quantifiable estimation of gene expression was expected with a minimum chance of aberrations. Our DESeq data analysis with a single replicate was valuable to identify differentially expressed genes (Alaux et al., 2011; Wang et al., 2010; Sharma et al., 2015; Assefa et al., 2020), which was subsequently validated by large scale gene expression analysis by real-time PCR. Full technical details and reports of sequence analysis with representative sample data as well as DGE analysis validation are presented in supplemental data (see Data sheet1).
Transcriptional Profiling analysis: For spatial and temporal expression analysis of DGE selected transcripts of mosquito and/or P. vivax origin were monitored through Real-Time PCR analysis, as described previously (De et al., 2018; Thomas et al., 2016; Sharma et al., 2020). Briefly, the target tissues from ~15 to 20 uninfected or P. vivax infected (average infection level i.e. 50-130 oocysts/midgut) mosquitoes were collected in trizol and isolated ~1 μg total RNA was subjected to cDNA through Reverse-Transcription protocol (Verso cDNA synthesis kit@Thermo Scientific, USA). Prior transcriptional profiling, the quality of each cDNA reaction was evaluated by amplification of internal control gene Actin or rpS 7 through standard RT-PCR and agarose gel analysis. Only the qualified cDNA templates were further used for relative gene expression profiling by BIORAD CFX96 Real-time systems, using SYBR® FAST Universal 2X qPCR master mix. PCR cycles included initial denaturation at 95 °C for 5 min, then further 40 cycles of 95 °C for 10 s, 52 °C for 15 s, and 72 °C for 22 s. After each cycle, fluorescence was detected at the 72 °C step. Then further final steps include 65 °C for 05sec and 95 °C for 5sec. Each experiment was conducted in triplicate to minimize variation, except for hemocytes which were tested in duplicate, especially due to technical limitation of sample collection. The actin gene was used as an internal control for normalizing the relative expression data, and analysis was performed by 2–ΔΔCt method (Sharma et al., 2020).
Statistical analysis: For statistical analysis “test” sample data was compared with the “control” data set. Since, we compared each ‘test’ data with a single ‘control’, and therefore statistically analyzed using Student’s t-test only. Significant differences were considered at the P < 0.05 level.
3. Results
Working hypothesis development and RNA-Seq data generation: Previous studies targeting individual tissues, either midgut or salivary glands, have been valuable to understand the mosquito-parasite interactions. But still, there are several unresolved questions regarding how (i) each tissue viz. midgut, hemocytes, and salivary glands together coordinate and manage the challenge of Plasmodium infection; and (ii) how Plasmodium avoids the tissue-specific host immunity and quickly respond and adapt to environmental changes in different tissues for its survival and transmission. To partly answer and resolve tissue-specific molecular complexity, we developed a working hypothesis. We opined that when blood meal itself significantly alters mosquitoes ‘metabolic physiology’, Plasmodium infection may cause an additional burden of immune activation. Thus, mosquitoes may need to follow a dual management strategy for overcoming the challenge of a metabolic shift in response to fast gut engorgement (De et al., 2019), as well as limiting the parasite population. Alternatively, for its survival, parasites may either suppress or temper/modulate the host tissue responses.
To test and evaluate the hypothesis, we designed a strategy to capture a molecular snapshot of the three different tissues directly interacting with Plasmodium vivax. After the establishment of an artificial membrane feeding protocol, we fed mosquitoes with clinically diagnosed P. vivax-infected patient’s blood samples (0.5–2% gametocytaemia). In our regular experience, we noticed an average infection intensity of 50–130 oocysts/midgut 5–6 days post-infection. But in a few experimental studies, we noticed a super-infection of P. vivax, raising the average infection intensity to 300–380 oocysts/midgut (Fig. 1). Since, for our RNAseq project, facing a challenge of technical limitation of getting ‘superinfection’ mosquito samples, we cautiously followed a sample pool, and add-in SMART-cDNA technology, which not only advantageous to discover tissue-specific P. vivax transcripts but also found reliable to validate the DEG data using ‘average infection’ samples for transcriptional profiling in triplicates (Palomares et al., 2019; Assefa et al., 2020; see Supplemental data sheet-1/Table S9).
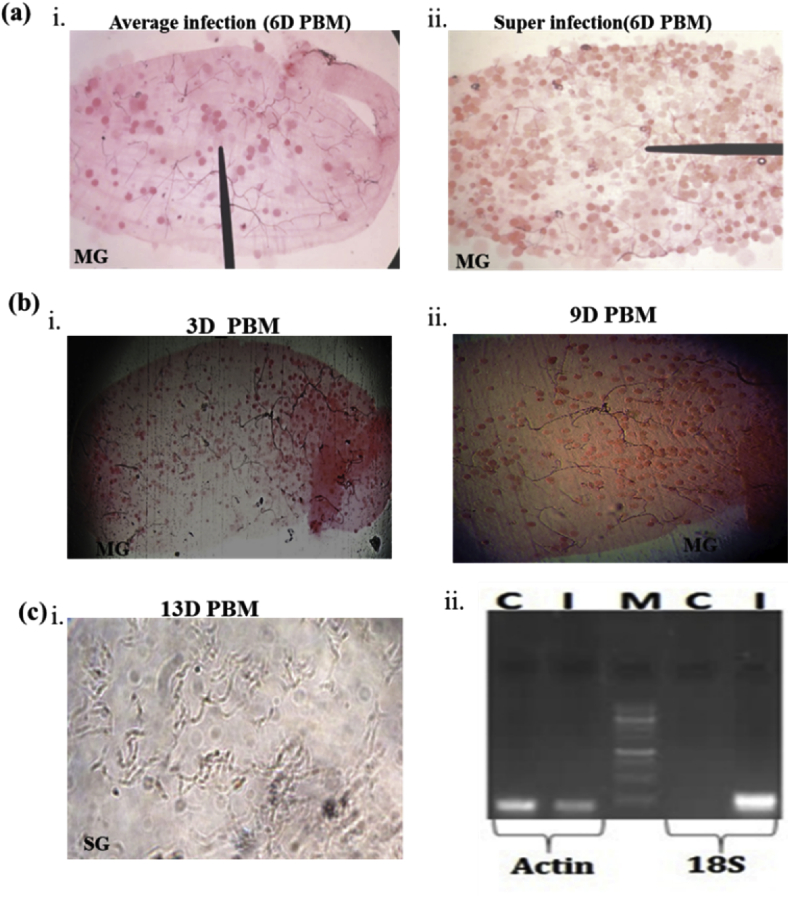
Fig. 1.
Technical presentation of mosquito infection validation and tissue collection strategy for tissue-specific RNA-Seq analysis: (a/i-ii) a comparative microscopic analysis of average infected and superinfected mosquito midgut, six days’ post Plasmodium vivax infection, (see “ruler needle pointer” of microscope eyepiece, highlighting the presence of gut oocysts as dark red circle); (b/i-ii) Microscopic view of early and late oocyst development in the mosquito midgut; (c): (i) Microscopic view of Plasmodium sporozoites in the mosquito salivary glands; (ii) Molecular confirmation of parasite infection in the mosquito by RT-PCR and agarose gel electrophoresis Uninfected (C) & Infected (I) midgut cDNA samples were subjected to 28 PCR cycle amplification using mosquito specific Actin & parasite-specific 18S primers. M = Molecular marker.
Accordingly, from infected 20–25 individual mosquitoes, we dissected and pooled targeted midgut, hemocytes, and salivary gland tissues, and performed a tissue-specific comparative RNA-Seq analysis. As per the technical design, we sequenced two midgut samples covering early (3–6 days) pre-mature to maturing oocysts, and late (8–10 days) fully matured/bursting oocysts; one hemocyte/hemolymph sample pooled from 9 to 12 DPI, a time window matching free circulatory sporozoites infection stage; and one sample of salivary glands 12–14 DPI (Fig. 1; see PanelA-B). For comparative study, we sequenced the naïve blood-fed mosquitoes’ tissues collected from the same aged mosquitoes. In the case of the midgut, we sequenced only one sample 3–4 days’ post blood meal (also see Sharma et al., 2020). From the seven RNA-Seq libraries we generated and analyzed a total of 28942203 reads (ST1). Table 1 represents the complete stat of the sequencing and analysis of each tissue-specific RNA-Seq data. Due to the unique nature of RNA-Seq sample i.e. ‘superinfection’, we mainly aimed to understand the genetic basis of parasite development in the mosquito, along with a very basic knowledge that how P. vivax infection may influence tissue-specific immune-physiological responses.
Table 1.
Complete sequencing stat of tissue-specific RNAseq libraries.
| Sl. No. | Sample Name | No. of Reads | No. of Contigs | Total transcripts | Transcripts with BLAST Hits (NR) | Annotated CDS analyzed |
|---|---|---|---|---|---|---|
| 1. | -MG-C | 4673408 | 74,548 | 11159 | 10219 (91%) | 6205 |
| 2. | MG-INF (3-6D) | 3625905 | 52,187 | 12065 | 11011 (91%) | 6813 |
| 3. | MG-INF (8-10D) | 5399936 | 42,212 | 7585 | 7222 (95%) | 7496 |
| 4. | HC-C | 3827903 | 30,959 | 6236 | 6044 (96%) | 2970 |
| 5. | HC-INF. | 3235689 | 18,462 | 3492 | 3333 (95%) | 3418 |
| 6. | SG-C | 3782999 | 64,960 | 7674 | 6840 (89%) | 6776 |
| 7. | SG-INF | 4396363 | 32,110 | 4492 | 4100 (91%) | 2743 |
Plasmodium vivax alters the molecular architecture of mosquito tissues: To understand that, how P. vivax infection influences tissue-specific molecular responses, we compared the global change in the transcript abundance of ‘uninfected’ and ‘infected’ blood-fed mosquito tissues. Our initial attempt of mapping of cleaned reads (filtered low quality, microbial, and P. vivax origin) to the available draft reference genome was unsuccessful. Alternatively, we mapped all the high-quality reads against the denovo assembled reference map, as described earlier (De et al., 2018; Sharma et al., 2015; Thomas et al., 2016). Though, a read density map analysis revealed that P. vivax infection causes a significant suppression in the salivary glands and midgut transcripts, but caused a greater shift in the read density of the infected hemocyte transcripts (Supplemental data sheet-2/Fig. S1). To further uncover the molecular and functional nature of the encoded proteins, we carried out a comprehensive GO annotation, cataloged, and extensively profiled tissues specific shortlisted genes altered in response to P. vivax infection.
Midgut response to Plasmodium oocysts development: A comparative catalog of biological process (L4) revealed that early maturing oocyst development coincides with the suppression of the majority of midgut proteins (ST1). Although oocyst development exceptionally induces the expression of proteins encoding cellular catabolic, organic substance catabolic, and organic acid metabolic processes, it also restricts the expression of proteins with regulatory functions (Fig. 2a). Interestingly, when compared to naïve midgut, late-stage oocysts showed a re-enrichment of the gut proteins with common functions (Fig. 2a). Venn diagram analysis of annotated transcripts showed that 877, 1036, and 1251 transcripts uniquely restricted their expression to naïve, early, and late oocysts in the infected mosquito guts, respectively, (Fig. 2c). A heat map analysis of differentially expressed common transcripts showed a significant alteration in response to early oocysts infection (Fig. 2e). Taken together, these observations suggest that the fast-developing oocysts of P. vivax may cause a biphasic modulation of gut metabolic machinery, allowing early suppression and late recovery for the maintenance of homeostasis.
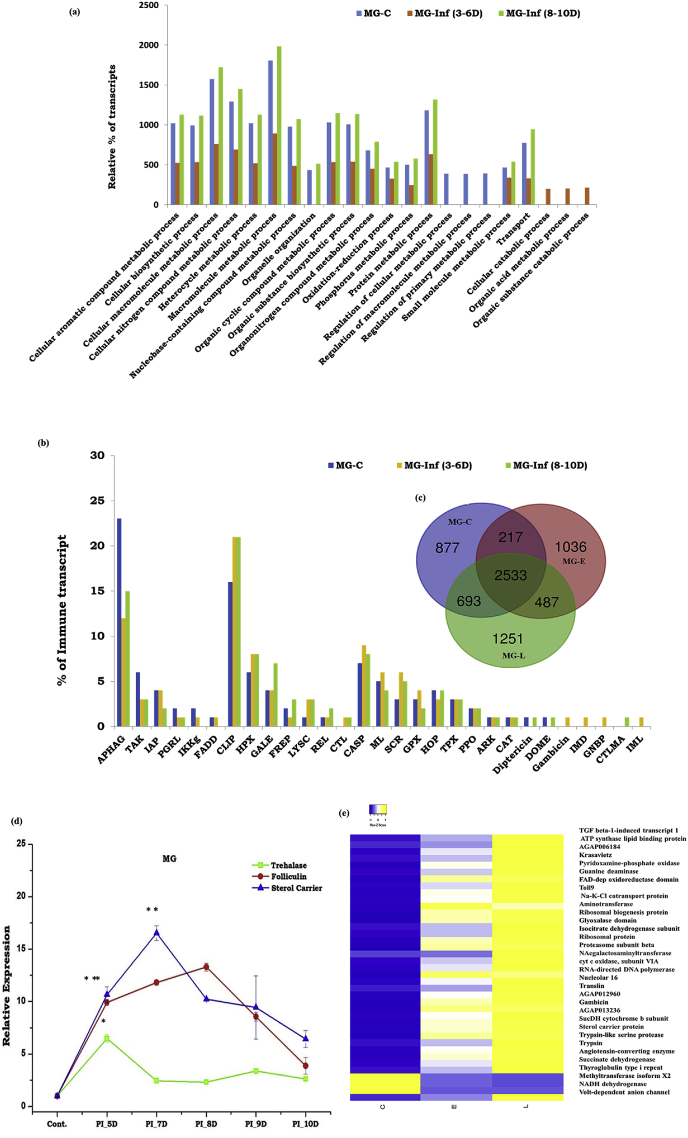
Fig. 2.
Detailed molecular cataloging of midgut transcriptome in response to P. vivax infection: (a) Comparative Gene Ontology (GO- biological process/Level 4) analysis of midgut transcriptome revealed an initial suppression/restricted expression of most of the midgut genes during early oocyst infection (MG-Inf 3-6D) followed by re-enrichment of the respective transcripts at later stages i.e. 8-10 DPI (MG-Inf 8-10D); (b) Comparative gut immunome analysis showing alteration of immune proteins of common function as a well exclusive increased percentage of immune proteins (CTL:C-type lectin; Gambicin; GNBP: Gram-negative bacteria-binding proteins; IMD: Immune deficiency pathway member; CTMLA: C-type lectin mannose binding group and IML: Immunolectins) in response to early oocyst infection; (c) Venn diagram analysis represents genes expressing uniquely in the midgut after a blood meal, during early oocyst infection as well as mature oocyst infection; (d) Time dependent relative gene expression analysis of selected gut transcripts in the mosquito gut (n = 20; N = 3): Trehalase p < 0.01; Folliculin p < 0.007 and Sterol carrier p < 0.002 (S2_primer list) encoding nutritional responsive proteins in response to P. vivax infection; (e) Heat map showing differential expression of the common midgut transcripts in response to P. vivax infection. Statistical significance was measured using the student’s t-test, where each test sample was compared with control blood-fed mosquito gut (∗P < 0.05;∗∗P < 0.001;∗∗∗P < 0.0001); (n represents the number of mosquito’s gut pooled for sample collection; N = number of replicates).
To further clarify how Plasmodium vivax infection alters gut immuno-physiological responses, we identified, cataloged, and compared the expression of selected transcripts in response to a blood meal and P. vivax infection. A comparative gut-immunome analysis showed an increased percentage of immune transcripts, except for a few classes such as Autophagy, TAK, IAP, PGRP, and IKKG (Fig. 2b). Additionally, we also observed an exclusive enrichment of several classes of immune proteins such as CTL, Gambicin, GNBP, IMD, CTMLA, and IML, in response to early infection (ST2). As expected, blood meal causes a transient change in immune gene expression (S1), but in the P. vivax-infected midgut samples, we observed a multifold enrichment of Gambicin expression after 48hr and late induction of other AMPs such as C1, C2, and D1 (see Sharma et al., 2020). Together, these data suggest that a time-dependent action of distinct AMPs against P. vivax is necessary to delimit the gut-specific oocysts’ development. Furthermore, the rapid induction of transcripts encoding Folliculin, Trehalase, and Sterol carrier indicated their possible role in managing nutritional imbalance during P. vivax development (Fig. 2d).
Hemocyte response to free circulatory sporozoites: Infected mosquito hemocytes showed a slight enrichment of transcripts (1408) encoding the diverse nature of proteins (ST1). The expression of 1128 transcripts linked to organelle organization and ribonucleoprotein complex biogenesis functions remains uniquely associated with infected hemocytes (Fig. 3a). Interestingly, we observed at least 966 transcripts encoding proteins of cellular catabolic and organic acid metabolic processes, which remained restricted to naïve blood-fed mosquito hemocytes. Surprisingly, contrary to this in the gut, similar categories of proteins were exclusively induced in response to early oocysts infection (see Fig. 2, Fig. 3). FPKM (Fragments Per Kilobase of transcript per Million mapped reads) based heat map analysis further showed a unique modulation of common transcripts, with restricted expression either in the naïve or infected mosquito hemocytes (Fig. 3e).
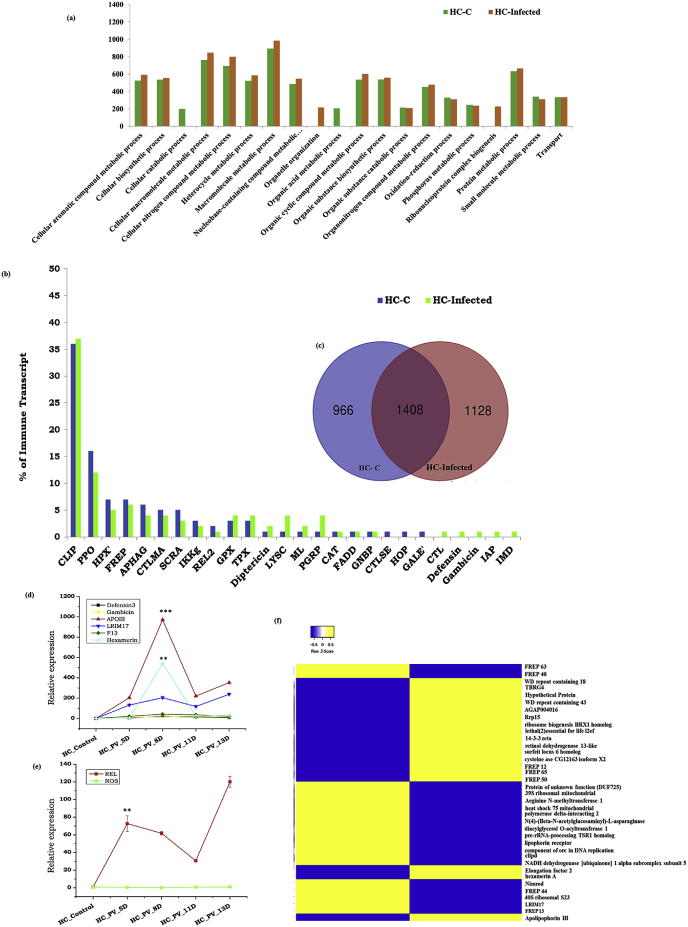
Fig. 3.
Comparative analysis of the molecular architecture of hemocyte transcriptomes during P. vivax infection: (a) Gene ontology (GO biological process/Level 4) analysis of hemocyte transcriptome showing exclusive enrichment of transcripts having a role in organelle organization and ribonucleoprotein complex biogenesis in response to P. vivax infection; (b) Comparative catalog of immune transcripts from uninfected and parasite-infected hemocytes revealing exclusive enrichment of unique protein families (CTL:C-type lectin; Gambicin; Defensin; IMD: Immune deficiency pathway member; and IAP: Inhibitors of apoptosis) in response to P. vivax infection; (c) Venn diagram analysis showing 1128 transcripts expressing exclusively in P. vivax infected hemocytes (HC-Infected), while 966 genes expression restricted to blood-fed hemocytes only (HC–C); (d) Transcriptional response of selected AMPs(Antimicrobial Peptides) and Non AMPs such as ApoIII (p < 0.0001), hexamerin (p < 0.0002) and FREP13: Fibrinogen related Protein 13) in the hemocytes after 5, 8, 11 and 13 DPI with P. vivax; (e) Transcriptional profiling of signaling molecules (REL: Relish p < 0.003; and NOS: Nitric oxide synthase) in P. vivax infected hemocyte samples (S2_primer list); (f) FPKM-based heat map expression analysis of common transcripts that were either restricted to blood fed or P. vivax infected hemocytes. Statistical significance was measured using the student’s t-test, where each test sample was compared with control blood-fed mosquito hemocytes (∗P < 0.05;∗∗P < 0.001;∗∗∗P < 0.0001); (n represents the number of mosquito’s hemocytes pooled for sample collection; N = number of replicates).
Since mosquito hemocytes mount a highly specific cellular immune response, we next targeted to decode the molecular nature of the immune interaction between mosquito hemocytes and free circulating sporozoites (fcSPZ) of P. vivax. A comparative immunome (ST2) analysis indicated that fcSPZ causes a greater suppression of many immune family proteins, except for the marked enrichment of GPX, TPX, Dipetricin, LYSC, PGRP, and ML family proteins (Fig. 3b). Interestingly, like the midgut, infected mosquito hemocytes also showed an exclusive induction of similar classes of immune proteins such as CTL, Gambicin, Defencin, IAP, and IMD. (Fig. 3d). Significant enrichment of REL than NOS coincided with early oocysts development in the gut (Fig. 3e). We also observed an exclusive induction of the physiologically active class of immune proteins such as ApoIII, Hexamerin, and FREP13 during the 8th day of P. vivax infection (Fig. 3d). While, in our recent study we further observed that the expression of FREP12 significantly upregulated after 10 days of P. vivax infection, highlighting its possible role in immunity against free circulating sporozoites (Chauhan et al., 2020).
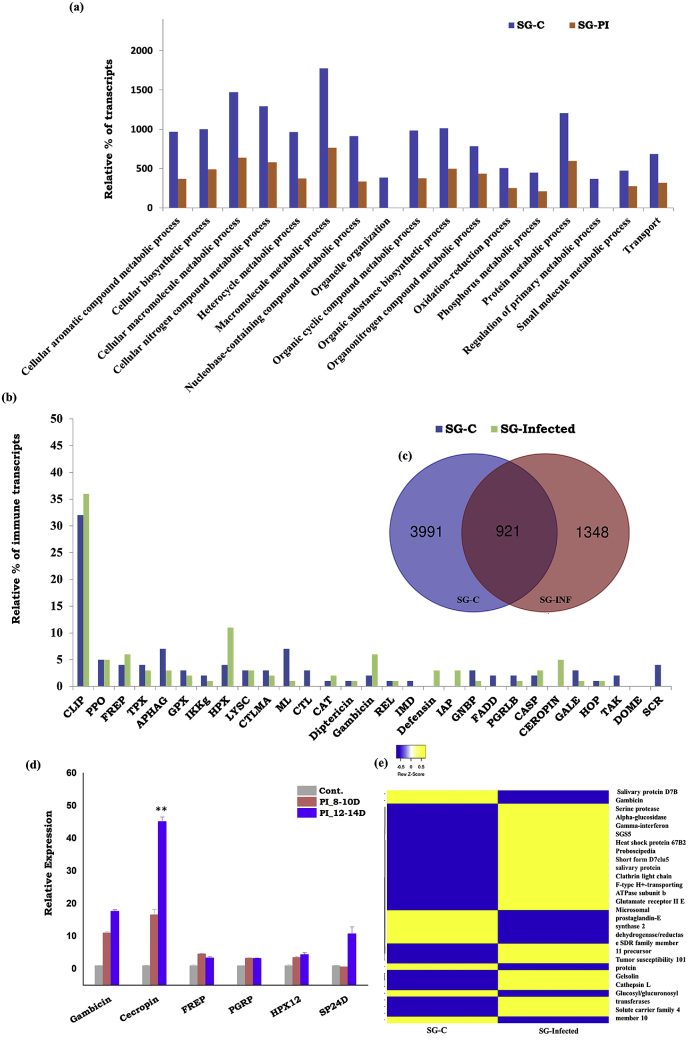
Salivary glands response to P. vivax infection: In contrast to midgut and hemocytes, P. vivax infection impaired the molecular architecture of the salivary glands by delimiting the expression of common salivary transcripts (ST1). Additionally, we also observed restricted expression of primary metabolic process proteins in uninfected mosquito salivary glands (Fig. 4a). Venn diagram analysis further indicated that only 1427 (28%) annotated salivary proteins shared a common function. However, a large pool of 3069 (60%) transcripts were restricted to uninfected mosquito salivary glands and the remaining 699 (12%) transcripts showed a unique appearance to sporozoites invaded salivary glands (Fig. 4c). A heat map data analysis further suggested that salivary sporozoites may keep a stronghold on the salivary metabolic machinery, possibly favoring its survival (Fig. 4e).
Fig. 4.
Detailed molecular map and comparative analysis of Blood fed (SG-C) and P. vivax infected (SG-Infected) salivary gland transcriptomes:(a) Gene ontology (GO- biological process/Level 4) based molecular cataloging and comparison of salivary gland transcriptome in response to P. vivax infection; (b) Molecular cataloging of the immune genes enriched in the salivary glands of Plasmodium-infected mosquitoes; (c) Venn diagram analysis showing several common and uncommon salivary transcripts altered in response to infection; (d)) Transcriptional profiling of selected salivary gland genes (Gambicin p < 0.0003; Cecropin p < 0.0004; FREP: Fibrinogen related Proteins p < 0.02; PGRP: Peptidoglycan Recognition Proteins p < 0.002; HPX12: Heme peroxidase 12 p < 0.001; SP24D: Serine Protease 24D p < 0.09) during P. vivax infection; (e) FPKM-based heat map analysis expression of common salivary genes. Statistical significance was measured using the student’s t-test, where each test sample was compared with control blood-fed mosquito salivary glands (∗P < 0.05;∗∗P < 0.001;∗∗∗P < 0.0001); (n represents the number of mosquito’s hemocytes pooled for sample collection; N = number of replicates).
Next, to test how the salivary immune system influences sporozoite development and survival, we cataloged and compared the relative percentage of immune transcripts (ST2). We noticed an exclusive appearance of four classes of immune family proteins, such as FADD, Gambicin, GNBP, and SCRC in the invaded salivary glands (Fig. 4b). The expression of TAK1, CASP, HOP, and STAT immune family proteins remained restricted to naïve mosquito salivary glands (Fig. 4d). Transcriptional profiling of selected immune transcripts such as Gambicin, Cecropin, and SP-24 D showed a high induction in the salivary glands (Fig. 4e). Surprisingly, several other salivary secretory proteins such as anopheline, D7 Family, ion transporter family proteins, and 53.7 kDa also showed significant up-regulation, suggesting their anti-Plasmodium role against P. vivax infection.
Deep sequencing identifies P. vivax transcripts: The above analysis allowed us to hypothesize that when Plasmodium sp. switches from one stage to another, it may follow a unique strategy to regulate tissue-specific metabolic and immuno-physiological responses. The surprising finding of a large pool of 4,449 transcripts of P. vivax origin, distinctly expressed in their respective infected mosquito tissues (Table 2), encouraged us to establish a genetic relationship of mosquito-parasite interaction. We noticed that 73.8% of P. vivax transcripts were associated with oocysts (early and late), while 74 transcripts are originated from free circulatory sporozoites, and 1,133 transcripts from salivary sporozoites (ST3).
Table 2.
Detailed information of P.vivax transcripts retrieved from mosquito transcriptomes.
| Target infection stage | Total P. vivax transcripts | The qualified Best match to P. vivax | Hypothetical/Unknown | Unique/stage-specific | The qualified Best match to other Plasmodium |
|---|---|---|---|---|---|
| MG(OOC-E) | 3208 | 1265 | 1943 | 2200 | 539 |
| MG(OOC-L) | 75 | 65 | 15 | 9 | 10 |
| HC(fcSPZ) | 74 | 57 | 17 | 6 | 7 |
| SG(sg-SPZ) | 1133 | 797 | 336 | 162 | 113 |
| TOTAL | 4449 | 2184 | 2311 | 2377 | 669 |
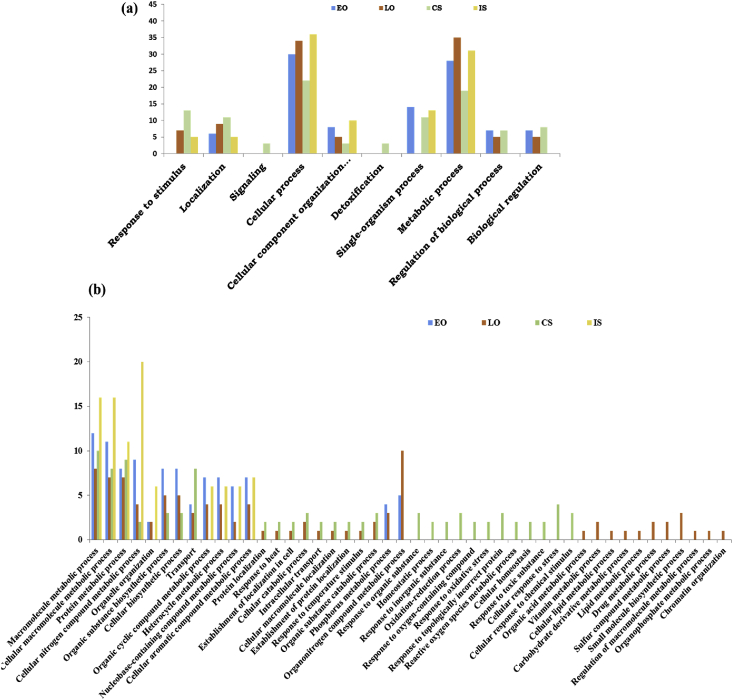
Molecular changes of P. vivax may facilitate its survival and transmission: To establish a functional correlation, we annotated, cataloged, and compared the relative abundances of the P. vivax-specific transcripts. Initially, putative transcripts encoding Plasmodium homolog proteins were filtered out from the pre-analyzed RNA-Seq dataset. Each developmental stage of P. vivax revealed a large pool of hypothetical proteins, especially in early oocysts and salivary sporozoites (Table- 2), suggesting complex biology is yet to unravel. Further, GO annotation revealed that each developmental stage carries an enriched transcript abundance of the genes linked to cellular as well as metabolic processes (Fig. 5a). Surprisingly, free circulatory sporozoites not only showed a selective enrichment for the family proteins encoding ‘Response to Stimulus’ and ‘Localization’ but also unraveled an exclusive appearance of signaling and detoxification linked genes in the hemocytes.
Fig. 5.
Molecular architecture of P. vivax transcripts during tissue-specific developmental stages within the mosquito An. stephensi: (a) BLAST2GO (GO- biological process Level 2 based annotation and functional classification of identified P. vivax transcripts; (b) detailed molecular and functional cataloging (GO- biological process Level 4) of P. vivax genes revealed that, despite having a limited number of proteins, Late oocyst (LO) and free circulatory sporozoite (CS) proteins are more diverse than early oocyst (EO) and salivary invaded sporozoite (IS) proteins.
Since the rupturing of late oocysts releases millions of fcSPZ, which are highly vulnerable to the hemocyte cellular immune response, we hypothesize that Plasmodium must have a unique strategy to defend its survival. An in-depth functional annotation analysis (BP-Level 4), clearly demonstrated that despite having a low number of transcripts, late oocysts and fcSPZ encoded a more diverse nature of proteins than early oocysts and salivary sporozoites (Fig. 5b). A Venn diagram analysis further reveals that the molecular nature of Plasmodium encoded proteins is significantly altered when it switches from one stage to another, particularly late oocysts (LO) to fcSPZ.
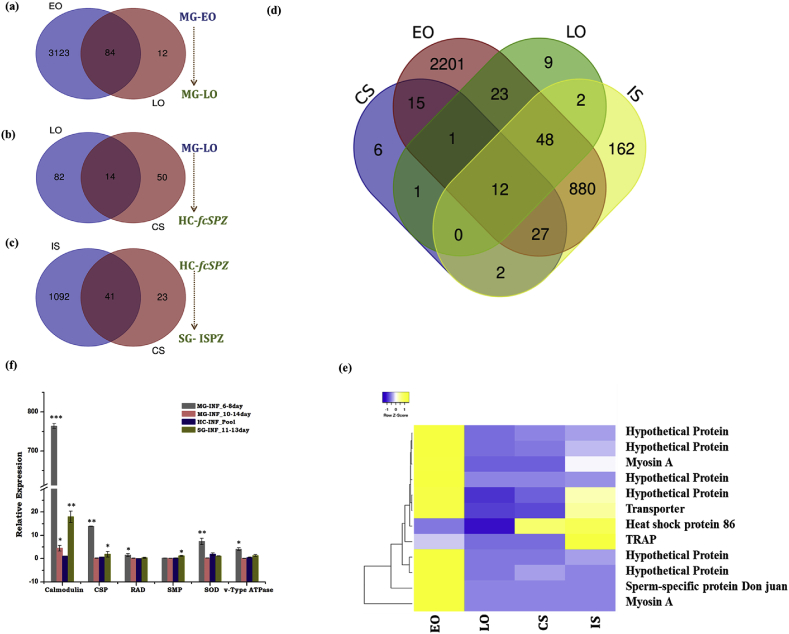
Remarkably, a cross tissue comparison showed a restricted expression of a large number of proteins in each developmental stage of P. vivax (Fig. 6a–c). Interestingly, a total of 2,200 transcripts showed restricted expression to early oocysts (EO), while 9 transcripts were exclusively expressed to the late oocysts (LO) stage. Out of the total 74 transcripts, at least 6 showed restricted expression to the fcSPZ stage and 162 transcripts expression exclusively remained in the salivary invaded sporozoites (Fig. 6d). Though the exact mechanism of this developmental transformation is yet unknown. But an FPKM-based relative expression analysis and transcriptional profiling of selected common transcripts further suggested that the ability to alter genetic makeup may enable P. vivax to misguide tissue-specific immuno-physiological responses and survival in the mosquito host (Fig. 6e–f).
Fig. 6.
Stage-specific comparative analysis of P. vivax transcripts: Venn diagrams showing P. vivax specific transcripts data comparison between (a) Midgut Early oocyst (MG-EO) and Late oocyst (MG-LO);(b) Midgut Late oocyst (MG-LO) and free circulatory Sporozoite (HC-fcSPZ) of hemocytes and (c) HC-fcSPZ and salivary gland sporozoite (sgSPZ); (d) Venn diagram showing the number of common and uncommon P. vivax transcripts expressed during distinct developmental stages i.e. Early oocyst (EO), Late oocyst (LO), free circulatory Sporozoite (CS) and salivary invaded sporozoites (IS); (e) FPKM-based comparative heat map of selected genes like myosin A, Transporter, TRAP (Throbospondin related adhesion protein) and some hypothetical proteins; (f) Tissue-specific relative expression analysis of P. vivax genes (Calmodulin; CSP: Circumsporozoite protein; RAD protein; SMP: Sporozoite microneme protein; SOD: superoxide dismutase; vATPase) in the infected mosquitoes (n = 20; N = 2); Statistical significance was measured using student’s t-test, (∗P < 0.05;∗∗P < 0.001;∗∗∗P < 0.0001); (n = represents the number of mosquito’s tissues pooled for sample collection; N = number of replicates).
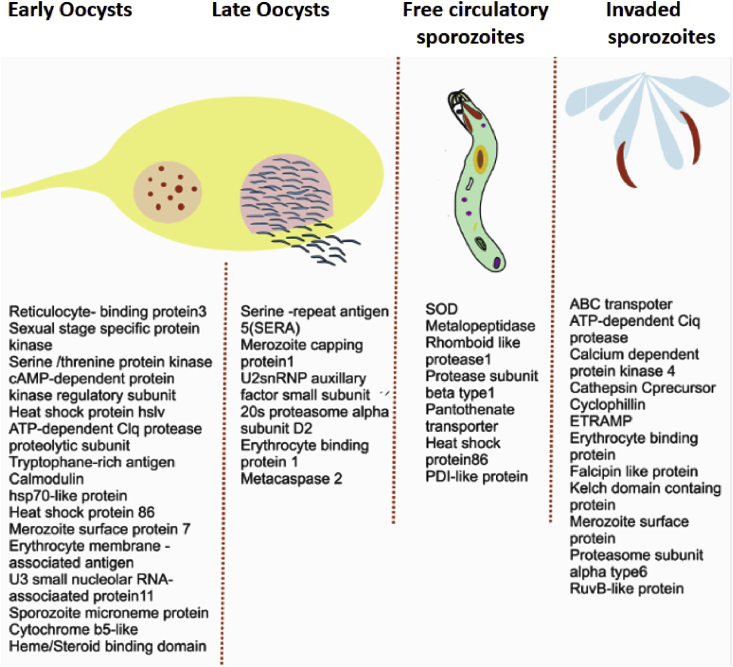
Finally, we targeted the catalog of well-annotated key genes that may have a potential role in parasite development and transmission. To do this, we performed a comprehensive literature search analysis and shortlisted transcripts with high FPKM, but a restricted expression to a particular tissue (Fig. 7). Though an ongoing independent detailed comparison of P. vivax transcript database with other parasites such as P. falciparum and P. berghei, yet to unravel genetic differences (unpublished), current findings provide a valuable resource for P. vivax transcripts, especially free circulatory sporozoites directly interacting with mosquito hemocytes, for future functional characterization.
Fig. 7.
Representative catalog of RNA-Seq identified P. vivax-specific transcripts dominantly expressing in distinct mosquito tissues.
4. Discussion
Evolution and adaptation to blood-feeding is an essential requirement for mosquitoes’ reproductive success (Attardo et al., 2005; Hansen et al., 2014; Kokoza et al., 2001). However, Plasmodium parasites take the benefit of mosquitoes’ blood-feeding behavior for the completion of the sexual cycle and disease transmission (Thomas, 2012; Lacroix et al., 2005; Schwartz and Koella, 2001). Thus, to win the developmental race, we opined that Plasmodium must have the ability to manipulate immuno-physiological responses of directly interacting tissues, such as the midgut, hemocytes, and salivary glands of mosquitoes (Cator et al., 2012). It has been well established that during its development inside susceptible mosquito hosts, the population dynamics of Plasmodium are significantly altered (Vézilier et al., 2012). But holistically the molecular nature of tissue-specific host-parasite interactions, covering gut-hemocyte-salivary gland-Plasmodium together in a single study, experimentally remains poorly understood (Molina-Cruz et al., 2012). Using a system-wide RNAseq analysis, here we now uncover the tissue-specific genetic basis of mosquito-Plasmodium interaction.
Initial profiling of AMPs/non-AMPs in response to uninfected blood-feeding showed a transient influence in the tested tissue, suggesting a general role in the maintenance of physiological homeostasis (Fig. S4). However, surprisingly, when compared to an uninfected counterpart, each tissue shows a unique relationship of metabolic alteration in response to P. vivax infection. Our functional annotation of gut-RNAseq data demonstrated that post-gut invasion, the mosquito can regulate and recover from the acute harms caused by fast-developing early oocysts. However, many naturally selected refractory anopheline species can cease the development of parasites through melanization (Molina-Cruz et al., 2012; Simões et al., 2017). It is plausible to predict that even in the susceptible strain, the controlled regulation of gut-parasite interplay is key for the survival of both mosquito and parasite (Dong et al., 2009). Studies also suggest that nutritional deprivation may have a direct impact on the gut oocysts development and mosquitoes’ reproductive outcome, although the mechanism is yet to unravel (Liu et al., 2013; Yang et al., 2019). Our observation of early induction of transcripts regulating gut-specific nutritional homeostasis, such as Folliculin, Trehalase, and Sterol carrier suggested that maturing oocysts may cause host nutritional resources imbalance (Baba et al., 2006; Schekman, 2013; Shukla et al., 2015). However, the delayed elevation of gut immune transcripts may counterbalance the negative impact of rapidly exiting sporozoites into the hemolymph. Thus, disrupting this relationship may favor the development of new tools to numb parasites either by delimiting the nutritional demand and/or enhancing gut immunity (Shea-Donohue et al., 2017).
Once left the gut epithelium, millions of fcSPZ directly encounter and are cleared by hemocytes, but the molecular nature of this interaction is not fully understood (Jull Hillyer et al., 2003; Jaramillo-Gutierrez et al., 2009). Our data suggest a major shift in transcript abundance, especially encoding structural and catabolic activity associated proteins, indicated the hyperactivity of hemocytes to control Plasmodium sporozoites population. The enriched number of hemocyte-specific immune transcripts and their time-dependent elevated expression together indicated that a hyper immunity of hemocytes is essential for the majority of sporozoite population clearance. Although the reason for how this hyper-response is achieved by hemocyte is unclear, however, our observation of increased expression of REL compared to NOS against early P. vivax oocysts supports the idea that pre-immune activation of hemocyte may exist (De et al., 2018; Kwon and Smith, 2018). Several previous studies suggested that both REL and NO are key to regulating tissue-specific immune regulation. Recently, we demonstrated that both REL and NO participate in inter-organ immune communication, however, each tissue specifically maintains the inter-organ flow of signals (Clayton et al., 2014; Simões et al., 2017; De et al., 2018). An indirect anti-Plasmodium response via hemocyte and REL immune signaling have also been suggested in the An. gambiae, P. berghei, and P. falciparum model (Hillyer and Estévez-Lao, 2010). Surprisingly, however, it remains unclear how a fraction of fcSPZ succeeds in avoiding the hyperimmune response of hemocytes and invade salivary glands.
Our RNA-Seq data analysis and annotation of salivary-parasite interactions indicated that Plasmodium sporozoites not only impair the metabolic machinery but also enrich nucleic acid binding transcriptional activities related proteins. For successful blood meal acquisition, salivary glands release a cocktail containing nucleic acid binding factors such as Apyrase (King et al., 2011), D7 family proteins (Calvo et al., 2006), and nucleotide transferase (Dhar and Kumar, 2003; Dixit et al., 2009; Vogt et al., 2018). Thus, it is plausible to hypothesize that Plasmodium infection may enhance host-seeking behavioral activities by stimulating the expression of nucleic acid binding factors (Chen et al., 2008). Additionally, increased expression of salivary immune transcripts indicated that an active local immune response is essential to restrict salivary invaded sporozoite (IS) populations.
As a proof of concept, our data suggested that even after mounting an effective tissue-specific immuno-physiological response, mosquitoes fail to disrupt the Plasmodium sporogonic cycle. Hence, we propose that Plasmodium parasites are clever enough to dodge the mosquito responses by wise manipulation of their own molecular architecture. Retrieval of a large pool of P. vivax transcripts originating from distinct developmental stages provided an opportunity to anticipate the molecular dynamics facilitating its survival. An initial observation of more than 50% transcripts encoding hypothetical proteins indicated that a deep understanding of the P. vivax sporogonic cycle is still obscure. However, an observation of a significant difference in the molecular repertoire of stage-specific P. vivax genes further strengthens our hypothesis that the Plasmodium parasite has the unique ability to misguide the track and trap system of mosquitoes.
5. Conclusion
For its successful survival and transmission, at every stage of development inside the mosquito host, Plasmodium vivax negotiates multiple tissues. Independent studies targeting individual tissues have been valuable (Roth et al., 2018; Boonkaew et al., 2020), but conceptually, we have unresolved major questions about how a mosquito’s tissue-specific actions manage the challenge of Plasmodium infection and/or how Plasmodium manages to avoid tissue-specific responses. For the first time, we demonstrate and establish that P. vivax follows a smart strategy of genetic makeup change to misguide and evade, even a highly sophisticated immune barrier of each tissue. We hypothesize and propose that an unharmed tissue-specific molecular wave of negotiations and actions by genetic changes benefits P. vivax successful development and transmission in the mosquito host. Further establishing a functional correlation may lead to the identification of mosquitoes as well as P. vivax specific crucial genetic factors for target selection and designing a new molecular strategy for malaria control.
Data deposition
The sequences of the individual tissue samples were submitted to the NCBI with accession numbers SRR8580009 (MG-Inf 8-10D); SRR8580010 (MG-C); SRR8580011 (MG-Inf 3-6D); SRR8501353 (HC–C); SRR8501352 (HC-PI); SRR8476334 (SG-PI); SRR8476333 (SG-C).
Grant Information
Work in the laboratory was supported by Department of Biotechnology (DBT), Government of India (BT/HRD/35/02/01/2009), and Indian Council of Medical Research (ICMR), Government of India (5/87 (301)v2011ECD-II), RKD is a recipient of a DBT sponsored Ramalingaswami Fellowship. SK, ST, and CC are recipients of CSIR (09/905 (0015)/2015-EMR-1), UGC (22/12/2013(II)EU-V), and DST (DST/INSPIRE/03/2014/003463) fellowships, respectively.
Authors’ contribution
ST, SK, CC, KCP, VP, RKD conceived scientific hypothesis and designed the experiments, ST, SK, CC performed tissue i.e. midgut, salivary gland, and hemocyte specific experiments, DS, PS, TT, JR, TDD, DSR contributed to design and performing the experiments, data acquisition, writing, and editing; ST, SK, CC, KCP, VP, RKD data analysis and interpretation, data presentation, contributed reagents/materials/analysis tools, wrote, reviewed, edited, and finalized MS. All authors read and approved the final manuscript.
Authors summary
Malaria transmission dynamics are heavily influenced by mosquito –parasite interactions. When passing through tissue-specific barriers, Plasmodium has to compromise by losing its population, but the genetic relationship is unknown. To win the developmental race, Plasmodium needs to overcome two important immuno-physiological barriers. The first one accounts for an indirect 24–30 h long pre-invasive gut-microbe-parasite interaction in the gut lumen. The second one follows a direct post-gut invasive 14–18 days long interaction with the midgut, hemocyte, and salivary glands. During the pre-invasive phase of interaction, we showed that Plasmodium vivax follows an immunosuppression strategy by restricting microbial growth in the gut lumen (Sharma et al., 2020). Here, we demonstrate that successful developmental progression by P. vivax is accompanied by the manipulation of tissue-specific metabolic responses and altering its genetic makeup. This strategy not only clears off the multifaceted mosquito’s tissue-specific immune responses but also favors Plasmodium’s survival and transmission. Comprehending this tissue-specific interaction between host and parasite at the molecular level could provide a new tool to intervene in the Plasmodium life cycle within a vector.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We would like to thank all the technical staff members of the central insectary facility for mosquito rearing and Kunverjeet Singh for lab assistance. The authors thank Dr. Neena Valecha, Dr. S. K. Sharma, and Dr. Mayur Kajla for reviewing and valuable comments on the manuscript. Authors thanks to NIMR-clinical facility support. Finally, we thank Xceleris Genomics, Ahmedabad for NGS sequencing. This manuscript has been released as a Pre-Print @www.Biorxiv.org; https://www.biorxiv.org/content/10.1101/774166v1.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crimmu.2021.02.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Aly A.S.I., Vaughan A.M., Kappe S.H.I. Malaria parasite development in the mosquito and infection of the mammalian host. Annu. Rev. Microbiol. 2009 doi: 10.1146/annurev.micro.091208.073403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaux C., Dantec C., Parrinello H., Le Conte Y. Nutrigenomics in honey bees: digital gene expression analysis of pollen’s nutritive effects on healthy and varroa-parasitized bees. BMC Genom. 2011;12:496. doi: 10.1186/1471-2164-12-496. Published 2011 Oct 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardo G.M., Hansen I.A., Raikhel A.S. Insect Biochemistry and Molecular Biology. 2005. Nutritional regulation of vitellogenesis in mosquitoes: implications for an autogeny. [DOI] [PubMed] [Google Scholar]
- Assefa A.T., Vandesompele J., Thas O. On the utility of RNA sample pooling to optimize cost and statistical power in RNA sequencing experiments. BMC Genom. 2020;21:312. doi: 10.1186/s12864-020-6721-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M., Hong S.-B., Sharma N., Warren M.B., Nickerson M.L., Iwamatsu A., Zbar B. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc. Natl. Acad. Sci. Unit. States Am. 2006;103(42):15552–15557. doi: 10.1073/pnas.0603781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baton L.A., Ranford-Cartwright L.C. Plasmodium falciparum ookinete invasion of the midgut epithelium of Anopheles stephensi is consistent with the Time Bomb model. Parasitology. 2004 doi: 10.1017/s0031182004005979. [DOI] [PubMed] [Google Scholar]
- Belachew E.B. Immune response and evasion mechanisms of Plasmodium falciparum parasites. Journal of Immunology Research. 2018 doi: 10.1155/2018/6529681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennink S., Kiesow M.J., Pradel G. The development of malaria parasites in the mosquito midgut. Cell Microbiol. 2016 doi: 10.1111/cmi.12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonkaew T., Mongkol W., Prasert S., Paochan P., Yoneda S., Nguitragool W., Kumpitak C., Sattabongkot J., Kubera A. Transcriptome analysis of Anopheles dirus and Plasmodium vivax at ookinete and oocyst stages. Acta Trop. 2020 doi: 10.1016/j.actatropica.2020.105502. (in press) [DOI] [PubMed] [Google Scholar]
- Calvo E., Mans B.J., Andersen J.F., Ribeiro J.M.C. Function and evolution of a mosquito salivary protein family. J. Biol. Chem. 2006 doi: 10.1074/jbc.M510359200. [DOI] [PubMed] [Google Scholar]
- Castillo J.C., Barletta Ferreira A.B., Trisnadi N., Barillas-Mury C. Activation of mosquito complement antiplasmodial response requires cellular immunity. Science Immunology. 2017 doi: 10.1126/sciimmunol.aal1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cator L.J., Lynch P.A., Read A.F., Thomas M.B. Do malaria parasites manipulate mosquitoes? Trends Parasitol. 2012 doi: 10.1016/j.pt.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.G., Mathur G., James A.A. Chapter 2 gene expression studies in mosquitoes. Adv. Genet. 2008 doi: 10.1016/S0065-2660(08)00802-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan C., De T.D., Kumari S., Rani J., Sharma P., Tevatiya S., Pandey K.C., Pande V., Dixit R. Hemocyte-specific FREP13 abrogates the exogenous bacterial population in the hemolymph and promotes midgut endosymbionts in Anopheles stephensi. Immunol. Cell Biol. 2020 doi: 10.1111/imcb.12374. [DOI] [PubMed] [Google Scholar]
- Cirimotich C.M., Dong Y., Garver L.S., Sim S., Dimopoulos G. Mosquito immune defenses against Plasmodium infection. Dev. Comp. Immunol. 2010 doi: 10.1016/j.dci.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton A.M., Dong Y., Dimopoulos G. The anopheles innate immune system in the defense against malaria infection. Journal of Innate Immunity. 2014 doi: 10.1159/000353602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A., Götz S., García-gómez J.M., Terol J., Talón M. Sequence analysis Blast2GO : a universal tool for annotation, visualization and analysis in functional genomics research, (October. 2005. 2-5. [DOI] [PubMed]
- De T., Das, Sharma P., Thomas T., Singla D., Tevatiya S., Kumari S., Dixit R. Interorgan molecular communication strategies of “Local” and “Systemic” innate immune responses in mosquito Anopheles stephensi. Front. Immunol. 2018 doi: 10.3389/fimmu.2018.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar R., Kumar N. Role of mosquito salivary glands. Curr. Sci. 2003 [Google Scholar]
- Dixit R., Rawat M., Kumar S., Pandey K.C., Adak T., Sharma A. Salivary gland transcriptome analysis in response to sugar feeding in malaria vector Anopheles stephensi. J. Insect Physiol. 2011 doi: 10.1016/j.jinsphys.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Dixit R., Sharma A., Mourya D.T., Kamaraju R., Patole M.S., Shouche Y.S. Salivary gland transcriptome analysis during Plasmodium infection in malaria vector Anopheles stephensi. Int. J. Infect. Dis. 2019;13(5):636–646. doi: 10.1016/j.ijid.2008.07.027. [DOI] [PubMed] [Google Scholar]
- Dong Y., Manfredini F., Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009 doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garver L.S., de Almeida Oliveira G., Barillas-Mury C. The JNK pathway is a key mediator of Anopheles gambiae antiplasmodial immunity. PLoS Pathog. 2013 doi: 10.1371/journal.ppat.1003622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouagna L.C., Mulder B., Noubissi E., Tchuinkam T., Verhave J.P., Boudin C. The early sporogonic cycle of Plasmodium falciparum in laboratory- infected Anopheles gambiae: an estimation of parasite efficacy. Tropical Medicine and International Health. 1998. [DOI] [PubMed]
- Han Y.S., Thompson J., Kafatos F.C., Barillas-Mury C. Memorias Do Instituto Oswaldo Cruz; 2000. MC8 - Molecular Interactions between Anopheles stephensi Midgut Cells and Plasmodium Berghei: the Time Bomb Theory of Ookinete Invasion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen I.A., Attardo G.M., Rodriguez S.D., Drake L.L. Four-way regulation of mosquito yolk protein precursor genes by juvenile hormone-, ecdysone-, nutrient-, and insulin-like peptide signaling pathways. Front. Physiol. 2014 doi: 10.3389/fphys.2014.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillyer Julián F., Estévez-Lao T.Y. Nitric oxide is an essential component of the hemocyte-mediated mosquito immune response against bacteria. Dev. Comp. Immunol. 2010 doi: 10.1016/j.dci.2009.08.014. [DOI] [PubMed] [Google Scholar]
- Hillyer Julián F., Schmidt S.L., Christensen B.M. Hemocyte-mediated phagocytosis and melanization in the mosquito Armigeres subalbatus following immune challenge by bacteria. Cell Tissue Res. 2003 doi: 10.1007/s00441-003-0744-y. [DOI] [PubMed] [Google Scholar]
- Hillyer Jullán F., Schmidt S.L., Christensen B.M. Rapid phagocytosis and melanization of bacteria and Plasmodium sporozoites by hemocytes of the mosquito Aedes aegypti. J. Parasitol. 2003 doi: 10.1645/0022-3395(2003)089[0062:RPAMOB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Jaramillo-Gutierrez G., Rodrigues J., Ndikuyeze G., Povelones M., Molina-Cruz A., Barillas-Mury C. Mosquito immune responses and compatibility between Plasmodium parasites and anopheline mosquitoes. BMC Microbiol. 2009 doi: 10.1186/1471-2180-9-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariu T., Yuda M., Yano K., Chinzei Y. MAEBL is essential for malarial sporozoite infection of the mosquito salivary gland. J. Exp. Med. 2002 doi: 10.1084/jem.20011876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J.G., Vernicks K.D., Hillyer J.F. Members of the salivary gland surface protein (SGS) family are major immunogenic components of mosquito saliva. J. Biol. Chem. 2011 doi: 10.1074/jbc.M111.280552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoza V.A., Martin D., Mienaltowski M.J., Ahmed A., Morton C.M., Raikhel A.S. Transcriptional regulation of the mosquito vitellogenin gene via a blood meal-triggered cascade. Gene. 2001 doi: 10.1016/S0378-1119(01)00602-3. [DOI] [PubMed] [Google Scholar]
- Kuehn A., Pradel G. The coming-out of malaria gametocytes. J. Biomed. Biotechnol. 2010 doi: 10.1155/2010/976827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Gupta L., Yeon S.H., Barillas-Mury C. Inducible peroxidases mediate nitration of Anopheles midgut cells undergoing apoptosis in response to Plasmodium invasion. J. Biol. Chem. 2004 doi: 10.1074/jbc.M409905200. [DOI] [PubMed] [Google Scholar]
- Kwon H., Smith R.C. Chemical depletion of phagocytic immune cells reveals dual roles of mosquito hemocytes in Anopheles gambiae anti-Plasmodium 2 immunity 3. 2018. [DOI] [PMC free article] [PubMed]
- Lacroix R., Mukabana W.R., Gouagna L.C., Koella J.C. Malaria infection increases attractiveness of humans to mosquitoes. PLoS Biol. 2005 doi: 10.1371/journal.pbio.0030298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Dong Y., Huang Y., Rasgon J.L., Agre P. Impact of trehalose transporter knockdown on Anopheles gambiae stress adaptation and susceptibility to Plasmodium falciparum infection. Proc. Natl. Acad. Sci. Unit. States Am. 2013 doi: 10.1073/pnas.1316709110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolajczak S.A., Silva-Rivera H., Peng X., et al. Distinct malaria parasite sporozoites reveal transcriptional changes that cause differential tissue infection competence in the mosquito vector and mammalian host. Mol. Cell Biol. 2008;28(20):6196–6207. doi: 10.1128/MCB.00553-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Cruz A., DeJong R.J., Ortega C., Haile A., Abban E., Rodrigues J., Barillas-Mury C. Some strains of Plasmodium falciparum, a human malaria parasite, evade the complement-like system of Anopheles gambiae mosquitoes. Proc. Natl. Acad. Sci. Unit. States Am. 2012 doi: 10.1073/pnas.1121183109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller A.K., Kohlhepp F., Hammerschmidt C., Michel K. Invasion of mosquito salivary glands by malaria parasites: prerequisites and defense strategies. Int. J. Parasitol. 2010 doi: 10.1016/j.ijpara.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musiime A.K., Okoth J., Conrad M., et al. Is that a real oocyst? Insectary establishment and identification of Plasmodium falciparum oocysts in midguts of Anopheles mosquitoes fed on infected human blood in Tororo, Uganda. Malar. J. 2019;18:287. doi: 10.1186/s12936-019-2922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomares M., Dalmasso C., Bonnet E., et al. Systematic analysis of TruSeq, SMARTer and SMARTer Ultra-Low RNA-seq kits for standard, low and ultra-low quantity samples. Sci. Rep. 2019;9:7550. doi: 10.1038/s41598-019-43983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramphul U.N., Garver L.S., Molina-Cruz A., Canepa G.E., Barillas-Mury C. Plasmodium falciparum evades mosquito immunity by disrupting JNK-mediated apoptosis of invaded midgut cells. Proc. Natl. Acad. Sci. Unit. States Am. 2015 doi: 10.1073/pnas.1423586112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A., Adapa S.R., Zhang M., Liao X., Saxena V., Goffe R., Adams J.H. Unraveling the Plasmodium vivax sporozoite transcriptional journey from mosquito vector to human host. Sci. Rep. 2018 doi: 10.1038/s41598-018-30713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Montagna G.N., Matuschewski K. Plasmodium berghei sporozoites acquire virulence and immunogenicity during mosquito hemocoel transit. Infection and Immunity Feb. 2014;82(3):1164–1172. doi: 10.1128/IAI.00758-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R. Discovery of the cellular and molecular basis of cholesterol control. Proc. Natl. Acad. Sci. Unit. States Am. 2013;110(37):14833–14836. doi: 10.1073/pnas.1312967110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz A., Koella J.C. Trade-offs, conflicts of interest and manipulation in Plasmodium-mosquito interactions. Trends Parasitol. 2001 doi: 10.1016/S1471-4922(00)01945-0. [DOI] [PubMed] [Google Scholar]
- Sharma P., Sharma S., Mishra A.K., Thomas T., Das De T., Rohilla S.L., Dixit R. Unraveling dual feeding associated molecular complexity of salivary glands in the mosquito Anopheles culicifacies. Biology Open. 2015 doi: 10.1242/bio.012294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Rani J., Rawal C., Seena Y. Altered gut microbiota and immunity defines Plasmodium vivax survival in Anopheles stephensi. Front. Immunol. 2020 doi: 10.3389/fimmu.2020.00609. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea-Donohue T., Qin B., Smith A. Parasites, nutrition, immune responses and biology of metabolic tissues. Parasite Immunol. 2017 doi: 10.1111/pim.12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiao S.H., Whitten M.M.A., Zachary D., Hoffmann J.A., Levashina E.A. Fz2 and Cdc 42 mediate melanization and actin polymerization but are dispensable for Plasmodium killing in the mosquito midgut. PLoS Pathog. 2006 doi: 10.1371/journal.ppat.0020133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla E., Thorat L.J., Nath B.B., Gaikwad S.M. Insect trehalase: physiological significance and potential applications. Glycobiology. 2015 doi: 10.1093/glycob/cwu125. [DOI] [PubMed] [Google Scholar]
- Simões M.L., Mlambo G., Tripathi A., Dong Y., Dimopoulos G. Immune regulation of Plasmodium is Anopheles species specific and infection intensity dependent. 2017. MBio. [DOI] [PMC free article] [PubMed]
- Smith R.C., King J.G., Tao D., Zeleznik O.A., Brando C., Thallinger G.G., Dinglasan R.R. Molecular profiling of phagocytic immune cells in Anopheles gambiae reveals integral roles for hemocytes in mosquito innate immunity. Mol. Cell. Proteomics. 2016 doi: 10.1074/mcp.M116.060723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T., De D.T., Sharma P., Sharma S., Lata S., Saraswat P., Pandey K.C., Dixit R. Hemocytome: deep sequencing analysis of mosquito blood cells in Indian malarial vector Anopheles stephensi. Gene. 2016 doi: 10.1016/j.gene.2016.02.031. [DOI] [PubMed] [Google Scholar]
- Talman A.M., Domarle O., McKenzie F.E., Ariey F., Robert V. Gametocytogenesis: the puberty of Plasmodium falciparum. Malar. J. 2004 doi: 10.1186/1475-2875-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderberg J.P. Development of infectivity by the Plasmodium berghei sporozoite. J. Parasitol. 1975;61(1):43–50. PMID: 1090717. [PubMed] [Google Scholar]
- Vézilier J., Nicot A., Gandon S., Rivero A. Proceedings of the Royal Society B: Biological Sciences. 2012. Plasmodium infection decreases fecundity and increases survival of mosquitoes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt M.B., Lahon A., Arya R.P., Kneubehl A.R., Spencer Clinton J.L., Paust S., Rico-Hesse R. Mosquito saliva alone has profound effects on the human immune system. PLoS Neglected Tropical Diseases. 2018 doi: 10.1371/journal.pntd.0006439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Luan J., Li J., et al. De novo characterization of a whitefly transcriptome and analysis of its gene expression during development. BMC Genom. 2010;11:400. doi: 10.1186/1471-2164-11-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Schleicher T.R., Dong T., Bong Park H., Lan J., Carwford J., Dimopoulos G. Disruption of mosGILT in Anopheles gambiae impairs ovarian development and Plasmodium infection. J. Exp. Med. 2019 doi: 10.1084/jem.20190682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.