Abstract
Tumor mass and its microenvironment alter host immune system in various ways to promote tumor growth. One of the modifications is evasion of immune surveillance by augmenting the number of Tregs in tumor vicinity. Elevated levels of Tregs are seen in peripheral circulation and tumor tissue of cancer patients. Cancer cells release several chemokines to attract Tregs in tumor-site. Infiltration of Tregs has clinical significance because being immunosuppressive infiltrating Tregs suppress other immune cells making the tumor microenvironment favorable for tumor growth. On the other hand, infiltrating Tregs show metabolic alteration in tumor microenvironment which allows their selective survival over the others. Persistence of Tregs in the tumor microenvironment and subsequent immunosuppression makes Tregs a potential therapeutic obstacle and the reason behind the failure of immunotherapy. In this review, we emphasize the recent development in the metabolic adaptation of tumor-infiltrating Tregs and the therapeutic approaches to boost immunity against cancer.
Keywords: T-regulatory cell, Metabolism, Immune-suppression, Anti-Tumor immunity, Tumor microenvironment
Abbreviations: Treg, T-regulatory cell; TME, Tumor microenvironment; CCR/CXCR, Chemokine receptor; CCL/CXCL, Chemokine ligand; NK cell, Natural killer cell; FOXP3, Forkhead box P3; TGFβ, Transforming growth factor-beta; IL, Interleukin
Graphical abstract
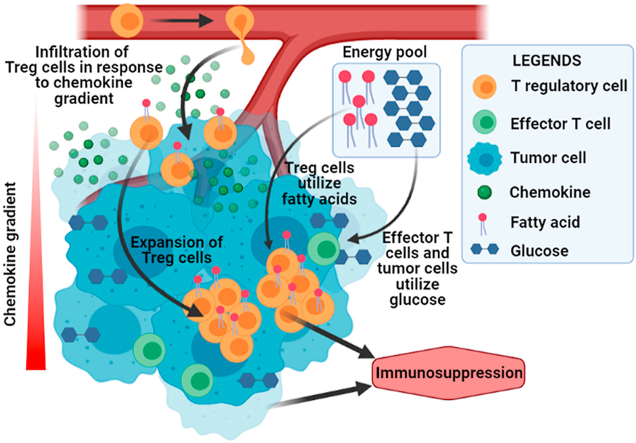
In tumor microenvironment, cancer cells show increased uptake of available glucose and metabolically change the environment. Effector T cells often cannot adapt to the glucose-depleted metabolic environment created by cancer cells and thus fail to survive. In the contrary, immunosuppressive Tregs which are infiltrated in the tumor by the tumor-derived chemokine gradient survive this inhospitable environment and facilitate tumor immune-evasion. The Tregs adapt to the glucose-depleted tumor microenvironment by shifting their metabolic preferences from glucose to fatty acid (Created in BioRender.com).
1. Introduction
Successful elimination of foreign pathogens depends upon the multifaceted interaction of innate and adaptive immune system in our body. The immune surveillance is also necessary to identify cancerous outgrowths in the host body (Smyth et al., 2001). But how tumor develops despite this immune surveillance mechanism has been a mystery till date. The relationship between immune system and tumorigenesis has long been established (Khong and Restifo, 2002; Pardoll, 2003). Tumorigenesis is an intricate process, consisting of three stages: (i) Elimination, (ii) Equilibrium and (iii) Escape; each step involves modification and alteration of immune system in such a way that favours tumor development. Tumor microenvironment (TME) plays a significant role in every step of tumorigenesis. During elimination phase antigen-processing cells (dendritic cells, macrophages etc.) recognize cancer antigens and eradicate emerging tumor cells thus protecting host against tumor. In equilibrium phase tumor cells and immune cells enter into a vigorous equilibrium. T cell mediated defence creates the barrier against excessive tumor cell proliferation during this stage. In escape phase cancer cells that escape from immune selection pressure develop into highly metastatic and invasive tumors by avoiding the immune surveillance (Dunn et al., 2002, 2004). Out of the various subsets of T cells, CD4+CD25+FOXP3+ T-regulatory cells (Tregs) are the key players that play vital role in this immune escape of tumor. Tregs are necessary for the orientation of T cell tolerance, which is one of the mechanisms of tumor-immune evasion (Elkord et al., 2010). Tregs are found in elevated amount in the peripheral circulation of cancer patients and a high density of Tregs is correlated with reduced survival of the patient. Being immunosuppressive, Tregs suppress effector T cell and NK cell responses, meddling with the immunity against tumor (Chaudhary and Elkord, 2016). To make the situation worse, Tregs infiltrate to the tumor-site and creates a tolerogenic microenvironment in the vicinity of tumor. Tumor-infiltration of Treg is facilitated by the chemokine gradient secreted from the growing tumor mass (Ondondo et al., 2013). The nutrients available in tumor microenvironment are almost totally used up by cancer cells for their proliferation and growth and as a result the tumor-infiltrating immune cells become deprived of the nutrients. This deprivation of nutrients as well as the presence of various metabolic by-products of cancer cells like lactate affects the function of the immune cells. The immune cells, most importantly the T cells are highly affected by this acidic tumor microenvironment in a way that favors tumor progression. Tumor-infiltrating Tregs show altered metabolic activity in TME compared to other effector T cells in order to survive. The collective result of infiltration and metabolic alteration allows rapid generation and sustenance of Tregs and makes the TME more tolerogenic.
Metabolic adaptation of infiltrated Tregs is a growing field of research and it has achieved clinical importance over time. In this manuscript, we focus on the recent development in the areas of Treg infiltration, their metabolic adaptations in different human malignancies, and the therapeutics targeting them.
2. Developmental origin of tumor Tregs
Tumor microenvironment is the surrounding non-tumor tissue and is made up of different heterogenic population of cells like lymphatic cells, immune cells, inflammatory cells, blood vessels, fibroblasts, extracellular matrix and the signaling molecules. Growing tumor mass constantly interacts with the TME for the availability of nutrients and growth signals. Immune cells share a major portion of TME. Among them T-regulatory cells is one of the crucial determinants of tumor growth. Tregs are a subset of T cells that mediate peripheral tolerance. Tregs are potent immune suppressors, playing important roles in autoimmunity and transplantation tolerance. There are two types of Tregs. Natural Tregs (nTregs), develop in the thymus by stimulation of self-antigens. On the other hand, Tregs can also develop from naïve T cells during specific stimulation in peripheral circulation. These Tregs are known as peripherally induced Tregs (pTregs) (Chen et al., 2003; Zheng et al., 2002). pTregs phenotypically resemble nTregs, and both of them have similar phenotypic characteristics (Lin et al., 2013a). Tregs suppress the immune response through different mechanisms. X-chromosome encoded transcription factor FOXP3 (Forkhead box P3) and its associated protein partners regulate the diverse functions of Tregs (Hori et al., 2003). The lineage-specification factor FOXP3 is essential for Treg functioning, which accomplishes its multiple actions through transcriptional regulation of its target genes. TGFβ signaling pathway is essential for FOXP3 expression which generates both nTregs and pTregs (Chen et al., 2003). IL2R (CD25) also plays a significant role in the generation of FOXP3+ Tregs. Various TME components also generate Tregs within the tumor-site (Chaudhary and Elkord, 2016). Tregs are essential for suppression of autoimmunity but this feature can be damaging in case of tumor condition. Numerous scientific discoveries suggest that both nTregs and pTregs are found to be elevated in various solid tumor tissues and tumor-infiltration depends upon the interaction of chemokine receptors of Tregs and tumor-secreted chemokines (Lin et al., 2013b; Hanahan and Coussens, 2012). Selective buildup of Tregs in TME can occur by two basic mechanisms; firstly by infiltration from peripheral circulation and secondly by the sustenance of these infiltrated Tregs in the TME. Tregs obtain their nourishment for growth by altered metabolic activity in the TME. Reformed metabolism of Tregs causes survival of them compared to other immune cells. All these eventually lead to the accumulation of immunosuppressive Tregs in the TME and subsequent tumor proliferation.
3. Tumor-derived chemokine gradient promotes Treg accumulation in TME
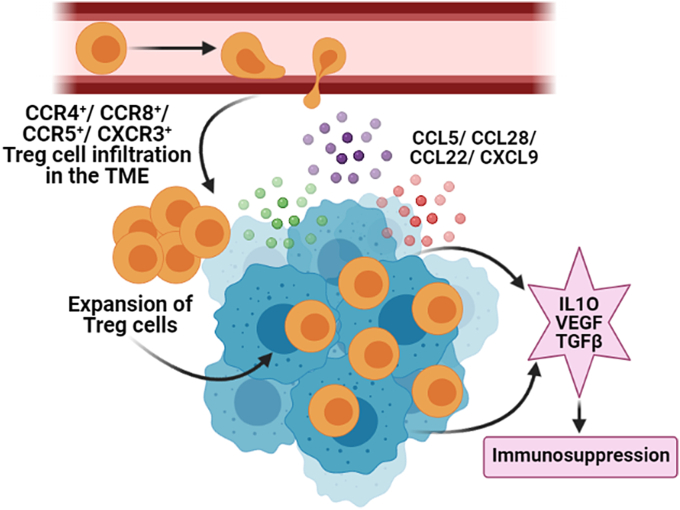
The selective buildup of Tregs within tumor tissue suggests that Tregs are preferentially recruited and infiltrated within the TME (Mailloux and Young, 2010). The mechanism of Tregs infiltration likely involves the interaction between chemokines and chemokine receptors. Chemokines are chemotactic cytokines that coordinate locomotion of different lymphocyte cell populations. Movement of immune cells in different cellular compartments is directed by the differential expression pattern of chemokines. Chemokines recruit leukocytes into the tumor microenvironment which ultimately determine the cancer progression (Fig. 1). This tumor-infiltration of leukocytes is multidimensional and consists of both pro-tumorigenic and anti-tumorigenic activities. Highly Treg infiltrated breast tumor has poor prognosis, so strategies to regulate Treg infiltration are widely scrutinized these days.
Fig. 1.
Tregs get recruited in tumor microenvironment by the interaction between chemokines and chemokine receptors: Tumor cells and tumor-associated cells-secreted chemokines attract Tregs. These immunosuppressive Tregs inhibits the activity of effector T cells and facilitates tumor growth. Chemokines and chemokine receptor-axis that are majorly involved in this chemotaxis of Tregs are CCL28-CCR10, CCL5-CCR5, CCL22-CCR4, and CXCL9/10/11-CXCR3. Effector T cells fail to infiltrate in the TME as they do not express these chemokine receptors. Even if effector T cells infiltrate in the TME they do not show immunogenicity due to high level of Tregs in the TME and their immunosuppressive effect (Created in BioRender.com).
The chemokine superfamily involves approximately 50 chemokine ligands and are divided into four subfamilies (CC, CXC, CX3C and XC) based on the position of the first two N-terminal cysteine residues. Likewise, chemokine receptors fall under the family of G protein–coupled seven-transmembrane signaling receptors (GPCRs) and till date approximately 20 chemokine receptors have been discovered. The role of chemokines and chemokine receptors has been widely studied in different human cancers. As studied extensively, Tregs express a variety of chemokine receptors depending upon the spatial and temporal nature of its suppressive action (Hanahan and Coussens, 2012; Mailloux and Young, 2010). During tumor-infiltration Tregs express CCR2-9, CXCR3/4 chemokine receptors and infiltrate in response to tumor-derived chemokines (CCL17, CCL22, CCL1, CCL28, and CCL9/10/11). Tumor cells release these chemokines to summon Tregs and make the TME favorable for its growth. It is also noted that the major interaction is facilitated by CCR4‐CCL17/22, CCR8‐ CCL1, CCR10‐CCL28, and CXCR3‐CCL9/10/11 axes and the underlying mechanism has been discussed below in breast cancer and several human malignancies (Table 1).
Table 1.
List of chemokine networks that play crucial role in the tumor-infiltration of Tregs: Major interaction is facilitated by CCR10‐CCL28, CCR5-CCL5/8, CXCR4-CXCL12, CCR4‐CCL17/22, CCR6-CCL20, CCR8‐CCL1, and CXCR3/CCR5‐CCL9/10/11 axes which are responsible for the tumor-infiltration of Tregs.
| Chemokine receptor on Treg | Tumor-derived chemokines | Tumor-type |
|---|---|---|
| CCR10 | CCL28 | Ovarian cancer, liver cancer (Facciabene et al., 2011) |
| CCR5 | CCL5/8 | Pancreatic ductal adenocarcinoma, mice metastatic tumor, skin squamous cell carcinoma, colorectal cancer (Halvorsen et al., 2016, Wang et al., 2017) |
| CXCR4 | CXCL12 | Basal-like breast cancer (Kryczek et al., 2005, Yan et al., 2011) |
| CCR4 | CCL22/17 | Lymphomas, lung, breast, ovarian, gastric, prostate cancer, esophageal squamous cell carcinoma (Gobert et al., 2009; Faget et al., 2011; Mizukami et al., 2008) |
| CCR6 | CCL20 | Colorectal cancer (Liu et al., 2011) |
| CCR8 | CCL1 | Breast cancer (Kuehnemuth et al., 2018) |
| CXCR3/CCR5 | CXCL9/10/11 | Sporadic human renal cell carcinoma, breast, ovarian, colorectal, hepatocellular carcinoma (Redjimi et al., 2012) |
Hypoxic tumor microenvironment not only promotes angiogenesis but also causes the expression of CCL28 in ovarian cancer and liver cancer, which promotes the infiltration of CCR10+ Tregs (Facciabene et al., 2011). Infiltrated CCR10+ Tregs secrete vascular endothelial growth factor A (VEGFA), promoting neo-angiogenesis, and metastasis at distant sites. Although FOXP3 is the master-regulator of Tregs, it is sometimes expressed by pancreatic ductal adenocarcinoma (PDAC) cells and secretes CCL5 to recruit CCR5+ Tregs in the tumor (Wang et al., 2017). Hypoxia also causes the recruitment of CCR5+ Tregs through CCL8/CCR5 signaling in mice metastatic tumors (Halvorsen et al., 2016). CCR5 is also responsible for recruitment of Tregs in skin squamous cell carcinoma (SSC) and colorectal cancer (CRC) (Ward et al., 2015). Basal-like breast cancer cells increase expression of CXCL12 during hypoxic environment which causes the infiltration of CXCR4+FOXP3+ Tregs (Kryczek et al., 2005, Yan et al., 2011). The picture that emerges from these reports is that hypoxia and subsequent angiogenesis drives the accumulation of CCR10/CXCR4/CCR5+ Tregs. Tumor-infiltrated Tregs in-turn secretes VEGFA which exert tumor proliferation. Reports suggest that accumulation of CCR10/CXCR4/CCR5+ Tregs in tumor tissue has been the reason for the immune evasion mechanism of growing tumor.
The CCL17/22-CCR4 axis is the requisite factor for recruitment of Tregs into lymphomas, lung, breast, ovarian, gastric and prostate cancers (Gobert et al., 2009; Faget et al., 2011; Moore et al., 2020). Augmented levels of CCL17 and/or CCL22 are known as poor prognostic markers in breast cancer. The same has been reported in cerebrospinal fluid of patients with lymphomatous and carcinomatous meningitis (Haas et al., 2008), gastric (Mizukami et al., 2008), and esophageal squamous cell carcinomas (Maruyama et al., 2010) because it causes infiltration of immunosuppressive Tregs. CCR4 is known to accumulate Tregs in mouse melanoma model. CCL22 is secreted by ovarian tumor cells and tumor-associated macrophages which attracts CCR4+ Tregs in tumor microenvironment from lymph nodes resulting in the destruction of anticancer immunity (Curiel et al., 2004). Therefore, tumor-derived CCL22 mediates tumor trafficking of Tregs. On the other hand, tumors that express little or no CCL22 are not infiltrated by Tregs, suggesting that Treg recruitment to the tumor occurs via the CCL22:CCR4 axis. Different tumor-associated cells (macrophages, dendritic cells, fibroblasts, myeloid-derived suppressor cells) help in the recruitment of Tregs by releasing cytokines and chemokines. For example, Tumor-associated macrophages (TAMs) release CCL20 which recruits CCR6+ Tregs in colorectal cancer (Liu et al., 2011). It should be noted that Treg infiltration can sometimes lead to a positive prognosis by monitoring inflammation associated with neoplastic transformation in some cancers, including colorectal and gastric cancers. CCL1 is shown to have a profound role in the accumulation of Tregs in breast tumor milieu as CCL1 is the functional ligand of CCR8. CCL1 remains up-regulated in breast cancer and it is positively correlated with CCR8+ Treg infiltration. CCL1 is secreted by some myeloid cells in TME that attract CCR8+ Tregs. CCR8+ Tregs express surface marker CD39 which causes ATP‐adenosine metabolism. They also release anti-inflammatory cytokine IL10 and apoptosis inducer granzyme-B. These features help Tregs to defeat effector T cell functions. High infiltration of CCR8+FOXP3+ Tregs is associated with poor prognosis and CCL1 may act as a therapeutic target in breast cancer (Kuehnemuth et al., 2018). Sporadic human renal cell carcinomas have increased expression of CXCL9, CXCL10, and CXCL11 which attracts CXCR3+CCR5+ T cells. On the other hand, human breast, ovarian, colorectal, and hepatocellular carcinomas recruit CXCR3+FOXP3+ Tregs in TME (Redjimi et al., 2012).
4. Altered energy expenditure determines the survival of Tregs in TME
4.1. Immunometabolism: integration of metabolism of immune cells with tumor environment
An emerging field “immunometabolism” indicates that there is a distinct relation between metabolic pathways and immune system (Smyth et al., 2001). Recent studies are trying to find out how immune cells perform their job according to their metabolism and how these metabolic pathways help in shaping the phenotype and activation of immune cells. Immunotherapy is considered to be effective even against drug-resistant cancers. And combining metabolism with immunotherapy targets the energy production required for the development and function of immune cells (Smyth et al., 2001; Kaymak et al., 2021). The efficacy of immunotherapies can be greatly increased by limiting metabolic competition in the tumor microenvironment (Zappasodi et al., 2021). Developing a therapeutic regimen combining conventional chemotherapy along with metabolic targets that can reduce the tumor-associated immune suppression might lead to a more robust immune system after the chemotherapy. This might be an important step towards dealing with the problem of cancer relapse since the immune system can recognize and eliminate resistant malignant cells that have survived chemotherapy (Wang et al., 2020).
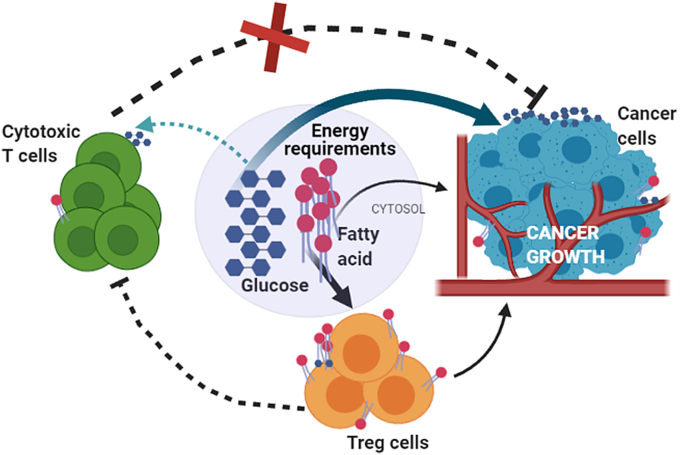
To mount any immune responses, cells need to rapidly propagate and produce cytokine and this involves different bio-energetic processes (DeBerardinis et al., 2007). Cell metabolism controls the signaling pathways that are involved in regulating immune cell activities. The pathways of glycolysis, tri-carboxylic acid (TCA) cycle and oxidative phosphorylation (OXPHOS) are interconnected to meet the energy requirements of these cells. Depending on oxygen availability, the pyruvate either generates CO2 and H2O or lactic acid. As tumor grows, the tumor cells take up most of the glucose available in that environment and generate lactate which gets accumulated in extracellular tissue. These lactate rich environments become non-suitable for the effector T cells, but Tregs by some means manage to survive in this inhospitable environment. Recent findings have suggested that the metabolic fitness of T cells can be achieved by modulating CD28 signaling as well as by blocking CTLA4 that in a way promotes the infiltration of immune cells, especially in glycolysis-low tumors (Zappasodi et al., 2021). Targeting Treg metabolism is currently of high-therapeutic interest (Parlo and Coleman, 1986) because these cells play an important role in maintaining immune tolerance to self-tissues, but become harmful to the body in case of tumor development. Glucose is the primary fuel source for most of the cells and T cells are no exception. They depend on glucose for the generation of ATP (Lu et al., 2014). The resting T cells which include naïve T cells, memory and anergic T cells have a low metabolic requirement and to meet that requirement they retain a metabolic steadiness that supports only basal energy production over biosynthesis (Chandran et al., 2015). Mitochondrial oxidative phosphorylation (OXPHOS) pathway generates almost 96% of the required ATP for naïve T cells (Bailey et al., 2014). Whereas, upon stimulation, the activated T cells divide and differentiate to generate various T cell subsets and each subset needs unique metabolic pathways for their energetics (Chang et al., 2013). To proliferate, these cells entail high amount of ATP and that requirement is fulfilled by shifting their metabolic preferences from catabolic mitochondrial OXPHOS to anabolic pathways like glycolysis. Although OXPHOS is more efficient than aerobic glycolysis in generating ATP per molecule of glucose, activated T cells and rapidly growing cancer cells utilize this pathway for their energetics which is known as “Warburg effect” (Wang et al., 2011; Zeng and Chi, 2014). Once differentiated, the different subsets of T cells exploit discrete biosynthetic and energetic pathways to sustain the functionality (Frauwirth et al., 2002; Frauwirth and Thompson, 2004). Recent studies specified that different subsets of T cell do not require identical metabolic programming. CD4+ effector T cells like, Th1, Th2, Th17, depends on aerobic glycolysis whereas the pTregs, which are predominantly effector Tregs have been shown to be less dependent on aerobic glycolysis and are different from the resting Tregs and their energy requirements (Carr et al., 2010; Sinclair et al., 2013). The tumor microenvironment is hostile for effector T cells whereas pTregs can survive in that situation and suppress the function of effector T cells and therefore help in the progression of the tumor (Fig. 2).
Fig. 2.
Overview of nutrient usage by different immune cells and cancer cells in tumor microenvironment: Rapidly growing tumor cells use most of the available glucose to proliferate and this makes the nutrient availability to effector T cells restricted. Tregs in tumor microenvironment use available fatty acids for their sustainability and growth and as a result further helps in tumor progression (Created in BioRender.com).
There are many studies that have already shown that Tregs use fatty acids as preferential metabolic substrates. Tregs favorably avoid glucose metabolism and that has a great functional importance and that might be mediated by CTLA4 overexpression as shown in the studies by Zappasodi et al. According to their studies, the CTLA4 overexpression blocks CD28 signaling towards glucose utilization and thus ensures the functional stability of Tregs (Zappasodi et al., 2021). In studies involving triple negative breast cancer cells when the utilization of fatty acid was hindered using etomoxir, a fatty acid uptake inhibitor, the maintenance of Tregs in tumor got affected as well as the tumor load got reduced. Understanding the metabolism of Tregs in tumor microenvironment will give the indication to manipulate these immunosuppressive pro-tumor Tregs to check tumor progression.
4.2. Distinct metabolic changes of immune cells in breast tumor condition
Breast cancer, a leading cause of death among women can be classified into Normal-like, Luminal A, Luminal B, HER2-enriched and Basal-like subtypes. High glycolytic rate in triple negative breast cancer are shown to promote macrophage inducing factors and in a way activate their pro-tumorigenic activities. All these factors help in the progression of the tumor in a glycolysis-dependent way. Mostly glycolysis related genes are highly expressed in breast cancer compared to other tissues (Chang et al., 2013). These glycolytic genes have direct positive impact on genes involved in immunological/inflammation functions of immune cells (Chang et al., 2015). As well as in breast cancer studies, it has been found that high glycolysis in these tumor conditions result in lower infiltration of immune cells such as NKT in tumor-sites. Rather they result in higher expression of immune checkpoints like CTLA4, FOXP3. In other words, the enhanced glycolytic activity associated with immune cells residing in breast cancer conditions results in pro tumorigenic immunity. These metabolic viewpoints of immune cells in breast cancer need to be analyzed thoroughly to get an idea of a way to manipulate these immune cells in breast tumor conditions so that we can generate a metabolic approach along with therapeutic approach to suppress the tumor growth.
4.3. Metabolic shifts of Tregs in tumor microenvironment
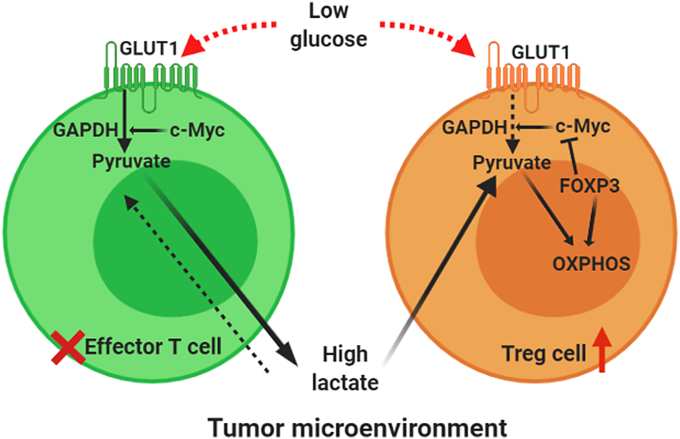
The extensive studies by Angelin et al., stated that the transcription factor FOXP3 plays an important role in modulating the metabolism of tumor-infiltrating CD4+CD25+ Tregs to survive and function in lactate rich environment (Angelin et al., 2017). Tregs are metabolically flexible and can withstand the high-lactate condition in the TME as evident from the works of Watson et al. (2021). Increased expression of FOXP3 results in inhibition of aerobic glycolysis and redirects the cellular metabolism to OXPHOS (Asano et al., 1996; Gerriets et al., 2016). In activated effector T cells c-Myc gets highly expressed and supports the glycolytic pathway whereas in Tregs FOXP3 blocks the expression of c-Myc and this can be one possible mechanism of this metabolic shift. Angelin et al. showed that the Complex 1 of electron transport chain (ETC) makes the Tregs capable of suppressing the proliferation of effector T cells (Angelin et al., 2017). In tumor microenvironment, in low glucose condition, Tregs oxidize NADH to NAD+ through the coupled action of ETC and TCA cycle. Whereas, effector T cells, in low glucose, high lactate condition face a redox imbalance as they solely depend on aerobic glycolysis to regenerate NAD+. And in absence of sufficient NAD+, GAPDH, a key regulator of glycolysis cannot function. And as a result, the effector T cells which are dependent on glucose for their growth and maintenance cannot survive in low glucose lactate rich tumor microenvironment whereas Tregs can easily sustain in that condition (Fig. 3).
Fig. 3.
Overview of metabolism of effector T cells and the reprogrammed metabolism of tumor-infiltrating Tregs: Under normal condition, activated effector T cells highly express GLUT1 (glucose transporter) and c-Myc which supports the glycolytic pathway through which these effector cells survive in normal conditions. In TME this dependency on glycolysis becomes fatal for effector T cells as in the high lactate low glucose tumor microenvironment, they face a redox imbalance and thus a key regulatory enzyme of glycolysis, GAPDH can not function. Whereas in Tregs FOXP3 blocks the expression of c-Myc and this can be one possible mechanism of this metabolic shift from glucose to fatty acid dependency. Tumor-infiltrating Tregs oxidize NADH to NAD+ through the coupled action of ETC and TCA cycle. And as a result, the effector T cells which are dependent on glucose for their growth and maintenance cannot survive in low glucose lactate rich tumor microenvironment. (Created in BioRender.com).
5. Suppressive activity of tumor-infiltrating Tregs
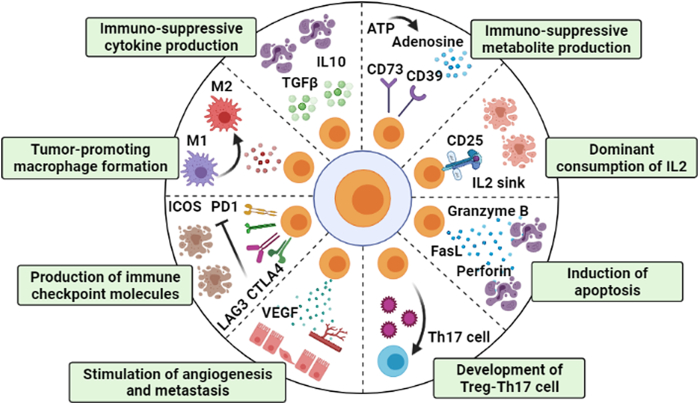
The critical balance between effector T cells versus Tregs is the most crucial factor that ultimately determines the progression of cancer. Tregs are the key players of the mechanisms of tumor-immune evasion (Motz and Coukos, 2013). Infiltrated Tregs suppress the action of tumor-specific effector T cells, promote immune evasion and develop a pro-tumorigenic TME and it has been seen in breast, ovarian, gastric, colorectal and pancreatic cancer (Pandiyan et al., 2007). There are several circumstances, by which Tregs increase their number and suppressive activity in the tumor milieu which are discussed below (Fig. 4).
Fig. 4.
Pictorial representation of suppressive activity of tumor-infiltrating Tregs: Tregs secrete immunosuppressive cytokines such as TGFβ, IL10 that transforms effector T cells into anergic T cells. Immunosuppressive CD39 and CD73 molecule makes the TME favorable for tumor progression. On the other hand, Tregs absorb most of the IL2 present in the TME by acting as IL2 sink, making IL2 less available to other effector T cells leading to their apoptosis. Tregs release apoptosis inducing molecules like FasL, granzyme, perforin. Development of immunosuppressive Treg-Th17 cell is also beneficial for tumor growth. Tregs stimulate metastasis and angiogenesis by several mechanisms. Checkpoint molecules like PD1, CTLA4, LAG3, ICOS production by Treg is also a sign for uncontrolled tumor progression. Tregs convert M1 macrophages (anti-tumorigenic) into M2 macrophages (pro-tumorigenic) which work with Treg to promote tumor growth (Created in BioRender.com).
Treg suppressive activity is dependent upon the master transcription factor FOXP3 and its associated protein complexes. Both nTreg and pTreg contributes to tolerogenic and immunosuppressive activity (Banerjee et al., 2013). Tregs inhibit the activation of CD4+ T cells, CD8+ T cells, NK cells, dendritic cells, promote tumor-associated M2 macrophage (TAMs) activity and induce the generation of suppressive Treg-Th17 cells. Treg induces effector T cell, NK cells, cytotoxic T lymphocytes (CTLs) and dendritic cells (DCs) apoptosis by secreting TNFα, perforin, granzyme A and B, Fas ligand. FOXP3 can perform both as transcriptional activator and repressor. FOXP3 activates the expression of inhibitory immune checkpoint molecules, such as, T-cell immunoglobulin and mucin-domain containing-3 (TIM- 3/HAVCR2), lymphocyte activation gene-3 (LAG-3), inducible T-cell co-stimulator (ICOS), programmed-death 1 (PD1), cytotoxic T-lymphocyte associated protein 4 (CTLA4), and glucocorticoid-induced TNFR family related gene (GITR); and T cell activation markers, CD25 and CD69 (Kakita et al., 2012; Schuler et al., 2012; Jie et al., 2013; Lin et al., 2013b; Han et al., 2014; Scurr et al., 2014). The functions of these proteins are to delay immune response and stop excessive T cell activation during physiological immune responses. LAG-3, TIM-3 and PD1 contribute to T cell cycle arrest and generation of immature APCs such as dendritic cells that are unable to induce effector immune responses against cancer (Nirschl and Drake, 2013; Buchbinder and Desai, 2016). LAG3 binds with MHC-II of DCs and prevents DC maturation and its effector function as APC. CTLA4 interacts with CD80 and CD86 ligand on DCs blocking the costimulatory protein CD28 and limits dendritic cell function. CTLA4+ Tregs also release Indoleamine 2,3-dioxygenase (IDO); which is a potent immunosuppressor and provides decreased costimulatory signal to DCs. Tregs isolated from the tissue of hepatocellular carcinoma (HCC), colorectal cancer (CRC) and pancreatic cancer patients show distinct expression pattern of CTLA4, ICOS, PD1, CD25 and CD69. Alternatively, the association of FOXP3 with NFATc2 down-regulates IL2 expression which causes scarcity of IL2 in tumor microenvironment and subsequent effector T cell death (Rudra et al., 2012; Rudensky et al., 2006). Tumor microenvironmental IL2 is consumed by Tregs through its receptor IL2Rα/CD25 that causes IL2 scarcity and induces BIM1-mediated apoptosis of adjacent effector cells (Sakaguchi et al., 2008, von Boehmer and Daniel, 2013).
A recent study has shown that FOXP3 acts as a co-transcription factor with STAT3 and up-regulates IL10 expression in pTregs (Bhattacharyya et al., 2010, Hossain et al., 2013, Hossain et al., 2015). Tregs-released IL10, IL35, TGFβ are vigorous anti-inflammatory and immune-suppressive cytokines that causes decreased activation of effector T cells, NK, NKT cells and APCs (Shevach, 2009; Lindau et al., 2013). Presence of high amounts of FOXP3+IL10+ Tregs in breast cancer patient may lead to adverse prognosis. IL10 is a potent immunosuppressive cytokine that suppresses the activity of Th17, Th1 cell. IL35 is an anti-inflammatory cytokine which blocks the development and function of Th1 and Th17 cells (Khong and Restifo, 2002). Treg-released IL35 suppresses inflammatory responses of effector immune cells which help in tumor proliferation. Prominent levels of IL35 have been found in several human tumors such as non-small cell lung cancer, breast cancer, acute myeloid leukemia, pancreatic ductal adenocarcinoma and colorectal cancer and are associated with poor survival of the patients (Wu et al., 2012; JCZhang et al., 2013; Jin et al., 2014). Tumor-associated cells like Plasmacytoid DCs, myeloid DCs and TAMs released TGFβ transform activated CD4+CD25+ T cells into FOXP3+ Tregs. Treg express ectonucleotidases CD39 and CD73 that hydrolyze exogenous ATP into AMP and immunosuppressive adenosine; both of them suppresses CD80 and CD86 costimulatory signals of DCs and makes them anergic. A high expression of CD39 has been observed in intra-tumoral Tregs of colon and head & neck cancers (HNC) (Sundström et al., 2016; Schuler et al., 2014; Dunne et al., 2016). NRP1 (neuropilin-1) is a non–tyrosine kinase receptor which is involved in tolerance-mediated responses, driving tumor growth. It is constitutively expressed on the surface of CD4+CD25+FOXP3+ Tregs during cancer condition and interacts with immature DCs and alters their functional activities (Bruder et al., 2004; Sarris et al., 2008). NRP1 restricts CD8+ T cell function and acts as an obstacle to the CD8+ T cell-mediated tumor immune surveillance. NRP1 is crucial for Treg suppressive nature in the TME because it acts as a coreceptor of VEGFR2 for VEGF ligands and Plexin for semaphorin (Sema) ligands, initiating numerous signaling pathways depending on the circumstances (Chuckran et al., 2020). Treg infiltration leads to secretion of VEGFA and subsequent angiogenesis in ovarian cancer and liver cancer. Latent TGFβ with the help of glycoprotein A repetitions predominant (GARP) gets anchored to the surface of T cells and the complex can be cleaved to release active TGFβ. Latency-associated peptide (LAP) binds with TGFβ in inactive latent TGFβ complexes and release active TGFβ. Highly suppressive LAP+ and GARP/LAP co-expressing Tregs are present in excess amount in the TILs of CRC patients and the peripheral blood of pancreatic, CRC and their suppressive activity is mediated by TGFβ and IL10 (Sun et al., 2012; Mahalingam et al., 2014; Abd Al Samid et al., 2016; Rifkin, 2005).
These tumor-infiltrating Tregs display heightened immunosuppressive activity in comparison with the Tregs isolated from peripheral blood (Bu et al., 2016; Strauss et al., 2007). This occurs because in TME Tregs are highly exposed to tumor-associated antigens (TAA) which causes their enhanced activation. Enhanced Treg proliferation and activation in TME have been seen in breast cancer and colorectal cancer patients. These Tregs express Ki67, a potent proliferation marker in comparison to Tregs from peripheral circulation.
6. Can targeting infiltration and metabolism of Tregs provide a better means of immunotherapy?
6.1. Targeting infiltration of Tregs in TME
Many treatment strategies these days focus on the ways to deplete Tregs from tumor microenvironment. Targeting chemokine receptors has become very famous in combinatorial immunotherapy along with chemotherapeutic drugs. Targeting CCL22 expressing tumor cells and TAMs by effector T cells can be a promising therapy so that CCR4+ Tregs fail to infiltrate in the tumor tissue thereby facilitating anticancer immunity. CCL22 epitopes can be added to cancer vaccines for this method. It has been seen that the signal sequence of CCL22 is the most probable epitope for its binding to the HLA-A2. This signal sequence is cleaved off before functional protein secretion. Hence, CCL22 expressing cells can be easily recognized by T cells even though CCL22 is secreted outside of the cell. Emerging studies have shown that using specific antibodies, antagonists, or siRNA, we can block CCL22/CCL17 – CCR4 axis, leading to decreased Treg accumulation in TME. Anti‐CCR4 mAb is very effective for depletion of Treg accumulation in TME and has been used in clinical trials in some solid tumor therapy; for example, breast cancer. On the other hand, anti-CCR4 mAb (Mogamulizumab) selectively depletes circulating CCR4+ Tregs in the adult T-cell leukemia-lymphoma patients (Moore et al., 2020). Blocking the interaction between chemokine and chemokine receptors has shown potential in immunotherapy. This way we can block Treg infiltration in the TME. For example, hypoxia makes the cancer cells express CCL28 that attracts CCR10+ Tregs. Intratumoral administration of anti‐CCR10 antibody can block the interaction between CCL28 and CCR10 and prevents Treg infiltration (Ohue and Nishikawa, 2019). CCR5 deficiency (CCR5−/− mice) or CCL5 blockade (using Met-RANTES) led to diminished Tregs numbers in TME and slow tumor growth (Kiss et al., 2009). CCR1 antagonist CCX721 inhibits metastasis in mouse Multiple Myeloma (MM) model (Dairaghi et al., 2012). CCR1 antagonist BL5923 inhibits liver metastasis in murine model of colon cancer. They are perfect candidates in combinatorial treatments. CCR2 inhibitor PF-04136309 along with chemotherapeutic drug Gemcitabine (GEM) has been used in preclinical model (Noel et al., 2020). Another CCR2 inhibitor, CCX872 along with anti-PD1 treatment has shown promising results in pancreatic tumors (Flores-Toro et al., 2020). CXCR4 antagonist AMD3100 (Plerixafor) is a promising therapeutic strategy in different preclinical data and clinical (Micallef et al., 2018). AMD3100 increases the efficiency of other treatments by inhibiting Treg infiltration and promotes anti-tumor T cell response. Selective blockade of CCL5 can be used to enhance antitumor immune response (Wang et al., 2017). Knockdown of CCR5 results in suppression of tumor growth and decreased infiltration of Tregs in the tumor microenvironment (Aldinucci and Casagrande, 2018). Disruption of CCR5/CCL5 signaling has also been shown in mouse models to reduce intra-tumoral Treg accumulation and slow tumor progression (Tan et al., 2009).
6.2. Preventing proliferation and survival of Tregs in TME by targeting metabolism
The tumor cells adapt their metabolism in a way so that they can sustain their growth in any condition and become resistant to any kind of treatments. The therapy resistant tumors have altered phenotype with respect to the naïve tumor cells. One of the immunotherapeutic approaches for treatment of pancreatic cancer targets the oncogenic KRASG12Dand the inactivation of this gene results in the regression of cancer (Viale et al., 2014). However, if KRASG12D is reactivated, the cancer reappears because some metabolically modified cancer cells endure in a quiescent state. These cells have improved respiration, reduced glycolysis, and when respiration is inhibited, they have an impaired ability to increase glycolysis. This renders existing cells susceptible to respiration inhibitors. Other studies are also there to suggest that mitochondrial respiration is empowering cells to bypass the therapies and thus indicating that targeting mitochondrial respiration may help in cancer regression (Navarro et al., 2016; Zhang et al., 2016).
Now to target metabolism for therapy, it is crucial to recognize the limiting metabolic processes sufficiently to target the process safely. Many chemotherapeutic drugs inhibit nucleotide metabolism. But literature proposes that tumors use various mechanisms to uphold nucleotide pools, and this may contribute to effectiveness. However, understanding differential sensitivity to metabolism-targeted chemotherapy in detail may help in using these therapies more effectively.
Apart from targeting cancer cells and their metabolism and infiltration, rejuvenation of the immune cells is a major goal for modern immunotherapy. A novel approach for immunotherapy is to generate functionally active tumor-specific effector T cells and memory T cells. The enhanced viability of memory T cells and their ability to differentiate upon re-exposure to cancer antigens will allow for long-lasting immune-mediated anti-tumor function. Since T cell differentiation and function highly depends on the metabolism, targeting metabolism of T cells with checkpoint inhibitors seems to be a promising indication that might shift the differentiation of tumor-specific T cells to generate potent effector T cells and memory T cells.
To combine metabolism-targeted therapies with immunotherapies we need to clearly understand communications between metabolism of cancer cells and immune cells. Cancer causes changes in the metabolism of whole-body that may influence tumor-nutrient availability. Cancer cells make the tumor microenvironment hypoxic and nutrient-deprived and these affect T cell and macrophage differentiation, raising the possibility that if tumor cell metabolisms are targeted it may promote anti-cancer immune responses. The excess use of available glucose by cancer cells restrict the availability of glucose to T cells and thus causes lymphocyte exhaustion (Chang et al., 2015; Ho et al., 2015). Thus, if glucose use by cancer cells can be decreased using therapies, it may make glucose available for T cells and promote immune-effector functions to further inhibit tumor growth. However, there lies the limitation that if the same therapies restrict the glucose metabolism of T cells, they may again lower anti-tumor immunity. Not only glucose, other nutrient levels also affect both immune and cancer cells. Modulating the diet to regulate the availability of amino acid can slow cancer growth.
To answer the ever more complex questions about how internal and external stimuli put together to create utilizable metabolic phenotypes in cancer, the metabolic preferences of cancer cells by tissue of origin, the communications between benign and malignant cells within the microenvironment, and effects of the diet and microbiome on the host need to be considered. Further investigations are required to explore the effect of diet on tumor growth and integrate the metabolism with therapeutic treatments.
6.3. Influence of environment and lifestyle factors on the functions of Tregs in cancer patients
There's a crucial link between diet and immune cells. How diet influences these immune cells remains largely unknown but recent evidence suggests a role for nutrients across the immune cell function as well as tumor progression. This means that the growth and maintenance of both immune cells and cancer cells can be affected by our dietary intake (Smith et al., 2013) i.e., what we eat—in particular, whether we have excess energy and nutrients. The regular diet not only affects these but also affect the microbiota of human body and as a whole exerts a great impact on cancer development and during cancer progression, there occurs a metabolic interaction between microbes and immune cells and that may shift the supporting microbiota to the threatening one, as microbes begin interacting with cancer cells rather than healthy human cells (Smith et al., 2013).
Not only diet but also there are other factors like the effect of individual's overall lifestyle, environmental, microbial and pathogenic exposures that affects the growth and function of immune cells. The Molecular pathological epidemiology (MPE) can generate data regarding how environmental, dietary, lifestyle, and microbial exposures influence host–tumor interactions (Herrington et al., 2019). It is increasingly becoming clear that nutrition, metabolic state, gut-microbiota, and the functions of Tregs are deeply interconnected. MPE which is an interdisciplinary study deals with the interaction of these factors and has been explored in breast, lung, prostate, and colorectal cancers.
Some studies have already linked obesity and cancer threats while stating the fact that, in addition to genetic factors, high caloric intake in the form of processed food enriched in protein, sugar, fat and salt, is extensively thought to contribute to the imbalance of Treg components. Dietary nutrients and microbial metabolites can influence the disease outcome by altering the anti-tumor immune response. Common dietary components like gluten, vitamins A and D, omega-3, omega-6 and probiotics are now widely accepted to be essential to protect against many diseases with an inflammatory nature. On the other hand, high-fat diets are documented to exert multiple deleterious effects, including fatty liver diseases. In the case of cancer, the cancer cells require a lot of energy and they derive it from glucose though they switch from an otherwise oxygen-requiring pathway to less efficient processes such as fermentation. This metabolic disruption, referred to as Warburg metabolism (Vander Heiden et al., 2009), results in the production of lactate which destroys the extracellular environment and facilitates invasion into new tissues and metastasis. Now that creates a lactate rich, glucose depleted micro-environment and to overcome these competitive conditions the Tregs use alternative metabolic pathway in TME i.e., they shift from glucose dependent to fatty acid dependent metabolic pathway. As Tregs use alternate metabolic pathway in the TME the effects of these dietary supplements must be considered for holistic approach.
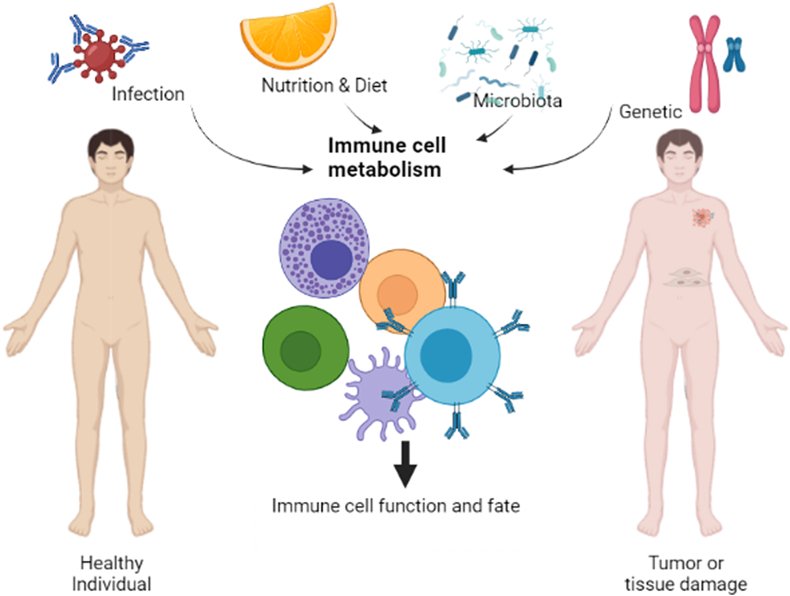
Fatty acids such as omega-3 and omega-6, which is supplemented in products such as bread and margarine, give them a health-promoting effect. Docosahexaenoic acid (DHA) is an omega-3 fatty acid which has been reported to reduce suppressive and migratory functions of Tregs leading to a proliferation of CD4+IL2RA− effector T cells (Song et al., 2016; Yessoufou et al., 2009). Fish oil or omega-3 polyunsaturated fatty acids (PUFAs) prevent cancer (Song et al., 2014; Shen et al., 2012) which has been demonstrated in an immuno-MPE study. Studies has been shown that higher intake of omega-3 PUFAs was associated with a lower risk of colorectal carcinomas with high infiltrating FOXP3+ Tregs (Song et al., 2016). DHA treatment causes increased expression of Treg-specific markers like FOXP3, CTLA4 and TGFβ mRNA, whereas decreases IL10 mRNA expression. Thus the effects of DHA treatment seem to be rather complex: on one hand, this fatty acid exerts an inhibitory effect on migratory and Treg function; then again, it promotes the expression of the typical Treg markers TGFβ and FOXP3. These discoveries led to the idea that omega-3 PUFAs can suppress the function of Tregs, thereby encouraging anti-tumor immune response (Song et al., 2016). Hence food components like vitamins and fatty acids supplements must be considered because they influence the development and character of Tregs. On the other hand, gut-associated microbiota influence the generation of CD4+CD25+FOXP3+ pTregs. Widespread research has established the crosstalk between commensal microbiota and immune cells. Several studies have shown that administration of probiotic bacteria containing members of the Lactobacillus, Streptococcus, and Bifidobacterium genera primed DCs to induce the development of FOXP3+ Tregs and IL10-secreting Tregs (Smits et al., 2005; Kwon et al., 2010). As functional food containing probiotics become more widely available and popular, understanding their mode of action on regulation of Treg could prove valuable to control cancer. Hence, integrative immuno-MPE research can provide new insights into the combined role of environmental factors and Tregs and possible therapeutic attributes to combat cancer (Fig. 5).
Fig. 5.
Pictorial representation of factors affecting the functions and fate of immune cells: The immune cells may have different fate in tumor conditions rather that the same in healthy individuals. Not only the tumor microenvironment but also other factors like infections (viral, bacterial), nutrition and diet of individuals, altered gut microbiota as a consequence of individual's diet and most importantly one's genetic makeup can affect and shape the functions and fate of immune cells (Created in BioRender.com).
On the other hand, gene-environment (G × E) interaction is when two people having two different genotypes respond to environmental factors in different ways. Environmental exposure such as various chemicals, duration of its exposure, sunlight, radiation, geographical location and cultural behaviors can initiate tumor formation by altering the genotypes. Risk of most cancers is determined by a multifaceted interaction of genetic and environmental factors. G × E interaction also shapes different components of immune system like effector T cells, Tregs during cancer, which inturn regulates the outcome of cancer (Ogino et al., 2018, 2019). Gene-environment interaction research has the potential to develop strategies for cancer prevention and control by modifying the immune system, so before developing efficacious therapeutics it should be given considerable thought.
7. Concluding remarks
Suppressive activity of Tregs in TME has gained clinical significance over the years. Different studies have focused on the ways of infiltration of Tregs in breast tumor and its subsequent consequences. Infiltrated Tregs show metabolic shift for the selective survival in the TME which ultimately leads to the proliferation of Tregs in the vicinity of tumor. The balance between Tregs and effector T cells disrupts which makes the TME more tolerogenic . Metabolic shift of infiltrated Tregs has been observed in breast cancer and various human malignancies. Recent advances in immunotherapy have focused on the blockade of the checkpoint molecules of Tregs or the adoptive transfer of effector T cells. Although these treatment strategies have generated considerable results in preclinical studies but not much success has been produced in clinical studies. TME plays a complex role in the survival of immunosuppressive Tregs which acts as a barrier for successful immunotherapy. Understanding the molecules involved in the infiltration and metabolic shift of Tregs can be very beneficial in this context. For the efficacy of the treatment the influence of diet, gut-microbiota, lifestyle and environmental factors must be considered thoroughly because these factors influence the behavior of Tregs in cancer patient. Removal of inhibitory signal is the key to the successful immunotherapy. Combining immunotherapy with traditional treatment strategies can also be used for selective depletion of Tregs which will help to reduce tumor volume.
Ethical approval and consent to participate
Not applicable.
Consent for publication
All authors read and approved the final manuscript and concur with the submission for the publication.
Availability of supporting data
Not applicable.
CRediT authorship contribution statement
Tania Sarkar: undertook the background literature study and prepared the initial draft of the manuscript and the figures. Subhanki Dhar: undertook the background literature study and prepared the initial draft of the manuscript and the figures. Gaurisankar Sa: supervised the entire project and made final corrections to the draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The study was funded by grants from Department of Biotechnology, Government of India.
References
- Abd Al Samid M., Chaudhary B., Khaled Y.S., Ammori B.J., Elkord E. Combining FoxP3 and Helios with GARP/LAP markers can identify expanded Treg subsets in cancer patients. Oncotarget. 2016 Mar 22;7(12):14083–14094. doi: 10.18632/oncotarget.7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldinucci D., Casagrande N. Inhibition of the CCL5/CCR5 Axis against the progression of gastric cancer. Int. J. Mol. Sci. 2018 May 16;19(5):1477. doi: 10.3390/ijms19051477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelin A., Gil-de-Gómez L., Dahiya S., Jiao J., Guo L., Levine M.H., Wang Z., Quinn W.J., 3rd, Kopinski P.K., Wang L., Akimova T., Liu Y., Bhatti T.R., Han R., Laskin B.L., Baur J.A., Blair I.A., Wallace D.C., Hancock W.W., Beier U.H. Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metabol. 2017 Jun 6;25(6):1282–1293. doi: 10.1016/j.cmet.2016.12.018. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano M., Toda M., Sakaguchi N., Sakaguchi S. Autoimmune disease as a consequence of developmental abnormality of a T cell subpopulation. J. Exp. Med. 1996 Aug 1;184(2):387–396. doi: 10.1084/jem.184.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S.R., Nelson M.H., Himes R.A., Li Z., Mehrotra S., Paulos C.M. Th17 cells in cancer: the ultimate identity crisis. Front. Immunol. 2014 Jun 17;5:276. doi: 10.3389/fimmu.2014.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Vasanthakumar A., Grigoriadis G. Modulating T regulatory cells in cancer: how close are we? Immunol. Cell Biol. 2013 May;91(5):340–349. doi: 10.1038/icb.2013.12. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S., Md Sakib Hossain D., Mohanty S., Sankar Sen G., Chattopadhyay S., Banerjee S., Chakraborty J., Das K., Sarkar D., Das T., Sa G. Curcumin reverses T cell-mediated adaptive immune dysfunctions in tumor-bearing hosts. Cell. Mol. Immunol. 2010 Jul;7(4):306–315. doi: 10.1038/cmi.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruder D., Probst-Kepper M., Westendorf A.M., Geffers R., Beissert S., Loser K., von Boehmer H., Buer J., Hansen W. Neuropilin-1: a surface marker of regulatory T cells. Eur. J. Immunol. 2004 Mar;34(3):623–630. doi: 10.1002/eji.200324799. [DOI] [PubMed] [Google Scholar]
- Bu M., Shen Y., Seeger W.L., An S., Qi R., Sanderson J.A., Cai Y. Ovarian carcinoma-infiltrating regulatory T cells were more potent suppressors of CD8(+) T cell inflammation than their peripheral counterparts, a function dependent on TIM3 expression. Tumour Biol. 2016 Mar;37(3):3949–3956. doi: 10.1007/s13277-015-4237-x. [DOI] [PubMed] [Google Scholar]
- Buchbinder E.I., Desai A. CTLA-4 and PD-1 pathways: similarities, differences, and implications of their inhibition. Am. J. Clin. Oncol. 2016 Feb;39(1):98–106. doi: 10.1097/COC.0000000000000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr E.L., Kelman A., Wu G.S., Gopaul R., Senkevitch E., Aghvanyan A., Turay A.M., Frauwirth K.A. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J. Immunol. 2010 Jul 15;185(2):1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran S.S., Paria B.C., Srivastava A.K., Rothermel L.D., Stephens D.J., Dudley M.E., Somerville R., Wunderlich J.R., Sherry R.M., Yang J.C., Rosenberg S.A., Kammula U.S. Persistence of CTL clones targeting melanocyte differentiation antigens was insufficient to mediate significant melanoma regression in humans. Clin. Canc. Res. 2015 Feb 1;21(3):534–543. doi: 10.1158/1078-0432.CCR-14-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.H., Curtis J.D., Maggi L.B., Jr., Faubert B., Villarino A.V., O'Sullivan D., Huang S.C., van der Windt G.J., Blagih J., Qiu J., Weber J.D., Pearce E.J., Jones R.G., Pearce E.L. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013 Jun 6;153(6):1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.H., Qiu J., O'Sullivan D., Buck M.D., Noguchi T., Curtis J.D., Chen Q., Gindin M., Gubin M.M., van der Windt G.J., Tonc E., Schreiber R.D., Pearce E.J., Pearce E.L. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015 Sep 10;162(6):1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary B., Elkord E. Regulatory T cells in the tumor microenvironment and cancer progression: role and therapeutic targeting. Vaccines (Basel) 2016 Aug 6;4(3):28. doi: 10.3390/vaccines4030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Jin W., Hardegen N., Lei K.J., Li L., Marinos N., McGrady G., Wahl S.M. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 2003 Dec 15;198(12):1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuckran C.A., Liu C., Bruno T.C., Workman C.J., Vignali D.A. Neuropilin-1: a checkpoint target with unique implications for cancer immunology and immunotherapy. J Immunother Canc. 2020 Jul;8(2) doi: 10.1136/jitc-2020-000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curiel T.J., Coukos G., Zou L., Alvarez X., Cheng P., Mottram P., Evdemon-Hogan M., Conejo-Garcia J.R., Zhang L., Burow M., Zhu Y., Wei S., Kryczek I., Daniel B., Gordon A., Myers L., Lackner A., Disis M.L., Knutson K.L., Chen L., Zou W. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 2004 Sep;10(9):942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- Dairaghi D.J., Oyajobi B.O., Gupta A., McCluskey B., Miao S., Powers J.P., Seitz L.C., Wang Y., Zeng Y., Zhang P., Schall T.J., Jaen J.C. CCR1 blockade reduces tumor burden and osteolysis in vivo in a mouse model of myeloma bone disease. Blood. 2012 Aug 16;120(7):1449–1457. doi: 10.1182/blood-2011-10-384784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis R.J., Mancuso A., Daikhin E., Nissim I., Yudkoff M., Wehrli S., Thompson C.B. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. U. S. A. 2007 Dec 4;104(49):19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn G.P., Bruce A.T., Ikeda H., Old L.J., Schreiber R.D. Cancer immunoediting: from immunosurveillance to tumor escape. Nat. Immunol. 2002 Nov;3(11):991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- Dunn G.P., Old L.J., Schreiber R.D. The three Es of cancer immunoediting. Annu. Rev. Immunol. 2004;22:329–360. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- Dunne M.R., Ryan C., Nolan B., Tosetto M., Geraghty R., Winter D.C., O'Connell P.R., Hyland J.M., Doherty G.A., Sheahan K., Ryan E.J., Fletcher J.M. Enrichment of inflammatory IL-17 and TNF-α secreting CD4(+) T cells within colorectal tumors despite the presence of elevated CD39(+) T regulatory cells and increased expression of the immune checkpoint molecule, PD-1. Front Oncol. 2016 Mar 7;6:50. doi: 10.3389/fonc.2016.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkord E., Alcantar-Orozco E.M., Dovedi S.J., Tran D.Q., Hawkins R.E., Gilham D.E. T regulatory cells in cancer: recent advances and therapeutic potential. Expet Opin. Biol. Ther. 2010 Nov;10(11):1573–1586. doi: 10.1517/14712598.2010.529126. [DOI] [PubMed] [Google Scholar]
- Facciabene A., Peng X., Hagemann I.S., Balint K., Barchetti A., Wang L.P., Gimotty P.A., Gilks C.B., Lal P., Zhang L., Coukos G. Tumour hypoxia promotes tolerance and angiogenesis via CCL28 and T(reg) cells. Nature. 2011 Jul 13;475(7355):226–230. doi: 10.1038/nature10169. [DOI] [PubMed] [Google Scholar]
- Faget J., Biota C., Bachelot T., Gobert M., Treilleux I., Goutagny N., Durand I., Léon-Goddard S., Blay J.Y., Caux C., Ménétrier-Caux C. Early detection of tumor cells by innate immune cells leads to T(reg) recruitment through CCL22 production by tumor cells. Canc. Res. 2011 Oct 1;71(19):6143–6152. doi: 10.1158/0008-5472.CAN-11-0573. [DOI] [PubMed] [Google Scholar]
- Flores-Toro J.A., Luo D., Gopinath A., Sarkisian M.R., Campbell J.J., Charo I.F., Singh R., Schall T.J., Datta M., Jain R.K., Mitchell D.A., Harrison J.K. CCR2 inhibition reduces tumor myeloid cells and unmasks a checkpoint inhibitor effect to slow progression of resistant murine gliomas. Proc. Natl. Acad. Sci. U.S.A. 2020 Jan 14;117(2):1129–1138. doi: 10.1073/pnas.1910856117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauwirth K.A., Thompson C.B. Regulation of T lymphocyte metabolism. J. Immunol. 2004 Apr 15;172(8):4661–4665. doi: 10.4049/jimmunol.172.8.4661. [DOI] [PubMed] [Google Scholar]
- Frauwirth K.A., Riley J.L., Harris M.H., Parry R.V., Rathmell J.C., Plas D.R., Elstrom R.L., June C.H., Thompson C.B. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002 Jun;16(6):769–777. doi: 10.1016/s1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- Gerriets V.A., Danzaki K., Kishton R.J., Eisner W., Nichols A.G., Saucillo D.C., Shinohara M.L., MacIver N.J. Leptin directly promotes T-cell glycolytic metabolism to drive effector T-cell differentiation in a mouse model of autoimmunity. Eur. J. Immunol. 2016 Aug;46(8):1970–1983. doi: 10.1002/eji.201545861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert M., Treilleux I., Bendriss-Vermare N., Bachelot T., Goddard-Leon S., Arfi V., Biota C., Doffin A.C., Durand I., Olive D., Perez S., Pasqual N., Faure C., Ray-Coquard I., Puisieux A., Caux C., Blay J.Y., Ménétrier-Caux C. Regulatory T cells recruited through CCL22/CCR4 are selectively activated in lymphoid infiltrates surrounding primary breast tumors and lead to an adverse clinical outcome. Canc. Res. 2009 Mar 1;69(5):2000–2009. doi: 10.1158/0008-5472.CAN-08-2360. [DOI] [PubMed] [Google Scholar]
- Haas J., Schopp L., Storch-Hagenlocher B., Fritzsching B., Jacobi C., Milkova L., Fritz B., Schwarz A., Suri-Payer E., Hensel M., Wildemann B. Specific recruitment of regulatory T cells into the CSF in lymphomatous and carcinomatous meningitis. Blood. 2008 Jan 15;111(2):761–766. doi: 10.1182/blood-2007-08-104877. [DOI] [PubMed] [Google Scholar]
- Halvorsen E.C., Hamilton M.J., Young A., Wadsworth B.J., LePard N.E., Lee H.N., Firmino N., Collier J.L., Bennewith K.L. Maraviroc decreases CCL8-mediated migration of CCR5(+) regulatory T cells and reduces metastatic tumor growth in the lungs. OncoImmunology. 2016 Mar 10;5(6) doi: 10.1080/2162402X.2016.1150398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Yang Y., Chen Z., Jiang Z., Gu Y., Liu Y., Xu S., Lin C., Pan Z., Zhou W., Cao X. Human hepatocellular carcinoma-infiltrating CD4⁺CD69⁺Foxp3⁻ regulatory T cell suppresses T cell response via membrane-bound TGF-β1. J. Mol. Med. (Berl.) 2014 May;92(5):539–550. doi: 10.1007/s00109-014-1143-4. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Coussens L.M. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Canc. Cell. 2012 Mar 20;21(3):309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- Herrington C.S., Poulsom R., Coates P.J. Recent advances in pathology: the 2019 annual review issue of the journal of pathology. J. Pathol. 2019 Apr;247(5):535–538. doi: 10.1002/path.5255. [DOI] [PubMed] [Google Scholar]
- Ho P.C., Bihuniak J.D., Macintyre A.N., Staron M., Liu X., Amezquita R., Tsui Y.C., Cui G., Micevic G., Perales J.C., Kleinstein S.H., Abel E.D., Insogna K.L., Feske S., Locasale J.W., Bosenberg M.W., Rathmell J.C., Kaech S.M. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell. 2015 Sep 10;162(6):1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori S., Nomura T., Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003 Feb 14;299(5609):1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- Hossain D.M., Panda A.K., Manna A., Mohanty S., Bhattacharjee P., Bhattacharyya S., Saha T., Chakraborty S., Kar R.K., Das T., Chatterjee S., Sa G. FoxP3 acts as a cotranscription factor with STAT3 in tumor-induced regulatory T cells. Immunity. 2013 Dec 12;39(6):1057–1069. doi: 10.1016/j.immuni.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Hossain D.M., Panda A.K., Chakrabarty S., Bhattacharjee P., Kajal K., Mohanty S., Sarkar I., Sarkar D.K., Kar S.K., Sa G. MEK inhibition prevents tumour-shed transforming growth factor-β-induced T-regulatory cell augmentation in tumour milieu. Immunology. 2015 Apr;144(4):561–573. doi: 10.1111/imm.12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jc, Zhang Z., Li T.Y., Liang Y.F., Wang H.M., Bao J.J., Zhang J.A., Wang W.D., Xiang W.Y., Kong B., Wang Z.Y., Wu B.H., Chen X.D., He L., Zhang S., Wang C.Y., Xu J.F. Assessing the role of IL-35 in colorectal cancer progression and prognosis. Int. J. Clin. Exp. Pathol. 2013 Aug 15;6(9):1806–1816. [PMC free article] [PubMed] [Google Scholar]
- Jie H.B., Gildener-Leapman N., Li J., Srivastava R.M., Gibson S.P., Whiteside T.L., Ferris R.L. Intratumoral regulatory T cells upregulate immunosuppressive molecules in head and neck cancer patients. Br. J. Canc. 2013 Nov 12;109(10):2629–2635. doi: 10.1038/bjc.2013.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P., Ren H., Sun W., Xin W., Zhang H., Hao J. Circulating IL-35 in pancreatic ductal adenocarcinoma patients. Hum. Immunol. 2014 Jan;75(1):29–33. doi: 10.1016/j.humimm.2013.09.018. [DOI] [PubMed] [Google Scholar]
- Kakita N., Kanto T., Itose I., Kuroda S., Inoue M., Matsubara T., Higashitani K., Miyazaki M., Sakakibara M., Hiramatsu N., Takehara T., Kasahara A., Hayashi N. Comparative analyses of regulatory T cell subsets in patients with hepatocellular carcinoma: a crucial role of CD25(-) FOXP3(-) T cells. Int. J. Canc. 2012 Dec 1;131(11):2573–2583. doi: 10.1002/ijc.27535. [DOI] [PubMed] [Google Scholar]
- Kaymak I., Williams K.S., Cantor J.R., Jones R.G. Immunometabolic interplay in the tumor microenvironment. Canc. Cell. 2021 Jan 11;39(1):28–37. doi: 10.1016/j.ccell.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong H.T., Restifo N.P. Natural selection of tumor variants in the generation of "tumor escape" phenotypes. Nat. Immunol. 2002 Nov;3(11):999–1005. doi: 10.1038/ni1102-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss D.L., Longden J., Fechner G.A., Avery V.M. The functional antagonist Met-RANTES: a modified agonist that induces differential CCR5 trafficking. Cell. Mol. Biol. Lett. 2009;14(4):537–547. doi: 10.2478/s11658-009-0017-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I., Lange A., Mottram P., Alvarez X., Cheng P., Hogan M., Moons L., Wei S., Zou L., Machelon V., Emilie D., Terrassa M., Lackner A., Curiel T.J., Carmeliet P., Zou W. CXCL12 and vascular endothelial growth factor synergistically induce neoangiogenesis in human ovarian cancers. Canc. Res. 2005 Jan 15;65(2):465–472. [PubMed] [Google Scholar]
- Kuehnemuth B., Piseddu I., Wiedemann G.M., Lauseker M., Kuhn C., Hofmann S., Schmoeckel E., Endres S., Mayr D., Jeschke U., Anz D. CCL1 is a major regulatory T cell attracting factor in human breast cancer. BMC Canc. 2018 Dec 20;18(1):1278. doi: 10.1186/s12885-018-5117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H.K., Lee C.G., So J.S., Chae C.S., Hwang J.S., Sahoo A., Nam J.H., Rhee J.H., Hwang K.C., Im S.H. Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc. Natl. Acad. Sci. U. S. A. 2010 Feb 2;107(5):2159–2164. doi: 10.1073/pnas.0904055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Chen M., Liu Y., et al. Advances in distinguishing natural from induced Foxp3(+) regulatory T cells. Int. J. Clin. Exp. Pathol. 2013 Jan 15;6(2):116–123. [PMC free article] [PubMed] [Google Scholar]
- Lin Y.C., Mahalingam J., Chiang J.M., Su P.J., Chu Y.Y., Lai H.Y., Fang J.H., Huang C.T., Chiu C.T., Lin C.Y. Activated but not resting regulatory T cells accumulated in tumor microenvironment and correlated with tumor progression in patients with colorectal cancer. Int. J. Canc. 2013 Mar 15;132(6):1341–1350. doi: 10.1002/ijc.27784. [DOI] [PubMed] [Google Scholar]
- Lindau D., Gielen P., Kroesen M., Wesseling P., Adema G.J. The immunosuppressive tumour network: myeloid-derived suppressor cells, regulatory T cells and natural killer T cells. Immunology. 2013 Feb;138(2):105–115. doi: 10.1111/imm.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhang N., Li Q., Zhang W., Ke F., Leng Q., Wang H., Chen J., Wang H. Tumor-associated macrophages recruit CCR6+ regulatory T cells and promote the development of colorectal cancer via enhancing CCL20 production in mice. PloS One. 2011 Apr 29;6(4) doi: 10.1371/journal.pone.0019495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.C., Yao X., Crystal J.S., Li Y.F., El-Gamil M., Gross C., Davis L., Dudley M.E., Yang J.C., Samuels Y., Rosenberg S.A., Robbins P.F. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin. Canc. Res. 2014 Jul 1;20(13):3401–3410. doi: 10.1158/1078-0432.CCR-14-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingam J., Lin C.Y., Chiang J.M., Su P.J., Chu Y.Y., Lai H.Y., Fang J.H., Huang C.T., Lin Y.C. CD4⁺ T cells expressing latency-associated peptide and Foxp3 are an activated subgroup of regulatory T cells enriched in patients with colorectal cancer. PloS One. 2014 Sep 30;9(9) doi: 10.1371/journal.pone.0108554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailloux A.W., Young M.R. Regulatory T-cell trafficking: from thymic development to tumor-induced immune suppression. Crit. Rev. Immunol. 2010;30(5):435–447. doi: 10.1615/critrevimmunol.v30.i5.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., Kono K., Izawa S., Mizukami Y., Kawaguchi Y., Mimura K., Watanabe M., Fujii H. CCL17 and CCL22 chemokines within tumor microenvironment are related to infiltration of regulatory T cells in esophageal squamous cell carcinoma. Dis. Esophagus. 2010 Jul;23(5):422–429. doi: 10.1111/j.1442-2050.2009.01029.x. [DOI] [PubMed] [Google Scholar]
- Micallef I.N., Stiff P.J., Nademanee A.P., Maziarz R.T., Horwitz M.E., Stadtmauer E.A., Kaufman J.L., McCarty J.M., Vargo R., Cheverton P.D., Struijs M., Bolwell B., DiPersio J.F. Plerixafor plus granulocyte colony-stimulating factor for patients with non-hodgkin lymphoma and multiple myeloma: long-term follow-up report. Biol. Blood Marrow Transplant. 2018 Jun;24(6):1187–1195. doi: 10.1016/j.bbmt.2018.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y., Kono K., Kawaguchi Y., Akaike H., Kamimura K., Sugai H., Fujii H. CCL17 and CCL22 chemokines within tumor microenvironment are related to accumulation of Foxp3+ regulatory T cells in gastric cancer. Int. J. Canc. 2008 May 15;122(10):2286–2293. doi: 10.1002/ijc.23392. [DOI] [PubMed] [Google Scholar]
- Moore D.C., Elmes J.B., Shibu P.A., Larck C., Park S.I. Mogamulizumab: an anti-CC chemokine receptor 4 antibody for T-cell lymphomas. Ann. Pharmacother. 2020 Apr;54(4):371–379. doi: 10.1177/1060028019884863. [DOI] [PubMed] [Google Scholar]
- Motz G.T., Coukos G. Deciphering and reversing tumor immune suppression. Immunity. 2013 Jul 25;39(1):61–73. doi: 10.1016/j.immuni.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P., Bueno M.J., Zagorac I., Mondejar T., Sanchez J., Mourón S., Muñoz J., Gómez-López G., Jimenez-Renard V., Mulero F., Chandel N.S., Quintela-Fandino M. Targeting tumor mitochondrial metabolism overcomes resistance to antiangiogenics. Cell Rep. 2016 Jun 21;15(12):2705–2718. doi: 10.1016/j.celrep.2016.05.052. [DOI] [PubMed] [Google Scholar]
- Nirschl C.J., Drake C.G. Molecular pathways: coexpression of immune checkpoint molecules: signaling pathways and implications for cancer immunotherapy. Clin. Canc. Res. 2013 Sep 15;19(18):4917–4924. doi: 10.1158/1078-0432.CCR-12-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel M., O'Reilly E.M., Wolpin B.M., Ryan D.P., Bullock A.J., Britten C.D., Linehan D.C., Belt B.A., Gamelin E.C., Ganguly B., Yin D., Joh T., Jacobs I.A., Taylor C.T., Lowery M.A. Phase 1b study of a small molecule antagonist of human chemokine (C-C motif) receptor 2 (PF-04136309) in combination with nab-paclitaxel/gemcitabine in first-line treatment of metastatic pancreatic ductal adenocarcinoma. Invest. N. Drugs. 2020 Jun;38(3):800–811. doi: 10.1007/s10637-019-00830-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S., Nowak J.A., Hamada T., Phipps A.I., Peters U., Milner D.A., Jr., Giovannucci E.L., Nishihara R., Giannakis M., Garrett W.S., Song M. Integrative analysis of exogenous, endogenous, tumour and immune factors for precision medicine. Gut. 2018 Jun;67(6):1168–1180. doi: 10.1136/gutjnl-2017-315537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S., Nowak J.A., Hamada T., Milner D.A., Jr., Nishihara R. Insights into pathogenic interactions among environment, host, and tumor at the crossroads of molecular pathology and epidemiology. Annu. Rev. Pathol. 2019 Jan 24;14:83–103. doi: 10.1146/annurev-pathmechdis-012418-012818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohue Y., Nishikawa H. Regulatory T (Treg) cells in cancer: can Treg cells be a new therapeutic target? Canc. Sci. 2019 Jul;110(7):2080–2089. doi: 10.1111/cas.14069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondondo B., Jones E., Godkin A., Gallimore A. Home sweet home: the tumor microenvironment as a haven for regulatory T cells. Front. Immunol. 2013 Jul 16;4:197. doi: 10.3389/fimmu.2013.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandiyan P., Zheng L., Ishihara S., Reed J., Lenardo M.J. CD4+CD25+Foxp3+ regulatory T cells induce cytokine deprivation-mediated apoptosis of effector CD4+ T cells. Nat. Immunol. 2007 Dec;8(12):1353–1362. doi: 10.1038/ni1536. [DOI] [PubMed] [Google Scholar]
- Pardoll D. Does the immune system see tumors as foreign or self? Annu. Rev. Immunol. 2003;21:807–839. doi: 10.1146/annurev.immunol.21.120601.141135. [DOI] [PubMed] [Google Scholar]
- Parlo R.A., Coleman P.S. Continuous pyruvate carbon flux to newly synthesized cholesterol and the suppressed evolution of pyruvate-generated CO2 in tumors: further evidence for a persistent truncated Krebs cycle in hepatomas. Biochim. Biophys. Acta. 1986 Apr 29;886(2):169–176. doi: 10.1016/0167-4889(86)90134-5. [DOI] [PubMed] [Google Scholar]
- Redjimi N., Raffin C., Raimbaud I., Pignon P., Matsuzaki J., Odunsi K., Valmori D., Ayyoub M. CXCR3+ T regulatory cells selectively accumulate in human ovarian carcinomas to limit type I immunity. Canc. Res. 2012 Sep 1;72(17):4351–4360. doi: 10.1158/0008-5472.CAN-12-0579. [DOI] [PubMed] [Google Scholar]
- Rifkin D.B. Latent transforming growth factor-beta (TGF-beta) binding proteins: orchestrators of TGF-beta availability. J. Biol. Chem. 2005 Mar 4;280(9):7409–7412. doi: 10.1074/jbc.R400029200. [DOI] [PubMed] [Google Scholar]
- Rudensky A.Y., Gavin M., Zheng Y. FOXP3 and NFAT: partners in tolerance. Cell. 2006 Jul 28;126(2):253–256. doi: 10.1016/j.cell.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Rudra D., deRoos P., Chaudhry A., Niec R.E., Arvey A., Samstein R.M., Leslie C., Shaffer S.A., Goodlett D.R., Rudensky A.Y. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat. Immunol. 2012 Oct;13(10):1010–1019. doi: 10.1038/ni.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S., Yamaguchi T., Nomura T., Ono M. Regulatory T cells and immune tolerance. Cell. 2008 May 30;133(5):775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Sarris M., Andersen K.G., Randow F., Mayr L., Betz A.G. Neuropilin-1 expression on regulatory T cells enhances their interactions with dendritic cells during antigen recognition. Immunity. 2008 Mar;28(3):402–413. doi: 10.1016/j.immuni.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler P.J., Schilling B., Harasymczuk M., Hoffmann T.K., Johnson J., Lang S., Whiteside T.L. Phenotypic and functional characteristics of CD4+ CD39+ FOXP3+ and CD4+ CD39+ FOXP3neg T-cell subsets in cancer patients. Eur. J. Immunol. 2012 Jul;42(7):1876–1885. doi: 10.1002/eji.201142347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler P.J., Saze Z., Hong C.S., Muller L., Gillespie D.G., Cheng D., Harasymczuk M., Mandapathil M., Lang S., Jackson E.K., Whiteside T.L. Human CD4+ CD39+ regulatory T cells produce adenosine upon co-expression of surface CD73 or contact with CD73+ exosomes or CD73+ cells. Clin. Exp. Immunol. 2014 Aug;177(2):531–543. doi: 10.1111/cei.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scurr M., Ladell K., Besneux M., Christian A., Hockey T., Smart K., Bridgeman H., Hargest R., Phillips S., Davies M., Price D., Gallimore A., Godkin A. Highly prevalent colorectal cancer-infiltrating LAP⁺ Foxp3⁻ T cells exhibit more potent immunosuppressive activity than Foxp3⁺ regulatory T cells. Mucosal Immunol. 2014 Mar;7(2):428–439. doi: 10.1038/mi.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X.J., Zhou J.D., Dong J.Y., Ding W.Q., Wu J.C. Dietary intake of n-3 fatty acids and colorectal cancer risk: a meta-analysis of data from 489 000 individuals. Br. J. Nutr. 2012 Nov 14;108(9):1550–1556. doi: 10.1017/S0007114512003546. [DOI] [PubMed] [Google Scholar]
- Shevach E.M. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity. 2009 May;30(5):636–645. doi: 10.1016/j.immuni.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Sinclair L.V., Rolf J., Emslie E., Shi Y.B., Taylor P.M., Cantrell D.A. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 2013 May;14(5):500–508. doi: 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P.M., Howitt M.R., Panikov N., Michaud M., Gallini C.A., Bohlooly Y.M., Glickman J.N., Garrett W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013 Aug 2;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits H.H., Engering A., van der Kleij D., de Jong E.C., Schipper K., van Capel T.M., Zaat B.A., Yazdanbakhsh M., Wierenga E.A., van Kooyk Y., Kapsenberg M.L. Selective probiotic bacteria induce IL-10-producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin. J. Allergy Clin. Immunol. 2005 Jun;115(6):1260–1267. doi: 10.1016/j.jaci.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Smyth M.J., Godfrey D.I., Trapani J.A. A fresh look at tumor immunosurveillance and immunotherapy. Nat. Immunol. 2001 Apr;2(4):293–299. doi: 10.1038/86297. [DOI] [PubMed] [Google Scholar]
- Song M., Chan A.T., Fuchs C.S., Ogino S., Hu F.B., Mozaffarian D., Ma J., Willett W.C., Giovannucci E.L., Wu K. Dietary intake of fish, ω-3 and ω-6 fatty acids and risk of colorectal cancer: a prospective study in U.S. men and women. Int. J. Canc. 2014 Nov 15;135(10):2413–2423. doi: 10.1002/ijc.28878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M., Nishihara R., Cao Y., Chun E., Qian Z.R., Mima K., Inamura K., Masugi Y., Nowak J.A., Nosho K., Wu K., Wang M., Giovannucci E., Garrett W.S., Fuchs C.S., Ogino S., Chan A.T. Marine ω-3 polyunsaturated fatty acid intake and risk of colorectal cancer characterized by tumor-infiltrating T cells. JAMA Oncol. 2016 Sep 1;2(9):1197–1206. doi: 10.1001/jamaoncol.2016.0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss L., Bergmann C., Szczepanski M., Gooding W., Johnson J.T., Whiteside T.L. A unique subset of CD4+CD25highFoxp3+ T cells secreting interleukin-10 and transforming growth factor-beta1 mediates suppression in the tumor microenvironment. Clin. Canc. Res. 2007 Aug 1;13(15 Pt 1):4345–4354. doi: 10.1158/1078-0432.CCR-07-0472. [DOI] [PubMed] [Google Scholar]
- Sun J., Tang D.N., Fu T., Sharma P. Identification of human regulatory T cells in the setting of T-cell activation and anti-CTLA-4 immunotherapy on the basis of expression of latency-associated peptide. Canc. Discov. 2012 Feb;2(2):122–130. doi: 10.1158/2159-8290.CD-11-0236. [DOI] [PubMed] [Google Scholar]
- Sundström P., Stenstad H., Langenes V., Ahlmanner F., Theander L., Ndah T.G., Fredin K., Börjesson L., Gustavsson B., Bastid J., Quiding-Järbrink M. Regulatory T cells from colon cancer patients inhibit effector T-cell migration through an adenosine-dependent mechanism. Cancer Immunol Res. 2016 Mar;4(3):183–193. doi: 10.1158/2326-6066.CIR-15-0050. [DOI] [PubMed] [Google Scholar]
- Tan M.C., Goedegebuure P.S., Belt B.A., Flaherty B., Sankpal N., Gillanders W.E., Eberlein T.J., Hsieh C.S., Linehan D.C. Disruption of CCR5-dependent homing of regulatory T cells inhibits tumor growth in a murine model of pancreatic cancer. J. Immunol. 2009 Feb 1;182(3):1746–1755. doi: 10.4049/jimmunol.182.3.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009 May 22;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A., Pettazzoni P., Lyssiotis C.A., Ying H., Sánchez N., Marchesini M., Carugo A., Green T., Seth S., Giuliani V., Kost-Alimova M., Muller F., Colla S., Nezi L., Genovese G., Deem A.K., Kapoor A., Yao W., Brunetto E., Kang Y., Yuan M., Asara J.M., Wang Y.A., Heffernan T.P., Kimmelman A.C., Wang H., Fleming J.B., Cantley L.C., DePinho R.A., Draetta G.F. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014 Oct 30;514(7524):628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Boehmer H., Daniel C. Therapeutic opportunities for manipulating T(Reg) cells in autoimmunity and cancer. Nat. Rev. Drug Discov. 2013 Jan;12(1):51–63. doi: 10.1038/nrd3683. [DOI] [PubMed] [Google Scholar]
- Wang R., Dillon C.P., Shi L.Z., Milasta S., Carter R., Finkelstein D., McCormick L.L., Fitzgerald P., Chi H., Munger J., Green D.R. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011 Dec 23;35(6):871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Lang M., Zhao T., Feng X., Zheng C., Huang C., Hao J., Dong J., Luo L., Li X., Lan C., Yu W., Yu M., Yang S., Ren H. Cancer-FOXP3 directly activated CCL5 to recruit FOXP3+Treg cells in pancreatic ductal adenocarcinoma. Oncogene. 2017 May 25;36(21):3048–3058. doi: 10.1038/onc.2016.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Franco F., Tsui Y.C., Xie X., Trefny M.P., Zappasodi R., Mohmood S.R., Fernández-García J., Tsai C.H., Schulze I., Picard F., Meylan E., Silverstein R., Goldberg I., Fendt S.M., Wolchok J.D., Merghoub T., Jandus C., Zippelius A., Ho P.C. CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat. Immunol. 2020 Mar;21(3):298–308. doi: 10.1038/s41590-019-0589-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S.T., Li K.K., Hepburn E., Weston C.J., Curbishley S.M., Reynolds G.M., Hejmadi R.K., Bicknell R., Eksteen B., Ismail T., Rot A., Adams D.H. The effects of CCR5 inhibition on regulatory T-cell recruitment to colorectal cancer. Br. J. Canc. 2015 Jan 20;112(2):319–328. doi: 10.1038/bjc.2014.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M.J., Vignali P.D.A., Mullett S.J., Overacre-Delgoffe A.E., Peralta R.M., Grebinoski S., Menk A.V., Rittenhouse N.L., DePeaux K., Whetstone R.D., Vignali D.A.A., Hand T.W., Poholek A.C., Morrison B.M., Rothstein J.D., Wendell S.G., Delgoffe G.M. Metabolic support of tumour-infiltrating regulatory T cells by lactic acid. Nature. 2021 Mar;591(7851):645–651. doi: 10.1038/s41586-020-03045-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Li P., Shao N., Ma J., Ji M., Sun X., Ma D., Ji C. Aberrant expression of Treg-associated cytokine IL-35 along with IL-10 and TGF-β in acute myeloid leukemia. Oncol Lett. 2012 May;3(5):1119–1123. doi: 10.3892/ol.2012.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Jene N., Byrne D., Millar E.K., O'Toole S.A., McNeil C.M., Bates G.J., Harris A.L., Banham A.H., Sutherland R.L., Fox S.B. Recruitment of regulatory T cells is correlated with hypoxia-induced CXCR4 expression, and is associated with poor prognosis in basal-like breast cancers. Breast Cancer Res. 2011 Apr 26;13(2):R47. doi: 10.1186/bcr2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yessoufou A., Plé A., Moutairou K., Hichami A., Khan N.A. Docosahexaenoic acid reduces suppressive and migratory functions of CD4+CD25+ regulatory T-cells. J. Lipid Res. 2009 Dec;50(12):2377–2388. doi: 10.1194/jlr.M900101-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappasodi R., Serganova I., Cohen I.J., Maeda M., Shindo M., Senbabaoglu Y., Watson M.J., Leftin A., Maniyar R., Verma S., Lubin M., Ko M., Mane M.M., Zhong H., Liu C., Ghosh A., Abu-Akeel M., Ackerstaff E., Koutcher J.A., Ho P.C., Delgoffe G.M., Blasberg R., Wolchok J.D., Merghoub T. CTLA-4 blockade drives loss of Treg stability in glycolysis-low tumours. Nature. 2021 Mar;591(7851):652–658. doi: 10.1038/s41586-021-03326-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Chi H. mTOR signaling and transcriptional regulation in T lymphocytes. Transcription. 2014;5(2) doi: 10.4161/trns.28263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Frederick D.T., Wu L., Wei Z., Krepler C., Srinivasan S., Chae Y.C., Xu X., Choi H., Dimwamwa E., Ope O., Shannan B., Basu D., Zhang D., Guha M., Xiao M., Randell S., Sproesser K., Xu W., Liu J., Karakousis G.C., Schuchter L.M., Gangadhar T.C., Amaravadi R.K., Gu M., Xu C., Ghosh A., Xu W., Tian T., Zhang J., Zha S., Liu Q., Brafford P., Weeraratna A., Davies M.A., Wargo J.A., Avadhani N.G., Lu Y., Mills G.B., Altieri D.C., Flaherty K.T., Herlyn M. Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J. Clin. Invest. 2016 May 2;126(5):1834–1856. doi: 10.1172/JCI82661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Sg, Gray J.D., Ohtsuka K., Yamagiwa S., Horwitz D.A. Generation ex vivo of TGF-beta-producing regulatory T cells from CD4+CD25- precursors. J. Immunol. 2002 Oct 15;169(8):4183–4189. doi: 10.4049/jimmunol.169.8.4183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.