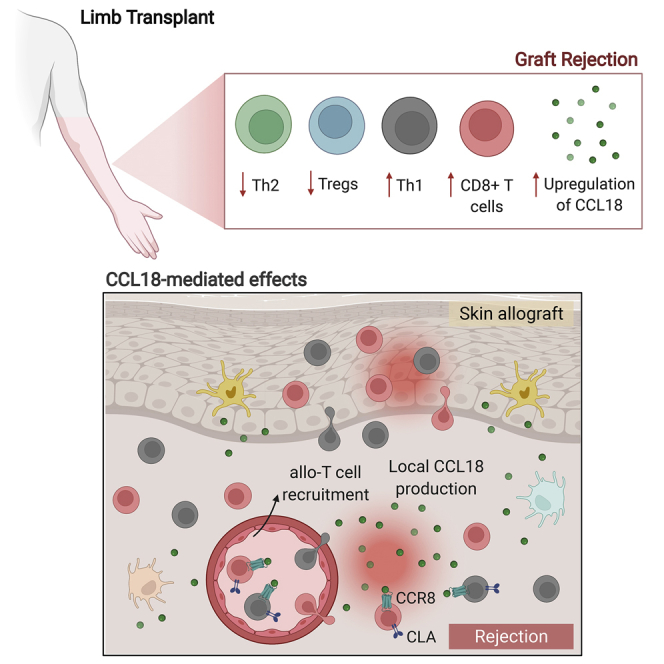
Summary
Limb transplantation is a life-changing procedure for amputees. However, limb recipients have a 6-fold greater rejection rate than solid organ transplant recipients, related in part to greater immunogenicity of the skin. Here, we report a detailed immunological and molecular characterization of individuals who underwent bilateral limb transplantation at our institution. Circulating Th17 cells are increased in limb transplant recipients over time. Molecular characterization of 770 genes in skin biopsies reveals upregulation of T cell effector immune molecules and chemokines, particularly CCL18. Skin antigen-presenting cells primarily express the chemokine CCL18, which binds to the CCR8 receptor. CCL18 treatment recruits more allo-T cells to the skin xenograft in a humanized skin transplantation model, leading to signs of accelerated graft rejection. Blockade of CCR8 remarkedly decreases CCL18-induced allo-T cell infiltration. Our results suggest that targeting the CCL18:CCR8 pathway could be a promising immunosuppressive approach in transplantation.
Keywords: upper extremity transplantation, limb transplantation, chemokines, CCL18, CCR8, extremity, rejection, vascular composite allograft
Graphical abstract
Highlights
-
•
Increased T cell infiltration in the stable graft compared with native skin
-
•
The chemokine CCL18 is upregulated during rejection in limb transplantation
-
•
CCL18 recruits more allo-T cells to the skin and leads to accelerated xenograft loss
-
•
CCR8 blockade halts CCL18-mediated T cell infiltration
Borges et al. provide a comprehensive immune characterization of limb transplant recipients, demonstrating that the chemokine CCL18 is a dominant signal during rejection. Local CCL18 mediates recruitment of pathogenic allo-T cells into grafts, which is abrogated by CCR8 blockade. Targeting the CCL18:CCR8 pathway is a promising complementary immunosuppressive approach in transplantation.
Introduction
Vascularized composite allotransplantation (VCA), including limb transplantation, is a life-changing procedure for individuals who have suffered severe traumatic injuries. Unlike solid organ transplantation, VCA involves transplantation of multiple tissues with different immunogenicity levels, including skin, muscle, bones, and nerves. Among these, the skin has the highest immunogenicity of all1 because of the presence of the following components: a dense population of antigen-presenting cells (APCs) and resident T cells, a rich microbiota, and continuous exposure to environmental threats, both physical and chemical. These unique skin features may explain the 6-fold greater rejection rate of VCA recipients compared with solid organ transplant recipients.2 More than 85% of individuals undergoing VCA experience acute cellular rejection in the first year after transplantation, and many have multiple episodes of rejection, leading to a higher burden of immunosuppressive therapy over time.3 It is reported that the number of rejection episodes may portend forthcoming chronic rejection events and graft loss.2 A better understanding of key distinctions between VCA and solid organ transplantation immune responses is essential for discovery of novel markers of rejection and predictive biomarkers.
Here we identified unique molecular signatures in skin graft biopsies at times of rejection that were dominated by T-effector molecules and chemokines, particularly CCL18. The CCL18 chemokine is present only in primates, with no murine ortholog, and has been identified as a key chemokine in the skin. CCL18 has been described to bind to the CCR8 receptor4 and induce homing of T cells in inflammatory skin conditions.5, 6, 7 Based on the unique rejection signal on graft biopsies, we sought to investigate the role of CCL18 in skin transplantation. By using a humanized skin transplantation model, we observed that CCL18 treatment recruited more allo-T cells to the skin and led to accelerated damage of the xenograft. This effect was abrogated markedly by treating the recipients with an anti-CCR8 blocking antibody. We characterized immunological changes in the graft microenvironment compared with proximal native skin tissue. Our data suggest an increase in T cell infiltration and action in the graft microenvironment compared with native tissue. We demonstrated a role of the chemokine CCL18 in attracting inflammatory allo-T cells to skin grafts, identifying the CCL18-CCR8 pathway as a potential therapeutic target in transplantation.
Results
Expansion of circulating Th17 cells after transplantation
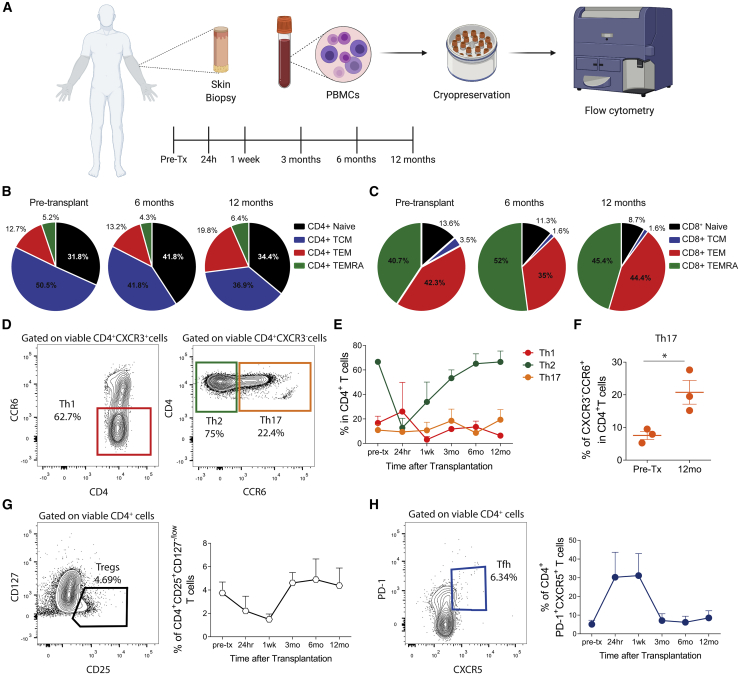
Th17 cells have a pathogenic role in different skin inflammatory disorders.8,9 We reasoned that Th17 cells would be enriched over time upon limb transplantation. Three individuals who received limb transplants in our institution between October 2011 and August 2016 were included in this analysis, with a mean follow-up of 5.2 years. The clinical characteristics of these individuals are detailed in Table 1. The individuals’ pre- and post-operative appearance is shown in Figure S1. Detailed information about the individuals’ immunosuppression is given in the STAR methods. We initially characterized T cell subsets from peripheral blood collected over time after transplantation according to our protocol (pre-transplantation, 24 h, 1 week, and 3, 6, and 12 months; Figure 1A). Analyses of the effector and memory T cell subsets (Figure S2) revealed that CD4+ central memory T cells (TCM cells; CD45RA–CCR7+) were the predominant T cell phenotype in the pool of CD4+ T cells, although they decreased after transplantation, whereas CD4+ effector memory T cells (TEMs; CD45RA–CCR7–) increased over time (Figure 1B). TEMs and effector memory CD45RA+ T cells (TEMRA cells; CD45RA+CCR7–) were the main subsets presented in the pool of CD8+ T cells with a decrease in naive CD8+ T cells (CD45RA+CCR7+) after transplantation (Figure 1C). Next, we assessed the T helper (Th) phenotypes based on CXCR3 and CCR6 expression (Figure 1D). Circulating Th2 cells were the predominant phenotype in individuals with upper extremity transplantation over time (Figure 1E). Th17 cells were increased markedly after transplantation (Figure 1F), whereas Th1 cells were stable over time (Figure 1E). The percentage of circulating regulatory T (Treg) cells (CD4+CD25+CD127−/low cells; Figure 1G) and T follicular helper (Tfh) cells (CD4+CXCR5+PD-1+ cells; Figure 1H) had no significant expansion after transplantation, other than transient changes early after transplantation, likely related to use of depletion induction therapy. These findings indicate that Th2 cells were the dominant phenotype over time after transplantation, with a significant expansion of circulating Th17 cells.
Table 1.
Baseline characteristics of upper extremity transplant recipients and donors
| Individual 1 | Individual 2 | Individual 3 | |
|---|---|---|---|
| Recipient characteristics | |||
| Age at transplantation (years) | 65 | 40 | 30 |
| Sex | male | male | male |
| Ethnicity | white | white | white |
| Cause of injury | septic shock | septic shock | ballistic trauma |
| Surgery | bilateral forearm | bilateral upper extremity | bilateral upper extremity |
| PRA (%) | 0 | 69 | 0 |
| Donor-specific antibodies | negative | positive | negative |
| HLA mismatch (A, B, DR) | 5/6 | 5/6 | 4/6 |
| CMV status | negative | negative | negative |
| EBV status | positive | positive | positive |
| Induction agent | thymoglobulin | thymoglobulin | thymoglobulin |
| Follow-up (years) | 9 | 6 | 4 |
| Donor characteristics | |||
| Age (years) | 44 | 23 | 27 |
| Sex | male | male | male |
| CMV status | negative | negative | negative |
| EBV status | positive | positive | positive |
| Total ischemia time (hours) | 4 | 4 | 4 right/5 left |
Figure 1.
Analysis of circulating CD4+ and CD8+ T cell subsets from upper extremity recipients over time
(A) Skin biopsies and peripheral blood were collected over time (pre-transplantation, 24 h, 1 week, and 3, 6, and 12 months after transplantation) from upper extremity transplant recipients. PBMCs were isolated for posterior flow cytometry analysis. The cartoon was created with BioRender.
(B and C) Mean percentages of blood CD4+ (B) and CD8+ (C) naive cells (CCR7+CD45RA+), central memory T cells (TCM cells; CCR7+CD45RA−), effector memory T cells (TEM cells; CCR7−CD45RA−), and effector memory RA cells (TEMRA cells; CCR7−CD45RA+) before transplantation and 6 and 12 months after transplantation, represented as pie charts. Data are from all three individuals.
(D) Representative contour plots of gating of T helper (Th) 1 (CD4+CXCR3+CCR6−), Th2 (CD4+CXCR3−CCR6−), and Th17 (CD4+CXCR3−CCR6+) cells.
(E) Mean percentages of Th1, Th2, and Th17 cells from all three individuals over time.
(F–H) Mean percentages of circulating Th17 cells before transplantation and 12 months after transplantation. Statistic by paired t test, ∗p < 0.05. (G and H) Representative contour plots and mean percentages of (G) regulatory T (Treg) cells (CD4+CD25+CD127−/low) and (H) T follicular helper (Tfh) cells (CD4+CXCR5+PD-1+) from all three individuals over time.
Graphs are displayed as mean ± SD at each time point examined.
See also Figure 1, Figure 2, Figure 3, Figure 4, Figure 5, Figure 6and S2.
Increased T cell infiltration and activation in the non-rejecting graft microenvironment compared with native skin
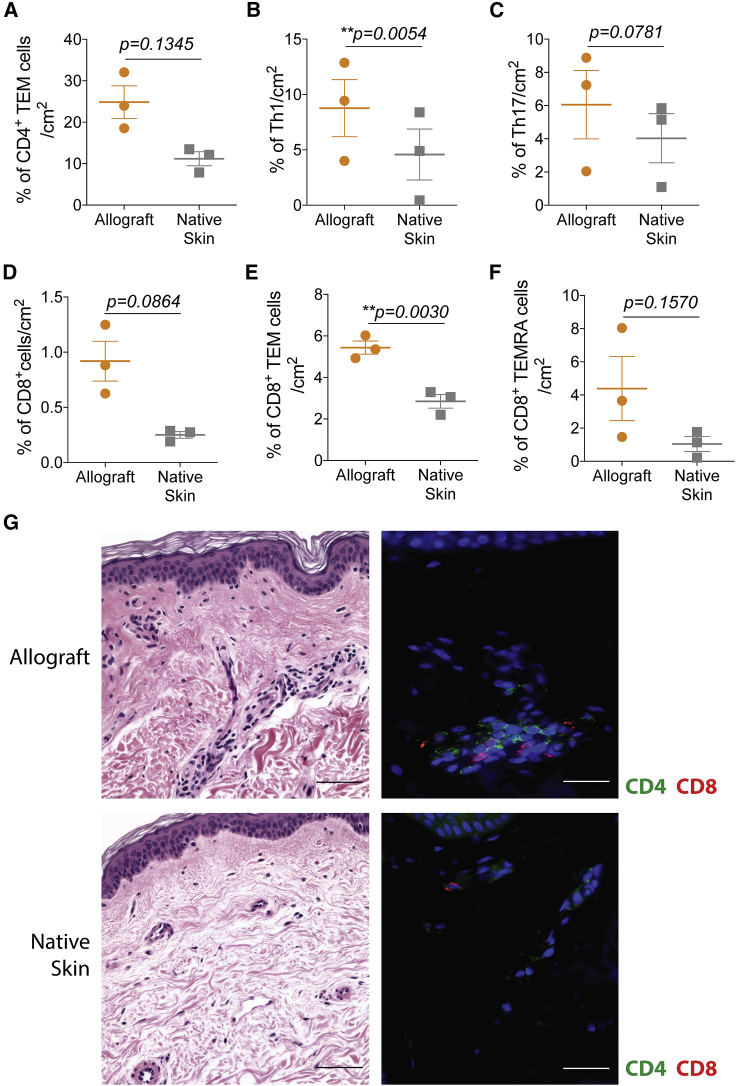
In transplantation, characterization of the T cells infiltrating human allografts compared with native tissues is technically limited by the small sample sizes collected in punch biopsies. We took advantage of debulking surgeries performed in our limb transplant recipients to examine T cells in the graft microenvironment at nonrejection time points and compared it with recipients’ adjacent native skin removed during the procedure. Allografts and native skins were processed, and infiltrating cells were isolated and stained by flow cytometry (Figure S3). Allografts had higher frequencies of activated CD4+ T cells, including CD4+ TEM cells (Figure 2A), Th1 cells (CD4+CXCR3+CCR6−; Figure 2B), and Th17 cells (CD4+CXCR3−CCR6+; Figure 2C) compared with native skin. Similarly, total CD8+ cells (Figure 2D), CD8+ TEM (Figure 2E) and TEMRA cells (Figure 2F) were also increased markedly compared with native skin. Immunofluorescence analyses of skin biopsies confirmed an increased influx of CD4+ and CD8+ cells in the allograft tissue compared with recipients’ adjacent native skin (Figure 2G), with approximately 5–10 positively labeled cells per vascular profile (CD8:CD4 ratio approximately 1:1) in allografts versus only rare ones (1–2 T cells per vascular profile) in adjacent native skin. Our data suggest that the graft microenvironment with alloantigens favors infiltration, expansion, and activation of T cells locally.
Figure 2.
Characterization of CD4+ and CD8+ T cell subsets from allografts and native skin of upper extremity recipients
(A–F) Mean percentages per skin area of infiltrating (A) CD4+ TEM cells (CCR7−CD45RA−), (B) Th1 cells (CD4+CXCR3+CCR6−), (C) Th17 cells (CD4+CXCR3−CCR6+), (D) total CD8+ cells, (E) CD8+ TEM cells, and (F) CD8+ TEMRA cells (CCR7−CD45RA+) from the allografts and adjacent native skins. Data are from all three individuals and represented as mean ± SD; statistics by paired t test.
(G) Representative H&E staining (left) and immunofluorescence of CD4+ and CD8+ cells (right) from the allograft and adjacent native skin of the same upper extremity transplant recipient; 200× (scale bars, 100 μm).
See also Figure S3.
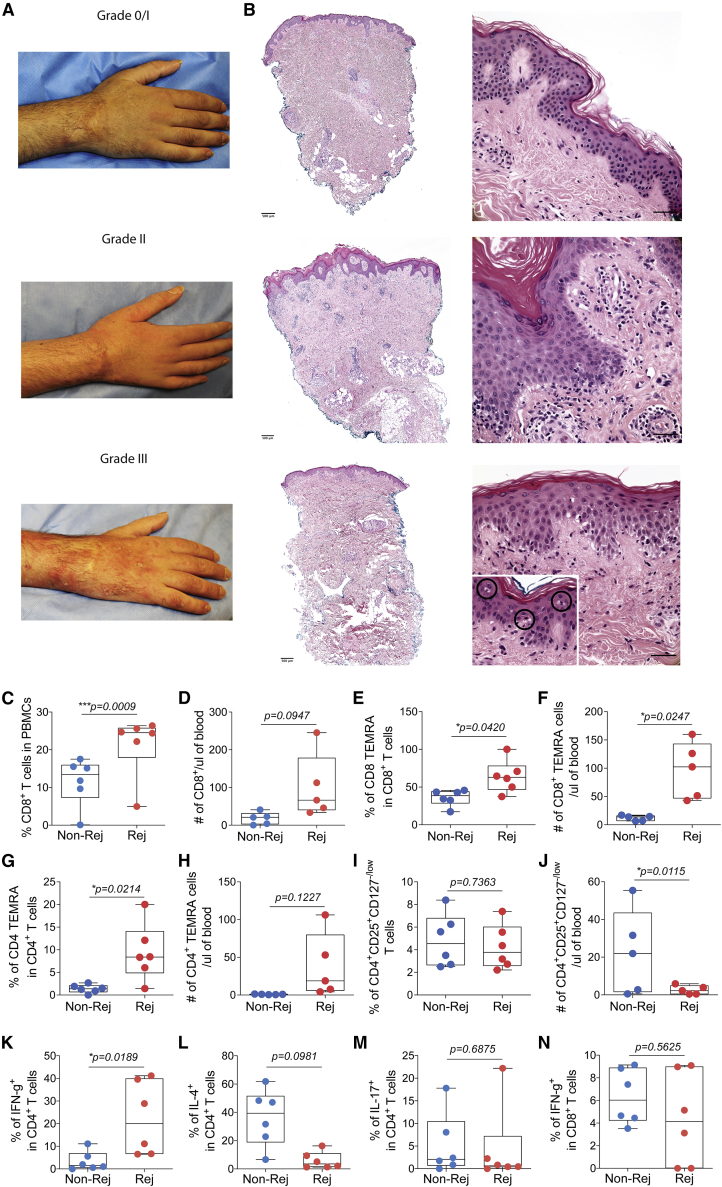
Expansion of circulating Th1 and TEM cells during rejection
In our cohort, all individuals developed at least one episode of acute cellular rejection within the first 3 years of transplantation (a total of 9 episodes; range, 2–4); about half occurred during the first 3 months after transplantation, whereas the remaining occurred later (>1 year after transplantation). Clinical aspects of the rejection included a maculopapular rash and edema (Figure 3A). Acute cellular rejection was assessed using Banff grading of skin-containing composite tissue,10 and most clinical rejection episodes were classified as between grades 2 and 3 (Figure 3B). There were no graft failures or recipient deaths. To characterize the unique features of rejection compared with nonrejection time points in limb transplantation, we characterized circulating T cells during those time points. For the rejection events, we selected blood samples corresponding to a respective Banff grading of 2, 2/3, or 3 skin biopsies from all three individuals. For nonrejection time points, the samples revealed grade 0 or 1 (grade 1 biopsy findings are regarded as non-specific for rejection and are not treated at our institution but monitored closely over time11). Compared with nonrejection time points, rejection episodes were characterized by an increase in circulating total CD8+ cells (Figures 3C and 3D), CD8+ TEMRA (Figures 3E and 3F) and CD4+ TEMRA cells (Figures 3G and 3H). Percentages of circulating Treg cells did not change during rejection (Figure 3I), whereas absolute numbers tended to be decreased at rejection time points compared with nonrejection (Figure 3J). In the peripheral blood, we observed an increase in interferon (IFN)-γ-producing CD4+ T cells (Figure 3K) and a decrease in interleukin-4 (IL-4) production by CD4+ T cells during rejection (Figure 3L). No changes were observed in production of IL-17 by circulating CD4+ T cells (Figure 3M) or IFN-γ production by CD8+ T cells (Figure 3N) during rejection events compared with nonrejection.
Figure 3.
Clinical and histopathological aspects in upper extremity allograft rejection with correlation to peripheral T cell populations
(A and B) Clinical photographs of an upper extremity transplant recipient and (B) corresponding H&E graft staining during clinical cellular rejection episodes with graft erythema and edema (grades 2–3) compared with mild rejection on surveillance biopsy (grade 1) without significant erythema or edema. Grade 3 rejection retains lymphocytic vasculopathy (bottom right panel) but also shows epithelial apoptosis associated with lymphoid exocytosis (circled in higher magnification, 400×). Left images, 40×; right images, 400× (scale bars, 50 μm).
(C–I) Percentages and (D, F, H, and J) absolute numbers of circulating total CD8+, CD8+ TEMRA, CD4+ TEMRA, and Treg cells at rejection (grades 2–3, n = 6) and nonrejection (grade 0, n = 6) events.
Percentages of (K) IFN-γ+, (L) IL-4+, (M) IL-17+ CD4+ T cells, and (N) IFN-γ+ CD8+ T cells from peripheral blood at rejection and nonrejection time points.
(C–N) Data are from all three individuals and represented as mean ± SD; statistics by paired t test. Rejection time points included samples from 1 week to 3 years after transplantation and nonrejection time points from 1 month to 4 years after transplantation.
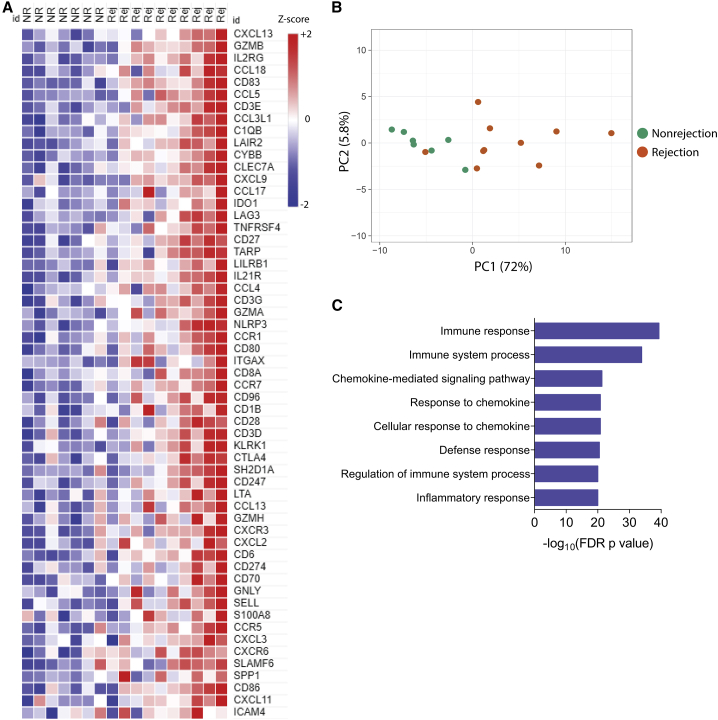
Molecular characterization of the graft microenvironment during rejection
To determine the molecular characteristics of the skin tissue associated with rejection, we compared the gene expression profiles of rejection (grades 2–3) and nonrejection (grade 0) events using NanoString technology. Among 770 genes analyzed, 57 genes were differentially expressed during rejection (log2 fold change > 2; unadjusted p < 0.01). The differentially expressed genes (DEGs) are displayed in Figure 4A. A principal-component analysis (PCA) was performed for the top 57 DEGs and demonstrated separate clustering of samples with rejection compared with nonrejection events, except for one resolving rejection that clustered with the nonrejection samples (Figure 4B). We used Gene Ontology analysis (GO) to better assess the biological processes that occurred during rejection in the allograft microenvironment. Immune, inflammatory, and chemokine-related responses were strongly associated with the DEGs during the rejection events (Figure 4C). Skin biopsies with rejection had a distinct gene signature compared with nonrejection biopsies.
Figure 4.
Unique gene expression signature in human upper extremity transplant rejection is associated with chemokines-related genes
(A) Heatmap of the 57 differentially expressed genes (DEGs) in rejection (grades 2–3, n = 10) compared with nonrejection (grades 0–1, n = 7) skin biopsies (log2 fold change of genes assessed were transformed into Z scores).
(B) Unsupervised principal-component analysis of the top 57 DEGs, clustering the samples with rejection compared with nonrejection events, except by one resolving rejection.
(C) Top 8 Gene Ontology (GO) biological process terms enriched among the 57 DEGs in rejection biopsies compared with nonrejection.
Statistics used Fisher’s one-tailed test with Benjamini-Hochberg false discovery rate (FDR; p value) for multiple testing correction. For all analyses, rejection biopsies included samples from 1 month to 3 years after transplantation and nonrejection biopsies from 1 month to 4.5 years after transplantation.
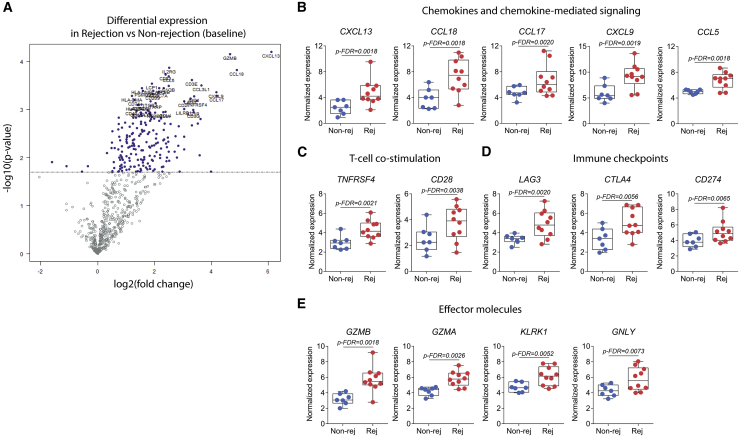
Rejection is characterized by expression of T cell-recruiting chemokines
From the distinctive gene signature during rejection (Figure 5A), the single most upregulated gene was CXCL13 (Figure 5B; log2 fold change = 6.1 compared with nonrejection). Following this, many of the top upregulated genes encoded for proteins associated with chemokines and chemokine-mediated signaling (CCL18, CCL17, CXCL9, and CCL5; Figure 5B), with CCL18 having a 2-fold increase during rejection. We also found increased gene expression in association with T cell co-stimulation (TNFRSF4 and CD28; Figure 5C) and effector immune molecules (GZMB, GZMA, KLRK1, and GNLY; Figure 5E). On the other hand, rejection biopsies also showed increased expression of inhibitory immune checkpoints (LAG3, CTLA4, and CD274; Figure 5D), suggesting that regulatory pathways may be triggered during rejection to counterbalance strong inflammatory responses.
Figure 5.
Chemokine-mediated signaling, T cell effector molecules, and immune checkpoints are upregulated in the limb transplant skin microenvironment during rejection
(A) Volcano plot showing DEGs in rejection in relation to nonrejection. Log2 fold change is represented on the x axis, and the y axis displays −log10 of each gene’s p value.
(B–E) Normalized expression of genes associated with (B) chemokines and chemokine-mediated signaling, (C) T cell co-stimulation, (D) immune checkpoints, and (E) effector molecules. Boxplots represent mean values with whiskers of maximum and minimum values.
Statistics are represented by FDR p values. For all analyses, rejection biopsies (n = 10; 1 month to 3 years after transplantation) and nonrejection biopsies (n = 7; 1 month to 4.5 years after transplantation).
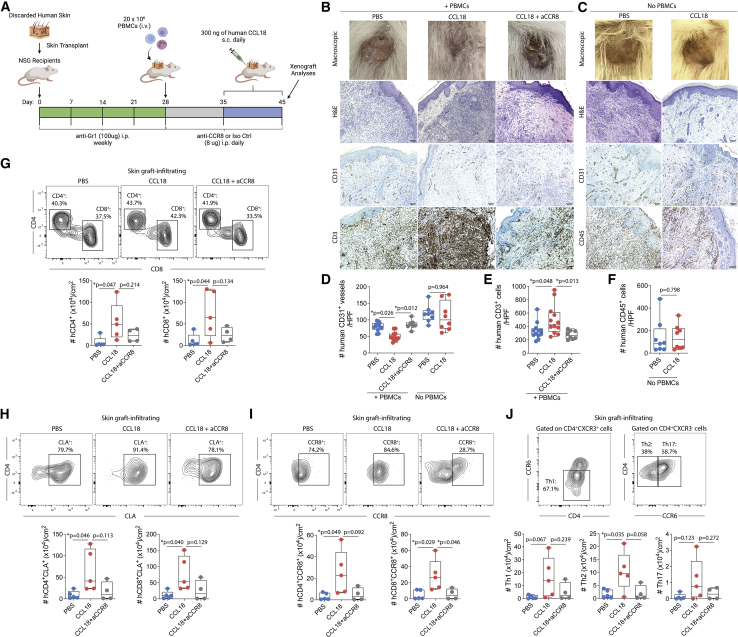
CCL18 enhances recruitment of allogeneic T cells to human skin xenografts
Among the chemokines upregulated during rejection, CCL18 was particularly interesting, based on its primary expression in skin dendritic cells, Langerhans cells, and macrophages12, 13, 14 and its role in recruiting T cells in inflammatory skin conditions such as atopic dermatitis.5 Therefore, we evaluated the in vivo effect of CCL18 and its receptor CCR8 in a humanized skin transplantation model because CCL18 has no murine ortholog (Figure 6A). In this model, we transplanted human foreskins onto genetically immunosuppressed NSG mice and then administered allogeneic PBMCs 4 weeks after the skin transplantation. One week later, animals received daily subcutaneous injections of recombinant human CCL18 or PBS 1× into human skin xenografts for 10 days (Figure 6A). In a subgroup, CCL18-treated animals were administered an anti-CCR8 blocking antibody or isotype control daily starting on the day of PBMCs transfer (Figure 6A). Among the NSG recipients that had received PBMCs, subcutaneous injections of CCL18 into human skin xenografts led to significant macroscopic changes, including tissue shrinkage, discoloration, and dry appearance (Figure 6B). Anti-CCR8 treatment reduced the macroscopic signs of graft rejection induced by CCL18 (Figure 6B). To evaluate whether CCL18 deleterious effects on skin xenografts were dependent on the recipient’s immune system, a subgroup of animals was treated with subcutaneous injections of CCL18 or PBS 1× in the absence of PBMCs (Figure 6A). Interestingly, CCL18 treatment did not induce macroscopic changes in skin xenografts of animals that did not receive PBMCs, suggesting that CCL18 effects are dependent on the recipient’s immune system (Figure 6C). Histologically, xenografts treated with CCL18 demonstrated reduced CD31+ (vascular endothelium) staining (Figures 6B and 6D) and increased presence of CD3+ cells (Figures 6B and 6E) compared with PBS-injected animals. The anti-CCR8 treatment restored the presence of CD31+ structures (Figures 6B and 6D) and decreased CD3+ cells (Figures 6B and 6E). NSG mice that did not receive PBMCs presented the highest levels of CD31+ vessels that were not affected by CCL18 treatment (Figures 6C and 6D). Xenograft-resident human CD45+ cells were unchanged in CCL18-treated animals that did not receive PBMCs (Figures 6C and 6F). Thus, our data suggest that CCL18 can have major deleterious effects on skin xenografts, which is dependent on the presence of human immune cells.
Figure 6.
CCL18 attracts human allo-T cells to human skin xenografts
(A and B) Discarded human skin was transplanted into NSG recipient mice. Mice were injected weekly with anti-Gr1 to reduce local inflammation and establish an intact vasculature (A). Twenty-eight days after transplantation, recipients were injected with 20 × 106 allo-PBMCs. Seven days thereafter, 300 ng of CCL18 or PBS 1× was injected subcutaneously into the skin xenografts for 10 consecutive days. Recipient animals were treated with anti-CCR8 or isotype control daily. Skin xenografts and peripheral blood were analyzed by histology and flow cytometry on day 45 after transplantation.
(B–D) Representative macroscopic images of skin xenografts, H&E graft staining, and immunohistochemical staining for human CD31, human CD3, or human CD45 after the CCL18 injections into animals (A) injected (B) or not injected (C) with human PBMCs. Shown are absolute counts of human CD31+ vessels.
(E and F) Human CD3+ cells (E) and human CD45+ cells (F) in the skin xenografts, 100×. The number of positive cells was quantified from two representative 400× fields from each transplanted xenograft. Statistics by one-way ANOVA with Tukey’s post-test.
(G–J) Representative contour plot and absolute numbers of skin-infiltrating (G) CD4+ and CD8+ cells, (H) skin CD8+CLA+ and CD4+CLA+ cells, (I) skin CD8+CCR8+ and CD4+CCR8+ cells, and (J) Th1, Th17, and Th2 in PBS-, CCL18−, or CCL18+ anti-CCR8-treated groups that received human PBMCs. All data were normalized by square centimeter of tissue. Data represent a pool of two independent experiments (n = 4–6 animals per group) and are represented as mean ± SD. Statistics by one-way ANOVA with Tukey’s post-test.
See also Figure S4.
We next assessed and quantified recruitment of T cells to the skin using T cell extraction protocols and flow cytometry. CCL18 significantly increased the numbers of CD4+ and CD8+ T cells per skin area (Figure 6G and S4). The anti-CCR8 treatment markedly decreased CCL18-induced recruitment of CD4+ and CD8+ T cells (Figure 6G). CCL18 has been shown to recruit cutaneous lymphocyte-associated antigen (CLA)+ T cells to the skin microenvironment.5 We observed enhanced numbers of CLA+CD4+ and CLA+CD8+ T cells (Figure 6H) as well as CCR8+CD4+ and CCR8+CD8+ T cells (Figure 6I) in skin allografts treated with CCL18. In contrast, CCL18-induced recruitment of CLA+ and CCR8+ T cells was reduced markedly in anti-CCR8-treated animals (Figures 6H and 6I). Last, we found that CCL18-treated skin was associated with an increased number of Th1, Th2, and Th17 CD4+ T cells compared with vehicle-treated skin, whereas anti-CCR8 treatment inhibited this recruitment (Figure 6J). Our data suggest that CCL18 is an important local chemokine that can increase recruitment of CLA+ and CCR8+ allo-T cells to human skin xenografts and accelerate graft rejection. CCR8 blockade can substantially abrogate CCL18-induced T cell recruitment and pathogenic effects in skin xenografts.
Discussion
This study demonstrates that circulating immune cells from limb transplant recipients with no significant graft rejection have predominant Th2 and Treg cell phenotypes that are shifted to Th1 and CD8 responses during rejection. A similar peripheral immune profile was observed in face transplant recipients, as described previously by our group.15 However, despite the increased circulating Th17 cells over time, we did not observe an increase in Th17 cells infiltrating the allograft during rejection, as demonstrated in face transplant recipients.15 On one hand, these data suggest that, although skin is the main organ targeted by the immune system in both cohorts of individuals, the dominant effector response appears to differ. On the other hand, chronic rejection has been reported for VCA patients,16 and this increase in circulating Th17 cells has been associated with chronic graft injury in kidney transplant recipients, whereas reduced Th17 cells have been linked to allograft tolerance.17,18 In our previous work, focused on face transplant rejection,19 we found that effector cells represented contributions from recipient and donor immune pools. Moreover, donor T cells in rejecting grafts exhibited resident memory phenotypes, implicating local expansion in the transplanted tissue. Targeting events appeared to involve primarily cutaneous venules as well as epithelial domains in the epidermis and hair follicles, where keratinocyte stem cells normally reside.19 Whether key differences will also emerge in the evolutionary immunopathology between limb and face transplant rejection is a topic in need of further study. However, it is intriguing that Th17 cell pathways are implicated in limb transplantation in the context of data showing that IL-17 can target and activate skin epithelial stem cells through the TRAF4-ERK5 axis.20
The skin is an immunologically rich tissue with more than ∼1 × 106 resident T cells/cm2 21 and a diverse and dynamic population of APCs.22 Seminal studies by Murray1 have suggested that the skin is the most immunogenic organ. Different skin locations are exposed to diverse external physical and chemical insults that may affect the skin microbiome and the local immune response.2,23,24 This exposure could cause local non-specific inflammation and mimic alloimmune injury. Through debulking surgeries, we were able to evaluate the local immune profile in the allograft microenvironment and compared it with the adjacent native skin, demonstrating higher numbers of activated T cells in non-rejecting allograft skin. The comparison with adjacent native tissue is crucial because skin from different body areas has significant variation in its immunological content.25 In agreement with our data, an independent study demonstrated that the cellular infiltrates in skin biopsies from hand transplant recipients were predominantly composed of T cells.26 Because allograft and adjacent native skin tissues were exposed to the same external factors, our data suggest a more immunologically active environment in the allograft, likely related to continuous local shedding of alloantigens that primarily trigger the adaptive immunity and potentially the innate memory alloresponse.27 Alternatively, it is possible that leukocyte-endothelium interactions responsible for T cell trafficking and accumulation are altered even in a homograft setting, and this important control situation requires further scrutiny to define this issue.
CCL18 is a chemokine produced by APCs from the dermis and epidermis as well as by keratinocytes, and skin-homing human T cells express CCR8.28 Increased levels of CCL18 have been linked to atopic dermatitis,5 psoriasis,29 allergic contact hypersensitivity,30 and other human chronic inflammatory diseases.31 In this report, we found an increase in CCL18 in skin tissue during rejection of VCA. In a humanized skin transplantation model, we demonstrated that local CCL18 injection led to a higher infiltration of T cells in the xenografted human skin. Our results suggest that CCL18 may contribute to VCA rejection by promoting binding of CLA+ T cells and increasing homing of human memory T cells to the skin.5 Supporting this possibility, we also observed an increase in skin-infiltrating CLA+ T cells following CCL18 treatment. These cells have been described to mainly have a memory Th1 cell phenotype28,32 and collaborate in immune surveillance of healthy skin. When activated T cells infiltrate the skin allograft, the local inflammatory response may potentiate their effector function and further drive allograft rejection. Among other chemokines present during rejection of extremity transplants, CCL18 may have a unique role in T cell recruitment to the skin in comparison with other organ transplants and thus is a potentially promising selective target for downmodulation of the alloimmune response in VCA transplantation.
Identification of potential biomarkers of rejection is of paramount importance to provide additional tools to diagnose rejection, help physicians in their decision-making about treatment, and develop new therapies. Our group has previously identified serum MMP3 protein as a potential biomarker to stratify VCA recipients according to the severity of rejection.33,34 Here, besides the increase in CCL18, we also observed an increase in different chemokines, including CXCL9 and CCL5. Along the same lines as our findings, Hautz et al.26 have demonstrated that markers related to lymphocyte trafficking correlated with the severity of skin rejection in a cohort of five limb transplant recipients. Thus, limb transplant rejection is characterized by upregulation of lymphocyte-attracting chemokines and trafficking markers. These markers comprise potential targets for immunosuppressive drugs. Different chemokines have been reported to be upregulated and have a role during skin allograft rejection responses. CXCL9 and CCL5 have been demonstrated to be upregulated in skin allografts, but not isografts, a few days before rejection.35 CXCL9, CXCL10, and CCL5 are associated with kidney rejection, and their presence in the urine of transplant recipients is being explored as a rejection biomarker.36, 37, 38 This highlights the potential of chemokines to be used as biomarkers.
Our small number of individuals reflects, in part, the novelty and challenges of extremity transplantation in humans. Despite this, our study employed prospective blood and skin graft collection in combination with high-throughput technologies like NanoString to undercover unique aspects of the rejection process in extremity transplant recipients that may account for its relatively high rejection rate. This comprehensive report of limb transplant recipients is a result of the assembly and curation of a unique biobank with more than 45 time points involving surveillance and rejection episodes. It is now important to validate the pathogenic role of CCL18 and other potential T cell-attracting chemokines in other limb and VCA transplantation cohorts.
Limb transplantation is a clinically feasible procedure for amputees, and use of a solid organ transplantation-based immunosuppressive regimen has yielded good short/medium-term graft outcomes. Nonetheless, the high frequency of cellular rejection is a concerning long-term barrier. Development of novel biomarkers in larger cohorts and less toxic, narrowly targeted skin-specific immunosuppression strategies are critical to advance the field.
Limitations of the study
Limitations of our study include its single-center nature and the small number of individuals evaluated, limiting major extrapolations. Our findings need to be validated in other limb and VCA cohorts from different centers. We also acknowledged that, although NanoString is a useful tool, its detection capacity is limited to fewer than 1,000 transcripts. More comprehensive analyses, like single-cell RNA sequencing, could have identified other transcriptional pathways unique to the rejection process in limb transplant recipients. Finally, the results from the humanized skin transplantation are limited by the variability found in this model and the inability to fully recapitulate the complexity of the human immune system.
STAR★Methods
Key resources table
| REAGENT | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| PBMCs from limb transplant patients | This study (BWH) | N/A |
| Punch skin biopsies from limb transplant patients | This study (BWH) | N/A |
| Human discarded foreskin specimens | This study (BWH) | N/A |
| PBMCs from healthy volunteers | This study (BWH and MGH) | N/A |
| Experimental models | ||
| NSG (M. musculus) | Jackson Lab | NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ |
| Antibodies | ||
| APC/Cyanine7 anti-human CD4 | Biolegend | Clone OKT4; RRID: AB_2687202 |
| PerCP/Cyanine5.5 anti-human CD4 | Biolegend | Clone OKT4; RRID: AB_1186122 |
| PerCP/Cy5.5 anti-human CD127 | Biolegend | Clone A019D5; RRID: AB_10900253 |
| FITC anti-human CD185 (CXCR5) | Biolegend | Clone J252D4; RRID: AB_2561896 |
| PE/Cyanine7 anti-human CD279 (PD-1) | Biolegend | Clone EH12.2H7; RRID: AB_2159325 |
| APC anti-human FOXP3 | ThermoFisher | Clone 236A/E7; RRID: AB_10804651 |
| Brilliant Violet 510™ anti-human CD8 | Biolegend | Clone SK1; RRID: AB_2564623 |
| BUV737 anti-human CD8 | BD Biosciences | Clone SK1; RRID: AB_2870085 |
| APC anti-human CD45RA | BD Biosciences | Clone HI100; RRID: AB_314416 |
| APC anti-human CD45RA | Biolegend | Clone HI100; RRID: AB_314416 |
| PE anti-human CD45RA | Biolegend | Clone HI100; RRID: AB_314412 |
| FITC anti-human CD183 (CXCR3) | Biolegend | Clone G025H7; RRID: AB_10983066 |
| PerCP/Cyanine5.5 anti-human CD197 (CCR7) | Biolegend | Clone G043H7; RRID: AB_10915275 |
| PE/Cyanine7 anti-human CD196 (CCR6) | Biolegend | Clone G034E3; RRID: AB_10916518 |
| APC anti-human CD196 (CCR6) | Biolegend | Clone G034E3; RRID: AB_10915987 |
| PE anti-mouse/human B220 | Biolegend | Clone RA3-6B2; RRID: AB_312992 |
| APC-eFluor 780 anti-human IFN-gamma | Thermo Fisher | Clone 4S.B3; RRID: AB_10853011 |
| PE anti-human IL-17A | Thermo Fisher | Clone eBio64DEC17; RRID: AB_1724136 |
| PE anti-human CD25 | BD Biosciences | Clone M-A251; RRID: AB_2561860 |
| PE/Cy7 anti-human CD45 Antibody | BD Biosciences | Clone HI30; RRID: AB_314403 |
| PerCP/Cyanine5.5 anti-human/mouse CLA | Biolegend | Clone HECA-452; RRID: AB_2565765 |
| APC anti-human CD198 (CCR8) | Biolegend | Clone L263G8; RRID: AB_2820018 |
| Pacific Blue anti-human CD19 | Biolegend | Clone HIB19; RRID: AB_2073118 |
| Brilliant Violet 605™ anti-human CD3 | Biolegend | Clone OKT3; RRID: AB_2565824 |
| PE anti-human FOXP3 | Biolegend | Clone 206D; RRID: AB_492986 |
| Purified anti-human CCR8 | Biolegend | Clone L263G8; RRID: AB_2562613 |
| Purified mouse IgG2a, κ | Biolegend | Clone MOPC-173; RRID: AB_326546 |
| InVivoMAb anti-mouse Ly6G/Ly6C (Gr-1) | Bio X Cell | Clone RB6-8C5 |
| FcR Blocking Reagent, human | Miltenyi | Cat # 130-059-901; RRID: AB_2892112 |
| Rabbit-anti-human CD4, polyclonal | Novus Biologicals | Cat # NBP1-19371; RRID: AB_1641682 |
| Mouse anti-human CD8 alpha | Abcam | Clone C8/144B; RRID: AB_1280806 |
| Goat anti-rabbit IgG Antibody (H+L), Biotinylated | Vector Labs | Cat # BA-1000-1.5 |
| Horse anti-mouse IgG Antibody (H+L), Biotinylated | Vector Labs | Cat # BA-2000-1.5 |
| Rabbit anti-CD3 | Roche | Clone 2GV6 |
| Rabbit Polyclonal Anti-CD31 | Abcam | Cat # ab28364; RRID: AB_726362 |
| Rabbit anti-human CD45 | Cell Signaling | Cat # 13917S; RRID: AB_2750898 |
| Rabbit anti-mouse CD45 | Cell Signaling | Cat # 70257S; RRID: AB_2799780 |
| Critical commercial assays | ||
| nCounter® PanCancer Immune Profiling Panel | NanoString | Cat # XT-CSO-HIP1-12 |
| Chemicals, peptides, and recombinant proteins | ||
| Fixable Viability Dye eFluor 780 | Thermo Fisher | Cat # 65-0865-14 |
| Zombie NIR Fixable Viability Kit | Biolegend | Cat # 423106 |
| LIVE/DEAD™ Fixable Blue Dead Cell Stain Kit | Thermo Fisher | Cat # L34961 |
| Foxp3/Transcription Factor Staining Buffer Set | ThermoFisher | Cat # 00-5523-00 |
| Phorbol 12-myristate 13-acetate (PMA) and Ionomycin | Biolegend | Cat # 423301 |
| GolgiStop | BD Biosciences | Cat # 554724 |
| Recombinant human CCL18 | Peprotech | Cat # 300-34 |
| RPMI 1640 Medium with L-Glutamine | Lonza | Cat # 12-702Q |
| BenchMark Fetal Bovine Serum | GeminiBio | Cat # 100-106 |
| Penicillin-Streptomycin Solution | Corning | Cat # 30-002-CI |
| Collagenase type I | Thermo Fisher | Cat # 17100017 |
| DNase I | Thermo Fisher | Cat # 18047019 |
| AccuCheck Counting Beads | Thermo Fisher | Cat # PCB100 |
| Target Retrieval Solution, Citrate pH 6 | Agilent Dako | Cat # S236984-2 |
| Streptavidin, Alexa Fluor 546 conjugate | Thermo Fisher | Cat # S11225 |
| Streptavidin, Alexa Fluor 647 conjugate | Thermo Fisher | Cat # S21374 |
| ProLong Gold Antifade Mountant with DAPI | Thermo Fisher | Cat # P36931 |
| RNeasy FFPE Kit | Qiagen | Cat # 73504 |
| nCounter Standard Master Kit | NanoString | Cat # NAA-AKIT-01 |
| Software and algorithms | ||
| FlowJo v 10.7.1 | FlowJo | N/A |
| Graphpad Prism v9.0 | GraphPad Software | N/A |
| nSolver Analysis Software v4.0.70 | Nanostring | N/A |
| Morpheus | https://software.broadinstitute.org/morpheus | N/A |
| ClustVis v2.0 | https://biit.cs.ut.ee/clustvis/ | Metsalu et al., 2015 |
| PANTHER v16.0 | http://pantherdb.org | Mi et al., 2019 |
| ZEN 2012 v1.1.2.0 | ZEISS | N/A |
| Illustrator v26.0.1 | Adobe | N/A |
| Photoshop v23.1.0 | Adobe | N/A |
| ImageJ v2.0.0-rc-69/1.52p | https://imagej.net | N/A |
| Other | ||
| Axio Imager.M2 | ZEISS | N/A |
| Fortessa X-20 | BD Biosciences | N/A |
| VENTANA BenchMark Stain System | Roche | N/A |
| nCOUNTER FLEX | NanoString | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact (lriella@mgh.harvard.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Upper extremities transplant subjects and study approval
Three patients who received upper extremities transplants at the Brigham and Women’s Hospital were included in the study. All patients provided written informed consent to participate in the clinical trial (ClinicalTrials.gov NCT01293214) for upper extremities transplantation, as approved by the Human Research Committee at Brigham and Women’s Hospital (2009P001719). Before participation, patients were evaluated by our multidisciplinary team. Donors and recipients were matched according to sex, skin color, and ABO compatibility, in addition to a negative T and B cell cytotoxic crossmatch. Demographic details are displayed in Table 1. Patients were followed weekly during the first 4–6 weeks after transplantation and if stable, clinical visits were further spaced to every 2 weeks, every month, and then every 3 months. After the first year of transplantation, the clinical visits were scheduled semiannually.
Patients’ immunosuppression
All patients received mycophenolate mofetil (1,000 mg), methylprednisolone (500 mg), and rabbit anti-thymocyte globulin (1.5 mg/kg/day × 4 days) for induction therapy starting at the time of transplantation. Maintenance immunosuppression consisted of mycophenolate mofetil (initially 1,000 mg twice daily, and reduced to 500–750 mg twice daily in long-term), tacrolimus (adjusted to achieve target levels of 10–15 ng/mL in the first 6 months, followed by 8–12 ng/mL up to 1 year and 6–10 ng/mL thereafter), and prednisone taper (down to 20 mg on day 5, and 5–7.5 mg in long-term) (Table 1). Prednisone withdrawal was attempted in Patient 1, but due to the higher occurrence of acute rejection episodes during winter months, low dose prednisone (5 mg) was seasonally reinitiated.24 Perioperative antibacterial prophylaxis consisted of vancomycin and cefazolin and was modified according to perioperative findings. All patients received trimethoprim–sulfamethoxazole and valganciclovir prophylaxis against Pneumocystis jirovecii and cytomegalovirus, respectively, for ≥6 months. In the presence of clinical acute cellular rejection, patients were treated with pulse solumedrol 500 mg/day for 3 days, followed by a taper. Acute rejection episodes with no clinical signs were treated with an increase in maintenance immunosuppression and closely followed. Topical steroids or tacrolimus were also used in a few cases as adjuvant therapy. In case of no response, further T cell–depletion therapy (rabbit anti-thymocyte globulin, alemtuzumab) was attempted.
Humanized skin transplant model
Six-to-eight weeks-old NSG recipient mice were transplanted with a full-thickness (1 cm2 section) human foreskin xenograft on dorsum using a sterile monofilament, non-absorbable suture, as previously described by a member of our group (GFM39). Foreskins were used because they have fewer resident T cells.40 Human skin tissues were obtained as discarded tissue from plastic surgery (MGB IRB 2016P001844 and 2019P002424). Transplanted animals were treated weekly with 100 μg of an anti-Gr1 antibody (clone RB6-8C5, Bio X Cell) to reduce local cellular infiltration, improve wound healing, and establish an intact human vasculature.41 Four weeks later, each mouse was i.v. injected with 20 × 106 PBMCs from a different donor (allo-PBMCs). One week later, 300 ng of CCL18 (in 100 μL of sterile PBS 1×; purchased from Peprotech) was subcutaneously injected, under the skin allograft for ten consecutive days. The control group received sterile PBS 1× alone. Some animals were intraperitoneally treated with 8 μg anti-human CCR8 (clone L263G8, Biolegend) or mouse IgG2a, κ isotype control (clone MOPC-173, Biolegend). Skin xenografts were daily monitored for signs of rejection, primarily change in color and necrosis. Skin allografts and peripheral blood were harvested for analyses on day 17 after the PBMCs adoptive transfer. All animals were housed following the Institutional Animal Care and Use Committee (IACUC) and National Institutes of Health (NIH) Animal Care guidelines. The Mass General Brigham IACUC approved all experiments (protocol number 2016N000250 and 2020N000125).
Methods details
Isolation and quantification of skin cells
Immune cells from debulking surgeries or skin xenografts were isolated, as described previously.42 After harvesting, skin tissues were recovered overnight in RPMI media (Lonza) supplemented with 20% FBS (GeminiBio), 100 mM L-glutamine and ×1 penicillin/streptomycin at 4°C. After that, the tissues were minced into small pieces in 10% FCS-supplemented RPMI, followed by incubation in Collagenase type I (Thermo Fisher, 0.2%) and DNase I (Thermo Fisher, 30 Kunitz Units/mL) at 37°C for 2 h with shaking (350 rpm). Cells were passed through 70 μm cell-strainer, washed and recovered in RPMI media (Lonza) supplemented with 20% FBS, 100 mM L-glutamine and penicillin/streptomycin for 4h or overnight at 37°C. We quantified the total skin cell numbers using fluorescent AccuCheck Counting Beads (Invitrogen) by flow cytometry. Each skin sample had its area calculated and all data were normalized by skin area (in cm2).
Flow cytometry
We stained PBMCs, skin cells from debulking surgeries or xenografts for flow cytometry. PBMCs from different time points from the same patient were thawed, washed and stained on the same day to avoid variability. Over time analysis displayed in Figure 1 included samples from pre-transplant, 24 h, 1 week, 3-, 6- and 12-months post-transplant. Rejection time points included samples from 1 week to 3 years post-transplantation, and nonrejection time points from 1 month to 4 years post-transplantation. Thawed PBMCs and recovered skin cells were Fc-blocked (Miltenyi) for 20 min before staining for surface markers for 30 min in FACS buffer (2% FBS in PBS 1x) on ice. Intracellular staining was performed using the Fixation/Permeabilization Kit (Thermo Fisher). For PBMC analyses over time and during rejection episodes, we used the following anti-human antibodies: anti-CD4 (1:100), anti-CD8 (1:100), anti-CD45RA (1:100), anti-CCR7 (1:20), anti-CD25 (1:66), anti-CD127 (1:50), anti-CXCR5 (1:100), anti-PD-1 (1:400), anti-CXCR3 (1:50), anti-CCR6 (1:40), anti-IFN-γ (1:40), anti-IL-4 (1:40) and IL-17A (1:40). For cytokine detection, cell suspensions were incubated for 6 h with 50 ng/mL of phorbol 12-myristate 13-acetate (PMA) plus 500 ng/mL ionomycin (Biolegend), and GolgiStop (BD Biosciences) in 10% FBS-RPMI, followed by surface staining, permeabilization, and intracellular staining. For the experiments with NSG mice, we used the following anti-human antibodies: anti-CD45 (1:100), anti-CD3 (1:100), anti-CD4 (1:100), anti-CD8 (1:50), anti-CD19 (1:200), anti-CLA (1:33), anti-CCR8 (1:33), anti-CXCR3 (1:50) and anti-CCR6 (1:40). Stained cells were analyzed on a FACS Canto II flow cytometer (BD Biosciences) with FACSDiva software (BD Biosciences). Data were analyzed with FlowJo software (TreeStar). Viable cells were selected based on the staining with Fixable Viability Dye eFluor 780 (Thermo Fisher), Zombie NIR Fixable Viability Kit (Biolegend) or LIVE/DEAD Fixable Blue Dead Cell Stain Kit (Thermo Fisher). Gating strategies for PBMCs analyzes were as previously described15 and can be found in Figures S2 and S3.
Histology and immunofluorescence staining
All specimens were sectioned into 5 μm sections from formalin-fixed, paraffin-embedded tissue. Sections were dewaxed in xylene and rehydrated using serial ethanol baths in decreasing concentrations. Specimens were histopathologically evaluated with conventional hematoxylin and eosin (H&E) staining. Further evaluation was performed via immunofluorescence staining. Antigen retrieval was performed with citrate buffer pH 6 (Agilent Dako) in a pressure cooker (BioCare Medical) programmed at 110°C for 15 min. Sections were incubated with primary antibodies rabbit-anti-human CD4 (Novus Biologicals) at 1:200 and mouse-anti-human CD8 (Abcam) at 1:50 overnight at 4°C. Afterward, biotinylated secondary antibodies goat-anti-rabbit (Vector) and horse-anti-mouse (Vector) were both incubated at 1:200 for 1 h at room temperature followed by Streptavidin-AF546 conjugate (Invitrogen) and Streptavidin-AF647 conjugate (Invitrogen), respectively, for 30 min at room temperature. Sections were mounted with ProLong Gold Antifade Reagent with DAPI (Invitrogen) and coverslipped. Immunofluorescence-stained sections were photographed using a fluorescence microscope (EVOS FL Auto 2, Invitrogen) and processed with ImageJ software.
Immunohistochemistry
All specimens were sectioned into 5 μm sections from formalin-fixed, paraffin-embedded tissue. Immunohistochemical staining was performed using an automated Ventana BenchMark Stain System (Roche). Antibodies used were ready-to-use rabbit anti-human CD3 (Roche), rabbit polyclonal anti-CD31 (Abcam) at 1:50, rabbit anti-human CD45 (Cell Signaling) at 1:500 and rabbit anti-mouse CD45 (Cell Signaling) at 1:500. Stained sections were photographed using an Axio Imager M2 microscope (Zeiss) and processed with ImageJ and Photoshop software.
RNA extraction
We obtained six consecutive 10 μm sections from formalin-fixed paraffin-embedded (FFPE) skin punch biopsies taken at different time points. Deparaffinization with Xylene and RNA extraction were performed in sterile 1.5 mL microcentrifuge tubes with the RNeasy FFPE Kit (Qiagen), according to the manufacturer’s instructions. The concentration and purity of the isolated total RNA were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher) at the Center for Advanced Molecular Diagnostics (CAMD) Research Core of the Brigham and Women’s Hospital. The absorbance ratio at 260/280 was used to determine RNA quality.
NanoString nCounter assay for mRNA gene expression assay
To investigate intragraft gene expression changes during rejection episodes, 18 limb allograft biopsies taken between December 2013 and November 2018 were retrieved from the pathology archive at Brigham and Women’s Hospital, Boston. We obtained a total of ten biopsies from rejection and eight from nonrejections episodes. From the 18 samples analyzed, one did not pass quality control (nonrejection) and was excluded from the analysis. For nonrejection episodes, these included five biopsies with Banff grades 0 and two samples with Banff grades 0/1. For rejection episodes, samples included two biopsies with Banff grades 2, two samples with Banff grade 2/3, and six biopsies with Banff grades 3. We then analyzed 770 genes with the NanoString nCounter PanCancer Immune Profiling Gene Expression (GX) Codeset. Gene expression was measured on 100–200 ng of extracted RNA. Samples were processed on the NanoString nCounter Analysis System (NanoString Technologies) following the manufacturer’s instructions at the CAMD Research Core of the Brigham and Women’s Hospital. Images were processed into RCC files from two batches of two different lots of reagents.
We normalized raw gene expression counts, batch effect, background correction, data quality control and analyzed the data with the nSolver Analysis Software (Version 4.0.70). Twenty-seven reference genes (EIF2B4, PRPF38A, DDX50, MRPS5, AMMECR1L, CNOT4, COG7, TLK2, ZNF143, DHX16, SAP130, TBP, SDHA, NOL7, ZC3H14, TMUB2, EDC, FCF1, PPIA, AGK, HDAC3, POLR2A, SF3A3, USP39, ZNF346, GUSB and MTMR14) were used for normalization. We used the quality control parameters recommended by the manufacturer.
Quantification and statistical analysis
For flow cytometry data, we used paired t-test for paired two group comparations. In animal experiments, we used unpaired Student’s t-test for comparison of the two independent groups. All statistical tests were two-sided with a type 1 error rate of 0.05 to determine statistical significance. Prism software was used for data analysis and drawing graphs (GraphPad Software, Inc., San-Diego, CA).
For the NanoString analyzes, differentially expressed genes (DEGs) between rejection and nonrejections samples were analyzed using the nSolver Analysis Software (Version 4.0.70). Samples were not paired in this analysis. Because of the low number of samples, a gene was considered differently expressed when the comparison between groups reached a log2 fold change > 2 and an unadjusted p-value < 0.01. The log2 fold change of genes assessed was transformed into Z-scores and a heatmap was created using Morpheus matrix visualization and analysis tool from the Broad Institute (https://software.broadinstitute.org/morpheus). Unsupervised principal component analysis of the top 57 DEGs clustering the samples with rejection compared to nonrejection events as generated using the web tool ClustVis43 using their default configurations. Gene Ontology terms were identified using the PANTHER tool (Protein Analysis Through Evolutionary Relationships, http://pantherdb.org).44 All the 57 DEGs were used as an input with Homo sapiens as the organism and enriched for GO (biological processes) terms only. Fisher’s one-tailed test with Benjamini-Hochberg False Discovery Rate (FDR p value) for multiple testing corrections was used as statistics. A Volcano plot showing differentially expressed genes (DEGs) in rejection in relation to nonrejection was generated using nSolver Analysis Software (Version 4.0.70). Log2 fold change is represented in the X axis, and the Y axis displays −log10 of each gene's p value.
Additional resources
The study was registered at ClinicalTrials.gov (NCT01293214).
Acknowledgments
L.V.R. received support from the Department of Defense (RT190059, award W81XWH-20-1-0758). S.G.T. receives salary support from the Office of the Assistant Secretary of Defense for Health Affairs under the Reconstructive Transplant Research Program – Qualitative Research Award under award W81XWH-17-1-0400 and the Joint Program Committee 8/Clinical and Rehabilitative Medicine Research Program Extremity Regeneration Technology/Therapeutic Development Award under award W81XWH-16-2-0067. R.A.C. was supported by the Department of Defense (awards RT150073 and RT170025). Opinions, interpretations, conclusions, and recommendations are those of the author and are not necessarily endorsed by the Department of Defense. T.J.B. was the recipient of an American Heart Association postdoctoral fellowship (20POST35210659). B.K. was the recipient of the Plastic Surgery Foundation research fellowship grant.
Author contributions
T.J.B. and L.V.R. conceived the study and wrote the manuscript. T.J.B. performed flow cytometry, NanoString, animal experiments, and analyses. P.A., G.F.M., and G.C.L. performed the patient histology experiments and analyses. B.K. collected the clinical data. R.B.G., M.L.-F., and B.T.A. assisted T.J.B. with the animal transplants. D.G. performed the xenograft histology. B.P., G.F.M., R.A.C., S.A.I., G.C.L., and S.G.T. helped interpret the results and edited the manuscript. All authors reviewed the manuscript critically for important intellectual content and gave final approval of the version to be submitted.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community.
Published: March 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100559.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. This paper does not report the original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Murray J.E. Organ transplantation (skin, kidney, heart) and the plastic surgeon. Plast. Reconstr. Surg. 1971;47:425–431. doi: 10.1097/00006534-197105000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Kollar B., Pomahac B., Riella L.V. Novel immunological and clinical insights in vascularized composite allotransplantation. Curr. Opin. Organ Transpl. 2019;24:42–48. doi: 10.1097/MOT.0000000000000592. [DOI] [PubMed] [Google Scholar]
- 3.Hein R.E., Ruch D.S., Klifto C.S., Leversedge F.J., Mithani S.K., Pidgeon T.S., Richard M.J., Cendales L.C. Hand transplantation in the United States: a review of the organ procurement and transplantation network/united network for organ sharing database. Am. J. Transpl. 2019;20:1417–1423. doi: 10.1111/ajt.15704. [DOI] [PubMed] [Google Scholar]
- 4.Islam S.A., Ling M.F., Leung J., Shreffler W.G., Luster A.D. Identification of human CCR8 as a CCL18 receptor. J. Exp. Med. 2013;210:1889–1898. doi: 10.1084/jem.20130240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Günther C., Bello-Fernandez C., Kopp T., Kund J., Carballido-Perrig N., Hinteregger S., Fassl S., Schwärzler C., Lametschwandtner G., Stingl G., et al. CCL18 is expressed in atopic dermatitis and mediates skin homing of human memory T cells. J. Immunol. 2005;174:1723–1728. doi: 10.4049/jimmunol.174.3.1723. [DOI] [PubMed] [Google Scholar]
- 6.Park C.O., Lee H.J., Lee J.H., Wu W.H., Chang N.S., Hua L., Lee M.G., Lee K.H. Increased expression of CC chemokine ligand 18 in extrinsic atopic dermatitis patients. Exp. Dermatol. 2008;17:24–29. doi: 10.1111/j.1600-0625.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- 7.Vieyra-Garcia P., Crouch J.D., O’Malley J.T., Seger E.W., Yang C.H., Teague J.E., Vromans A.M., Gehad A., Win T.S., Yu Z., et al. Benign T cells drive clinical skin inflammation in cutaneous T cell lymphoma. JCI Insight. 2019;4:e124233. doi: 10.1172/jci.insight.124233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weaver C.T., Elson C.O., Fouser L.A., Kolls J.K. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu. Rev. Pathol. Mech. Dis. 2013;8:477–512. doi: 10.1146/annurev-pathol-011110-130318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu T., Li S., Ying S., Tang S., Ding Y., Li Y., Qiao J., Fang H. The IL-23/IL-17 pathway in inflammatory skin diseases: from bench to bedside. Front. Immunol. 2020;11:2971. doi: 10.3389/fimmu.2020.594735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cendales L.C., Kanitakis J., Schneeberger S., Burns C., Ruiz P., Landin L., Remmelink M., Hewitt C.W., Landgren T., Lyons B., et al. The Banff 2007 working classification of skin-containing composite tissue allograft pathology. Am. J. Transplant. 2008;8:1396–1400. doi: 10.1111/j.1600-6143.2008.02243.x. [DOI] [PubMed] [Google Scholar]
- 11.Tasigiorgos S., Kollar B., Turk M., Perry B., Alhefzi M., Kiwanuka H., Nizzi M.-C., Marty F.M., Chandraker A., Tullius S.G., et al. Five-Year follow-up after face transplantation. N. Engl. J. Med. 2019;380:2579–2581. doi: 10.1056/NEJMc1810468. [DOI] [PubMed] [Google Scholar]
- 12.Günther C., Zimmermann N., Berndt N., Grosser M., Stein A., Koch A., Meurer M. Up-regulation of the chemokine CCL18 by macrophages is a potential immunomodulatory pathway in cutaneous T-cell lymphoma. Am. J. Pathol. 2011;179:1434–1442. doi: 10.1016/j.ajpath.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He H., Suryawanshi H., Morozov P., Gay-Mimbrera J., Del Duca E., Kim H.J., Kameyama N., Estrada Y., Der E., Krueger J.G., et al. Single-cell transcriptome analysis of human skin identifies novel fibroblast subpopulation and enrichment of immune subsets in atopic dermatitis. J. Allergy Clin. Immunol. 2020;145:1615–1628. doi: 10.1016/j.jaci.2020.01.042. [DOI] [PubMed] [Google Scholar]
- 14.Reynolds G., Vegh P., Fletcher J., Poyner E.F.M., Stephenson E., Goh I., Botting R.A., Huang N., Olabi B., Dubois A., et al. Developmental cell programs are co-opted in inflammatory skin disease. Science. 2021;371:eaba6500. doi: 10.1126/science.aba6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borges T.J., O’Malley J.T., Wo L., Murakami N., Smith B., Azzi J., Tripathi S., Lane J.D., Bueno E.M., Clark R.A., et al. Codominant role of interferon-γ– and interleukin-17–producing T cells during rejection in full facial transplant recipients. Am. J. Transpl. 2016;16:2158–2171. doi: 10.1111/ajt.13705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krezdorn N., Lian C.G., Wells M., Wo L., Tasigiorgos S., Xu S., Borges T.J., Frierson R.M., Stanek E., Riella L.V., et al. Chronic rejection of human face allografts. Am. J. Transpl. 2019;19:1168–1177. doi: 10.1111/ajt.15143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung B.H., Kim K.W., Kim B.-M., Doh K.C., Cho M.-L., Yang C.W. Increase of Th17 cell phenotype in kidney transplant recipients with chronic allograft dysfunction. PLoS One. 2015;10:e0145258. doi: 10.1371/journal.pone.0145258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nova-Lamperti E., Romano M., Christakoudi S., Runglall M., McGregor R., Mobillo P., Kamra Y., Tsui T.-L., Norris S., John S., et al. Reduced TCR signaling contributes to impaired Th17 responses in tolerant kidney transplant recipients. Transplantation. 2018;102:e10–e17. doi: 10.1097/TP.0000000000001920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lian C.G., Bueno E.M., Granter S.R., Laga A.C., Saavedra A.P., Lin W.M., Susa J.S., Zhan Q., Chandraker A.K., Tullius S.G., et al. Biomarker evaluation of face transplant rejection: association of donor T cells with target cell injury. Mod. Pathol. 2014;27:788–799. doi: 10.1038/modpathol.2013.249. [DOI] [PubMed] [Google Scholar]
- 20.Wu L., Chen X., Zhao J., Martin B., Zepp J.A., Ko J.S., Gu C., Cai G., Ouyang W., Sen G., et al. A novel IL-17 signaling pathway controlling keratinocyte proliferation and tumorigenesis via the TRAF4-ERK5 axis. J. Exp. Med. 2015;212:1571–1587. doi: 10.1084/jem.20150204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark R.A., Chong B., Mirchandani N., Brinster N.K., Yamanaka K.-I., Dowgiert R.K., Kupper T.S. The vast majority of CLA+ T cells are resident in normal skin. J. Immunol. 2006;176:4431–4439. doi: 10.4049/jimmunol.176.7.4431. [DOI] [PubMed] [Google Scholar]
- 22.Heath W.R., Carbone F.R. The skin-resident and migratory immune system in steady state and memory: innate lymphocytes, dendritic cells and T cells. Nat. Immunol. 2013;14:978–985. doi: 10.1038/ni.2680. [DOI] [PubMed] [Google Scholar]
- 23.Etra J.W., Shores J.T., Sander I.B., Brandacher G., Lee W.P.A. Trauma-induced rejection in vascularized composite allotransplantation. Ann. Surg. 2020;271:e113–e114. doi: 10.1097/SLA.0000000000003365. [DOI] [PubMed] [Google Scholar]
- 24.Lopdrup R.G., Turk M., Win T.S., Marty F.M., Molway D., Tullius S.G., Pomahac B., Talbot S.G. Seasonal variability precipitating hand transplant rejection? Transplantation. 2017;101:e313. doi: 10.1097/TP.0000000000001877. [DOI] [PubMed] [Google Scholar]
- 25.Tong P.L., Roediger B., Kolesnikoff N., Biro M., Tay S.S., Jain R., Shaw L.E., Grimbaldeston M.A., Weninger W. The skin immune atlas: three-dimensional analysis of cutaneous leukocyte subsets by multiphoton microscopy. J. Invest. Dermatol. 2015;135:84–93. doi: 10.1038/jid.2014.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hautz T., Zelger B., Grahammer J., Krapf C., Amberger A., Brandacher G., Landin L., Pratschke J., Margreiter R., Schneeberger S. Molecular markers and targeted therapy of skin rejection in composite tissue allotransplantation. Am. J. Transpl. 2010;10:1200–1209. doi: 10.1111/j.1600-6143.2010.03075.x. [DOI] [PubMed] [Google Scholar]
- 27.Dai H., Lan P., Zhao D., Abou-Daya K., Liu W., Chen W., Friday A.J., Williams A.L., Sun T., Chen J., et al. PIRs mediate innate myeloid cell memory to nonself MHC molecules. Science. 2020;368:1122–1127. doi: 10.1126/science.aax4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaerli P., Ebert L., Willimann K., Blaser A., Roos R.S., Loetscher P., Moser B. A skin-selective homing mechanism for human immune surveillance T Cells. J. Exp. Med. 2004;199:1265–1275. doi: 10.1084/jem.20032177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim H.O., Cho S.I., Chung B.Y., Ahn H.K., Park C.W., Lee C.H. Expression of CCL1 and CCL18 in atopic dermatitis and psoriasis. Clin. Exp. Dermatol. 2012;37:521–526. doi: 10.1111/j.1365-2230.2011.04295.x. [DOI] [PubMed] [Google Scholar]
- 30.Goebeler M., Trautmann A., Voss A., Bröcker E.B., Toksoy A., Gillitzer R. Differential and sequential expression of multiple chemokines during elicitation of allergic contact hypersensitivity. Am. J. Pathol. 2001;158:431–440. doi: 10.1016/s0002-9440(10)63986-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schutyser E. Involvement of CC chemokine ligand 18 (CCL18) in normal and pathological processes. J. Leukoc. Biol. 2005;78:14–26. doi: 10.1189/jlb.1204712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colantonio L., Iellem A., Sinigaglia F., D’Ambrosio D. Skin-homing CLA+ T cells and regulatory CD25+ T cells represent major subsets of human peripheral blood memory T cells migrating in response to CCL1/I-309. Eur. J. Immunol. 2002;32:3506–3514. doi: 10.1002/1521-4141(200212)32:12<3506::AID-IMMU3506>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 33.Kollar B., Shubin A., Borges T.J., Tasigiorgos S., Win T.S., Lian C.G., Dillon S.T., Gu X., Wyrobnik I., Murphy G.F., et al. Increased levels of circulating MMP3 correlate with severe rejection in face transplantation. Sci. Rep. 2018;8:14915. doi: 10.1038/s41598-018-33272-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kollar B., Uffing A., Borges T.J., Shubin A.V., Aoyama B.T., Dagot C., Haug V., Kauke M., Safi A.F., Talbot S.G., et al. MMP3 is a non-invasive biomarker of rejection in skin-bearing vascularized composite allotransplantation: a multicenter validation study. Front. Immunol. 2019;10:2771. doi: 10.3389/fimmu.2019.02771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watarai Y., Koga S., Paolone D.R., Engeman T.M., Tannenbaum C., Hamilton T.A., Fairchild R.L. Intraallograft chemokine RNA and protein during rejection of MHC-matched/multiple minor histocompatibility-disparate skin grafts. J. Immunol. 2000;164:6027–6033. doi: 10.4049/jimmunol.164.11.6027. [DOI] [PubMed] [Google Scholar]
- 36.Hricik D.E., Nickerson P., Formica R.N., Poggio E.D., Rush D., Newell K.A., Goebel J., Gibson I.W., Fairchild R.L., Riggs M., et al. Multicenter validation of urinary CXCL9 as a risk-stratifying biomarker for kidney transplant injury. Am. J. Transpl. 2013;13:2634–2644. doi: 10.1111/ajt.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho J., Schaub S., Wiebe C., Gao A., Wehmeier C., Koller M.T., Hirsch H.H., Hopfer H., Nickerson P., Hirt-Minkowski P. Urinary CXCL10 chemokine is associated with alloimmune and virus compartment-specific renal allograft inflammation. Transplantation. 2018;102:521–529. doi: 10.1097/TP.0000000000001931. [DOI] [PubMed] [Google Scholar]
- 38.Kaminski M.M., Alcantar M.A., Lape I.T., Greensmith R., Huske A.C., Valeri J.A., Marty F.M., Klämbt V., Azzi J., Akalin E., et al. A CRISPR-based assay for the detection of opportunistic infections post-transplantation and for the monitoring of transplant rejection. Nat. Biomed. Eng. 2020;4:601–609. doi: 10.1038/s41551-020-0546-5. [DOI] [PubMed] [Google Scholar]
- 39.Christofidou-Solomidou M., Longley B.J., Whitaker-Menezes D., Albelda S.M., Murphy G.F. Human skin/SCID mouse chimeras as an in vivo model for human cutaneous mast cell hyperplasia. J. Invest. Dermatol. 1997;109:102–107. doi: 10.1111/1523-1747.ep12276733. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe R., Gehad A., Yang C., Scott L.L., Teague J.E., Schlapbach C., Elco C.P., Huang V., Matos T.R., Kupper T.S., et al. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci. Transl. Med. 2015;7:279ra39. doi: 10.1126/scitranslmed.3010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Racki W.J., Covassin L., Brehm M., Pino S., Ignotz R., Dunn R., Laning J., Graves S.K., Rossini A.A., Shultz L.D., et al. NOD-scid IL2rgamma(null) mouse model of human skin transplantation and allograft rejection. Transplantation. 2010;89:527–536. doi: 10.1097/TP.0b013e3181c90242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borges T.J., Murakami N., Machado F.D., Murshid A., Lang B.J., Lopes R.L., Bellan L.M., Uehara M., Antunes K.H., Pérez-Saéz M.J., et al. March1-dependent modulation of donor MHC II on CD103+ dendritic cells mitigates alloimmunity. Nat. Commun. 2018;9:3482. doi: 10.1038/s41467-018-05572-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Metsalu T., Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47:D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. This paper does not report the original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.